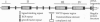Abstract
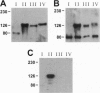
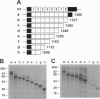
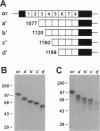
Megalin (gp330), an epithelial endocytic receptor, is a major target antigen of Heymann nephritis (HN), an autoimmune disease in rats. To elucidate the mechanisms of HN, we have mapped a pathogenic epitope in megalin that binds anti-megalin antibodies. We focused our attention on four clusters of cysteine-rich, low density lipoprotein receptor (LDLR) ligand binding repeats in the extracellular domain of megalin because they represent putative ligand binding regions and therefore would be expected to be exposed in vivo and to be able to bind circulating antibodies. Rat megalin cDNA fragments I through IV encoding the first through fourth clusters of ligand-binding repeats, respectively, were expressed in a baculovirus system. All four expression products were detected by immunoblotting with two antisera capable of inducing passive HN (pHN). When antibodies eluted from glomeruli of rats with pHN were used for immunoblotting, only the expression product encoded by fragment II was detected. This indicates that the second cluster of LDLR ligand binding repeats is directly involved in binding anti-megalin antibodies and in the induction of pHN. To narrow the major epitope in this domain, fragment II was used to prepare proteins sequentially truncated from the C- and N-terminal ends by in vitro translation. Analysis of the truncated translation products by immunoprecipitation with anti-megalin IgG revealed that the fifth ligand-binding repeat (amino acids 1160-1205) contains the major epitope recognized. This suggests that a 46-amino acid sequence in the second cluster of LDLR ligand binding repeats contains a major pathogenic epitope that plays a key role in pHN. Identification of this epitope will facilitate studies on the pathogenesis of HN.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachinsky D. R., Zheng G., Niles J. L., McLaughlin M., Abbate M., Andres G., Brown D., McCluskey R. T. Detection of two forms of GP330. Their role in Heymann nephritis. Am J Pathol. 1993 Aug;143(2):598–611. [PMC free article] [PubMed] [Google Scholar]
- Biemesderfer D., Dekan G., Aronson P. S., Farquhar M. G. Biosynthesis of the gp330/44-kDa Heymann nephritis antigenic complex: assembly takes place in the ER. Am J Physiol. 1993 Jun;264(6 Pt 2):F1011–F1020. doi: 10.1152/ajprenal.1993.264.6.F1011. [DOI] [PubMed] [Google Scholar]
- Falcone D., Andrews D. W. Both the 5' untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol Cell Biol. 1991 May;11(5):2656–2664. doi: 10.1128/mcb.11.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar M. G., Saito A., Kerjaschki D., Orlando R. A. The Heymann nephritis antigenic complex: megalin (gp330) and RAP. J Am Soc Nephrol. 1995 Jul;6(1):35–47. doi: 10.1681/ASN.V6135. [DOI] [PubMed] [Google Scholar]
- Huang J., Makker S. P. Role of receptor-associated 39/45 kD protein in active Heymann nephritis. Kidney Int. 1995 Feb;47(2):432–441. doi: 10.1038/ki.1995.56. [DOI] [PubMed] [Google Scholar]
- Kerjaschki D., Farquhar M. G. Immunocytochemical localization of the Heymann nephritis antigen (GP330) in glomerular epithelial cells of normal Lewis rats. J Exp Med. 1983 Feb 1;157(2):667–686. doi: 10.1084/jem.157.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Farquhar M. G. The pathogenic antigen of Heymann nephritis is a membrane glycoprotein of the renal proximal tubule brush border. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5557–5561. doi: 10.1073/pnas.79.18.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Miettinen A., Farquhar M. G. Initial events in the formation of immune deposits in passive Heymann nephritis. gp330-anti-gp330 immune complexes form in epithelial coated pits and rapidly become attached to the glomerular basement membrane. J Exp Med. 1987 Jul 1;166(1):109–128. doi: 10.1084/jem.166.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Noronha-Blob L., Sacktor B., Farquhar M. G. Microdomains of distinctive glycoprotein composition in the kidney proximal tubule brush border. J Cell Biol. 1984 Apr;98(4):1505–1513. doi: 10.1083/jcb.98.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Ullrich R., Diem K., Pietromonaco S., Orlando R. A., Farquhar M. G. Identification of a pathogenic epitope involved in initiation of Heymann nephritis. Proc Natl Acad Sci U S A. 1992 Dec 1;89(23):11179–11183. doi: 10.1073/pnas.89.23.11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerjaschki D., Ullrich R., Exner M., Orlando R. A., Farquhar M. G. Induction of passive Heymann nephritis with antibodies specific for a synthetic peptide derived from the receptor-associated protein. J Exp Med. 1996 May 1;183(5):2007–2015. doi: 10.1084/jem.183.5.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen A., Törnroth T., Tikkanen I., Virtanen I., Linder E. Heymann nephritis induced by kidney brush border glycoproteins. Lab Invest. 1980 Dec;43(6):547–555. [PubMed] [Google Scholar]
- Orlando R. A., Kerjaschki D., Farquhar M. G. Megalin (gp330) possesses an antigenic epitope capable of inducing passive Heymann nephritis independent of the nephritogenic epitope in receptor-associated protein. J Am Soc Nephrol. 1995 Jul;6(1):61–67. doi: 10.1681/ASN.V6161. [DOI] [PubMed] [Google Scholar]
- Orlando R. A., Kerjaschki D., Kurihara H., Biemesderfer D., Farquhar M. G. gp330 associates with a 44-kDa protein in the rat kidney to form the Heymann nephritis antigenic complex. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6698–6702. doi: 10.1073/pnas.89.15.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietromonaco S., Kerjaschki D., Binder S., Ullrich R., Farquhar M. G. Molecular cloning of a cDNA encoding a major pathogenic domain of the Heymann nephritis antigen gp330. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1811–1815. doi: 10.1073/pnas.87.5.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychowdhury R., Niles J. L., McCluskey R. T., Smith J. A. Autoimmune target in Heymann nephritis is a glycoprotein with homology to the LDL receptor. Science. 1989 Jun 9;244(4909):1163–1165. doi: 10.1126/science.2786251. [DOI] [PubMed] [Google Scholar]
- Saito A., Pietromonaco S., Loo A. K., Farquhar M. G. Complete cloning and sequencing of rat gp330/"megalin," a distinctive member of the low density lipoprotein receptor gene family. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):9725–9729. doi: 10.1073/pnas.91.21.9725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada Y., Hemler M. E. The primary structure of the VLA-2/collagen receptor alpha 2 subunit (platelet GPIa): homology to other integrins and the presence of a possible collagen-binding domain. J Cell Biol. 1989 Jul;109(1):397–407. doi: 10.1083/jcb.109.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Bachinsky D. R., Stamenkovic I., Strickland D. K., Brown D., Andres G., McCluskey R. T. Organ distribution in rats of two members of the low-density lipoprotein receptor gene family, gp330 and LRP/alpha 2MR, and the receptor-associated protein (RAP). J Histochem Cytochem. 1994 Apr;42(4):531–542. doi: 10.1177/42.4.7510321. [DOI] [PubMed] [Google Scholar]