Abstract
A voluminous polymer coat adorns the surface of many eukaryotic cells. Although the pericellular matrix (PCM) often extends several microns from the cell surface, its macromolecular structure remains elusive. This massive cellular organelle negotiates the cell’s interaction with surrounding tissue, influencing important processes such as cell adhesion, mitosis, locomotion, molecular sequestration, and mechanotransduction. Investigations of the PCM’s architecture and function have been hampered by the difficulty of visualizing this invisible hydrated structure without disrupting its integrity. In this work, we establish several assays to noninvasively measure the ultrastructure of the PCM. Optical force probe assays show that the PCM of rat chondrocyte joint (RCJ-P) cells easily reconfigures around optically manipulated microparticles, allowing the probes to penetrate into rather than compress the matrix. We report distinct changes in forces measured from PCMs treated with exogenous aggrecan, illustrating the assay’s potential to probe proteoglycan distribution. Measurements reveal an exponentially increasing osmotic force in the PCM arising from an inherent concentration gradient. With this result, we estimate the variation of the PCM’s mesh size (correlation length) to range from ∼100 nm at the surface to 500 nm at its periphery. Quantitative particle exclusion assays confirm this prediction and show that the PCM acts like a sieve. These assays provide a much-needed tool to study PCM ultrastructure and its poorly defined but important role in fundamental cellular processes.
Introduction
The pericellular matrix (PCM) lies at the nebulous interface of the extracellular matrix (ECM) and the cell surface, but unlike the ECM, the PCM is defined by its direct anchorage to the plasma membrane (1). This matrix, or cell coat, decorates the surface of a variety of cells in vivo and in tissue culture including fibroblasts (2), smooth muscle cells (3), prostate cancer cells (4), epithelial cells (5), chondrocytes (6), and mesothelial cells (7). Studies have shown that the cell coat can extend outward by anywhere from a few microns up to 20 μm (1,8) (Fig. 1 a) depending on the cell type and cell state. The underlying scaffold of the PCM is comprised of hyaluronan (HA; Fig. 1 b), a negatively charged linear polysaccharide found in situ with contour lengths as long as 2–25 μm (9). Anchored to the cell surface via HA-binding receptors and HA synthase, HA densely assembles bottlebrush-shaped proteoglycans (PGs) such as aggrecan and versican along its chain, the major constituent of the PCM. These PGs are extremely large and negatively charged, with semirigid cylindrical geometries of ∼80 × 350 nm (10). Their attachment increases HA’s persistence length dramatically (11) to form extended configurations that give rise to the cell coat (12,13).
Figure 1.
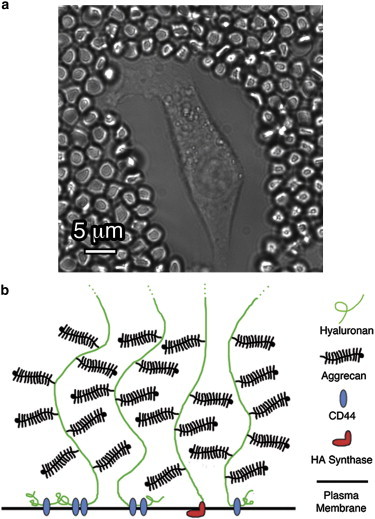
(a) Traditional PEA of chondrocyte RCJ-P cells, which uses fixed RBCs to visualize the extent of the PCM. Measurement of chondrocytes at the center of the flat, elongated side of the cell yields an average PCM thickness of L0,PEA = 7.0 ± 0.5 μm (n = 110 cells). In vivo, there is an ambiguous transition between the surface-bound PCM and the ECM, with the ECM being partially intertwined with the PCM. In the in vitro assays, there is no significant ECM component. (b) Schematic of the PCM. HA polymer chains decorated with PGs (e.g., aggrecan) are bound to surface receptors.
The functions of the PCM are poorly defined, but studies have shown that the matrix influences a diverse range of fundamental cell processes and disease states. These include cell adhesion (5,14–17), proliferation (3,18), migration (3,19), embryonic development (20), wound healing (21,22), mechanotransduction (23,24), protection from viral infections (25), sequestration of growth factors (26), osteoarthritis (27), and various forms of cancer (9,28–31). Recent work has also emphasized the importance of the PCM in drug delivery applications (32). Improved characterization methods are needed to clarify the mechanisms by which the PCM contributes to these cellular functions.
Particle exclusion and biochemical assays of the PCM contents provide some clues, suggesting that the supramolecular organization of the matrix is dynamically rearranged, especially when a cell’s adhesion to the surroundings is altered. For example, during mitosis and migration, changes occur in the length of the cell coat’s constituent HA chains (19), the distribution and type of PGs (14), and the HA grafting density and spatial organization at the cell surface (33). In mitosis, the PCM swells as the cell releases its attachments to the ECM (34). Similarly, in migrating cells, which rely on a delicate coordination of attachment and detachment of focal adhesions (35), the cell coat is reorganized into a distinct asymmetric distribution around the cell’s exterior, with little PCM at the leading edge and an accumulation at the rear. Further, it has been shown that removal or reduction of the PCM significantly diminishes proliferation rates and migration speeds (3). Increasing the cell coat thickness by adding exogenous HA or PGs increases the rate of migration (4,22). Far more work is needed to explain the role of the PCM in these processes, in particular using methods that can dynamically characterize the state and macromolecular organization of the cell coat on living cells.
The cell coat has remained a somewhat obscure structure in part because of its invisibility to phase contrast and differential interference contrast microscopy, and the difficulty of preparing samples for conventional histochemistry and electron microscopy without collapsing the matrix. This invisibility and fragility is due to the high water content resulting from the hydration of the highly charged PGs. Until now, the PCM’s spacious swelling has typically been studied using the classical particle exclusion assay (PEA; Fig. 1 a). In this approach, fixed red blood cells (RBCs) are added to the sample, and subsequent visualization of the empty space between the RBCs and the adherent cell reveals the presence and extent of the PCM. A handful of studies have introduced strategies to characterize the structural or mechanical properties of the cell coat (7,12,36–41). Nevertheless, characterizations of the ultrastructure and the physicochemical properties of this extensive cell matrix structure are still lacking.
Little is understood about the polymer physics of these supramolecular HA-PG assemblies or their resultant collective properties when they are tethered at the cell surface (42,43). Even less is known about how they come to regulate cell function. The ability to monitor the ultrastructure of the PCM of a living single cell should help delineate the mechanistic roles of the PCM in the numerous physiological processes correlated with its presence. To meet this need, we present an optical trap (OT)-based single-cell measurement force assay and a novel (to our knowledge) quantitative PEA (qPEA) that together move beyond the traditional PEA in determining the mechanics and spatial organization of the PCM. Our measurements establish the true perimeter of the PCM on rat chondrocyte joint (RCJ-P) cells, demonstrate its malleability and robustness, and for the first time (to our knowledge) illustrate and quantify the existence of a spatially varying mesh size throughout the cell coat. Comparative measurements of cell coats before and after their transformation by exogenous aggrecan show distinct changes in the extracted force curves, illustrating the optical force probe assay’s sensitivity to PG distribution. These methods provide a new avenue for investigating the structural and mechanical changes that occur during dynamic cell processes such as division and migration.
Materials and Methods
Cell culture
RCJ-P cells (fetal calvaria, batch 15.01.98; Prochon Biotech, Rehovot, Israel) were cultured under 5% CO2 with α-MEM, 15% FBS, 2% L-glutamine (Mediatech, Manassas, VA). The cells were plated for 18–20 h at low density on 78.5 mm2 (6 × 104 cells), passages 15–35. During optical force probe assays, a stage-top microscope incubator (LiveCell, Pathology Devices, Westminster, MD) maintained the cells at 37°, 5% CO2, 80% humidity. For qPEAs, the samples were supplemented with 10 mM HEPES (Cellgro, Manassas, VA) and held at 37°C with a stage-top incubator (N. E-MSI 07-3156; Okolab, Ottaviano, Italy). Fluorescent labeling of the HA in the PCM was achieved using neurocan-green fluorescent protein (GFP) expressed by HEK cells, and purified according to our published protocol (44,45).
For the exogenous aggrecan experiments, aggrecan (Invitrogen, Grand Island, NY) was diluted in PBS (2 mg/ml) and then further diluted in cell media (200 μg/mL) and incubated with the cells for 2 h. In a control, we verified that fluorescently tagged aggrecan (Atto) binds to the pericellular coat. To that end, we showed that replacing the fluorescent aggrecan solution with media after incubation does not destroy the fluorescent halo associated with the bound aggrecan. If the aggrecan were not strongly bound to the cell coat, the clear difference in fluorescence would decay over time, diffusing away from the cell coat area, but this did not occur even after a 15 min period.
Passivation of microspheres
Carboxylated polystyrene microspheres (3 μm) were purchased from Invitrogen. In the qPEAs, we used fluorescently labeled beads (FluoSpheres, 580/605, 505/515 nm; Invitrogen). Passivation was achieved using two schemes. For larger particles (>200 nm), Pluronic F127 was absorbed to the surfaces and anchored to the particles through physical entanglement achieved by reversible swelling with toluene as described previously (46). Smaller microspheres were covalently modified with methoxypolyethylene glycol amine (Fluka, St. Louis, MO) as described previously (47).
PEAs
Traditional PEA
Fixed sheep RBCs (R3378; Sigma Aldrich, St. Louis, MO) were used for PEAs. The erythrocytes were washed twice and resuspended in PBS (100 mg/ml), and then 20 μl was added to 300 μl of media.
qPEA
Passivated, fluorescent polystyrene spheres (FluoSpheres; Invitrogen) of varying size (40–3000 nm) were used to characterize the PCM. Beads (50 μL) were added to 170 μL media (in a Teflon ring) and allowed to settle for 10 min. To facilitate image analysis, the cell surface was fluorescently labeled with wheat germ agglutinin (WGA-Alexa Fluor 633 conjugate; Sigma), which was added to the sample (125 μg/ml) 5 min before measurements were obtained. The samples were imaged with a confocal microscope (FV1000; Olympus, Tokyo, Japan). We surveyed the microsphere distribution perpendicular to the cell surface (at a location similar to where the optical force probe assays were performed) by measuring the intensity distribution (averaged over 2 μm). We extracted the effective thickness, deff, by identifying the plateau in the average bead intensity. Controls showed that using multiple bead sizes simultaneously or exchanging the particles altered the observed particle distribution and deff. Therefore, to avoid artifacts, we performed the qPEAs using a fixed bead size for a given cell sample.
Optical trapping and calibration
Optical trapping was achieved with a 1064 nm laser (YLR-10-1064-LP; IPG, Oxford, MA). A pair of Keplerian relay telescopes, combined with steering mirrors, were used to deliver the beam (280 mW) to the back aperture of the microscope objective (60x, 1.4 NA oil; Nikon, Melville, NY). A dichroic mirror coupled the beam to the microscope, to enable imaging and trapping. Samples were imaged in bright field using a Nikon TE-2000 microscope equipped with a high-speed camera (Phantom v7.1; Vision Research, Wayne, NJ), or a Nikon DQC-FS camera (Nikon) for fluorescence imaging of the PCM. Movies were taken at 500 fps during optical force probe assays and 2000 fps for calibration. OTs, both conventional and holographic, were calibrated by evaluating the probability distribution of a trapped particle in the trap (48). A nonlinear least-squares method was used to fit a Gaussian to the data and to extract the trap stiffness (typically ∼50 pN/μm). We verified that the OT was linear for the range of relevant forces using Stokes drag measurements.
We implemented holographic optical trapping by replacing the first of the two mirrors with a reflective phase-only liquid crystal spatial light modulator (HEO 1080P; HOLOEYE, Carlsbad, CA). We created the two holographic OTs using kinoforms displayed on the spatial light modulator, which we calculated using a direct search optimization (49). During holographic trapping experiments, beads were moved into the PCM in 0.25 μm steps via a sequence of precalculated kinoforms. The average update time between kinoforms was 0.07 s. We calculated the forces on the beads at each position by subtracting the theoretical position of the bead in a force-free environment from its measured position (50). The stiffness of each trap was approximated to be constant with respect to different kinoforms, an assumption that we verified for these short translation distances.
Optical force probe assays
We performed optical force probe assays using a static calibrated OT and a programmable stage (ProScan H117; Prior Scientific, Rockland, MA). Cells with a phenotype like that shown in Fig. 1 a were selected. In a typical experiment, an OT holding a 3 μm probe was positioned 20 μm outside of the PCM, where it could be translated toward and away (orthogonally) to the cell surface at 8 μm/s. The OT was paused for 5 s at the cell surface (always at a distance of 3 μm ) and outside of the matrix. We extracted the forces on the probe by using a standard subpixel particle-tracking algorithm (51) to find the bead position in the OT and hence the force. The bead height above the coverslip was ∼5 μm. We approximated the detectable width of the coat, L0,dyn, by observing the location where the smoothed dynamic curve (20 point moving average) surpassed 0.5 pN, the noise level in the system.
Results
RCJ-P cells have a malleable and robust PCM
The optical force probe assay introduced in this work interrogates the dynamic and equilibrium properties of the PCM. Data from a typical assay are shown in Fig. 2 a. In stage I, a pegylated microsphere is carried orthogonally toward the cell surface at a fixed speed. The force on the probe increases once the edge of the PCM is encountered. At a distance of a few microns from the cell surface to the bead, the movement is paused. This distance is maintained to avoid contact and possible tethering to the cell surface (52), which is often decorated by microvilli, particularly on cells with HA-rich cell coats (53). During this pause, as shown in stage II, the force on the probe decays due to rearrangement of the PCM around the microsphere. Remarkably, even after full matrix relaxation, a nonzero equilibrium force remains on the halted probe particle. The bead is then moved away from the cell surface, during which its deflection in the OT reveals a decreasing dynamic force (stage III). Outside of the PCM, at the original starting location, a pause is taken to allow possible matrix entanglement with the bead to relax (stage IV) before the assay is repeated.
Figure 2.
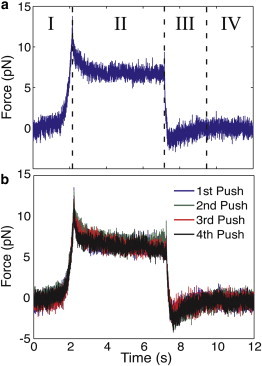
(a) A typical force curve from the optical force probe assay. In stage I, the dynamic force, Fdyn, is measured as the bead is pushed into the cell coat. During stage II, the force decays to a nonzero equilibrium force, Feq, while the bead motion pauses for 5 s at a distance of 3 μm from the cell’s surface. Stage III shows the dynamic force on the bead as it is retracted. Stage IV shows the force on the bead when it is at rest, outside of the matrix. (b) A comparison of consecutive optical force probe assays (stages I–IV) reveals no significant alteration in the force curves.
The large PCM of RCJ-P cells makes them ideal for refining the optical force probe assay measurements. Traditional PEAs show that the average thickness of these cells’ PCM is L0,PEA = 7.0 ± 0.5 μm (n = 114 cells). The optical force studies should be optimized for specific cell types and possibly for changes in cell state, such as occur during the cell cycle. In the work reported here, the probe diameter was 3 μm, the probe speed was 8 μm/s, and the duration of pauses inside and outside of the PCM was 5 s.
The optical force probe measurements typically yield a smooth force profile (stage I), indicating no detectable disruption of a network structure in the PCM even when recording at high frame rates (500 Hz). A subfraction of experiments (∼10%) resulted in jagged force profiles, with apparent snapping events. These data corresponded with clear evidence of poor probe passivation (e.g., particles sticking to the sample substrate) and were not included in the final analyses. A comparison of the force profiles from consecutive measurements of the same cell reveals no change in the force signature (Fig. 2 b). This result was consistent for the >50 measurements made. The unchanged, smooth force profile suggests that the optical force probe assay is nondestructive, leaving the structure of the pericellular coat intact and undisturbed. The reproducibility also indicates that on these short experimental timescales (<60 s), no transformation due to an active cell response is apparent.
The interaction of the probe particle with the PCM is critical to establish a model for the quantitative analysis of the force curves. Several complementary experiments provided evidence that the probe particle penetrates the PCM rather than compressing it. In one study, we combined imaging of the fluorescently labeled PCM (using neurocan-GFP, which binds to HA) with optical manipulation of a probe bead into the matrix. We observed no deformation of the PCM as particles were moved toward and away from the cell surface (see Movie S1 in the Supporting Material). Rather, it appears that the bead penetrates the PCM with the matrix recovering around the bead.
To test the validity of this qualitative observation, we examined the recovery of the PCM behind a probe bead using two holographic OTs (see inset in Fig. 3). To do this, we measured the force on a single bead translated to the cell surface with a holographic trap (50). We then translated the same bead parallel to the cell surface (5 μm) to accommodate a second probe particle within the PCM. Lastly, we moved the second particle along the same path as the first one while monitoring the force on both probes. Nearly a dozen such measurements confirmed that the forces exerted on the second particle were similar to those on the first particle (typical force curves on both probes are shown in Fig. 3; n = 10). We interpret this observation as a clear indication that much of the PCM material responsible for the measured forces recovers behind the first probe bead as it is pushed inward. Interestingly, we also observed that the final equilibrium force on the first bead remained constant when the second bead was translated into the matrix (Fig. S1). This indicates that there is little mechanical coupling between the beads via the PCM, despite their final separation from edge to edge of just 2 μm. This preliminary evidence that the RCJ-P cell coat is not a cross-linked network is in contradiction to the frequent depictions in the literature of the PCM as a gel-like network.
Figure 3.
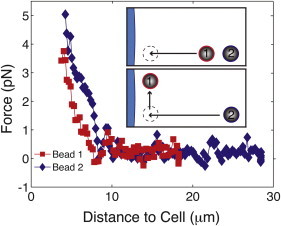
Probing the PCM with two probe particles reveals that particles penetrate rather than compress the matrix as they move toward the cell surface. The inset summarizes the experiment. The first bead is translated toward the cell surface (indicated in blue) with a holographic OT while the force is measured. Bead 1 is then moved slightly to the side to make room in the PCM for a second probe, which is translated along the same path. Determination of the force on the second bead shows that a similar environment must be present around bead 2, because the force curve is nearly identical (n = 10). This would be unlikely to occur if the PCM did not recover behind the first bead or if the matrix were cross-linked, coupling the two beads. Measurement on the first bead as the second bead moves inward also shows no change in force despite the close proximity of the two beads, another indication that the PCM is not cross-linked (see Fig. S1).
Dynamic force probe measurements are ultrasensitive and detect differences in PG distribution
The dynamic measurement made during the optical force probe studies (Fig. 2 a, stage I) is far more sensitive to the pericellular coat’s extent than the traditional PEAs used by researchers who study the PCM. Unlike PEAs, which give the average position of RBCs sitting in equilibrium with the PCM, the translating bead in the OT experiences a force arising from a combination of elastic and viscous components. The inset in Fig. 4 a illustrates how the sign of their relative contributions depends on the direction of motion. This observation explains in part why the magnitude and spatial variation of the force depend on the direction of the motion. Fig. 4 a compares inward and outward dynamic force curves acquired from the same cell. During the inward motion, the viscous and elastic forces are complementary, working together to push the optically trapped bead away from the cell surface. During the reverse motion, the elastic force maintains the same directionality, whereas the viscous component reverses direction, reducing the overall force.
Figure 4.
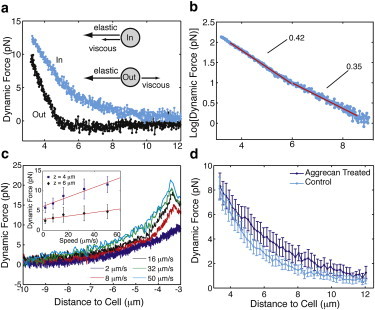
(a) Dynamic force curves measured from a single cell’s PCM. The blue force curve represents the force on the bead as it moves inward toward the cell surface (stage I). The black force curve is the dynamic force upon retraction (stage III). The difference in the value at a position z = 3 μm arises from the time decay to the equilibrium force. The inset illustrates the direction of the elastic and viscous forces on a translating bead. (b) Semi-log plot of the average inward dynamic curve. Two distinct regions, each of which is well fit by an exponential, appear. (c) Investigation of the speed dependence of the dynamic force. The inset shows that the force is roughly linear with speed at the distances z = 4 μm, 6 μm from the cell surface. (d) Comparison of average inward dynamic force curves for control cells (n = 30) and cells conditioned with exogenous aggrecan (200 μg/mL, n = 17). The curves are clearly different, showing the sensitivity of the analysis to changes in PG distribution.
The inward dynamic measurements indicate that the PCM extends to much greater distances than are normally appreciated (Fig. 4 a). The average distance at which a nonzero force is discernible on a 3 μm bead moving toward the cell surface is L0,dyn = 11.5 ± 1.1 μm, where the data are averaged from measurements of n = 30 cells (see Table 1) and the subscript dyn refers to the ingoing dynamic force measurement (the error reported is twice the standard error (SE) unless otherwise stated). This is nearly 40% larger than can be discerned with either a traditional PEA, which yields an effective thickness of L0,PEA = 7.0 ± 0.5 (n = 114), or fluorescent staining with neurocan-GFP, which for RCJ-P cells falls off exponentially so that it is barely visible beyond 6 μm from the surface, as we previously reported (38). Interestingly, the ranges of the standard deviation (SD) of L0,dyn and L0,PEA are similar, reflecting consistency in our observation of a sizable heterogeneity in the cells’ PCMs.
Table 1.
Summary of the physical parameters measured in optical force probe assays of the PCM on the RCJ-P cell line for both control cells and cells modified with exogenous aggrecan
| Average values | Control (n = 30) | Aggrecan-treated (n = 17) |
|---|---|---|
| Fdyn peak (pN) | 9.1 ± 0.9 | 9.7 ±1.2 |
| Feq (pN) | 5.5 ± 0.7 | 3.9 ± 0.9 |
| L0,PEA (μm) | 7.0 ± 0.5 (n = 114) | 17.6 ± 1.3 (n = 86) |
| Lo,dyn (μm) | 11.5 ± 1.1 | 14.1 ± 1.2 |
| Lo,eq(μm) | 8.0 ± 0.6 | 13.1 ± 2.1 (n = 11) |
The error reported is twice the SE.
Analysis of the dynamic force curve averaged from single-cell measurements (v = 8 μm/s, n = 30) shows that the inward force increases toward the cell surface with an exponential dependence, Fdyn ∼ exp(-bz), but where that dependence has two unique values in the cell coat (Fig. 4 b). For the outer regime of the coat, in the range of 6–9 μm, the value is b = 0.35 ± 0.07 μm−1. In the inner region, where measurements take place from 3 μm to 6 μm, the force increases more rapidly with b = 0.42 ± 0.04 μm−1. Due to the naturally limited extent of the PCM, the fitting range is small; however, two separate exponentials were used in lieu of a single exponential (b = 0.39 ± 0.04 μm−1) because the two exponentials provide a better fit to the data according to χ2 analysis. A separate analysis shows that the outward dynamic force is a single exponential that decays more rapidly with b = 1.7 ±0.1 μm−1 in the range of 3 μm < z < 5 μm (although a fraction of the data (n = 9/30) were not fit well by any curve). Beyond 5 μm, the outward dynamic force is negligible, likely due to the competition of elastic and viscous forces.
Next, we investigated how changing the probe speed alters the dynamic response of the pericellular coat. In these measurements, the same cell was probed consecutively at five different speeds (v = 2, 8, 16, 32, and 50 μm/s; n = 7). We observed that the dynamic force relaxed to the same equilibrium value independently of the speed used during the probe. This suggests that probing at higher speeds does not damage or alter the cell coat, and justifies analysis of the scaling of the five speeds. Predictably, the higher-speed probes encountered more resistance as they were moved through the coat, as shown in Fig. 4 c. The inset in Fig. 4 c shows the variation in force with speed at two fixed positions in the cell coat (z = 4 μm, 6 μm). Interestingly, the force is approximately linear with speed at both positions, and the drag on the bead, indicated by the slopes of the inset lines, is higher closer to the cell surface.
Dynamic force curve measurements are particularly useful for quickly registering changes in the pericellular coat. The speed of their acquisition (∼10 s) makes them a preferable readout for time-sensitive situations such as diagnostic applications or scenarios in which the PCM is rapidly modified by the cell (e.g., cell mitosis or migration, during which changes occur on the timescale of tens of minutes). To demonstrate the sensitivity of this approach, we compared typical force signatures from cell coats before and after they were incubated with exogenously added aggrecan. This led to a dramatic swelling of the PCM that was dependent on the PG concentration. When 200 μg/mg aggrecan was added, the PEAs gave an average thickness of L0,PEA=17.6 ± 1.3 μm (n = 86), compared with 7.0 ±0.5 μm (n = 114) for the untreated RCJ-P cells.
Fig. 4 c compares the average dynamic force curve from measurements of the aggrecan-modified cells (n = 17) with that of untreated control cells. The inward dynamic force curve is well fit by a single exponential function with b = 0.31+/0.10 μm−1. Forces from the modified PCMs are detected farther from the cell surface (L0,dyn = 14.1 ± 1.2 μm) as compared with the control cells (L0,dyn =11.5 ± 1.1 μm). Despite this change in coat size, the peak dynamic force is similar between the control cells (9.1 ± 0.9 pN) and the aggrecan-treated cells (9.7 ± 1.2 pN). This similarity continues for the inner region of the coat (∼3–4.5 μm), where the two curves agree to within 1 pN. The similarity of the curves suggests that the exogenous aggrecan is not able to diffuse into the matrix due to steric and/or charge repulsion, or possibly because there are limited available binding sites along the HA chain. The increased forces beyond this region imply that aggrecan does find available binding sites at distances farther out from the cell.
More comprehensive optical force studies using comparative experiments that are supplemented by a more thorough theoretical analysis might ultimately lead to methods to estimate the distribution of PG throughout the PCM. Our approaches also provide opportunities to gain a deeper understanding of the biophysical mechanisms involved in PCM regulation of cellular function.
Static equilibrium force in PCM increases exponentially toward the cell surface
We observed that the PCM relaxes around a probe particle after it is translated into the matrix and stopped near the cell surface (region III, Fig. 2). The typical relaxation time is on the order of ∼300 ms. During relaxation, the dynamic force decays to an equilibrium force, Feq, that is sufficient to eject the bead out of the PCM when the OT is turned off. We investigated whether the equilibrium force persists at other locations throughout the PCM. To that end, we repeated the full optical force probe assay (stages I–IV) sequentially, with the final position varying by steps of 0.3 μm. We found that the equilibrium force is detectable in a smaller range compared with the dynamic force (a typical equilibrium force curve from a single cell is shown in Fig. 5 a). Only the inner regions of the PCM, at distances of z < 8 μm, exert a static equilibrium force on the probe particle. The equilibrium force data are reliably fit by a single exponential profile:
| (1) |
where c = 0.50 ± 0.04 μm−1 and a = 27.3 ± 6.4 pN. From the fits (n = 30), we determined the average distance at which we can detect the equilibrium force with an optical force probe assay to be L0,eq = 8.0 ± 0.6 μm. The difference between L0,dyn and L0,eq of several microns (∼3.5 μm) is expected given the absence of viscous forces for these stationary measurements. Interestingly, the observation that L0,eq > L0,PEA provides evidence that the densely arranged erythrocytes used in the PEAs may exert an osmotic pressure that compresses the PCM. This result reinforces the observation that equilibrium measurements, including the traditional PEA and passive microrheology (38,39), are less sensitive to the full extent of the pericellular coat than dynamic measurements.
Figure 5.
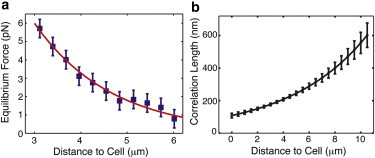
(a) The PCM exerts a force on optically trapped stationary beads. We infer, based on our data, that the equilibrium force arises from molecular concentration gradients in the cell coat. This equilibrium force, Feq, increases exponentially toward the cell surface with an exponent c = 0.50 ± 0.04 μm−1 (red curve). The plot shows the equilibrium force curve from a single-cell measurement, with SD error bars. (b) The correlation length, or mesh size, in the PCM increases exponentially away from the cell surface (the prefactor is set to one). This result relies on our experimental assessment that the probe particles penetrate rather than compress the PCM, and that the measured equilibrium force arises from an osmotic pressure gradient.
Osmotic pressure gradient gives rise to the equilibrium force in the PCM
By a process of elimination, we determine that the equilibrium force in the PCM predominantly arises from osmotic effects. Other possible sources for the observed force, such as elastic compression of the PCM or binding to the matrix, are unlikely given our reported experimental observations, which indicate that these effects are negligible. However, a probe particle will experience an osmotic force if the osmotic pressure is unequal throughout the matrix, creating a pressure gradient across the particle probe. We show in a derivation (see Supporting Material) that the approximate force on a bead of radius R generated by a pressure gradient is
| (2) |
where R is the bead radius, P is the pressure, and z is the perpendicular distance to the cell surface from the center of the bead (see Fig. S3). We hypothesize that molecular concentration gradients in the PCM give rise to the measured force, because osmotic pressure is linearly proportional to concentration. A spatially varying concentration in the PCM will arise from natural variations in HA length as well as the nonuniform decoration by PGs of the HA strands (our comparative studies with exogenous aggrecan support this picture). Additionally, the basic physics of polymers bound to a surface dictates a varying concentration perpendicular to the attachment plane. Even for a monodisperse uniform layer of densely grafted polymer strands to a surface (a polymer brush), the concentration is predicted to decrease parabolically for linear neutral polymers in a good solvent (54). Similar theoretical approaches have been used to predict the spatial variation in concentration for different configurations of surface-bound polymers (55). We therefore surmise that large probe particles that penetrate (rather than compress) the PCM experience an inherent molecular gradient due in part to the configuration of the HA-PG strands and in part to a nonuniform distribution of PGs, molecules whose size and minimal spacing are one to two orders of magnitude smaller than those of the microsphere probe.
The PCM has a spatially varying mesh size
We can estimate the correlation length of the PCM if the equilibrium force arises predominantly from natural concentration variations in the PCM. The correlation length (more loosely defined as the mesh size of a polymer network) is a statistical measure of the distance between segments on neighboring chains (56). Never before, to the best of our knowledge, have the magnitude and variation of the correlation length throughout the PCM been reported, and such information should provide useful insights into the role of the PCM in mediating transport and the interactions of the cell with its surrounding environment.
To find the correlation length, ξ, we employ a useful result from polymer physics that gives the dependence of the osmotic pressure on the correlation length as P ∼ kBT/ξ3, where kB is the Boltzmann constant and T is the temperature (56). Our calculation of the osmotic pressure from the measured equilibrium force (see Supporting Material, Fig. S3 and, Fig. S4) shows that the pressure increases exponentially toward the cell surface and peaks at a value of a few pascals:
| (3) |
where a and c are the fitting parameters from Eq. 1. Equipped with this experimental result, we can estimate the correlation length up to a prefactor of order unity and find that it is equal to
| (4) |
where c is the exponent from the fit to the equilibrium force curve. Fig. 5 b plots the outcome of this analysis, the mesh size prediction calculated from 30 independent cell measurements. Strikingly, the results indicate that the correlation length varies throughout the matrix, decreasing exponentially toward the cell surface, with a typical size on the scale of ∼100 nm (assuming the prefactor is one). This value is consistent with the dimensions of the PGs in the PCM such as aggrecan (∼80 × 350 nm), whose size and distribution along the HA are likely to impact the minimum mesh size (see Fig. 1 b).
To corroborate this result, we developed a complementary assay that we refer to as the qPEA. In this approach, we use passivated microspheres of well-defined sizes to probe the accessibility of the PCM for objects of different sizes. These assays show that particles become nonuniformly distributed throughout the matrix in a size-dependent fashion. To quantify the variation and size dependence, we measured the intensity profiles of fluorescent microspheres as a function of distance to the cell. Fig. 6 a shows typical intensity profiles associated with different particle sizes. A typical intensity profile for particle sizes < 500 nm consists of a nonzero intensity at the cell surface followed by a gradient of increasing intensity until it plateaus. Larger-sized beads show more abrupt changes in concentration with no transition zone.
Figure 6.
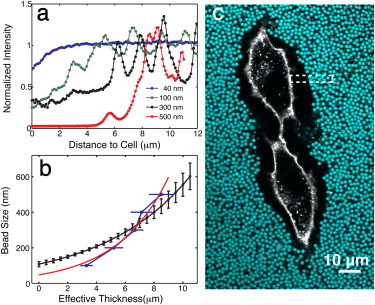
(a) qPEAs using monodisperse passivated beads of different sizes (40–3000 nm) reveal that the cell coat acts like a sieve, with the final spatial distributions of the particles dependent on the size. Intensity profiles of fluorescent beads reveal differences in particle distribution perpendicular to surface (40, 100, 300, and 500 nm). Particles ≥ 500 nm are excluded completely from the inner region of the cell coat z < 8 μm, whereas smaller particles show different intensity profiles versus distance (and hence concentrations), consistent with predictions of mesh size variation. (b) Bead size versus effective thickness, deff, is a rough measure for the correlation length versus position in the PCM. The correlation length estimated from the optical force probe study is included for comparison (the prefactor is set to one). Both assays indicate that mesh size scales exponentially. (c) qPEA with 2 μm diameter particles. The box indicates a typical location on a cell from which both optical force and qPEA data are extracted.
These data show that the PCM acts as a sieve for passivated particles of different sizes. The profiles show that the distribution of particles roughly plateaus at a distance deff that depends on the particle size. We extracted the average effective thickness for particle diameters of 40 nm, 100 nm, 200 nm, 300 nm, 400 nm, 500 nm, 2000 nm, and 3000 nm, as shown in Fig. 6 b and summarized in Table S1 and Fig. S6. The data are consistent with the optical force measurements, suggesting that the ability of particles to penetrate the PCM decreases as the particle size increases, confirming that the mesh size is indeed spatially varying. Interestingly, particle sizes of ≥500 nm have the same average effective thickness, deff,500 = 8.5 ± 0.8 μm, suggesting that this is the edge of an inner domain within the PCM. It is also consistent with the equilibrium thickness measured by the optical force studies, L0,eq = 8.0 ± 0.6 μm.
Next, we seek to directly compare the correlation length prediction from the force probe experiments with the size-dependent particle distributions found in the qPEA (57). To do this, we assume that the bead size corresponds roughly to the correlation length in the PCM at the effective thickness where that bead type shows constant concentration. We note that 40 nm bead data are excluded because the plateau is at deff,40 = 1.4 ± 0.3 μm, a distance where microvilli complicate the PCM structure on RCJ-P cells (38). With this assumption, the qPEA data can be recast as correlation length versus position in the pericellular coat. Fitting the qPEA data to an exponential yields an exponent of 0.29 μm−1, a value for the correlation length scaling that is almost twice that obtained from the optical force measurements, c/3 = 0.17 ± 0.01 μm−1. Overlaying the optical force data with the ad hoc qPEA curve provides a visible comparison as shown in Fig. 6 b. Although the agreement is crude due to the large error that arises from cell heterogeneity, the two assays are consistent. In the future, optimizing experiments to perform both assays on single cells to avoid averaging over measurements will greatly improve the comparison. So could a full theoretical treatment of the qPEA data, which we expect will provide guidance for a more precise comparison between the two techniques.
The data extracted from the optical force probe and qPEA assays provide strong evidence that the PCM has a spatially varying mesh size. The penetration of 40 nm particles to the surface and their uniform distribution at deff,40 = 1.4 μm suggest that the mesh size ranges from a <100 nm length scale near the surface to ∼500 nm at a distance of ∼8 μm from the cell surface. As might be expected, the observed mesh size variation strongly affects the transport of objects through the matrix. This is illustrated by the intensity profiles, which reflect distinct spatial distributions of particles in the PCM as a function of size.
Discussion
The PCM is a neglected but important construct of single cells. A growing body of evidence suggests that the mechanical and structural properties of this cell-associated matrix influence numerous physiological processes. Meanwhile, researchers working in bioengineering (26) and drug delivery (32) are increasingly concerned with the influence of the PCM, and possible opportunities to manipulate it to realize their applications. In similarity to other passive and active microrheology approaches used to interrogate the viscoelastic properties of cells (39,58–62), in this work we introduce several complementary assays to nondestructively interrogate the mechanics and organization of the PCM. Optical force probe measurements show that the PCM is a robust yet malleable structure. The PCM tolerates repeated probing with a 3 μm particle without any measurable changes. Fluorescence imaging of the PCM during optical manipulation of probe particles shows that the PCM appears to rearrange around entrant particles. This is further supported by dual holographic optical-tweezers experiments that verify that the matrix recovers behind a particle such that it exerts similar forces on a secondary bead. The observed rearrangement of matrix around the second probe particle, as well as the observation that the first probe does not feel a force as the second moves inward, provides evidence in support of the ultrastructural insight that the PCM of this chondrocyte cell line is not cross-linked.
The speed of experimentation enabled by dynamic force assays allows for real-time measurements of PCM changes during quick processes, such as cell migration and cell mitosis, in which the PCM is rapidly modified. Our dynamic force probe measurements of PCMs altered with exogenous aggrecan illustrate how the optical force probe approach will be useful for future time-dependent studies of PCM transformation. Although studies of individual PCMs probed both before and after exogenous aggrecan treatment will provide the most precise understanding of the nature of these changes, the averaged data presented here already provide useful information about how aggrecan modifies the PCM and yields clues concerning the availability of binding sites along the HA chains. In the future, the full battery of experiments and analyses developed in this work can be applied to investigate exogenously treated RCJ-P cells or other cells undergoing PCM transformations.
A combined analysis of the dynamic and equilibrium measurements suggests that the PCM on RCJ-P cells has two distinct spatial regimes, consistent with other studies of the same cell line (5). There is an inner region that is capable of exerting an equilibrium force on external objects and a more diffuse outer region that is easily penetrated by objects with diameters at least as large as 3 μm. The inner region is delimited by an edge located at ∼6–8 μm, as detected by both the dynamic and equilibrium force experiments. Our qPEAs also identify this distance (∼8.5 μm) as a unique position in the PCM, a position that particles (diffusing in solution) with diameters of ≥500 nm do not penetrate. The outer region of the PCM is detectable only by dynamic optical force probe measurements, and our measurements consistently confirmed its presence. Indeed, our dynamic force measurements reveal that the PCM extends much farther (40%) from the cell surface of RCJ-P cells than was previously detectable. The ultrasensitivity of dynamic force probe measurements stems from the viscous component that is not present in equilibrium measurements.
Our analysis of the equilibrium force measurements led to the indirect and interesting implication that the PCM must possess a spatially varying correlation length. We further investigated this prediction by developing a novel qPEA to look at the size-dependent diffusion of (passivated) particles into the PCM. Those measurements corroborated the optical force probe data, roughly confirming that the length scale and exponential increase of the mesh size are accurate. Hence, to the best of our knowledge, our assays provide the first quantitative evidence that the mesh size of the PCM varies with distance to the cell surface. The spatial and chemical variations in the cell coat will greatly influence the transport of objects and molecules to and from the cell surface. In these studies, we minimized the role of chemical interactions to focus on steric exclusion. The outcome demonstrates that that even without the electrostatic repulsion or chemical interactions that are expected to occur in the highly negatively charged matrix due to PGs, access to the cell surface is affected by the cell coat ultrastructure. This has direct implications for cell defense against viral and bacterial infections, and drug delivery applications.
Conclusions
In conclusion, this work introduces several novel (to our knowledge) biophysical strategies to interrogate the PCM. Our investigations uncovered valuable ultrastructural details about the PCM on RCJ-P cells. Experiments showed that the PCM is extremely malleable and easily penetrated by passivated 3 μm probe particles. There is no supporting evidence that the PCM of RCJ-P cells is cross-linked; rather, several experiments suggested that there is little coupling between the PG-decorated HA strands grafted to the surface. We observed two unique regions within the PCM. The inner region extends out to ∼8 μm and is capable of exerting osmotic pressure on 3 μm beads and restraining diffusive particles ≥500 nm from entering. The outer region extends to an average of ∼11.5 μm, but its diffuse structure is only detectable by the ultrasensitive dynamic optical force probe assay. The assays revealed a significant heterogeneity in PCM between cells, pointing to the importance of single-cell experiments, especially for future work focused on dynamic changes in the PCM. Two independent methods separately confirmed and provided quantitative data about the exponentially increasing mesh size in the PCM. When compared with molecular scales, the mesh size is large, ranging from ∼100 nm near the cell surface to ∼500 nm at the edge of the inner PCM region. These insights and others that will be gained in future work based on these tools should provide researchers with useful clues to unravel the mechanisms by which the PCM influences so many fundamental biological processes.
Acknowledgments
We thank M. Rubinstein and L. Cai for extremely helpful discussions about the PCM and its osmotic pressure. We are also grateful to A. Souslov and P. Goldbart for insightful discussions. J.E.C. gratefully acknowledges J. Spatz, in whose laboratory the preliminary work was performed, and C. Boehm. We also thank B. Geiger and U. Rauch for their respective gifts of the RCJ-P cell line and the neurocan-GFP (HEK cell line).
This work was supported by the National Science Foundation (CAREER grant 0848797 to J.E.C., L.T.M., and P.C.), the Human Frontiers in Science Program (RGP0013/2010 to J.S. and J.E.C.), the VINNOVA Foundation (A.G.), and the Max Planck Society (H.B.).
Supporting Material
References
- 1.Evanko S.P., Tammi M.I., Wight T.N. Hyaluronan-dependent pericellular matrix. Adv. Drug Deliv. Rev. 2007;59:1351–1365. doi: 10.1016/j.addr.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedman K., Kurkinen M., Höök M. Isolation of the pericellular matrix of human fibroblast cultures. J. Cell Biol. 1979;81:83–91. doi: 10.1083/jcb.81.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evanko S.P., Angello J.C., Wight T.N. Formation of hyaluronan- and versican-rich pericellular matrix is required for proliferation and migration of vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 1999;19:1004–1013. doi: 10.1161/01.atv.19.4.1004. [DOI] [PubMed] [Google Scholar]
- 4.Ricciardelli C., Russell D.L., Horsfall D.J. Formation of hyaluronan- and versican-rich pericellular matrix by prostate cancer cells promotes cell motility. J. Biol. Chem. 2007;282:10814–10825. doi: 10.1074/jbc.M606991200. [DOI] [PubMed] [Google Scholar]
- 5.Cohen M., Klein E., Addadi L. Organization and adhesive properties of the hyaluronan pericellular coat of chondrocytes and epithelial cells. Biophys. J. 2003;85:1996–2005. doi: 10.1016/S0006-3495(03)74627-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knudson C.B. Hyaluronan receptor-directed assembly of chondrocyte pericellular matrix. J. Cell Biol. 1993;120:825–834. doi: 10.1083/jcb.120.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldin P., Suzuki M., Pertoft H. Chondroitin sulfate proteoglycan modulates the permeability of hyaluronan-containing coats around normal human mesothelial cells. J. Cell. Physiol. 1995;165:54–61. doi: 10.1002/jcp.1041650107. [DOI] [PubMed] [Google Scholar]
- 8.Clarris B.J., Fraser J.R.E. On the pericellular zone of some mammalian cells in vitro. Exp. Cell Res. 1968;49:181–193. doi: 10.1016/0014-4827(68)90530-2. [DOI] [PubMed] [Google Scholar]
- 9.Toole B.P. Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 10.Ng L., Grodzinsky A.J., Ortiz C. Individual cartilage aggrecan macromolecules and their constituent glycosaminoglycans visualized via atomic force microscopy. J. Struct. Biol. 2003;143:242–257. doi: 10.1016/j.jsb.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Hellmann M., Weiss M., Heermann D.W. Monte Carlo simulations reveal the straightening of an end-grafted flexible chain with a rigid side chain. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2007;76:021802. doi: 10.1103/PhysRevE.76.021802. [DOI] [PubMed] [Google Scholar]
- 12.Lee G.M., Johnstone B., Caterson B. The dynamic structure of the pericellular matrix on living cells. J. Cell Biol. 1993;123:1899–1907. doi: 10.1083/jcb.123.6.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranova N.S., Attili S., Richter R.P. The sweet coat of living cells-from supramolecular structure and dynamics to biological function. Int. J. Mater. Res. 2011;102:3. [Google Scholar]
- 14.Yamagata M., Saga S., Kimata K. Selective distributions of proteoglycans and their ligands in pericellular matrix of cultured fibroblasts. Implications for their roles in cell-substratum adhesion. J. Cell Sci. 1993;106:55–65. doi: 10.1242/jcs.106.1.55. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerman E., Geiger B., Addadi L. Initial stages of cell-matrix adhesion can be mediated and modulated by cell-surface hyaluronan. Biophys. J. 2002;82:1848–1857. doi: 10.1016/S0006-3495(02)75535-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen M., Kam Z., Geiger B. Dynamic study of the transition from hyaluronan- to integrin-mediated adhesion in chondrocytes. EMBO J. 2006;25:302–311. doi: 10.1038/sj.emboj.7600960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen M., Joester D., Geiger B. Hyaluronan in the pericellular coat: an additional layer of complexity in early cell adhesion events. Soft Matter. 2007;3:327–332. doi: 10.1039/b613770a. [DOI] [PubMed] [Google Scholar]
- 18.Tammi R., Tammi M. Correlations between hyaluronan and epidermal proliferation as studied by [3H]glucosamine and [3H]thymidine incorporations and staining of hyaluronan on mitotic keratinocytes. Exp. Cell Res. 1991;195:524–527. doi: 10.1016/0014-4827(91)90405-j. [DOI] [PubMed] [Google Scholar]
- 19.Itano N., Atsumi F., Kimata K. Abnormal accumulation of hyaluronan matrix diminishes contact inhibition of cell growth and promotes cell migration. Proc. Natl. Acad. Sci. USA. 2002;99:3609–3614. doi: 10.1073/pnas.052026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toole B.P. Hyaluronan in morphogenesis. J. Intern. Med. 1997;242:35–40. doi: 10.1046/j.1365-2796.1997.00171.x. [DOI] [PubMed] [Google Scholar]
- 21.Chen W.Y.J., Abatangelo G. Functions of hyaluronan in wound repair. Wound Repair Regen. 1999;7:79–89. doi: 10.1046/j.1524-475x.1999.00079.x. [DOI] [PubMed] [Google Scholar]
- 22.Savani R.C., Wang C., Turley E.A. Migration of bovine aortic smooth muscle cells after wounding injury. The role of hyaluronan and RHAMM. J. Clin. Invest. 1995;95:1158–1168. doi: 10.1172/JCI117764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han Y., Cowin S.C., Weinbaum S. Mechanotransduction and strain amplification in osteocyte cell processes. Proc. Natl. Acad. Sci. USA. 2004;101:16689–16694. doi: 10.1073/pnas.0407429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vincent T.L., McLean C.J., Saklatvala J. FGF-2 is bound to perlecan in the pericellular matrix of articular cartilage, where it acts as a chondrocyte mechanotransducer. Osteoarthritis Cartilage. 2007;15:752–763. doi: 10.1016/j.joca.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Clarris B.J., Fraser J.R., Rodda S. Effect of cell-bound hyaluronic acid on infectivity of Newcastle disease virus for human synovial cells in vitro. Ann. Rheum. Dis. 1974;33:240–242. doi: 10.1136/ard.33.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macri L., Silverstein D., Clark R.A.F. Growth factor binding to the pericellular matrix and its importance in tissue engineering. Adv. Drug Deliv. Rev. 2007;59:1366–1381. doi: 10.1016/j.addr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 27.Alexopoulos L.G., Williams G.M., Guilak F. Osteoarthritic changes in the biphasic mechanical properties of the chondrocyte pericellular matrix in articular cartilage. J. Biomech. 2005;38:509–517. doi: 10.1016/j.jbiomech.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Itano N., Sawai T., Kimata K. Relationship between hyaluronan production and metastatic potential of mouse mammary carcinoma cells. Cancer Res. 1999;59:2499–2504. [PubMed] [Google Scholar]
- 29.Simpson M.A., Wilson C.M., McCarthy J.B. Manipulation of hyaluronan synthase expression in prostate adenocarcinoma cells alters pericellular matrix retention and adhesion to bone marrow endothelial cells. J. Biol. Chem. 2002;277:10050–10057. doi: 10.1074/jbc.M110069200. [DOI] [PubMed] [Google Scholar]
- 30.Hayen W., Goebeler M., Nehls V. Hyaluronan stimulates tumor cell migration by modulating the fibrin fiber architecture. J. Cell Sci. 1999;112:2241–2251. doi: 10.1242/jcs.112.13.2241. [DOI] [PubMed] [Google Scholar]
- 31.Zhang L., Underhill C.B., Chen L. Hyaluronan on the surface of tumor cells is correlated with metastatic behavior. Cancer Res. 1995;55:428–433. [PubMed] [Google Scholar]
- 32.Zhou R., Zhou H., Yeung E.S. Pericellular matrix enhances retention and cellular uptake of nanoparticles. J. Am. Chem. Soc. 2012;134:13404–13409. doi: 10.1021/ja304119w. [DOI] [PubMed] [Google Scholar]
- 33.Turley E.A., Torrance J. Localization of hyaluronate and hyaluronate-binding protein on motile and non-motile fibroblasts. Exp. Cell Res. 1985;161:17–28. doi: 10.1016/0014-4827(85)90486-0. [DOI] [PubMed] [Google Scholar]
- 34.Brecht M., Mayer U., Prehm P. Increased hyaluronate synthesis is required for fibroblast detachment and mitosis. Biochem. J. 1986;239:445–450. doi: 10.1042/bj2390445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horwitz A.R., Parsons J.T. Cell migration—movin’ on. Science. 1999;286:1102–1103. doi: 10.1126/science.286.5442.1102. [DOI] [PubMed] [Google Scholar]
- 36.Bader D.L., Ohashi T., Sato M. Deformation properties of articular chondrocytes: a critique of three separate techniques. Biorheology. 2002;39:69–78. [PubMed] [Google Scholar]
- 37.Alexopoulos L.G., Setton L.A., Guilak F. The biomechanical role of the chondrocyte pericellular matrix in articular cartilage. Acta Biomater. 2005;1:317–325. doi: 10.1016/j.actbio.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Boehm H., Mundinger T.A., Curtis J.E. Mapping the mechanics and macromolecular organization of hyaluronan-rich cell coats. Soft Matter. 2009;5:4331–4337. [Google Scholar]
- 39.Nijenhuis N., Mizuno D., Schmidt C.F. High-resolution microrheology in the pericellular matrix of prostate cancer cells. J. R. Soc. Interface. 2012;9:1733–1744. doi: 10.1098/rsif.2011.0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ng L., Hung H.-H., Grodzinsky A. Nanomechanical properties of individual chondrocytes and their developing growth factor-stimulated pericellular matrix. J. Biomech. 2007;40:1011–1023. doi: 10.1016/j.jbiomech.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Iyer S., Gaikwad R.M., Sokolov I. Atomic force microscopy detects differences in the surface brush of normal and cancerous cells. Nat. Nanotechnol. 2009;4:389–393. doi: 10.1038/nnano.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Attili S., Richter R.P. Combining colloidal probe atomic force and reflection interference contrast microscopy to study the compressive mechanics of hyaluronan brushes. Langmuir. 2012;28:3206–3216. doi: 10.1021/la204602n. [DOI] [PubMed] [Google Scholar]
- 43.Baranova N.S., Nilebäck E., Richter R.P. The inflammation-associated protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6 oligomers. J. Biol. Chem. 2011;286:25675–25686. doi: 10.1074/jbc.M111.247395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Baader S.L., Rauch U. Neurocan-GFP fusion protein: a new approach to detect hyaluronan on tissue sections and living cells. J. Histochem. Cytochem. 2004;52:915–922. doi: 10.1369/jhc.3A6221.2004. [DOI] [PubMed] [Google Scholar]
- 45.Scrimgeour J., Cho J.K., Curtis J. Microfluidic dialysis cell for characterization of macromolecule interactions. Soft Matter. 2011;7:4762–4767. [Google Scholar]
- 46.Kim A.J., Manoharan V.N., Crocker J.C. Swelling-based method for preparing stable, functionalized polymer colloids. J. Am. Chem. Soc. 2005;127:1592–1593. doi: 10.1021/ja0450051. [DOI] [PubMed] [Google Scholar]
- 47.Valentine M.T., Perlman Z.E., Weitz D.A. Colloid surface chemistry critically affects multiple particle tracking measurements of biomaterials. Biophys. J. 2004;86:4004–4014. doi: 10.1529/biophysj.103.037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuman K.C., Block S.M. Optical trapping. Rev. Sci. Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Polin M., Ladavac K., Grier D. Optimized holographic optical traps. Opt. Express. 2005;13:5831–5845. doi: 10.1364/opex.13.005831. [DOI] [PubMed] [Google Scholar]
- 50.McLane L.T., Carroll K.M., Curtis J.E. Force measurements with a translating holographic optical trap. Proc. SPIE. 2010;7762:77610. [Google Scholar]
- 51.Crocker J.C., Grier D.G. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 52.Dai J., Sheetz M.P. Mechanical properties of neuronal growth cone membranes studied by tether formation with laser optical tweezers. Biophys. J. 1995;68:988–996. doi: 10.1016/S0006-3495(95)80274-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kultti A., Rilla K., Tammi M.I. Hyaluronan synthesis induces microvillus-like cell surface protrusions. J. Biol. Chem. 2006;281:15821–15828. doi: 10.1074/jbc.M512840200. [DOI] [PubMed] [Google Scholar]
- 54.Milner S.T., Witten T.A., Cates M.E. Theory of the grafted polymer brush. Macromolecules. 1988;21:2610–2619. [Google Scholar]
- 55.de Gennes P.G. Polymers at an interface; a simplified view. Adv. Colloid Interface Sci. 1987;27:189–209. [Google Scholar]
- 56.Rubinstein M., Colby R.H. Oxford University Press; New York: 2003. Polymer Physics. [Google Scholar]
- 57.Button B., Cai L.-H., Rubinstein M. A periciliary brush promotes the lung health by separating the mucus layer from airway epithelia. Science. 2012;337:937–941. doi: 10.1126/science.1223012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selhuber-Unkel C., Yde P., Oddershede L.B. Variety in intracellular diffusion during the cell cycle. Phys. Biol. 2009;6:025015. doi: 10.1088/1478-3975/6/2/025015. [DOI] [PubMed] [Google Scholar]
- 59.Daniels B.R., Masi B.C., Wirtz D. Probing single-cell micromechanics in vivo: the microrheology of C. elegans developing embryos. Biophys. J. 2006;90:4712–4719. doi: 10.1529/biophysj.105.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deng L., Trepat X., Fredberg J.J. Fast and slow dynamics of the cytoskeleton. Nat. Mater. 2006;5:636–640. doi: 10.1038/nmat1685. [DOI] [PubMed] [Google Scholar]
- 61.Lau A.W.C., Hoffman B.D., Lubensky T.C. Microrheology, stress fluctuations, and active behavior of living cells. Phys. Rev. Lett. 2003;91:198101. doi: 10.1103/PhysRevLett.91.198101. [DOI] [PubMed] [Google Scholar]
- 62.Wei M.-T., Zaorski A., Ou-Yang H.D. A comparative study of living cell micromechanical properties by oscillatory optical tweezers. Opt. Express. 2008;16:8594–8603. doi: 10.1364/oe.16.008594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


