Abstract
Spinal muscular atrophy (SMA) is a neurodegenerative disorder that is characterized by progressive loss of motor neuron function. It is caused by the homozygous loss of the SMN1 (survival of motor neuron 1) gene and a decrease in full-length SMN protein. SMN2 is a nearly identical homolog of SMN1 that, due to alternative splicing, expresses predominantly truncated SMN protein. SMN2 represents an enticing therapeutic target. Increasing expression of full-length SMN from the SMN2 gene might represent a treatment for SMA. We describe a newly designed cell-based reporter assay that faithfully and reproducibly measures full-length SMN expression from the SMN2 gene. This reporter can detect increases of SMN protein by an array of compounds previously shown to regulate SMN2 expression and by the overexpression of proteins that modulate SMN2 splicing. It also can be used to evaluate changes at both the transcriptional and splicing level. This assay can be a valuable tool for the identification of novel compounds that increase SMN2 protein levels and the optimization of compounds already known to modulate SMN2 expression. We present here preliminary data from a high-throughput screen using this assay to identify novel compounds that increase expression of SMN2.
Keywords: spinal muscular atrophy, survival of motor neuron, SMN1, SMN2, cell-based assay, high-content screening, HTS
Introduction
Spinal muscular atrophy (SMA) is a neurological disorder that results from loss of function of the anterior horn cells in the spinal cord,1 manifesting as progressive motor weakness, muscle wasting, and paralysis. SMA is caused by insufficient levels of the survival motor neuron (SMN) protein. The SMN locus on chromosome 5q13 contains two inverted copies of SMN called SMN1 and SMN2.2,3 Most cases of SMA harbor homozygous deletions of the SMN1 gene and retain at least one copy of SMN2 (reviewed in4,5). With a carrier rate of about 1 in 40, SMA is estimated to be the most frequent genetic cause of infant mortality.6,7
SMN2 is a gene duplication of SMN1 with the same predicted amino acid coding capacity. The nucleotide sequences of SMN1 and SMN2 are nearly identical. A critical difference is a C to T transition at the +6 position in exon 7, which dramatically influences the splicing pattern in these genes. Greater than 90% of SMN1 transcripts include exon 7, whereas there is less than 15% exon 7 inclusion in SMN2 transcripts.8 This alternatively spliced product produces a truncated and unstable form of the SMN protein.9 Any increase in the inclusion of exon 7 in SMN2 transcripts would result in higher levels of full-length SMN protein. In fact, any treatment that increases the amount of full-length SMN2 mRNA should result in increased levels of SMN protein. Based on this premise, we developed an in vivo screen that can detect increases in full-length exon 7–included SMN2 transcripts.
We previously constructed a splicing reporter that fused SMN exons 6, 7, and 8 and their introns in frame with firefly luciferase and was expressed from a cytomegalovirus (CMV) promoter in C33a cells.10 We found that this reporter could recapitulate changes in splicing observed with overexpression of the splicing factor Tra2β.10,11 This assay was used successfully to identify compounds that increase the amount of full-length transcript produced by the SMN2 gene and SMN protein in fibroblasts isolated from an SMA patient.12,13 Another study used an SMN2 promoter–based reporter to screen a library of small molecules for the ability to increase SMN expression levels in NSC34 cells.14 This reporter measured only SMN2-specific transcription and lacked any SMN gene sequence.
It has been reported that histone deacetylase (HDAC) inhibitors such as sodium butyrate,15 trichostatin A (TSA),16 valproic acid (VPA),17 suberoylanilide hydroxamic acid (SAHA),18 and LBH58919 increase SMN transcription and inclusion of exon 7. For many of these HDACs, relatively high (micromolar or millimolar) concentrations of these compounds are necessary. These activators are nonspecific and will alter transcription of many genes, so long-term safety has been questioned. However, type I severe SMA is fatal, and short-term administration of such compounds may be beneficial. These results serve as proof of principle that SMN reporters can be used as tools for identifying and characterizing protein factors and chemical compounds that increase expression of full-length SMN2 transcripts. Although these successes are promising, there is a clear need for more drug candidates.
Our first-generation splicing assay had low signal intensity, high basal expression of SMN-luciferase, and became less responsive with serial cell passage.10 On this basis, we concluded that these cells were unsuitable for high-throughput screening (HTS). We redesigned our reporter system and built a more stable and reproducible assay that could be used for HTS. Here, we report a clonal second-generation SMN-luciferase reporter cell line that combines the strengths of both the promoter-based assay and our previous splicing reporter. We also present preliminary results from an HTS to identify novel compounds that increase SMN2 expression using this cell-based SMN-luciferase reporter assay.
This assay is much more robust, has lower well-to-well variation, and displays more stable luciferase expression that does not change with serial passage. It also faithfully reproduces the reported activity of an array of drug-like compounds that have been shown to increase SMN expression levels. This reporter can detect changes in SMN2 levels in response to overexpression of splicing factors such as Tra2β. This assay is a vast improvement on the previous generation of reporters and represents a valuable tool for further identification and characterization of compounds that increase expression of full-length SMN protein from the SMN2 gene.
Materials and Methods
Cloning
The luciferase minigene from our previous reporter vectors SMN1-luc (T-luc) or SMN2-luc (C-luc)10 was shortened by digestion with Sma I and Swa I to remove 2 kB from intron 6. The SMN exon 1–5 fragment was generated by PCR from human cDNA (exon 1 forward: ccacaaatgtgggagggcgataacc and exon 6 reverse: tatctcgagtggtccagaaggaaatggaggcagcc). The SMN promoter elements were from p3.4T and p3.4C SMN.20 These were combined into pIRES cloning vector (BD Clontech, Mountain View, CA) at the multiple cloning site. The entire reporter fragment was excised from pIRES and ligated into a pCEP4 (Invitrogen, Carlsbad, CA) plasmid that also expressed renilla luciferase from nucleotides 299–1259 of phRL-null (Promega, Madison, WI) from the CMV promoter.
Cell Culture
Cells were incubated at 37 °C with 5% CO2. HEK-293 cells were cultured in D-MEM (Gibco 11995) with 10% fetal bovine serum (FBS; Atlas) and 1× pen-strep (Gibco 15140). Reporter cell lines containing SMN1, SMN2, or control luciferase reporter were selected and maintained in D-MEM with 10% FBS and 1× pen-strep with 200 µg/mL hygromycin B (Invitrogen 10687-010). 3813 and 3814 fibroblasts were cultured in D-MEM (Gibco 11995) with 15% fetal bovine serum (FBS; Atlas) and 1× pen-strep (Gibco 15140).
Luciferase Reporter Assay
The reporter cell lines were plated at 50 000 cells per well in 96-well plates and incubated overnight. Compounds were added to each well and incubated at 37 °C overnight. The final DMSO concentration was 0.1%. Luciferase activity was assayed with either SteadyGlo (Promega E2510) or DualGlo (Promega E2920) luciferase using the Perkin Elmer Envision multilabel reader. For detailed assay conditions, see Table 1. All data points were transformed from counts per second (CPS) to percentage increase over basal expression in the treated control wells (DMSO or H2O as appropriate).
Table 1.
SMN-Luciferase Standard Conditions: 96-Well Format
| Sequence | Parameter | Value | Description |
|---|---|---|---|
| 1 | Cells | 100 µL | 50 000 cells/well 96 TC–treated white plate |
| 2 | Time | 24 h | 37 °C 5% CO2 |
| 3 | Compound | 100 µL | With compound 2× concentration |
| 4 | Time | 24 h | 37 °C 5% CO2 |
| 5 | Remove media from wells | ||
| 6 | Reagent | 30 µL | SteadyGlo or DualGlo reagent (Promega) |
| 7 | Time | 30 s | Room temperature |
| 8 | Detector | 1.0 s integration | Perkin Elmer Envision |
Protein Detection
For analysis of SMN-luciferase fusion, cells were treated with compound or DMSO for 24 h. Cells were lysed with protein lysis buffer (100 mM Tris pH 8.0, 100 mM NaCl, 0.1% NP-40, 8.0 M urea, and protease inhibitor). Each sample was separated on a 10% SDS-page gel, transferred to Immobilon-P membrane (Millipore IVPH00010), and blotted for the SMN-luciferase fusion with anti-luciferase antibody (Promega, G7541), SMN with the 4f11 mouse monoclonal antibody,21 HA-tag with 12CA5 monoclonal, actin (Sigma A2066), or a-tubulin (DM1a; Sigma T6199).
For detection of SMN protein in patient fibroblasts, 8000 cells per cm2 were plated 24 h prior to drug addition. Fresh media and compound were added every 24 h. After 72 h, cells were harvested, washed with cold phosphate-buffered saline, and lysed as above. We have determined that 10 µg total protein per lane is within the linear range for immunoblot detection of SMN and a-tubulin. Western blots were probed for SMN with the 4f11 mouse monoclonal antibody and a-tubulin.
Quantification of protein was performed with the Fujifilm LAS-4000 Multifunctional Imaging System. The signal intensity was measured for each band on an immunoblot, normalized to the loading control, and the fold increase was determined in relation to the appropriate DMSO-treated control.
Overexpression Assays
Cells were plated at a density of 2 × 106 per 60 cm dish and incubated overnight. Cells were transfected with up to 6 µg of HA-tagged expression vector using Fugene 6 at a 3:1 fugene:DNA ratio and incubated overnight. After 24 h, cells were replated at 1 × 105 cells per well in a 96-well plate or 1 × 106 cells per well in 6-well dishes. Cells were tested 24 to 48 h later. Luciferase was assayed using DualGlo luciferase as described above. Protein lysates and RNA samples were collected using protein lysis buffer or Trizol, respectively. Protein was analyzed by Western blot, and RNA was analyzed by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR).
PCR and RT-PCR
Compounds were tested at three concentrations that display maximal activity in the luciferase assay. Cells were treated as described above for the luciferase assay. Cells were harvested by trypsinization, neutralized with trypsin inhibitor, and washed. RNA was isolated from the cells using Trizol Reagent (Invitrogen 15596-026). cDNA was generated using the Improm-II Reverse Transcription System (Promega A3801).
The forward primer pair recognizes the exon 5–6 junction, which includes a restriction site that was engineered into the reporter and will exclude amplification of endogenous SMN mRNA. The reverse primers recognize either exon 7 or luciferase for detection of full-length or total SMN-luciferase transcripts, respectively. For a reference control, we amplified cDNA from the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primers used include SMN exon5-forward (catttccttctggaccactcgag), luciferase-reverse (atagcttctgccaaccgaacgg), exon7-reverse (taaggaatgtgagcaccttccttc), GAPDH-reverse (G3A) (tccaccaccctgttgctgta), and GAPDH-forward (G3S) (accacagtccatgccatcac). qPCR was performed as described in the protocol for iQ SybrGreen Supermix (BioRad 170–8882) using an Eppendorf Mastercycler ep realplex4 Thermo Cycler. Reactions were incubated for a 10-min 94 °C hot start followed by 45 cycles of the following: 94 °C for 45 s, 60 °C for 15 s, and 72 °C for 45 s. Melting curves for each reaction were obtained. Each sample was assayed in triplicate, and every plate contained a five-point cDNA dilution course to calculate amplification efficiency for each primer pair. The Pfaffl method was used to determine the change in transcript levels relative to the DMSO and normalized to GAPDH.22
Screening Protocol
Cells were plated in phenol red-free D-MEM (Gibco 21063) with sodium pyruvate (Gibco 11360) and 10% FBS in the absence of hygromycin B and pen-strep and allowed to adhere 1 h prior to addition of compound. Each compound was added to a single well with a Beckman Coulter Biomek FX 384 to a final concentration of approximately 2 µM from compound stocks, based on an average molecular weight of 500. The final DMSO concentration in test and control wells was 0.13%. Plates were sealed with porous paper tape and incubated for 24 h. Luciferase expression was measured using the SteadyLite luciferase (Perkin Elmer 6016981) substrate and the Molecular Devices LJL Analyst HT and read for CPS at an integration time of 100 000 µS. For comparison, data points were transformed from CPS to percentage activation over basal expression in the DMSO-treated control wells. This is summarized in Table 2.
Table 2.
SMN2-Luciferase Conditions for High-Throughput Screening: 384-Well Format
| Sequence | Parameter | Value | Description |
|---|---|---|---|
| 1 | Cells | 40 µL | 10 000 cells/well 384 TC–treated white plate |
| 2 | Time | 1 h | 37 °C 5% CO2 |
| 3 | Compound | 2.2 mM | Final of 0.13% DMSO |
| 4 | Time | 25 h | 37 °C 5% CO2 |
| 5 | Reagent | 25 µL | Perkin Elmer SteadyLite |
| 6 | Time | 30 min | Room temperature |
| 7 | Detector | 0.1 s integration | Molecular Devices LJL Analyst HT |
Library Composition
The compound library consisted of 115 000 small molecules, including compounds approved by the Food and Drug Administration, a purified natural products library, compounds purchased from Peakdale (High Peak, UK), Maybridge Plc. (Cornwall, UK), Cerep (Paris, France), Bionet Research Ltd. (Cornwall, UK), Prestwick (Ilkirch, France), Specs and Biospecs (CP Rijswijk, the Netherlands), ENAMINE (Kiev, Ukraine), I.F. Lab LTD (Burlington, Canada), and Chemical Diversity Labs (San Diego, CA), and small molecules from academic institutions. Compounds were selected from vendors by applying a series of filters including clogP and predicted solubility. All of the small molecules generally adhere to Lipinski’s rules23 and contain a low proportion of known toxicophores (i.e., Michael acceptors and alkylating agents) and unwanted functionalities (i.e., imines, thiols, and quarternary amines) and have been optimized for molecular diversity. Compound source plates for the assay were prepared by spotting 0.4 µL of 1.67 mM compound in DMSO in each well of a Greiner 384-well plate, with columns 23 and 24 spotted with neat DMSO for positive and negative controls. These were then sealed with aluminum plate seals and stored at −20 °C.
Results
Development of a New Reporter
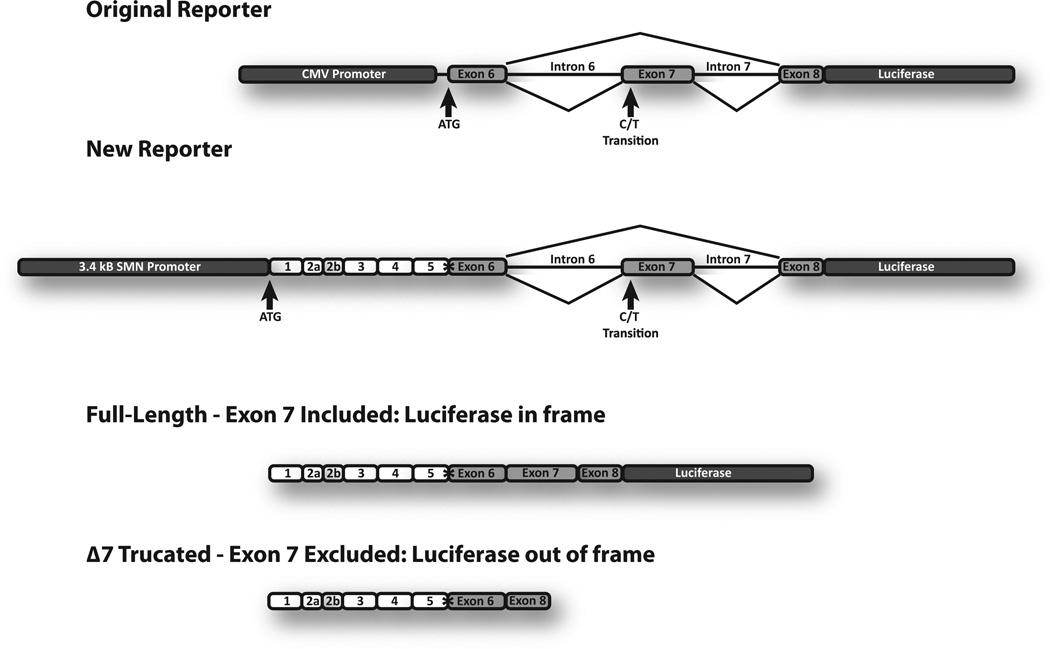
Over time, the original C33a reporter cell lines10 displayed a decrease in the difference in luciferase signal from SMN1 and SMN2 (Supplemental Figure 1) and showed inconsistent responses to treatment with drugs known to increase SMN expression (data not shown). These original reporters were also driven by the CMV promoter and displayed much higher basal levels of SMN1- and SMN2-luciferase activity in the absence of treatment (Supplemental Figure 1), which could diminish the reporters’ ability to detect compounds that are less potent and more selective. To address this, a new reporter assay was designed that would be more responsive to molecular cues that regulate the levels of SMN expression through multiple pathways. The SMN promoter and exon 7 splicing cassette were combined into a single construct to simultaneously identify compounds that increase SMN transcription or exon 7 inclusion (Fig. 1). The presence of the native SMN promoter may also influence recruitment of splicing factors to the transcript and better reproduce the context in which the endogenous gene is processed and expressed. Because a subset of compounds might stabilize the SMN RNA or protein, for example, by interfering with its metabolism or ubiquitination, exons 1 to 5 were included in the new reporter. This reporter produces a full-length SMN-luciferase fusion protein that should be regulated and metabolized in a manner that is more consistent with the endogenous SMN protein. The entire reporter was cloned into an Epstein Barr virus (EBV) vector that is maintained autonomously as stable episomes in some human cell lines.
Figure 1.
SMN-reporter mini-genes. The top diagram illustrates the changes made to the new reporter (see text). The asterisk denotes a restriction site included during cloning that is unique to the SMN reporter gene. These reporters produce an SMN-luciferase fusion protein when exon 7 is included (third diagram) but not when exon 7 is skipped (bottom diagram); ~90% of mRNA from the SMN2-reporter mini-gene skips exon 7.
To summarize, our new assay design: (1) replaced the CMV promoter with 3.4 kb of the SMN1 or SMN2 promoters with an intact transcription start site; (2) included the cDNA for exons 1 to 5 following the authentic promoter and translational ATG; (3) deleted a portion of intron 6 since the size of the entire 6 kb intron complicated cloning; (4) included the exon 7 splicing cassette with the firefly luciferase reporter; (5) cloned renilla luciferase expressed from the CMV promoter into the construct for monitoring copy number, cell viability, and specificity of the transcriptional effects; and (6) transferred the 9 kb reporter SMN1 and SMN2 cassettes into EBV ori-based pCEP4. The result is reporters with sequences from SMN1 and SMN2 in the context of their respective promoters. These constructs, each ~19 kb, will produce increased SMN-luciferase fusion protein if there is an induction of (1) transcription from the SMN promoter, (2) inclusion of exon 7, or (3) stabilization of the SMN-luciferase mRNA or fusion protein. Not only will this reporter be able to identify potential positive modifiers of SMN expression, but the design of the screen and the renilla control will also reduce the likelihood of selecting compounds that are toxic or cause nonspecific increases in transcription. These reporters were transfected into HEK 293 cells. These human cells are easy to culture and expand, highly transfectable, and maintain EBV-based plasmids extrachromosomally. Stable clonal cell lines were isolated that express the full-length SMN-luciferase protein as appropriate in both the SMN1-luc or SMN2-luc reporters.
Analysis of SMN1-luc and SMN2-luc Clonal Cell Lines Population
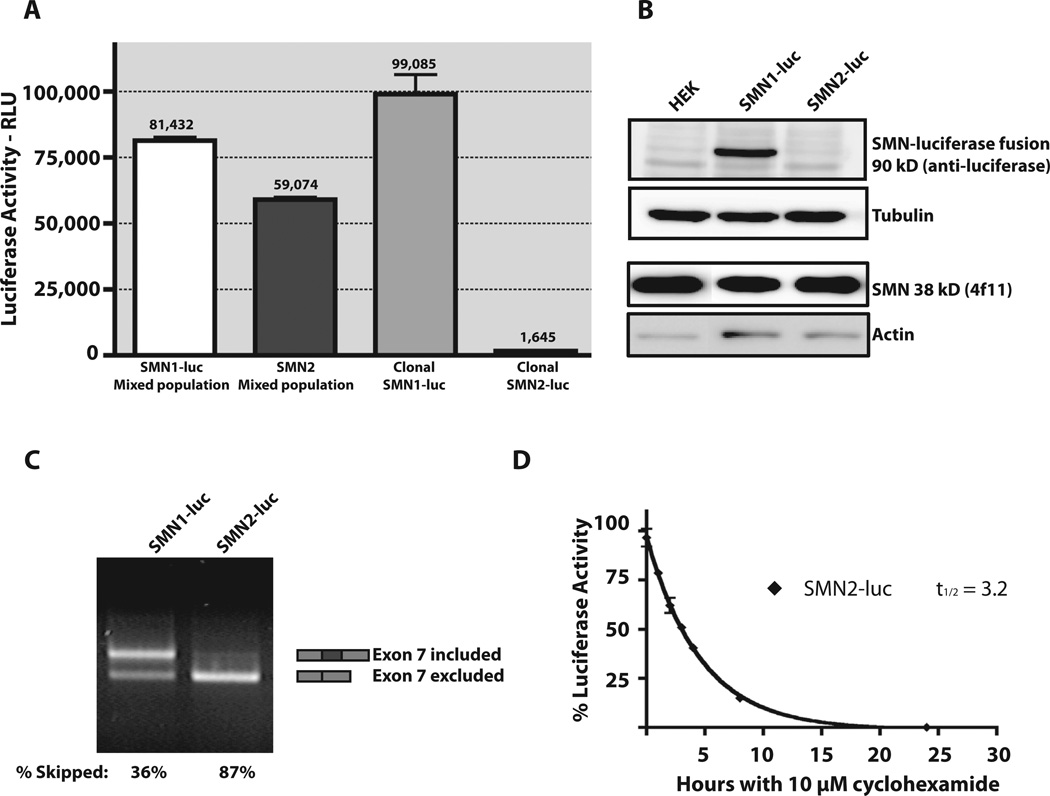
Luciferase expression was analyzed in the stable HEK293 cell lines containing the SMN1- and SMN2-luciferase reporters. In the initial stable mixed cell populations, mixed SMN1-luc cells displayed 30% more luciferase activity than the mixed SMN2-luc cells. A pair of clonal cell lines was isolated, clonal SMN1-luc and clonal-SMN2-luc, in which the clonal-SMN1-luc cell line has 50-fold higher luciferase activity when compared with clonal-SMN2-luc cell line (Fig. 2A). This range of expression provides a large window for potential activation of SMN2 expression with drug treatment. Because these reporters were designed for use in either high- or low-throughput screens, the dynamic range of activation is very important. The selection of SMN2-luc clones that maintain low levels of basal SMN-luciferase expression was instrumental in establishing this range of activation and was a dramatic improvement over not only the original C33a–based reporter cell lines but also the mix population HEK293 SMN1-luc and SMN2-luc cell lines (Supplemental Figure 1).
Figure 2.
Reporter assay validation. (A) Comparison of luciferase activity for equivalent numbers of mixed population SMN1-luc, mixed population SMN2-luc, clonal SMN1-luc, and clonal SMN2-luc cells. Luciferase activity, scored as relative light units (RLU). (B) Detection of SMN-luciferase fusion protein in HEK parent, SMN1-luc, and SMN2-luc reporter cells. Lysates were blotted for the fusion protein with an anti-luciferase antibody and endogenous SMN and compared with actin and tubulin. (C) End point RT-PCR was used to compare mRNA species of SMN1-luc and SMN2-luc cells. Primer pairs were designed to amplify both full-length and exon 7–excluded SMN-luciferase mRNA. Percentage inclusion was determined by comparing band intensities using Quantity One software. (D) SMN-luciferase fusion stability was determined by treating cells with 10 µM cycloheximide and measuring luciferase activity at various time points. Data were plotted using Prism4 (GraphPad Software Inc.), and nonlinear regression was used to determine protein half-life.
Expression of the SMN-luciferase protein fusion was confirmed by Western blot (Fig. 2B). All cells express endogenous SMN (38 kD), but only the SMN1-luc cell line expresses detectable levels of the SMN-luciferase fusion protein. The SMN-luciferase fusion protein could be detected only in the SMN2-luc cell line upon induction with compounds that increase SMN expression (SAHA and sodium butyrate in Fig. 3B).
Figure 3.
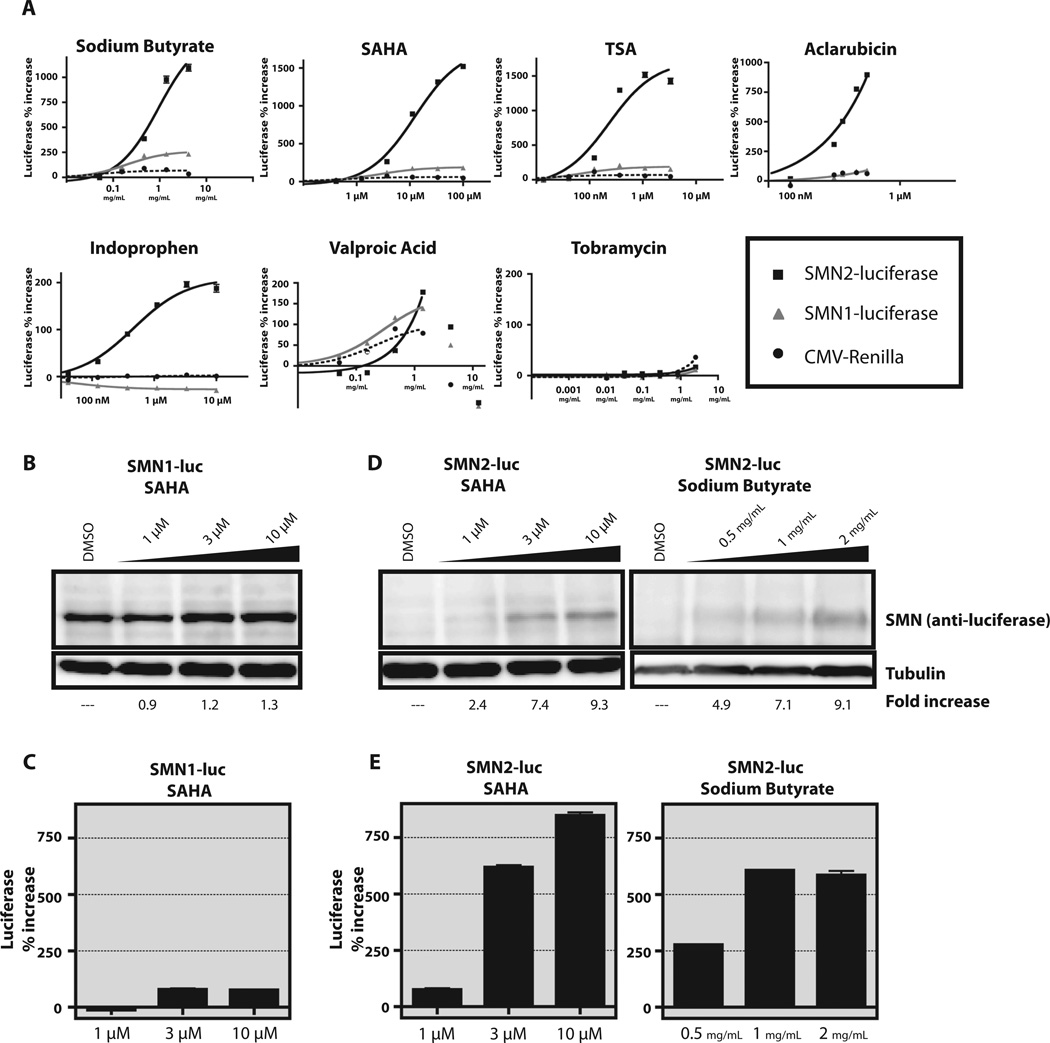
Activity of known compounds in the reporter cells. (A) Sodium butyrate, SAHA, TSA, aclarubicin, indoprofen, valproic acid, and tobramycin were tested in six-point dose-response experiments. Each compound was tested at concentrations previously reported to increase SMN protein levels. Black square with solid line, SMN2-luc cells; gray triangle, SMN1-luc cells; black circle with dotted line, SV40 control. Y-axis represents percentage activation relative to DMSO control. All points were tested in quadruplicate and plotted as mean ± SEM. Curves were created by linear regression using Prism4 (GraphPad Software Inc.). (B, C) SMN1-luc cells were treated for 24 h at increasing concentrations of SAHA. The amount of SMN-luciferase fusion protein detected by anti-luciferase antibody (B) is similar to the percentage increase in luciferase activity in the same samples (C). (D, E) SMN2-luc cells were treated for 24 h with increasing amounts of SAHA or sodium butyrate. The increase in SMN-luciferase fusion protein (D) correlates with luciferase activity (E). Experiments were performed three times with similar results. Blots shown are representative.
These clonal cell lines display the expected patterns of exon 7 inclusion in the reporter transcripts for both the SMN1-luc and SMN2-luc cell lines. Published studies have determined that 90% of transcripts from SMN1 include intron 7, whereas only 10% to 20% of transcripts from SMN2 include exon 7.8 In Figure 2C, end-point RT-PCR for each cell line is shown, and inclusion of exon 7 was calculated as 64% for SMN1-luc and 13% for SMN2-luc. By qRT-PCR using the primers illustrated in Figure 4, the percentage inclusion for transcripts in each reporter was calculated more precisely; 95.1% ± 6.7% for SMN1-luc and 10.2% ± 0.9% for SMN2-luc. Using these primers, we estimated the number of copies of each reporter in these clonal cell lines. SMN1-luc has one copy of the episomal reporter per cell, and the SMN2-luc cell line has about 10 copies of the reporter per cell.
Figure 4.
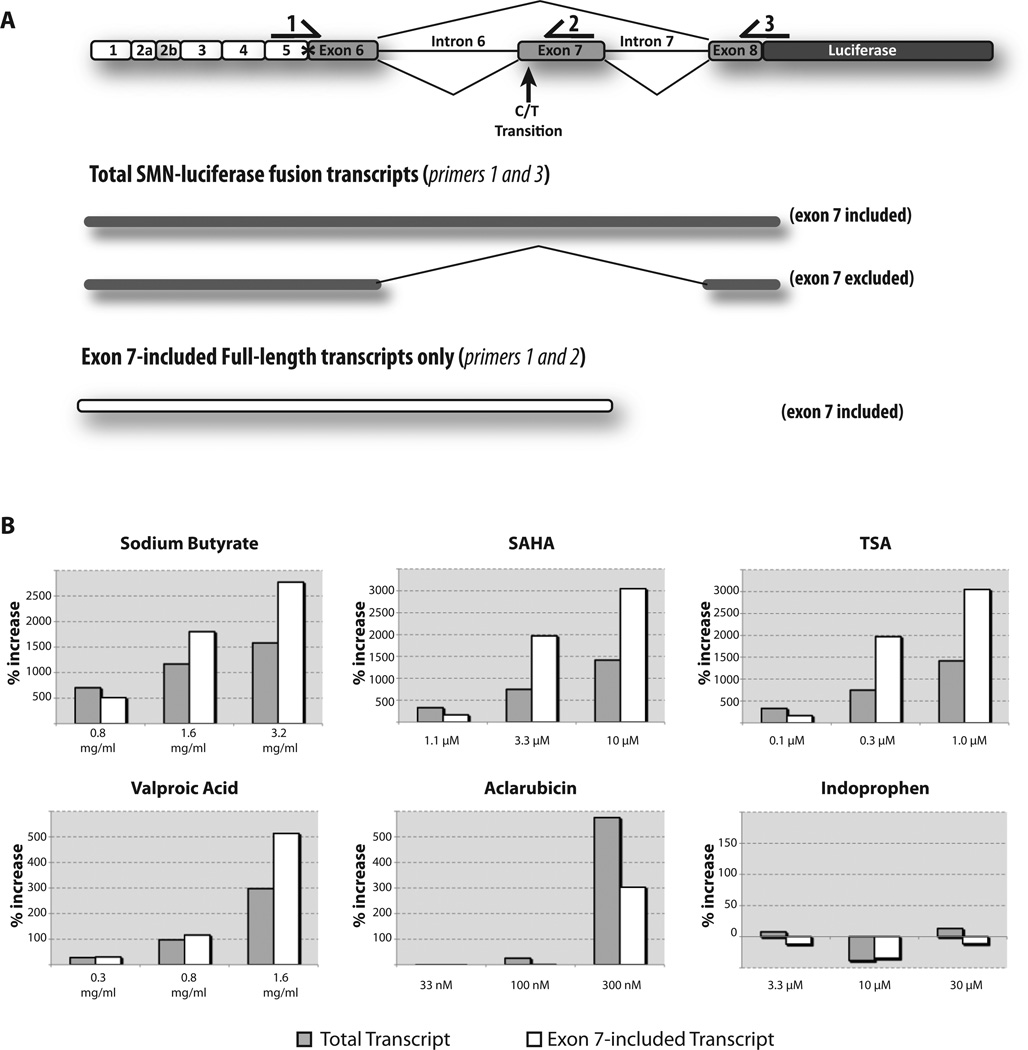
Analysis of SMN-luciferase fusion transcripts by qRT-PCR. (A) A schematic of the primer design for qRT-PCR. Primer pairs were chosen to amplify only the SMN-luciferase fusion transcripts but not endogenous SMN. Primer 1 overlaps a unique Xho I site (*). Primers 1 and 2 can amplify only full-length SMN-luc transcripts that contain exon 7. Primers 1 and 3 amplify SMN-luc reporter transcripts (both exon 7 included and excluded). (B) Comparison of increases in amount of total reporter transcripts (gray bars) and amount of exon 7–included reporter transcripts (white bars). Cells were treated at increasing concentrations of compound for 24 h. Percentage increase was calculated in relation to treatment with DMSO and normalized to GAPDH.
To determine the half-life of the SMN-fusion protein in these cell lines, cells were treated with cycloheximide and assayed for residual luciferase activity for 24 h. The luciferase activity in the SMN2-luc cell line had a t1/2 of 3.2 h (Fig. 2D). This matches well with published data for endogenous SMN protein.24 These data suggest that any changes in protein stability for the SMN-luciferase fusion protein would be easily detected within 24 to 48 h.
When the cell lines were tested for tolerance to DMSO (data not shown), it was found the luciferase expression was virtually unchanged when the final DMSO concentration in the reaction ranged from 0% to 1%. Concentrations greater than 1% decreased luciferase activity and were very likely detrimental to cell viability. Compounds are routinely screened at a final DMSO concentration equal to or less than 0.1%. Basal activity and response to compounds did not vary with serial cell passage and were very reproducible after the clones were thawed from liquid nitrogen storage (data not shown).
SMN-Luciferase Reporter Cell Response to Compound Treatment
In this assay, cells were treated with compounds previously shown to increase SMN2 protein levels and luciferase expression was read for counts per second (CPS) at an integration time of 100 000 µS (see Table 1 for detailed assay conditions). For comparison, all data points are expressed as percentage increase over basal luciferase expression in the control cells.
Compounds that were previously shown to increase SMN expression including SAHA, sodium butyrate, aclarubicin, TSA, indoprofen, and tobramycin were tested in our cell lines (Fig. 3A). These compounds were screened in both the SMN1-luc and SMN2-luc cell lines, and percentage increase of luciferase activity was plotted for SMN1-luciferase, SMN2-luciferase, and the CMV-driven internal renilla luciferase control. The activity for each compound, except tobramycin, matched or exceeded the published activities for these compounds (Fig. 3A and Supplemental Table 1). Tobramycin displayed no response in these cell lines. Tobramycin is an aminoglycoside that increases stable SMN protein levels from the SMN2 gene through transcriptional read-through of the termination codon in exon 8. By design, our reporter requires the inclusion of exon 7 and the frameshift mutation therein to restore the reading frame for luciferase. In the absence of exon 7, luciferase is out of frame. Read-through will not correct the frame shift, so tobramycin cannot and did not increase SMN-luciferase protein levels in the SMN-luciferase reporter cell lines.
Changes in the SMN-luciferase fusion protein levels were confirmed with SAHA in both the SMN1-luc and SMN2-luc cell lines and with sodium butyrate in the SMN2-luc cell line. There was a moderate increase in SMN-luciferase protein in the SMN1-luc cell line with increasing SAHA (Fig. 3B), which corresponds to the increase in luciferase activity (Fig. 3C). We observed a greater than ninefold increase in SMN-luciferase protein when the SMN2-luc cell line was treated with either SAHA or sodium butyrate (Fig. 3B). This protein increase corresponds to an increase in luciferase activity (Fig. 3C). These data confirm the correlation between luciferase activity and levels of full-length SMN-luciferase protein.
Analysis of mRNA in SMN2-Luciferase Reporter by qRT-PCR
Based on the new reporter design, we predicted that this screen could detect increased SMN-luciferase protein levels caused by compounds that stimulate SMN2 transcription or exon 7 inclusion or by increasing the half-life of the SMN mRNA or protein. qRT-PCR was used to analyze SMN-luciferase mRNAs. All assays used RNA from control and compound-treated SMN2-luc cells, and primer pairs were chosen to amplify only the SMN-luciferase–derived transcripts (Fig. 4A). We used a pair of primers that specifically detects both full-length (exon 7 included) and Δ7 (exon 7 excluded) SMN-luciferase transcripts (primers 1 and 3). To measure changes in exon 7 splicing efficiency, we used a primer pair that detects only the full-length (exon 7 included) SMN-luciferase transcript (primers 1 and 2).
For each sample, the percentage increase in the amount of total SMN-luciferase transcripts (gray bar) and exon 7–included full-length SMN-luciferase transcripts (white bar) was plotted (Fig. 4). Compounds that increase transcription should show increased total SMN-luciferase transcripts (gray bar) with a proportional increase for exon 7–included transcripts (white bar), assuming that proper splicing of exon 7 is not rate limiting. Compounds that stimulate exon 7 inclusion should increase the amount of exon 7–included full-length transcripts (white bar) detected with little to no change in the expression of total SMN-luciferase transcripts (gray bar). In these experiments, cells were treated with compound and harvested, and each cell pellet was divided for isolation of RNA and to assay luciferase activity. Each qRT-PCR sample was normalized to the housekeeping gene GAPDH. The Pfaffl method was used to calculate the change in the amount of total SMN mRNA and to determine whether compound treatments increased SMN transcripts.22
With the pan HDAC inhibitor compounds SAHA, TSA, valproic acid, and sodium butyrate, we observed increases in both the total and exon 7–included full-length SMN-luciferase transcripts. The increase in total SMN-luciferase mRNA suggests that these compounds are increasing transcription, as would be expected for HDAC inhibitors. These compounds also display a dramatic increase in exon 7–included full-length transcripts that was greater in magnitude than the increase of total SMN-luciferase transcripts. This suggests that the HDAC inhibitors stimulate both SMN transcription and exon 7 recognition and inclusion. With aclarubicin at 300 nM, there was a potent increase in total SMN-luciferase transcript. This increase was accompanied by a lesser increase in exon 7–included transcript, indicating that aclarubicin increases transcription of SMN2. In a cell in which transcription is increased and splicing efficiency is unchanged, the percentage increase in exon 7–included transcripts (gray bar) would be equal to the increase in total transcripts (white bar). In this case, the amount of exon 7–included transcripts has decreased even as the number of total transcripts increased. We conclude that although aclarubicin does increase transcription, it antagonizes exon 7 recognition and inclusion. We did not observe any consistent change in transcript levels with indoprofen.
Genetic Modulation of SMN-luciferase Protein Expression
To confirm that the reporter could also respond to protein-induced changes in SMN expression, we overexpressed the splicing factors SF2/ASF and Tra2β. SF2/ASF recognizes a splicing enhancer that includes C in the +6 position at the 5′ end of exon 7 in SMN1 to promote exon 7 inclusion.25 This interaction is antagonized by hnRNPA1, which binds at an overlapping splicing silencer that is created by the C to T transition at the +6 position in exon 7 in the SMN2 transcripts.25 Little to no increase in luciferase activity or change in RNA was observed in the SMN2-luciferase reporter cell-line (Fig. 5A, C). This was expected, because hnRNPA1 has a high affinity for the silencing element in exon 7 of SMN2. Even at its highest levels of overexpression, SF2/ASF was unable to effectively stimulate the weakened enhancer element (Fig. 5B).
Figure 5.
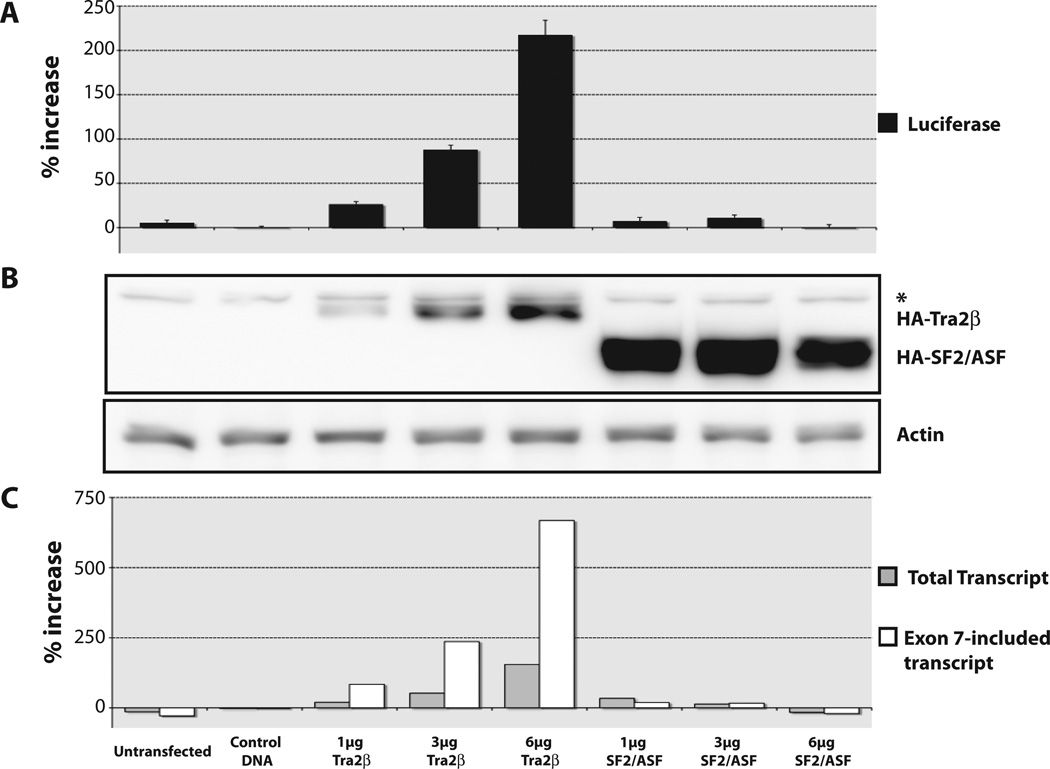
Overexpression of the splicing factors hTra2β and SF2/ASF. SMN2-luciferase reporter cells were transfected with increasing amounts of HA-tagged hTra2β or SF2/ASF. Cells were incubated for 48 h and assayed for luciferase activity. (A) Percentage increase was calculated in relation to treatment with control DNA transfection and normalized to internal renilla control. (B) Transfected protein expression was confirmed using anti-HA antibody. The asterisk denotes a background HA band. Actin was used as a loading control. (C) Comparison of increases in amount of total reporter transcripts (gray bars) and amount of exon 7–included reporter transcripts (white bars). Percentage increase was calculated in relation to treatment with DMSO and normalized to GAPDH.
Tra2β is known to increase exon 7 inclusion in SMN2.11 Despite low levels of Tra2β expression (Fig. 5B), transfected Tra2β stimulated luciferase activity twofold to threefold greater than the negative control (Fig. 5A). With increasing amounts of Tra2β expressed, there was up to a sixfold increase in exon 7–included transcripts (white bar) with only a 1.4-fold increase in total transcripts (gray bar; Fig. 5C). Analysis of the reporter mRNA confirmed that this SMN2-luciferase reporter detects increased inclusion of exon 7. From these data, we calculated the efficiency of exon 7 splicing of the reporter construct. Inclusion of exon 7 was 6.5% in the negative control sample, and no increase was observed with heterologous SF2/ASF expression. Exon 7 inclusion increased to 20% with the highest amount of transfected Tra2β. This validates the use of the SMN2-luciferase reporter cells to detect and quantify changes in exon 7 inclusion as well as changes in SMN expression in response to drug treatment and protein overexpression.
Assay Validation for High Throughput
To evaluate the suitability of the SMN2-luc cells for HTS, test plates in both 96-well and 384-well format were prepared to determine signal strength, well-to-well percentage coefficient of variation (%CV), amplitude of drug response, optimal cell density, and time of treatment. We observed that these HEK-derived cell lines displayed signal variability at low cell densities. As can be seen in Table 3, the correlation between luciferase signal and cell number was not linear. This is most obvious in the 384-well format. At low cell densities, these cells grow slowly and are less responsive to treatment. A cell density of 50 000 or 10 000 per well was found to be optimal in the 96- and 384-well plates, respectively. To assess responsiveness, 500 µg/mL sodium butyrate was selected as a stimulus. Although sodium butyrate is an HDAC inhibitor that can cause non-specific transcriptional activation, it has been reported to stimulate SMN2 transcription and SMN2 exon 7 inclusion.15,26 Cells were treated for 24 or 48 h, and although we observed increases in overall signal intensity at the 48-h time point, there was no difference in the percentage activation with sodium butyrate over control. However, there was an increase in the well-to-well %CV. Therefore, 24 hours was chosen as the optimal time point for treatment. In the 96-well format, luciferase activity in the SMN2-luc cell increased by more than threefold with sodium butyrate, and well-to-well %CV was 5% for both treated and control cells. In 384-well format, the %CV for control cells was 11% and 3% for treated. The z′ scores were calculated as 0.74 in 96-well and 0.78 in the 384-well formats, confirming that these assay conditions are suitable for HTS. These data are summarized in Table 3.
Table 3.
SMN2-Luciferase Reporter Validation with 0.5 mg/mL Sodium Butyrate
| Format | 96-Well Validation | 384-Well Validation | 384-Well LDDN Screen | |||||
|---|---|---|---|---|---|---|---|---|
| Cells per well | 25 000 | 50 000 | 25 000 | 50 000 | 5000 | 7500 | 10 000 | 10 000 |
| Time point | 24 h | 24 h | 48 h | 48 h | 24 h | 24 h | 24 h | 24 h |
| Na-But RLUa ave | 2046 | 4798 | 3696 | 7505 | 1332 | 2832 | 39 287 | 13 894 |
| Na-But stdev | 138 | 234 | 536 | 756 | 131 | 329 | 1337 | 1272.55 |
| CV Na-But | 0.07 | 0.05 | 0.15 | 0.10 | 0.10 | 0.12 | 0.03 | 0.09 |
| DMSO RLUa ave | 684 | 1357 | 1425 | 2626 | 521 | 1008 | 8054 | 4677 |
| DMSO stdev | 44 | 64 | 61.7 | 248 | 98 | 123 | 910 | 387 |
| CV DMSO | 0.06 | 0.05 | 0.04 | 0.09 | 0.19 | 0.12 | 0.11 | 0.08 |
| S/B | 3.0 | 3.5 | 2.6 | 2.9 | 2.6 | 2.8 | 4.9 | 3.0 |
| Z′ | 0.60 | 0.74 | 0.21 | 0.38 | 0.15 | 0.26 | 0.78 | 0.46 |
Relative light units (RLU) vary based on luciferase substrate, plate reader, and protocol.
HTS and Hit Selection
An 115 000 compound library was screened using the screening protocol outlined in Table 2. Each compound was added to a single well to a final concentration of ~2.2 µM. The final DMSO concentration in test and control wells was 0.13%. Each plate included negative controls of 0.13% DMSO (n = 16) and positive controls of 500 mg/mL sodium butyrate with 0.13% DMSO (n = 16). For the entire screen, the average %CV was 9.1% for DMSO alone and 9.8% for sodium butyrate–treated wells. The average increase with 500 mg/mL was 3.1-fold or 210%. A hit was defined by activation of greater than six times the %CV (60%). A total of 462 hits were identified for an overall 0.4% hit rate.
Hit Confirmation
The 462 hits were replated from the screening library into master plates and then rescreened at 0.1, 1, and 5 µM in quadruplicate under conditions identical to the original HTS. Each compound was counterscreened against an unrelated luciferase control cell line that expresses luciferase from the minimal SV40 promoter and lacks an intron. This reporter should not respond to compounds that specifically affect the SMN promoter or compounds that change regulation of splicing. This allowed us to exclude compounds that may cause nonspecific increases in luciferase expression. Of the 462 initial hits, 168 failed to reproduce at least 60% activation with the SMN2-luciferase reporter clone and were categorized as false-positives (34% false-positive rate). The remaining 294 compounds were limited to a high-priority group of 19 scaffolds based on potency, strength of activation, dose dependency, specificity against luciferase control, and favorable chemical properties. All 19 compounds showed greater than 100% increase in reporter expression in the rescreen and stimulated the control reporter less than 40%. All 19 lacked overtly toxic functional groups and had chemical scaffolds that were tractable to chemical modification.
Fresh lots of 17 commercially available compounds were ordered and then rescreened in quadruplicate using a 12-point dose-response under the same conditions as the first two rounds of screening with both the SMN2-luc and SMN1-luc cell lines and using the SV40 luciferase control cell line. Because the SMN1 and SMN2 promoters are nearly identical,20,27 compounds that increased transcription of the SMN2-luc reporter would also increase transcription of the SMN1-luc reporter. The SMN1-luc cells should therefore be responsive to compounds that increase transcription from the SMN promoter. However, because the SMN gene in the SMN1 cells includes exon 7 efficiently (>90%), only a small increase in percentage activation is possible with compounds that stimulate exon 7 inclusion.
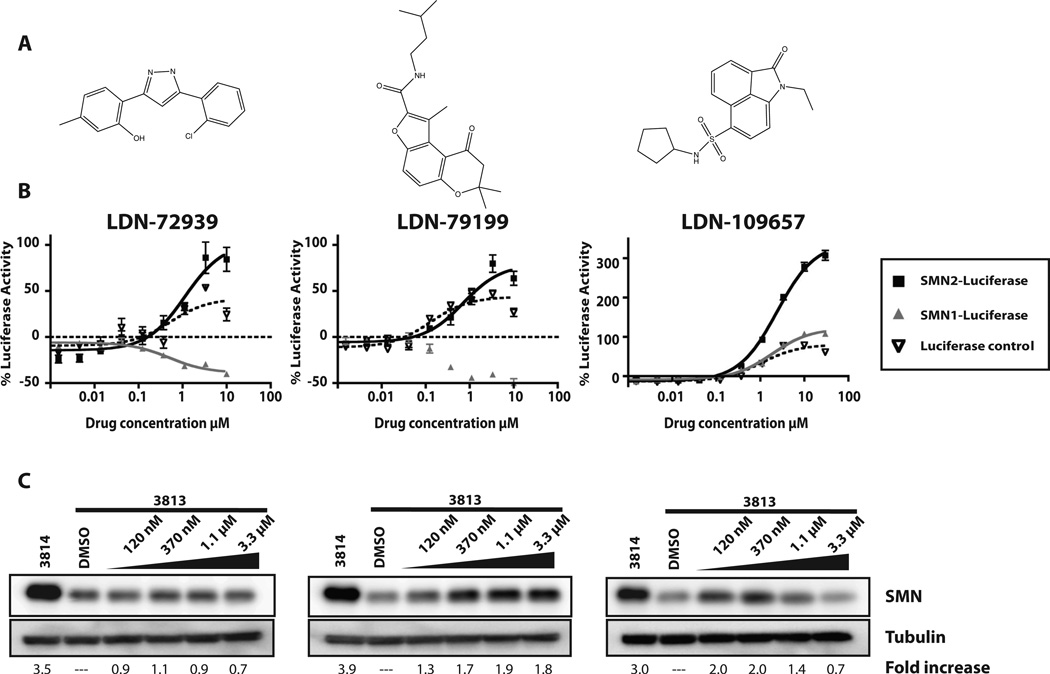
Overall, this panel of reporter cell lines offers the ability to discriminate between compounds that increase SMN expression through promoter and splicing specific mechanisms and rule out nonspecific activators. For example, LDN-109657 (Fig. 6B) increased SMN2-luciferase levels up to 300% (fourfold) but also increased luciferase expression in the SMN-luc1 reporter and, to a lesser extent, the luciferase control cells. These data suggest that this compound acts by either increasing SMN transcription or protein stabilization. Several compounds decreased SMN1-luciferase expressed in the SMN1-luc reporter cell line but still increased SMN2-luciferase expression in the SMN2-luc cell line (LDN-72939 and LDN-79199; Fig. 6B). Of the 17 compounds tested in the 12-point dose response, 13 increased luciferase expression by >60% and four did not. Preliminary confirmation and characterization of three of these compounds, LDN-72939, LDN-79199, and LDN-109657, are reported here. Overall, the amplitude of SMN-luciferase increase varied, but all three compounds displayed an EC50 in the low micromolar range: 1.1 µM, 750 nM, and 2.4 µM, respectively.
Figure 6.
Hit confirmation in the reporter cells. (A) Structures of three hits from the high-throughput screen. (B) Twelve-point dose response experiments. Each compound was tested with the reporter cell lines at 12 concentrations (0.17, 0.5, 1.5, 4.5, 13.7, 41, 123, 370, 1111, 3333, 10 000, 30 000 nM). Black, SMN2-luc cells; gray, SMN1-luc cells; dotted line, SV40 control. Y-axis represents percentage activation over DMSO control. All points were tested in quadruplicate and plotted as mean ± SEM. Curves were created by linear regression using Prism4 (GraphPad Software Inc.). (C) Primary human fibroblast lysates from carrier (3814; SMN1+/−; SMN2+/+) and SMA (3813; SMN1−/−; SMN2+/+) cells were blotted with antibodies to SMN and a-tubulin. Cells were treated for 48 h with increasing concentrations of compound. Fold increase was calculated in relation to DMSO-treated 3813 and normalized to tubulin levels. Experiments were performed three times. Blots shown are representative.
To further evaluate the preliminary lead compounds, their effects on endogenous SMN protein levels were examined. Any cell containing even one copy of the SMN1 gene will produce enough SMN protein from SMN1 to mask the effects of the compounds on protein expression from the SMN2 gene. SMN null cells are not viable. The current standard is to test primary human fibroblast cells derived from a severe type 1 SMA patient. Most commonly used are the 3813 cells (SMN1−/−; SMN2+/+). These cells express very low basal levels of full-length SMN protein. For comparison, 3814 primary fibroblasts from the carrier parent (SMN1+/−; SMN2+/+) of this SMA infant are available. By quantitative immunoblot for full-length SMN protein, the heterozygous 3814 cells express three to five times more full-length SMN protein than in the 3813 cells28 (Fig. 6C).
The selected compounds were tested for their effect on total endogenous SMN protein levels. 3813 fibroblasts were treated with varying concentrations of each compound for 72 h (Fig. 6C). We observed that these primary fibroblasts were sensitive to lower concentrations of compound than those used in the immortal reporter cell lines. Not all compounds increase endogenous SMN levels in the fibroblasts as well as others, regardless of their activity in the reporter cell lines. LDN-72939 was more active in the reporter cell line but showed little activity in the fibroblasts. LDN-79199 was less active in the reporter cells but increased endogenous SMN level twofold at 370 nM. LDN-109657 displayed high activity in both the reporter assay and in fibroblasts. It increased endogenous SMN levels twofold at 120 nM. The decrease in SMN at higher concentrations (1.1 µM and 3.3 µM) is likely due to either poor solubility or a decrease in cell proliferation in the fibroblasts.
Discussion
Spinal muscular atrophy is caused by an insufficient amount of functional, full-length SMN protein, usually resulting from deletion or disruption of both SMN1 alleles. All SMA patients retain at least one SMN2 gene, and disease severity inversely correlates with SMN2 copy number.29 Because the SMN2 gene has the potential to express functional full-length SMN protein, it is an attractive therapeutic target for the treatment for SMA.30,31
SMN2 expression can be increased through upregulation of transcription, promotion of exon 7 inclusion, stabilization of SMN2 mRNA and protein, or elevation of translation efficiency. HDAC inhibitors are known to influence transcription through histone deacetylation, which causes chromatin remodeling. We observed that the HDAC inhibitors sodium butyrate, TSA, VPA, and SAHA all increased the amount of total and exon 7 included SMN2-luc reporter transcripts (Fig. 4B). Not only do these pan-HDAC inhibitors increase the quantity of SMN-luciferase transcripts, but a higher percentage of those transcripts are correctly spliced (Fig. 4B). This bimodal mechanism for pan-HDAC inhibitor regulation of SMN2 expression may explain the high potency and efficacy observed with these compounds but may also predict other potential and possibly detrimental off-target effects.
Splicing of exon 7 is tightly regulated in both the SMN1 and SMN2 mRNAs. Conserved splicing enhancer and splicing silencing elements within exon 7 recruit splicing factors to the mRNA to regulate exon 7 recognition.32 We tested two of these splicing factors, hTra2β and SF2/ASF, in the SMN2-luc reporter cell line. As expected, SF2/ASF had no impact on SMN2-luciferase expression (Fig. 5). hTra2β binds to a splicing enhancer downstream of the hnRNP A1 site and can stabilize U1 snRNP binding at the 5′ splice site of exon 7. This should dramatically increase exon 7 recognition and increase its splicing efficiency. We observe an increase in SMN-luciferase activity and a considerable increase in exon 7–included full-length SMN2-luciferase transcripts with hTra2β overexpression (Fig. 5). The small increase in total reporter mRNA that accompanies this effect may be due to changes in the stability of the full-length, exon 7–included mRNA and not an actual increase in transcription of the reporter.
Disruption or regulation of the mRNA or protein turn-over machinery could also increase steady-state SMN levels. This could be the mechanism for the activity of indoprofen, the aminoglycosides, and the quinazoline compounds.14,21,33 Indoprofen, a nonsteroidal anti-inflammatory drug, was identified using our first-generation reporter assay.13 Its mechanism of action has not been determined, but recent evidence suggests that it has antiterminator activity that may stabilize the SMN Δ7 protein. It is possible that the quinazoline compounds could increase the translation efficiency of SMN mRNA, as suggested by the recent report of interaction with the RNA decapping factor DcpS.34
Our new SMN2 reporter cell line combines the benefits of previous reporters and expands their potential to identify new regulatory circuitry for SMN2 expression. This assay has proven to be stable and reliable in 96-, 384-, and 1536-well formats and has been used in three independent screening centers to identify novel modulators of SMN2 protein expression. Interestingly, the HTS at the NIH Chemical Genomics Center identified chemically distinct activators.35 In addition to novel compounds, these screens also identified compounds known to modify SMN2 protein expression, including indoprofen, resveratrol, hydroxyurea, and aclarubicin.
In this report, we show preliminary characterization of LDN-72939, LDN-79199, and LDN-109657. Each of these compounds increases SMN-luciferase expression by at least 60%. LDN-79199 and LDN-109657 increased the levels of endogenous SMN protein in SMA-derived primary fibroblasts. Further characterization of these compounds as well as others is ongoing.
One of the drawbacks of the previous C33a reporter cell line was its high basal level of SMN-luciferase expression. This made detection of small increases in SMN-luciferase difficult. We initially encountered high basal level expression with the new HEK293 mixed-population reporter cells as well. By isolating and expanding clonal cell lines, we were able to select for cells with much lower levels of basal SMN2-luciferase expression. Another issue with the previous C33a reporter was a progressive loss of luciferase signal intensity and decreased responsiveness to drug treatment. This variability in signal strength and drug response made the previous generation reporter unsuitable for HTS. The new HEK293 cell lines have maintained constant luciferase expression and reproducible induction with drug treatment over hundreds of population doublings. This consistency was also observed by our collaborators at the other screening centers. The improvement in this reporter assay is likely due to a combination of factors, including the new reporter design, the episomal nature of the vector, isolation of clonal cell lines, and use of HEK293 cells.
Spinal muscular atrophy is fatal for approximately 5000 children each year in the United States, whereas tens of thousands of others live with limited mobility and progressive muscle weakness. There is no approved drug for the treatment of spinal muscular atrophy. It has been established that the amount of SMN protein expressed has an inverse relationship to the severity to the disease. Compounds that safely increase SMN protein levels would dramatically improve the quality of life for individuals with SMA. Our new assay represents a valuable tool for identifying new compounds, characterizing existing compounds, and driving medicinal chemistry programs on active chemical scaffolds.
Supplementary Material
Acknowledgments
We thank Mickey Huang and Eli Schuman for their help with screening.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: J.J.C., M.C.E., and E.J.A. were supported by FightSMA and NICHD R21HD57402. J.N., G.D.C., and M.A.G. thank the Harvard NeuroDiscovery Center for financial support.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening’s Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Crawford TO, Pardo CA. The Neurobiology of Childhood Spinal Muscular Atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre S, Burglin L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, LePaslier D, Frezal F, Cohen D, Weissenbach J, Munnich A, Melki J. Identification and Characterization of a Spinal Muscular Atrophy Determining Gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Burglen L, Lefebvre S, Clermont O, Burlet P, Viollet L, Cruaud C, Munnich A, Melki J. Structure and Organization of the Human Survival Motor Neuron (SMN) Gene. Genomics. 1996;32:479–482. doi: 10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- 4.Melki J. Spinal Muscular Atrophy. Curr. Opin. Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Jablonka S, Sendtner M. Molecular and Cellular Basis of Spinal Muscular Atrophy. Amyotroph. Lateral. Scler. Other Motor Neuron. Disord. 2003;4:144–149. doi: 10.1080/14660820310011296. [DOI] [PubMed] [Google Scholar]
- 6.DiDonato CJ, Ingraham SE, Mendell JR, Prior TW, Lenard S, Moxley RT, Florence J, Burghes AHM. Deletion and Conversion in Spinal Muscular Atrophy Patients: Is There a Relationship to Severity? Ann. Neurol. 1997;41:230–237. doi: 10.1002/ana.410410214. [DOI] [PubMed] [Google Scholar]
- 7.Wirth B, Schmidt T, Hahnen E, Rudnik-Schöneborn S, Krawczak M, Müller MB, Schönling J, Zerres K. De Novo Rearrangements Found in 2% of Index Patients with Spinal Muscular Atrophy: Mutational Mechanisms, Parental Origin, Mutation Rate, and Implications for Genetic Counseling. Am. J. Hum. Genet. 1997;61:1102–1111. doi: 10.1086/301608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A Single Nucleotide in the SMN Gene Regulates Splicing and Is Responsible for Spinal Muscular Atrophy. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorson CL, Androphy EJ. An Exonic Enhancer Is Required for Inclusion of an Essential Exon in the SMA-Determining Gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 10.Zhang ML, Lorson CL, Androphy EJ, Zhou J. An In Vivo Reporter System for Measuring Increased Inclusion of Exon 7 in SMN2 mRNA: Potential Therapy of SMA. Gene Ther. 2001;8:1532–1538. doi: 10.1038/sj.gt.3301550. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B. Htra2-beta 1 Stimulates an Exonic Splicing Enhancer and Can Restore Full-Length SMN Expression to Survival Motor Neuron 2 (SMN2) Proc. Natl. Acad. Sci. U. S. A. 2000;97:9618–9623. doi: 10.1073/pnas.160181697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreassi C, Jarecki J, Zhou J, Coovert DD, Monani UR, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, Burghes AH. Aclarubicin Treatment Restores SMN Levels to Cells Derived from Type I Spinal Muscular Atrophy Patients. Hum. Mol. Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 13.Lunn MR, Root DE, Martino AM, Flaherty SP, Kelley BP, Coovert DD, Burghes AH, Man NT, Morris GE, Zhou J, Androphy EJ, Sumner CJ, Stockwell BR. Indoprofen Upregulates the Survival Motor Neuron Protein through a Cyclooxygenase-Independent Mechanism. Chem. Biol. 2004;11:1489–1493. doi: 10.1016/j.chembiol.2004.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarecki J, Chen X, Bernardino A, Coovert DD, Whitney M, Burghes A, Stack J, Pollok BA. Diverse Small-Molecule Modulators of SMN Expression Found by High-Throughput Compound Screening: Early Leads towards a Therapeutic for Spinal Muscular Atrophy. Hum. Mol. Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- 15.Chang JG, Hsieh-Li HM, Jong YJ, Wang NM, Tsai CH, Li H. Treatment of Spinal Muscular Atrophy by Sodium Butyrate. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Avila AM, Burnett BG, Taye AA, Gabanella F, Knight MA, Hartenstein P, Cizman Z, Di Prospero NA, Pellizzoni L, Fischbeck KH, Sumner CJ. Trichostatin A Increases SMN Expression and Survival in a Mouse Model of Spinal Muscular Atrophy. J. Clin. Invest. 2007;117:659–671. doi: 10.1172/JCI29562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumner CJ, Huynh TN, Markowitz JA, Perhac JS, Hill B, Coovert DD, Schussler K, Chen X, Jarecki J, Burghes AH, Taylor JP, Fischbeck KH. Valproic Acid Increases SMN Levels in Spinal Muscular Atrophy Patient Cells. Ann. Neurol. 2003;54:647–654. doi: 10.1002/ana.10743. [DOI] [PubMed] [Google Scholar]
- 18.Hahnen E, Eyupoglu IY, Brichta L, Haastert K, Trankle C, Siebzehnrubl FA, Riessland M, Holker I, Claus P, Rom-stock J, Buslei R, Wirth B, Blumcke I. In Vitro and Ex Vivo Evaluation of Second-Generation Histone Deacetylase Inhibitors for the Treatment of Spinal Muscular Atrophy. J. Neurochem. 2006;98:193–202. doi: 10.1111/j.1471-4159.2006.03868.x. [DOI] [PubMed] [Google Scholar]
- 19.Garbes L, Riessland M, Holker I, Heller R, Hauke J, Trankle C, Coras R, Blumcke I, Hahnen E, Wirth B. LBH589 Induces up to 10-fold SMN Protein Levels by Several Independent Mechanisms and Is Effective Even in Cells from SMA Patients Non-responsive to Valproate. Hum. Mol. Genet. 2009;18:3645–3658. doi: 10.1093/hmg/ddp313. [DOI] [PubMed] [Google Scholar]
- 20.Monani UR, McPerson JD, Burghes AHM. Promoter Analysis of the Human Centromeric and Telomeric Survival Motor Neuron Genes (SMNC and SMNT) Biochim. Biophys. Acta. 1999;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 21.Mattis VB, Butchbach ME, Lorson CL. Detection of Human Survival Motor Neuron (SMN) Protein in Mice Containing the SMN2 Transgene: Applicability to Preclinical Therapy Development for Spinal Muscular Atrophy. J. Neurosci. Methods. 2008;175:36–43. doi: 10.1016/j.jneumeth.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfaffl MW. A New Mathematical Model for Relative Quantification in Real-Time RT-PCR. Nucleic Acids Res. 2001;29:e45–000. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 24.Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN Protein Stability. Mol. Cell. Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nlend Nlend R, Meyer K, Schumperli D. Repair of PremRNA Splicing: Prospects for a Therapy for Spinal Muscular Atrophy. RNA Biol. 2010;7:430–440. doi: 10.4161/rna.7.4.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andreassi C, Angelozzi C, Tiziano FD, Vitali T, De Vincenzi E, Boninsegna A, Villanova M, Bertini E, Pini A, Neri G, Brahe C. Phenylbutyrate increases SMN expression in vitro: relevance for treatment of spinal muscular atrophy. Eur J Hum Genet. 2004;12(1):59–65. doi: 10.1038/sj.ejhg.5201102. [DOI] [PubMed] [Google Scholar]
- 27.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The Promoters of the Survival Motor Neuron Gene (SMN) and Its Copy (SMNc) Share Common Regulatory Elements. Am. J. Hum. Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coovert D, Le T, McAndrew P, Strasswimmer J, Crawford T, Mendell J, Coulson S, Androphy EJ, Prior T, Burghes AHM. The Survival Motor Neuron Protein in Spinal Muscular Atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 29.Lorson CL, Rindt H, Shababi M. Spinal Muscular Atrophy: Mechanisms and Therapeutic Strategies. Hum. Mol. Genet. 2010;19:R111–R118. doi: 10.1093/hmg/ddq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirth B, Brichta L, Hahnen E. Spinal Muscular Atrophy and Therapeutic Prospects. Prog. Mol. Subcell. Biol. 2006;44:109–132. doi: 10.1007/978-3-540-34449-0_6. [DOI] [PubMed] [Google Scholar]
- 31.Sumner CJ. Therapeutics Development for Spinal Muscular Atrophy. NeuroRx. 2006;3:235–245. doi: 10.1016/j.nurx.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh NN, Singh RN, Androphy EJ. Modulating Role of RNA Structure in Alternative Splicing of a Critical Exon in the Spinal Muscular Atrophy Genes. Nucleic Acids Res. 2006;35:371–389. doi: 10.1093/nar/gkl1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heier CR, DiDonato CJ. Translational Readthrough by the Aminoglycoside Geneticin (G418) Modulates SMN Stability In Vitro and Improves Motor Function in SMA Mice In Vivo. Hum. Mol. Genet. 2009;18:1310–1322. doi: 10.1093/hmg/ddp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh J, Salcius M, Liu SW, Staker BL, Mishra R, Thurmond J, Michaud G, Mattoon DR, Printen J, Christensen J, Bjornsson JM, Pollok BA, Kiledjian M, Stewart L, Jarecki J, Gurney ME. DcpS as a Therapeutic Target for Spinal Muscular Atrophy. ACS Chem. Biol. 2008;3:711–722. doi: 10.1021/cb800120t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao J, Marugan JJ, Zheng W, Titus SA, Southall N, Cherry JJ, Evans M, Androphy EJ, Austin CP. Discovery, Synthesis and Biological Evaluation of Novel SMN Protein Modulators. J. Med. Chem. 2011;54:6215–6233. doi: 10.1021/jm200497t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.