Abstract
RNA interference (RNAi), the sequence-specific suppression of gene expression, offers great opportunities for insect science, especially to analyze gene function, manage pest populations, and reduce disease pathogens. The accumulating body of literature on insect RNAi has revealed that the efficiency of RNAi varies between different species, the mode of RNAi delivery, and the genes being targeted. There is also variation in the duration of transcript suppression. At present, we have a limited capacity to predict the ideal experimental strategy for RNAi of a particular gene/insect because of our incomplete understanding of whether and how the RNAi signal is amplified and spread among insect cells. Consequently, development of the optimal RNAi protocols is a highly empirical process. This limitation can be relieved by systematic analysis of the molecular physiological basis of RNAi mechanisms in insects. An enhanced conceptual understanding of RNAi function in insects will facilitate the application of RNAi for dissection of gene function, and to fast-track the application of RNAi to both control pests and develop effective methods to protect beneficial insects and non-insect arthropods, particularly the honey bee (Apis mellifera) and cultured Pacific white shrimp (Litopenaeus vannamei) from viral and parasitic diseases.
Keywords: antiviral therapy, dsRNA, insect pest control, RNA interference, siRNA, systemic RNAi
1. Introduction
RNA interference (RNAi) has transformed insect science research because it enables the researcher to suppress a gene of interest and thereby link a phenotype to gene function. For basic research purposes, RNAi offers a route to functional genetics in all insects, including those for which transgene resources do not exist (Belles, 2010). RNAi also has enormous potential for applied entomology (Price and Gatehouse, 2008; Xue et al., 2012). For example, RNAi can be used for insect pest control by suppressing essential genes leading to reduced fitness and/or mortality. Furthermore, by priming the antiviral RNAi response with innocuous viral sequences, beneficial insect species, such as honey bee (Apis mellifera) and silkworm (Bombyx mori), can be protected from highly pathogenic viral infections. However, the reality is not yet matching the envisioned potential of RNAi. Practitioners are increasingly aware that RNAi in insects can be capricious; efficacy varies across insect taxa, among genes, with mode of delivery, and even between different laboratories (Terenius et al., 2011). All too often, the application of RNAi technology is an empirical exercise: “try it, for it might work”.
The goal of this article is to promote the use of practical principles to design and interpret insect RNAi studies. We know that no single protocol can be applied for any gene in any insect. Therefore, the specific purpose of this article is to provide a roadmap for the application of RNAi for experimental analysis of gene function, management of pests and protection of beneficial arthropods. The article is divided into three sections. First, current knowledge of the mechanisms and function of RNAi in insects is reviewed, highlighting the known variation among insect taxa. This information offers a guide to the most appropriate strategy for different insect systems, and provides the springboard for much-needed future innovation in RNAi technology. Second, the design of RNAi studies is addressed, using both empirical data and conceptual understanding to identify successful experimental designs, effective methods for RNAi delivery, and informative indices of RNAi efficacy. Importantly, there is no single protocol for the perfect RNAi experiment, partly because the efficacy of RNAi strategies varies among insect groups. In the third section, we turn to the application of RNAi for the management of pest and beneficial insects, and discuss the unique opportunities and challenges associated with each of these applications.
2. Mechanisms of RNAi
RNAi refers to the suppression of gene expression by small noncoding RNA molecules, predominantly by the cleavage of a target mRNA in a sequence-specific manner (Fire et al., 1998), and the general steps involved in this process are shown in Fig. 1. Upon cell entry and recognition, double stranded RNA (dsRNA) is cleaved by the RNase III Dicer into 20–25 bp fragments with a two base overhang at the 3’ end. These fragments are incorporated into the multi-protein RNA-induced silencing complex (RISC), where one strand (the “passenger” strand) is eliminated and the other “guide” strand is retained. The catalytic component of RISC is the RNase H-like domain of an Argonaute protein, which cleaves single-stranded RNA molecules having sequence complementary to the guide RNA. Most eukaryotes, including animals and plants, have Dicer and Argonaute proteins, and possess the RNAi machinery (Shabalina and Koonin, 2008).
Fig. 1.
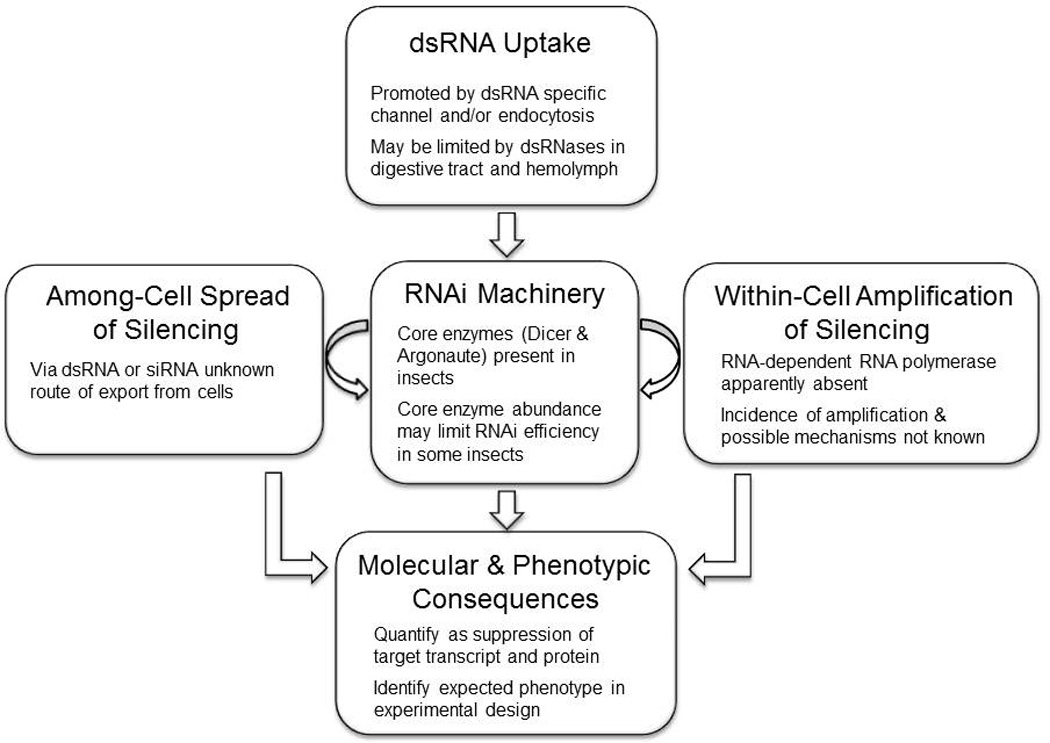
The RNAi process in insects. See text for additional details
The 20–25 bp RNAs generated by Dicer comprise two groups (Ghildiyal and Zamore, 2009; Matranga and Zamore, 2007; Asgari, 2013): microRNAs (miRNAs), which are processed from endogenous gene transcripts and function in the regulation of gene expression, and small interfering RNAs (siRNAs), which are derived from dsRNA molecules and provide defense against viruses and transposable elements. The experimental use of RNAi exploits the siRNA pathway, specifically the capacity of cells to degrade a single-stranded RNA (ssRNA) (including mRNAs) with sequence identity to the administered dsRNA molecules.
Three processes determine what can be achieved by RNAi: cellular uptake of the RNAi molecule (usually dsRNA), the production of secondary dsRNA molecules in the cell, and the transfer of these molecules to other cells (Fig. 1). Where the RNAi-mediated silencing is transmitted widely throughout the treated organism, RNAi is described as systemic (Whangbo and Hunter, 2008; Huvenne and Smagghe, 2010). In principle, the success of an RNAi experiment could be predicted from the level of these activities in the insect of interest, and strategies that increase these activities might enhance RNAi-mediated knockdown of the target gene expression. The difficulty is that we have little or no understanding of whether or how dsRNAs are amplified within insect cells or disseminated among insect cells.
In C. elegans, systemic spread of RNAi is optimal for dsRNA molecules ≥ 50 bp long, and is independent of Argonaute function (Feinberg and Hunter, 2003; Tabara et al., 1999), suggesting that the molecules moving between cells are dsRNAs, and not amplified siRNA. Uptake of dsRNA by somatic cells requires the protein SID-1 (systemic interference defective-1), which is inferred to function as a dsRNA channel (Winston et al., 2002). Other C. elegans proteins, SID-2 and SID-5, have been implicated in dsRNA uptake by gut cells and dsRNA export from cells, respectively (Hinas et al., 2012; Winston et al., 2007). Putative insect orthologs of the C. elegans sid genes have been described (e.g. Dong and Friedrich, 2005; Xu and Han, 2008), but these reports deserve careful evaluation. The sid-1-like genes in various insects have a greater sequence identity with the C. elegans gene chup-1, also known as tag-130, than to sid-1. CHUP-1 is a cholesterol transporter and has no known involvement in RNAi (Valdes et al., 2012). As an illustration, a gene in Locusta migratoria initially identified as sid-1 -like is not required for systemic RNAi (Luo et al., 2012), and is the ortholog of C. elegans CHUP-1 (as determined by top reciprocal BLASTp hit), and not SID-1.
Plants and the nematode Caenorhabditis elegans possess an RNA-dependent RNA polymerase (RdRP) (Pak and Fire, 2007; Xie et al., 2001) that facilitates the within cell amplification of silencing. The fragments of the target ssRNA released from the RISC act as a template for RdRP-dependent dsRNA synthesis, yielding more substrate for RISC-mediated degradation of the target ssRNA. Efficient amplification of RNAi by RdRP can drive the abundance of the target ssRNA molecule to undetectable levels, and RdRP is essential for RNAi in C. elegans (Sijen et al., 2001). RdRP has been identified in a few animal species beyond Caenorhabditis nematodes, including the cephalochordate Branchiostoma floridae (Vienne et al., 2003), but no verified RdRP homolog is evident in any insect genome sequenced to date (Tomoyasu et al., 2008). It is unclear whether or how the RNAi triggered by the acquisition of dsRNA molecules is sustained in insect cells (Fig. 1).
Analysis of gene orthology between C. elegans and insects has been a productive approach to identify the core RNAi machinery in insects, but far less informative for understanding the molecular basis of intracellular amplification and systemic spread of RNAi. The alternative discovery-based strategy of a genetic screen, using rescue from RNAi-lethality, has great potential. For example, Ulvila et al. (2006) demonstrated that uptake of dsRNA by Drosophila S2 cells is strongly endocytosis-dependent, and mediated principally by the scavenger receptors Eater and SR-CI. We should, however, be cautious in extrapolating from these data to organismal RNAi because S2 cells, which are hemocyte-like, display high rates of endocytosis as compared to the majority of cell types in the intact insect.
The physiological role of insect RNAi could be informative in predicting RNAi efficacy in different insect taxa and cell types and, by extension, the development of sustainable strategies for RNAi applications in field conditions. There is a strong consensus that RNAi contributes to insect immunity against viruses with a dsRNA genome or dsRNA replicative intermediates in the cytoplasm of infected cells (Blair, 2011; Schnettler et al., 2012). We could, therefore, expect greatest success with RNAi in insect species and cell types that utilize RNAi as a primary antiviral immune response. Some insects or cell types may have low responsiveness to exogenously-applied dsRNA because they utilize alternative anti-viral defenses (e.g. apoptosis of infected cells, symbiont-mediated protection) (Merkling and van Rij, 2013). It should also be noted that certain insect viruses suppress RNAi. For example, flock house virus (FHV) codes for a protein, known as the B2 protein, that binds to dsRNA, including the FHV replication intermediate, preventing cleavage by insect Dicer and incorporation into RISC (Chao et al., 2005). An insect infected with an asymptomatic, persistent virus that codes for an RNAi suppressor would display limited responsiveness to experimental RNAi (Berry et al., 2009).
3. Designing a RNAi experiment
As described above, RNAi application and efficacy remains variable between genes, organisms and life stages, despite the tremendous utility that RNAi presents for improving our understanding of fundamental biological questions and for pest control. In addition, in insect species where RNAi is predominantly environmental with little evidence for systemic propagation, interference can vary widely between tissues due to differences in the efficacy of dsRNA uptake. Extreme examples are D. melanogaster and Manduca sexta where transcript knockdown by injection of dsRNA has only been achieved in hemocytes, which are capable of endocytosis (Miller et al., 2008; Terenius et al., 2011). In mosquitoes, most tissues can be reached by the injection of dsRNA, however the success of knockdown in the central nervous system varies highly between genes and may be dose-dependent (Lycett et al., 2006; Biessmann et al., 2010). Tissue differences in RNAi efficacy may be overcome by the design of new delivery methods, including transgenesis or viral transduction, which eliminate the requirement for cellular uptake of the RNAi trigger. Development of such technologies is lacking for the majority of species (Fig. 2 and Section 3.2 below).
Fig. 2. Current methodologies for RNAi delivery in insects: a guide on the performance of different delivery methods.
All approaches yield transient RNAi other than the tran sgenic method. ? = lack of data prevents evaluation. RNAi efficacy in Drosophila is limited to hemocytes if delivered by injection (Miller et al., 2008). Delivery by feeding can be highly effective (Whyard et al., 2009), but transgenesis is the preferred methodology. In Aedes aegypti, RNAi has been most successful by injection (e.g. Campbell et al., 2008), transgenesis (e.g. Bian et al., 2005), and viral transduction (e.g. Adelman et al., 2001), with a few reported instances of success by feeding (Coy et al., 2012) or topical application (Pridgeon et al., 2008). Most cases of successful RNAi in Tribolium castaneum have been by injection (e.g. Arakane et al., 2005). RNAi efficacy in Manduca sexta is most effective by injection for immune-related genes, and there is evidence to support successful gene suppression by delivery by feeding (Terenius et al., 2010). RNAi in A. pisum has been successful by both injection (Mutti et al., 2006) and feeding (Shakesby et al., 2009). RNAi in Apis mellifera is effective for a variety of genes and life stages both by injection and feeding (Amdam et al., 2003; Hunter et al., 2010). Even where a specific method of application has shown success for RNAi of certain genes, the effective suppression of all genes by that technique is not guaranteed.
The aforementioned biological variables, including presence/absence of the core RNAi machinery, cellular uptake and propagation of signal (Roignant et al., 2003; Miller et al., 2008), and dsRNA degrading enzymes (Arimatsu et al., 2007), as well as other differences in genetic backgrounds (Kitzmann et al., 2013), greatly affect the success of RNAi experiments in different species. Often, these challenges can be mitigated by experimental factors including the design of the RNAi molecule, the mode of delivery and the dose of the dsRNA molecule.
3.1. The RNAi molecule
The success of an RNAi experiment hinges on the production of a specific RNAi molecule (in the form of dsRNA, siRNA, or a hairpin RNA) for a target gene of interest (GOI). Experiments should include an RNAi molecule against a heterologous sequence absent from the target insect’s genome (typically green fluorescent protein (GFP) or LacZ), to control for both the administration of the experimental dsRNA and the physiological impact of triggering the RNAi cascade. In some cases, a positive control can be incorporated into the experimental design. For example, in Tribolium, the use of RNAi against vermillion (white) or Lac-2 provides rapid phenotypic evidence of RNAi success manifest in white compound eyes or white pupae, respectively (Arakane et al., 2005; Arakane et al., 2011).
A crucial consideration is the choice of sequence for dsRNA preparation, especially its length and sequence identity to the target transcript of the insect. Huvenne and Smagghe (2010) provide a comprehensive survey of the length range of dsRNAs used in early studies: from 134–1842 bp, with most studies using 300–520 bp. Comparisons among gene regions (e.g., 5’ end) to which RNAi molecules are designed have yielded variable results. For example, RNAi against hunchback (hb) in Acyrthosiphon pisum resulted in similar mortality whether the RNAi trigger was designed against the 5´ or 3´ end of the gene (Mao and Zeng 2012), but 3’ portion of the inhibitor of apoptosis gene in Aedes aegypti yielded a greater effect on mosquito mortality than dsRNA targeting the 5’ or central region of the gene (Pridgeon et al., 2008), and the most effective antiviral RNAi molecule against infectious myonecrosis virus (which infects the Pacific white shrimp, Litopenaeus vannamei) was at the extreme 5’ end of the genome (Loy et al., 2012). These varied results illustrate the importance of screening multiple RNAi sequences for a gene of interest.
Generally speaking, greater success with insect RNAi has been obtained with dsRNA molecules ≥50–200 bp in length (Huvenne and Smagghe, 2010), although the minimal length required to obtain maximal biological activity varies among insect species (Bolognesi et al., 2012). Suppression of gene expression has been achieved with siRNAs (either synthesized directly or obtained by “dicing” the dsRNA in vitro before administration to the insect), for example in the lepidopteran Helicoverpa armigera (Kumar et al., 2012), aphid A. pisum (Mutti et al., 2006) and tsetse (Attardo et al., 2012). It may, sometimes, be necessary to design the RNAi molecule of shorter length than ideal to obtain specificity, especially where one member in a gene family that has high sequence similarity is being targeted. Regardless of the desired size of the RNAi molecule, the design process can be aided by software that is informed by genome sequence and RNA folding kinetics to optimize effectiveness; for example, E-RNAi currently offers dsRNA and siRNA design suggestions for A. mellifera, Tribolium castaneum, A. pisum, Anopheles gambiae and Ae. aegypti (Horn and Boutros, 2010).
A further issue to be considered in the design of RNAi molecules is the exquisite specificity of RNAi. In the context of field applications of RNAi, this property facilitates design of insect-lethal sequences that are highly species-specific. For example, feeding four species of Drosophila with species-specific vATPase dsRNA resulted in reduced vATPase mRNA and significant mortality in conspecific, but not heterospecific flies (Whyard et al., 2009). In basic research pursuits, this property affords researchers the capacity to silence alleles (using short dsRNA) of the same gene specifically, e.g. TEP1 alleles in An. gambiae (Blandin et al., 2009). Conversely, two alleles of a heterozygous individual, as well as genetically-distinct members within an insect population, whether in the laboratory or field, may differ in their susceptibility to RNAi. This concern is amply justified by studies on the effect of mismatches between the dsRNA and its intended target (i.e. mRNA) using synthetic siRNAs administered to mammalian cells in culture. Most single mismatches impair the RNAi effect (Birmingham et al., 2006; Jackson et al., 2003; Joseph and Osman, 2012a, b; Wu et al., 2011); some mismatches, however, alter the cellular response from one of transcript loss (siRNA) to translational repression, (Hu et al., 2010; Tomari et al., 2007). The advantages of using longer >200bp dsRNA for RNAi strategies in pest management is the production of many siRNAs against the targeted mRNA transcript; potentially maximizing the RNAi response. Further studies will be necessary to clarify the extent to which the responses to mismatches in dsRNA and target mRNA in whole insects differ from the siRNA studies which used a single construct conducted on cultured mammalian cells.
3.2. RNAi delivery
Efficacy of an RNAi experiment can be influenced strongly by the mode of delivery of the RNAi trigger (Fig. 2 and references within). The most widely used routes for administering RNAi to insects are injection into the hemolymph and feeding. Microinjection was used in the first successful application of RNAi in an insect, to obtain knockdown of frizzled in Drosophila melanogaster (Kennerdell and Carthew, 1998). This method was quickly transferred to T. castaneum (Brown et al., 1999) and subsequently applied to adult insects in An. gambiae (Blandin et al., 2002). Microinjection has been applied to all life stages in hemi- and holometabolous insects in a rapidly growing number of orders; indeed routine protocols are now in place for injection for various taxa, including Tribolium, B. mori, several genera of Diptera, the honey bee, cockroaches and orthopterans [for a list of references, see Belles (2010)].
An important barrier to the use of microinjection in some insects is non-specific damage caused by mechanical damage, which is most often pronounced when targeting embryos. Experimental variables that influence survivorship include methods of immobilization (cold, CO2, adherence to a substrate), injection volume, site of injection, and dilutants. Although water or physiologic saline work well for most species, the diluent may require adjustment to the particular osmotic pressure of the hemolymph.
Oral delivery is a less-invasive and potentially a high-throughput method for RNAi delivery. It has particular value for insects that are intolerant of injection (Fig. 2) and for field applications for RNAi-mediated pest control (see section 4). Protocols for administration of dsRNA synthesized in vitro and incorporated into the diet are now available for honey bees, aphids, whiteflies and psyllids (Aronstein et al., 2006; Wuriyanghan et al., 2011; Ghanim et al., 2007; Whyard et al., 2009). RNAi delivery to phytophagous insects can also be achieved by engineering plants to express dsRNAs in plant systems for which transgene introduction technologies are available (Fig. 2). Two complementary methods are in use: stable transformation by hairpin dsRNAs that target insect genes (Baum et al., 2007) and transient virus-induced gene silencing (VIGS), in which engineered viral vectors carrying the gene sequence of interest are transformed into Agrobacterium tumefaciens and infiltrated into the plant tissue (Burch-Smith et al., 2004). Both approaches have been exploited, to achieve transcript suppression in Coleoptera (Baum et al., 2007), Lepidoptera (Baum et al., 2007; Kumar et al., 2012) and Hemiptera (Pitino et al. 2011; Zha et al., 2011). In some species, notably dipterans, oral delivery of RNAi triggers has yielded less consistent results than microinjection (Zhang et al., 2011). Further, in Lepidoptera, feeding as a mode of delivery necessitates the provision of high doses of RNAi trigger (Terenius et al., 2011). This can be attributed to a variety of factors. The efficacy of RNAi of midgut transcripts may be reduced due to low or inconsistent doses taken up by individual insects, frequency and size of feeding, plus GI tract morphology and physiology will affect the actual dose of RNAi that reaches the midgut epithelium. In addition, there is evidence for production of mRNAs that encode putative secretory dsRNA-degrading enzymes in insects, notably B. mori, that can interfere with the RNAi response (Arimatsu et al., 2007; Liu et al., 2012). Establishing protocols for consistent RNAi induction by feeding in different species may, therefore, prove challenging. In addition, oral delivery of RNAi molecules in species where systemic RNAi cannot be achieved limits its application to genes expressed in gut cells (Fig. 2).
A minority of studies have exploited alternative routes for dsRNA delivery, including electroporation, soaking or ectopic application, incorporation into nanoparticles, expression in bacteria, topical application, injection into woody plants, direct absorption of dsRNA in water solution into plant cuttings, or rooted seedlings and trees and solubilization using transfection agents, such as Lipofectamine™ (Wang et al., 2011; Karim et al., 2010; Zhang et al., 2010; Pridgeon et al., 2008; Lopez-Martinez et al., 2012; Hunter et al., 2012). For additional consideration of this topic, the reader is referred to Yu et al. (2013), which provides a comprehensive review of the history and current practice.
3.3. RNAi dosage
The requisite dose of RNAi molecules varies with insect species, life stage, the target gene transcript abundance and its spatial and temporal expression profiles, and according to the delivery method of choice. The viscosity of high dsRNA concentrations limits the injectable concentrations to 6 µg µl−1 (K. Michel, unpub data), and the cost of synthesizing large amounts of dsRNA presents a challenge for high concentrations in artificial diets. Species- and tissue-specific biological factors, including the degradation of dsRNA, and weak activity of the RNAi machinery, can influence the efficacy of RNAi, often requiring relatively high dosage of RNAi molecules. There is now persuasive evidence for dsRNase activity in various extracellular fluids of insects, including the digestive juices of B. mori (Arimatsu et al., 2007), the saliva of the hemipteran Linus lineolaris (Allen et al., 2012) and the hemolymph of M. sexta (Garbutt et al., 2013). Although, to our knowledge, this has not been reported in insects, the difficulties in achieving RNAi of genes expressed in neurons of the nematode C. elegans has been attributed to the high expression of a nuclease (eri-1, enhanced RNAi-1) in these cells (Kennedy et al., 2004).
The mode of uptake, ability to spread RNAi molecules and ability to process the RNAi molecules are other important considerations that no doubt strongly influences the requisite dose required to induce a RNAi response. In D. melanogaster larvae, cell autonomous RNAi can be induced readily by the expression of short hairpin RNAs from a transgene; however, injected dsRNAs fail to trigger RNAi in most tissues with the exception of hemocytes (Miller et al., 2008). A higher dose is usually required when the RNA molecule is delivered orally as compared to injection. Multiple introductions of dsRNA can enhance the efficacy of RNAi in the salivary glands of Rhodnius prolixus (Araujo et al., 2006), and although the basis for this effect is not fully understood, one attractive hypothesis is that elements of the RNAi machinery may be expressed at low levels in some tissues (Chintapalli et al., 2007; Rinkevich and Scott, 2013), but can be induced in response to the RNAi molecule (Garbutt and Reynolds, 2012; Liu et al., 2013). Pertinently, Ae. aegypti, mounts an antiviral RNAi response to Sindbis virus infection, but transcript levels for Dicer and Argonaute do not change appreciably; only Tudor staphylococcal nuclease, an element of the RISC, shows moderate increase in transcript abundance during an active RNAi response (Campbell et al., 2008). Further research is required to establish the incidence and significance of inducibility in RNAi function.
3.4. Choice of gene: transcript abundance and protein stability
In principle, the ideal gene target for RNAi produces an mRNA pool with high turnover that codes for a protein with a short half-life. The use of RNAi for phenotypic analysis of gene function in any life stage could be more difficult if the protein product of the target gene has a long half-life. For example, nicotinic acetylcholine receptors (nAChRs) can be stable for ≥ 2 weeks (Lomazzo et al., 2011) and this protein stability may explain the weak phenotypic response associated with RNAi-mediated knockdown of Dα6 (nicotinic acetylcholine receptor subunit) expression in both D. melanogaster and T. castaneum (Rinkevich and Scott, 2013). However, for the great majority of genes, mRNA turnover and protein half-life are not known.
3.5. Evaluation of RNAi experiments
The desired result of an RNAi experiment varies with the purpose of the study. High insect mortality is a successful outcome for investigations designed to identify novel RNAi-based strategies to control an insect pest, but a hindrance to many experimental investigations of gene function. For many analyses of gene function, physiological indices of predicted function should be central to the analysis. For example, if a gene under study has a predicted role in protein digestion, osmoregulation or olfaction, then analyses of gut protease activity, hemolymph osmotic pressure and electroantennogram data, respectively, may be useful physiological indices. For some experiments, it may be necessary to reduce the RNAi dose to obtain a reliable physiological signal of gene function obtained by an intermediate expression knockdown, because strong knockdown could result in secondary, deleterious effects on insect fitness that obscure the primary lesion. It is, therefore, important to define the appropriate physiological and fitness assays as an integral part of the experimental design.
The successful reduction of transcript levels as a result of RNAi is most commonly measured by RT-qPCR and expressed as a percent reduction of the relevant transcript in the treatment group versus the negative control group (in which animals were subjected to an RNAi molecule for a heterologous gene). Although this methodology is widely accepted, the choice of reference or housekeeping genes for calculating relative transcript levels is challenging. Even if reference genes for RT-qPCR have been described and validated on the species level, the expression of a reference gene may vary with the physiology and the tissue being targeted [e.g. Ponton et al., 2011 (Drosophila), Scharlaken et al., 2008 (honey bees), Majerowicz et al., 2011 (Rhodnius)].
Ultimately the phenotypic result of an RNAi experiment hinges on the reduction of protein levels for the gene of interest, and it is highly desirable to determine relative protein concentration. The effect of RNAi on the protein may not be well-correlated to the level of transcript suppression. For example, following dsRNA injection targeting An. gambiae SRPN2, SRPN2 protein is not detectable by western blot in the hemolymph, but transcript levels remain at 40–60% compared to controls (Michel et al., 2005). Finally, it is possible that RNAi could lead to suppression of transcript (and protein), but not yield a phenotype, particularly where redundancy is built into a specific biological function. For example, deletion of one of the most abundant nAChRs in the insect nervous system results in flies that are “normal” (Perry et al., 2007). Whether redundancy will present a limitation for a significant number of other genes remains to be established.
4 Application of RNAi for the management of insect populat
The potential of RNAi for the management of pest insects and protection of domesticated beneficial insects, especially the honey bee, is widely recognized (Xue et al., 2012). In principle, the sequence used in RNAi can be tailored to any taxonomic scale, from a single genotype to a family or even order of insects; and the identity of the target sequence can be manipulated at will, enabling the practitioner to respond rapidly to novel pest taxa or to diminishing efficacy (due to the evolution of resistance, for example) of one target sequence or combination of sequences. In other words, RNAi offers exquisite specificity and flexibility that cannot be matched by traditional chemical insecticides, biological control by natural enemies, or plants bearing protein-coding transgenes.
4.1. RNAi and the control of insect pests
Proof of principle for the application of RNAi in insect crop pest control comes from early studies conducted on the western corn rootworm, Diabrotica virgifera virgifera (WCRW) (Baum et al., 2007), and cotton bollworm Helicoverpa armigera (CBW) (Mao et al., 2007). Baum et al. (2007) fed larval WCRW on 290 dsRNAs, from which they identified 14 genes that reduced larval performance, and one of these, vacuolar ATPase subunit A (V-ATPase), was carried forward for detailed analysis. Low concentrations of orally-delivered dsRNA against V-ATPase in artificial diet suppressed the corresponding WCRW mRNA. Importantly, larvae reared on transformed corn plants that express V-ATPase dsRNA also displayed reduced expression of the V-ATPase gene and caused much reduced plant root damage (Baum et al., 2007). In the study of Mao et al. (2007) on CBW, the target gene was a cytochrome P450, CYP6AE14, which is expressed in the larval midgut and detoxifies gossypol, a secondary metabolite common to cotton plants. When CBW was exposed to either Arabidopsis thaliana or Nicotiana tobacum expressing CYP6AE14 dsRNA, levels of this transcript in the insect midgut decreased, larval growth was retarded, and both effects were more dramatic in the presence of gossypol (Mao et al., 2007). Transgenic cotton plants expressing CYP6AE14 dsRNA also support drastically retarded growth of the CBW larvae, and suffered less CBW damage than control plants (Mao et al., 2011). The research on both WCRW and CBW has been extended to additional genes. The Snf7 gene, which is involved in trafficking of membrane receptors, has been reported to be effective against both D. v. virgifera and D. v. howardi larvae (Bolognesi et al., 2012; Ramaseshadri et al., 2013); and cotton plants engineered to express cysteine proteases attenuated the peritrophic matrix of CBW, resulting in increased uptake of the dsRNA (Mao et al., 2013). Importantly, cotton plants expressing both the dsCYP6AE14 and cysteine protease were more protected from bollworm than either of the single-transgene lines (Mao et al., 2013).
The studies of Baum et al. (2007) and Mao et al. (2007) illustrate two key issues for successful RNAi of insect crop pests: choice of the target sequence(s) for RNAi; and mode of delivery. The target gene must be an essential insect gene that is consistently expressed through the relevant life-stages and yields reliable RNAi-induced depression of insect performance. As the technology moves from proof of principle to application, very careful consideration of the design of the target sequence(s) is required. The prerequisites for success are perfect sequence identity between at least some of the 21–25 bp siRNAs derived from the dsRNA and the cognate mRNA of the insect pests; and sufficient sequence divergence between all the siRNAs and protein-coding genes of non-target organisms. These analyses can be conducted in silico, by comparing a 21–25 bp moving window along the candidate dsRNA sequence to both the target gene in all target insect taxa, and to all predicted protein-coding genes in all other publicly-available genomes. It may be appropriate to obtain genomic or transcriptomic data for other non-target taxa that currently lack genomic resources, so that the in silico analysis of the proposed dsRNA sequences includes ecologically-relevant organisms. Any proposed dsRNA that fails to yield multiple siRNAs with perfect match to the sequence in all pest insects, or that yields a single siRNA that matches the sequence in any relevant non-target organism should be discarded. Less certain is the degree of sequence mismatch between dsRNA-derived siRNAs and a non-target organism that can be tolerated. Because siRNA molecules can inhibit translation of transcripts with less than perfect sequence identity, the threshold for concern about non-target effects could be less than 100% sequence identity. Further work is required to determine the relative amount of mismatch between the target and effector that causes lack of efficacy. Such results would inform our understanding of how to optimize pest management while minimizing effects to non-target organisms and slowing the evolution of resistance (Section 4.3).
The second issue important for the success of RNAi is delivery at an effective dose, while maintaining acceptable production costs. Recent breakthroughs in dsRNA production methods, which can produce kilogram quantities, continues to reduce the cost associated with dsRNA production and makes it feasible to start discussing strategies which will apply dsRNA products as baits, sprays, or through irrigation systems (Hunter et al., 2010; Hunter et al., 2012). In planta RNAi has great potential not only against chewing insect pests [such as the WCRW and CBW studied by Baum et al. (2007) and Mao et al. (2007)], but also against plant sap feeding pests. Transgenic technologies involving expression of toxins from Bacillus thuringiensis (Bt) in crop plants have, contributed little to the control of sucking insect pests, because Bt endotoxins have yet to be identified with activity against these pests (Li et al., 2011). In addition, these insects are becoming increasingly prevalent in Bt crops, as a result of ecological release due to reduced use of broad-spectrum insecticide treatments previously used to control lepidopteran and coleopteran pests (Faria et al., 2007; Lu et al., 2010). Pyramiding RNAi technologies against sap feeders with Bt (or other technologies) against chewing insects could resolve these difficulties. The experimental demonstrations of in planta RNAi against the rice plant hopper Nilaparvata lugens (Zha et al., 2011) and the aphid Myzus persicae (Pitino et al., 2011) provide a proof of principle for this technology.
Alternative approaches are being developed for RNAi delivery as a conventional pesticide, for example as insecticidal baits for urban pests, such as ants, cockroaches and termites (Zhou et al., 2008), or for the aquatic larval stages of mosquitoes (see below). The commercial potential of these methods depends critically on the ability to deliver dsRNA to the target insect, which is in part determined by stability of the dsRNA in the environment, its concentration in the baits and take-up rates by the insects, as offset against the production costs for dsRNA. These objectives will be facilitated by formulations that enhance the uptake of dsRNA into insect cells and its protection against insect dsRNases. For example, dsRNA forms stable 100–400 nm particles in association with chitosan, through the electrostatic forces between the positive charges of the amino group in chitosan and the negatively-charged RNA (McCarroll and Kavallaris, 2012). Zhang et al. (2010) used the chitosan nanoparticle-based RNAi technology to suppress the expression of two chitin synthase genes (AgCHS1 and AgCHS2) in African malaria mosquito (An. gambiae) larvae. Although this treatment did not kill the larvae, it did reduce the larval chitin content and increased larval susceptibility to the insecticide diflubenzuron.
4.2. RNAi and the protection of insects against parasites and pathogens
The susceptibility of many eukaryotic parasites to RNAi offers a novel strategy to enhance the health of beneficial insects. Of course, this strategy does not apply to bacterial pathogens or the various eukaryotes (e.g. trypanosomes and Plasmodium species) which lack any known capacity for RNAi. The opportunity is vividly illustrated by the microsporidian parasite Nosema of the honey bee. Nosema causes high morbidity and mortality of honey bees (Martin-Hernandez et al., 2011). Two aspects of the biology of Nosema make it an especially suitable target for RNAi-based strategies: it has the molecular machinery for RNAi, and it colonizes midgut epithelial cells, a site readily accessed by ingested dsRNA. When fed honey infected with N. ceranae plus dsRNA specific to the Nosema ADP/ATP transporter gene, which is essential for Nosema energy metabolism, honey bees had a reduced Nosema load and lower mortality, together with suppressed transcript abundance of the target genes (Paldi et al., 2010).
Eukaryotic parasites that exploit insect organs other than the gut would be susceptible to RNAi only where the insect host displays systemic spread of the RNAi signal. This has been demonstrated for the ectoparasitic mite, Varroa destructor, which feeds on the blood of honey bees (Garbian et al., 2012). When bees were fed on dsRNA specific to a panel of Varroa genes, the density of Varroa mites on the bees was reduced by up to 50%, with no apparent deleterious effect on the honey bees. The pattern of spread of the RNAi was tested by allowing honey bees to feed on sucrose solution containing dsRNA-GFP (green fluorescent protein; because the genomes of both the insect and mite lack the GFP gene, the distribution of GFP-RNA could be monitored without interference from sequence of endogenous origin). When Varroa-infested bees were fed on the test solution, the GFP-RNA was recovered in the Varroa. Moreover, when these Varroa were subsequently transferred to bees feeding on sugar solution without dsRNA-GFP, the recipient bees acquired the GFP-RNA. These experiments demonstrate that the RNAi can be amplified and spread not only at the level of the individual insect, but also at the colony level in honey bees. Further research is required to establish the frequency and dose of RNAi applications required to sustain protection of colonies, and whether this approach offers a cost-effective strategy for the control of Varroa mite, which is of first-order importance in compromising the health of honey bee colonies.
RNAi also holds potential to clear insect vector species from parasites, that themselves are not susceptible to RNAi. A prime example is parasites of the genus Plasmodium, the causative agent of malaria (Baum et al., 2009). Conceptually, depletion of proteins required for parasite entry or survival within the insect vector by means of RNAi could be used to create refractory mosquitoes (Brown and Catteruccia, 2006). Proof-of-principle successes have been achieved in the laboratory (Dong et al., 2011). Effective RNAi delivery methodologies that are field-deployable involve oral exposure or transgenic population replacement strategies, and are currently under development.
The natural function of RNAi is protection against viruses, and RNAi has enormous potential in anti-viral therapy. There are opportunities for RNAi-mediated suppression of viral infections in insects, including vectors of socio-economically important viral diseases of humans, livestock and crop plants. Exogenously-applied or ingested dsRNA can be considered as a boost to the native RNAi machinery of the host, conferring protection both by prophylaxis and direct treatment. The value of such boosting is illustrated by research on the titer of various mosquito-vectored arboviruses. RNAi-mediated antiviral immunity contributes to the suppression of viruses, including dengue virus and Sindbis virus, in the mosquito Ae. aegypti, as demonstrated by the increased titer and transmission of these viruses in mosquitoes in which the RNAi machinery was experimentally silenced (Franz et al., 2006; Campbell et al., 2008; Khoo et al., 2010). Furthermore, viral suppression is promoted by enhancing the RNAi pathway, achieved by engineering the insects to express an inverted-repeat RNA that triggers production of dsRNA specific to the virus sequence (Franz et al., 2011; Mathur et al., 2010). There is some evidence for viral-mediated suppression of RNAi, for example by the Sindbis virus and West Nile virus, in mosquito cells (Cirimotich et al., 2009; Schnettler et al., 2012). Such suppression may be the reason why supplementary dsRNA is required to achieve RNAi-mediated elimination of viral infection from the insect host.
Another insect system demanding urgent solutions to viral infections is the honey bee, especially in the context of evidence that viruses, including the Israeli acute paralysis virus (IAPV), may contribute to the ongoing decline of honey bees, including colony collapse disorder (Evans and Schwarz, 2011). Evidence that exogenous dsRNA can supplement the endogenous RNAi machinery comes from the demonstration that IAPV infection of honey bees can be eliminated by orally-delivered dsRNA corresponding to two different sequences of the IAPV genome (Maori et al., 2009). Among colonies inoculated with IAPV, mortality was reduced in those treated with IAPV-dsRNA relative to those that were not treated or that were treated with non-IAPV dsRNA. These results led to large-scale field test in the USA, in which honey bees were fed a dsRNA product, Remebee-I, in the presence of the IAPV (Hunter et al., 2010). Honey bee survival, colony size and honey production were all increased in the Remebee-I treatment. Ingested IAPV-specific dsRNA successfully reduced the negative effects of IAPV infection in 160 honey bee hives in two states (Florida and Pennsylvania) with very different climates and seasons. These results provide the first successful field demonstration of the use of RNAi as a large scale preventative treatment for an insect disease.
The antiviral effect of RNAi has also been successfully augmented for disease control in a non-insect arthropod, the cultured shrimp, L. vannamei. Diseases caused by viruses are economically devastating to the shrimp industry, and induced RNAi provides protection from a number of different viruses, including single- and double-stranded RNA viruses and a DNA virus (Bartholomay et al., 2012). This strategy enhances RNAi-based antiviral immunity, providing long-term, highly specific protection and a route for vaccination of cultured shrimp against viral diseases. For example, dsRNA designed to target the 5’ end of ORF1 in the genome of Infectious myonecrosis virus (IMNV) provides significant disease protection even 52 days after vaccination (Loy et al., 2012). More recently, the same RNAi trigger was used to provide therapeutic effect such that disease pathology resolved and 50% of animals survived if the RNAi trigger was provided within 48 hours post-infection (Loy et al., 2012). The outstanding challenge is a viable delivery strategy, because shrimp culture involves hundreds of thousands of animals in hectare-sized ponds.
4.3. The evolutionary stability of RNAi-based management of insect populations
The relationship between viruses and RNAi-based insect immunity is evolutionarily dynamic. This is indicated by both the presence of viral suppressors of RNAi (see above) and the positive selection on the genes contributing to RNAi-machinery interacting with siRNAs, but not the endogenous miRNAs (Obbard et al., 2006). We can, therefore, anticipate that insects, viruses and eukaryotic parasites will respond to strong selection exerted by RNAi-based control strategies. For example, insects that carry viruses with RNAi suppressors would be at a selective advantage on RNAi-protected crops, and RNAi-based prophylactics for honey bee colonies would select for viral pathogens with RNAi suppression. The RNAi suppression mechanisms that have evolved are not specific to a particular target sequence. This implies that resistance to a dsRNA specific to one gene cannot be prevented by pyramiding multiple genes with different function, nor overcome by switching to a different gene or gene set.
The genetic variation that exists within and among insect populations could also present a challenge to the application of RNAi for pest control, depending on the amount of mismatch present between the dsRNA and the target transcript. Furthermore, single nucleotide polymorphisms (SNPs) that result in lower effectiveness of the RNAi, could potentially be selected for and lead to the evolution of resistance. If such SNPs were synonymous they would be expected to have little or no fitness cost in the absence of the selecting agent (dsRNA), and resistance could evolve rapidly. However, the degree of mismatch (i.e. the number of SNPs) that would be needed to prevent RNAi from controlling a pest is not known. The long-term benefits of RNAi-based applications in insect pest management will require new and independent thought on effective resistance management strategies designed to minimize selective pressures and delay the evolution of resistance.
4.4. RNAi risks and regulation
The above examples offer clear evidence for potential applications for RNAi for the control of insect pests, manipulation of insect disease vectors, and management of beneficial insects, together with concerns about the stability of RNAi strategies in the face of selection for resistance. Overlying these considerations is a very real uncertainty regarding the environmental and ecological risks posed by these technologies. The Federal regulatory framework for estimating the ecological risks associated with RNAi technologies is still in development, and a number of critical gaps remain including potential toxicity to non-target organisms (see Section 4.1) , environmental fate, and importantly, the risk of resistance evolution in target pests (Section 4.3). Documenting efficacy of the technology is ongoing and regulatory considerations for RNAi-based insecticidal traits, such as the development of standardized environmental risk assessment, are still being developed (Auer and Frederick, 2009; http://cera-gmc.org/docs/cera_publications/pub_08_2011.pdf). Considerations of how to evaluate sequence specificity, environmental fate, and exposure of non-target organisms are still being developed. However, U.S. regulatory agencies such as the Environmental Protection Agency and the Department of Agriculture have provided preliminary assessments (http://cera-gmc.org/docs/cera_publications/pub_08_2011.pdf) suggesting that data requirements for RNAi traits may be reduced based primarily on the lack of a plant incorporated protectant, such as a protein toxin. There is also a lack of information on the risk of insect resistance to RNAi-mediated control that is a critical impediment to the development of an insect resistance management plan aimed at promoting a responsible and sustainable use of the technology. Insecticide resistance presents a major challenge for the sustainable control of pests. In the case of insects, pest species have found ways to evolve resistance to nearly every control strategy that has been used. Predictions that resistance could not develop to a new control strategy (e.g., Williams, 1967) have proven to be wrong time and time again.
5. Concluding comments
A decade of research on RNAi in insects has demonstrated the great power of the technology for discovery-led science and potential for improved management of insect populations. As the science has matured, it has equally become evident that RNAi is no panacea, but introduces a range of new conceptual and technological challenges for insect scientists. Insects vary widely in their amenability to RNAi, and no single protocol is suitable for all species. Against the backdrop of this functional diversity, it is unfortunate that there has been a dearth of systematic investigation of the mechanisms of RNAi in insects. We still have only a weak understanding of whether and how the RNAi signal is amplified in individual cells and disseminated between cells in insects. It is increasingly recognized that the caveats in our understanding of insect-specific mechanism are a major limitation to the implementation of RNAi. A priority for the future is for the insect research community to apply their persistence and ingenuity to solve the fundamentals of how insect RNAi works, in the context of the physiology of the insect body, and apply that to the pressing problems posed by pests and beneficial insects.
Highlights.
RNAi is a highly valuable tool for understanding gene function.
RNAi holds great potential for pest management.
RNAi efficiency varies among insect species and genes.
Development of RNAi protocols is a highly empirical process.
Systematic analysis of RNAi mechanisms in insects will facilitate future research.
Acknowledgements
We thank all of the speakers and participants at the 2012 ESA symposium “RNAi: The Power, the Promise and the Frustration”, from which the idea for this paper originated. This work was supported by the following grants: NIH R01 AI095842 to K.M., NSF 1114370 to L.C.B, and AFRI-NIFA NYW-2011-04650 (to A.E.D).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Molecular Biology. 2001;10:265–273. doi: 10.1046/j.1365-2583.2001.00267.x. [DOI] [PubMed] [Google Scholar]
- Allen ML, Walker WB., 3rd Saliva of Lygus lineolaris digests double stranded ribonucleic acids. Journal of Insect Physiology. 2012;58:391–396. doi: 10.1016/j.jinsphys.2011.12.014. [DOI] [PubMed] [Google Scholar]
- Amdam GV, Simões ZLP, Guidugli KR, Norberg K, Omholt SW. Disruption of vitellogenin gene function in adult honeybees by intra-abdominal injection of double-stranded RNA. BMC Biotechnology. 2003;3:1–000. doi: 10.1186/1472-6750-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakane Y, Baguinon MC, Jasrapuria S, Chaudhari S, Doyungan A, Kramer KJ, Muthukrishnan S, Beeman RW. Both UDP N-acetylglucosamine pyrophosphorylases of Tribolium castaneum are critical for molting, survival and fecundity. Insect Biochemistry and Molecular Biology. 2011;41:42–50. doi: 10.1016/j.ibmb.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, Pereira MH. RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochemistry and Molecular Biology. 2006;36:683–693. doi: 10.1016/j.ibmb.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimatsu Y, Kotani E, Sugimura Y, Furusawa T. Molecular characterization of a cDNA encoding extracellular dsRNase and its expression in the silkworm, Bombyx mori . Insect Biochemistry and Molecular Biology. 2007;37:176–183. doi: 10.1016/j.ibmb.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Aronstein K, Pankiw T, Saldivar E. SID-1 is implicated in systemic gene silencing in the honey bee. Journal of Apicultural Research. 2006;45:20–24. [Google Scholar]
- Asgari S. MicroRNA functions in insect. Insect Biochemistry and Molecular Biology. 2013;43:388–397. doi: 10.1016/j.ibmb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Attardo GM, Benoit JB, Michalkova V, Yang G, Roller L, Bohova J, Takác P, Aksoy S. Analysis of lipolysis underlying lactation in the tsetse fly, Glossina morsitans . Insect Biochemistry and Molecular Biology. 2012;42:360–370. doi: 10.1016/j.ibmb.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer C, Frederick R. Crop improvement using small RNAs: applications and predictive ecological risk assessments. Trends in Biotechnology. 2009;27:644–651. doi: 10.1016/j.tibtech.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Bartholomay LC, Loy DS, Dustin Loy J, Harris DL. Nucleic-acid based antivirals: augmenting RNA interference to ‘vaccinate’ Litopenaeus vannamei . Journal of Invertebrate Pathology. 2012;110:261–266. doi: 10.1016/j.jip.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, Waters AP, Cowman AF, Crabb BS, de Koning-Ward TF. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Research. 2009;37:3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, et al. Control of coleopteran insect pests through RNA interference. Nature Biotechnology. 2007;25:1322–1326. doi: 10.1038/nbt1359. [DOI] [PubMed] [Google Scholar]
- Belles X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annual Review of Entomology. 2010;55:111–128. doi: 10.1146/annurev-ento-112408-085301. [DOI] [PubMed] [Google Scholar]
- Berry B, Deddouche S, Kirschner D, Imler JL, Antoniewski C. Viral suppressors of RNA silencing hinder exogenous and endogenous small RNA pathways in Drosophila . PLoS ONE. 2009;4:e5866. doi: 10.1371/journal.pone.0005866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G, Shin SW, Cheon HM, Kokoza V, Raikhel AS. Transgenic alteration of Toll immune pathway in the female mosquito Aedes aegypti. Proc Natl Acad Sci U S A. 2005;102:13568–13573. doi: 10.1073/pnas.0502815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biessmann H, Andronopoulou E, Biessmann MR, Douris V, Dimitratos SD, Eliopoulos E, Guerin PM, Iatrou K, Justice RW, Kröber T, et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS One. 2010;5:e9471. doi: 10.1371/journal.pone.0009471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorow Y, Baskerville S, Maksimova E, Robinson K, Karpilow J, et al. 3' UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nature Methods. 2006;30:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiology. 2011;6:267–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandin S, Moita LF, Köcher T, Wilm M, Kafatos FC, Levashina EA. Reverse genetics in the mosquito Anopheles gambiae: targeted disruption of the Defensin gene. EMBO Reports. 2002;3:852–856. doi: 10.1093/embo-reports/kvf180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blandin SA, Wang-Sattler R, Lamacchia M, Gagneur J, Lycett G, Ning Y, Levashina EA, Steinmetz LM. Dissecting the genetic basis of resistance to malaria parasites in Anopheles gambiae . Science. 2009;326:147–150. doi: 10.1126/science.1175241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R, Ilagan O, Lawrence C, Levine S, Moar W, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte) PLoS ONE. 2012;7:e47534. doi: 10.1371/journal.pone.0047534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AE, Bugeon L, Crisanti A, Catteruccia F. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Research. 2003;31:e85. doi: 10.1093/nar/gng085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AE, Catteruccia F. Toward silencing the burden of malaria: progress and prospects for RNAi-based approaches. Biotechniques. 2006:38–44. doi: 10.2144/000112117. [DOI] [PubMed] [Google Scholar]
- Brown SJ, Mahaffey JP, Lorenzen MD, Denell RE, Mahaffey JW. Using RNAi to investigate orthologous homeotic gene function during development of distantly related insects. Evolution and Development. 1999;1:11–15. doi: 10.1046/j.1525-142x.1999.99013.x. [DOI] [PubMed] [Google Scholar]
- Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. The Plant Journal. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Keene KM, Brackney DE, Olson KE, Blair CD, Wilusz J, Foy BD. Aedes aegypti uses RNA interference in defense against Sindbis virus infection. BMC Microbiology. 2008;8:47. doi: 10.1186/1471-2180-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao JA, Lee JH, Chapados BR, Debler EW, Schneemann A, Williamson JR. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nature Structural and Molecular Biology. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nature Genetics. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiology. 2009;9:49. doi: 10.1186/1471-2180-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Friedrich M. Nymphal RNAi: systemic RNAi mediated gene knockdown in juvenile grasshopper. BMC Biotechnology. 2005;5:25. doi: 10.1186/1472-6750-5-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Das S, Cirimotich C, Souza-Neto JA, McLean KJ, Dimopoulos G. Engineered anopheles immunity to Plasmodium infection. PLoS Pathogens. 2011;7:e1002458. doi: 10.1371/journal.ppat.1002458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Schwarz RS. Bees brought to their knees: microbes affecting honey bee health. Trends in Microbiology. 2011;19:614–620. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Faria CA, Wackers FL, Pritchard J, Barrett DA, Turlings TC. High susceptibility of Bt maize to aphids enhances the performance of parasitoids of lepidopteran pests. PLoS ONE. 2007;2:e600. doi: 10.1371/journal.pone.0000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Franz AW, Jasinskiene N, Sanchez-Vargas I, Isaacs AT, Smith MR, Khoo CC, Heersink MS, James AA, Olson KE. Comparison of transgene expression in Aedes aegypti generated by mariner Mos1 transposition and PhiC31 site-directed recombination. Insect Molecular Biology. 2011;20:587–598. doi: 10.1111/j.1365-2583.2011.01089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz AWE, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, James AA, Olson KE. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti . Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbian Y, Maori E, Kalev H, Shafir S, Sela I. Bidirectional transfer of RNAi between honey bee and Varroa destructor: Varroa gene silencing reduces Varroa population. PLoS Pathogens. 2012;8:e1003035. doi: 10.1371/journal.ppat.1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbutt JS, Belles X, Richards EH, Reynolds SE. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica . Journal of Insect Physiology. 2013;59:171–178. doi: 10.1016/j.jinsphys.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Garbutt JS, Reynolds SE. Induction of RNA interference genes by double-stranded RNA; implications for susceptibility to RNA interference. Insect Biochemistry and Molecular Biology. 2012;42:621–628. doi: 10.1016/j.ibmb.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Ghanim M, Kontsedalov S, Czosnek H. Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius) Insect Biochemistry and Molecular Biology. 2007;37:732–738. doi: 10.1016/j.ibmb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nature Reviews Genetics. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A, Wright AJ, Hunter CP. SID-5 is an endosome-associated protein required for efficient systemic RNAi in C. elegans . Current Biology. 2012;22:1938–1943. doi: 10.1016/j.cub.2012.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn T, Boutros M. E-RNAi: a web application for the multi-species design of RNAi reagents--2010 update. Nucleic Acids Research. 2010;38:W332–W339. doi: 10.1093/nar/gkq317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liu J, Corey DR. Allele-selective inhibition of Huntingtin expression by switching to an miRNA-like RNAi mechanism. Chemical Biology. 2010;17:1183–1188. doi: 10.1016/j.chembiol.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter W, Ellis J, Vanengelsdorp D, Hayes J, Westervelt D, Glick E, Williams M, Sela I, Maori E, Pettis J, et al. Large-scale field application of RNAi technology reducing Israeli acute paralysis virus disease in honey bees (Apis mellifera, Hymenoptera: Apidae) PLoS Pathogens. 2010;6:e1001160. doi: 10.1371/journal.ppat.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter WB, Glick E, Paldi N, Bextine BR. Advances in RNA interference: dsRNA treatment in trees and grapevines for insect pest population suppression. Southwestern Entomologist. 2012;37:85–87. [Google Scholar]
- Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. Journal of Insect Physiology. 2010;56:227–235. doi: 10.1016/j.jinsphys.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Jackson A, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation of RNAi. Nature Biotechnology. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Joseph TT, Osman R. Convergent transmission of RNAi guide-target mismatch information across argonaute internal allosteric network. PLOS Computational Biology. 2012a;8:e1002693. doi: 10.1371/journal.pcbi.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph TT, Osman R. Thermodynamic basis of selectivity in guide-target-mismatched RNA interference. Proteins: Structure, Function, Bioinformatics. 2012b;80:1283–1298. doi: 10.1002/prot.24025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Troiano E, Mather TN. Functional genomics tool: gene silencing in Ixodes scapularis eggs and nymphs by electroporated dsRNA. BMC Biotechnology. 2010;10:1. doi: 10.1186/1472-6750-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans . Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- Khoo CC, Piper J, Sanchez-Vargas I, Olson KE, Franz AW. The RNA interference pathway affects midgut infection- and escape barriers for Sindbis virus in Aedes aegypti . BMC Microbiology. 2010;10:130. doi: 10.1186/1471-2180-10-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzmann P, Schwirz J, Schmitt-Engel C, Bucher G. RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genomics. 2013 doi: 10.1186/1471-2164-14-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Pandit SS, Baldwin IT. Tobacco rattle virus vector: A rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS ONE. 2012;7:e31347. doi: 10.1371/journal.pone.0031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chougule NP, Bonning BC. Interaction of the Bacillus thuringiensis delta endotoxins Cry1Ac and Cry3Aa with the gut of the pea aphid, Acyrthosiphon pisum (Harris) Journal of Invertebrate Pathology. 2011;107:69–78. doi: 10.1016/j.jip.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Liu J, Swevers L, Iatrou K, Huvenne H, Smagghe G. Bombyx mori DNA/RNA nonspecific nuclease: expression of isoforms in insect culture cells, subcellular localization and functional assays. Journal of Insect Physiology. 2012;58:1166–1176. doi: 10.1016/j.jinsphys.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Liu J, Smagghe G, Swevers L. Transcriptional response of BmToll9-1 and RNAi machinery genes to exogenous dsRNA in the midgut of Bombyx mori . Journal of Insect Physiology. 2013;59:646–654. doi: 10.1016/j.jinsphys.2013.03.013. [DOI] [PubMed] [Google Scholar]
- Lomazzo E, Hussmann GP, Wolfe BB, Yasuda RP, Perry DC, Kellar KJ. Effects of chronic nicotine on heteromeric neuronal nicotinic receptors in rat primary cultured neurons. Journal of Neurochemistry. 2011;119:153–164. doi: 10.1111/j.1471-4159.2011.07408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Martinez G, Meuti M, Denlinger DL. Rehydration driven RNAi: a novel approach for effectively delivering dsRNA to mosquito larvae. Journal of Medical Entomology. 2012;49:215–218. doi: 10.1603/me11122. [DOI] [PubMed] [Google Scholar]
- Loy DL, Mogler MA, Loy DS, Janke B, Kamrud K, Scura ED, Harris DLH, Bartholomay LC. Double-stranded RNA provides sequence dependent protection against infectious myonecrosis virus in Litopenaeus vannamei . Journal of General Virology. 2012;93:880–888. doi: 10.1099/vir.0.038653-0. [DOI] [PubMed] [Google Scholar]
- Lu Y, Wu K, Jiang Y, Xia B, Li P, Feng H, Wyckhuys KA, Guo Y. Mirid bug outbreaks in multiple crops correlated with wide-scale adoption of Bt cotton in China. Science. 2010;328:1151–1154. doi: 10.1126/science.1187881. [DOI] [PubMed] [Google Scholar]
- Luo Y, Wang X, Yu D, Kang L. The SID-1 double-stranded RNA transporter is not required for systemic RNAi in the migratory locust. RNA Biology. 2012;9:663–671. doi: 10.4161/rna.19986. [DOI] [PubMed] [Google Scholar]
- Lycett GJ, McLaughlin LA, Ranson H, Hemingway J, Kafatos FC, Loukeris TG, Paine MJ. Anopheles gambiae P450 reductase is highly expressed in oenocytes and in vivo knockdown increases permethrin susceptibility. Insect Molecular Biology. 2006;15:321–327. doi: 10.1111/j.1365-2583.2006.00647.x. [DOI] [PubMed] [Google Scholar]
- Majerowicz D, Alves-Bezerra M, Logullo R, Fonseca-de-Souza AL, Mey-erFernandes JR, Braz GR, Gondim KC. Looking for reference genes for real-time quantitative PCR experiments in Rhodnius prolixus (Hemiptera: Reduviidae) Insect Molecular Biology. 2011;20:713–722. doi: 10.1111/j.1365-2583.2011.01101.x. [DOI] [PubMed] [Google Scholar]
- Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature Biotechnology. 2007;25:1307–1313. doi: 10.1038/nbt1352. [DOI] [PubMed] [Google Scholar]
- Mao YB, Tao XY, Xue XY, Wang LJ, Chen XY. Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Research. 2011;20:665–673. doi: 10.1007/s11248-010-9450-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YB, Xue XY, Tao XY, Yang CQ, Wang LJ, Chen XY. Cysteine protease enhances plant-mediated bollworm RNA interference. Plant Molecular Biology. 2013 doi: 10.1007/s11103-013-0030-7. [Epub 4 March] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Zeng F. Feeding-based RNA interference of a gap gene is lethal to the pea aphid, Acyrthosiphon pisum . PLoS ONE. 2012;7:e48718. doi: 10.1371/journal.pone.0048718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maori E, Paldi N, Shafir S, Kalev H, Tsur E, Glick E, Sela I. IAPV, a bee-affecting virus associated with Colony Collapse Disorder can be silenced by dsRNA ingestion. Insect Molecular Biology. 2009;18:55–60. doi: 10.1111/j.1365-2583.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- Martin-Hernandez R, Botias C, Barrios L, Martinez-Salvador A, Meana A, Mayack C, Higes M. Comparison of the energetic stress associated with experimental Nosema ceranae and Nosema apis infection of honeybees (Apis mellifera) Parasitology Research. 2011;109:605–612. doi: 10.1007/s00436-011-2292-9. [DOI] [PubMed] [Google Scholar]
- Mathur G, Sanchez-Vargas I, Alvarez D, Olson KE, Marinotti O, James AA. Transgene-mediated suppression of dengue viruses in the salivary glands of the yellow fever mosquito, Aedes aegypti . Insect Molecular Biology. 2010;19:753–763. doi: 10.1111/j.1365-2583.2010.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matranga C, Zamore PD. Small silencing RNAs. Current Biology. 2007;17:R789–R793. doi: 10.1016/j.cub.2007.07.014. [DOI] [PubMed] [Google Scholar]
- McCarroll J, Kavallaris M. Nanoparticle delivery of siRNAs as a novel therapeutic for human disease. Australian Biochemist. 2012;43:9–20. [Google Scholar]
- Merkling SH, van Rij RP. Beyond RNAi: Antiviral defense strategies in Drosophila and mosquito. Journal of Insect Physiology. 2013;59:159–170. doi: 10.1016/j.jinsphys.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Michel K, Budd A, Pinto S, Gibson TJ, Kafatos FC. Anopheles gambiae SRPN2 faciliates midgut invasion by the malaria parasite Plasmodium berghei . EMBO Reports. 2005;6:891–897. doi: 10.1038/sj.embor.7400478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SC, Brown SJ, Tomoyasu Y. Larval RNAi in Drosophila? Developmental Genes and Evolution. 2008;218:505–510. doi: 10.1007/s00427-008-0238-8. [DOI] [PubMed] [Google Scholar]
- Mutti NS, Park Y, Reese JC, Reeck GR. RNAi knockdown of a salivary transcript leading to lethality in the pea aphid, Acyrthosiphon pisum . Journal of Insect Science. 2006;6:1–7. doi: 10.1673/031.006.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Current Biology. 2006;16:580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans . Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Paldi N, Glick E, Oliva M, Zilberberg Y, Aubin L, Pettis J, Chen Y, Evans JD. Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Applied and Environmental Microbiology. 2010;76:5960–5964. doi: 10.1128/AEM.01067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry T, McKenzie JA, Batterham P. A Dalpha6 knockout strain of Drosophila melanogaster confers a high level of resistance to spinosad. Insect Biochemistry and Molecular Biology. 2007;37:184–188. doi: 10.1016/j.ibmb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Pitino M, Coleman AD, Maffei ME, Ridout CJ, Hogenhout SA. Silencing of aphid genes by dsRNA feeding from plants. PLoS ONE. 2011;6:e25709. doi: 10.1371/journal.pone.0025709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster . Journal of Insect Physiology. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
- Price DR, Gatehouse JA. RNAi-mediated crop protection against insects. Trends in Biotechnology. 2008;26:393–400. doi: 10.1016/j.tibtech.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Pridgeon JW, Zhao L, Becnel JJ, Strickman DA, Clark GG, Linthicum KJ. Topically applied AaeIAP1 double-stranded RNA kills female adults of Aedes aegypti . Journal of Medical Entomology. 2008;45:414–420. doi: 10.1603/0022-2585(2008)45[414:taadrk]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ramaseshadri P, Segers G, Flannagan R, Wiggins E, Clinton W, Ilagan O, McNulty B, Clark T, Bolognesi R. Physiological and cellular responses caused by RNAi-mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS ONE. 2013;8:e54270. doi: 10.1371/journal.pone.0054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich FD, Scott JG. Limitations of RNAi of a6 nicotinic acetylcholine receptor subunits for assessing the in vivo sensitivity to spinosad. Insect Science. 2013;20:101–108. doi: 10.1111/j.1744-7917.2012.01523.x. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Carré C, Mugat B, Szymczak D, Lepesant JA, Antoniewski C. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA. 2003;9:299–308. doi: 10.1261/rna.2154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharlaken B, de Graff D, Goossens K, Brunain M, Peelman L, Jacobs F. Reference gene selection for insect expression studies using quantitative real-time PCR: the head of the honeybee, Apis mellifera, after a bacterial challenge. Journal of Insect Science. 2008;8:223–237. [Google Scholar]
- Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. Journal of Virology. 2012;86:13486–13500. doi: 10.1128/JVI.01104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends in Ecology and Evolution. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakesby AJ, Wallace IS, Isaacs HV, Pritchard J, Roberts DM, Douglas AE. A water-specific aquaporin involved in aphid osmoregulation. Insect Biochemistry and Molecular Biology. 2009;39:1–10. doi: 10.1016/j.ibmb.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans . Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Terenius O, Papanicolaou A, Garbutt JS, Eleftherianos I, Huvenne H, Kanginakudru S, Albrechtsen M, An C, Aymeric JL, Barthel A, et al. RNA interference in Lepidoptera: an overview of successful and unsuccessful studies and implications for experimental design. Journal of Insect Physiology. 2011;57:231–245. doi: 10.1016/j.jinsphys.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G. Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium . Genome Biol. 2008;9:R10. doi: 10.1186/gb-2008-9-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, Ramet M. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. Journal of Biological Chemistry. 2006;281:14370–14375. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- Valdes VJ, Athie A, Salinas LS, Navarro RE, Vaca L. CUP-1 is a novel protein involved in dietary cholesterol uptake in Caenorhabditis elegans . PLoS ONE. 2012;7:e33962. doi: 10.1371/journal.pone.0033962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vienne A, Shiina T, Abi-Rached L, Danchin E, Vitiello V, Cartault F, Inoko H, Pontarotti P. Evolution of the proto-MHC ancestral region: more evidence for the plesiomorphic organisation of human chromosome 9q34 region. Immunogenetics. 2003;55:429–436. doi: 10.1007/s00251-003-0601-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Li H, Miao X. Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS ONE. 2011;6:e18644. doi: 10.1371/journal.pone.0018644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whangbo JS, Hunter CP. Environmental RNA interference. Trends in Genetics. 2008;24:297–305. doi: 10.1016/j.tig.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Whyard S, Singh AD, Wong S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochemistry and Molecular Biology. 2009;39:824–832. doi: 10.1016/j.ibmb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Winston WM, Molodowitch C, Hunter CP. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science. 2002;295:2456–2459. doi: 10.1126/science.1068836. [DOI] [PubMed] [Google Scholar]
- Winston WM, Sutherlin M, Wright AJ, Feinberg EH, Hunter CP. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10565–10570. doi: 10.1073/pnas.0611282104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Ma H, Ye C, Ramirez D, Chen S, Montoya J, Phankar P, Wang XA, Manjunath N. Improved siRNA/shRNA functionality by mismatched duplex. PLoS ONE. 2011;6:e28580. doi: 10.1371/journal.pone.0028580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuriyanghan H, Rosa C, Falk BW. Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli . PLoS ONE. 2011;6:e27736. doi: 10.1371/journal.pone.0027736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Fan B, Chen C, Chen Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:6516–6521. doi: 10.1073/pnas.111440998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Han Z. Cloning and phylogenetic analysis of sid-1-like genes from aphids. Journal of Insect Science. 2008;8:1–6. doi: 10.1673/031.008.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue XY, Mao YB, Tao XY, Huang YP, Chen XY. New approaches to agricultural insect pest control based on RNA interference. Advances in Insect Physiology. 2012;42:73–117. [Google Scholar]
- Yu N, Christiaens O, Liu JM, Niu J, Cappelle K, Caccia S, Huvenne H, Smagghe G. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Science. 2013;20:4–14. doi: 10.1111/j.1744-7917.2012.01534.x. [DOI] [PubMed] [Google Scholar]
- Zha W, Peng X, Chen R, Du B, Zhu L, He G. Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens . PLoS ONE. 2011;6:e20504. doi: 10.1371/journal.pone.0020504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang J, Zhu KY. Chitosan/double-stranded RNA nanoparticle-mediated RNA interference to silence chitin synthase genes through larval feeding in the African malaria mosquito (Anopheles gambiae) Insect Molecular Biology. 2010;19:683–693. doi: 10.1111/j.1365-2583.2010.01029.x. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wheeler MM, Oi FM, Scharf ME. RNA interference in the termite Reticulitermes flavipes through ingestion of double-stranded RNA. Insect Biochemistry and Molecular Biology. 2008;38:805–815. doi: 10.1016/j.ibmb.2008.05.005. [DOI] [PubMed] [Google Scholar]



