Significance
Voltage-gated sodium (Nav) channels are members of a large complex that plays a crucial role in rapid electrical signaling throughout the human body. As prominent members of this complex, β-subunits modify Nav channel function and cause debilitating disorders when mutated. Collectively, the functional and crystallographic results reported in this work uncover intricate interactions of these elements within the Nav-channel signaling complex and establish a key role for β-subunits in shaping Nav1.2 pharmacology. An important concept emerging from our results is that β-subunits provide exciting opportunities for designing new therapeutic strategies to correct their abnormal behaviors.
Keywords: voltage-gated sodium channel, beta4 subunit, ProTx-II, X-ray structure, disease mutations
Abstract
Voltage-gated sodium (Nav) channels are embedded in a multicomponent membrane signaling complex that plays a crucial role in cellular excitability. Although the mechanism remains unclear, β-subunits modify Nav channel function and cause debilitating disorders when mutated. While investigating whether β-subunits also influence ligand interactions, we found that β4 dramatically alters toxin binding to Nav1.2. To explore these observations further, we solved the crystal structure of the extracellular β4 domain and identified 58Cys as an exposed residue that, when mutated, eliminates the influence of β4 on toxin pharmacology. Moreover, our results suggest the presence of a docking site that is maintained by a cysteine bridge buried within the hydrophobic core of β4. Disrupting this bridge by introducing a β1 mutation implicated in epilepsy repositions the 58Cys-containing loop and disrupts β4 modulation of Nav1.2. Overall, the principles emerging from this work (i) help explain tissue-dependent variations in Nav channel pharmacology; (ii) enable the mechanistic interpretation of β-subunit–related disorders; and (iii) provide insights in designing molecules capable of correcting aberrant β-subunit behavior.
Voltage-gated sodium (Nav) channels play a key role in cellular communication by manipulating the transmembrane voltage gradient to encode and propagate vital information rapidly over long distances (1). Consequently, mutations that modify Nav channel activity underlie debilitating neurological diseases, muscular disorders, and pain syndromes (2, 3). Typically, Nav channels are part of a membrane-embedded signaling complex that involves various integral membrane proteins (4). The significance of this environment for proper channel function is highlighted by divergent Nav channel responses to changes in membrane voltage when expressed in native tissues or in heterologous systems. β-Subunits are prominent members of the Nav channel signaling complex but do not contribute to the ion-conducting pore (5). Instead, they are multifunctional single-transmembrane segment glycoproteins that (i) modulate the gating properties of voltage-gated ion channels; (ii) regulate Nav channel trafficking and expression levels; and (iii) promote cell adhesion and migration (5–14). Of the four known β-subunits and their splice variants (15–20), β4 is unique in that it enables resurgent current, a feature that renders certain Nav channel isoforms capable of high-frequency firing in excitable tissues (21). Moreover, aberrant behavior of the ubiquitously expressed β4 subunit has been implicated in long-QT syndrome (LQTS) (22), LQTS-associated Sudden Infant Death Syndrome (23), atrial fibrillation (24), Huntington’s disease (25), and prostate cancer (26), possibly through dysregulation of the Nav channel signaling complex. β4 also is targeted by β- and γ-secretase enzymes from the amyloidogenic pathway, a recent observation that suggests a potential contribution of this particular subunit to the development of Alzheimer's disease (27).
Despite accumulating evidence supporting an important contribution to neuronal excitability and various health disorders (28), fundamental questions about the molecular mechanisms underlying β4 interaction with Nav channels remain unanswered. Moreover, the notion that the β4 subunit shapes the overall pharmacological sensitivities of the Nav channel signaling complex remains unexplored. However, altered ligand interactions may be exploited to detect the presence of β4 in normal or pathological conditions (29). Here, we investigated whether β-subunits influence Nav channel sensitivity to molecules isolated from animal venom and discovered that β4 can drastically alter the response of the neuronal Nav1.2 isoform to spider and scorpion toxins that target paddle motifs within Nav channel voltage sensors. To elucidate the machinery underlying this observation, we solved the crystal structure of the extracellular β4 domain and found a 58Cys-containing binding interface that is involved in Nav channel modulation of toxin pharmacology by β4. Remarkably, dismantling the strictly conserved internal cysteine bridge in β4 by introducing a β1 mutation implicated in epilepsy (30) does not preclude protein folding and trafficking to the membrane. However, conformational changes induced by the mutation perturb the 58Cys-containing loop and disrupt β4 interaction with Nav1.2, in turn altering the functional and pharmacological properties of the larger Nav channel signaling complex.
Results
β-Subunits Shape Nav Channel Pharmacology.
Although the influence of β-subunits on ion channel gating is well documented (5), little is known about their ability to manipulate the pharmacological sensitivities of Nav channel signaling complexes (29, 31, 32). To investigate the extent to which these versatile glycoproteins modify ligand interactions, we applied seven toxins from spider, scorpion, sea anemone, and wasp venom (ProTx-I, ProTx-II, TsVII, AaHII, LqqIV, ATX-II, and β-PMTX) as well as two drugs (lidocaine and ambroxol) to Xenopus oocytes expressing the neuronal Nav1.2 isoform and determined potential changes in ligand susceptibility induced by the presence of each of the four β-subunits. We uncovered multiple conditions in which Nav1.2’s sensitivity to a particular toxin was modified by β-subunits, whereas sensitivity to neither drug was significantly affected (Fig. 1, Fig. S1, and Table S1). For example, the sea anemone toxin ATX-II interacts exclusively with the Nav1.2 domain IV voltage sensor to inhibit fast inactivation, resulting in a large increase in inward sodium ion flow (33). When β2 is present, 100 nM ATX-II still prevents Nav1.2 from inactivating rapidly; however, the peak sodium current increases only marginally (Fig. S1). A similar effect is seen when 100 nM of the domain IV-targeting scorpion toxin LqqIV is applied to Nav1.2 coexpressed with β1 [binding site identification is given in Fig. S2 (34)]. In this instance, however, LqqIV also shifts the steady-state inactivation curve to more positive potentials (V1/2 from −57 mV to −48 mV; P ≤ 0.001), thereby increasing channel availability to open in response to membrane depolarizations (Fig. S1). In contrast, the related scorpion toxin AaHII (34) decreases Nav1.2 availability when coexpressed with β2 (V1/2 from −42 mV to −61 mV; P ≤ 0.001) or β4 (V1/2 from −45 mV to −63 mV; P ≤ 0.001).
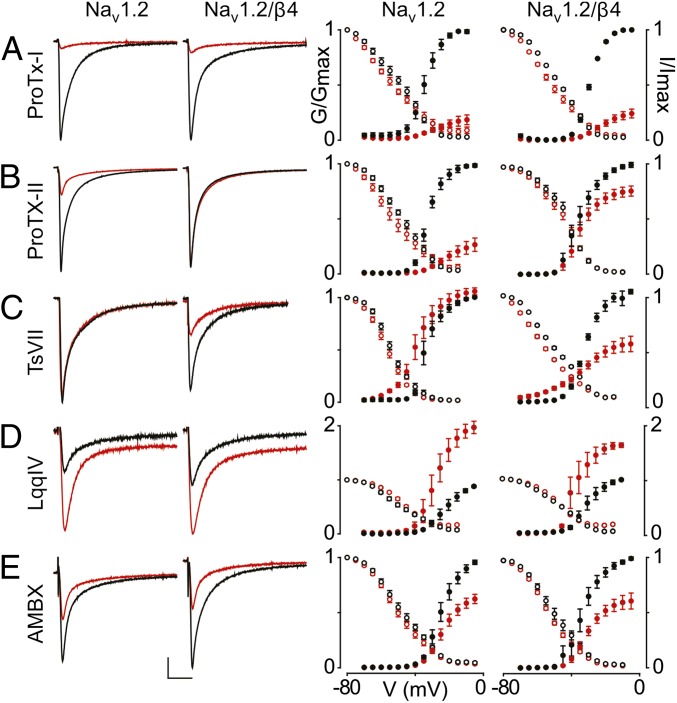
Fig. 1.
Influence of β4 on the ligand susceptibility of Nav1.2. (A–E) Effect of saturating concentrations [100 nM ProTx-I (A), 100 nM ProTx-II (B), 500 nM TsVII (C), 100 nM LqqIV (D), and 500 μM ambroxol (AMBX) (E)] (34, 77) on Nav1.2 and Nav1.2/β4. (Left) Representative sodium currents are elicited by a depolarization to −20 mV before (black) and after (red) addition of toxin or drug from a holding potential of −90 mV. The x-axis is 10 ms; the y-axis is ∼0.5 μA. (Right) Normalized conductance–voltage relationships (G/Gmax; black filled circles) and steady-state inactivation relationships (I/Imax; black open circles) of the WT Nav1.2 channel with or without β4 coexpression are compared before (black circles) and after (red circles) toxin or drug application. β4 alters Nav1.2 susceptibility to ProTx-II and TsVII, whereas ProTx-I, LqqIV, and AMBX are not affected. Channel-expressing oocytes were depolarized in 5-mV steps from a holding potential of −90 mV. Boltzmann fit values are reported in Table S1. n = 3–5; error bars represent S.E.M.
Interestingly, the most striking effects on toxin susceptibility are observed when Nav1.2 is expressed together with the β4 subunit. First, the tarantula toxin ProTx-II has been shown to interact with the voltage sensors in domains I, II, and IV of Nav1.2 and to inhibit channel opening (34, 35). However, in the presence of β4, Nav1.2 is dramatically less inhibited, suggesting that β4 may prevent ProTx-II from interacting with one or more of its receptor sites (Fig. 1B). Second, the structurally unrelated scorpion toxin TsVII promotes Nav channel opening by preferentially interacting with the voltage sensor in domain II and stabilizing it in an activated state (34, 36). Without β-subunits present, 500 nM TsVII causes Nav1.2 to open at more negative voltages, although the maximal conductance of the channel is not affected. In contrast, TsVII greatly decreases Nav1.2 maximal conductance with a smaller shift in activation voltage when β4 is present (Fig. 1C), raising the possibility that this β-subunit exerts an influence on the domain II voltage sensor. Next, we crystallized the extracellular β4 domain to explore the mechanisms underlying the influence of this particular β-subunit on Nav1.2 pharmacology.
The β4 Subunit Structure Reveals an Exposed Cysteine.
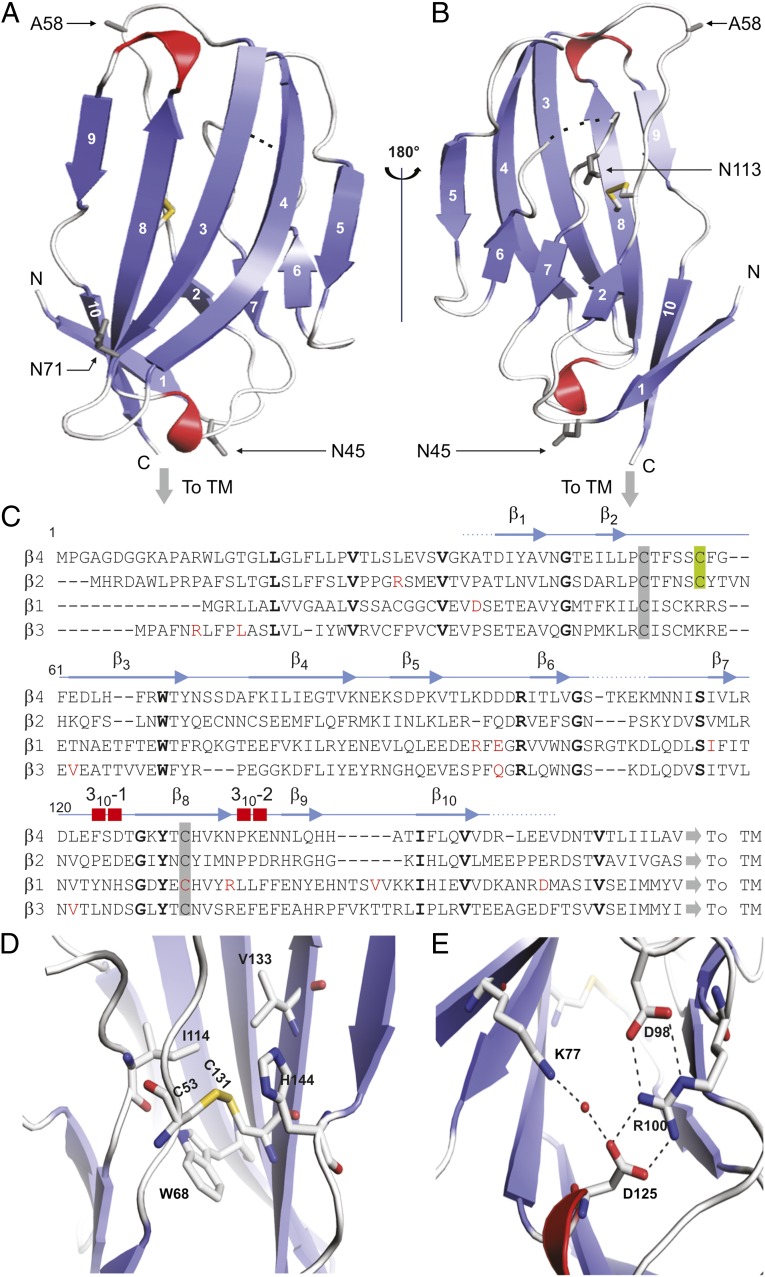
Despite their impact on the functional and pharmacological properties of Nav channels, tertiary structural information about β-subunits has been lacking. To address this inadequacy, we solved the crystal structure of the extracellular β4 domain (residues 32–157) at 1.7-Å resolution (Fig. 2, Fig. S3, and Table 1) and found that this subunit has a compact fold, similar to that of immunoglobulins, that consists of 10 β-strands and two 310 helices (37, 38). A low sequence complexity suggests that the upstream region of this domain (residues 1–31) is likely to be disordered in the absence of a binding partner, because constructs containing this particular segment result in protein aggregation. Moreover, a large portion of this region also may serve as a membrane-targeting signal sequence that is proteolytically cleaved during cellular processing (19). Overall, the structure is held together by a buried disulfide bridge connecting the β2–β3 loop with the β8-strand which is flanked by 68Trp, a residue conserved among all β-subunit isoforms. On the other side, it packs against 114Ile and 133Val, both of which correspond to the similar Ile, Val, or Leu residues in other β-subunits (Fig. 2C). Because of the strictly conserved nature of the protein core (Fig. 2D), it is likely that the buried disulfide bond has a major effect on the stability of the protein. Another stabilizing element in the β4 structure is an ion-pair network involving two hydrogen bonds between 98Asp and 100Arg, which forms two additional bonds with 125Asp, as well as a hydrogen bond between 77Lys and 125Asp that is mediated by a water molecule (Fig. 2E). Such networks of ionic pairs are more stabilizing than individual salt bridges because of lower desolvation penalties (39). Next, we examined the surface of β4 for unique structural features that may contribute to an interaction with Nav1.2.
Fig. 2.
Crystal structure of the β4 extracellular domain. (A and B) Two views of the hβ4 extracellular domain, rotated by 180° around a vertical axis. β-Strands are in blue; 310 helices are in red. Dotted lines indicate a flexible region that is invisible in the electron-density map. The conserved cysteine bridge is shown in stick format, with the sulfur atoms colored yellow. The C58A mutation is indicated as A58, and three potential glycosylation sites are shown in as 45Asn (N45), 71Asn (N71), and 113Asn (N113). “To TM” indicates the position at which the single-transmembrane helix starts; N and C specify the N- and C-terminal ends of the structured part of the extracellular domain, respectively. (C) Sequence alignment of the extracellular regions of hβ1–4 with the secondary structure of β4 shown above. Amino acids conserved between β4 and other β-subunits are in bold, and known β-subunit–related disease mutations are in red. Conserved cysteines are highlighted by a gray background and the C58A locus by a green background. (D) Close-up view of the disulfide bond showing the nearby conserved hydrophobic core. (E) Close-up view of the ion-pair network formed by hydrogen bonds between 98Asp and 100Arg, which forms two additional bonds with 125Asp as well as a water-mediated hydrogen bond between 77Lys and 125Asp.
Table 1.
Data collection and refinement statistics
| Parameter | β4 C58A | β4 C131W |
| Data collection | ||
| Space group | P32 2 1 | P 1 21 1 |
| Cell dimensions | ||
| a, b, c, in Å | 43.44, 43.44, 108.38 | 31.50, 42.50, 89.20 |
| α, β, γ, in degrees | 90.0, 90.0, 120.0 | 90.0, 91.5, 90.0 |
| Resolution, in Å | 27.09–1.72 (1.77–1.72)*,† | 29.51–1.74 (1.77–1.74)*,† |
| Rsym or Rmerge | 6.0 (70.1) | 6.2 (53.8) |
| I/σI | 21.4 (2.9) | 13.6 (2.6) |
| Completeness, % | 99.9 (98.5) | 98.8 (91.4) |
| Redundancy | 10.4 (8.9) | 3.6 (3.1) |
| Refinement | ||
| Resolution, in Å | 26.0–1.72 | 26.0–1.74 |
| No. reflections | 13,164 | 24,173 |
| Rwork/Rfree | 18.7/ 22.8 | 17.5/21.9 |
| No. atoms | ||
| Protein | 907 | 1,961 |
| Water | 143 | 279 |
| B-factors | ||
| Protein | 29.4 | 20.8 |
| Water | 43.8 | 32.3 |
| rmsd | ||
| Bond lengths, in Å | 0.007 | 0.007 |
| Bond, in degrees | 1.189 | 1.172 |
For each structure, one crystal was used for data collection and structure determination.
Values in parentheses indicate the highest-resolution shell.
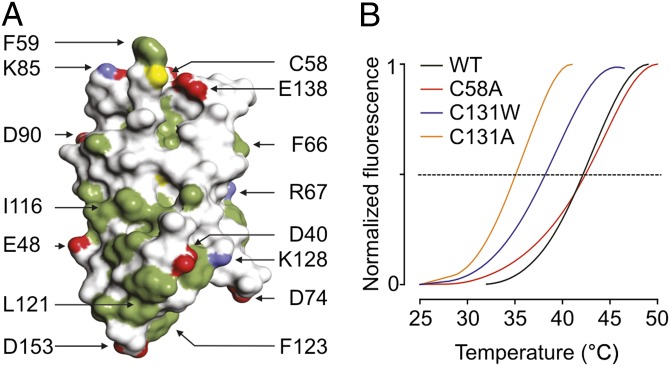
The β4 extracellular domain consists of solvent-accessible surface area of ∼6,600 Å2 with one side exposing multiple hydrophobic side chains as well as residue 58, a Cys in WT protein but replaced by Ala to facilitate crystallization (Fig. 3A). 58Cys is located in a loop between the β2- and β3-strands and is lined by two hydrophobic pockets, thereby placing it in an ideal position to form a disulfide bond with another free Cys (Fig. 2A). Nonetheless, conformational changes resulting from the C58A mutation are unlikely, because the residue is located at the protein surface, and the mutant has a thermal stability similar to that of WT β4 (Fig. 3B). Strikingly, the position of 58Cys in β4 corresponds to that of the Cys (26Cys) thought to be involved in linking β2 to Nav1.1 (40). Here, removing the covalent bond with Nav1.1 by mutating 26Cys to Ala results in an altered subcellular localization pattern of β2. Given the prominent position of 58Cys in the β4 crystal structure and the functional importance of the corresponding residue in β2, we asked whether 58Cys plays a role in determining the influence of β4 on Nav1.2 toxin pharmacology.
Fig. 3.
Surface representation and thermal stability of the β4 extracellular domain. (A) Surface representation of the β4 extracellular domain. Hydrophobic side chains are indicated in green, carboxyl groups of Asp and Glu are shown in red, and the positively charged nitrogen groups of Lys and Arg are in blue. The position of 58Cys is shown in yellow; other select residues are labeled for reference purposes. (B) ThermoFluor experiments showing average melting curves for WT β4 and three mutants under reducing conditions (14 mM 2-ME). The melting temperatures are WT: 42.2 ± 0.1 °C; C58A: 42.2 ± 0.2 °C; C131W: 38.3 ± 1.1 °C; and C131A: 35.1 ± 0.4 °C (SDs are the results of three measurements). The melting curves for C131W and C131A are identical in the absence or presence of 2-ME.
Mutating 58Cys in β4 Restores Nav1.2 Sensitivity to ProTx-II.
To examine whether the ability of β4 to alter Nav1.2 toxin susceptibility depends on a disulfide bond involving 58Cys, we initially incubated Nav1.2/β4-expressing oocytes with a reducing agent (DTT) for 60 min. Subsequently, we transferred the oocyte to a physiological recording solution and tested whether Nav1.2 regains sensitivity to ProTx-II (Fig. S4). Even though all exposed cysteines may have been reduced, channel-gating behavior is not affected significantly; however, we observe that the toxin now inhibits Nav1.2 as if no β4 is present, suggesting that the disulfide bond between 58Cys and Nav1.2 is crucial in modifying channel susceptibility to ProTx-II in the presence of β4.
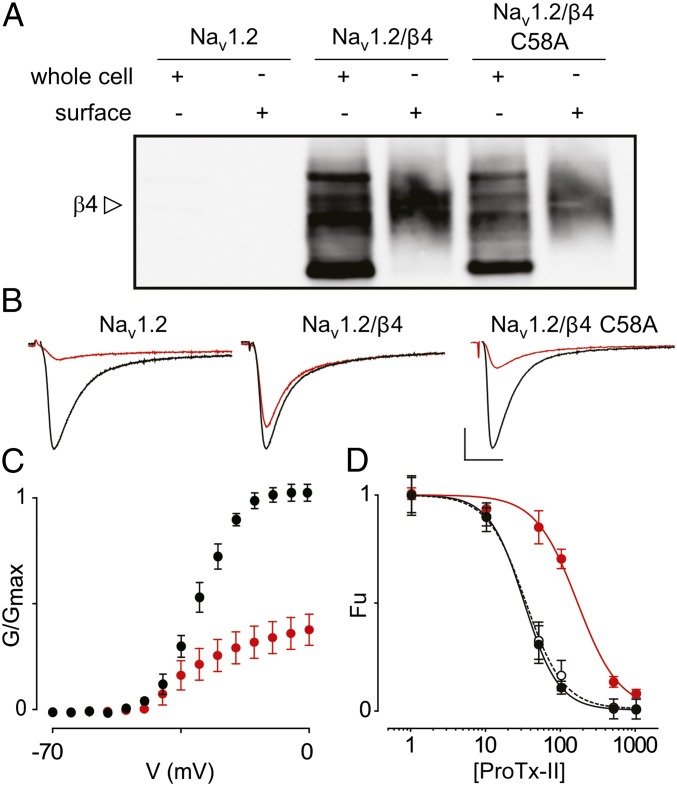
Next, we replaced 58Cys in β4 with Ala and exploited the presence of an intracellular Myc-tag to determine whether cellular trafficking of this mutant is altered. By doing so, we discovered that the WT β4 protein and the C58A mutant are produced in large quantities (Fig. 4A). Moreover, biotinylation experiments reveal the presence of both variants within the lipid membrane, most likely in a glycosylated form (41). When β4 glycosylation is removed using peptide-N-glycosidase F (PNGase F), an amidase that releases N-linked oligosaccharides, the molecular weight of the protein on the membrane surface corresponds to the predicted mass of β4 (28 kDa) (Fig. S5) (19). Having established the membrane insertion of β4 and the C58A mutant (without or in the presence of Nav1.2; Fig. S5), we next applied 100 nM ProTx-II to cells expressing Nav1.2/β4 C58A and observed a level of inhibition similar to that obtained when no β4 is present (Fig. 4 B and C). When comparing the affinities of ProTx-II for Nav1.2 in more detail, we note an approximately fivefold decrease in the presence of β4 compared with control conditions without β4 (IC50 from 32 ± 1 nM to 164 ± 33 nM with slopes of 1.9 ± 0.1 and 1.6 ± 0.3, respectively). Moreover, the affinity of ProTx-II for Nav1.2 coexpressed with the C58A mutant (IC50 = 33 ± 2 nM with a slope of 1.7 ± 0.1) is comparable to the IC50 obtained on cells lacking β4 (Fig. 4D). Taken together, our results identify 58Cys as a reactive residue that, when mutated, eliminates the influence of β4 on Nav1.2 toxin pharmacology by perturbing a unique disulfide bond with the Nav channel.
Fig. 4.
Influence of β4 C58A on ProTx-II susceptibility of Nav1.2. (A) Correct cellular trafficking of WT β4 and the C58A mutant in oocytes is shown using Western blot analyses by probing for an intracellular β4 Myc-tag combined with primary amine biotinylation of surface proteins. The gel demonstrates that the oocytes produce WT β4 as well as the C58A mutant and that the membrane-inserted protein is heavily glycosylated. The open arrow indicates glycosylated β4. A more detailed gel showing deglycosylated β4 is shown in Fig. S5. (B) The effect of 100 nM ProTx-II on Nav1.2, Nav1.2/β4, and Nav1.2/β4 C58A. Representative sodium currents are elicited by a depolarization to −20 mV before (black trace) and after (red trace) the addition of ProTx-II from a holding potential of −90 mV. The x-axis is 10 ms; the y-axis is ∼0.5 μA. (C) Normalized conductance–voltage relationship (G/Gmax) of the Nav1.2/β4 C58A channel without (black filled circles) and in the presence (red filled circles) of ProTx-II. (D) Affinity (IC50) of ProTx-II interacting with WT Nav1.2 (black filled circles), Nav1.2/β4 (red filled circles), and Nav1.2/β4 C58A (black open circles connected by the dashed line). The concentration dependence for toxin inhibition is plotted as the fraction of uninhibited channels (Fu). Lines represent a fit with the Hill equation, and IC50 values are mentioned in the text. n = 3–5 for each toxin concentration; error bars represent S.E.M.
Mapping Clinically Relevant β-Subunit Mutations onto the β4 Structure.
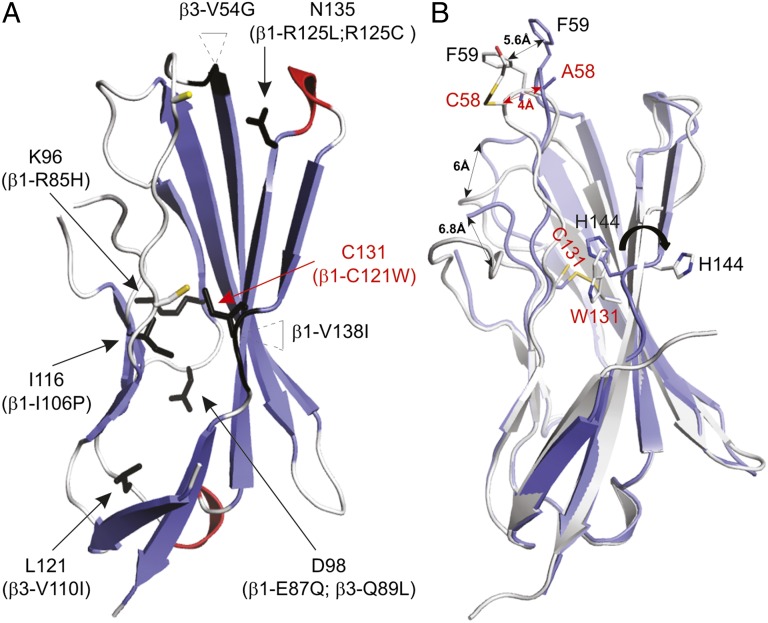
Reflecting its medical importance, atypical β-subunit behavior has been implicated in various epilepsy syndromes and cardiac disorders (5). However, little is known about the relationship between the structural and functional consequences of mutations and a particular clinical phenotype. Our results presented here provide a unique opportunity to map abnormalities within the extracellular domain onto a high-resolution β4 crystal structure and to explore their mode of action (Fig. 5 and Table S2). For example, R85H and E87Q in β1 have been associated with atrial fibrillation and Brugada syndrome, respectively (42, 43). Both residues are located near or within the β5–β6 loop and form a patch of solvent-accessible surface area (Fig. 5A), suggesting that these amino acids are part of an important functional interface that may be targeted by therapeutics (28). Similarly, the R125L variant has been identified in patients who have generalized epilepsy plus febrile seizures plus (GEFS+) (44), whereas R125C may contribute to the occurrence of Dravet syndrome (45). Because this residue lines a pocket near the conserved 58Cys, the addition of a Cys may interfere with the formation of a proper disulfide bond with the Nav channel (Fig. 5A).
Fig. 5.
Disease-related mutations mapped onto the β4 extracellular domain structure and crystal structure of the β4 C131W variant. (A) Arrows indicate the positions of known β1 and β3 disease mutations (summarized in Table S2) mapped onto the β4 extracellular domain. The corresponding residue substitutions are shown in parentheses, and the positions of 53Cys and 58Cys are indicated by a yellow stick for reference purposes. Two mutations occur within inserted regions, and the main chains of amino acids next to these insertions are also colored black. (B) The crystal structure of the C131W mutant (white) superimposed onto the WT β4 crystal structure (blue). Shifts in the positions of several regions are indicated by double-headed arrows, and select side chains are shown for reference purposes. 58Cys in the C131W structure appears to have a 2-ME molecule attached to it, further highlighting its reactivity. In both A and B, mutants discussed in this work are displayed in red.
Surface-exposed mutations may hint at a disruption of a functional interface, whereas buried substitutions are likely to affect overall protein folding. For instance, in patients who suffer from Dravet syndrome (46) a Pro residue replaces a concealed Ile at position 106 in β1, where it is likely to interfere with protein folding (Fig. 5A). The V110I mutation in β3 has been linked to Brugada syndrome and also affects a residue buried within a hydrophobic core, resulting in reduced channel expression (47). Interestingly, the first epileptogenic mutation attributed to Nav channel β-subunits is a Cys-to-Trp substitution that disrupts the disulfide bridge within the extracellular domain of β1 (30). Although the protein traffics to the membrane, the precise mechanism relating this C121W variant to the resulting GEFS+ disease phenotype is still a matter of debate (48–50). We chose to mutate the corresponding residue in β4 (C131W and C131A) to determine whether disrupting this strictly conserved internal disulfide bond repositions 58Cys to such an extent that it cannot interact with the Nav channel.
Even though a lack of space for a Trp side chain at the 131Cys position suggests that this mutant should be unable to fold properly and aggregate, both the C131W and the C131A construct can be purified to obtain nonaggregated protein, as indicated by size-exclusion chromatography (Fig. S6). Moreover, thermal stability measurements in the presence of reducing agent [14 mM 2-mercaptoethanol (2-ME)] only show ∼4-°C and ∼7-°C decreases in melting temperatures for C131W and C131A, respectively, compared with WT (Fig. 3B). The presence or absence of 2-ME did not affect the melting curves for the two mutants, and we did not observe an interpretable melting curve for WT β4 without reducing agent, an observation that is consistent with the major stabilizing effect of a disulfide bond. Taken together, these results demonstrate that, although the disulfide bond adds to stability, it is not strictly required for folding.
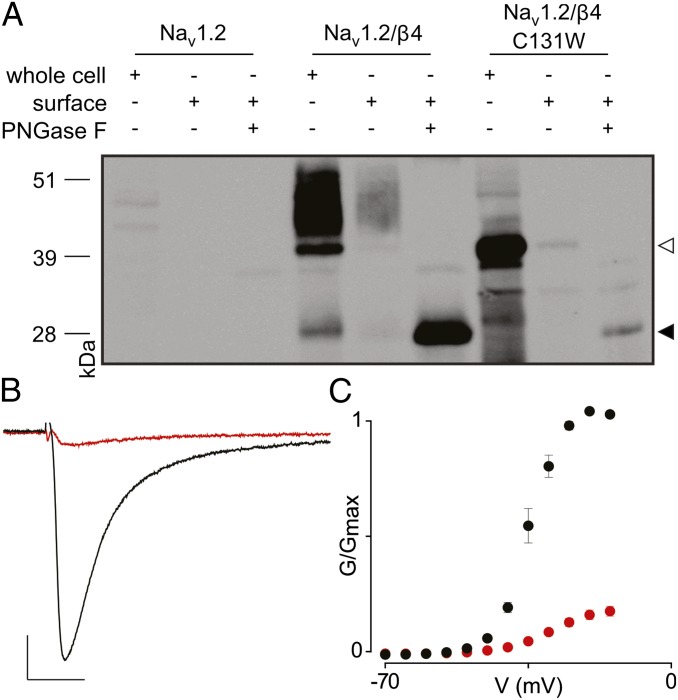
To explore this surprising result further, we next solved the crystal structure of the C131W construct at a resolution of 1.7 Å and found that the mutation results in conformational changes at multiple positions (Fig. 5B). Because the location of 144His in the β9–β10 loop would clash with the bulky Trp residue, this residue has swung away completely, and the β9–β10 loop adopts a β-strand conformation that results in a merged β9–β10 strand. In addition, 53Cys no longer is constrained by the disulfide bond, resulting in a dramatic shift of the β2–β3 loop (∼5.2 Å) within the main chain. This shift of the β2–β3 loop in turn shifts the positions of the neighboring β4–β5 loop (∼6 Å) and the β6–β7 loop (∼6.8 Å), resulting in complete remodeling of the surface. Because the β2–β3 loop contains the functionally important 58Cys residue, the C131W mutation remodels the Nav channel-binding interface, potentially altering the functional and pharmacological properties of the Nav channel signaling complex. To substantiate this notion, we tested whether the C131W mutant indeed loses its ability to modify Nav1.2 sensitivity to ProTx-II.
The C131W Mutation Alters the Influence of β4 on Nav1.2 Toxin Pharmacology.
To examine the effect of the C131W substitution on Nav1.2 toxin susceptibility, we first determined whether cellular trafficking of this mutant is altered. Similar to WT β4, C131W is produced in large amounts (Fig. 6A); however, a nonquantitative Western analysis of total cell lysate uncovers potential alterations in glycosylation patterns that may complicate protein separation from the endoplasmatic reticulum. Nonetheless, biotinylation experiments reveal the presence of C131W within the oocyte membrane (Fig. 6A and Fig. S5). Indeed, when glycosylation is removed using PNGase F, the molecular mass of the C131W mutant on the membrane surface closely matches the predicted mass of β4 (19). These results are consistent with previously reported observations with the β1 C121W mutant in a mouse model for epilepsy (51) and in human embryonic kidney cells stably expressing Nav1.1 (6). Subsequently, we applied 100 nM ProTx-II to cells expressing Nav1.2/β4 C131W and observed a level of inhibition over a wide voltage range similar to that obtained when neither β4 nor the C58A mutant is present (Fig. 6 B and C). Because the C131W mutant traffics to the membrane surface in oocytes, results from the ProTx-II experiment suggest that 58Cys no longer may be able to interact with Nav1.2. It is worth noting that our Western blot analysis (Fig. 6A and Fig. S5) does not rule out the possibility that impaired C131W trafficking in oocytes may contribute, at least in part, to the restoration of Nav1.2 toxin sensitivity. Together with our crystallographic data, these functional results show that, although the conserved cysteine bond is not strictly required to produce folded protein, it does dictate the overall conformation, including the position of the important 58Cys-containing bioactive surface.
Fig. 6.
Influence of β4 C131W on ProTx-II susceptibility of Nav1.2. (A) Cellular trafficking of WT β4 and the C131W mutant in oocytes is shown using nonquantitative Western blot analyses by probing for an intracellular Myc-tag combined with primary amine biotinylation of surface proteins. The gel demonstrates that WT β4, as well as the C131W mutant, is produced and that the membrane-inserted protein is glycosylated. The open arrowhead indicates glycosylated protein, whereas the closed arrowhead represents deglycosylated protein. (B) Effect of 100 nM ProTx-II on Nav1.2/β4 C131W. Representative sodium current is elicited by a depolarization to −20 mV before (black trace) and after (red trace) the addition of ProTx-II from a holding potential of −90 mV. The x-axis is 10 ms; the y-axis is ∼0.5 μA. (C) Normalized conductance–voltage relationship (G/Gmax) of the Nav1.2/β4 C131W channel without (black filled circles) and in the presence of (red filled circles)100 nM ProTx-II. n = 3–5; error bars represent S.E.M.
Discussion
β-Subunits are vital members of the larger Nav channel signaling complex in which they modify channel function to fine-tune the electrical excitability of native tissues (5–7, 9, 52). The goal of the present study was to investigate the interaction of β-subunits with Nav channels on a molecular level and the role of these ancillary proteins in shaping the pharmacological sensitivities of the neuronal Nav1.2 isoform. Our experiments with an extensive collection of animal toxins establish that β-subunits can alter Nav1.2 pharmacology drastically and that the resulting effects vary among β-subunit–toxin pairs (Fig. 1, Fig. S1, and Table S1). For example, β4 coexpression results in an approximately fivefold decrease in the affinity of ProTx-II for Nav1.2, whereas the same subunit reduces sodium influx upon TsVII application, an effect that is not observed with the WT channel or in the presence of β1–3. It is worth considering potential principles underlying β-subunit modulation of Nav channel pharmacology. For example, a toxin may compete with a β-subunit for binding to a particular region within Nav1.2. For instance, LqqIV interacts with the domain IV voltage sensor (Fig. S2), and its effect is influenced by β1, suggesting a direct or allosteric interaction of this particular subunit with domain IV within Nav1.2 (53–55). Alternatively, ligands may bind directly to β-subunits (56), an intriguing concept that also has been observed with the voltage-gated calcium channel inhibitor gabapentin, which acts through the transmembrane α2δ subunit (57). Interestingly, β3 is unable to modulate Nav1.2 susceptibility to any of the toxins we tested, but this particular subunit influences lidocaine binding to the neonatal Nav1.3 and cardiac Nav1.5 isoform (58, 59). This result highlights the unique character of the interaction between a particular β-subunit and a given Nav channel isoform (60).
Because the most striking effects on Nav1.2 toxin susceptibility are observed with the β4 subunit, we crystallized its extracellular domain and identified 58Cys as a surface-exposed residue that, when mutated, abolishes the impact of β4 on ProTx-II binding to Nav1.2 (Figs. 2 and 3). Because this Cys is conserved in β2 and β4, it likely belongs to a universal docking site for multiple Nav1 channel isoforms. The two other conserved cysteines (53Cys and 131Cys) are buried within a hydrophobic core where they form a disulfide bond. When 131Cys is mutated to Trp or Ala (Fig. 3B), we surprisingly find that the extracellular domain is still folded, thus demonstrating that the cysteine bridge is not strictly required to produce functional protein. However, the crystal structure of the C131W mutant, which mimics a well-established epileptogenic mutation found in β1, reveals conformational changes at multiple locations, including the loop that contains the conserved 58Cys (Fig. 5B). Subsequently, an important role of the buried cysteine bridge may be to maintain a specific local structure that allows an interaction with the Nav channel and that, when disrupted (Fig. 6), creates a loss-of-function phenotype that relates to disorders such as GEFS+ (30).
Collectively, the functional and crystallographic results reported here shed light on the intricate interactions of β4 within the Nav channel signaling complex and establish a key role for β-subunits in shaping Nav1.2 pharmacology. As such, an important concept emerging from our work is that β-subunits provide exciting opportunities for designing new therapeutic strategies to correct their abnormal behaviors.
Materials and Methods
Toxin Acquisition and Purification.
ProTx-I and ProTx-II (61) were acquired from Peptides International, β-PMTX (62) was obtained from Alomone Laboratories, and ATX-II (63), lidocaine, and ambroxol were from Sigma-Aldrich. AaHII from Androctonus australis hector venom, TsVII from Tityus serrulatus venom, and LqqIV from Leiurus quinquestriatus were purified as described previously (64–66). Toxins were kept at −20 °C, and aliquots were dissolved in appropriate solutions containing 0.1% BSA.
Two-Electrode Voltage-Clamp Recording from Xenopus Oocytes.
The DNA sequences of rNav1.2a (67), rβ1–4 (acquired from Origene and modified for oocyte expression), and of the C58A and C131W mutants were confirmed by automated DNA sequencing, and cRNA was synthesized using T7 polymerase (mMessage mMachine kit; Ambion) after linearizing the DNA with appropriate restriction enzymes. LqqIV binding site experiments on chimeric voltage-gated potassium channels were carried out as previously described (34). Channels were expressed together with a β-subunit (1:5 molar ratio) in Xenopus oocytes that were incubated at 17 °C in 96 mM NaCl, 2 mM KCl, 5 mM Hepes, 1 mM MgCl2, 1.8 mM CaCl2, and 50 g/mL gentamycin (pH 7.6) with NaOH for 1–2 d after cRNA injection, and then were studied using two-electrode voltage-clamp recording techniques (OC-725C; Warner Instruments) with a 150-μL recording chamber. Data were filtered at 4 kHz and digitized at 20 kHz using pClamp 10 software (Molecular Devices). Microelectrode resistances were 0.5–1 MΩ when filled with 3 M KCl. The external recording solution (ND100) contained 100 mM NaCl, 5 mM Hepes, 1 mM MgCl2, and 1.8 mM CaCl2 (pH 7.6) with NaOH. All experiments were performed at room temperature (∼22 °C). Leak and background conductances, identified by blocking the channel with tetrodotoxin (Alomone Laboratories), have been subtracted for all Nav channel currents. All chemicals used were obtained from Sigma-Aldrich unless otherwise indicated.
Analysis of Channel Activity and Toxin–Channel Interactions.
Voltage–activation relationships were obtained by measuring steady-state currents and calculating conductance (G). In representative cases a Boltzmann function was fitted to the data according to: 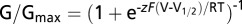 where G/Gmax is the normalized conductance, z is the equivalent charge, V1/2 is the half-activation voltage, F is Faraday's constant, R is the gas constant, and T is temperature in Kelvin. Occupancy of closed or resting channels by ProTx-II and other toxins was examined using negative holding voltages when open probability was very low, and the fraction of uninhibited channels (Fu) was estimated using depolarizations that are too weak to open toxin-bound channels, as described previously (34). After the toxin was added to the recording chamber, the equilibration between the toxin and the channel was monitored using weak depolarizations elicited at 5-s intervals. Concentration dependence for ProTx-II inhibition of Nav channels is plotted as Fu measured at negative voltages versus toxin concentration. A Hill equation was fitted the data to obtain affinity values. Off-line data analysis was performed using Clampfit 10 (Molecular Devices) and Origin 8 (OriginLab).
where G/Gmax is the normalized conductance, z is the equivalent charge, V1/2 is the half-activation voltage, F is Faraday's constant, R is the gas constant, and T is temperature in Kelvin. Occupancy of closed or resting channels by ProTx-II and other toxins was examined using negative holding voltages when open probability was very low, and the fraction of uninhibited channels (Fu) was estimated using depolarizations that are too weak to open toxin-bound channels, as described previously (34). After the toxin was added to the recording chamber, the equilibration between the toxin and the channel was monitored using weak depolarizations elicited at 5-s intervals. Concentration dependence for ProTx-II inhibition of Nav channels is plotted as Fu measured at negative voltages versus toxin concentration. A Hill equation was fitted the data to obtain affinity values. Off-line data analysis was performed using Clampfit 10 (Molecular Devices) and Origin 8 (OriginLab).
Nonquantitative Biochemical Assessment of β4 Production in Xenopus Oocytes.
Batches of 20 oocytes expressing Nav1.2, Nav1.2/β4, Nav1.2/β4 C58A, and Nav1.2/β4 C131W were washed with ND100 and incubated with 0.5 mg/mL Sulfo-NHS-LC-biotin (Pierce) for 30 min. Oocytes were thoroughly washed again (by pipetting up and down) in ND100 before lysis in 400 μL buffer H (1% Triton X-100, 100 mM NaCl, 20 mM Tris⋅HCl, pH 7.4) plus protease inhibitors (Clontech). All subsequent steps were performed at 4 °C. Lysates were shaken gently for 15 min and then were centrifuged at 16,200 × g for 3 min. The pellet was discarded, and the supernatant was transferred to a fresh 1.5-mL Eppendorf tube; 40 μL of supernatant was stored at −80 °C for later use as the whole-cell protein aliquot. Then 200 μL of hydrophilic streptavidin magnetic beads (New England Biolabs) was added, and the sample was shaken gently at 4 °C overnight. Beads were washed six times with buffer H and were resuspended in 40 μL buffer H, after which biotinylated protein was dissociated from the beads by the addition of 40 uL 1× LDS loading buffer plus reducing agent [final concentration: 10% (vol/vol) 2-ME, 50 mM DTT] and boiling at 95 °C for 5 min. Deglycosylated samples were prepared by incubating the surface protein aliquot with PNGase-F and 1% Nonidet P-40 at 37 °C for I h. All samples were diluted appropriately in buffer H to give roughly equal protein concentrations, as measured by absorbance at 280 nM. Then 7.5 μL of the supernatant was run on a 10% (wt/vol) Bis-Tris NuPAGE Novex Mini-Gel (Invitrogen) with 3-(N-morpholino)propanesulfonic acid running buffer and were analyzed by Western blot analysis. Nitrocellulose membranes were probed with 1:1,000 mouse anti-Myc antibody (Cell Signaling Technologies) as the primary antibody and 1:10,000 goat anti-mouse HRP-conjugated antibody (Thermo-Fisher Scientific) as the secondary antibody. Membranes were incubated for 5 min with an enhanced chemiluminescent substrate (68) before imaging.
Production of the β4 Extracellular Domain.
Human β4 (hβ4) (32-157) acquired from OriGene was cloned into pET28HMT (69). Mutations were introduced using the QuikChange kit from Stratagene according to the manufacturer’s instructions. Proteins were expressed at 18 °C in Escherichia coli Rosetta (DE3) pLacI strains (Novagen), were induced at an OD600 of ∼0.6 with 0.3 mM isopropyl β-d-1-thiogalactopyranoside, and were grown overnight before harvesting. Cells were lysed via sonication in buffer A (250 mM KCl and 10 mM Hepes, pH 7.4) supplemented with 25 µg/mL DNaseI and 25 µg/mL lysozyme. After centrifugation, the supernatant was applied to a PorosMC column (Tosoh Biosep), washed with buffer A plus 10 mM imidazole, and eluted with buffer B (250 mM KCl plus 500 mM imidazole, pH 7.4). The protein was dialyzed overnight against buffer A and cleaved simultaneously with recombinant TEV protease. Next, the samples were run on another PorosMC column in buffer A, and the flowthrough was collected and dialyzed against buffer C (50 mM KCl plus 20 mM Tris⋅Cl, pH 8.0), applied to a HiloadQ column (GE Healthcare), and eluted with a gradient from 0–30% buffer D (2 M KCl plus 20 mM Tris⋅Cl, pH 8.0). Finally, the samples were run on a Superdex200 gel-filtration column (GE Healthcare) in buffer A. The protein samples were exchanged with 25 mM KCl plus 10 mM Hepes (pH 7.4), concentrated to 10–20 mg/mL using Amicon concentrators (3 K molecular weight cut off; Millipore), and stored at −80 °C.
Crystallization, Data Collection, and Structure Solution.
Crystals were grown using the sitting-drop method at 4 °C. β4 (32-157) C58A was crystallized in 0.1 M Hepes (pH 7) and 15% (wt/vol) PEG 20000. The C131W mutant was crystallized in 0.2 M ammonium formate, 20% (wt/vol) PEG formate, and 20% (wt/vol) PEG 3350 at 4 °C. Crystals were flash-frozen after transfer to the same solution supplemented with 30% (vol/vol) glycerol. Diffraction experiments were performed at the Advanced Photon Source (Chicago) beamline 23-ID-D-GM/CA, and datasets were processed using XDS (70). A search model was created by using only β-strands from Protein Data Bank (PDB) ID code 1NEU and with all side chains truncated to Ala. Molecular replacement was performed using Phaser (71), yielding poor initial phases which were improved via autobuilding in ARP/wARP (72). The model was completed by successive rounds of manual model building in COOT (73) and refinement using Refmac5.5 (74). A simulated annealing composite omit map was calculated with CNS (75) to verify the absence of residual model bias. No residues were found to be in disallowed regions of the Ramachandran plot. All structure figures were prepared using PYMOL (DeLano Scientific). Coordinates are available in the PDB database (ID codes 4MZ2 and 4MZ3).
Thermal Melting Experiments.
The protein melting curves were measured by means of ThermoFluor experiments (69, 76). Samples for melting curves contained 50 μL of 0.1 mg/mL protein and 1× SYPRO Orange solution (Invitrogen) using the manufacturer’s instructions. The curves were measured in a DNA engine Opticon 2 real-time PCR machine (Bio-Rad), using the SYBR green filter option. The temperature was ramped from 25–95 °C in 0.5-°C steps, with 15 s at each step. The melting temperatures were taken as the midpoint of each transition.
Supplementary Material
Acknowledgments
We thank Al Goldin for sharing the rNav1.2a clone; Marie-France Martin-Eauclaire and Pierre Bougis for sharing TsVII, AaHII, and LqqIV; YaWen Lu for assistance with biochemistry; the staff of the Advanced Photon Source (Chicago) GM/CA-CAT beamline 23-iD-D; the staff at the SSRL beamlines (Menlo Park); and the members of the F.B. and F.V.P. laboratories for helpful discussions. This work was supported in part by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award R00NS073797 (to F.B.) and by Human Frontier Science Program Grant RGY0064/2013 (to F.B. and F.V.P.). S.D. is supported by a postdoctoral fellowship of the Heart and Stroke Foundation of Canada. F.V.P. is a Michael Smith Foundation of Health Research Career Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4MZ2 and 4MZ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314557110/-/DCSupplemental.
References
- 1.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57(4):397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 2.Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: From molecule to man. Nat Rev Neurosci. 2013;14(1):49–62. doi: 10.1038/nrn3404. [DOI] [PubMed] [Google Scholar]
- 3.George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115(8):1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15(1):35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Brackenbury WJ, Isom LL. Na channel beta subunits: Overachievers of the ion channel family. Front Pharmacol. 2011;2:53. doi: 10.3389/fphar.2011.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aman TK, et al. Regulation of persistent Na current by interactions between beta subunits of voltage-gated Na channels. J Neurosci. 2009;29(7):2027–2042. doi: 10.1523/JNEUROSCI.4531-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buffington SA, Rasband MN. Na+ channel-dependent recruitment of Navβ4 to axon initial segments and nodes of Ranvier. J Neurosci. 2013;33(14):6191–6202. doi: 10.1523/JNEUROSCI.4051-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chioni AM, Brackenbury WJ, Calhoun JD, Isom LL, Djamgoz MB. A novel adhesion molecule in human breast cancer cells: Voltage-gated Na+ channel beta1 subunit. Int J Biochem Cell Biol. 2009;41(5):1216–1227. doi: 10.1016/j.biocel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho C, Zhao J, Malinowski S, Chahine M, O’Leary ME. Differential expression of sodium channel β subunits in dorsal root ganglion sensory neurons. J Biol Chem. 2012;287(18):15044–15053. doi: 10.1074/jbc.M111.333740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marionneau C, et al. The sodium channel accessory subunit Navβ1 regulates neuronal excitability through modulation of repolarizing voltage-gated K⁺ channels. J Neurosci. 2012;32(17):5716–5727. doi: 10.1523/JNEUROSCI.6450-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen HM, et al. Modulation of voltage-gated K+ channels by the sodium channel β1 subunit. Proc Natl Acad Sci USA. 2012;109(45):18577–18582. doi: 10.1073/pnas.1209142109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreibmayer W, Wallner M, Lotan I. Mechanism of modulation of single sodium channels from skeletal muscle by the beta 1-subunit from rat brain. Pflugers Arch. 1994;426(3-4):360–362. doi: 10.1007/BF00374796. [DOI] [PubMed] [Google Scholar]
- 13.Zhang ZN, et al. The voltage-gated Na+ channel Nav1.8 contains an ER-retention/retrieval signal antagonized by the beta3 subunit. J Cell Sci. 2008;121(Pt 19):3243–3252. doi: 10.1242/jcs.026856. [DOI] [PubMed] [Google Scholar]
- 14.McCormick KA, et al. Molecular determinants of Na+ channel function in the extracellular domain of the beta1 subunit. J Biol Chem. 1998;273(7):3954–3962. doi: 10.1074/jbc.273.7.3954. [DOI] [PubMed] [Google Scholar]
- 15.Isom LL, et al. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256(5058):839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 16.Isom LL, et al. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83(3):433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 17.Morgan K, et al. Beta 3: An additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci USA. 2000;97(5):2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patino GA, et al. Voltage-gated Na+ channel β1B: A secreted cell adhesion molecule involved in human epilepsy. J Neurosci. 2011;31(41):14577–14591. doi: 10.1523/JNEUROSCI.0361-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu FH, et al. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci. 2003;23(20):7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazen-Gillespie KA, et al. Cloning, localization, and functional expression of sodium channel beta1A subunits. J Biol Chem. 2000;275(2):1079–1088. doi: 10.1074/jbc.275.2.1079. [DOI] [PubMed] [Google Scholar]
- 21.Bant JS, Raman IM. Control of transient, resurgent, and persistent current by open-channel block by Na channel beta4 in cultured cerebellar granule neurons. Proc Natl Acad Sci USA. 2010;107(27):12357–12362. doi: 10.1073/pnas.1005633107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medeiros-Domingo A, et al. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116(2):134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan BH, et al. Sudden infant death syndrome-associated mutations in the sodium channel beta subunits. Heart Rhythm. 2010;7(6):771–778. doi: 10.1016/j.hrthm.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li RG, et al. Mutations of the SCN4B-encoded sodium channel β4 subunit in familial atrial fibrillation. Int J Mol Med. 2013;32(1):144–150. doi: 10.3892/ijmm.2013.1355. [DOI] [PubMed] [Google Scholar]
- 25.Oyama F, et al. Sodium channel beta4 subunit: Down-regulation and possible involvement in neuritic degeneration in Huntington’s disease transgenic mice. J Neurochem. 2006;98(2):518–529. doi: 10.1111/j.1471-4159.2006.03893.x. [DOI] [PubMed] [Google Scholar]
- 26.Diss JK, et al. Beta-subunits of voltage-gated sodium channels in human prostate cancer: Quantitative in vitro and in vivo analyses of mRNA expression. Prostate Cancer Prostatic Dis. 2008;11(4):325–333. doi: 10.1038/sj.pcan.4501012. [DOI] [PubMed] [Google Scholar]
- 27.Wong HK, et al. beta Subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J Biol Chem. 2005;280(24):23009–23017. doi: 10.1074/jbc.M414648200. [DOI] [PubMed] [Google Scholar]
- 28.Brackenbury WJ, Isom LL. Voltage-gated Na+ channels: Potential for beta subunits as therapeutic targets. Expert Opin Ther Targets. 2008;12(9):1191–1203. doi: 10.1517/14728222.12.9.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson MJ, et al. Navβ subunits modulate the inhibition of Nav1.8 by the analgesic gating modifier μO-conotoxin MrVIB. J Pharmacol Exp Ther. 2011;338(2):687–693. doi: 10.1124/jpet.110.178343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace RH, et al. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19(4):366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 31.Zhang MM, et al. Co-expression of Na(V)β subunits alters the kinetics of inhibition of voltage-gated sodium channels by pore-blocking μ-conotoxins. Br J Pharmacol. 2013;168(7):1597–1610. doi: 10.1111/bph.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uebachs M, et al. Efficacy loss of the anticonvulsant carbamazepine in mice lacking sodium channel beta subunits via paradoxical effects on persistent sodium currents. J Neurosci. 2010;30(25):8489–8501. doi: 10.1523/JNEUROSCI.1534-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rogers JC, Qu Y, Tanada TN, Scheuer T, Catterall WA. Molecular determinants of high affinity binding of alpha-scorpion toxin and sea anemone toxin in the S3-S4 extracellular loop in domain IV of the Na+ channel alpha subunit. J Biol Chem. 1996;271(27):15950–15962. doi: 10.1074/jbc.271.27.15950. [DOI] [PubMed] [Google Scholar]
- 34.Bosmans F, Martin-Eauclaire MF, Swartz KJ. Deconstructing voltage sensor function and pharmacology in sodium channels. Nature. 2008;456(7219):202–208. doi: 10.1038/nature07473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiao Y, Blumenthal K, Jackson JO, 2nd, Liang S, Cummins TR. The tarantula toxins ProTx-II and huwentoxin-IV differentially interact with human Nav1.7 voltage sensors to inhibit channel activation and inactivation. Mol Pharmacol. 2010;78(6):1124–1134. doi: 10.1124/mol.110.066332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcotte P, Chen LQ, Kallen RG, Chahine M. Effects of Tityus serrulatus scorpion toxin gamma on voltage-gated Na+ channels. Circ Res. 1997;80(3):363–369. doi: 10.1161/01.res.80.3.363. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z, et al. Crystal structure of the extracellular domain of human myelin protein zero. Proteins. 2012;80(1):307–313. doi: 10.1002/prot.23164. [DOI] [PubMed] [Google Scholar]
- 38.Yereddi NR, et al. The immunoglobulin domain of the sodium channel β3 subunit contains a surface-localized disulfide bond that is required for homophilic binding. FASEB J. 2013;27(2):568–580. doi: 10.1096/fj.12-209445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karshikoff A, Ladenstein R. Ion pairs and the thermotolerance of proteins from hyperthermophiles: A “traffic rule” for hot roads. Trends Biochem Sci. 2001;26(9):550–556. doi: 10.1016/s0968-0004(01)01918-1. [DOI] [PubMed] [Google Scholar]
- 40.Chen C, et al. Identification of the cysteine residue responsible for disulfide linkage of Na+ channel α and β2 subunits. J Biol Chem. 2012;287(46):39061–39069. doi: 10.1074/jbc.M112.397646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou TT, Zhang ZW, Liu J, Zhang JP, Jiao BH. Glycosylation of the sodium channel β4 subunit is developmentally regulated and involves in neuritic degeneration. Int J Biol Sci. 2012;8(5):630–639. doi: 10.7150/ijbs.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheffer IE, et al. Temporal lobe epilepsy and GEFS+ phenotypes associated with SCN1B mutations. Brain. 2007;130(Pt 1):100–109. doi: 10.1093/brain/awl272. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe H, et al. Sodium channel β1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118(6):2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fendri-Kriaa N, et al. New mutation c.374C>T and a putative disease-associated haplotype within SCN1B gene in Tunisian families with febrile seizures. Eur J Neurol. 2011;18(5):695–702. doi: 10.1111/j.1468-1331.2010.03216.x. [DOI] [PubMed] [Google Scholar]
- 45.Patino GA, et al. A functional null mutation of SCN1B in a patient with Dravet syndrome. J Neurosci. 2009;29(34):10764–10778. doi: 10.1523/JNEUROSCI.2475-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogiwara I, et al. A homozygous mutation of voltage-gated sodium channel β(I) gene SCN1B in a patient with Dravet syndrome. Epilepsia. 2012;53(12):e200–e203. doi: 10.1111/epi.12040. [DOI] [PubMed] [Google Scholar]
- 47.Ishikawa T, et al. Novel SCN3B mutation associated with brugada syndrome affects intracellular trafficking and function of Nav1.5. Circ J. 2013;77(4):959–967. doi: 10.1253/circj.cj-12-0995. [DOI] [PubMed] [Google Scholar]
- 48.Baroni D, Barbieri R, Picco C, Moran O. Functional modulation of voltage-dependent sodium channel expression by wild type and mutated C121W-β1 subunit. J Bioenerg Biomembr. 2013;45(4):353–368. doi: 10.1007/s10863-013-9510-3. [DOI] [PubMed] [Google Scholar]
- 49.Egri C, Vilin YY, Ruben PC. A thermoprotective role of the sodium channel β1 subunit is lost with the β1 (C121W) mutation. Epilepsia. 2012;53(3):494–505. doi: 10.1111/j.1528-1167.2011.03389.x. [DOI] [PubMed] [Google Scholar]
- 50.Meadows LS, et al. Functional and biochemical analysis of a sodium channel beta1 subunit mutation responsible for generalized epilepsy with febrile seizures plus type 1. J Neurosci. 2002;22(24):10699–10709. doi: 10.1523/JNEUROSCI.22-24-10699.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wimmer VC, et al. Axon initial segment dysfunction in a mouse model of genetic epilepsy with febrile seizures plus. J Clin Invest. 2010;120(8):2661–2671. doi: 10.1172/JCI42219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cusdin FS, et al. The sodium channel beta3-subunit induces multiphasic gating in NaV1.3 and affects fast inactivation via distinct intracellular regions. J Biol Chem. 2010;285(43):33404–33412. doi: 10.1074/jbc.M110.114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chanda B, Asamoah OK, Bezanilla F. Coupling interactions between voltage sensors of the sodium channel as revealed by site-specific measurements. J Gen Physiol. 2004;123(3):217–230. doi: 10.1085/jgp.200308971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makita N, Bennett PB, George AL., Jr Molecular determinants of beta 1 subunit-induced gating modulation in voltage-dependent Na+ channels. J Neurosci. 1996;16(22):7117–7127. doi: 10.1523/JNEUROSCI.16-22-07117.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qu Y, et al. Functional roles of the extracellular segments of the sodium channel alpha subunit in voltage-dependent gating and modulation by beta1 subunits. J Biol Chem. 1999;274(46):32647–32654. doi: 10.1074/jbc.274.46.32647. [DOI] [PubMed] [Google Scholar]
- 56.Jover E, Massacrier A, Cau P, Martin MF, Couraud F. The correlation between Na+ channel subunits and scorpion toxin-binding sites. A study in rat brain synaptosomes and in brain neurons developing in vitro. J Biol Chem. 1988;263(3):1542–1548. [PubMed] [Google Scholar]
- 57.Gee NS, et al. The novel anticonvulsant drug, gabapentin (Neurontin), binds to the alpha2delta subunit of a calcium channel. J Biol Chem. 1996;271(10):5768–5776. doi: 10.1074/jbc.271.10.5768. [DOI] [PubMed] [Google Scholar]
- 58.Lenkowski PW, Shah BS, Dinn AE, Lee K, Patel MK. Lidocaine block of neonatal Nav1.3 is differentially modulated by co-expression of beta1 and beta3 subunits. Eur J Pharmacol. 2003;467(1-3):23–30. doi: 10.1016/s0014-2999(03)01595-4. [DOI] [PubMed] [Google Scholar]
- 59.Makielski JC, Limberis J, Fan Z, Kyle JW. Intrinsic lidocaine affinity for Na channels expressed in Xenopus oocytes depends on alpha (hH1 vs. rSkM1) and beta 1 subunits. Cardiovasc Res. 1999;42(2):503–509. doi: 10.1016/s0008-6363(99)00024-3. [DOI] [PubMed] [Google Scholar]
- 60.Farmer C, et al. Splice variants of Na(V)1.7 sodium channels have distinct β subunit-dependent biophysical properties. PLoS ONE. 2012;7(7):e41750. doi: 10.1371/journal.pone.0041750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Middleton RE, et al. Two tarantula peptides inhibit activation of multiple sodium channels. Biochemistry. 2002;41(50):14734–14747. doi: 10.1021/bi026546a. [DOI] [PubMed] [Google Scholar]
- 62.Konno K, et al. Isolation and structure of pompilidotoxins, novel peptide neurotoxins in solitary wasp venoms. Biochem Biophys Res Commun. 1998;250(3):612–616. doi: 10.1006/bbrc.1998.9299. [DOI] [PubMed] [Google Scholar]
- 63.Béress L, Béress R, Wunderer G. Isolation and characterisation of three polypeptides with neurotoxic activity from Anemonia sulcata. FEBS Lett. 1975;50(3):311–314. doi: 10.1016/0014-5793(75)80517-5. [DOI] [PubMed] [Google Scholar]
- 64.Céard B, De Lima ME, Bougis PE, Martin-Eauclaire MF. Purification of the main beta-toxin from Tityus serrulatus scorpion venom using high-performance liquid chromatography. Toxicon. 1992;30(1):105–110. doi: 10.1016/0041-0101(92)90506-z. [DOI] [PubMed] [Google Scholar]
- 65.Martin MF, Rochat H. Large scale purification of toxins from the venom of the scorpion Androctonus australis Hector. Toxicon. 1986;24(11-12):1131–1139. doi: 10.1016/0041-0101(86)90139-x. [DOI] [PubMed] [Google Scholar]
- 66.Kopeyan C, Martinez G, Rochat H. Primary structure of toxin IV of Leiurus quinquestriatus quinquestriatus: Characterization of a new group of scorpion toxins. FEBS Lett. 1985;181(2):211–217. [Google Scholar]
- 67.Auld VJ, et al. A rat brain Na+ channel alpha subunit with novel gating properties. Neuron. 1988;1(6):449–461. doi: 10.1016/0896-6273(88)90176-6. [DOI] [PubMed] [Google Scholar]
- 68.Diaz AN, Sanchez FG, Garcia JAG. Hydrogen peroxide assay by using enhanced chemiluminescence of the luminol-H2O2-horseradish peroxidase system: Comparative studies. Anal Chim Acta. 1996;327(2):161–165. [Google Scholar]
- 69.Lobo PA, Kimlicka L, Tung CC, Van Petegem F. The deletion of exon 3 in the cardiac ryanodine receptor is rescued by β strand switching. Structure. 2011;19(6):790–798. doi: 10.1016/j.str.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 70.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3(7):1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 74.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 75.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 76.Nettleship JE, Brown J, Groves MR, Geerlof A. Methods for protein characterization by mass spectrometry, thermal shift (ThermoFluor) assay, and multiangle or static light scattering. Methods Mol Biol. 2008;426:299–318. doi: 10.1007/978-1-60327-058-8_19. [DOI] [PubMed] [Google Scholar]
- 77.Weiser T, Wilson N. Inhibition of tetrodotoxin (TTX)-resistant and TTX-sensitive neuronal Na(+) channels by the secretolytic ambroxol. Mol Pharmacol. 2002;62(3):433–438. doi: 10.1124/mol.62.3.433. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.