Abstract
Folding of membrane proteins begins in the ribosome as the peptide is elongated. During this process, the nascent peptide navigates along 100 Å of tunnel from the peptidyltransferase center to the exit port. Proximal to the exit port is a ‘folding vestibule’ that permits the nascent peptide to compact and explore conformational space for potential tertiary folding partners. The latter occurs for cytosolic subdomains, but has not yet been shown for transmembrane segments. We now demonstrate, using an accessibility assay and an improved, intramolecular crosslinking assay, that the helical transmembrane S3b-S4 hairpin (‘paddle’) of a voltage-gated potassium (Kv) channel, a critical region of the Kv voltage sensor, forms in the vestibule. S3-S4 hairpin interactions are detected at an early stage of Kv biogenesis. Moreover, this vestibule hairpin is consistent with a closed-state conformation of the Kv channel in the plasma membrane.
Keywords: Folding of nascent peptides, potassium channel voltage-sensor biogenesis, transmembrane helical hairpin, PEG-MAL, intramolecular crosslinking
INTRODUCTION
A tunnel in the ribosome is home to the nascent peptide as it is elongated and wends its way from the peptidyl transferase center (PTC) to the exit port, a distance of approximately 80-100 Å. The width of this tunnel is 10-20Å, the widest region being near the exit port in the last 20Å of the tunnel (Figure 1a). This region is an operational extension of the tunnel with distinct physico-chemical properties1 that are permissive for secondary folding2-8. We refer to this region as the ‘folding vestibule’ 4. It is likely comprised of ribosomal RNA and proteins, chaperones, and other factors9-12. Based on our previous studies, this vestibule appears to begin at a tunnel location approximately 80Å from the PTC, but its terminal border has not yet been defined. This vestibule hosts tertiary interactions between small fragments within soluble domains of the nascent peptide13-15. Proteins that acquire tertiary structures while tethered to the ribosome may begin to fold in this vestibule16, 17, an event that could, in the case of membrane proteins, influence membrane insertion efficiency and the ultimate fate of transmembrane segments. To our knowledge, no studies yet directly assess whether helical transmembrane segments in the vestibule undergo tertiary folding, e.g., hairpin formation.
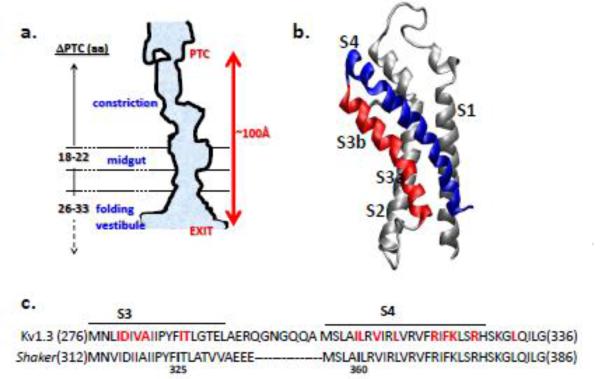
Figure 1. Biogenic participants.
(a) Cartoon of ribosome tunnel (after Wilson & Beckmann 201152). The constriction is a region of narrower dimensions approximately 25-30Å from the PTC; the midgut is a region that discriminates side chain volumes53; the folding vestibule begins at a tunnel location that is approximately 80Å from the PTC, near the exit port4. As indicated by the dashed arrow, its terminal border has not yet been defined. Numbers for ΔPTC are estimates derived from previous studies of modification of nascent chain cysteines in the tunnel4, 53. This angstrom distance is an estimate based on the observation that engineered cysteines located 26-33 residues from the PTC are accessible and that an extended peptide displaces 3-3.4 Å/amino acid. (b) Voltage-sensor domain (VSD) of a Kv channel. The S3 and S4 are red and blue, respectively. S1 and S2 are shown in gray. Model drawn in DS Viewer Pro using coordinates of the Kv1.2/2.1 crystal structure 20. (c) Comparison of the primary sequences of the S3-S4 hairpin from Kv1.3 and Shaker channels. The amino acid sequence is indicated by a single-letter code. Shaker residues 336-355 are indicated by dashes. Cysteine engineered in Kv1.3 for crosslinking experiments are bolded in red. In Shaker, residues I325 and I360 (bolded in black) are crosslinked in the closed state when substituted with cysteines25.
To explore this hypothesis, we focused on the voltage-sensor domain (VSD), comprised of four transmembrane segments, S1-S4, of a tetrameric voltage-gated potassium (Kv) channel. The VSD contains a functionally important tertiary fold, the S3b-S4 hairpin (Figure 1b), the so-called ‘paddle’ of the voltage-sensor18. A Kv test peptide offers two major advantages over other polytopic membrane proteins: it has been studied functionally in detail, and several high-resolution crystal structures exist. Thus, folded states identified in the vestibule and the ER (early biogenic stages) can be compared with specific states of the mature channel in the plasma membrane.
Despite some controversy about the voltage-dependent movement of S3b relative to S4 as the channel responds to changes of membrane potential, its existence and role as a folded subdomain is supported by 3 lines of evidence. First, the crystal structures of Kv channels19, 20 all show a helix-turn-helix motif. Second, this subdomain is conserved and can be transplanted not only from archaebacterial KvAP to eukaryotic Kv channels, but also from distinctly diverse voltage-sensing proteins like Hv121 and Ci-VSP22, with retention of function 23. Third, S3b and S4 appear to be in close proximity24, 25 and residues in these two helices in the Shaker Kv channel can be crosslinked in a functionally competent state in the plasma membrane25. There is high sequence conservation between Shaker and the channel we study here, Kv1.3, albeit with slight shifts in alignment (Figure 1c). Thus, we used S3-S4 as a model to explore whether tertiary interactions occur in the folding vestibule, and therefore early in Kv biogenesis, and to identify requirements for formation of this helix-turn-helix structure.
Upstream transmembrane segments may serve as retention signals inside the translocon26-28, thereby enabling sequential transmembrane helix bundles to fold correctly and insert into the ER membrane. Moreover, helical transmembrane segments of polytopic proteins are well known to accumulate in the translocon (e.g., 29-32), where they may even require cooperative intra-segment interactions for bilayer insertion. Our investigation of Kv S3-S4 hairpin formation bears on these hypotheses. In this paper, using an improved intramolecular crosslinking assay, we show that transmembrane segments are capable of tertiary prefolding inside the vestibule prior to targeting of the nascent chain-ribosome complex to the membrane. We draw three conclusions. First, S3-S4 hairpin interactions in the Kv monomer are detected in the folding vestibule, thus, at an early stage of biogenesis. Second, the entire S4 segment forms a compact structure, likely helical, before exiting the ribosomal tunnel. Third, the S3-S4 hairpin in the vestibule is consistent with a closed-state conformation of the Kv channel in the plasma membrane.
Prefolding of transmembrane segments inside the vestibule may facilitate or even promote folding of transmembrane segments inside the translocon and influence both the timing and efficiency of insertion events. In the case of a voltage-gated channel, misfolding will result in a lack of trafficking of the channel and/or a disease phenotype 33, 34sometimes with disastrous consequences for cell excitability.
RESULTS
To determine whether, and in which compartment, tertiary interactions occur during Kv biogenesis, we examined the S3-S4 transmembrane segment of Kv1.3 inside the folding vestibule, which appears to be large enough to hold a tertiary helical hairpin. Any nascent chain candidate for helical hairpin formation must fulfill two criteria. Each arm of the hairpin must have a helical secondary structure, and the putative interaction surfaces of a hairpin must be in close proximity in the vestibule. With these two criteria in mind, we set out to evaluate each of them for the nascent S3-S4 segment.
S4 location and secondary structure
To determine the location and secondary structure of S4, we used an accessibility assay that reliably estimates compaction in the tunnel. For each S4 construct, we made mRNA from DNA that lacked a stop codon so that translation in a cell-free rabbit reticulocyte lysate would yield the designed nascent peptide attached to tRNA and the ribosome. A cysteine was engineered, one at a time, along the transmembrane segment of S4 (Figure 2a) and covalently modified by a high molecular-weight maleimide (polyethylene glycol maleimide, PEGMAL). The adduct can be detected as a shift in mobility of the peptide on a protein gel 2-4, and labeling is compared to an all-extended molecular tape measure2 (black circles, Figure 2c). To completely emerge from the ribosomal exit tunnel, a given residue in an extended conformation is ~33 amino acids from the PTC, which corresponds to a distance of ~99-112Å, the operational length of the tunnel, in excellent agreement with anatomical dimensions derived from structural studies35 36. Residues that are ≥ 33 amino acids from the PTC are ≥ 80% pegylated in a fully extended peptide. If a nascent test peptide is cysteine-scanned, pegylated, and then compared with the extended tape measure (Figure 2c), a decreased slope indicates the presence of secondary structure. Specifically, a fractional slope of ≤0.5, relative to that of the extended tape measure, is consistent with the presence of an α-helix (1.5Å/amino acid for an α-helix versus 3-3.4Å/amino acid for an extended peptide37).
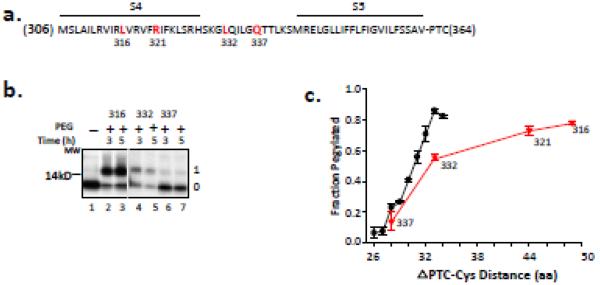
Figure 2. Accessibility of S4 residues.
(a) Primary sequence of S4 and flanking residues. The amino acid sequence is indicated by a single-letter code. Residues 316, 321, 332 and 337 (bolded, red) were mutated to cysteines, one at a time, and used in the pegylation assay. The N-terminus of the peptide is M306 and a restriction enzyme (Acc1) was used to generate the terminal amino acid, V364, at the peptide’s C-terminus. The peptide thus includes S4-S5, attached to the PTC at residue 364. (b) Pegylation of selected S4 residues of Kv1.3 indicated in (a). Nascent peptides were translated from mRNA engineered from DNA that lacked a stop codon, pegylated (1 mM PEG-MAL) for the times indicated, and fractionated using SDS-PAGE as described previously4. The unpegylated protein is shown in lane 1 and is identical for all peptides substituted with cysteines at 316, 332, and 337. Gels were 12% NuPAGE Bis-Tris gels with MES running buffer. The number to the left of the gel is a molecular weight standard; numbers to the right of the gel indicate unpegylated (0) and singly pegylated (1) protein. (c) Fraction pegylated for the S4 residues (red triangles) indicated along the x-axis, which indicates the number of amino acids from the PTC to (and including) the labeled cysteine. Pegylation of a known all-extended nascent peptide is shown by the black circles and represents data taken from Lu and Deutsch, 20052. Symbols are mean ± SEM (n= 3).
To avoid any potentially complicating influence of tertiary structure from the preceding transmembrane segments S1-S3 (see below), we made a construct containing only the S4-S5-S6 C-terminus of Kv1.3 (Figure 2a, PTC 364), positioning S4 in the vestibule. A cysteine scan of S4 residues 316, 332, and 337 using pegylation gave the results shown in Figure 2b. The lower band (band 0) represents the unpegylated protein, whereas the upper band (band 1) is the pegylated protein. Residue 337, located a distance of 28 amino acids from the PTC (ΔPTC 28), is mostly unpegylated (lanes 6 and 7). Residue 316, ΔPTC 50, is mostly pegylated (lanes 2 and 3), whereas residue 332, ΔPTC 33, is intermediate between these two extremes (lanes 4 and 5). Quantification of these data gives the fraction pegylated (Fpeg). A plot of Fpeg versus distance of the cysteine from the PTC (ΔPTC-Cys) yields a relationship (red triangles; Figure 2c) with Fpeg at ΔPTC 33 that is 0.55 that of the tape measure and an additional increased Fpeg for residue 321. These results are consistent with those obtained for pegylation of canonical helices from different transmembrane segments (S1-S62-4), and suggest that scanned regions in the S4-S5 linker and S4 are compact, thereby situating the most C-terminal residues of S4 in the tunnel. Moreover, these results showing C-terminal S4 compaction, combined with previous findings demonstrating N-terminal S4 compaction4, indicate that S4 folding occurs vectorially upon entry of the given segment into the vestibule. S3 is also compact in the tunnel vestibule4. Thus, both S3 and S4 are likely helical in the vestibule (also see below), thereby fulfilling the first criterion for helical hairpin formation.
Tertiary paddle-like structure
The second criterion for helical hairpin formation stipulates a tertiary fold. If a tertiary structure forms in the vestibule, then this predicts a decrease in the accessibility of residues at the putative hairpin interface. To evaluate this hypothesis, we first used the pegylation accessibility assay. We cysteine scanned and pegylated S4 in the full-length T1− Kv1.34, which contains the complete voltage sensor domain, S1-S4 (Figure 3a). This construct is identical to the construct used in Figure 2 except that it contains S1-S3 at its N terminus. In both cases, the C terminus of S4 is positioned in the folding vestibule. Residue 298 (ΔPTC 68) is ~ 80% pegylated (Figure 3b, top gel), whereas 321(ΔPTC 44) is only ~30% pegylated. Residue 307 (ΔPTC 58) is intermediate between the two. Fpeg for all the scanned residues indicated in Figure 3a was calculated (red circles, Figure 3c). Compared to the accessibility curve of S4 lacking S1-S3 (Figure 2c), accessibility is markedly reduced in the construct containing the complete VSD, eliminating the possibility that S4 is independently responsible for the low Fpeg. Rather, the low Fpeg may be due to the presence of tertiary interactions between S4 and N-terminal regions of the peptide (see below) in the confinement of the folding vestibule, but not to non-specific block. Three lines of evidence support this conclusion. First, the presence of upstream N-terminal domains per se does not lead to a decrease in Fpeg2-4. Second, residue 298, in the exposed linker segment between S3 and S4, should be accessible, even in a hairpin fold, and is a good control for non-specific hindrance(top gel). . Fpeg for 298 is ~0.8 (Figure 3b). Third, release of peptides containing engineered cysteines at positions 316, 321, or 323 increased pegylation (70, 50, and 65%, respectively; data not shown), suggesting that the intrinsic reactivity of these cysteines is not responsible for the reduced pegylation in the vestibule, but rather the folded confinement of the peptide is. These results are consistent with a specific tertiary fold for the VSD, including a S3-S4 hairpin.
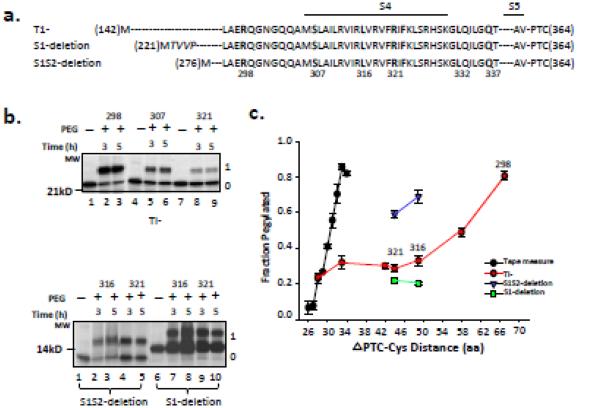
Figure 3. Accessibility of S4 residues in Kv1.3T1− and deletion constructs.
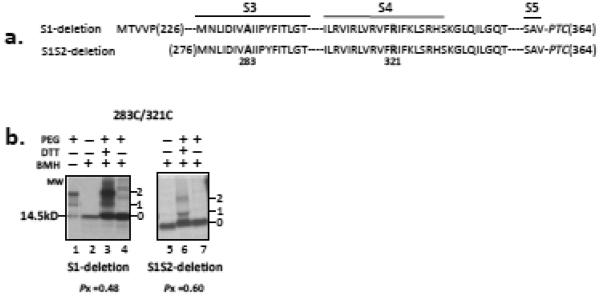
(a) Schematic of S4 in the T1- construct (N-terminal residue is M142; construct contains S1S2S3S4S5), S1-deletion construct (N-terminal residue is M221; italicized residues are non-native residues due to the engineered restriction site and do not affect measurements of crosslinking; construct contains S2S3S4S5), and S1S2-deletion construct (N-terminal residue is M276; construct contains S3S4S5). Bars represent the indicated transmembrane segments S1-S4. Dashes represent residues in the native sequence but not explicitly shown. All constructs terminate at V364 (derived from an Acc1-cut DNA template as in Figure 2) at the PTC. Engineered cysteines are bolded. (b) Pegylation of 298C, 307C, and 321C in the T1-construct, and 316C and 321C in S1-deletion and S1S2-deletion constructs. Methods and gels as described in Figure 2b. The first lane for each deletion construct is the unpegylated peptide. (c) Fraction pegylated, as described in Figure 2c, for the 7 engineered cysteines in the T1- construct (red circles) and for the 316C and 321C in S1-deletion (green squares) and in S1S2-deletion (blue triangles) constructs. Symbols are mean ± SEM (n= 3).
For Kv1.3, S2-S3-S4 forms a biogenic unit that acts cooperatively to ensure correct topological architecture of the voltage sensor in the membrane 38 suggesting a role (including facilitation of S3-S4 interactions) for upstream VSD segments in Kv biogenesis. If so, then we expect deletion of these segments to diminish the tertiary folding of the S3-S4 hairpin and increase Fpeg. To test this hypothesis, we made sequential deletions of S1 and S2 in the T1- construct and assayed for accessibility of 316C and 321C, residues located in the middle of S4. In the complete S1-S4 VSD, these residues give Fpeg of ~0.3. An S1-deleted construct (contains S2-S3-S4) shows a similarly low Fpeg whereas an S1S2-deleted construct (contains S3-S4) manifests a dramatically increased Fpeg (Figure 3b, bottom gel; Figure 3c, green squares and blue triangles, respectively). These results are consistent with retention of compact secondary structure for S4 in the S1S2-deleted construct and a tertiary conformation that differs from that of the full-length T1− Kv1.3. Only when S2 is re-instated (in the absence of S1 and the T1-S1 linker) is the tertiary conformation rescued (green squares). Moreover, these results suggest that S1 may not be involved in S3-S4 hairpin formation in the vestibule and the presence of S3 alone permits some tertiary folding (Fpeg for residue 321 in S1S2-deleted construct is 0.6 versus 0.7 for S4 alone (Figure 2c)). It is important to note that deletion of transmembrane segments might allosterically rearrange the nascent peptide in the tunnel and increase Fpeg.
To more directly test the hypothesis that S3 and S4 form a tertiary helical hairpin in the folding vestibule of the tunnel, we used an intramolecular crosslinking strategy (Figure 4a). We engineered selected cysteine pairs in the S3-S4 segment in the full-length Kv1.3 T1- construct in which S4 was situated in the vestibule. The nascent peptide-ribosome complex was isolated and treated with a bifunctional crosslinker, either phenyldimaleimide (PDM; intermaleimide distance of 6 Å) or bis-maleidyl-hexane (BMH; spacer arm of 13Å). BMH has a relatively flexible hydrocarbon chain. Any reaction of our peptide with either PDM or BMH will result in a mixture of five different species (Figure 4a), each of which would be indistinguishable on a protein gel and appear at the parent molecular weight. To distinguish the fraction of crosslinked peptide, we developed a modified strategy of the tertiary folding assay39, as depicted in Figure 4a, which increases the sensitivity and accuracy of our determination. The bifunctional reagent was quenched, the complex solubilized in lithium dodecyl sulfate (LDS), and then treated with dithiothreitol (DTT), which efficiently converts all non-crosslinked species to a dithiol peptide. This protocol reduces the reaction mixture from 5 peptide species to only two, an advantage over crosslinking strategies previously developed 39. These two species can easily be distinguished by treating the mixture of crosslinked peptide and dithiol peptide with PEG-MAL. Pegylation of the dithiol peptide produces gel-shifted products, leaving the crosslinked peptide at the original parent molecular weight. The predicted gel phenotypes for the extreme cases of 100% and 0% crosslinked peptide are depicted as schematic cartoons in Figure 4a (right). The bands labeled “0”, “1”, and “2” represent unpegylated, singly and doubly pegylated protein, respectively. A 1:1 mixture of crosslinked and uncrosslinked peptide will manifest a bimodal pattern. Note that a pegylation efficiency of <100% will result in a finite fraction of singly pegylated peptide. Intermediate mixtures of crosslinked/uncrosslinked will alter the intensity distribution of the three bands. This is quantified in our analysis (see Methods) and exemplified by the experiments shown in Figure 5. The pegylation efficiency (Ppeg) and crosslinking probability (Pxlink) were the two free parameters estimated simultaneously by maximum likelihood (see Methods, Analysis of Data). Under LDS conditions, Ppeg ranges from 0.8-0.9 (Table 1 In only 2 out of 15 cases, Ppeg is slightly lower than 0.8 (~ 0.7), likely due to incomplete denaturation in LDS. Lower Ppeg values will shift the apparent bimodality (see below, Figures 6 and 7). The efficiency of BMH and DDT labeling is >95% (Figure 4b), consistent with reported labeling efficiencies for other small bifunctional maleimides39.
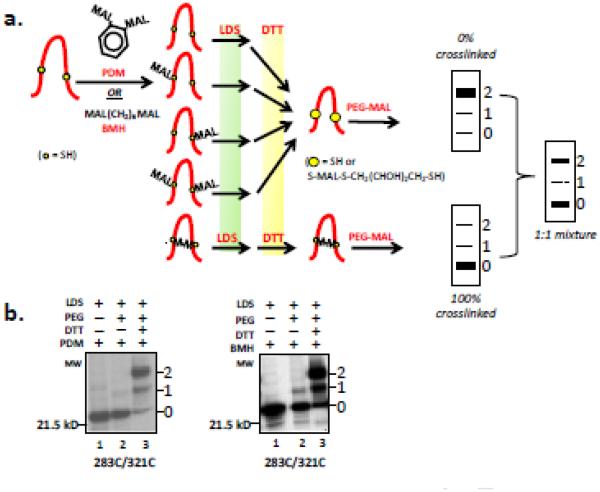
Figure 4. Crosslinking strategy.
(a) Crosslinking Protocol. The nascent peptide (derived from an Acc1-cut DNA template as in Figure 2), while attached to the ribosome PTC, was treated with bifunctional crosslinking reagent (PDM or BMH) to yield the indicated 5 species. Subsequent denaturation and solubilization with lithium dodecylsulfate (LDS) and treatment with dithiothreitol (DTT) converts peptidyl maleimides to a free thiol. Thus, all species except for the crosslinked species (bottom) can be efficiently pegylated upon addition of PEG-MAL, the final step in the protocol. To the right of the protocol are cartoons of protein gels indicating the predicted pattern for the 0%, (top), 100% crosslinking (bottom), or a mixture of the two possibilities (middle). The numbers to the right of each cartoon gel indicate unpegylated (0) and singly pegylated (1) and doubly pegylated (2) protein. (b) Efficiency for overall PDM- (left) and BMH-pathway (right) shown in (a), for the cysteine pair 283C/321C. Lane 1 was derived from a translation product that was denatured in LDS and then treated with bifunctional reagent. Lane 2 was derived from an LDS-denatured translation product, subsequently treated with bifunctional reagent, then with β-mercaptoethanol (βME), and pegylated. Lane 3 shows the same peptide released with LDS but treated with DTT prior to pegylation. The number to the left of the gel is a molecular weight standard; numbers to the right of the gel indicate unpegylated (0) and singly pegylated (1) and doubly pegylated (2) protein.
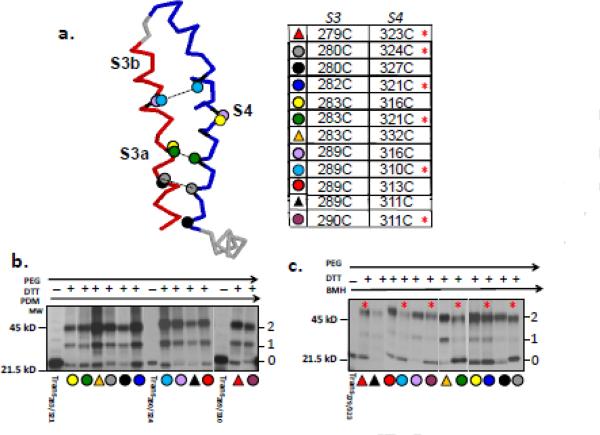
Figure 5. Crosslinking of selected pairs in the S3-S4 hairpin of Kv1.3T1−.
(a) Cartoon of the S3-S4 segment (left) indicating selected cysteine pairs in S3-S4 hairpin for crosslinking experiments. Dashed lines indicate selected crosslinked pairs. The table (right) is more extensive and indicates all cysteine pairs tested as the same color and symbol. (b) PDM-treatment of pairs indicated in (a). according to the protocol shown in Figure 4a. Lanes marked “trans” are translation products for the construct with the indicated cysteine pair, treated with PDM, then released and treated with β-mercaptoethanol, and then with PEG-MAL. These lanes confirm complete labeling by bifunctional reagent and no crosslinking in the absence of DTT. All other lanes represent products from crosslinking experiments for the pairs indicated by the colored symbols, which are keyed to the table shown in (a). (c) BMH-treatment of pairs indicated in A. according to the protocol shown in Figure 4a. Lanes as described in (b). Numbers as described in Figure 4. Quantification of these gels gives the probability of crosslinking (Pxlink), which is given in Table 1. Red asterisks in (a) and (c) indicate crosslinked pairs, i.e., Pxlink >0.4.
Table 1. Efficiency of crosslinking.
Engineered cysteine pairs are listed for the Kv1.3T1-constructs studied as nascent peptides attached to the ribosome tunnel as described in Figure 5c. As described in the Methods, Pxlink is the probability that the cysteines are crosslinked and PPeg is the efficiency of pegylation for the indicated cysteine pair. Bolded data indicate crosslinked pairs. Values for the last pair in the table, A283C/R321C, were determined subsequent to integration of the peptide into microsomal membranes. Data represent means ± SD for triplicate experiments.
| Cysteine pairs | Pxlink | P peg |
|---|---|---|
| I279C/F323C | 0.41+0.02 | 0.8910.01 |
| D280C/K324C | 0.43±0.02 | 0.83+0.05 |
| D280C/R327C | 0.1910.03 | 0.8810.03 |
| V282C/R321C | 0.41+0.02 | 0.88+0.03 |
| A283C/L316C | 0.1910.05 | 0.8310.04 |
| A283C/R321C | 0.52+0.03 | 0.83+0.05 |
| A283C/S332C | 0.11±0.02 | 0.8610.01 |
| I289C/I310C | 0.40+0.05 | 0.9010.01 |
| I289C/L311C | 0.18±0.01 | 0.86+0.03 |
| I289C/V313C | 0.24±0.06 | 0.9210.02 |
| I289C/L316C | 0.14±0.06 | 0.8510.02 |
| T290C/L311C | 0.45±0.03 | 0.8010.05 |
| A283C/R321C | 0.6110.01 | 0.71+0.01 |
Figure 6. Crosslinking of the S3-S4 hairpin in deletion constructs.
(a) Schematic of S1-deletion (contains S2-S3-S4-S5, lanes 1-4) and S1S2-deletion (contains S3-S4-S5, lanes 5-7) constructs starting at native residue 226 and 276, respectively. Each deletion peptide terminates at residue 364. Dashes represent residues in the native sequence but not explicitly shown. (b) Crosslinking of pair 283C/321C. Methods as described in Figure 5b. Lane 1 is a pegylation control lane showing nascent peptide pegylation subsequent to denaturation to verify quality of PEG-MAL. Lanes 2 and 5 are derived from a translation product that was treated with BMH, indicating the unpegylated (0 band) protein. Lanes 3 and 6 exhibit a band pattern indicative of crosslinking (see Figure 4a). Lanes 4 and 7 represent reactions in which β-mercaptoethanol, but not DTT, was present, i.e., no conversion of peptidyl maleimide to a free modifiable peptide (see Figure 4a). The number under the gels indicates the Pxlink for S1-deletion and S1S2-deletion (0.48±0.02 and 0.60±0.01, respectively), which have Ppeg of 0.85± 0.01 and 0.72±0.02 (n=3), respectively.
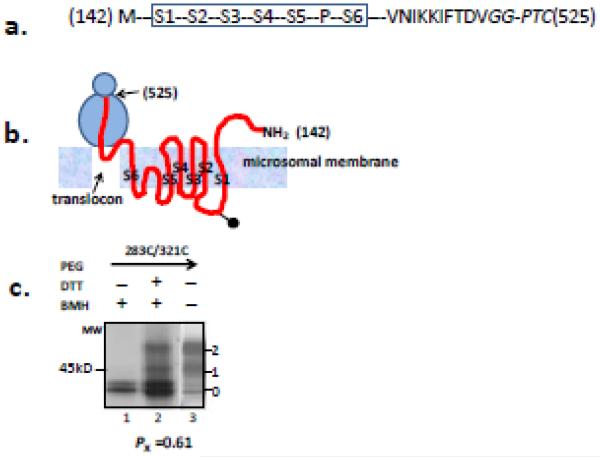
Figure 7. Crosslinking of Kv1.3 T1- integrated into microsomal membranes.
(a) Schematic of Kv1.3 T1- (S1-S2-S3-S4-S5-P-S6-C-terminus) with a C-terminal residue at 525 (NotI-cut in the DNA template, see Methods), which locates S1-S6 in the membrane38 and the cytosolic C terminus in the ribosomal tunnel. The N-terminal residue is 142. The engineered cysteine pair in this construct is 283C/321C. The GG residues (italicized) adjacent to the PTC are non-native due to the engineered restriction site and do not affect measurements of crosslinking. Dashes represent residues in the native sequence but not explicitly shown. (b) Cartoon of the putative topology and locations of Kv1.3 segments (red line) in the tunnel, translocon, and membrane. This represents an approximation according to transmembrane orientation and insertion of Kv1.3 described in Tu et al., 200038. Transmembrane segments are labeled as S1-S6. The ball-and-stick in the linker between S1 and S2 represents a glycosylation moiety. (c) Crosslinking of pair 283C/321. Lanes and bands are as described in Figure 6b. Lane 1 represents a reaction in which β-mercaptoethanol, but not DTT, was present, i.e., no conversion of peptidyl maleimide to a free modifiable peptide (see Figure 4a). Lane 2 shows a band pattern indicative of crosslinking (see Figure 4a). Lane 3 represents a control experiment showing nascent peptide pegylation subsequent to denaturation that verifies the efficacy of PEG-MAL. The number under the gels indicates the Pxlink, which has a Ppeg of 0.71±0.01 (n=4). The doublet at band 0 is unpegylated protein that is unglycosylated (lower band) or ER-glycosylated (upper band).
Based on a survey of homologous x-ray crystal structures19, 20 (open state) and available functional data25, 40, we engineered a series of constructs with paired cysteine mutations (Figure 5a). Although residue proximities implicated by functional and crystallographic studies of a mature Kv channel in the plasma membrane may not be identically manifest at early stages of Kv biogenesis, they nonetheless served to guide our choice of putative neighborhoods for pairwise interactions. We recognize that pairs that are far apart in the crystal structure of the channel’s open state might be re-located in a different state of the channel (e.g., a closed state). The engineered constructs were translated (no membranes) peptides crosslinked according to our crosslinking strategy (Figure 4a) using either PDM (Figure 5b) or BMH (Figure 5c). PDM-crosslinking was not detected for any of the chosen pairs, however, BMH crosslinked (red asterisk) several, but not all, pairs. Constructs containing paired cysteines at residues 279/323, 280/324, 282/321, 283/321, 289/310, and 290/311 gave a distinct bimodal pattern (as predicted in Figure 4a) of unpegylated (band 0), singly (band 2) and doubly (band 3) pegylated peptide.
Using maximum likelihood estimation (see Methods, Analysis of Data), we fit the gel intensities of all three bands to simultaneously solve for Pxlink and Ppeg for each experiment (Table 1). Constructs containing paired cysteines at positions 279/323, 280/324, 282/321, 283/321, 289/310, and 290/311 averaged a Pxlink close to 0.4-0.5. In contrast, a control pair engineered with distant cysteines at positions 283C/332C, gave a Pxlink of only ~0.11. Of 12 pairs investigated, only 6 exhibited a Pxlink of 0.4-0.5 . Not all of the crosslinked pairs reside within S3b. A few are present in the C-terminal part of S3a. For three different locations along the hairpin, crosslinked pairs (280C/324C, 283C/321C, 289C/310C) are shown connected by a dashed line in Figure 5a. Although the nascent peptide need not resemble any state of maturity or gating of the tetrameric protein, we note that a cysteine pair in Shaker, corresponding to 289C/310C in Kv1.3, is crosslinkable in the closed state of the mature functional Shaker channel in the plasma membrane 25. Moreover, Shaker residues 316 and 374, corresponding to 280/324 in Kv1.3, have been implicated to form a salt bridge in the closed state 40. Cysteine pairs 280C/324C and 289C/310C crosslink, whereas 280C/327C, 289C/313C, and 289C/316C, whose analogs in Shaker are in proximity only in the open state25, 40, do not. These crosslinking results are thus consistent with monitoring a closed-state facsimile in the folding vestibule (see Discussion).
These first attempts to track hairpin formation may only capture certain features of the incipient structure. It is significant that PDM, a bifunctional crosslinker with an intermaleimide distance of only 6Å labeled individual cysteines completely (>95%), but did not crosslink any of the more than 12 pairs tested. These results demonstrate that impaired cysteine reactivity is not the cause for lack of crosslinking, but rather the orientation/distance/accessibility of the cysteines within the pair is the primary determinant.
Finally, we asked whether S3 and S4 form a hairpin fold under two circumstances: first, in the absence of the N-terminal region, including S1and S2, and second, in the ER membrane following integration of the voltage sensor into the bilayer. In the first case, we translated and crosslinked 283C/321C, a strongly crosslinkable pair (Pxlink = 0.52), engineered into a S1-deleted construct or into a construct lacking both S1 and S2 (Figure 6a). Comparable Pxlink values of 0.48±0.02 and 0.60±0.01, respectively, were obtained (Figure 6b). Therefore, both constructs permit crosslinking of 283C/321C. We interpret the crosslinking and accessibility results for deletion constructs to suggest that S2 hinders accessibility of S4 residues either by decreasing PEG-MAL access or by altering S4’s surroundings, but does not interfere with S3-S4 hairpin interactions.
To test whether full-length monomeric T1− nascent chain manifests an S3-S4 hairpin conformation in the ER membrane, we introduced a Not1- restriction site at the end of the C terminus of the construct (Figure 7a). This places the voltage sensor on a long tether with the Kv1.3 cytosolic C-terminus in the tunnel and the last residue of S6 > 90 amino acids from the PTC (Figure 7b). Transmembrane segments S1-S6 are thus integrated into the bilayer38. Crosslinking of 283C/321C in this construct (Figure 7c) gives a Pxlink of 0.61 ± 0.01 (Ppeg =0.71 ± 0.01, n = 3), consistent with a tertiary S3-S4 hairpin in a monomer embedded in the ER membrane.
DISCUSSION
Hairpin interactions may be characterized as an equilibrium between folded and unfolded species. The equilibrium constant (affinity) will be different for different peptide sequences at different locations within and without the ribosomal tunnel. If the affinity is high, then the dwell time of the peptide in the hairpin configuration will be longer, leading to a higher intra-hairpin Pxlink. Additionally, conformational flexibility and length of the intervening hairpin loop may be factors in hairpin formation in the vestibule. An entropic window, in which a nascent peptide explores conformational space and samples potential tertiary partners, has been described for some cytosolic hairpin interactions in the ribosomal folding vestibule13. We now show that transmembrane helical hairpins can also form in this vestibule. The hairpin investigated herein is part of the voltage sensor (S1-S4), a modular and functionally critical unit of voltage-sensing proteins21, 22, 41.
Secondary structure of VSD segments
The C-terminal region of S4, in the folding vestibule, is compact and likely helical, even in the absence of S3 and all other upstream elements of Kv1.3. Both accessibility and crosslinking results support this conclusion. Transmembrane segments S1, S2, S3, and S4 each acquire their individual secondary structures before completely emerging from the ribosomal exit and likely retain this structure in the translocon3, 4(this paper). These preformed secondary structures may facilitate tertiary folding of the VSD, whether inside the vestibule or in a more distal compartment such as the translocon or lipid bilayer, assisting membrane insertion. Moreover, such folding capacity might underlie coupled secondary-tertiary folding for helical hairpins. Vectorial (N- to C-terminus) secondary folding of an entire transmembrane segment occurs in the absence of microsomal membranes when C-terminal residues reside in the folding vestibule4(this paper). Upon emergence from the vestibule, a transmembrane helix has been suggested to be stabilized by the SRP, even in the absence of microsomal membranes 11.
Tertiary Interactions of VSD segments
S3-S4 hairpin interactions in the Kv monomer are detected in the folding vestibule, i.e., at an early stage in Kv biogenesis. Although precedents exist, both experimental and theoretical, for tertiary interactions within soluble domains in the vestibule13-16, our results are the first demonstrating that transmembrane tertiary interactions can occur in the vestibule. To do this, we used both accessibility and crosslinking assays. In the latter case, three important considerations should be noted: 1) apparent side-chain distances between cysteines on opposite hairpin arms are distinct from backbone distances between the hairpin arms, 2) a hairpin may be either parallel, or skewed in a crossed configuration, and 3) a lack of crosslinking with PDM (6Å) may be due to the rigidity of the probe, as well as its short linker length. Crosslinking will depend on the position of the hairpin backbones (arms), the orientations of the cysteine side chains, and consequently on the linker length and flexibility of the bifunctional maleimide. Whereas PDM does not crosslink residues within S3-S4, BMH, a longer (13 Å) and more flexible molecule, can crosslink specific residues within S3-S4. We therefore suggest the helix-turn-helix has distance constraints for the paired cysteine side chains that limit the use of PDM. Four cysteine pairs, 279C/323C, 280C/324C, 289C/310C, and 290C/311C crosslink, which is not consistent with predictions derived from a parallel orientation of the S3-S4 hairpin and may implicate a skewed hairpin arrangement. Bimodal crosslinking implies two populations, one that is crosslinked and one that is not. Each could contain several conformational states. Based on the high efficiency of maleimide labeling (Ppeg, Table 1), 0.4-0.5 crosslinking from a bimodal distribution indicates that 40-50% of the peptide is in a conformation(s) characterized by near proximity of the specified cysteine pair, as in a hairpin conformation. This hairpin in the vestibule resembles the conformation in the ER membrane (Figure 7), suggesting the vestibule hairpin is a bonafide conformation along the biogenic pathway, however, the precise orientation of such a hairpin may eventually be fine-tuned in the context of the fully assembled tetramer. The cysteine pairs I289C/I310C and D280C/K324C in Kv1.3 (≥ 0.4 Pxlink) correspond to Shaker residues that are crosslinkable in the closed state of the mature channel in the plasma membrane25 and produce functional consequences when mutated 40. Thus, the nascent Kv paddle structure in the vestibule may represent a physiologically relevant functional state of the mature channel.
Although S3-S4 have an intrinsic affinity and crosslink in the absence of S2 (Figure 6), S2 may nonetheless serve a role in biogenesis of the VSD. At least one important role is targeting the Kv nascent peptide to the ER membrane38. But S2 may serve additional roles. First, S2 may interact with chaperones in the vicinity of the tunnel’s exit port and/or translocon to align the nascent VSD for efficient integration. Facilitation of VSD insertion/integration into the bilayer could also be mediated by S2’s sequence and consequent integration propensity. S2 integrates well into the ER membrane38. In contrast, the isolated S3 segment is a secretory peptide and does not integrate into the membrane 38. S4 manifests only 50% integration efficiency38, 44 and S3-S4 does not integrate efficiently38. Thus, a strong integrator such as S2 would confer the required driving force for insertion of the VSD.
Yet another possible role for S2 is as a translocon retention signal 26-28. An increased dwell time of S2 in the translocon would allow the C-terminal segments of the VSD (S3-S4) to partner correctly (prefold) and form a cooperative biogenic unit (S2-S3-S4) for efficient membrane integration of Kv VSD transmembrane helices38, 42, 43. It also participates in electrostatic interactions between charged side chains within S2, S3, and S4 that drive the gating of mature Kv channels at the plasma membrane in response to membrane potential4020, 45, 46. While S2 plays a role in both these functions, the roles need not be the same. These salt bridges may not be critical for promotion of initial folding of the hairpin in the ribosome-nascent chain complex, as charge-neutralizing (e.g., D280S, R321C, R260A) and charge-reversing (e.g., D280R) mutations failed to alter Fpeg (Tu and Deutsch, data not shown). Nonetheless, salt bridges within the VSD may exist later in the translocon and, if so, their formation could be facilitated by prior hairpin formation in the adjacent folding vestibule of the ribosome.
Why does early hairpin biogenesis matter?
Hairpin formation in the tunnel raises an interesting conundrum, namely, how will such a bulky unit proceed into the translocon and thereafter into the lipid bilayer of the ER? Moreover, we may ask if there is an advantage for the nascent peptide to acquire a helix-turn-helix motif at this stage of biogenesis. The cytosolic entryway to the translocon (20-25Å; 47) appears to be dynamic and large enough to accommodate a hairpin. It widens upon engagement of the translocon with a ribosome-nascent chain complex48, 49. This expansion also includes the narrowest passage (5-8Å), a ring of bulky hydrophobic residues. Secondary folds are maintained as a nascent Kv chain transits from folding vestibule to translocon4, and specific crosslinking of S3-S4 is similar in the folding vestibule and ER membrane. Thus, we suggest that for a nascent chain entering the upper cavity of the translocon, between the putative canaliculus and the ribosomal exit port, the prefolded hairpin signals expansion of the canaliculus. This could accommodate efficient passage of the nascent chain through the translocon. As such, the folding vestibule would constitute an operational antechamber where reorientation and refolding can be optimized for efficient integration. Moreover, successive transmembrane segments, e.g., S2, might serve to keep the translocon canaliculus patent throughout the peptide’s journey from translocon to bilayer. These provocative hypotheses require further experimental investigation.
Efficient protein folding and packing is key to understanding not only biogenesis but also trafficking mechanisms, because ER export and degradation signals are modulated by how the protein is folded and packed and will alter levels of expression of Kv channels at the cell surface, which leads to pathology. In addition to defining fundamental steps in cotranslational folding, these studies begin to address folding events in Kv formation of a critical region of the voltage sensor.
MATERIALS AND METHODS
Constructs and In Vitro Translation
Standard methods of bacterial transformation, plasmid DNA preparation, and restriction enzyme analysis were used. The nucleotide sequences of all mutants were confirmed by automated cycle sequencing performed by the DNA Sequencing Facility at the School of Medicine on an ABI 377 Sequencer using Big dye terminator chemistry (A0BI). pSP/ΔT1)Kv1.3 was constructed by religation of the HindIII/NcoI-digested, blunt-ended pSP/Kv1.3/cysteine-free 2. Engineered cysteines and restriction enzyme sites were introduced into pSP/ΔT1Kv1.3/cysteine-free using QuikChange Site-Directed Mutagenesis Kit. All mutant DNAs were sequenced throughout the entire coding region. Capped cRNA was synthesized in vitro from linearized templates using Sp6 RNA polymerase (Promega, Madison, WI). Linearized templates for Kv1.3 translocation intermediates were generated using several restriction enzymes to produce different length DNA constructs lacking a stop codon and to position the nascent peptide segments at the specified locations. N-terminal deletion constructs were made as follows (see4). pSp/S2-S3-S4-S5-COOH was constructed by relegation of SmaI/PpumI-digested pSp/Kv1.3 cysteine-free DNA. pSp/S3-S4-S5-COOH constructs were made by relegation of HindIII/NruI-digested pSp/Kv1.3 cysteine-free DNA, and pSp/S4-S5-COOH constructs were made by PCR-amplification as described previously38. We mutated native cysteine to serine and introduced cysteines according to the needs of the experiment. Proteins were translated in vitro with [35S]Methionine (4 μl/50 μl translation mixture; ~10 μCi/μl Express, Dupont/NEN Research Products, Boston, MA) for 60-90 min at 22-25°C in a rabbit reticulocyte lysate (2 mM final (DTT), typically in the absence of microsomal membranes, according to the Promega Protocol and Application Guide. When microsomal membranes were included in the translation reaction, they were added at a final concentration of 1.0 μl/25 μl translation reaction according to the Promega Protocol and Application Guide.
Pegylation Measurements
As described previously 2, translation reaction (40 μl) was added to 500 μl phosphate-buffered saline (PBS, Ca-free, containing 4 mM MgCl2 (pH 7.3)) with 2 mM DTT. The suspension was centrifuged (Beckman Optima TLX Ultracentrifuge, Beckman TLA 100.3 rotor) through a sucrose cushion (100 μl; 0.5 M sucrose, 100 mM KCl, 5 mM MgCl2, 50 mM Hepes, and 1 mM DTT (pH 7.5)) for 20 min at 70,000rpm (~245,000xg) at 4° C to isolate ribosome-bound peptide. The pellet was resuspended on ice in 90 μl of PBS Ca-free buffer containing 4 mM Mg2+ and 100 μM DTT (pH 7.3). 10μl buffer containing 10 mM PEG-MAL (5 kDa, SunBio Inc.) was added (final PEG-MAL concentration of 1 mM) and incubated at 0°C for two consecutive time points, 3h and 5 h. Reactions were terminated by addition of 100 mM DTT, followed by incubation of the mixture at room temperature for 10-15 min. Samples were collected either by centrifugation at 70,000 rpm at 4° C for 20 min and/or by precipitation with a 90% final volume of cold acid-acetone (900 μl; a stock acid-acetone solution was made by adding 10 μl of HCl to 120 ml acetone). The final sample was mixed with NuPAGE sample buffer and subject to SDS-NuPAGE analysis. The fraction of protein pegylated for each residue was calculated as the ratio of counts per minute in the pegylated band (band 1) to the sum of the counts per minute in the pegylated and unpegylated (band 0) bands. If incompletely translated products were present, then the ratio of the full-length to incompletely translated products from the control lane (first lane) was used to correct the amount of pegylation of the full-length nascent peptide.
Crosslinking Assay
Translation reaction (50μl) in the absence of microsomal membranes was added to 500μl of PBS buffer with 2mM DTT. The suspension was centrifuged through a sucrose cushion (120μl containing 0.5 M sucrose, 100 mM KCl, 5 mM MgCl2, 50 mM HEPES (pH 7.3) and 1mM DTT) at 4°C for 20 min at 70,000 rpm using a TLA 100.3 Beckman ultra-centrifuge rotor to isolate ribosome-bound peptide. The pellet was re-suspended at 4°C in 500μl of PBS for 30-60mins in the presence of either 0.5 mM phenyldimaleimide (PDM, SigmaAldrich) or 1-2mM bismaleimide hexane (BMH, ThermoScientific). The reaction was terminated by adding DTT (a final DTT concentration that is 50x the concentration of the bifunctional crosslinker ) and incubated first at room temperature for 5 mins, then at 4°C 10 mins. Samples were centrifuged again through a sucrose cushion (120μl, but minus DTT) for 20 min at 70,000 rpm at 4°C for 20 min at 70,000 rpm using a TLA 100.3 Beckman ultra-centrifuge rotor to isolate ribosome-bound peptide. Pellets were re-suspended at room temperature for 30 mins in 80 μl PBS buffer containing 2% LDS (to denature the peptide) and 2mM DTT. PEG-MAL (10 mM; 150μl) was added to give a final PEG-MAL concentration of 6.5 mM and the sample was incubated at room temperature for 2 hr.
Translation reaction (50μl) in the presence of microsomal membranes (2 μl) was added to 500μl of PBS buffer with 2mM DTT. The suspension was centrifuged through a sucrose cushion (120μl; see above) at 4°C for 7 min at 50,000 rpm using a TLA 100.3 Beckman ultra-centrifuge rotor to isolate ribosome-bound peptide. The pellet was re-suspended on ice in 500 μl of PBS containing 0.5% dodecyl maltoside (C12M), incubated for 30mins, followed by incubation with 1-2mM BMH for 60 mins at 4°C and worked up as described above.
Gel Electrophoresis and Fluorography
All final samples were treated with ~2μl of 500 μg/ml RNase for 20 min at room temperature to digest tRNA and remove contaminating peptidyl-tRNA bands. Samples were mixed with NuPAGE sample buffer (1 M glycerol, 0.5 mM EDTA, 73 mM LDS, 141 mM Tris base, and 106 mM Tris-HCl) and heated at 70° C for 10 min before being loaded onto the gel. Electrophoresis was performed using the NuPAGE system and precast Bis-Tris gels and MES (50 mM) or Mops (50 mM) running buffer (50 mM Tris base, 3.5 mM SDS, and 1 mM EDTA). Gels were soaked in Amplify (Amersham Corp., Arlington Heights, IL) to enhance 35S fluorography, dried, and exposed to Kodak X-AR film at -70° C. Typical exposure times were 16-30 h. Quantitation of gels was carried out directly using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager.
Analysis of Data
Monomeric constructs having engineered pairs of cysteines were subjected to a cross-linking strategy using bifunctional maleimide reagents and PEG-MAL (Figure 4a). We employed a maximum likelihood algorithm50 to simultaneously estimate the probability (Pxlink) that the cysteine pairs were cross-linked, and the probability (Ppeg) that available cysteine residues were labeled by PEG-MAL after denaturation in LDS. The protein gels had 3 bands. The fractional weight of these bands (Fx), represents the distribution of the number of PEG-MAL labels (x=0,1,2) on the nascent peptides. If the two cysteine residues on each peptide are identical and independent with respect to labeling, we can estimate Pxlink and Ppeg using the binomial distribution, as follows. Let
The log-likelihood for the data, LL(Fx | yx), given the parameters Pxlink and Ppeg, can be expressed as
We used a variable metric algorithm51to minimize the negative of LL(Fx | yx) as a function of the two free parameters. Simulations showed these estimates to be highly accurate as well as robust. In this formulation the average number of pegylated cysteines per molecule is F1 +2F2.
Highlights.
Transmembrane segments undergo tertiary prefolding in the folding vestibule
S3-S4 hairpin in the Kv monomer is detected in the folding vestibule
Entire S4 segment forms a compact structure before exiting the ribosomal tunnel
S3-S4 hairpin in the vestibule resembles a closed-state conformation of Kv channels
Improved intramolecular crosslinking assay is introduced
ACKNOWLEDGMENTS
We thank Dr. Richard Horn, for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM 52302 to CD.
Abbreviations
- PTC
peptidyl transferase center
- Kv
voltage-gated K+
- PEG-MAL
polyethylene glycol maleimide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005; 44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 3.Tu L, Wang J, Deutsch C. Biogenesis of the T1-S1 linker of voltage-gated K+ channels. Biochemistry. 2007; 46:8075–8084. doi: 10.1021/bi700319f. [DOI] [PubMed] [Google Scholar]
- 4.Tu LW, Deutsch C. A folding zone in the ribosomal exit tunnel for Kv1.3 helix formation. J. Mol. Biol. 2010; 396:1346–1360. doi: 10.1016/j.jmb.2009.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004; 116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 6.Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010; 17:313–317. doi: 10.1038/nsmb.1756. [DOI] [PubMed] [Google Scholar]
- 7.Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Progress in Nucleic Acid Research & Molecular Biology. 2001; 66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 8.Mingarro I, Nilsson I, Whitley P, von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biology [Computer File] 2000; 1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier T, Ferbitz L, Deuerling E, Ban N. A cradle for new proteins: trigger factor at the ribosome. Curr. Opin. Struct. Biol. 2005; 15:204–212. doi: 10.1016/j.sbi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 10.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for cotranslational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009; 16:589–597. doi: 10.1038/nsmb.1614. [DOI] [PubMed] [Google Scholar]
- 11.Robinson PJ, Findlay JE, Woolhead CA. Compaction of a prokaryotic signal-anchor transmembrane domain begins within the ribosome tunnel and is stabilized by SRP during targeting. J. Mol. Biol. 2012; 423:600–612. doi: 10.1016/j.jmb.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Pechmann S, Willmund F, Frydman J. The ribosome as a hub for protein quality control. Mol. Cell. 2013; 49:411–421. doi: 10.1016/j.molcel.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kosolapov A, Deutsch C. Tertiary interactions within the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2009; 16:405–411. doi: 10.1038/nsmb.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elcock AH. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLoS. Comput. Biol. 2006; 2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Brien EP, Hsu ST, Christodoulou J, Vendruscolo M, Dobson CM. Transient tertiary structure formation within the ribosome exit port. J. Am. Chem. Soc. 2010; 132:16928–16937. doi: 10.1021/ja106530y. [DOI] [PubMed] [Google Scholar]
- 16.Kowarik M, Kung S, Martoglio B, Helenius A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Molecular Cell. 2002; 10:769–778. doi: 10.1016/s1097-2765(02)00685-8. [DOI] [PubMed] [Google Scholar]
- 17.Kelkar DA, Khushoo A, Yang Z, Skach WR. Kinetic analysis of ribosome-bound fluorescent proteins reveals an early, stable, cotranslational folding intermediate. J. Biol. Chem. 2012; 287:2568–2578. doi: 10.1074/jbc.M111.318766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Lee A, Chen J, Ruta V, Cadene M, Chait BT, MacKinnon R. X-ray structure of a voltage-dependent K+ channel. Nature. 2003; 423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 19.Long SB, Campbell EB, MacKinnon R. Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science. 2005; 309:903–908. doi: 10.1126/science.1116270. [DOI] [PubMed] [Google Scholar]
- 20.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007; 450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 21.Murata Y, Iwasaki H, Sasaki M, Inaba K, Okamura Y. Phosphoinositide phosphatase activity coupled to an intrinsic voltage sensor. Nature. 2005; 435:1239–1243. doi: 10.1038/nature03650. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki M, Takagi M, Okamura Y. A voltage sensor-domain protein is a voltage-gated proton channel. Science. 2006; 312:589–592. doi: 10.1126/science.1122352. [DOI] [PubMed] [Google Scholar]
- 23.Alabi AA, Bahamonde MI, Jung HJ, Kim JI, Swartz KJ. Portability of paddle motif function and pharmacology in voltage sensors. Nature. 2007; 450:370–375. doi: 10.1038/nature06266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Ramu Y, Lu Z. A shaker K+ channel with a miniature engineered voltage sensor. Cell. 2010; 142:580–589. doi: 10.1016/j.cell.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broomand A, Elinder F. Large-scale movement within the voltage-sensor paddle of a potassium channel-support for a helical-screw motion. Neuron. 2008; 59:770–777. doi: 10.1016/j.neuron.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Hou B, Lin PJ, Johnson AE. Membrane protein TM segments are retained at the translocon during integration until the nascent chain cues FRET-detected release into bulk lipid. Mol. Cell. 2012; 48:398–408. doi: 10.1016/j.molcel.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cross BC, High S. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J. Cell Sci. 2009; 122:1768–1777. doi: 10.1242/jcs.046094. [DOI] [PubMed] [Google Scholar]
- 28.Pitonzo D, Yang Z, Matsumura Y, Johnson AE, Skach WR. Sequence-specific retention and regulated integration of a nascent membrane protein by the endoplasmic reticulum Sec61 translocon. Mol. Biol. Cell. 2009; 20:685–698. doi: 10.1091/mbc.E08-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skach W, Lingappa VR. Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actrions of two internal sequences. Journal of Biological Chemistry. 1993; 268:23552–23561. [PubMed] [Google Scholar]
- 30.Borel AC, Simon SM. Biogenesis of polytopic membrane proteins: membrane segments of P-glycoprotein sequentially translocate to span the ER membrane. Biochemistry. 1996; 35:10587–10594. doi: 10.1021/bi960950q. [DOI] [PubMed] [Google Scholar]
- 31.Sadlish H, Pitonzo D, Johnson AE, Skach WR. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005; 12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- 32.Kida Y, Morimoto F, Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 2007; 179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang FD, Chen J, Lin M, Keating MT, Sanguinetti MC. Long-QT syndrome-associated missense mutations in the pore helix of the HERG. 2001 doi: 10.1161/hc3501.093815. [DOI] [PubMed] [Google Scholar]
- 34.Anderson CL, Delisle BP, Anson BD, Kilby JA, Will ML, Tester DJ, Gong Q, Zhou Z, Ackerman MJ, January CT. Most LQT2 mutations reduce Kv11.1 (hERG) current by a class 2 (trafficking-deficient) mechanism. Circulation. 2006; 113:365–373. doi: 10.1161/CIRCULATIONAHA.105.570200. [DOI] [PubMed] [Google Scholar]
- 35.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution.[comment] Science. 2000; 289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 36.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. [see comments.] Science. 2000; 289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 37.Creighton TE. Proteins. W.H. Freeman and Company; New York: 1993. Conformational Properties of Polypeptide Chains; pp. 171–199. [Google Scholar]
- 38.Tu L, Wang J, Helm A, Skach WR, Deutsch C. Transmembrane biogenesis of Kv1.3. Biochemistry. 2000; 39:824–836. doi: 10.1021/bi991740r. [DOI] [PubMed] [Google Scholar]
- 39.Kosolapov A, Tu L, Wang J, Deutsch C. Structure Acquisition of the T1 Domain of Kv1.3 During Biogenesis. Neuron. 2004; 44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Papazian DM, Shao XM, Seoh S-A, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 voltage sensor in Shaker K+ channel. Neuron. 1995; 14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 41.Bezanilla F. The voltage sensor in voltage-dependent ion channels. Physiol Rev. 2000; 80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 42.Sato Y, Sakaguchi M, Goshima S, Nakamura T, Uozumi N. Integration of Shaker-type K+ channel, KAT1, into the endoplasmic reticulum membrane: synergistic insertion of voltage-sensing segments, S3-S4, and independent insertion of pore-forming segments, S5-P-S6. Proceedings of the National Academy of Sciences of the United States of America. 2002; 99:60–65. doi: 10.1073/pnas.012399799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang L, Sato Y, Hessa T, von HG, Lee JK, Kodama I, Sakaguchi M, Uozumi N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. U. S. A. 2007; 104:8263–8268. doi: 10.1073/pnas.0611007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005; 307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 45.Tiwari-Woodruff SK, Schulteis CT, Mock AF, Papazian DM. Electrostatic interactions between transmembrane segments mediate folding of Shaker K+ channel subunits. 1997:1489–1500. doi: 10.1016/S0006-3495(97)78797-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nat. Chem. Biol. 2011; 7 doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van den Berg B, Clemons WM, Jr., Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004; 427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 48.Hamman BD, Chen JC, Johnson EE, Johnson AE. The aqueous pore through the translocon has a diameter of 40-60 A during cotranslational protein translocation at the ER membrane. Cell. 1997; 89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 49.Egea PF, Stroud RM. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. U. S. A. 2010; 107:17182–17187. doi: 10.1073/pnas.1012556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehmann EL, Casella G. Theory of Point Estimation. Springer-Verlag; 1998. New. [Google Scholar]
- 51.Powell MJD. A fast algorithm for nonlinearity constrained optimization calculations. In: Watson GA, editor. Numerical Analysis. Berlin: 1978. [Google Scholar]
- 52.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 2011; 21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 53.Lu J, Hua Z, Kobertz WR, Deutsch C. Nascent peptide side chains induce rearrangements in distinct locations of the ribosomal tunnel. J. Mol. Biol. 2011; 411:499–510. doi: 10.1016/j.jmb.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]