KNL1 is essential for efficient kinetochore–microtubule attachment dynamics during mitosis, in part through promotion of Bub1-mediated targeting of Aurora B to the kinetochore.
Abstract
Aurora B kinase phosphorylates kinetochore proteins during early mitosis, increasing kinetochore–microtubule (MT) turnover and preventing premature stabilization of kinetochore–MT attachments. Phosphorylation of kinetochore proteins during late mitosis is low, promoting attachment stabilization, which is required for anaphase onset. The kinetochore protein KNL1 recruits Aurora B–counteracting phosphatases and the Aurora B–targeting factor Bub1, yet the consequences of KNL1 depletion on Aurora B phospho-regulation remain unknown. Here, we demonstrate that the KNL1 N terminus is essential for Aurora B activity at kinetochores. This region of KNL1 is also required for Bub1 kinase activity at kinetochores, suggesting that KNL1 promotes Aurora B activity through Bub1-mediated Aurora B targeting. However, ectopic targeting of Aurora B to kinetochores does not fully rescue Aurora B activity in KNL1-depleted cells, suggesting KNL1 influences Aurora B activity through an additional pathway. Our findings establish KNL1 as a requirement for Aurora B activity at kinetochores and for wild-type kinetochore–MT attachment dynamics.
Introduction
Congression and proper segregation of mitotic chromosomes is critically dependent on the interaction between spindle microtubules (MTs) and kinetochores. The strength of kinetochore–MT attachments must be precisely regulated to prevent the accumulation of attachment errors and to facilitate proper activation and silencing of the spindle assembly checkpoint (SAC). Aurora B kinase influences the binding affinity between kinetochores and MTs (Biggins et al., 1999; Tanaka et al., 2002; Cimini et al., 2006), in part through phosphorylation of the NDC80 complex component Hec1 (Cheeseman et al., 2006; DeLuca et al., 2006). During early mitosis, Hec1 is highly phosphorylated by Aurora B, reducing its MT binding activity and preventing premature stabilization of kinetochore–MT attachments. Conversely, during late mitosis, Hec1 phosphorylation levels are low, increasing kinetochore–MT binding affinity and promoting stable attachments and SAC silencing (DeLuca et al., 2011). A current model to explain the marked change in Aurora B kinase-mediated phosphorylation posits that phosphorylation efficiency depends on the distance between kinetochore substrates and the permanently activated kinase that emanates as a gradient from the inner centromere, such that increased inter-kinetochore distance results in decreased levels of substrate phosphorylation (Liu et al., 2009; Welburn et al., 2010; Wang et al., 2011). Although protein gradients exist and can be sustained in the cytoplasm (Lipkow and Odde, 2008), the mechanism by how a steep, nanometer-scale gradient of active Aurora B arises and regulates the interaction between kinetochores and MTs is not well understood. Alternatively, Aurora B activity at the kinetochore may be modulated during mitotic progression as a result of biochemical and physical changes occurring at the kinetochore. Indeed, several kinetochore proteins have been demonstrated to influence Aurora B activity. However, little is known about the interplay between Aurora B regulators and the mechanisms by which they modulate Aurora B activity.
The kinetochore-associated phosphatases PP1 and PP2A have been implicated in counteracting Aurora B activity to facilitate kinetochore–MT stabilization (Liu et al., 2010; Foley et al., 2011; Suijkerbuijk et al., 2012; Kruse et al., 2013). The kinetochore protein KNL1 mediates recruitment of PP1 (directly) and PP2A (indirectly through BubR1) to kinetochores, as well as recruitment of the SAC proteins BubR1 and Bub1, which are known to down-regulate Aurora B kinase activity and promote Aurora B localization, respectively (Lampson and Kapoor 2005; Kiyomitsu et al., 2007; Liu et al., 2010; Suijkerbuijk et al., 2012; Kruse et al., 2013). Although KNL1 is a scaffold for both positive and negative regulators of Aurora B activity, the consequences of KNL1 depletion in mammalian cells and its effect on Aurora B–mediated regulation of kinetochore–MT attachment have not been studied. Here we find that depletion of KNL1 or perturbation of the KNL1 N terminus abolishes Aurora B–mediated phosphorylation of outer kinetochore proteins, including Hec1 and Dsn1, and prevents cells from properly regulating kinetochore–MT attachments. This lack of phosphorylation is correlated to a significant decrease in Aurora B activity at kinetochores. We show that the N terminus of KNL1 also facilitates kinase activity of Bub1, a SAC protein known to promote Aurora B recruitment by phosphorylating histone H2A. However, bypassing the requirements of Bub1-mediated Aurora B recruitment in KNL1-depleted cells by direct targeting of Aurora B does not rescue wild-type levels of Aurora B activity or substrate phosphorylation, suggesting that Bub1 and KNL1 may act in an alternative pathway to regulate Aurora B activity. Together, our results demonstrate that KNL1 is essential for Aurora B kinase activity at kinetochores and for proper regulation of kinetochore–MT attachments.
Results
KNL1 facilitates phosphorylation of the outer kinetochore protein Hec1
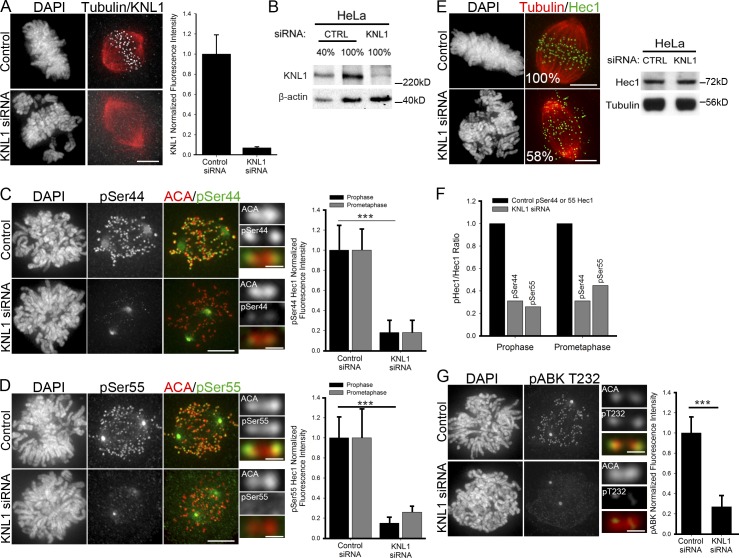
The kinetochore protein KNL1 has been described as a scaffold for both SAC proteins and phosphatase activities. PP1 is recruited to kinetochores by an N-terminal RVSF motif, and PP2A is recruited by BubR1, a well-known KNL1 interacting partner (Liu et al., 2010; Suijkerbuijk et al., 2012; Kruse et al., 2013). Both PP1 and PP2A are proposed to counteract Aurora B activity. In support of this, depletion of BubR1 (which recruits PP2A to kinetochores) or Sds22 (a PP1 regulatory subunit) results in hyperactivation of Aurora B (Lampson and Kapoor 2005; Posch et al., 2010). Based on these findings, we predicted that loss of KNL1, a hub for kinetochore Aurora B antagonists, would result in high levels of Hec1 phosphorylation. To test this, we depleted KNL1 from HeLa and RPE-1 cells (Fig. 1, A and B and Fig. S1 A) and quantified Hec1 phosphorylation at kinetochores. Quantification was performed in early mitotic cells, when Hec1 phosphorylation levels at kinetochores are highest (DeLuca et al., 2011). Surprisingly, KNL1 depletion caused a significant decrease in Hec1 phosphorylation (>80%; Fig. 1, C and D; Fig. S1 B). We measured a 42% decrease in total Hec1 levels at kinetochores after KNL1 depletion, but measured no change in total Hec1 protein levels (Fig. 1 E). The reduction in Hec1 phosphorylation after KNL1 depletion was significant after accounting for the 42% loss of total Hec1 at kinetochores (Fig. 1 F). Thus, KNL1 is required for Hec1 phosphorylation in early mitosis.
Figure 1.
KNL1 is required for Aurora B kinase-mediated Hec1 phosphorylation at the kinetochore. (A) Control and KNL1-depleted HeLa cells immunostained with KNL1 and tubulin antibodies. Kinetochore fluorescence intensities were measured after KNL1 depletion using a KNL1 antibody. Error bars represent SD from independent experiments (n = 3). For each experiment n ≥ 100 kinetochores were measured from at least 10 cells. (B) Western blot of HeLa cell extracts from control and KNL1 siRNA-treated cells immunostained with KNL1 and β-actin antibodies. Percentage lysate loaded in the gel is indicated. Quantification of band intensities indicates that KNL1 levels were decreased by 96%. (C–G) Control and KNL1-depleted HeLa cells were immunostained with Hec1 phosphospecific antibodies (C and D), a non-phosphospecific Hec1 antibody (E, 9G3), a phosphospecific Aurora B antibody (G, pT232), and kinetochore fluorescence intensities were quantified. Error bars in graphs represent SD from independent experiments (n = 3 for Aurora B pT232 and Hec1 pSer44; n = 2 for Hec1 pSer55). For each experiment n ≥ 100 kinetochores were measured from at least 10 cells. ***, P < 0.001 (Mann-Whitney rank sum test). Fluorescence intensities are indicated as percentages in (E, left). (E, right) Western blot of control and KNL1-depleted HeLa extracts. Blots were probed with antibodies to Hec1 and tubulin as a loading control. (F) Levels of phospho-Hec1 remaining at kinetochores calculated as a ratio of phospho-Hec1 fluorescence intensities from C and D to total Hec1 remaining at kinetochores after KNL1 depletion (58%) in E. See C and D above for n values and statistics. (A–G) Error bars in all controls represent SD between cells. Bars: (cell panels) 5 µm; (kinetochore pair insets) 0.5 µm.
KNL1 is required for Aurora B activation
To determine if the observed defects in Hec1 phosphorylation upon KNL1 depletion were due to perturbation of Aurora B kinase activity, we evaluated levels of active Aurora B kinase in KNL1-depleted cells. Active Aurora B has been detected using antibodies to either auto-phosphorylated Thr232 (pT232; Yasui et al., 2004) or phosphorylated Ser331 (pS331; Petsalaki et al., 2011). Compared with control cells, kinetochores of KNL1-depleted cells exhibited a marked reduction of Aurora B pT232 and pS331 localization (Fig. 1 G; Fig. S1, C and E), suggesting that KNL1 is involved in the regulation of Hec1 phosphorylation by facilitating recruitment and/or activation of Aurora B. Contrary to the bulk of centromeric Aurora B (between sister kinetochores) observed with pan-Aurora B antibodies, pT232 and pS331 Aurora B antibodies localize at the kinetochore region in early mitosis. Despite the different localization patterns, the Aurora B phosphospecific antibodies are not detected in cells when Aurora B is chemically inhibited (DeLuca et al., 2011; Petsalaki et al., 2011), and Western blotting with the pT232 antibody suggests that it primarily recognizes Aurora B (Fig. S1 D). Whether phosphorylated Aurora B is recruited to specific sites at the kinetochore or whether Aurora B localizes to multiple sites between sister kinetochores and only those molecules near the kinetochore are phosphorylated at T232 (and S331) is unclear. Regardless, auto-phosphorylation of Aurora B (a requirement for kinase activity; Yasui et al., 2004) and phosphorylation of Aurora B substrates significantly decrease after KNL1 depletion. Thus, we conclude that KNL1 is essential for proper Aurora B activity at the kinetochore.
Using a non-phosphospecific Aurora B antibody (AIM1), we determined if KNL1 depletion had an effect on centromeric Aurora B localization. We found only a modest reduction of inner centromeric Aurora B (40%) in cells depleted of KNL1 (Fig. S1 F) compared with the more severe reduction of Aurora B pT232 (>80%). Strikingly, depletion of KNL1 resulted in a significant decrease in phosphorylation of the Aurora B substrate and chromosomal passenger complex (CPC) component INCENP (Fig. S1 G), indicating that activity of Aurora B at the inner centromere is also compromised upon KNL1 depletion. In contrast, global Aurora B activity, determined by phosphorylation of Ser10 histone H3 (Crosio et al., 2002), did not significantly change upon depletion of KNL1 (Fig. S1 H). Together, our data demonstrate that KNL1 promotes activity of Aurora B at the centromere and kinetochore region and acts as a mediator of outer kinetochore protein phosphorylation.
KNL1 is required for proper regulation of kinetochore–MT dynamics
Given its role in promoting Aurora B kinase activity at kinetochores, we next probed the phenotypic consequences of KNL1 depletion. Lack of Aurora B kinase activity in early mitosis results in premature stabilization of kinetochore–MT fibers, which persist after exposing cells to cold-induced MT depolymerization (Rieder, 1981; Liu et al., 2009). Both KNL1-depleted cells and cells treated with ZM447439, an Aurora B kinase inhibitor, exhibited higher spindle fluorescence intensity in early mitosis than control cells after cold-induced MT depolymerization (Fig. 2 A). This indicates that depletion of KNL1 results in premature stabilization of kinetochore–MT attachments and further supports the idea that KNL1 promotes Aurora B kinase activity. KNL1 depletion leads to a phenotype in which a population of chromosomes aligns at the spindle equator, while another population of chromosomes remains trapped near one or both poles (Fig. 1 A; Fig. S1 A; Cheeseman et al., 2008). Interestingly, chromosomes trapped at the poles in KNL1-depleted cells generally did not exhibit stable, end-on kinetochore–MT attachments after cold treatment (unpublished data). Although this may appear inconsistent with reduced Aurora B activity at kinetochores, the result is possibly due to kinetochore delocalization of proteins required to mediate attachment of chromosomes stranded in regions of low MT density (e.g., Cenp-E) that occurs upon KNL1 depletion. Similar to previous observations in cells with decreased Aurora B activity or impaired Hec1 phosphorylation (Cimini et al., 2006; DeLuca et al., 2006, 2011), aligned chromosomes in KNL1-depleted HeLa cells exhibited abnormal chromosome oscillations (Fig. 2, B–D). Specifically, oscillatory movements of kinetochores in KNL1-depleted cells were significantly dampened compared with those in control cells (Fig. 2, B–D; Videos 1 and 2). These results indicate that despite its MT-binding activity (Cheeseman et al., 2006), KNL1 is not absolutely essential for the formation of end-on kinetochore–MT attachments in human cells, but is instead required for their proper regulation. Consistent with this idea, KNL1 MT-binding activity does not contribute to chromosome biorientation in Caenorhabditis elegans (Espeut et al., 2012). Together, these results demonstrate that KNL1 contributes to the regulation of kinetochore–MT plus-end attachment dynamics likely through activation of Aurora B.
Figure 2.
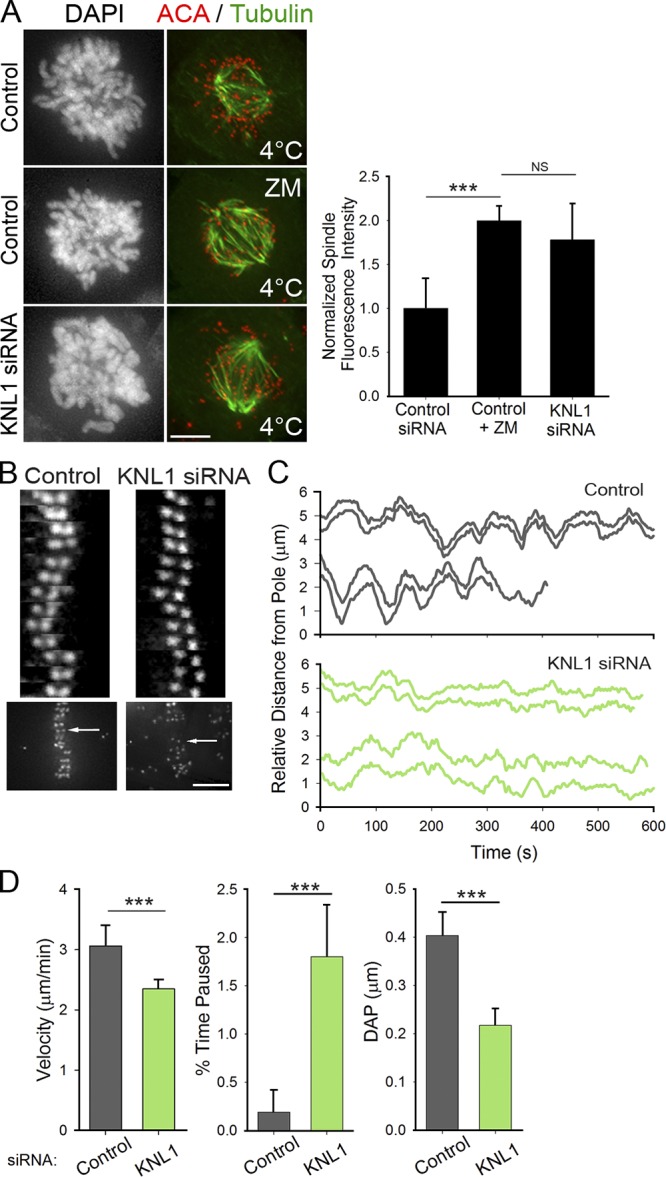
KNL1 is required for wild-type kinetochore–MT dynamics. (A) Representative images of control, ZM-treated, and KNL1-depleted early mitotic HeLa cells subjected to a cold-induced MT depolymerization assay and immunostained for tubulin and a kinetochore marker (ACA). Total spindle fluorescence intensities are shown for each condition. Error bars in graph represent SD from independent experiments (n = 2). For each experiment n ≥ 8 cells per condition. ***, P < 0.001 (Mann-Whitney rank sum test); NS, not statistically significantly different. (B) Kymographs of sister kinetochore pairs from live-cell time-lapse imaging sequences in control and KNL1-depleted HeLa cells. Kymographs were generated from the kinetochore pairs indicated in the panels below (arrows). (C) Plots of kinetochore tracks over time in control and KNL1-depleted HeLa cells. For each condition, two representative sister kinetochore pairs are shown. (D) Quantification of kinetochore oscillations in control and KNL1-depleted cells. DAP indicates deviation from average position, a measure of oscillation amplitude (Stumpff et al., 2008). Error bars represent SD from independent experiments (n = 3). For each experiment n ≥ 32 kinetochores were tracked from at least 5 cells. ***, P < 0.001 (t test). Bars, 5 µm.
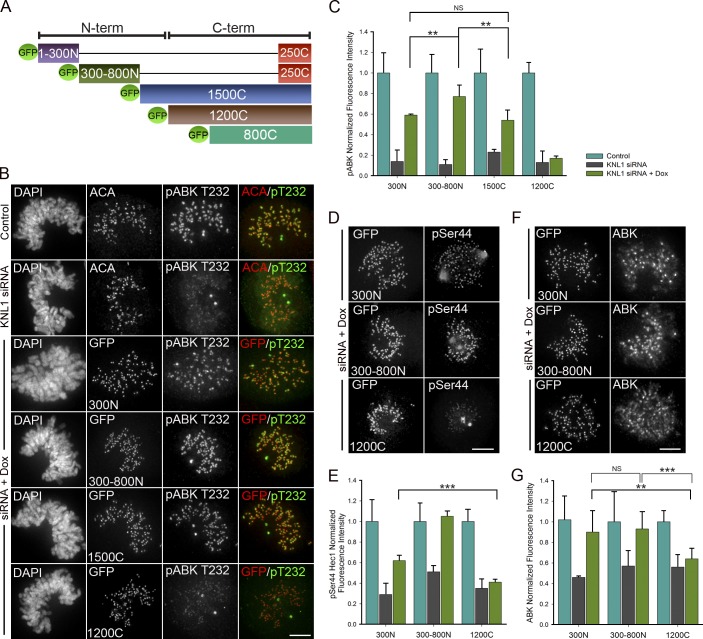
The N-terminal region of KNL1 is sufficient to promote Aurora B activity and phosphorylation of outer kinetochore proteins
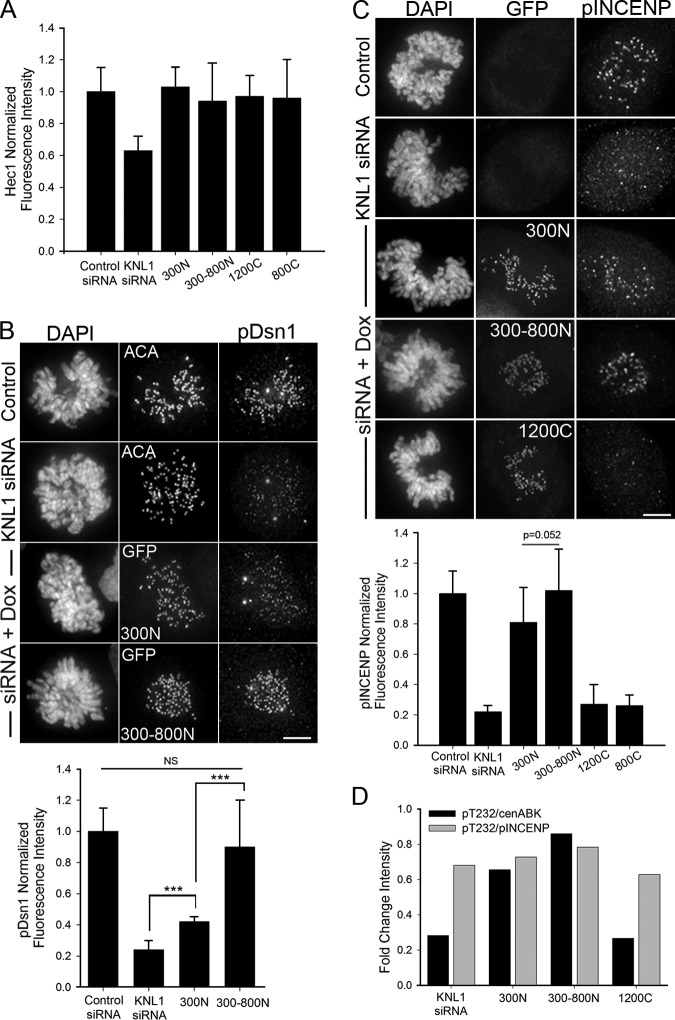
To define the domain in KNL1 required to promote Aurora B activity at kinetochores we generated isogenic doxycycline-inducible HeLa stable cell lines containing KNL1 fragments fused to GFP for use in silence and rescue experiments (Fig. 3 A). All fragments contained a small C-terminal region required to target KNL1 to kinetochores (aa 2056–2316, “250C”), which on its own was not sufficient to rescue Aurora B pT232 localization (Fig. S3 A). KNL1 fragments containing portions of the N terminus (300N, 300–800N, 1500C) significantly restored Aurora B pT232, whereas the fragments containing only the C terminus (1200C, 800C) did not (Fig. 3, B and C). Similar results were obtained by transient transfection with KNL1 fragments (Fig. S2, A–D). In all cases, GFP fluorescence intensity levels of the mutant constructs at kinetochores were comparable, as well as the penetrance of KNL1 depletion (Fig. S2, D–F). As expected, the KNL1 N terminus coordinately rescued phosphorylation of Hec1 and phosphorylation of the Aurora B kinetochore substrate Dsn1, whose phosphorylation was also inhibited after KNL1 depletion (Fig. 3, D and E; Fig. 4 B). KNL1 fragments lacking the N-terminal half of the protein were not able to significantly restore Hec1 phosphorylation, while all KNL1 fragments, including the short 250C fragment, facilitated wild-type levels of Hec1 recruitment to kinetochores as detected by a non-phosphospecific Hec1 antibody (Fig. 4 A; Fig. S3 C). These results reveal a previously undescribed role for the KNL1 N-terminal region in promoting activity of Aurora B and phosphorylation of Aurora B substrates.
Figure 3.
The N terminus of KNL1 promotes Aurora B activity. (A) Schematic of GFP-KNL1 constructs stably incorporated into Flp-In T-REx HeLa cell lines; the exact amino acids of all KNL1 constructs shown are as follows: aa 1–300 (300N), 300–818 (300-800N), 1174–2316 (1200C), 1519–2316 (800C), 819–2316 (1500C), and 2056–2316 (250C). (B–G) Flp-In T-REx HeLa cells were depleted of endogenous KNL1, rescued with the indicated GFP-KNL1 fragment upon doxycycline addition, and immunostained with the indicated antibodies. Kinetochore fluorescence intensities were quantified as indicated in the graphs. Error bars represent SD from independent experiments; (B and C) Aurora B pT232 (n = 3), (D and E) Hec1 pSer44 (n = 2), (F and G) Aurora B (n = 3). For each experiment n ≥ 100 kinetochores were measured from at least 8 cells. ***, P < 0.001; **, P < 0.05; NS, not statistically significantly different (Mann-Whitney rank sum test). Bars, 5 µm.
Figure 4.
The N-terminal region of KNL1 is sufficient to recover Aurora B activity at kinetochores. (A) Quantification of kinetochore fluorescence intensities measured using the Hec1 9G3 antibody in stable HeLa FlpIn T-Rex cell lines expressing KNL1 fragments; n ≥ 100 kinetochores and n ≥ 5 cells. No statistical differences were found between control and HeLa stable cell lines expressing KNL1 fragments. (B) Quantification of phospho-Dsn1 at kinetochores in cells expressing KNL1 fragments; n ≥ 100 kinetochores and n ≥ 5 cells, n = 3 independent experiments. For control and KNL1 siRNA panels, cells were stained with ACA and pDsn1 antibodies. For doxycycline-induced cell lines, cells were stained with pDsn1 antibodies and the GFP fluorescence is shown. The 300N cell line is shown in the panel for control and KNL1 siRNA. (C) Quantification of phospho-INCENP at kinetochores in cells expressing KNL1 fragments. The data shown are from a single representative experiment out of two repeats. For the experiment shown, n ≥ 100 kinetochores and n ≥ 5 cells. The 300N cell line is shown in the panel for control and KNL1 siRNA. Error bars represent SD between cells (Mann-Whitney rank sum test). (A and B) In all cases error bars represent SD between independent experiments. ***, P < 0.001; NS, not statistically significantly different (Mann-Whitney rank sum test). (D) Ratios between the average Aurora B pT232 and pINCENP fluorescence intensities and between Aurora B pT232 and AIM1 fluorescence intensities for each KNL1 mutant cell line are shown as bar graphs. The pT232/pINCENP ratio suggests that pT232 and pINCENP levels are highly correlated between cell lines. The pT232/cenABK(AIM1) ratio suggests that pT232 and cenABK are not highly correlated. See Fig. 3, C and G, and panel C above for n values and statistics. Bars, 5 µm.
We next examined the requirements of the different KNL1 regions on the localization of inner centromeric Aurora B. Similar to the results observed upon KNL1 depletion, cells lacking the N-terminal half of KNL1 (1200C, 800C) exhibited a moderate reduction in centromeric Aurora B accumulation (∼30–40%; Fig. 3, F and G). Conversely, fragments containing KNL1 N-terminal regions (300N, 300–800N, 1500C) almost completely reestablished centromeric Aurora B targeting (∼90%; Fig. 3, F and G), indicating that the N terminus of KNL1 is also sufficient for wild-type levels of centromeric Aurora B enrichment. Importantly, the KNL1 300N region recovered high levels of centromeric Aurora B (∼90%) but only partial levels of Aurora B pT232 (∼60%). Thus, although the KNL1 N terminus promotes both enrichment of centromeric Aurora B and Aurora B auto-phosphorylation (pT232) at kinetochores, we did not observe a close quantitative correlation between the levels of these two populations (Fig. 4 D). Instead, levels of phosphorylated INCENP (phospho-Ser893/Ser894; Wang et al., 2011), a substrate and enhancer of Aurora B activity that also localizes to the inner centromere (Honda et al., 2003), correlated well with levels of Aurora B pT232 in all KNL1 mutant stable cell lines (Fig. 4, C and D; Fig. S3 B). Thus, it is possible that KNL1 differentially influences centromeric Aurora B recruitment and Aurora B activity.
The N-terminal region of KNL1 rescues phenotypic defects observed upon KNL1 depletion
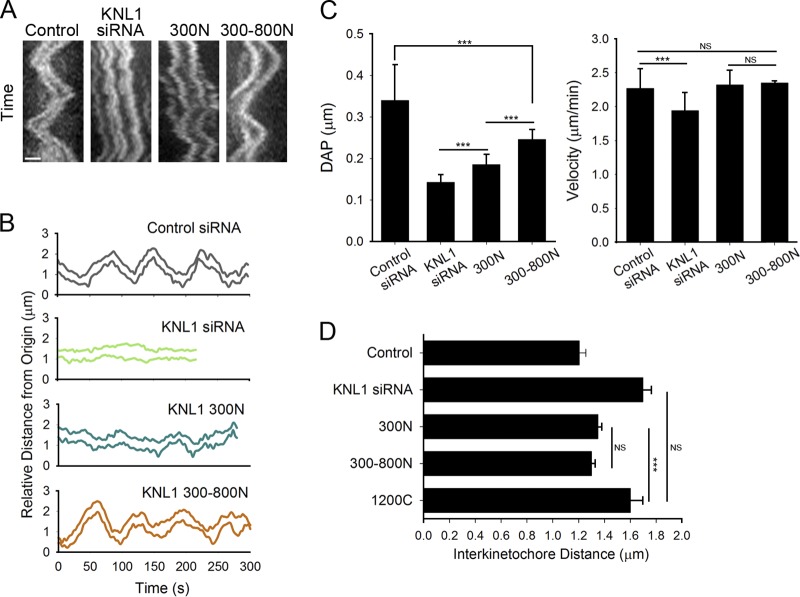
In agreement with our results demonstrating different contributions of the KNL1 N terminus to Aurora B activity at the kinetochore, 300–800N KNL1 significantly restored kinetochore oscillations of aligned chromosomes, whereas cells expressing 300N only partially restored oscillatory movements (Fig. 5, A–C). Additionally, we measured the effect of KNL1 depletion on the generation of tension across sister kinetochores. Contrary to a previous report (Liu et al., 2010), we found that inter-kinetochore distances of bi-oriented chromosomes were significantly increased in KNL1-depleted cells (Fig. 5 D), indicating either hyper-stabilized kinetochore–MT connections or weakened centromere cohesion (DeLuca et al., 2006; Stevens et al., 2011). Consistently, 300N and 300–800N KNL1 significantly restored inter-kinetochore distances of aligned chromosomes to control values (Fig. 5 D). Thus, the KNL1 N terminus is critical for Aurora B activity at the kinetochore and for normal kinetochore–MT dynamics in cells.
Figure 5.
The KNL1 N terminus partially rescues wild-type MT dynamics and kinetochore–MT attachment regulation. (A–D) Flp-In T-REx HeLa cells were depleted of endogenous KNL1, rescued with the indicated GFP-KNL1 fragment upon doxycycline addition and analyzed for oscillatory kinetochore movements and inter-kinetochore distances in metaphase. (A) Kymographs of sister kinetochore pairs from live-cell time-lapse imaging sequences. Bar, 1 µm. (B) Representative plots of kinetochore tracks over time in cells expressing the indicated fragments. (C) Quantification of kinetochore oscillations. DAP indicates deviation from average position; n ≥ 12 kinetochores per cell line. Error bars represent SD between cells. ***, P < 0.001 (t test). (D) Inter-kinetochore distances were measured on bi-oriented sister kinetochore pairs using ACA as a kinetochore marker. Error bars represent SD from independent experiments (n = 3). For each experiment n ≥ 100 kinetochores were measured from at least 5 cells. ***, P < 0.001 (t test). NS, not statistically significantly different (Mann-Whitney rank sum test).
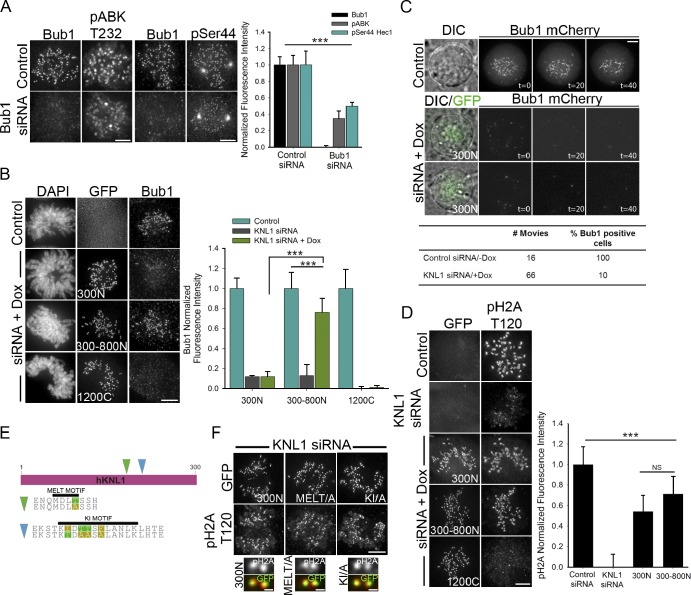
KNL1 N terminus facilitates Bub1 kinase activity at kinetochores
The N terminus of KNL1 serves to recruit multiple outer kinetochore proteins implicated in the modulation of Aurora B activity (Kiyomitsu et al., 2007; Liu et al., 2010; Suijkerbuijk et al., 2012). We therefore examined if KNL1 promotes Aurora B activity by mediating the recruitment of these regulatory factors. Silence and rescue with a KNL1 construct in which the PP1-binding motif (RVSF) was disrupted (Liu et al., 2010) did not affect kinetochore levels of Aurora B pT232, nor did depletion of BubR1 (Fig. S4, A and B). Bub1 depletion, in contrast, led to reduced Aurora B pT232 levels, and this resulted in decreased Hec1 phosphorylation (Fig. 6 A). These data are in agreement with recent findings suggesting that Bub1 kinase promotes Aurora B activation (Ricke et al., 2011; Ricke et al., 2012). Thus, the ability of KNL1 to promote Aurora B recruitment and activation could depend on its function as a kinetochore scaffold for Bub1. The KNL1 N terminus, which we find indispensable for Aurora B activity at the kinetochore, contains multiple MELT motifs that upon Mps1-mediated phosphorylation allow for direct recruitment of Bub1 (Fig. S2 A; London et al., 2012; Shepperd et al., 2012; Yamagishi et al., 2012). Interestingly, although all KNL1 constructs capable of recovering significant levels of Aurora B pT232 localization contain at least 1 MELT repeat (Fig. S2 A), not all are able to mediate Bub1 kinetochore accumulation (Fig. 6, B and C). Fragments 300–800N (4 MELT repeats) and 1500C (5 MELT repeats) highly restored Bub1 localization to kinetochores (Fig. 6 B; unpublished data). In contrast, 300N (1 MELT repeat) was unable to mediate Bub1 kinetochore localization, even upon nocodazole treatment (Fig. 6, B and C; Fig. S4 C). Thus, our data suggest that Bub1 accumulation at kinetochores enhances Aurora B activation and inner centromere Aurora B targeting, but it is not a requisite for these processes to occur.
Figure 6.
KNL1 N terminus is required for Bub1 kinase activity at the kinetochore. (A) Control and Bub1-depleted HeLa cells were immunostained with antibodies to Aurora B pT232 (left panels) or Hec1 pSer44 (right panels). Kinetochore fluorescence intensities are shown. Error bars represent SD from independent experiments (n = 2). For each experiment n ≥ 50 kinetochores were measured from at least 6 cells. (B–D) Flp-In T-REx HeLa cells were depleted of endogenous KNL1, rescued with the indicated GFP-KNL1 fragment upon doxycycline addition, and immunostained with Bub1 (B), histone H2A pT120 (D), or overexpressing hBub1-mCherry (C). Kinetochore fluorescence intensities were quantified. In B and D, error bars represent SD from independent experiments (n = 3 in B; n = 2 in D). For each experiment n ≥ 100 kinetochores, n ≥ 10 cells in B; n ≥ 50 kinetochores, n ≥ 5 cells in D. ***, P < 0.001; NS, not statistically significantly different (Student’s t test). (C) Live-cell imaging in control and 300N KNL1 cells expressing Bub1-mCherry and treated with nocodazole. Time is indicated in minutes. Upon KNL1 depletion 90% of cells had no detectable Bub1 at kinetochores, while 10% of cells showed high levels of Bub1, presumably due to incomplete knockdown. (E) Schematic of GFP-KNL1 300N fragments containing mutations in the MELT or KI motifs. The wild-type (top) and mutated (bottom) sequences are shown. (F) HeLa cells depleted of endogenous KNL1 were rescued with N-terminal GFP-tagged 300N, 300N-KI (300N-KI/A), or 300N-MELT (300N-MELT/A) mutants and immunostained for histone H2A pT120. Bars: (cell panels) 5 µm; (kinetochore pair insets) 0.5 µm.
Because the 300 N-terminal amino acids of KNL1 are not necessary or sufficient for accumulation of Bub1 at kinetochores, but mediate centromeric Aurora B accumulation and partial Aurora B activity, we asked if this region on KNL1 could facilitate Bub1 kinase activity. By analyzing the levels of histone H2A phosphorylation (T120), a well-described Bub1 kinetochore substrate (Kawashima et al., 2010), we found evidence of significant Bub1 kinase activity at kinetochores in cells expressing 300N KNL1 or 300–800N KNL1 but negligible activity when KNL1 was depleted or replaced with C-terminal fragments. Specifically, cells expressing the 300–800N fragment rescued pH2A-T120 to 70%, and those expressing the 300N fragment to 55% (Fig. 6 D). Thus, the N-terminal region of KNL1 facilitates both Aurora B and Bub1 kinase activities at the kinetochore. Importantly, the 300–800N fragment recruits significantly higher levels of Bub1 to kinetochores than the 300N fragment, and this corresponds to a high level of both Aurora B and Bub1 kinase activities. These results suggest that Bub1 accumulation at kinetochores is required for wild-type levels of Aurora B–mediated phospho-regulation of kinetochore–MTs in cells.
Previous studies mapped the binding of the Bub1 tetratricopeptide repeats (TPRs) domain to aa 200–250 in the N-terminal region of KNL1 (referred to as the KI motif; Kiyomitsu et al., 2011). Recently, however, and in agreement with our results, it was demonstrated that the Bub1 TPR domain is not sufficient for Bub1 localization to kinetochores (Krenn et al., 2012; Yamagishi et al., 2012). Furthermore, the catalytic activity of Bub1 measured in vitro was not affected by expression of a Bub1 mutant unable to interact with the KI motif of KNL1 (Krenn et al., 2012). Similarly, silence and rescue experiments in mammalian cells using a mutant of KNL1 lacking the KI motif confirmed that the TPR–KI interaction is dispensable for Bub1 localization to kinetochores (Yamagishi et al., 2012). Therefore, we hypothesized that a 300N KNL1 fragment containing mutations in the KI motif predicted to disrupt the KNL1–TPR interaction (Krenn et al., 2012) would not impair Bub1 kinase activity in cells. Indeed, silence and rescue experiments using a GFP-tagged 300N KNL1-KI mutant (Fig. 6 E) confirmed that the ability of the KNL1 300N fragment to mediate Bub1 kinase activity does not rely on its KI motif (Fig. 6 F). We next examined if disruption of the MELT motif contained in the 300N KNL1 region disrupted Bub1-mediated histone H2A phosphorylation. Surprisingly, a 300N KNL1 mutant containing an alanine substitution in the single MELT motif (MELA) was able to mediate histone H2A phosphorylation in cells (Fig. 6 F). Therefore, neither the KI nor the MELT motif in the 300N region of KNL1 are essential for Bub1 kinase activity at kinetochores. How the 300N-terminal region of KNL1 is able to mediate Bub1 and Aurora B kinase activities remains to be addressed. One possibility, however, is that Bub1 interaction with 300N KNL1, even transiently, results in close positioning of the kinase to its substrate (H2A). Alternatively, a transient interaction with KNL1 could promote allosteric changes in Bub1 that enhance its activation.
KNL1 is required for full Aurora B activity independent of Aurora B accumulation
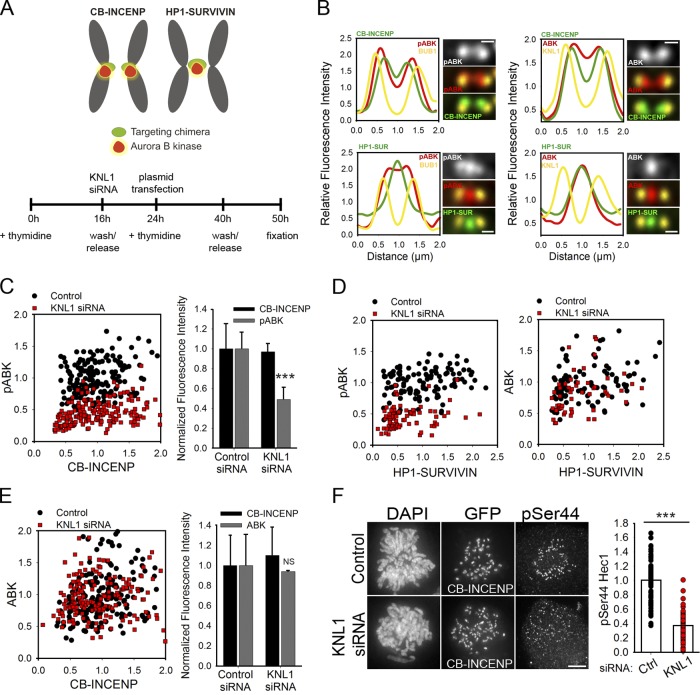
Bub1 promotes Aurora B recruitment to the inner centromere through phosphorylation of histone H2A, which results in targeting of Borealin, a CPC component (Tsukahara et al., 2010). It is proposed that accumulation at this inner centromeric site results in Aurora B activation and a gradient of active kinase emanating toward the kinetochore to regulate kinetochore–MT interactions (Wang et al., 2011). Therefore, it is possible that KNL1 contributes to Aurora B activity entirely by facilitating Bub1-mediated Aurora B recruitment. Our data, however, suggest distinct contributions of KNL1 to Aurora B localization and Aurora B activity (Figs. 1 G, 3 (C and G), 4 D, and S1). We therefore hypothesized that KNL1 not only contributes to Aurora B activity by promoting its recruitment but also through an alternative pathway. To directly test this, we targeted Aurora B to the kinetochore through a Cenp-B (CB)–INCENP chimera (Liu et al., 2009) and to the inner centromere through an HP1-Survivin chimera (Fig. 7 A). Endogenous Aurora B, detected using pan-antibodies and antibodies to its phosphorylated form (pT232), was redistributed to kinetochores or to the inner centromere in cells expressing CB-INCENP or HP1-Survivin, respectively (Fig. 7 B). We quantified the levels of both Aurora B and Aurora B pT232 colocalizing with these fusion proteins in control and KNL1-depleted cells (Fig. 7, C–E; Fig. S5, A–D). Although control and KNL1-depleted cells were able to recruit similar levels of Aurora B in the presence of the targeting chimeras (Fig. 7, D and E), Aurora B pT232 and substrate phosphorylation were not fully restored in KNL1-depleted cells (Fig. 7, C, D, and F). Although ectopic targeting of Aurora B to these regions may not reflect endogenous kinase activity, these results argue that kinase accumulation is not sufficient to achieve control levels of kinase activity in the absence of KNL1. Therefore, it is likely that the influence of KNL1 on Aurora B activity at kinetochores is not exclusively dependent on Bub1-mediated recruitment of the kinase.
Figure 7.
Aurora B accumulation is not sufficient for wild-type levels of Aurora B activity in KNL1-depleted cells. (A) Schematic of the localization of CB-INCENP and HP1-Survivin chimeras (top) and schematized experimental procedure (bottom). (B) Line scans of sister kinetochore pairs from HeLa cells expressing CB-INCENP or HP1-Survivin and immunostained with the indicated antibodies. Line scans show the fluorescence intensity across sister kinetochores for a pan-antibody to Aurora B (AIM1) or Aurora B pT232 antibody and the kinetochore markers Bub1 or KNL1. Line scans represent the average position of at least 30 kinetochores per condition. (C–E) Normalized kinetochore fluorescence intensities of control and KNL1-depleted HeLa cells expressing either CB-INCENP or HP1-Survivin and immunostained with antibodies to Aurora B pT232 (C and D) or Aurora B (AIM1, D and E). Scatter plots show intensities at individual kinetochores for Aurora B pT232 or Aurora B (AIM1) versus GFP intensities of the indicated chimera. C shows n ≥ 180 kinetochores from at least 10 cells and 3 independent experiments; D shows n ≥ 50 kinetochores from at least 8 cells; E shows n ≥ 180 kinetochores from at least 8 cells and 2 independent experiments. Graphs in C and E (right) represent the mean and SD from independent experiments. ***, P < 0.001; NS, not statistically significantly different (Mann-Whitney rank sum test). (F) Control and KNL1-depleted HeLa cells expressing CB-INCENP and immunostained with a Hec1 phosphospecific antibody to Ser44. Fluorescence intensity quantification is shown on the right. ***, P < 0.001 (t test). Bars: (cell panels) 5 μm; (kinetochore pair insets) 0.5 μm.
Discussion
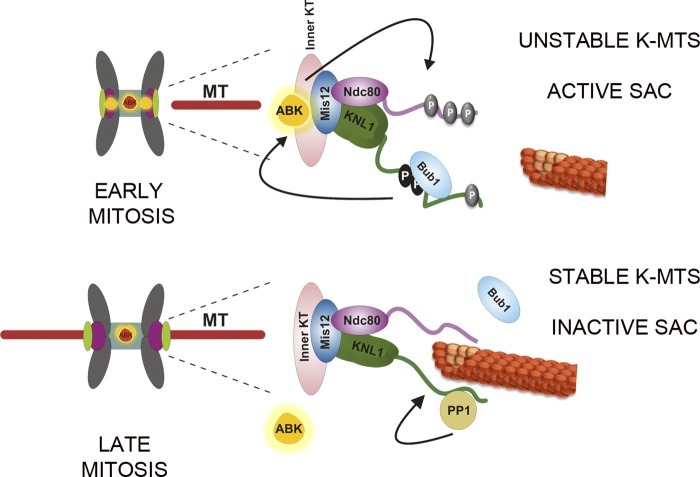
KNL1 is widely recognized as a kinetochore scaffolding protein that contributes to the formation of kinetochore–MT attachments and is required for accurate chromosome segregation during mitosis (Desai et al., 2003). Studies in C. elegans and mammalian cells have demonstrated its involvement in the recruitment of SAC proteins (Bub1, BubR1; Kiyomitsu et al., 2007) and phosphatases (PP1 and PP2A via BubR1) to kinetochores (Liu et al., 2010; Suijkerbuijk et al., 2012; Kruse et al., 2013) as well as its ability to bind MTs (Welburn et al., 2010; Espeut et al., 2012). KNL1 is proposed to be required for initial activation of the SAC (Kiyomitsu et al., 2007) and for checkpoint silencing (Meadows et al., 2011; Rosenberg et al., 2011; Espeut et al., 2012), yet how this large kinetochore protein of unknown structure contributes to different signaling pathways temporally throughout mitosis is not well understood. Adding more complexity, Bub1, BubR1, and the phosphatases PP1 and PP2A are regulators of Aurora B (Lampson and Kapoor 2005; Kiyomitsu et al., 2007; Liu et al., 2010; Suijkerbuijk et al., 2012; Kruse et al., 2013), the kinase considered the “master regulator” of kinetochore–MT attachments during mitosis. Based on its role in recruiting proteins that impact Aurora B function, we reasoned that KNL1 might be critical for the proper regulation of Aurora B activity. We found that KNL1 is required for Aurora B activity and for Aurora B–mediated phosphorylation of outer kinetochore proteins including Hec1, the primary kinetochore–MT attachment protein, and Dsn1, a member of the kinetochore-associated Mis12 complex, which also contributes to the formation of stable kinetochore–MT attachments (Kline et al., 2006). We identified the N-terminal half of KNL1 (aa 1–1200N) as sufficient to mediate such activities. In addition, the N-terminal region of KNL1 sufficient to promote Aurora B activity and substrate phosphorylation is also required for Bub1-mediated histone H2A phosphorylation, a known contributor to Aurora B centromeric localization (Yamagishi et al., 2010). Previously, it was unclear if Bub1 accumulation to kinetochores was required for Bub1 to perform its functions. Here we found that KNL1-mediated Bub1 accumulation to kinetochores is not a requirement for its activity at kinetochores. However, the N-terminal region of KNL1 is necessary and sufficient to facilitate Bub1 kinase activity. The levels of Bub1 kinase activity, measured by the levels of phosphorylation of its substrate, histone H2A, correlate with the levels of Aurora B activity. Thus, we propose that KNL1 promotes Aurora B activation and Aurora B–mediated phosphorylation by facilitating Bub1 kinase activity during early mitosis (Fig. 8). Importantly, although Bub1 functions upstream of Aurora B targeting, localization of Aurora B by ectopic targeting in the absence of KNL1 did not result in full Aurora B activity. Therefore, KNL1 and Bub1 may influence Aurora B activity in a manner that is distinct from their roles in targeting the kinase to centromeres. The interplay between Bub1 and Aurora B kinase activities at kinetochores and how Bub1 interaction with KNL1 could modulate Bub1 function during mitotic progression are issues that remain to be addressed.
Figure 8.
Model for KNL1-mediated Aurora B activation. In early mitosis, phosphorylation of outer kinetochore substrates relies on Aurora B kinase activity, which is dependent on the N terminus of KNL1. KNL1-mediated Bub1 kinase activity enhances Aurora B activation at both the inner centromere and the kinetochore. Upon generation of stable kinetochore–MT attachments and sister kinetochore bi-orientation, KNL1 is no longer able to mediate Bub1 kinase activity and Bub1 accumulation, resulting in decreased levels of active Aurora B. During metaphase, reduced levels of active Aurora B at the kinetochore region (DeLuca et al., 2011) lead to further stabilization of kinetochore–MT attachments and checkpoint silencing (mediated in part through PP1 binding; Meadows et al., 2011; Rosenberg et al., 2011).
The current model describing the regulation of kinetochore–MT attachment strength during mitosis proposes that Aurora B, emanating as a gradient from the inner centromere, selectively reaches kinetochore proteins by diffusion, depending on their distance from the origin of the gradient (Liu et al., 2009; Wang et al., 2011). However, it has been demonstrated both in chicken cells and in budding yeast that mitotic chromosomes are able to accurately bi-orient and segregate in absence of centromeric Aurora B (Yue et al., 2008; Campbell and Desai 2013). In agreement with these studies, we find that in the absence of KNL1, significant levels of centromeric Aurora B or ectopic targeting of Aurora B do not result in control levels of Aurora B activity as the previously described model proposes. We find that maintenance of Aurora B activity, promoted by the N terminus of KNL1, correlates with Bub1 activity at kinetochores (which is low during late mitosis, when checkpoint proteins no longer accumulate at kinetochores). Thus, an alternative model for the regulation of kinetochore–MT attachment suggests that levels of active Aurora B at kinetochores are modulated during mitotic progression as a result of changes in Bub1 kinase activity (Fig. 8). Specifically, it is possible that changes at the kinetochore during late mitosis result in decreased KNL1-mediated Bub1 kinase activity and Aurora B–mediated phosphorylation of kinetochore substrates. Our results demonstrating Bub1’s contribution to Aurora B activity at the kinetochore support the notion that a single sensory mechanism may be used for both MT turnover/error correction and checkpoint signaling (Fig. 8; Musacchio, 2011). A previously demonstrated role for Bub1 in chromosome congression (Klebig et al., 2009) is likely to be linked to the maintenance of Aurora B activity at the kinetochore. Conversely, a role for Aurora B in checkpoint signaling may be to not only prevent premature checkpoint silencing (by stalling PP1 recruitment) but also to maintain high levels of Bub1 kinetochore activity through the Mps1–KNL1 pathway.
Materials and methods
Cell culture and transfection
HeLa (ATCC) and RPE1 cells (hTERT-RPE1; ATCC) were cultured in DMEM and DMEM/F12 (Invitrogen), respectively, supplemented with 10% FBS and 1% penicillin/streptomycin at 37°C in 5% CO2. For immunostaining experiments after KNL1 depletion, cells were synchronized in early mitosis using a double thymidine block and release. Cells were treated with 2.5 mM thymidine for 16 h, washed with PBS, replaced with fresh medium, transfected with siRNA, and incubated for 8 h. For the second block, cells were transfected with siRNA and treated with 2.5 mM thymidine for 16 h. Cells were then washed with PBS, replaced with fresh medium, and fixed after 10 h for immunostaining. For all experiments, cells were plated at 50% confluency 24 h before transfection or thymidine treatment on acid-washed glass coverslips for immunofluorescence or on glass-bottomed dishes for live-cell imaging (MatTek Corporation). For experiments with ZM447439, cells were treated with 2 µM ZM447439 for 2 h before fixing or harvesting. For cold-induced depolymerization assays, cells were incubated in ice-cold DMEM for 10 min, fixed with paraformaldehyde, and prepared for immunofluorescence as described below. siRNA transfections were performed using Oligofectamine (Invitrogen), according to the manufacturer’s instructions, and cells were analyzed 48 h after transfection. For KNL1 silence and rescue experiments, cells were transfected with siRNA, 8 h later with plasmid DNA using Effectene (QIAGEN), and analyzed 48 h after DNA transfection. siRNAs targeting KNL1 were purchased from Invitrogen. The sequences for “stealth” siRNAs were 5′-GAACACAUUGCUUUCUGCUCCCAUU-3′, 5′-GGGCAGGAUGACAUGGAGAUCACUA-3′, and 5′-AAGAUCUGAUUAAGGAUCCACGAAA-3′. The siRNA used for KNL1 silence and rescue experiments for wild-type and RVSF/AAAA plasmids (Liu et al., 2010) was synthesized by QIAGEN (5′-AAGGAAUCCAAUGCUUUGAGA-3′). Bub1 siRNA was 5′-CAGCUUGUGAUAAAGAGUCAA-3′ and BubR1 siRNA was 5′-ACGAGAAUACCUAAUAUGUGA-3′, both purchased from QIAGEN. Luciferase siRNA (QIAGEN) was used as control in all experiments displayed in Figs. 1 and 2 and Figs. S1–S3.
The FlpIn T-REx HeLa host cell line was a gift from S. Taylor (University of Manchester, Manchester, England, UK). To generate FlpIn T-REx HeLa cells containing KNL1 fragments fused to GFP at the N terminus, Flp-In T-REx HeLa host cells were cotransfected with a ratio of 9:1 (wt/wt) pOG44:pcDNA5/FRT/TO expression plasmid using Fugene HD (Promega). 48 h after transfection, cell lines were placed under selection with 100 µg/ml hygromycin (EMD Millipore) for 2 wk. Hygromycin-resistant foci were chosen, expanded, and tested for GFP expression. Gene expression was induced with 1 µg/ml doxycycline (Sigma-Aldrich) for 24 h. For silence and rescue experiments, stable cell lines were doubly blocked with thymidine and depleted of endogenous KNL1 using the corresponding siRNAs described above.
Plasmids
The siRNA-resistant KNL1 wild-type and RVSFAAAA mutant plasmids were a gift from M. Lampson (University of Pennsylvania, Philadelphia, PA). The GFP-tagged KNL1 fragments were as follows: amino acids 1–300 (300N), 300–818 (300–800N), 819–1518 (800–1500N), 1174–2316 (1200C), 1519–2316 (800C), 819–2316 (1500C), 2056–2316 (250C), and 1834–2316 (400C), and were generated by PCR using KNL1 wild-type as the template (Liu et al., 2009) and cloned into the pEGF-C2 vector and pcDNA5/FRT/TO vector using In-Fusion cloning (Takara Bio Inc.). The KNL1 fragments KNL1 300N, 300–800N, and 800–1500N were cloned into pEGFP-C2 and/or pcDNA5/FRT/TO containing the KNL1 kinetochore-binding region (aa 2056–2316). Constructs 800–1500N and 1500C differ from the sequence published in Bolanos-Garcia et al. (2011), in that aa 910–1120 are not contained. The GFP-tagged CB-INCENP plasmid containing hCenp-B aa 1–158 and hINCENP aa 47–920 was a gift from S. Lens (University Medical Center, Utrecht, Netherlands). The GFP-HP1-Survivin construct was generated as follows: full-length Survivin and HP1 were obtained by RT-PCR from HeLa cells. Survivin was cloned into pEGFP-C2 to generate GFP-Survivin, and HP1 was cloned into GFP-Survivin to generate GFP-HP1-Survivin. All plasmid DNAs were purified using an Endo-free Maxi kit (QIAGEN) before transfection.
Immunofluorescence
Fixation and immunostaining of HeLa and RPE-1 cells were performed as described previously (DeLuca et al., 2011). In brief, cells were rinsed in 37°C PHEM buffer (60 mM Pipes, 25 mM Hepes, 10 mM EGTA, and 4 mM MgSO4, pH 6.9), fixed in in 4% paraformaldehyde for 20 min, and extracted in PHEM buffer + 0.5% Triton X-100 for 5 min. Staining of phospho-Ser331 Aurora B was performed as described in Petsalaki et al. (2011). In brief, cells were rinsed in PHEM buffer, extracted in PHEM buffer + 0.5% CHAPS for 5 min, and fixed with cold methanol for 5 min at 20°C. Immunostaining was performed using the following antibodies: rabbit polyclonal anti-phosphorylated Ser44-Hec1 (pSer44 against antigen PTFGKL(pS)INKPTSE) and anti-phosphorylated Ser55-Hec1 (pSer55 against antigen KPTSERKV(pS)LFGKR) were used at 1:2,000; mouse anti-Hec1 9G3 at 1:2,000 (GeneTex); rabbit anti-Dsn1 at 1:500 (GeneTex); human anti-centromere antibody at 1:500 (ACA; Antibodies, Inc.); mouse anti-tubulin at 1:200 (Sigma-Aldrich); sheep anti-tubulin at 1:300 (Cytoskeleton, Inc.); rabbit anti-phosphorylated Aurora B (pThr232) at 1:2,000 (Rockland); rabbit anti-phosphorylated Aurora-B (pSer331, against peptide CPWVRAN(pS)RRVLPPS); a gift from G. Zachos [University of Crete, Heraklion, Greece]) at 1:100; mouse anti-BubR1 at 1:500 (EMD Millipore); rabbit anti-phosphorylated INCENP (Ser893/Ser894, against peptide RYHKRT(pS)(pS)AVWNSPC); a gift from M. Lampson) at 1:1,000; rabbit anti-phosphorylated Dsn1 (Ser109 against peptide TNRRK(pS)LHPIH); a gift from I. Cheeseman [Whitehead Institute, Cambridge, MA]) at 1:1,000; rabbit anti-phosphorylated histone H2A (Thr120) at 1:1,000 (Active Motif); mouse anti-Bub1 at 1:200 (Abcam); mouse anti–Aurora B monoclonal (AIM1) at 1:300 (BD); and mouse anti-phosphorylated histone H3 (Ser10; Cell Signaling Technology) at 1:1,000. Affinity-purified antibodies against KNL1 were generated at 21st Century Biochemicals; rabbits were immunized with the following KNL1 peptide: MDGVSSEANEENDNIERPVRRR, and serum was double affinity purified. Secondary antibodies conjugated to either Alexa 647, Alexa 488, or Rhodamine Red-X (Jackson ImmunoResearch Laboratories, Inc.) were used at 1:300.
Image acquisition and analysis
Image acquisition was performed on an imaging system (DeltaVision Personal DV; Applied Precision) equipped with a camera (CoolSNAP HQ2; Photometrics/Roper Scientific), a 60×/1.42 NA PlanApochromat objective (Olympus), and SoftWoRx acquisition software (Applied Precision). For fixed cell experiments, images were acquired at room temperature as Z-stacks at 0.2-µm intervals and kinetochore fluorescence intensity measurements were performed using MetaMorph software (Molecular Devices). The integrated fluorescence intensity minus the calculated background was determined for each kinetochore in control and treated samples and normalized to the average value obtained from control cells (Hoffman et al., 2001). Inter-kinetochore distances were determined using SoftWorx software. For live-cell imaging of kinetochore oscillations, HeLa cells in 35-mm glass-bottomed dishes (MatTek Corporation) were transfected with KNL1 siRNA and 24 h later with 0.2 µg of GFP-CENP-B (a gift from K. Sullivan, NUI Galway, Galway, Ireland) and GFP-centrin (a gift from M. Bornens, Institut Curie, Paris, France) as described above. Cells were imaged 24 h after DNA transfection in Leibovitz’s L-15 media (Invitrogen) supplemented with 10% FBS, 7 mM Hepes, pH 7.0, and 4.5 g/L glucose. Stage temperature was maintained at 37°C with an environmental chamber (Precision Control). Fluorescence images of GFP-CENP-B/GFP-centrin–expressing cells were acquired every 3 s for 10 min. At each time point, 5 images were collected in a Z-series, using a 0.5-µm step size. Cells chosen for analysis expressed both GFP-CENP-B and centrin-GFP proteins and were in late prometaphase or metaphase with primarily bi-oriented chromosomes. Sister kinetochore pairs chosen for analysis were located within the middle of the spindle. Kinetochore and spindle pole movements were tracked using CENP-B-GFP fluorescence and centrin-GFP fluorescence, respectively, on maximum projection time-lapse sequences using MetaMorph software. The spindle pole marker centrin was used to correct for cell movement during the time-lapse imaging sequence. Kinetochore movements were tracked on maximum projection time-lapse sequences using the “track points” function in MetaMorph software. Tracking data were analyzed in Microsoft Excel. A “pause” event was recorded when a kinetochore did not move for two sequential time frames. Velocity was calculated by linear regression analysis of kinetochore distance versus time plots, and deviation from average position (DAP) was determined by subtracting the position of the kinetochore in the regression line from the original kinetochore position (Stumpff et al., 2008; DeLuca et al., 2011). Kinetochore oscillation analysis in HeLa TRex FlpIn stable cell lines was performed as described above, with the exception that cells were expressing mCherry tagged CenpB. For live-cell imaging with mCherry-Bub1, 300N KNL1 HeLa stable cells were transfected with 0.3 µg mCherry-Bub1 and imaged 48 h after DNA transfection in filming medium containing 330 nM nocodazole. All presented data were analyzed in SigmaPlot software v.11.0 (Systat Software) and the statistical tests performed are specified in figure legends.
Immunoprecipitation and immunoblotting
Cells were lysed in lysis buffer (75 mM Hepes, pH 7.5, 150 mM KCl, 1.5 mM EGTA, 1.5 mM MgCl2, 0.1% NP-40, Complete protease inhibitor cocktail [Roche], and PhosSTOP phosphatase inhibitors [Roche]. Western blotting was performed using the following antibodies: anti-Hec1 9G3 at 1:1,000 (GeneTex); rabbit anti-phosphorylated Aurora B (pThr232) at 1:100 (Rockland); rabbit anti–Aurora B at 1:1,000 (Abcam); rabbit anti-KNL1 at 1:500; mouse anti–β-actin at 1:500 (Abcam); and mouse anti-tubulin at 1:2,000 (Sigma-Aldrich). For immunoprecipitation assays, lysates were incubated with mouse anti–Aurora B (mouse AIM1; BD) for 2 h at 4°C, and with a mix of Sepharose protein A/G (Bio-Rad Laboratories) for an additional 2 h at 4°C. The unbound fraction was recovered and beads were washed with lysis buffer (3×). Beads were treated with 2× SDS sample buffer and boiled at 95°C for 5 min for elution.
Online supplemental material
Fig. S1 shows the effect of KNL1 depletion in RPE1 cells (Fig. 1). In addition, this figure demonstrates the specificity of the pT232 Aurora B kinase antibody and the effect of KNL1 depletion on Aurora B S331, INCENP, and histone H3 S10 phosphorylation (Fig. 1 G). Fig. S2 demonstrates that the results observed in HeLa Flp-In T-REx stable cell lines are mirrored using transient transfections (Fig. 3). Fig. S3 demonstrates that the C terminus of KNL1 is not sufficient to mediate Aurora B and INCENP phosphorylation in cells (Fig. 3). Fig. S4 shows that phosphorylation of Aurora B T232 does not depend on BubR1 and that pT232 phosphorylation is not affected in cells expressing a KNL1 mutant incapable of recruiting PP1 (Fig. 6). Finally, this figure shows that Bub1 does not accumulate to kinetochores in cells expressing GFP-KNL1-300N in nocodazole (Fig. 6). Fig. S5 shows immunofluorescence images supporting the results shown in Fig. 7, which demonstrate that ectopic accumulation of Aurora B does not result in full Aurora B activity in the absence of KNL1. Videos 1 and 2 show kinetochore oscillations in control and KNL1-depleted cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201306054/DC1.
Supplementary Material
Acknowledgments
We thank members of the DeLuca laboratory and the Cytoskeleton Group Meeting for helpful discussions; Tina Lynch and Jeanne Mick for experimental assistance; and O’Neil Wiggan, Arshad Desai, and Ted Salmon for providing insightful comments on the manuscript. We also thank Stephen Taylor, Mike Lampson, Susanne Lens, George Zachos, Iain Cheeseman, William Sullivan, and Michel Bornens for providing reagents.
This work was supported by National Institutes of Health grant R01GM088371 (to J.G. DeLuca). J.G. DeLuca is also supported by the Pew Scholars Program in the Biomedical Sciences.
Footnotes
Abbreviations used in this paper:
- CB
- Cenp-B
- MT
- microtubule
- SAC
- spindle assembly checkpoint
References
- Biggins S., Severin F.F., Bhalla N., Sassoon I., Hyman A.A., Murray A.W. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544 10.1101/gad.13.5.532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolanos-Garcia V.M., Lischetti T., Matak-Vinković D., Cota E., Simpson P.J., Chirgadze D.Y., Spring D.R., Robinson C.V., Nilsson J., Blundell T.L. 2011. Structure of a Blinkin-BUBR1 complex reveals an interaction crucial for kinetochore-mitotic checkpoint regulation via an unanticipated binding site. Structure. 19:1691–1700 10.1016/j.str.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.S., Desai A. 2013. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature. 497:118–121 10.1038/nature12057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., Chappie J.S., Wilson-Kubalek E.M., Desai A. 2006. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 127:983–997 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2008. KNL1 and the CENP-H/I/K complex coordinately direct kinetochore assembly in vertebrates. Mol. Biol. Cell. 19:587–594 10.1091/mbc.E07-10-1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D., Wan X., Hirel C.B., Salmon E.D. 2006. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr. Biol. 16:1711–1718 10.1016/j.cub.2006.07.022 [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G.M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C.D., Sassone-Corsi P. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874–885 10.1128/MCB.22.3.874-885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Gall W.E., Ciferri C., Cimini D., Musacchio A., Salmon E.D. 2006. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 127:969–982 10.1016/j.cell.2006.09.047 [DOI] [PubMed] [Google Scholar]
- DeLuca K.F., Lens S.M., DeLuca J.G. 2011. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 124:622–634 10.1242/jcs.072629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., Rybina S., Müller-Reichert T., Shevchenko A., Shevchenko A., Hyman A., Oegema K. 2003. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 17:2421–2435 10.1101/gad.1126303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeut J., Cheerambathur D.K., Krenning L., Oegema K., Desai A. 2012. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J. Cell Biol. 196:469–482 10.1083/jcb.201111107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E.A., Maldonado M., Kapoor T.M. 2011. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 13:1265–1271 10.1038/ncb2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., Salmon E.D. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R.K., Körner R., Nigg E.A. 2003. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol. Biol. Cell. 14:3325–3341 10.1091/mbc.E02-11-0769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. 2010. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 327:172–177 10.1126/science.1180189 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. 2007. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 13:663–676 10.1016/j.devcel.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Murakami H., Yanagida M. 2011. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31:998–1011 10.1128/MCB.00815-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebig C., Korinth D., Meraldi P. 2009. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185:841–858 10.1083/jcb.200902128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline S.L., Cheeseman I.M., Hori T., Fukagawa T., Desai A. 2006. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J. Cell Biol. 173:9–17 10.1083/jcb.200509158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenn V., Wehenkel A., Li X., Santaguida S., Musacchio A. 2012. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J. Cell Biol. 196:451–467 10.1083/jcb.201110013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T., Zhang G., Larsen M.S., Lischetti T., Streicher W., Kragh Nielsen T., Bjørn S.P., Nilsson J. 2013. Direct binding between BubR1 and B56-PP2A phosphatase complexes regulate mitotic progression. J. Cell Sci. 126:1086–1092 10.1242/jcs.122481 [DOI] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98 10.1038/ncb1208 [DOI] [PubMed] [Google Scholar]
- Lipkow K., Odde D.J. 2008. Model for protein concentration gradients in the cytoplasm. Cell Mol Bioeng. 1:84–92 10.1007/s12195-008-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M.J., Lampson M.A., Lens S.M. 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 323:1350–1353 10.1126/science.1167000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C.B., Hori T., Fukagawa T., Cheeseman I.M., Lampson M.A. 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188:809–820 10.1083/jcb.201001006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London N., Ceto S., Ranish J.A., Biggins S. 2012. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 22:900–906 10.1016/j.cub.2012.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J.C., Shepperd L.A., Vanoosthuyse V., Lancaster T.C., Sochaj A.M., Buttrick G.J., Hardwick K.G., Millar J.B. 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell. 20:739–750 10.1016/j.devcel.2011.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A. 2011. Spindle assembly checkpoint: the third decade. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:3595–3604 10.1098/rstb.2011.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E., Akoumianaki T., Black E.J., Gillespie D.A., Zachos G. 2011. Phosphorylation at serine 331 is required for Aurora B activation. J. Cell Biol. 195:449–466 10.1083/jcb.201104023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch M., Khoudoli G.A., Swift S., King E.M., Deluca J.G., Swedlow J.R. 2010. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191:61–74 10.1083/jcb.200912046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., Jeganathan K.B., van Deursen J.M. 2011. Bub1 overexpression induces aneuploidy and tumor formation through Aurora B kinase hyperactivation. J. Cell Biol. 193:1049–1064 10.1083/jcb.201012035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke R.M., Jeganathan K.B., Malureanu L., Harrison A.M., van Deursen J.M. 2012. Bub1 kinase activity drives error correction and mitotic checkpoint control but not tumor suppression. J. Cell Biol. 199:931–949 10.1083/jcb.201205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 1981. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 84:145–158 10.1007/BF00293368 [DOI] [PubMed] [Google Scholar]
- Rosenberg J.S., Cross F.R., Funabiki H. 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21:942–947 10.1016/j.cub.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepperd L.A., Meadows J.C., Sochaj A.M., Lancaster T.C., Zou J., Buttrick G.J., Rappsilber J., Hardwick K.G., Millar J.B. 2012. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 22:891–899 10.1016/j.cub.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D., Gassmann R., Oegema K., Desai A. 2011. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS ONE. 6:e22969 10.1371/journal.pone.0022969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J., von Dassow G., Wagenbach M., Asbury C., Wordeman L. 2008. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell. 14:252–262 10.1016/j.devcel.2007.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suijkerbuijk S.J., Vleugel M., Teixeira A., Kops G.J. 2012. Integration of kinase and phosphatase activities by BUBR1 ensures formation of stable kinetochore-microtubule attachments. Dev. Cell. 23:745–755 10.1016/j.devcel.2012.09.005 [DOI] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329 10.1016/S0092-8674(02)00633-5 [DOI] [PubMed] [Google Scholar]
- Tsukahara T., Tanno Y., Watanabe Y. 2010. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature. 467:719–723 10.1038/nature09390 [DOI] [PubMed] [Google Scholar]
- Wang E., Ballister E.R., Lampson M.A. 2011. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J. Cell Biol. 194:539–549 10.1083/jcb.201103044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J.P., Vleugel M., Liu D., Yates J.R., III, Lampson M.A., Fukagawa T., Cheeseman I.M. 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell. 38:383–392 10.1016/j.molcel.2010.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi Y., Honda T., Tanno Y., Watanabe Y. 2010. Two histone marks establish the inner centromere and chromosome bi-orientation. Science. 330:239–243 10.1126/science.1194498 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Yang C.H., Tanno Y., Watanabe Y. 2012. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 14:746–752 10.1038/ncb2515 [DOI] [PubMed] [Google Scholar]
- Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. 2004. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 279:12997–13003 10.1074/jbc.M311128200 [DOI] [PubMed] [Google Scholar]
- Yue Z., Carvalho A., Xu Z., Yuan X., Cardinale S., Ribeiro S., Lai F., Ogawa H., Gudmundsdottir E., Gassmann R., et al. 2008. Deconstructing Survivin: comprehensive genetic analysis of Survivin function by conditional knockout in a vertebrate cell line. J. Cell Biol. 183:279–296 10.1083/jcb.200806118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.