Significance
Virus infection can trigger profound alterations in host cell metabolism, but the role of individual enzymes in this process is relatively unstudied. Here, we show that argininosuccinate synthetase 1 (AS1) antagonizes the production of herpes simplex virus 1 (HSV-1) in cultured fibroblasts. Infection reduces the level of AS1 protein, and further reduction induced by siRNA treatment enhances the production of HSV-1. AS1 deficiency mimics many of the metabolic changes induced by HSV-1 infection, demonstrating that a decrease in the activity of a single cellular enzyme is responsible for much of the metabolic reprogramming induced by HSV-1. These results underscore the dependence of HSV-1 replication on altered host cell metabolism.
Keywords: metabolomics, herpesviruses
Abstract
Herpes simplex virus 1 (HSV-1) infection triggers specific metabolic changes in its host cell. To explore the interactions between cellular metabolism and HSV-1 infection, we performed an siRNA screen of cellular metabolic genes, measuring their effect on viral replication. The screen identified multiple enzymes predicted to influence HSV-1 replication, including argininosuccinate synthetase 1 (AS1), which consumes aspartate as part of de novo arginine synthesis. Knockdown of AS1 robustly enhanced viral genome replication and the production of infectious virus. Using high-resolution liquid chromatography-mass spectrometry, we found that the metabolic phenotype induced by knockdown of AS1 in human fibroblasts mimicked multiple aspects of the metabolic program observed during HSV-1 infection, including an increase in multiple nucleotides and their precursors. Together with the observation that AS1 protein and mRNA levels decrease during wild-type infection, this work suggests that reduced AS1 activity is partially responsible for the metabolic program induced by infection.
Many viruses, including herpes simplex virus 1 (HSV-1), human cytomegalovirus (HCMV), influenza, Dengue, hepatitis C, and hepatitis E (1–11), have been shown to significantly perturb metabolic homeostasis over the course of infection. HSV-1 is an intriguing case study, because the virus encodes several of its own metabolic enzymes, including a dUTPase, uracil-DNA glycosylase, ribonucleotide reductase, and thymidine kinase (12). Infection of host cells by multiple strains of HSV-1 induces alterations to central carbon metabolism, such as increased flux through upper glycolysis, and the anapleurotic feeding of carbons into the citric acid (TCA) cycle through the activity of pyruvate carboxylase (1). Infection also induces nucleotide synthesis, manifested by the increased flux of carbons through aspartate to nucleotides, as well as the increased pool levels of multiple nucleotides themselves (1).
Although the metabolic reprogramming that occurs during HSV-1 infection has been described, the importance of specific host cell metabolic enzymes to infection is relatively unknown. Here we sought to further resolve the relationship between infection and metabolism by screening for cellular metabolic enzymes that modulate the efficiency of viral infection in primary human fibroblasts. The screen identified several dozen enzymes that modulate HSV-1 growth, including argininosuccinate synthetase (AS1), which antagonized viral replication.
AS1 acts as a homotetramer to catalyze the synthesis of argininosuccinate from aspartate and citrulline (13, 14). The enzyme plays a role in the production of urea in the kidney and liver, but functions primarily as the limiting step in the citrulline-nitric oxide (NO) cycle in other cell types (15). As a constituent of the citrulline-NO cycle, AS1 participates in the de novo production of the nonessential amino acid arginine as well as the soluble factor NO.
We found that AS1 knockdown by siRNA increased the yield of HSV-1 and that overexpression of the enzyme can reduce virus yield in healthy fibroblasts. AS1-deficient cells have the capacity to make more viral genomes and express higher levels of certain viral proteins and transcripts, while exhibiting a metabolic program reminiscent of that observed during infection. This study demonstrates that decreasing the level of a single metabolic enzyme is sufficient to mimic key aspects of the virus-induced metabolome and significantly improve the replicative capacity of HSV-1.
Results
A Subset of Human Metabolic Enzymes Modulates HSV-1 Replication.
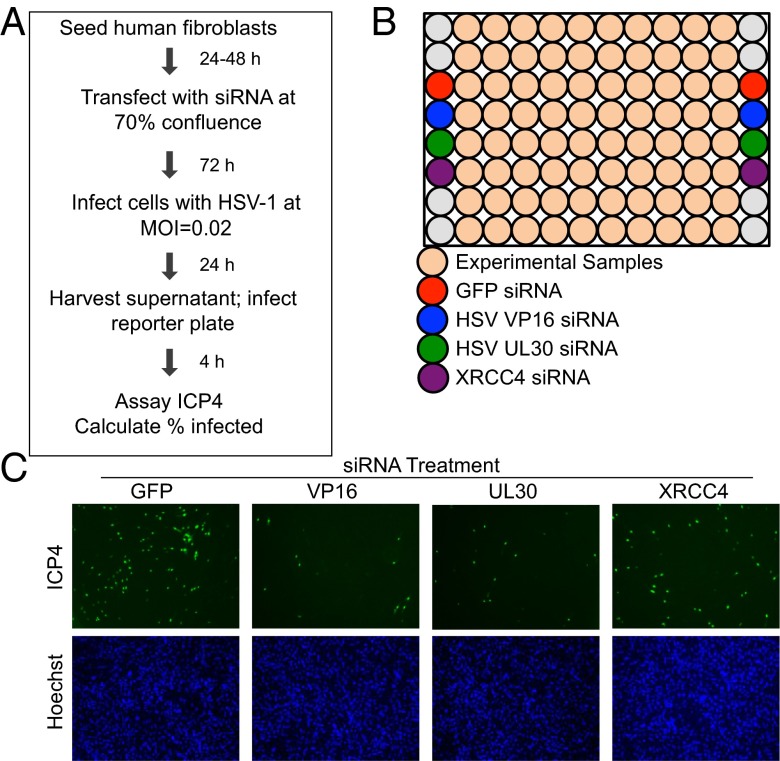
It has been well documented that HSV-1 infection triggers large-scale changes to host cell metabolism (1, 4), but the role played by individual cellular metabolic enzymes during the viral life cycle is less understood. To identify host metabolic enzymes that modulate the production of infectious progeny, we performed an siRNA screen (7) (Fig. 1A). Primary human fibroblasts were transfected with siRNAs targeting 401 mRNAs encoding human metabolic enzymes. Each mRNA was targeted by three separate siRNAs, which were tested independently. Transfected cells were incubated for 3 d before being infected at a multiplicity of 0.02 infectious units (IU) per cell with HSV-1. Infection at this low input multiplicity allowed for the identification of phenotypes that might otherwise be masked by multiplicity-dependent leakiness and guaranteed that the read-out of the screen remained in the linear range for all siRNAs tested. The infectious supernatant was collected at 24 h after infection (hpi) and then used to infect a fresh plate of untransfected fibroblasts. At 4 hpi, these cells were stained for the presence of the viral immediate-early infected cell protein 4 (ICP4) and host cell nuclei. Images were quantified and used to calculate the percentage of infected cells in each well. This technique allowed us to capture changes in viral replication efficiency at any step from attachment through assembly and egress.
Fig. 1.
Protocol for siRNA screen. (A) Subconfluent human fibroblasts were transfected with individual siRNAs and infected with HSV-1 72 h later at 0.02 IU per cell. At 24 hpi, supernatants were used to infect a fresh plate of fibroblasts, which were fixed 4 hpi and stained for the viral immediate-early protein ICP4 and nuclei. (B) Each plate in the screen included wells containing siRNAs targeting two viral proteins (VP16 and UL30) and one cellular protein (XRCC4) as positive controls, whereas a GFP siRNA was used as a negative control. (C) Representative images of control wells after staining the secondary infection plates for ICP4.
Each screen plate also included several experimental controls (Fig. 1B). An siRNA targeting GFP was used as a negative control, and siRNAs targeting the viral proteins VP16 and pUL30 (16) were used as positive controls. An siRNA targeting the cellular protein XRCC4, known to be important during HSV-1 infection (17), also served as a positive control. Sample images from the ICP4-stained secondary infection plates show that each of the three positive control siRNAs effectively decreased virus yield (Fig. 1C).
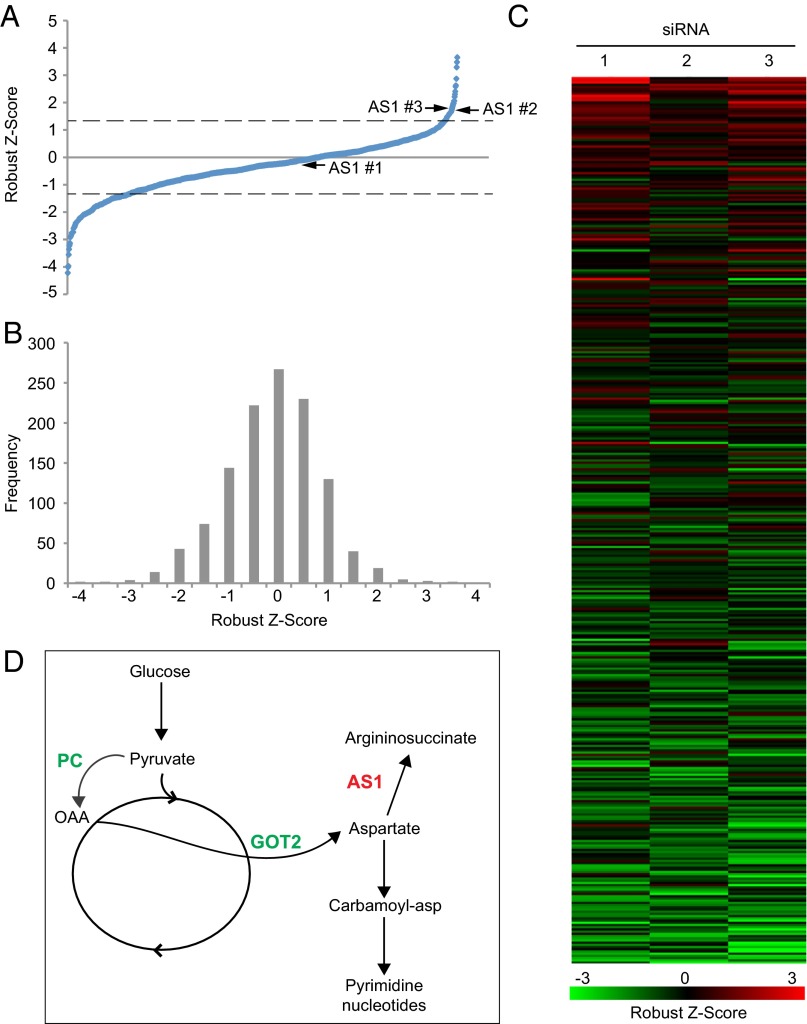
After collecting percent infected values for all siRNAs, data were normalized by robust z score (18) and plotted in ascending order (Fig. 2A and Dataset S1). A negative score indicates that knockdown of a cellular gene reduces virus yield, whereas a positive score indicates that knockdown increases yield. When z-score frequency was plotted, a leftward shift to the negative values was observed, suggesting that more cellular metabolic enzymes support HSV-1 replication than are inhibitory (Fig. 2B). The three independent scores for each cellular enzyme were grouped together (Fig. 2C), and the threshold for hit determination was set as those enzymes that had two or more siRNAs with absolute z scores ≥1.25. This ranking resulted in the discovery of 35 enzymes that contribute to viral replication (negative z score), and 3 that have an inhibitory effect (positive z score) (Table 1). Several functional groupings of enzymes appear, including two, heparan sulfate 3-O-sulfotransferase 1 and heparan sulfate 3-O-sulfotransferase 1 (HS3ST1 and HS6ST1), which are involved in synthesis of heparan sulfate, which is an HSV-1 cellular receptor (19), and two, glycerol-3-phosphate acyltransferase 1 and 2 (GPAM and GPAT2), which are involved in glycerolipid biosynthesis (Dataset S1). Interestingly, when the screen hits were plotted to a global metabolic map, one cluster of enzymes was identified as important to infection. This cluster included pyruvate carboxylase [PC; Enzyme Commission (EC) 6.4.1.1] and glutamic-oxaloacetic acid transaminase 2 (GOT2; EC 2.6.1.1), which have been shown to support HSV-1 replication (1), as well as argininosuccinate synthetase I (AS1; EC 6.3.4.5), whose normal expression inhibited the production of infectious progeny (Fig. 2D).
Fig. 2.
Knockdown of cellular metabolic enzymes alter HSV-1 replication. (A) Robust z score values for all individual siRNAs from the screen, plotted in ascending order. Dashed line indicates significance cutoff of |z score| > 1.25, arrows indicate z scores for three siRNAs targeting AS1. (B) The frequency of robust z scores for all individual siRNAs from the screen. (C) Robust z-score values for each of the three independent siRNAs targeting a single cellular gene. Genes are organized by descending average robust z score for all three siRNAs. (D) Metabolic pathway map showing the relationship between two siRNAs that decrease viral yield (PC and GOT2) and one that increases viral yield (AS1). OAA, oxaloacetate.
Table 1.
Entrez gene identification numbers and full gene names for the 38 hits from the siRNA screen, categorized by their effect on HSV-1 replication
| Gene ID | Gene name |
| Decrease virus yield | |
| 113451 | ADC: Arginine decarboxylase |
| 440138 | ALG11: Asparagine-linked glycosylation 11 homolog |
| 12035 | BCAT1: Branched chain aminotransferase 1, cytosolic |
| 760 | CA2: Carbonic anhydrase II |
| 766 | CA7: Carbonic anhydrase VII |
| 9085 | CDY1: Chromodomain protein, Y-linked, 1 transcript variant 1 |
| 203611 | CDY2B: Chromodomain protein, Y-linked, 2B |
| 854057 | COQ3: Coenzyme Q3 homolog, methyltransferase |
| 1629 | DBT: Dihydrolipoamide branched chain transacylase E2 |
| 12686 | ELOVL3: Elongation of very long chain fatty acids-like 3 |
| 51303 | FKBP11: FK506 binding protein 11 |
| 14417 | GAD2: Glutamate decarboxylase 2 |
| 26912 | GCAT: Glycine C-acetyltransferase |
| 2648 | GCN5L2: GCN5 general control of amino acid synthesis 5-like 2 |
| 2651 | GCNT2: Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| 2806 | GOT2: Glutamic-oxaloacetic transaminase 2, mitochondrial |
| 57678 | GPAM: Glycerol-3-phosphate acyltransferase, mitochondrial |
| 2765094 | HEMK2: HemK methyltransferase family member 2 |
| 15476 | HS3ST1: Heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| 50785 | HS6ST1: Heparan sulfate 6-O-sulfotransferase 1 |
| 51451 | LCMT1: Leucine carboxyl methyltransferase 1 |
| 150763 | GPAT2: Glycerol-3-phosphate acyltransferase 2, mitochondiral |
| 4141 | MARS: Methionine-tRNA synthetase |
| 4258 | MGST2: Microsomal GST 2 |
| 4507 | MTAP: Methylthioadenosine phosphorylase |
| 25902 | MTHFD1L: Methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| 5091 | PC: Pyruvate carboxylase |
| 10999 | SLC27A4: Solute carrier family 27 (fatty acid transporter), member 4 |
| 8677 | STX10: Syntaxin 10 |
| 415117 | STX19: Syntaxin 19 |
| 6861 | SYT5: Synaptotagmin V |
| 6916 | TBXAS1: Thromboxane A synthase 1 |
| 80745 | THUMPD2: THUMP domain containing 2 |
| 259230 | SGMS1:Sphingomyelin synthase 1 |
| 7453 | WARS: Tryptophanyl-tRNA synthetase |
| Increase virus yield | |
| 445 | AS1: Argininosuccinate synthetase 1 |
| 55251 | PCMTD2: Protein-l-isoaspartate- O-methyltransferase domain containing 2 |
| 5229 | PGGT1B: Protein geranylgeranyltransferase type I, β subunit |
Cellular metabolic enzymes can alter HSV-1 replication. Those genes which had two or more siRNAs with absolute robust z scores ≥1.25 were identified as hits in the screen. Gene identification numbers and full enzyme named are listed in alphabetical order by the phenotype they induced.
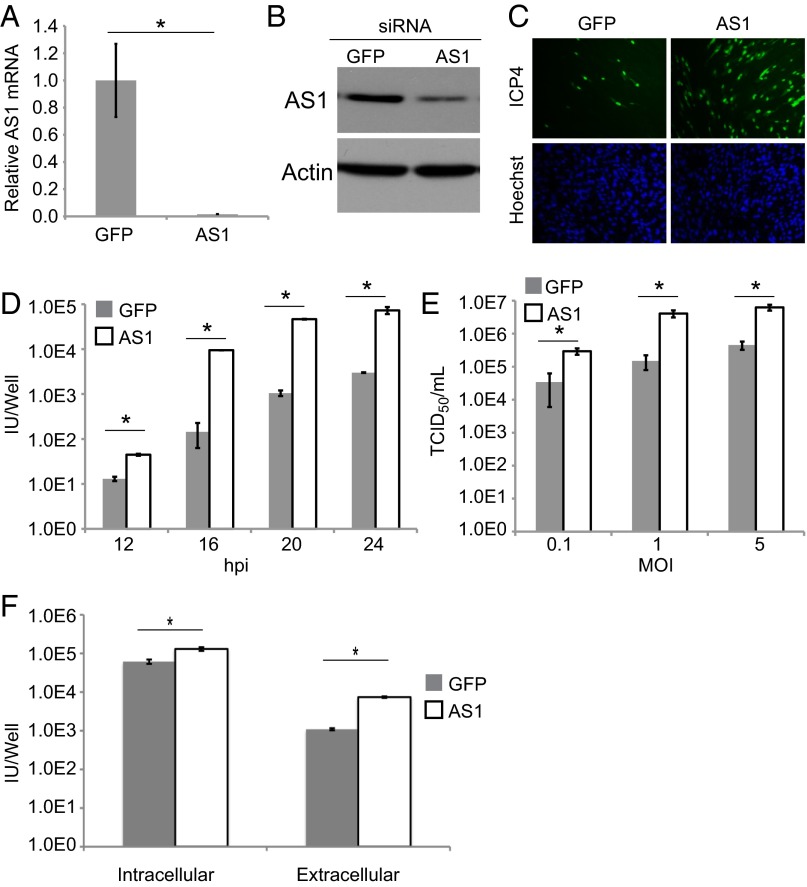
Knockdown of AS1 Increases Virus Yield.
PC catalyzes the conversion of pyruvate to oxaloacetate, and the virus uses this enzyme to anapleurotically feed carbons into the citric acid cycle. GOT2 can then convert oxaloacetate to aspartate, which contributes to the synthesis of nucleotides. AS1 is a cytosolic enzyme that condenses aspartate and citrulline into argininosuccinate. It represents the limiting step in the production of the nonessential amino acid arginine and the soluble messenger NO (15). When AS1 transcript and protein levels were decreased by siRNA (Fig. 3 A and B), virus yield increased significantly (Fig. 3C). This phenotype was true at multiple time points, where the yield of infectious virus in the supernatant increased by a factor of 10 to nearly 100 (Fig. 3D), and the phenotype was multiplicity independent (Fig. 3E). The phenotype was significant, but less robust (twofold increase), when only intracellular virus was measured (Fig. 3F).
Fig. 3.
AS1 knockdown increases the yield of HSV-1. (A and B) Reverse transcription (RT)-qPCR and Western blot showing knockdown of AS1 mRNA and protein 3 d after siRNA treatment. (C) Immunofluorescence images showing the increase in viral titer from cells treated with AS1 versus GFP siRNA. Transfected cells were infected (0.02 IU per cell) 3 d after siRNA addition. Supernatants were collected 24 hpi and used to infect fresh fibroblasts. Cells were fixed 4 hpi and stained for nuclei and the immediate-early viral protein ICP4. (D) HSV-1 titers (ICP4-positive cells per well) over the course of infection (0.1 IU per cell) after treatment with GFP or AS1 siRNA as described above. (E) HSV-1 titers after low, medium, or high MOI infections. Supernatants were collected 18 hpi. (F) Intracellular and extracellular virus titers (ICP4-positive cells per well) at 18 hpi (0.1 IU per cell). Data are derived from two independent experiments, and error bars represent ±1 SD. *P < 0.05.
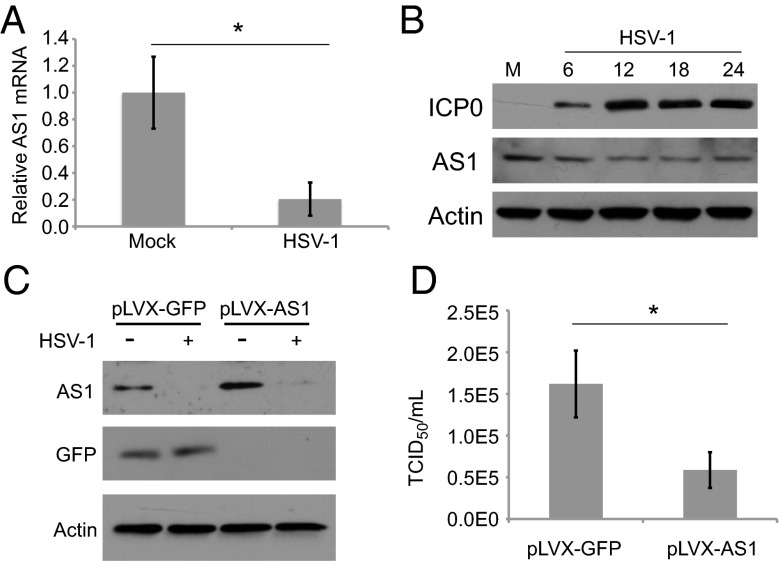
Activity of AS1 Limits Viral Replication.
During the course of a wild-type infection, HSV-1 replication reduces AS1 RNA levels by a factor of approximately 5 (Fig. 4A). Similarly, AS1 protein levels were decreased following infection, and the magnitude of the reduction reflected the portion of cells infected in the culture (0.1 IU per cell, Fig. 4B, compared with 1.0 IU per cell, Fig. 4C). Consistent with our previous conclusion that AS1 activity limits viral replication (Fig. 3), overexpression of AS1 (Fig. 4C) reduced the accumulation of HSV-1 progeny in the supernatant (Fig. 4D).
Fig. 4.
Wild-type HSV-1 infection triggers a decrease in AS1 mRNA and protein levels. (A) RT-qPCR showing levels of AS1 mRNA in cells 18 h after infection or mock infection (0.1 IU per cell). (B) Levels of AS1 protein during a time course of viral infection (0.1 IU per cell). (C) Levels of AS1 in fibroblasts infected with a lentivirus vector expressing AS1 versus GFP. Cells were treated with lentivirus expressing AS1 or GFP and puromycin selected. Confluent cells were infected with HSV-1 (1.0 IU per cell) and lysates were collected 18 hpi. (D) HSV-1 titers from lentivirus-infected cells assayed in C. Data are derived from two independent experiments, and error bars represent ±1 SD. *P < 0.05.
AS1 Knockdown Increases Production of Viral Proteins and Genomes.
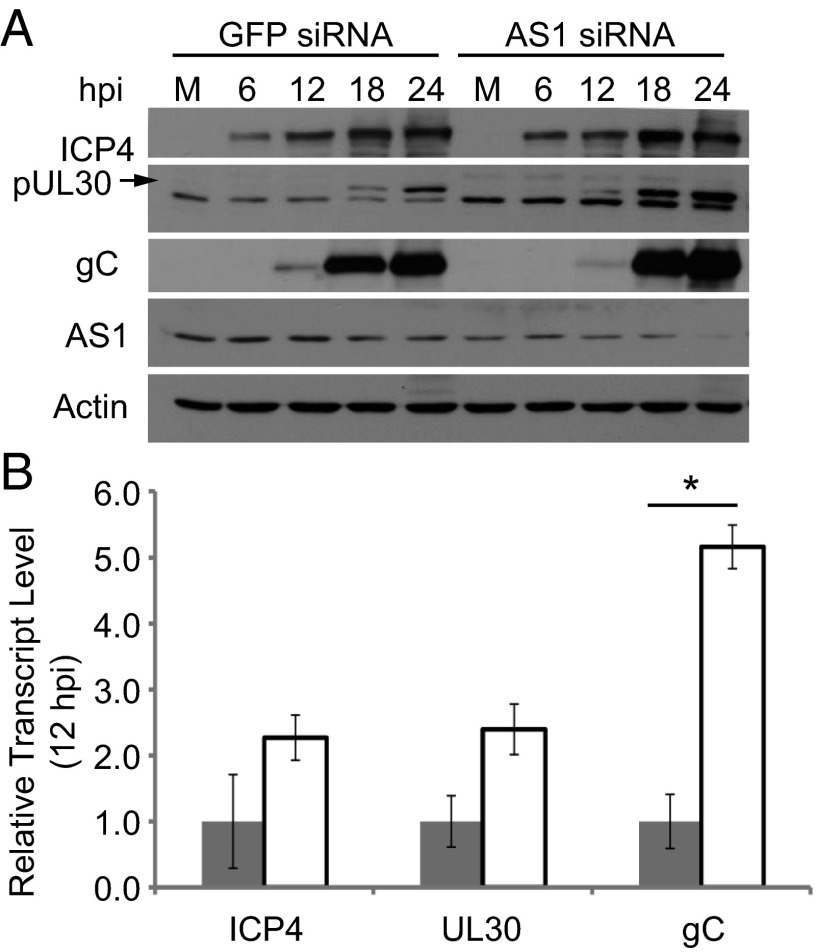
To determine whether the enhanced production of HSV-1 following knockdown of AS1 (Fig. 3) was accompanied by altered expression of viral proteins, representative proteins were assayed by Western blot at various times after infection (Fig. 5A). Knockdown of AS1 did not appear to alter the levels of the immediate-early protein ICP4. However, the amount of the early protein pUL30, which is the catalytic subunit of the viral DNA polymerase holoenzyme required for the replication of the viral genome (20), increased in cells containing less AS1. Expression of the true-late viral protein gC also increased in AS1 knockdown cells, as did its transcript levels by 12 hpi (Fig. 5B). True-late viral genes require viral genome replication for their maximal expression (21, 22).
Fig. 5.
AS1 knockdown increases the protein and transcript levels of viral genes. (A) Western blot showing levels of viral proteins over the course of infection (MOI = 0.1 IU per cell) of AS1 or GFP siRNA-treated cells. pUL30 is represented by upper band on blot, denoted by arrow. (B) Relative levels of viral transcripts in GFP or AS1 knockdown cells at 12 hpi (MOI = 0.1). *P < 0.05.
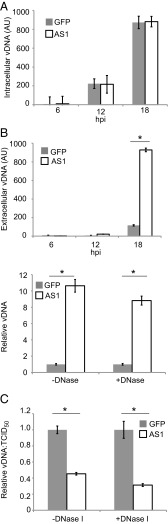
Because AS1 knockdown increased the transcript and protein levels of a viral gene that requires viral genome replication for its production, we next tested whether knockdown altered the accumulation of viral DNA (vDNA) (Fig. 6A). No difference in the amount of intracellular vDNA was evident when quantitative PCR (qPCR) assays were performed at various times after infection. However, when extracellular vDNA was assayed, more genomes were produced by the knockdown cells, e.g., at 18 hpi the medium from AS1-deficient cells contained approximately ninefold more vDNA (Fig. 6B, Upper). A similar result was obtained whether or not the samples from the medium were treated with DNase I (Fig. 6B, Lower), consistent with the interpretation that vDNA in the medium was present within intact virions. To determine whether AS1 knockdown altered the infectivity of HSV-1 virions, we measured the ratio of DNase I-resistant extracellular viral genomes (a measure of vDNA within released virions) to TCID50 titers (Fig. 6C). Although both extracellular genome production and viral titer increased in AS1 knockdown cells, the ratio of these two values was decreased by a factor of 2–3 compared with virions produced in control cells, regardless of DNase I treatment. This result indicates that virions from AS1 knockdown cells are somewhat more infectious than those produced from cells with normal AS1 expression.
Fig. 6.
AS1 knockdown increases extracellular viral DNA and improves infectivity. (A) Intracellular viral DNA (vDNA) accumulation over the course of infection. Cells were infected (0.1 IU per cell) 3 d after siRNA treatment. At appropriate times, cells were lysed and DNA was collected. Primers against the viral gene UL30 were used as a proxy for viral genomes. (B, Upper) Extracellular vDNA accumulation over the course of infection. Samples were infected as in A, infectious supernatants were collected, and vDNA was isolated. (B, Lower) Infectious supernatants were gathered as in Left and were treated with DNase I or PBS before vDNA isolation. (C) Particle infectivity as measured by vDNA:TCID50/mL ratios. Infectious supernatants were collected as in A, and aliquots were used to quantify vDNA or to determine titers by TCID50. AU, arbitrary units. Data are derived from two independent experiments, and error bars represent ±1 SD. *P < 0.05.
Perturbing AS1 Product Levels Does Not Phenocopy the Increased Virus Titers Observed Following AS1 Knockdown.
The increase in HSV-1 yield observed after AS1 knockdown could have been due to decreased production of an (HSV-1 inhibiting) AS1 product or maintenance of an (HSV-1 promoting) AS1 substrate. To explore the former possibility, we investigated the role of two key metabolites that require AS1 for their production: arginine and NO.
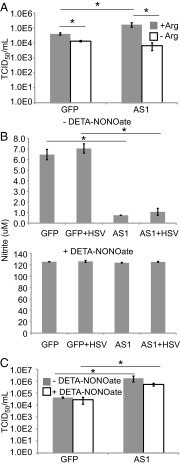
Arginine is a nonessential amino acid that is present in excess in the cell-culture medium used to propagate HSV-1. When AS1 knockdown was performed and cells were grown in medium lacking exogenous arginine, virus yields were significantly decreased regardless of AS1 knockdown status (Fig. 7A), suggesting that the potential loss of arginine in knockdown cells was not responsible for the increased production of HSV-1. This result also suggests that the increased virus titers seen upon AS1 knockdown depend on the presence of at least some arginine in the medium, consistent with earlier studies showing that the amino acid is necessary for productive infection (23, 24).
Fig. 7.
Altering the levels of AS1 products does not mimic the increased HSV-1 replication induced by AS1 knockdown. (A) HSV-1 titers from cells treated with siRNA and replaced with media containing 0.084 or 0.00 g/L of arginine at the time of infection (0.1 IU/cell). Supernatants were collected 18 hpi. (B, Upper) Extracellular concentration of the NO breakdown product nitrate. Cells were siRNA transfected and replaced with serum-free media at the time of infection (1 IU per cell). Supernatants were gathered 18 hpi. (B, Lower) Cells were transfected and infected as in Upper. Extracellular nitrite concentration at 18 hpi was measured following application of 200 μM DETA-NONOate at 1 hpi. (C) Titers of infectious supernatants from B. Data are derived from two independent experiments, and error bars represent ±1 SD. *P < 0.05.
NO is a soluble signaling molecule released by cells in response to specific triggers such as inflammatory cytokines or bacterial lipopolysaccharides (25, 26). NO can also exhibit antiviral activity under certain conditions (27–30). NO is unstable, but reliable measurements can be made of its breakdown product, nitrite (31). Upon knockdown of AS1, nitrite levels in the medium decreased by approximately 5- to 10-fold, and this trend was maintained during infection (Fig. 7B, Upper). When a NO donor, DETA-NONOate, was added to the medium, such that nitrite levels in AS1 knockdown cultures were equal to that of control cells (Fig. 7B, Lower), the virus yield was still significantly higher in AS1 knockdown cells (Fig. 7C). This experiment again argues that the loss of a product downstream of AS1 in knockdown cells is not solely responsible for the increased production of virus. It is likely that the failure to consume the AS1 substrate, aspartate, favors viral replication by maintaining aspartate pools for the production of nucleotides.
The Metabolome Induced by AS1 Deficiency Mimics That Seen During Infection.
To explore the possibility that reduced AS1 activity in HSV-1–infected cells contributed to elevated nucleotide production, we performed a metabolomic analysis of cells that were transfected with AS1 or GFP siRNA, and then mock infected or infected with HSV-1. Metabolites were analyzed at two times after infection: 6 hpi, approximately when viral DNA replication begins, and 18 hpi, when the output of virions is near its maximum level. At both of these times, metabolite pool levels were normalized to mock-infected cells transfected with GFP siRNA.
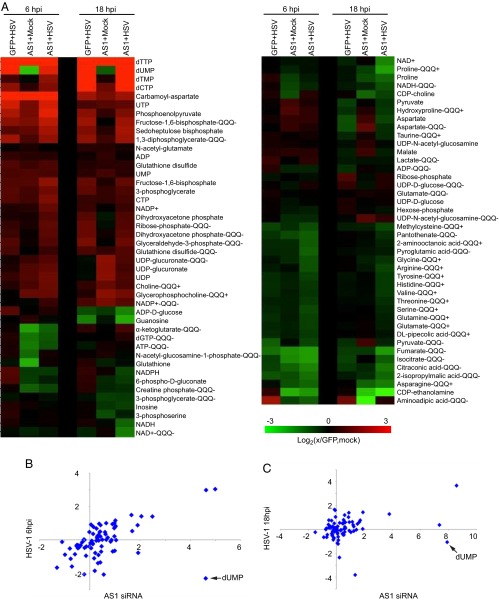
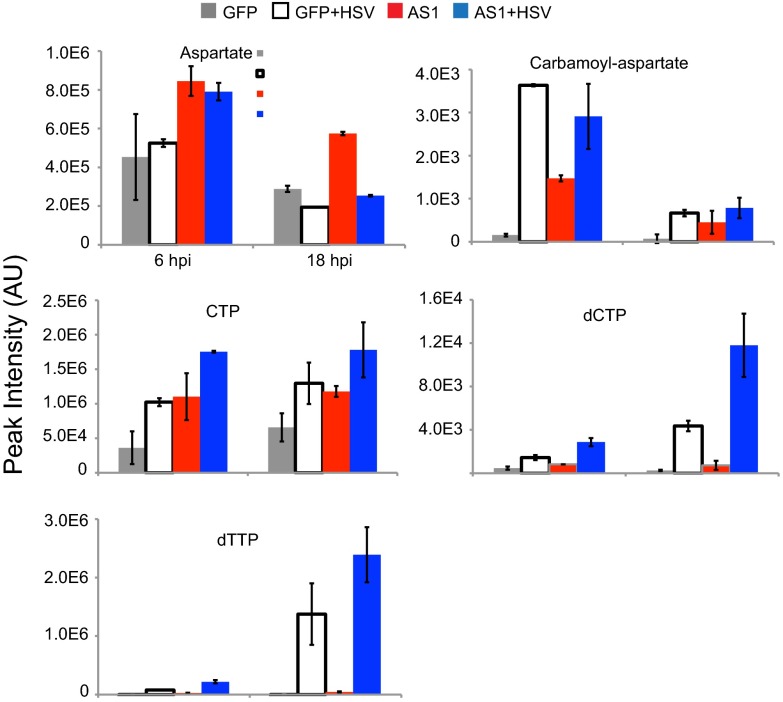
HSV-1 infection induces major metabolic changes in host cells including increased levels of multiple intermediates of upper glycolysis, as well as many nucleotides (1). Similar changes were also seen in uninfected AS1 knockdown cells (Fig. 8A). The correlation between the metabolic profiles of infected and AS1 knockdown cells is stronger at earlier time points following infection (6 hpi, P < 0.001) but remains significant late in infection (18 hpi, P < 0.05) (Fig. 8 B and C). Most significantly, following AS1 knockdown, the levels of aspartate and carbamoyl-aspartate, one of the first committed metabolites on the pathway to nucleotide synthesis, are increased, as are multiple nucleotides themselves (Fig. 9). This result demonstrates that AS1 knockdown induces a metabolic program similar to that seen during infection, suggesting that decreased levels of this enzyme help create a cellular environment more conducive to HSV-1 replication.
Fig. 8.
AS1 knockdown induces a metabolic profile similar to that observed during HSV-1 infection. (A) Metabolite abundances after transfection with siRNA and subsequent infection or mock infection with HSV-1 in serum-free medium (1 IU per cell). Metabolites were harvested at 6 or 18 hpi and normalized to packed cell volume. Values are expressed relative to GFP-transfected, mock-infected cells at the appropriate time point and are log2 transformed. Measurements were taken by using the Exactive instrument unless noted. (Metabolites measured by the triple-quadrupole machines are noted by QQQ+ or QQQ−). (B) Correlation between normalized metabolite levels for HSV-1 infection (x axis) and AS1 knockdown alone (y axis) at 6 hpi. R = 0.50 and P < 0.001. (C) Correlation between normalized metabolite levels for HSV-1 infection (x axis) and AS1 knockdown alone (y axis) at 18 hpi. R = 0.27 and P < 0.05.
Fig. 9.
AS1 knockdown increases pool sizes of metabolites in nucleotide synthesis pathways. Individual levels of metabolites involved in nucleotide synthesis were taken from the heat map in Fig. 7. Values are averages of duplicate biological experiments ±1 SD.
Effects of AS1 Activity on Replication Is Virus Specific.
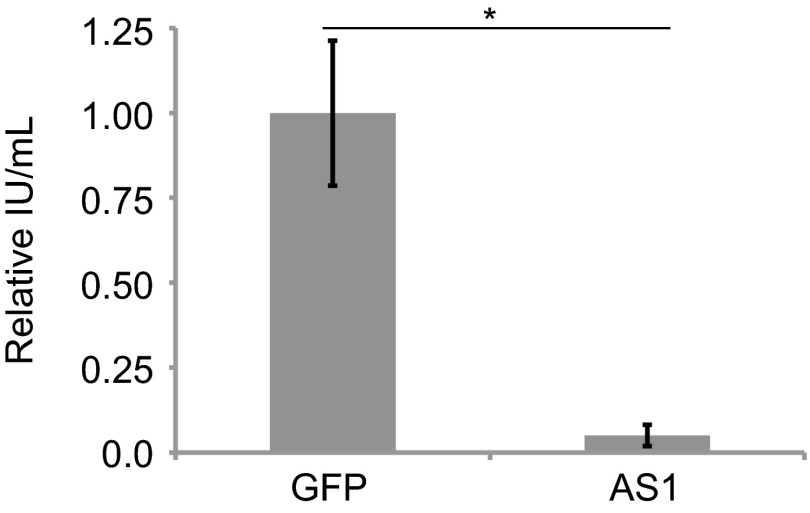
Human cytomegalovirus (HCMV), like HSV-1, is also known to reprogram metabolism during infection. However, the two herpes viruses have a dissimilar effect on specific metabolic pathways (e.g., HSV-1 uses carbons from glucose for pyrimidine biosynthesis, whereas HCMV uses glucose-derived carbons primarily for fatty acid metabolism) (1–3, 7). We predicted, based on our previous metabolic analysis of HCMV, that knockdown of AS1 would not increase the yield of HCMV progeny. In contrast to its effect on HSV-1, AS1 deficiency decreased the yield of HCMV by a factor of approximately 10 (Fig. 10). We conclude that inhibition of AS1 is a strategy specific to HSV-1, but not all herpes viruses. Furthermore, this observation reinforces our previous conclusion that the two herpes viruses have divergent, and sometimes opposite, effects on metabolism.
Fig. 10.
AS1 knockdown decreases the replication of a different herpesvirus, HCMV. HCMV titers (IE1-positive cells) taken from cells treated with AS1 or GFP siRNA. siRNA was added to cells, and they were infected with HCMV (0.1 IU per cell) 24 h later. Infectious supernatants were gathered 96 hpi. Experiments are representative of two independent experiments, and all error bars represent ±1 SD. *P < 0.05.
Discussion
Knockdown of a single enzyme, AS1, mimics key aspects of the HSV-1–induced perturbation of cellular metabolism and is sufficient to increase the titers, genome production, and infectivity of HSV-1.
We completed an siRNA-based screen that independently knocked down the expression of 401 metabolic enzymes (Fig. 1). Of these enzymes, 35 (8.7%) reduced viral yield and 3 (0.7%) increased virus yield (Fig. 2, Table 1, and Dataset S1). The bias toward hits that decrease viral titers, as well as the shift toward negative robust z scores for the screen as a whole, suggests that the virus has evolved to use numerous host cell metabolic enzymes for its own benefit. This trend is similar to that observed with HCMV infection (7) and is not surprising given that pharmacological inhibitors of multiple metabolic enzymes have been shown to posses antiherpesvirus properties (1, 3, 7). Several of the hits identified by this screen agree with previously published data, including PC and GOT2, which are known to play a role in the metabolic program induced by HSV-1 infection (1).
Unlike PC and GOT2, knockdown of AS1 increased extracellular viral titers by at least a factor of 10 at each time tested (Fig. 3D). Intracellular virus yields were also increased, but to a lesser extent (Fig. 3F). Overexpression of AS1 had the opposite effect and reduced the yield of progeny virus (Fig. 4D). Given these results, it is likely that AS1 activity presents a bottleneck to HSV-1 replication. It had been shown that HSV-1 infection induces flux through aspartate to nucleotides, presenting a drain on aspartate pools (1). Because AS1 consumes aspartate, we hypothesized that knockdown of this enzyme may increase viral titers by increasing pools of aspartate and allowing it to serve as a substrate for further synthesis of the nucleotides needed for DNA replication and mRNA transcription. The increased level of RNA encoding a late viral gene product supports this idea (Fig. 5). This metabolic rerouting model was also supported by the increased pools of metabolites of nucleotide synthesis in knockdown cells with replenished aspartate (Figs. 8 and 9).
AS1 knockdown increases the accumulation of extracellular vDNA but does not alter intracellular genome number. Additionally, the increase in viral titers after AS1 knockdown is less robust when intracellular virus is measured (∼twofold increase) than when extracellular virus is measured (∼10-fold increase) (Fig. 3F). Perhaps the environment induced by AS1 knockdown allows for the production of more infectious virus particles by supplying nucleotides to support enhanced viral gene mRNA production later in infection (Fig. 5), when the host cell’s transcriptional capacity is particularly stressed. This change would allow for the increased and/or earlier accumulation of virion proteins such as gC to support the production of fully mature virions (Fig. 5). Conceivably, the increased production of virion constituents generates virions with enhanced infectivity (Fig. 6C) and facilitates their release from the cell (Fig. 3F).
Polyamines are metabolites of arginine, and it has long been established that arginine is essential during HSV-1 replication because of the requirement for polyamines to neutralize the viral genome for packaging (32, 33). In a cell culture system, where excess arginine is provided, the loss of AS1 has a beneficial effect; however, further work under conditions of limiting arginine is needed to determine whether this enzyme is required for optimal nucleic acid packaging.
The fact that HSV-1 infection resulted in reduced AS1 transcript (Fig. 4A) and protein levels (Fig. 4 B and C), combined with the decrease in viral titers upon AS1 overexpression (Fig. 4D), argues that loss of AS1 activity during infection, whether directly or indirectly triggered by the virus, creates a cellular environment more conducive to replication. Given the decrease in AS1 mRNA, it is possible that the virus inhibits AS1 mRNA transcription, processing, or stability, all of which have been described for cellular transcripts during infection (34–36).
There are multiple striking similarities in the global metabolic profile of AS1 knockdown cells and HSV-1–infected cells, especially early in infection (Fig. 8). Under both conditions, there is an accumulation of multiple intermediates of upper glycolysis, a decrease in TCA cycle compounds, and increased nucleotides (Figs. 8 and 9). Because these changes result from AS1 knockdown alone, which also rescues aspartate pools, it makes sense that AS1 activity typically limits viral infection. One possible explanation for the decreased correlation between infected and knockdown cells late after infection is that early metabolic changes induced by the virus are the results of increased aspartate availability, whereas late changes require additional perturbations.
A major difference between knockdown-induced AS1 deficiency and infected cell metabolism was the decreased pools of dUMP in knockdown cells. This difference may be present because the virus encodes a dUTPase that is able to convert dUTP to dUMP to support the production of thymine nucleotides, which increase substantially during infection (37). This viral enzyme would be absent in uninfected cells. AS1 knockdown cells also appear to have an altered glutathione:glutathione disulfide ratio compared with infected cells, suggesting a change in redox state. It has been shown, however, that the HSV-1–induced metabolic reprogramming is maintained even when redox state is purposefully perturbed (4).
The autosomal recessive human disease citrullinemia type I is associated with decreased AS1 activity and results in hyperammonemia and hypercitrullinemia (38, 39). Multiple mutation sites have been linked with the disease, and treatments include low-protein diets and arginine supplementation (40). Clinically, this work suggests that individuals suffering from citrullinemia type I may be at risk for more severe HSV-1 infection. There has been success ameliorating citrullinemia symptoms in animal models through the rescue of AS1 activity (41–43), and it would be interesting to determine whether treated citrullinemic animals have altered susceptibility to HSV-1 infection.
One of the significant conclusions to come from recent work on herpesvirus metabolomics is the difference in pathway preferences between the two related viruses HSV-1 and HCMV. Whereas HSV-1 places a high priority on rapid nucleotide synthesis, HCMV depends more on the production of very long chain fatty acids (1, 3, 7). One reason for this divergence may be due to the difference in replication cycle length: HCMV takes approximately 4 d to reach maximal titers compared with the 1-d cycle of HSV-1. This longer time period, together with the induction of an S phase phenotype, may mean that nucleotide metabolism does not present a bottleneck in HCMV replication. HSV-1 replicates in a much shorter time frame, perhaps owing to the evolution of viral genes that aid in nucleic acid metabolism. It is also interesting to consider that HSV-1 has the ability to infect peripheral neurons, which do not replicate, and, thus, have little need to generate their own nucleotides. Both of these observations may point to why the virus has evolved to place such a high priority on the generation of nucleotides.
Given this information, it is not surprising that knockdown of AS1 had different effects for these two viruses (Fig. 10). AS1 knockdown decreases pools of multiple amino acids, which may hinder the overall protein translation that HCMV infection induces. Additionally, AS1 knockdown decreases multiple intermediates in the TCA cycle, and HCMV uses the TCA intermediate citrate to feed fatty acid synthesis.
In sum, this work reveals a mechanism, AS1 depletion, which mimics major elements of the HSV-1–induced metabolome and increases virus production. By decreasing AS1 protein levels, HSV-1 induces a metabolic environment conducive to its own replication by increasing nucleotide precursors like aspartate. Together, this work supplies a context for further investigation of the causes and effects of metabolic perturbations during infection.
Methods
Cells, Viruses, and Drugs.
Primary human fibroblasts were used between passages 8 and 15. All cells were grown in Dulbecco’s modified Eagle medium (DMEM) supplemented with 4.5g/L glucose, 10% (vol/vol) FBS, and 100 μg/mL streptomycin and penicillin (Invitrogen). HSV-1 (F strain) was kindly provided by Bernard Roizman (University of Chicago, Chicago) (44) and prepared by pooling extracellular supernatants from infected Vero cells (American Type Culture Collection). HCMV (strain AD169) was prepared as reported (45). Viral titers were determined by tissue-culture infectious dose 50/mL (TCID50/mL) or IU per well (4). Intracellular viral titers were determined by collecting cell pellets, sonicating these pellets in 1 mL of media, and removing cell debris.
DETA-NONOate (Cayman Chemical) was dissolved in PBS and used at a final concentration of 200 μM. Arginine-deficient DMEM was supplemented with l-arginine•HCl (Sigma) at a final concentration of 0.084 g/L before filter sterilization. Puromycin (Sigma) was dissolved in water and used as a final concentration of 2 μg/mL.
siRNA Screen.
Human fibroblasts were grown to 75% confluence in 96-well plates before being transfected with 10 pmol of a single siRNA by using Lipofectamine RNAiMAX (Invitrogen), following the manufacturer’s instructions. In larger well formats, reagents were scaled up based on growth area. Two siRNA libraries were used: Mission Metabolism and Cell Trafficking Panel and Mission Transferase Panel (Sigma) for a total of 401 cellular enzymes (7). Three days after transfection, cells were infected with HSV-1 at a multiplicity of 0.02 IU per cell. Infectious supernatants were collected 24 hpi and diluted before being applied to a fresh plate of fibroblasts. At 4 hpi, these cells were methanol fixed and stained for the viral immediate-early protein ICP4 and cell nuclei (4).
Percent-infected values for each well were normalized to the row median of the corresponding screen plate to negate evaporation-based inequalities across a single plate (46). Robust z scores were calculated for the entire screen as described (18). Candidates were considered hits if two or more of the siRNAs targeting that enzyme gave scores greater than or equal to |1.25|.
In subsequent siRNA experiments, fibroblasts were grown in 24-well plates and transfected with 10 pmol of two AS1-specific siRNAs (siRNA 1: 5′-CAAAUAGACCCGUGUACAA-3′, siRNA 2: 5′-CCAAAUAGACCCGUGUACA-3′) or 20 pmol of an siRNA targeting GFP (5′-GCAAGCUGACCCUGAAGUUCAU-3′). Experiments followed the same basic protocol as above. For infections with HCMV, transfections were completed in the same way, but infections occurred 24 h after transfection, to accommodate for the slower replication cycle of the virus (7). Infectious supernatants were gathered at 96 hpi and applied to fresh fibroblasts, which were methanol fixed at 24 hpi and stained for IE1.
Metabolite Analysis.
Fibroblasts treated with GFP or AS1 siRNA were serum starved 2 d after transfection to synchronize cells in G0 (47). At the time of infection, virus was diluted in serum-free medium and added to the cells at the appropriate multiplicity of infection (MOI). Mock-infected cells were treated with media alone. After the 1-h adsorption period, the cells were washed and new serum-free media was applied. At various times after infection, metabolites were quenched and harvested as described (1, 4). Briefly, medium was rapidly removed from cells and cold 80% methanol was added to quench any further metabolic processes. After harvesting, extracted metabolites were dried under nitrogen gas and resuspended in HPLC-grade water.
Following sample separation by liquid chromatography, mass spectrometric analysis was completed by using three Thermo Scientific instruments. The two triple-quadrupole machines, a Finnigan TWQ Quantum Ultra mass spectrometer operated in the positive ionization mode (labeled QQQ+), and a TSQ Quantum Discovery MAX spectrometer used in the negative ionization mode (labeled QQQ-), were used for MS/MS analysis. The Exactive instrument was used for high-resolution untargeted analysis from 85 to 1,000 m/z. Metabolite peaks were identified by using Metabolomic Analysis and Visualization Engine based on retention times of standards (48). All metabolite values are the result of multiple technical and biological replicates.
Protein Analysis.
For Western blot analysis, cells were transfected and/or infected as described above. At the indicated time after infection, cells were washed with a PBS solution and harvested. Cells were then lysed in buffer (50 mM Tris⋅HCl at pH 8.0, 1% Nonidet P-40, 0.1% SDS, 150 mM NaCl, 0.1% Triton X-100, and 5 mM EDTA) with protease inhibitors (Roche Applied Science). Protein concentration was determined by using the Bradford assay before being loaded on to 8 or 10% polyacrylamide gels. After separation, proteins were transferred to nitrocellulose membranes, which were blocked in 5% nonfat dry milk in PBS + 0.1% Tween. All primary and secondary antibodies were also diluted in the same buffer. Anti-mouse primary antibodies included anti-AS1 (BD Transduction Labs 611700), anti-ICP4 (hybridoma supernatant), anti-gC (Virusys P1104), and anti-actin (HRP conjugated, Abcam ab4990). The anti-rabbit primary antibody used was a kind gift of Paul Boehmer (University of Arizona College of Medicine, Phoenix) (49). HRP-conjugated secondary antibodies were raised in goat (Jackson ImmunoResearch).
Nucleic Acid Analysis.
For quantification of RNAs, total RNA was isolated with TRIzol (Invitrogen) by following the manufacturer’s instructions. RNA was reverse transcribed to cDNA by using a TaqMan RT kit (Invitrogen) before being prepared for qPCR. Primers were developed to target actin [forward (FWD): 5′-TCCTCCTGAGCGCAAGTACTC-3′; reverse (REV): 5′-CGGACTCGTCATACTCCTGCTT-3′], AS1 (FWD: 5′-TGGGCTTGAAATTTGCTGAGCTG-3′; REV: 5′-CGATGC AGTGGCGGACAAATTC-3′), HSV-1 UL30 (FWD: 5′-CATCACCGA CCCGGAGAGGGAC-3′; REV: 5′-GGGCCAGGCGCTTGTTGGTGTA-3′) and HSV-1 gC (FWD: 5′-GTTCACCACAGTCTCTACCG-3′; REV: 5′-ATTGCGTCGCGAGAA CGTCA-3′).
For quantification of viral DNA, a segment within the UL30 region was assayed. Infectious supernatants were collected and pretreated with DNase I (NEB) for 10 min at 37 °C before inactivating the enzyme for 10 min at 75 °C. A 100-μL aliquot of infectious supernatant was mixed with 400-μL qPCR resuspension buffer (400 mM NaCl, 10 mM Tris at pH 7.5–8.0, and 10 mM EDTA), 2 μL of 10 mg/mL Proteinase K (Roche), and 4 μL of 20% SDS. After incubation overnight at 37 °C, samples were treated with RNase A before extraction with phenol:chloroform. Each sample was then incubated overnight at −20 °C with 10% vol/vol 3M NaOAc and 200% vol/vol cold 100% ethanol, and then viral DNA was pelleted by centrifugation and resuspended in nuclease-free water. qPCR was performed as above.
Nitrite Analysis.
Cells were transfected and infected as described above. At the time of harvesting, infectious supernatants were collected and spun through a 10-kDa filter (Amicon) for 10 min at 10,000 × g. Equal volumes of flow-through and resolubilized Griess reagent (Sigma) were combined and incubated for 5–10 min. The absorbance of the resulting mixture at 540 nm was read in a spectrophotometer.
Lentivirus Production and Infection.
The human AS1 cDNA was cloned into pLVX-EF1a-acGFP (Clontech), which was transfected into 293FT cells by using XtremeGene 9 (Roche), following the manufacturer’s instructions. Two days later, replication-deficient lentivirus was harvested at 12-h intervals and filtered before being used to infect subconfluent fibroblasts (three applications). Infected cultures were puromycin treated to select for transduced cells.
Statistical Analysis.
All statistical tests were performed by using a two-tailed, unpaired t test. For metabolomic data, correlation coefficients were used to determine the significance of similarities between metabolic profiles.
Supplementary Material
Acknowledgments
We thank L. Terry, J. Hwang, and E. Koyuncu for technical advice and critical commentary. This work was supported by National Institutes of Health Grants CA82396 and AI78063. S.L.G. is supported by National Science Foundation Graduate Research Fellowship DGE-0646086, and J.G.P. is supported by American Heart Association Postdoctoral Fellowship 12POST9190001.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321305110/-/DCSupplemental.
References
- 1.Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS Pathog. 2011;7(7):e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2(12):e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26(10):1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grady SL, Hwang J, Vastag L, Rabinowitz JD, Shenk T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J Virol. 2012;86(15):8259–8268. doi: 10.1128/JVI.00495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabinowitz JD, Purdy JG, Vastag L, Shenk T, Koyuncu E. Metabolomics in drug target discovery. Cold Spring Harb Symp Quant Biol. 2011;76:235–246. doi: 10.1101/sqb.2011.76.010694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces adipocyte-like lipogenesis through activation of sterol regulatory element binding protein 1. J Virol. 2012;86(6):2942–2949. doi: 10.1128/JVI.06467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T. Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog. 2013;9(5):e1003333. doi: 10.1371/journal.ppat.1003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munshi SU, et al. Metabonomic analysis of hepatitis E patients shows deregulated metabolic cycles and abnormalities in amino acid metabolism. J Viral Hepat. 2011;18(10):e591–e602. doi: 10.1111/j.1365-2893.2011.01488.x. [DOI] [PubMed] [Google Scholar]
- 9.Birungi G, Chen SM, Loy BP, Ng ML, Li SF. Metabolomics approach for investigation of effects of dengue virus infection using the EA.hy926 cell line. J Proteome Res. 2010;9(12):6523–6534. doi: 10.1021/pr100727m. [DOI] [PubMed] [Google Scholar]
- 10.Diamond DL, et al. Temporal proteome and lipidome profiles reveal hepatitis C virus-associated reprogramming of hepatocellular metabolism and bioenergetics. PLoS Pathog. 2010;6(1):e1000719. doi: 10.1371/journal.ppat.1000719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roe B, Kensicki E, Mohney R, Hall WW. Metabolomic profile of hepatitis C virus-infected hepatocytes. PLoS ONE. 2011;6(8):e23641. doi: 10.1371/journal.pone.0023641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehmer PE, Lehman IR. Herpes simplex virus DNA replication. Annu Rev Biochem. 1997;66:347–384. doi: 10.1146/annurev.biochem.66.1.347. [DOI] [PubMed] [Google Scholar]
- 13.Ratner S. Argininosuccinate synthetase of bovine liver: Chemical and physical properties. Proc Natl Acad Sci USA. 1982;79(17):5197–5199. doi: 10.1073/pnas.79.17.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kobayashi K, Jackson MJ, Tick DB, O’Brien WE, Beaudet AL. Heterogeneity of mutations in argininosuccinate synthetase causing human citrullinemia. J Biol Chem. 1990;265(19):11361–11367. [PubMed] [Google Scholar]
- 15.Xie L, Gross SS. Argininosuccinate synthetase overexpression in vascular smooth muscle cells potentiates immunostimulant-induced NO production. J Biol Chem. 1997;272(26):16624–16630. doi: 10.1074/jbc.272.26.16624. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YQ, Lai W, Li H, Li G. Inhibition of herpes simplex virus type 1 by small interfering RNA. Clin Exp Dermatol. 2008;33(1):56–61. doi: 10.1111/j.1365-2230.2007.02543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muylaert I, Elias P. Knockdown of DNA ligase IV/XRCC4 by RNA interference inhibits herpes simplex virus type I DNA replication. J Biol Chem. 2007;282(15):10865–10872. doi: 10.1074/jbc.M611834200. [DOI] [PubMed] [Google Scholar]
- 18.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Donnell CD, Kovacs M, Akhtar J, Valyi-Nagy T, Shukla D. Expanding the role of 3-O sulfated heparan sulfate in herpes simplex virus type-1 entry. Virology. 2010;397(2):389–398. doi: 10.1016/j.virol.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boehmer PE. Expression, purification, and characterization of the herpes simplex virus type-1 DNA polymerase. Methods Enzymol. 1996;275:16–35. doi: 10.1016/s0076-6879(96)75004-8. [DOI] [PubMed] [Google Scholar]
- 21. Fields BN, Knipe DM, Howley PM (2007) Fields Virology (Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia) 5th Ed.
- 22.Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14(1):8–19. doi: 10.1128/jvi.14.1.8-19.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tankersley RW., Jr Amino acid requirements of herpes simplex virus in human cells. J Bacteriol. 1964;87:609–613. doi: 10.1128/jb.87.3.609-613.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roizman B, Spring SB, Roane PR., Jr Cellular compartmentalization of herpesvirus antigens during viral replication. J Virol. 1967;1(1):181–192. doi: 10.1128/jvi.1.1.181-192.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seyffarth G, et al. Lipopolysaccharide induces nitric oxide synthase expression and platelet-activating factor increases nitric oxide production in human fetal membranes in culture. Reprod Biol Endocrinol. 2004;2:29. doi: 10.1186/1477-7827-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, Ghahary A, Shen YJ, Scott PG, Tredget EE. Human dermal fibroblasts produce nitric oxide and express both constitutive and inducible nitric oxide synthase isoforms. J Invest Dermatol. 1996;106(3):419–427. doi: 10.1111/1523-1747.ep12343428. [DOI] [PubMed] [Google Scholar]
- 27.Mehta DR, Ashkar AA, Mossman KL. The nitric oxide pathway provides innate antiviral protection in conjunction with the type I interferon pathway in fibroblasts. PLoS ONE. 2012;7(2):e31688. doi: 10.1371/journal.pone.0031688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Regev-Shoshani G, et al. Gaseous nitric oxide reduces influenza infectivity in vitro. Nitric Oxide. 2013;31:48–53. doi: 10.1016/j.niox.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bi Z, Reiss CS. Inhibition of vesicular stomatitis virus infection by nitric oxide. J Virol. 1995;69(4):2208–2213. doi: 10.1128/jvi.69.4.2208-2213.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croen KD. Evidence for antiviral effect of nitric oxide. Inhibition of herpes simplex virus type 1 replication. J Clin Invest. 1993;91(6):2446–2452. doi: 10.1172/JCI116479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hart CM, Kleinhenz DJ, Dikalov SI, Boulden BM, Dudley SC., Jr The measurement of nitric oxide production by cultured endothelial cells. Methods Enzymol. 2005;396:502–514. doi: 10.1016/S0076-6879(05)96042-4. [DOI] [PubMed] [Google Scholar]
- 32.Gibson W, Roizman B. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc Natl Acad Sci USA. 1971;68(11):2818–2821. doi: 10.1073/pnas.68.11.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roizman B, et al. Human herpersviruses I: A model for molecular organization and regulation of herpesviruses-a review. IARC Sci Publ. 1975;(11 Pt 1):3–38. [PubMed] [Google Scholar]
- 34.Spencer CA, Dahmus ME, Rice SA. Repression of host RNA polymerase II transcription by herpes simplex virus type 1. J Virol. 1997;71(3):2031–2040. doi: 10.1128/jvi.71.3.2031-2040.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraser KA, Rice SA. Herpes simplex virus immediate-early protein ICP22 triggers loss of serine 2-phosphorylated RNA polymerase II. J Virol. 2007;81(10):5091–5101. doi: 10.1128/JVI.00184-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hardy WR, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68(12):7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caradonna SJ, Cheng YC. Induction of uracil-DNA glycosylase and dUTP nucleotidohydrolase activity in herpes simplex virus-infected human cells. J Biol Chem. 1981;256(19):9834–9837. [PubMed] [Google Scholar]
- 38.McMurray WC. Biochemical genetics and mental retardation. Can Med Assoc J. 1962;87(9):486–490. [PMC free article] [PubMed] [Google Scholar]
- 39.Beaudet AL, O’Brien WE, Bock HG, Freytag SO, Su TS. The human argininosuccinate synthetase locus and citrullinemia. Adv Hum Genet. 1986;15:161–196, 291–292. doi: 10.1007/978-1-4615-8356-1_3. [DOI] [PubMed] [Google Scholar]
- 40.Engel K, Höhne W, Häberle J. Mutations and polymorphisms in the human argininosuccinate synthetase (ASS1) gene. Hum Mutat. 2009;30(3):300–307. doi: 10.1002/humu.20847. [DOI] [PubMed] [Google Scholar]
- 41.Ye X, Whiteman B, Jerebtsova M, Batshaw ML. Correction of argininosuccinate synthetase (AS) deficiency in a murine model of citrullinemia with recombinant adenovirus carrying human AS cDNA. Gene Ther. 2000;7(20):1777–1782. doi: 10.1038/sj.gt.3301303. [DOI] [PubMed] [Google Scholar]
- 42.Lee B, et al. Hepatocyte gene therapy in a large animal: A neonatal bovine model of citrullinemia. Proc Natl Acad Sci USA. 1999;96(7):3981–3986. doi: 10.1073/pnas.96.7.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patejunas G, et al. Evaluation of gene therapy for citrullinaemia using murine and bovine models. J Inherit Metab Dis. 1998;21(Suppl 1):138–150. doi: 10.1023/a:1005322010854. [DOI] [PubMed] [Google Scholar]
- 44.Ejercito PM, Kieff ED, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 45.Yu D, Smith GA, Enquist LW, Shenk T. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J Virol. 2002;76(5):2316–2328. doi: 10.1128/jvi.76.5.2316-2328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7(7):R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: Viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75(24):12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC-MS data. Anal Chem. 2010;82(23):9818–9826. doi: 10.1021/ac1021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogani F, et al. Association between the herpes simplex virus-1 DNA polymerase and uracil DNA glycosylase. J Biol Chem. 2010;285(36):27664–27672. doi: 10.1074/jbc.M110.131235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.