Abstract
The nuclear pore complex proteins SonA and SonB, the orthologs of mammalian RAE1 and NUP98, respectively, were identified in Aspergillus nidulans as cold-sensitive suppressors of a temperature-sensitive allele of the essential mitotic NIMA kinase (nimA1). Subsequent analyses found that sonB1 mutants exhibit temperature-dependent DNA damage sensitivity. To understand this pathway further, we performed a genetic screen to isolate additional conditional DNA damage-sensitive suppressors of nimA1. We identified two new alleles of SonA and four intragenic nimA mutations that suppress the temperature sensitivity of the nimA1 mutant. In addition, we identified SonC, a previously unstudied binuclear zinc cluster protein involved with NIMA and the DNA damage response. Like sonA and sonB, sonC is an essential gene. SonC localizes to nuclei and partially disperses during mitosis. When the nucleolar organizer region (NOR) undergoes mitotic condensation and removal from the nucleolus, nuclear SonC and histone H1 localize in a mutually exclusive manner with H1 being removed from the NOR region and SonC being absent from the end of the chromosome beyond the NOR. This region of chromatin is adjacent to a cluster of nuclear pore complexes to which NIMA localizes last during its progression around the nuclear envelope during initiation of mitosis. The results genetically extend the NIMA regulatory system to include a protein with selective large-scale chromatin location observed during mitosis. The data suggest a model in which NIMA and SonC, its new chromatin-associated suppressor, might help to orchestrate global chromatin states during mitosis and the DNA damage response.
Keywords: NIMA, DNA damage, Zn2Cys6 domain, Mag1, nucleolar organizer region (NOR)
THE NIMA kinase is an essential conserved regulator of mitotic events in the filamentous fungus Aspergillus nidulans (Osmani et al. 1988). NIMA was first discovered through a genetic screen that defined several different temperature-sensitive alleles of NIMA that cause a Never in Mitosis phenotype (Morris 1975). Subsequent studies showed that NIMA is essential for mitotic entry but not for the activation of the Cdk1 mitotic kinase (Oakley and Morris 1983; Osmani et al. 1988, 1991; Morris et al. 1992; Ye et al. 1995). Not only is NIMA essential for initiating mitosis, but also its overexpression can prematurely induce mitotic events including DNA condensation in A. nidulans, fission yeast, Xenopus, and human cells (O’Connell et al. 1994; Lu and Hunter 1995), indicating the existence of conserved mitotic substrates as recently confirmed in mammalian cells (Laurell et al. 2011). NIMA is subject to complex regulation at both the mRNA and protein levels, leading to maximum activity during mitosis (Osmani et al. 1987; Pu and Osmani 1995; Ye et al. 1995, 1996, 1998).
One of the key roles for NIMA at the onset of mitosis is its regulation of nuclear pore complexes. This insight came from a genetic screen aimed at identifying suppressors of the temperature-sensitive nimA1 allele. This genetic screen identified nimA1 suppressor mutations in two genes encoding nuclear pore complex (NPC) proteins (Wu et al. 1998; De Souza et al. 2003), which were named “SonA” and “SonB” for suppressors of nimA1. Further studies of SonA, SonB, and additional NPC proteins revealed that in A. nidulans the nuclear pore complex undergoes complex rearrangements during mitosis, with 13 core NPC proteins remaining associated with the nuclear envelope and 14 peripheral NPC proteins becoming dispersed or even targeted to other locations to presumably fulfill mitotic functions (De Souza et al. 2004; Osmani et al. 2006a; De Souza and Osmani 2009; Liu et al. 2009). The partial disassembly of NPCs allows nuclear access for tubulin and other important proteins required for mitosis (De Souza et al. 2004).
In addition to nuclear pores undergoing mitotic disassembly, the nucleolus of A. nidulans, as in higher eukaryotes, also undergoes mitotic disassembly (Dimario 2004; Hernandez-Verdun 2011). Nucleoli are assembled around nucleolar organizer regions (NORs) and are the sites of ribosome biogenesis (Boisvert et al. 2007). In mammalian cells, the disassembly of nucleoli involves shutdown of ribosomal DNA (rDNA) transcription, and regeneration of nucleoli is initiated by the reassembly of the rDNA transcription machinery onto the NORs (Leung et al. 2004; Boisvert et al. 2007). In A. nidulans, nucleolar proteins are separated from nuclei via a double restriction of the nuclear envelope after the NORs are removed from the nucleolus at anaphase (Ukil et al. 2009). The nucleolar structure is then disassembled in a stepwise manner and reassembled onto the NORs in the daughter nuclei (Ukil et al. 2009). The mechanisms by which these dynamic processes are regulated are largely unknown.
Although NIMA is essential for mitotic entry, there is also evidence to suggest that NIMA has functions later in mitosis. In cells arrested at pseudo-metaphase by the addition of the microtubule inhibitor nocodazole, NIMA remains in a hyperphosphorylated and active state (Ye et al. 1995) and during metaphase, NIMA localizes to the spindle and then later to the spindle pole bodies during mitotic exit (De Souza et al. 2000). The degradation of NIMA at the end of mitosis is also necessary for mitotic exit. NIMA contains two PEST sequences in its C-terminal regulatory domain that are important for its degradation and a C-terminal truncation allele stabilizes NIMA and prevents cells from exiting mitosis (Pu and Osmani 1995). More recently, cell biological and genetic analysis (Govindaraghavan et al. 2013; Shen and Osmani 2013) have provided further direct evidence that NIMA plays sequential roles during all phases of the mitotic process.
NIMA is the founding member of the NIMA-related kinase (Nek) family identified in organisms ranging from plants to humans. This family of kinases has been implicated in regulation of mitosis and cilia and may coordinate microtubule-dependent processes in dividing and nondividing cells (reviewed in O’Connell et al. 2003; Quarmby and Mahjoub 2005). There are 11 known Neks in mammals and several of these have roles in cell cycle progression and cilia functions (reviewed in Malumbres 2011; Fry et al. 2012). In addition, some Neks play roles in the cellular responses to different stresses, at least in part by contributing to cell cycle checkpoints (reviewed in Moniz et al. 2011; Fry et al. 2012). In Arabidopsis, Nek6 has been implicated in the response to salt stress (Zhang et al. 2011). Evidence has also been accumulating that some Neks function in the DNA damage response. For example, budding yeast Kin3 is important for cell cycle arrest in response to genotoxic agents (Moura et al. 2010), and mammalian Nek1, Nek2, Nek10, and Nek11 are involved in checkpoints activated in response to ionizing and ultraviolet radiation, the DNA-damaging drug methyl methanesulfonate (MMS), oxidative damage-inducing hydrogen peroxide, and the DNA-alkylating agent cisplatin (Noguchi et al. 2002, 2004; Polci et al. 2004; Mi et al. 2007; Chen et al. 2008; Melixetian et al. 2009; Pelegrini et al. 2010; Sorensen et al. 2010; Moniz and Stambolic 2011; Fry et al. 2012; Liu et al. 2013). Mutations in several Nek kinase genes have also been associated with cancer (reviewed in Moniz et al. 2011; Fry et al. 2012), and knockdown of Nek2 or Nek6 in several cancer cell lines inhibits proliferation or induces apoptosis (Tsunoda et al. 2009; Nassirpour et al. 2010). Due to their upregulation in cancers and their involvement in cell cycle progression and cell cycle checkpoint activation, Neks have been proposed as potential targets of anticancer drugs (Sorensen et al. 2010; Fry et al. 2012). However, the mechanisms by which Neks contribute to oncogenesis are unknown, and it is therefore important to identify other proteins involved in NIMA-regulated pathways.
Histone H3 and NUP98 (SonB in A. nidulans) have been identified as substrates for A. nidulans NIMA, and NUP98 phosphorylation is a rate-limiting step for NPC disassembly in human cells (De Souza et al. 2000; Laurell et al. 2011). Intriguingly, NUP98 has also been implicated in oncogenesis (for review see Gough et al. 2011) as many different NUP98 translocation events induce myeloproliferative disease, with some leading to leukemia (Moore et al. 2007). Introduction of the NUP98-HOXA9 chimera has been used in a mouse model of leukemia to induce the disease (Kroon et al. 2001). Interestingly, all NUP98 translocations result in fusions of the NUP98 N-terminus, which contains the Gle2-binding sequence (GLEBS) domain, the motif that interacts with RAE1 (SonA in A. nidulans) (Bailer et al. 1998; Funasaka et al. 2011), but are upstream of the identified NIMA phosphorylation sites (Borrow et al. 1996; Gough et al. 2011; Laurell et al. 2011).
A link between SonB and genomic stability has previously been established. An in-depth genetic analysis of the SonB allele that suppresses the nimA1 temperature sensitivity, sonB1, found that sonB1 confers conditional temperature-dependent DNA damage sensitivity. Furthermore, sonB1 causes lethality at higher temperatures when combined with a deletion of the DNA damage response gene scaA, the A. nidulans homolog of human NBS1 (De Souza et al. 2006). The overall genetic analysis of this mutant allele suggested that SonB is involved in a novel DNA damage response pathway (De Souza et al. 2006). No other proteins have yet been linked to this pathway although NIMA is likely involved since the same mutation in SonB that causes DNA damage sensitivity also suppresses nimA1 temperature sensitivity.
To further characterize the regulatory pathways involved with NIMA and SonB in mitosis and the response to DNA damage, we designed a genetic screen to isolate suppressors of nimA1 that also cause conditional temperature-dependent DNA damage sensitivity. Our expectation was the identification of additional essential genes involved in the DNA damage response. One such essential gene is reported here encoding a binuclear zinc cluster protein that we have named SonC, which displays distinctive chromatin localization during mitosis.
Materials and Methods
General techniques
The A. nidulans strains used in this work are listed in Supporting Information, Table S1, and were generated using standard techniques as previously described (Pontecorvo et al. 1953; Yang et al. 2004; Nayak et al. 2006; Szewczyk et al. 2006), except as noted below. A. nidulans strains were grown on MAG (2% malt extract, 0.2% peptone, 1% dextrose, 2% agar, 1 ml/L Clive Roberts trace elements solution (trace elements solution: 1 g/L FeSO4·7H2O, 8.8 g/L ZnSO4·7H2O, 0.4 g/L CuSO4·5H2O, 0.15 g/L MnSO4·H2O, 0.1 g/L Na2B4O7·10H2O, 0.05 g/L (NH4)6MO7O2·4H2O, 0.3% chloroform), 1 ml/L vitamin solution (vitamin solution: 1 mg/ml p-aminobenzoic acid, 2 mg/ml nicotinic acid, 200 mg/ml choline, 0.02 mg/ml D-biotin, 0.5 mg/ml pyrodoxine HCl, 25mg/ml riboflavin HCl), 0.5 g/L arginine) or MAGUU (supplemented with 1.2 g/L uridine and 1.12 g/L uracil) media unless otherwise noted, and standard growth and genetic methodologies were applied (Pontecorvo et al. 1953; Liu et al. 2010).
Genetic screen
Sensitivity to the DNA-damaging agent 1,2,7,8-diepoxyoctane (DEO) (Ong and Serres 1975) was assessed as in De Souza et al. (2006) and Liu et al. (2009). Mutagenesis of strain JLA1 was carried out as described (Wu et al. 1998). Following mutagenesis, spores were plated out at 42° to select for suppressors of nimA1 temperature sensitivity. Spores from colonies able to grow at 42° were spotted onto MAGUU plates and incubated at 32° for 2 days. These plates were replica-plated to MAGUU media containing 0.01% DEO and incubated for 2–4 days at 32° and 42°. Colonies exhibiting sensitivity to DEO were isolated and backcrossed to strain C61 to determine if mutations were intragenic or extragenic to nimA and to strains CDS509 and CDS364 to determine if mutations were linked to sonA or sonB, respectively.
Cloning of sonC by complementation with a high-copy-number plasmid genomic DNA library was carried out as described (De Souza et al. 2003). The mutants were transformed with the genomic plasmid library DNA, overlaid with media containing 0.02% DEO, and incubated at 42°. Twenty transformants that were no longer sensitive to DEO at 42° were selected for further analysis. DNA was recovered from transformants using the Promega Wizard Plus DNA Purification System according to the manufacturer’s protocol (Promega, Madison, WI). To isolate the plasmids from this DNA, DH5α Escherichia coli were transformed with the isolated DNA, and transformants containing the plasmids were selected by growth on LB plates containing 100 mg/ml ampicillin (Maniatis et al. 1989). Plasmid DNA was isolated using the Zymo Research Zyppy Plasmid Miniprep Kit according to the manufacturer’s specifications (Zymo Research, Irvine, CA). Four plasmids were recovered and compared by double-restriction enzyme digestion using KpnI and NotI (NotI sites flank the genomic DNA insert in the library vector). One plasmid from each unique digest pattern was sequenced to obtain the sequence flanking the genomic DNA insert to identify the region of the genome present on each complementing plasmid. To sequence the plasmids isolated from the suppressor mutants, primers M13 forward (Life Technologies, Carlsbad, CA) and oMN33 (Park and Yu 2012), which prime on either side of the genomic DNA insert in the plasmids, were used. The resulting sequences were compared with the annotated Aspergillus Comparative Database (Broad Institute, http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html).
Primers used for amplifying and sequencing sonA, sonB, sonC, nimA, and mag1 are listed in Table S2. All DNA sequencing was carried out at The Ohio State University Plant-Microbe Genomics Facility.
sonC analysis
Primers used to generate constructs for the deletion and GFP-tagging of sonC are listed in Table S2. Endogenous GFP-tagging and gene deletion constructs were constructed using fusion PCR followed by transformation of a ΔnkuAku70 strain as previously described (Yang et al. 2004; Nayak et al. 2006; Szewczyk et al. 2006). To delete sonC, primers SP251-F/SP259-R and SP258-F/SP252-R were used to amplify the regions upstream and downstream of the gene, respectively. Primers SP251-F/SP252-R were used for the fusion PCR. To GFP-tag SonC, primers SP255-F/SP260-R and SP258-F/SP252-R were used to amplify the regions upstream and downstream of the stop codon, respectively. Primers SP255-F/SP252-R were used for the fusion PCR. To tag NIMA with GFP, primers HP15/HP16 and HP17/HP18 were used to amplify the regions upstream and downstream of the stop codon, and primers HP19/HP20 were used to generate the GFP fragment. Primers HP18/HP15 were used for the fusion PCR. Confirmations of endogenous tagging and gene deletion were carried out by diagnostic PCR and by immunoblot as described (De Souza et al. 2009). Generation of histone H1 tagged with the red fluorescent protein mCherry (H1-mCherry) and Nup170-GFP were previously described in Nayak et al. (2010) and Osmani et al. (2006a), respectively, and generation of Bop1-mCherry was as described in Ukil et al. (2009) with the exception that mCherry amplified from pHL86 (Liu et al. 2009) was used instead of GFP. Generation of Pol I-mCherry, Topo I-mCherry, Fib-mCherry, Bop1-GFP, Nup170-mCherry, and CgrA-GFP were described in Ukil et al. (2009).
Live-cell imaging in the presence or absence of benomyl was carried out as described (De Souza et al. 2011) or with an UltraVIEW VoX spinning disc confocal system (PerkinElmer, Inc.). Image analysis and 3D projection generations were carried out using ImageJ freeware (ImageJ, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/) or Volocity Image Analysis Software (PerkinElmer, Inc.). The percentage of SonC remaining in nuclei during mitosis was calculated by measuring the average pixel intensity within a region of interest (ROI) inside a G2 nucleus and comparing it to the average pixel intensity within an equally sized ROI once the same nucleus entered mitosis.
The heterokaryon rescue technique used to determine that sonC is an essential gene and the corresponding diagnostic PCR were performed as previously described (Osmani et al. 2006b).
Sequence alignments of SonC were carried out using Biology Workbench (http://workbench.sdsc.edu).
Results
A genetic screen to identify novel suppressors of nimA1
The previous genetic screen used to identify nimA1 suppressors isolated mutants that, in addition to suppressing the nimA1 temperature sensitivity, conferred cold sensitivity (Wu et al. 1998). Selecting for a second phenotype facilitates the molecular cloning of suppressors by complementation. We hypothesized that, since sonB1 mutants are also sensitive to DNA-damaging agents, we could use this drug sensitivity as a secondary phenotype instead of cold sensitivity to identify additional suppressors of nimA1. We therefore designed a screen similar to the one performed previously (Wu et al. 1998) to identify new extragenic nimA1 suppressors (Figure S1). In brief, nimA1 spores were mutagenized using 4-nitroquinoline 1-oxide and were then plated at 42° to select for mutations that allowed growth at the restrictive temperature for nimA1. The isolated colonies were spotted to make arrays, which were replica-plated to media containing DEO at both 32° and 42°. The nimA1 suppressors that exhibited sensitivity to DEO were selected for further study (Figure 1A and Figure S1). We were specifically interested in identifying nimA1 suppressors that conferred temperature-dependent DNA damage sensitivity, similar to sonB1, as the isolation of such conditional alleles might permit the identification of additional essential genes involved in the DNA damage response.
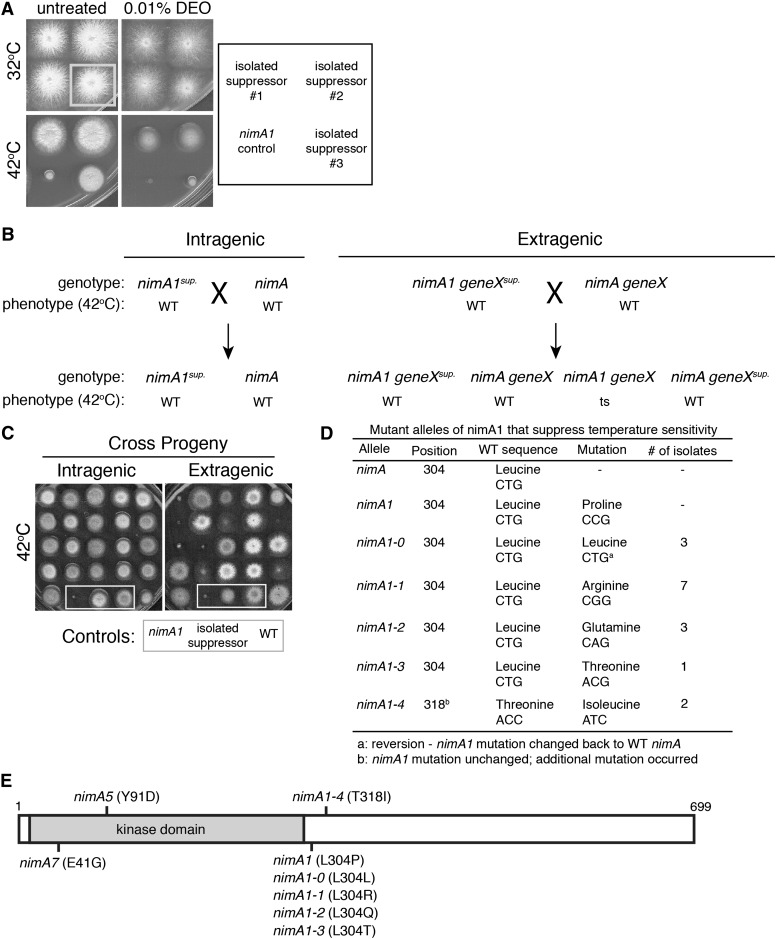
Figure 1.
Characterization of intragenic nimA1 suppressors identified new alleles of nimA. (A) Three different nimA1 suppressors isolated from the screen were replica-plated onto plates with or without 0.01% DEO and incubated at 32° or 42° for 2 days. (Bottom left) The unmutagenized nimA1 control strain (JLA1) was spotted for comparison. Although all three mutants suppressed the temperature sensitivity of nimA1, only one of them (#3) displayed temperature-dependent DNA damage sensitivity. (B) Diagram illustrating how intragenic suppressors were identified. Shown are the expected outcomes from crosses between isolated nimA1 suppressors and a wild-type (WT) strain to distinguish intragenic from extragenic suppressors. (C) The temperature sensitivity caused by nimA1 segregates independently from extragenic suppressors. Plates grown at 42° for 2 days show examples of progeny from crosses diagrammed in B. Extragenic suppressors were identified based on the isolation of temperature-sensitive progeny from a cross with a wild-type strain. In contrast, all progeny from a cross between a wild-type strain (C61) and intragenic suppressors (mutagenized strain JLA1) are not temperature-sensitive. Control strains are boxed by a shaded outline. Strain JLA1 was used as the nimA1 control. (D) Table of new intragenic NIMA alleles isolated as suppressors of nimA1 temperature sensitivity. (E) Diagram of NIMA with the positions of identified alleles marked. The catalytic domain is shaded.
Identification of intragenic nimA1 suppressors
While the goal of the screen was to identify new genes involved in the DNA damage response, we additionally identified several intragenic nimA1 suppressors by choosing 24 of the strongest suppressors that did not exhibit a drug-sensitive phenotype and determining if the nimA1 suppressor mutations were intragenic or extragenic by backcrossing to a wild-type strain. Progeny from the crosses were examined for temperature sensitivity (Figure 1, B and C). If none of the progeny exhibit temperature sensitivity, then the suppressor mutation must be co-segregating with the nimA1 mutation and is thus likely to be intragenic (Figure 1, B and C, left). If temperature-sensitive progeny are recovered, then the suppressor mutation is segregating independently of the nimA1 mutation and is therefore extragenic (Figure 1, B and C, right).
From the 24 mutants tested, 16 were determined to be intragenic based on the absence of temperature sensitivity of all the progeny from the backcross. The remaining 8 strong nimA1 suppressors were extragenic suppressors with no identified secondary phenotype. The nimA gene was PCR-amplified from the 16 intragenic suppressor strains and sequenced to identify the suppressor mutations. The nimA1 mutation is a single-base-pair change resulting in the codon change of leucine 304 to a proline (L304P) (Pu et al. 1995) (Figure 1D). Three of the intragenic suppressors were reversions of proline 304 changed to the wild-type leucine (P304L) designated as nimA1-0. Most of the other intragenic suppressors contained a different change in amino acid 304, resulting in the identification of three new nimA alleles. These alleles were named nimA1-1, nimA1-2, and nimA1-3 and had amino acid 304 changed from a proline to an arginine, glutamine, or threonine, respectively, with a majority being changed to an arginine (seven different isolates) (Figure 1D). Two other mutants still retained the nimA1 mutation (L304P) but had an additional single-base-pair substitution, which resulted in a change of amino acid 318 from a threonine to an isoleucine (T318I) termed nimA1-4 (Figure 1, D and E).
Mag1 is a copy-number suppressor of sonA1 involved in the DNA damage response
Two separate rounds of the screen were performed, and from the first we identified six mutants that exhibit DEO sensitivity to varying degrees (Table 1, mutants A–F). Importantly, for this first round of the screen we included any DEO-sensitive suppressors that were also cold-sensitive. Two of the six mutants (mutants A and B) were cold-sensitive (data not shown). To begin our characterization of the extragenic nimA1 suppressors, we first identified those that were linked to the previously identified DNA damage-sensitive nimA1 suppressor, sonB1, by crossing the isolated mutants to strains carrying the sonB1 mutation. This identified one suppressor linked to sonB (mutant A). Sequencing of the sonB gene from this mutant revealed that this suppressor was identical to sonB1, containing a point mutation resulting in an amino acid substitution (K193E).
Table 1. DEO-sensitive nimA1 suppressors isolated from the screen.
| Mutant | Linked to sonA | Linked to sonB | Gene/allele | Wild-type sequence | Mutation | Amino acid change |
|---|---|---|---|---|---|---|
| A | √ | sonB1 | GAG | AAG | K193E | |
| B | √ | sonA1 | CCT | CGT | P205R | |
| C | sonC1 | GCG | CCG | A113P | ||
| D | Unknown | Unknown | Unknown | Unknown | ||
| E | Unknown | Unknown | Unknown | Unknown | ||
| F | Unknown | Unknown | Unknown | Unknown | ||
| 7 | √ | sonA2 | CGT | CAT | R297H | |
| 55 | √ | sonA2 | CGT | CAT | R297H | |
| 72 | √ | sonA2 | CGT | CAT | R297H | |
| 89 | Unknown | Unknown | Unknown | Unknown | ||
| 118 | √ | sonA2 | CGT | CAT | R297H | |
| 136 | Unknown | Unknown | Unknown | Unknown | ||
| 143 | √ | sonA2 | CGT | CAT | R297H | |
| 146 | Unknown | Unknown | Unknown | Unknown | ||
| 147 | Unknown | Unknown | Unknown | Unknown | ||
| 155 | Unknown | Unknown | Unknown | Unknown | ||
| 244 | Unknown | Unknown | Unknown | Unknown | ||
| 270 | √ | sonA2 | CGT | CAT | R297H | |
| 299 | √ | sonA2 | CGT | CAT | R297H | |
| 309 | √ | sonA2 | CGT | CAT | R297H | |
| 314 | Unknown | Unknown | Unknown | Unknown | ||
| 331 | √ | sonA2 | CGT | CAT | R297H | |
| 332 | √ | sonA2 | CGT | CAT | R297H | |
| 393 | √ | sonA2 | CGT | CAT | R297H | |
| 411 | √ | sonA3 | CAT | TAT | H296Y | |
| 420 | √ | sonA2 | CGT | CAT | R297H | |
| 421 | √ | sonA2 | CGT | CAT | R297H | |
| 449 | Unknown | Unknown | Unknown | Unknown | ||
| 460 | √ | sonA2 | CGT | CAT | R297H | |
| 501 | Unknown | Unknown | Unknown | Unknown | ||
| 547 | Unknown | Unknown | Unknown | Unknown | ||
| 558 | Unknown | Unknown | Unknown | Unknown | ||
| 603 | √ | sonA2 | CGT | CAT | R297H |
We cloned the DNA damage-sensitive suppressor mutant B by complementation of its DNA damage sensitivity at 42° using a high-copy-number genomic DNA plasmid library (a kind gift from Greg May) (Osherov and May 2000). The region of the genome containing two genes designated AN4388 and AN4389 was identified from plasmids derived from two transformants, and the sonA gene was identified from plasmids from seven different transformants. AN4388 is a predicted subunit of ubiquinol-cytochrome-c reductase orthologous to Saccharomyces cerevisiae QCR7 (David et al. 2008; Arnaud et al. 2012). AN4389, hereafter referred to as Mag1, is the ortholog of S. cerevisiae Mag1 (Arnaud et al. 2012), a known DNA damage response protein (Chen et al. 1989). Since ubiquinol-cytochrome-c reductases are mitochondrial proteins involved in energy metabolism and mag1 is a DNA damage response gene, we concluded that mag1 was the gene of interest on these plasmids. To determine which gene was mutated to cause the nimA1 suppression, we sequenced mag1 and sonA from genomic DNA isolated from mutant B and compared their sequences to the sequencing results from the unmutagenized strain (JLA1). Mutant B does not contain any mutations in mag1 but carries the sonA1 mutation, indicating that Mag1 is a copy-number suppressor of sonA1. To confirm these results, we examined the ability of the multi-copy number plasmid containing mag1 to suppress the DNA damage sensitivity of a previously studied sonA1 mutant strain. High-copy mag1 is able to suppress the temperature-dependent DNA damage sensitivity of the sonA1 mutant similar to the wild-type sonA gene (Figure 2A) but not the DNA damage sensitivity of a sonB1 mutant (data not shown), likely because sonB1 mutants are much more sensitive to DNA damage than sonA1 mutants. High-copy mag1 is not able to suppress the temperature sensitivity of nimA1 mutants (data not shown).
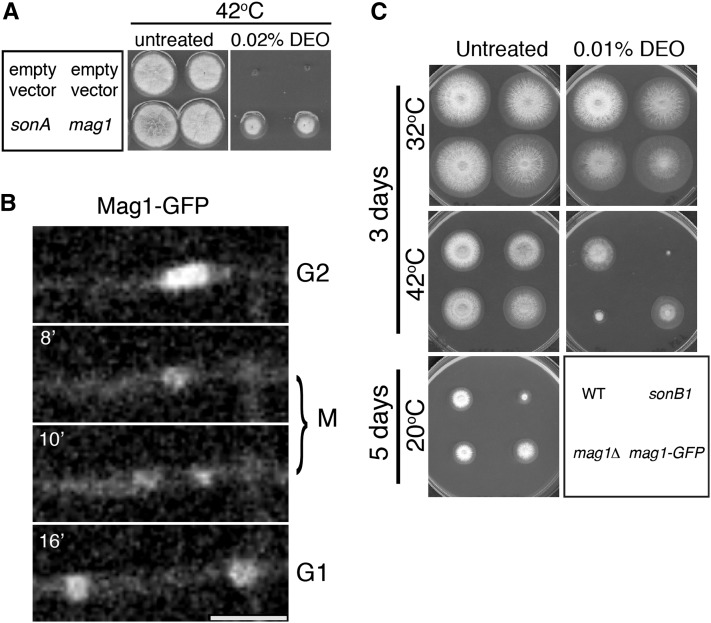
Figure 2.
Mag1 is a nuclear protein and is involved in the DNA damage response. (A) Strain SO369 (sonA1) was transformed with high-copy plasmids containing the pyrG selectable marker and genes for sonA or mag1. An empty vector containing only the selectable marker was used as a negative control. Transformants were spotted onto media selective for the plasmids with or without 0.02% DEO. Plates were incubated at 42° for 3 days. (B) Time-lapse confocal microscopy of Mag1-GFP (strain JLA263). Mag1-GFP is nuclear during interphase. During mitosis a majority of Mag1-GFP disperses from nuclei while a small percentage remains nuclear, potentially associated with chromatin. See File S1. Bar, 5 μm. (C) Strains R153 (WT), CDS364 (sonB1), JLA264 (mag1Δ), and JLA263 (mag1-GFP) were spotted onto MAGUU plates with or without 0.02% DEO and incubated at either 32° or 42° for 3 days or at 20° for 5 days.
To further characterize Mag1, we analyzed its cellular localization by endogenously tagging it with GFP. Mag1-GFP localizes to nuclei during interphase (Figure 2B and File S1). During mitosis, nearly all Mag1-GFP disperses into the cytoplasm with only a faint signal still present in the nuclei. Since the nuclear Mag1-GFP signal appears to condense and then divide into the two daughter nuclei, the structure that it is associating with during mitosis is likely chromatin. Cytoplasmic Mag1-GFP is then re-imported into nuclei following mitotic exit.
We next determined the phenotypes caused by deletion of mag1. Using the heterokaryon rescue technique, we determined that it is not an essential gene (data not shown) as haploid mag1-deleted strains were readily obtained. We examined both the mag1 deletion mutant and the Mag1-GFP strain for DEO and cold sensitivity. The Mag1-GFP strain does not exhibit sensitivity in any of the conditions assayed (Figure 2C). However, the mag1 deletion mutant showed temperature-dependent DNA damage sensitivity (Figure 2C). This sensitivity was also seen on media containing 0.025% MMS and was evident when conidia lacking mag1 were either treated directly or allowed to germinate first (data not shown). These results reveal that Mag1 plays a role in the DNA damage response in A. nidulans.
Identification and characterization of new SONA alleles
Since we recovered both the sonA1 and sonB1 mutants from the preliminary round of the genetic screen we performed the screen a second time using more stringent criteria, isolating only the stronger nimA1 suppressors and, to avoid re-isolating sonA1 and sonB1 again, we eliminated all cold-sensitive mutants from further analyses. We isolated an additional 603 nimA1 suppressors (numbered 1-603), 27 of which had a DNA damage-sensitive phenotype and were not cold-sensitive (Table 1, mutants designated by a number). The new mutants were crossed to sonA1 and sonB1 strains to identify any suppressors linked to these genes, which might identify new non-cold-sensitive alleles. We did not identify any new mutants linked to sonB, but did identify 16 nimA1 suppressor mutations linked to sonA (Table 1). Sequence analysis of these 16 identified two new alleles of sonA. The sonA1 allele contains a single C-to-G mutation, which results in a P205R change in the protein sequence (Wu et al. 1998). The two new mutations identified are downstream of the sonA1 mutation in the codons for amino acids 297 (R297H) and 296 (H296Y) (Figure 3, A and B), and we named them “sonA2” and “sonA3,” respectively. Fifteen mutants contain the sonA2 allele and 1 mutant contains the sonA3 allele.
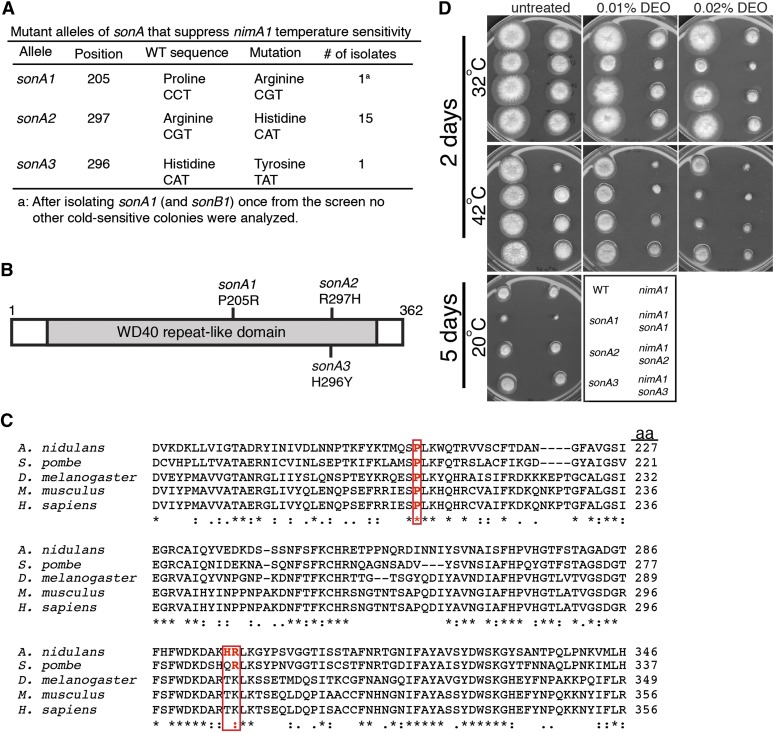
Figure 3.
Two new alleles of sonA were identified as nimA1 suppressors. (A) Table of SonA alleles that suppress the temperature sensitivity of nimA1. (B) Diagram of SonA (drawn to scale) showing the positions of the nimA1 suppressor mutations. Shaded area indicates the WD40 repeat-like domain. (C) Two of the amino acids changed in the different SonA alleles are highly conserved. Shown is the amino acid sequence alignment of sonA from A. nidulans aligned with its orthologs from other organisms. The changed amino acids present in the different sonA alleles are boxed in red. A. nidulans, Aspergillus nidulans; S. pombe, Schizosaccharomyces pombe; D. melanogaster, Drosophila melanogaster; M. musculus, Mus musculus; H. sapiens, Homo sapiens. (D) Strains R153 (WT), JLA1 (nimA1), CDS509 (sonA1), JLA227 (sonA2), JLA268 (sonA3), LPW29 (sonA1 nimA1), JLA185 (nimA1 sonA2), and JLA255 (nimA1 sonA3) were spotted onto YAGUU (0.5% yeast extract, 1% glucose, 2 ml/L vitamin mix, 1 ml/L trace elements, 10 mM MgSO4, 15 g/L agar, 1.2 g/L uridine, 1.12 g/L uracil) with or without DEO at 32° and 42° for 2 days or at 20° for 5 days.
Most of the SonA protein sequence consists of a WD40 repeat-like domain (amino acids 33–338) (Wu et al. 1998; Arnaud et al. 2012), and all three of the nimA1 suppressor mutations in SonA are within this WD40 domain (Figure 3B). SonA is 44% identical to human Rae1 and is therefore predicted to form a seven-bladed β-propeller structure similar to Rae1 (Ren et al. 2010). The sonA1 mutation occurs in a residue conserved from fungi to humans (Wu et al. 1998 and Figure 3C) and is located between two of the β-propellers. The sonA2 and sonA3 mutations are present in a highly conserved region of the protein, and the sonA2 mutation occurs in a conserved residue (Figure 3C). Both the sonA2 and sonA3 mutations reside in the β-sheet of the sixth β-propeller. None of the sonA mutations occur in regions predicted to interact with SonB/Nup98 (Ren et al. 2010).
The sonA1 mutation results in cold sensitivity, suppression of nimA1 temperature sensitivity (Wu et al. 1998), and mild DEO sensitivity. To compare these phenotypes with those caused by the newly identified sonA2 and sonA3 alleles, we spotted these mutants with or without the nimA1 mutation in the background onto media with and without DEO and incubated them at different temperatures (Figure 3D). The sonA2 and sonA3 alleles suppress the temperature sensitivity of nimA1 to the same level as sonA1. By this assay we confirmed that, of the three different sonA alleles, only sonA1 confers cold sensitivity. sonA1 and sonA2 mutants exhibit sensitivity to 0.02% DEO, while sonA3 mutants show mild sensitivity to this concentration of DEO only in the nimA1 background (Figure 3D).
Identification of SONC, a new nimA1 suppressor
In total, we isolated 15 DNA damage-sensitive nimA1 suppressors that were not linked to sonA or sonB and we selected the first 5 to begin our characterizations (Table 1, mutants C–F and mutant 89) by comparing their phenotypes in a nimA1 background with those of sonB1 (Figure 4). We found that mutants E and F are linked (data not shown), so we also did a comparison between these two mutants (Figure 4C). All five of the new mutants are able to suppress nimA1 temperature sensitivity to a degree similar to sonB1 and, in contrast to sonB1, only one of them confers cold sensitivity (mutant E, Figure 4C; note that mutants C–F were from the first version of the genetic screen in which we did not eliminate mutants with cold sensitivity). All mutants in combination with nimA1 show some degree of DNA damage sensitivity (Figure 4).
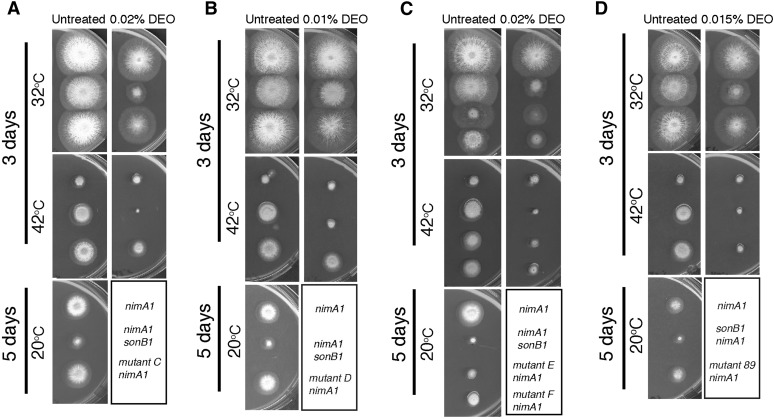
Figure 4.
Characterization of the isolated nimA1 suppressors. (A) Mutant C suppresses the temperature sensitivity of nimA1, and the double mutant shows mild DEO sensitivity. Strains JLA1 (nimA1), CDS36 (nimA1 sonB1), and JLA77 (nimA1 + mutant C) were spotted onto MAGUU plates with or without 0.02% DEO and incubated at either 32° or 42° for 3 days or at 20° for 5 days. (B) Mutant D suppresses nimA1 temperature sensitivity, and the double mutant shows temperature-dependent DEO sensitivity. Strains JLA1 (nimA1), CDS36 (nimA1 sonB1), and JLA73 (nimA1 + mutant D) were spotted onto MAGUU plates with or without 0.01% DEO and incubated as in A. (C) Mutants E and F are genetically linked and show the same degree of nimA1 suppression, and the double mutants show sensitivity to DEO. Strains JLA1 (nimA1), CDS36 (nimA1 sonB1), JLA78 (nimA1 + mutant E), and JLA75 (nimA1 + mutant F) were spotted onto MAGUU plates with or without 0.02% DEO and incubated as in A. (D) Mutant 89 suppresses nimA1 temperature sensitivity, and the double mutant shows temperature-dependent DNA damage sensitivity. Strains JLA1 (nimA1), CDS36 (nimA1 sonB1), and JLA184 (nimA1 + mutant 89) were spotted onto MAGUU plates with or without 0.015% DEO and incubated as in A.
We cloned the DNA damage-sensitive suppressor mutant C by complementation of its DNA damage sensitivity at 42° using the genomic DNA plasmid library as described above for mutant B. Two different regions of the genome were represented more than once from the sequenced plasmids. The same region of the genome that was identified from the sonA1 complementing plasmids and containing mag1 was identified from seven different transformants, and the gene designated as AN1232 was identified from another three. We sequenced mag1 and AN1232 from mutant C. Mutant C does not contain any mutations in mag1, indicating that Mag1 can act as a multi-copy number suppressor of at least two mutations (sonA1 and mutant C) that cause relatively minor DEO sensitivity. Mutant C, however, contains a single-base-pair substitution in AN1232, resulting in an A113P change (Figure 5A). We have named AN1232 sonC (Suppressor of nimA1 C) and the allele isolated sonC1.
Figure 5.
Mutant C contains a mutation in the locus designated as AN1232 and representing the sonC gene, which defines a protein containing a Zn2Cys6 domain. (A) Table showing the mutation found in mutant C. (B) Diagram of SonC, which contains a C6 zinc finger potential DNA-binding domain (gray box), and the mutation found in mutant C (sonC1), which is located in the N-terminal region. (C) ClustalW alignment (http://workbench.sdsc.edu) of SonC and its orthologs. Identical groups [indicated by an asterisk (*)], conserved strong groups [indicated by a colon (:)], and conserved weak groups [indicated by a period (.)] are noted. Accession numbers for the sequences are the following: A. nidulans (AN1232; Aspergillus Genome Database, http://www.aspgd.org); Penicillium chrysogenum (Pc16g03990); Coccidioides immitis (CIMG_10322); Bipolaris maydis (COCC4DRAFT_161203, with amino acids 642–689 removed prior to performing the alignment as this region is absent from the other orthologs and so is likely part of an intron); Sclerotinia sclerotiorum (SS1G_09964); Fusarium oxysporum (FOXB_15136, with amino acids 1–67 removed prior to performing the alignment as this region is likely part of the 5′ UTR); Trichoderma reesei (hypothetical protein, locus EGR51802); and Neurospora crassa (NCU07692). The C6 zinc finger domain is highly conserved, and the six cysteine residues with the spacing CX2CX6CX5CX2CX12C are found in all SonC orthologs (boxed in red). (D) SonC orthologs were found only in the Ascomycota, and the amino acid changed in sonC1 (boxed in red) occurs in a short, highly conserved region of the protein predicted to form an alpha-helix (JPred 3) (Cole et al. 2008). Sequences used are the same as in C. See Figure S2 for the alignment of the entire protein sequences.
sonC is essential
SonC contains a C6 zinc finger domain although the mutation that we identified occurs outside of this region (Figure 5B). SonC is found only in the filamentous Ascomycota, and the C6 zinc finger domain is highly conserved among these fungi with orthologs identified within the Dothideomycetes, Leotiomycetes, and Sordariomycetes as well as the Eurotiomycetes of the Pezizomycotina (Figure 5C and Figure S2), consisting at its core a cluster of six cysteine residues arranged with the following conserved spacing: CX2CX6CX5CX2CX12C. SonC is unique in that it is the only C6 zinc finger domain-containing protein in the Aspergillus genus that possesses 12 amino acids between the last two cysteines within the cluster (Wortman et al. 2009). In addition to the C6 zinc finger binding domain, a region of ∼70 amino acids at the N-terminus of the protein (Figure 5D) is also highly conserved (Figure S2). The presence of a C6 zinc finger domain suggests that these proteins could potentially bind to DNA, but neither SonC nor any of its orthologs have yet been characterized. The mutation identified occurs in a short stretch of highly conserved amino acids predicted to form an α-helix (Figure 5, A and D) and changes an alanine to a proline (A113P), which would predictably disrupt this secondary structure.
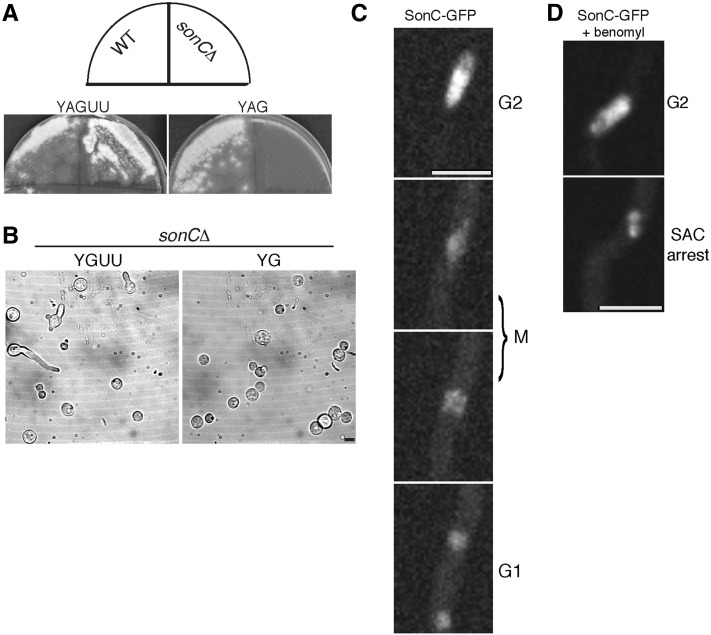
We first sought to determine if sonC is an essential gene using the heterokaryon rescue technique (Osmani et al. 2006b). We deleted sonC using the pyrG selection marker and found that sonC is essential (Figure 6A). To observe the terminal phenotype caused by sonC deletion, conidia from heterokaryons generated after the deletion were examined at the microscopic level (Figure 6B). The vast majority of conidia germinated under selective pressure (Figure 6B, “YG”) failed to undergo polarized growth, which is the phenotype displayed by pyrG89 mutant cells in the absence of complementation. This indicates either that deletion of sonC prevents polarity establishment or that the heterokaryon failed to generate conidia carrying the deleted sonC allele. The terminal growth arrest phenotype was therefore not unambiguously defined.
Figure 6.
SonC is an essential nuclear protein. (A) Heterokaryon rescue analysis showing that sonC is an essential gene. Conidia from a wild-type (WT) pyrG+ control strain (R153) or from ΔsonC::pyrG transformants were streaked onto media with (YAG, right) or without (YAGUU, left) selective pressure for the pyrG selective marker gene and grown for 2 days at 32°. If sonC were not essential, then haploid null strains would be generated and all conidia generated from the colony would contain the functional pyrG gene in place of sonC. If sonC were essential, then heterokaryons containing two different types of nuclei, those with the deletion (pyrG+ ΔsonC) and those with WT sonC (pyrG− sonC+), would be generated. The uninucleate conidia from heterokaryon colonies will contain either type of nuclei and can be distinguished by their ability to grow on pyrG+ selective media (Osmani et al. 2006b). If sonC were essential, no conidia from the heterokaryotic colonies would grow on selective YAG media, but those containing pyrG− sonC+ nuclei would grow and form colonies on nonselective YAGUU media. These are the growth phenotypes seen indicating that sonC is an essential gene. Heterokaryons were confirmed by diagnostic PCR (data now shown). (B) Bright-field images of conidia from a heterokaryon containing nuclei with the sonC deletion (sonCΔ::pyrG+) and pyrG89 nuclei with the wild-type allele of sonC (sonC+; pyrG−). Growth was for 24 h at 32° in nonselective (YAGUU) or selective (YAG) media for pyrG+. While 54% (n = 139) of cells sent out a germ tube in the nonselective media, only 4% (n = 357) germinated (not shown) in the selective media. (C) Time-lapse confocal microscopy of SonC-GFP. SonC-GFP is located within nuclei throughout interphase and partially disperses during mitosis. Strain JLA265 (sonC-GFP) was inoculated into minimal media and incubated at room temperature for 24 h prior to imaging. See corresponding File S2 and File S3. Bar, 5 μm. (D) Time-lapse confocal microscopy of SonC-GFP in cells treated with benomyl. Strain JLA265 (sonC-GFP) was inoculated into minimal media and incubated at room temperature for 22 h. Media was then exchanged with media containing 2.4 μg/ml benomyl to arrest cells in mitosis. See corresponding File S4 and File S5. Bar, 5 μm.
SonC locates to mitotic chromatin in a distinctive manner
A. nidulans undergoes partially open mitosis wherein the nuclear envelope remains intact but the nuclear pore complexes partially disassemble, allowing diffusion of nuclear proteins out of mitotic nuclei if they are not attached to nuclear structures such as DNA (De Souza et al. 2004; Osmani et al. 2006a; De Souza and Osmani 2007). If SonC is a DNA-binding protein, it would be expected to localize to nuclei and not be released from nuclei during mitosis. To investigate this, we tagged endogenous SonC with GFP and examined the localization of SonC-GFP by live-cell confocal microscopy during mitosis (Figure 6C; File S2; File S3). SonC-GFP localizes to nuclei during interphase, and an examination of nuclear levels in populations of cycling cells showed that these nuclear levels remain the same from G1 through G2 (data not shown). During mitosis SonC-GFP partially disperses into the cytoplasm; however, 50 ± 6% (n = 41 nuclei) remains within nuclei presumably through an association with DNA or chromatin (Figure 6C; File S2; File S3). Following mitosis, the cytoplasmic fraction becomes re-imported and SonC-GFP relocalizes back to the nucleoplasm in G1 (Figure 6C; File S2; File S3). Since some SonC remains in the nucleus during mitosis, we wanted to see if its mitotic localization might reflect association with condensing chromatin. We therefore monitored SonC-GFP during spindle assembly checkpoint (SAC)-mediated mitotic arrest using the microtubule inhibitor benomyl (Oakley and Morris 1980). During mitotic SAC arrest, some SonC-GFP remained within the nuclei and behaved similarly to chromatin, becoming condensed during entry into mitotic arrest (Figure 6D; File S4; File S5), further suggesting that some SonC is bound to DNA or chromatin during mitosis.
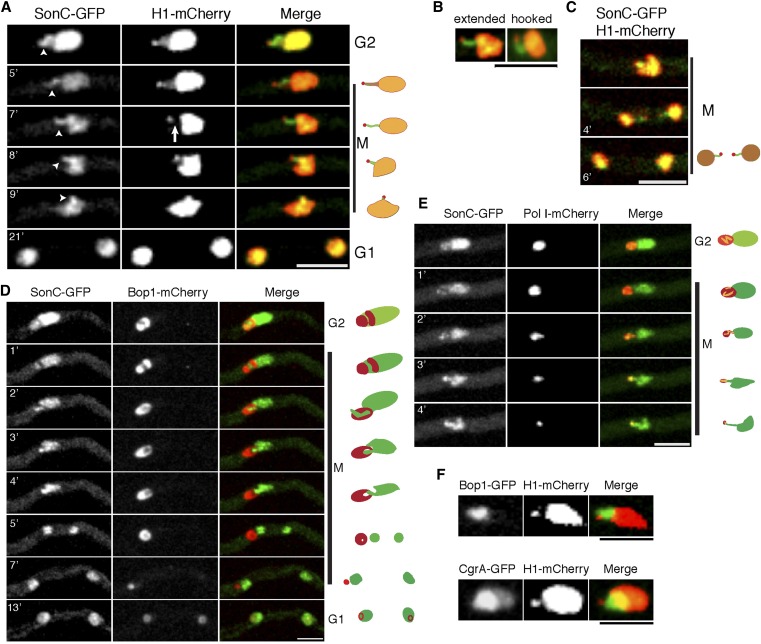
To investigate the relationship between SonC and chromatin, we examined its localization in a strain expressing H1-mCherry. Dual fluorescence imaging revealed several unexpected and intriguing results. The first is that, while we observed that histone H1 localizes to the main mass of chromatin as expected, we also noted that there is a less pronounced labeled projection of histone H1 that extends away from the main chromatin mass during G2 and the early stages of mitosis (Figure 7A; File S6; File S7). This projection ends with a small, brighter H1 focus that is separate from the main chromatin mass (Figure 7A; File S6; File S7). This distal focus was also detected in interphase nuclei. Importantly, the histone H1 projection and associated distal focus had been seen in several other studies but was not discussed [for examples, see figure 1E and figure 2B in Ukil et al. (2009) and figure 5B in Ukil et al. (2008)]. SonC colocalizes with histone H1 in the main chromatin mass (Pearson’s correlation coefficient 0.66 ± 0.16; n = 23) and along the projection but does not colocalize with H1 on the distal, brighter H1 focus (Pearson’s correlation coefficient = 0.136 ± 0.095; n = 25). As chromatin condensation progresses, SonC and histone H1 become localized in a mutually exclusive manner to distinct regions of the projection. Histone H1 becomes selectively evicted from the middle region of the DNA projection but remains at its distal end (Figure 7A, arrow in 7′ time point; File S6; File S7), whereas SonC is present along the projection but remains excluded from the distal H1 focus. In terms of its overall structure, at the onset of mitosis the SonC-coated projection starts as either a hooked-like or a fully extended structure (50% of nuclei have either form; n = 74) (Figure 7, A and B; File S7; File S8). The SonC-coated projection ending in the H1 focus is dynamic during mitosis and becomes progressively condensed into the main chromatin mass as nuclei approach metaphase when it is no longer easy to distinguish. During anaphase/telophase, the projection became more visible again in some nuclei (28% of nuclei) and was observed to separate as the last two portions of DNA to be segregated (Figure 7C and File S9). This pattern of segregation suggests that the extended portion of chromatin decorated with SonC, with histone H1 concentrated at its distal tip, represents one arm of a chromosome. This indicates that in Figure 7C the late-segregating chromatin decorated by SonC with H1 at the ends represents lagging chromosomes.
Figure 7.
SonC localizes to the NOR. (A) Time-lapse confocal microscopy of SonC-GFP in conjunction with histone H1-mCherry. Strain JLA319 (sonC-GFP H1-mCherry) was inoculated in minimal media and grown at room temperature for 24 h prior to imaging. The projection of SonC-GFP that corresponds to the NOR is marked by arrowheads. H1-mCherry is evicted from the middle region of the projection during mitotic chromosome condensation prior to anaphase (arrow) while SonC-GFP remains associated with the projection. Note that SonC-GFP does not locate to the distal H1-mCherry focus. Diagrams of the SonC and H1 localizations at the different time points are shown on the right. See corresponding File S6 and File S7. (B) The SonC projection can be visualized as an extended or a hooked structure. Strain is the same as in A. See the corresponding File S8 for hooked structure. (C) The SonC projection tipped by H1 was sometimes seen as lagging chromosome arms. Strain and growth conditions are the same as in A. See File S9. (D) Time-lapse confocal microscopy of SonC-GFP and the nucleolar marker Bop1-mCherry. The SonC-GFP projection is cradled within a portion of the nucleolar signal from Bop1-mCherry. Strain JLA324 (sonC-GFP bop1-mCherry) was treated as in A. Diagrams of the SonC and Bop1 localizations at different time points are shown on the right. See File S10 and File S11. (E) Time-lapse confocal microscopy of strain JLA325 (sonC-GFP pol I-mCherry). Strain was treated as in A. Diagrams of the SonC and Pol I localizations at different time points are shown on the right. See File S12. (F) The H1 focus is located outside of the nucleolus near the nuclear periphery. The position of the nucleolus is marked by the nucleolar proteins Bop1-GFP (strain LU193) or CgrA-GFP (strain LU178). The brightness and contrast of the images were adjusted using ImageJ software to highlight the relationship of the SonC projection with the other nuclear proteins more clearly. Bars, 5 μm.
In A. nidulans, the nucleolus resides in one-half of the nucleus that does not contain much chromatin decorated with H1 (Ukil et al. 2009), suggesting that the SonC projection is potentially associated with the nucleolus and represents part of the left arm of chromosome V, which contains the NOR (Brody et al. 1991; Clutterbuck and Farman 2008). To investigate this, we imaged a strain containing SonC-GFP and the nucleolar marker protein Bop1-mCherry, the ortholog of mammalian Bop1, a conserved protein involved in maturation of the 25S and 5.8S ribosomal RNAs, termed “Erb1” in yeast (Strezoska et al. 2000; Pestov et al. 2001; Miles et al. 2005). During A. nidulans mitosis, the NOR is removed from the nucleolus, which undergoes complete disassembly in the cytoplasm, and nucleolar proteins then are reimported into daughter nuclei during G1 to reform their nucleoli (Ukil et al. 2009). If the SonC-decorated projection contains the NOR, then the projection would be expected to be associated with the nucleolus during G2 and the early stages of mitosis and become separated from the nucleolus during mitotic progression. Supporting this possibility, the SonC-coated projection was found to be cradled by Bop1 in G2, prior to mitosis (Figure 7D; File S10; File S11). When examining the SonC-GFP and Bop1-mCherry channels individually, a shadow can be seen in the mCherry channel prior to and during the early stages of mitosis (Figure 7D; File S10; File S11), and the SonC projection was always located within this red fluorescent protein shadow, indicating that this region of the genome resides within the nucleolus. During mitosis, the SonC projection condenses away from its nucleolar cradle in a progressive fashion (Figure 7D; File S10; File S11) until the nucleolus associates with only the end of the SonC projection. The nucleolus then becomes detached as the SonC projection is condensed into the main SonC (chromatin) mass, just before DNA segregation (Figure 7D; File S10; File S11).
The RNA polymerase Pol I is responsible for transcribing most of the rRNAs and is therefore located primarily within the nucleolus at the NORs in most eukaryotes, including A. nidulans (Ukil et al. 2009). Pol I-mCherry also locates to the SonC projection during G2 (Figure 7E and File S12). During both G2 and at the beginning of mitosis, Pol I-mCherry surrounds the SonC projection (n = 30; Figure 7E and File S12). As mitosis progresses, Pol I-mCherry is progressively removed from the SonC projection until metaphase, when, in approximately one-half of the nuclei (54%, n = 147), no nuclear Pol I-mCherry signal was detected. Just prior to metaphase, when the majority of Pol I-mCherry is no longer nuclear, the final Pol I-mCherry signal is located toward the tip of the SonC projection. The cell cycle-regulated localizations of Pol I and SonC provide further evidence that the SonC-DNA projection corresponds to the NOR. To address the location of the distal H1 focus in relationship to the nucleolus, their relative locations were followed during entry into mitosis (Figure 7F). This analysis revealed that the H1 focus resides outside of the nucleolus near the nuclear periphery. This pattern was seen when either Bop1 or CgrA, which is involved in rRNA processing for the 60S ribosome subunit (Sun et al. 2001; Moy et al. 2002), was used as the nucleolar marker.
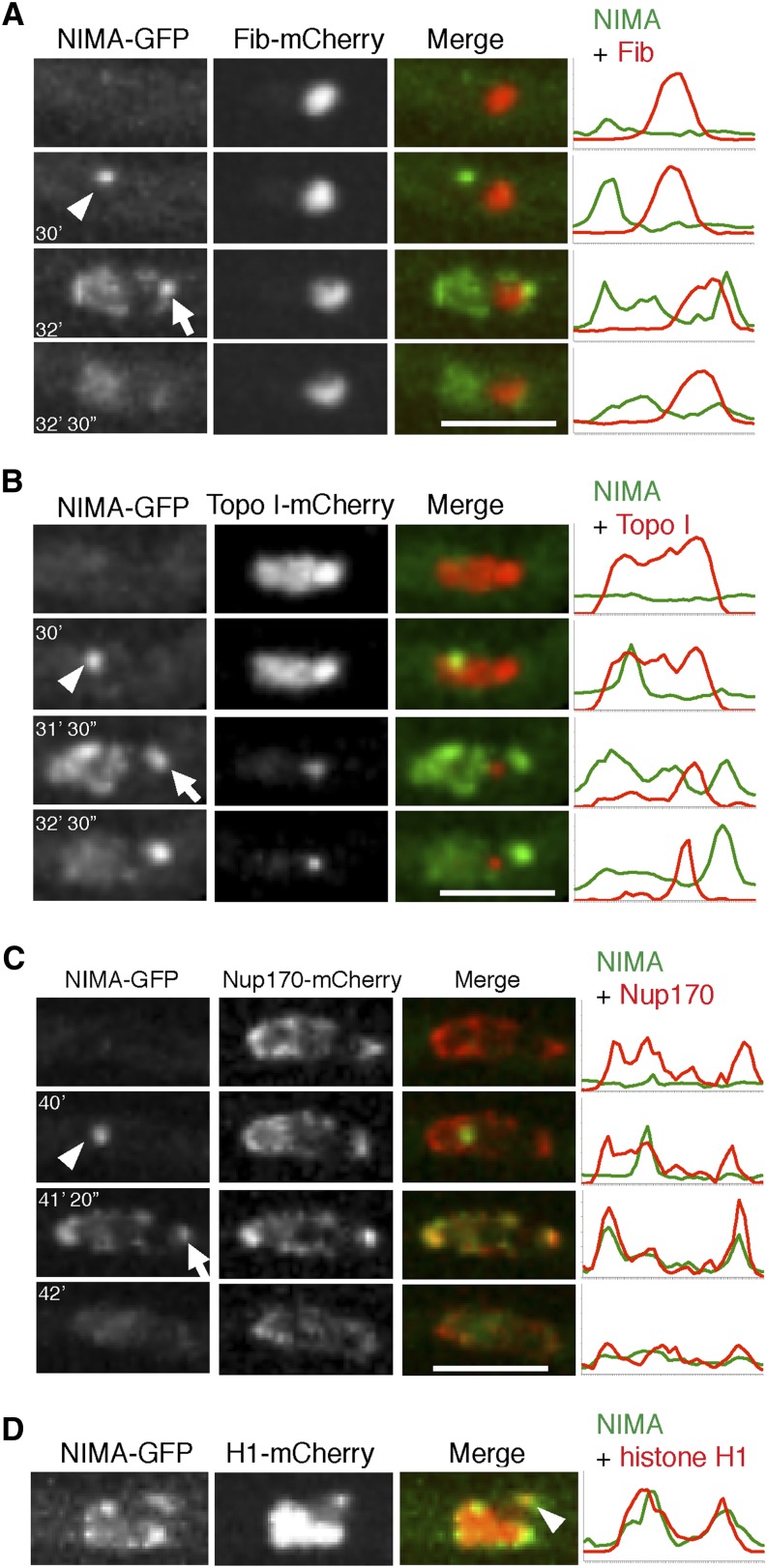
NIMA locates to a focus on the nuclear periphery adjacent to the nucleolus during its mitotic progression around the nuclear envelope
Live-cell imaging of functional endogenously tagged NIMA-GFP has shown that it first locates to spindle pole bodies (SPBs) during entry into mitosis (Shen and Osmani 2013) before progressively locating around the nuclear envelope in sequential association with NPCs (De Souza et al. 2003, 2004), promoting their disassembly. We also observed NIMA-GFP consistently locating at a second focal point after first locating to the SPBs and moving around the nuclear periphery. This NIMA-GFP focus appeared late in the process of it spreading around the mitotic nuclear envelope and was located in the opposite half of the nucleus from that containing the SPB. The A. nidulans nucleolus locates within the opposite half of the nucleus to the SPBs, suggesting that the location of the second NIMA focal point might be adjacent to the nucleolus. To test this, we followed the distribution of NIMA-GFP and Fib-mCherry, as a nucleolar marker, during mitosis. Fib, short for fibrillarin, is a component of a protein complex required for pre-18S rRNA processing (Tollervey et al. 1991). This demonstrated that the second NIMA focal point lies adjacent to the nucleolus (Figure 8A). Topo I is located throughout nuclei during interphase, but much of it disperses during mitotic entry, leaving a concentration previously defined as the mitotic NOR (Ukil et al. 2009). Colocalization studies indicate that the second NIMA focal point lies adjacent to the mitotic NOR rather than being associated with it (Figure 8B).
Figure 8.
NIMA locates to a NPC cluster adjacent to the nucleolus and the distal H1 focus during mitosis. (A) The location of NIMA-GFP in relation to the nucleolar marker Fib-mCherry was monitored in strain KF120 during mitosis. NIMA-GFP first locates to the SPBs (arrowhead), spreads around the nucleus, and locates to a second focus located on the nuclear envelope near the nucleolus (arrow). (B) The location of NIMA-GFP in relation to the NOR as defined by Topo I-mCherry was followed in strain KF122. After NIMA-GFP locates to the SPBs (arrowhead) during mitotic entry, it then locates to a second focus adjacent to the NOR as mitosis proceeds (arrow). (C) The localization of NIMA-GFP in relation to NPCs marked using Nup170-mCherry was followed in strain KF110. Only after initially locating to the SPBs (arrowhead) does NIMA-GFP then colocalize with a NPC cluster (arrow). (D) NIMA-GFP localization was followed in relation to the distal H1 focus in strain KF018. The second NIMA focus is located in the region of the nuclear envelope adjacent to the distal H1 focus (arrowhead). Pixel-intensity line profiles for A-D are shown to the right. Bars, 5 μm.
Although NPCs distribute quite evenly around the nuclear envelope, there is often an obvious separated cluster of NPCs near the nucleolus. To test if the second NIMA focal point colocalizes with this NPC cluster we followed the distribution of NIMA-GFP in relation to Nup170-mCherry, a core NPC protein that does not disperse during mitosis (Osmani et al. 2006a). The results indicate that the second NIMA focus represents the NPC cluster located next to the nucleolus (Figure 8C).
The mitotic locations of NIMA to a region of the nucleus next to the nucleolus late during its progression around the nuclear envelope indicates that this location might be near the histone H1 focus at the end of the SonC projection/NOR. Colocalization studies following histone H1-mCherry and NIMA-GFP confirmed that NIMA does locate late during mitosis to the region of the nuclear envelope populated by the H1 focus (Figure 8D).
Discussion
New alleles of nimA and sonA
There is evidence that NIMA and related kinases have functions in the DNA damage response. We therefore reasoned it would be informative to identify additional nimA1 suppressors that, like sonB1, are DNA damage-sensitive and that could be cloned by complementation of their DNA damage sensitivity. As part of the study, we selected several mutants that exhibited strong suppression of nimA1 temperature sensitivity and identified new alleles of NIMA (Figure 1). Most of the new alleles are the result of a substitution in the same codon as the nimA1 mutation, but one new allele causes an amino acid substitution just downstream of the nimA1 mutation. Secondary structure prediction analysis (JPred 3) (Cole et al. 2008) indicates that the nimA1 mutation and the downstream suppressor mutation are within a single α-helix, suggesting that the intragenic nimA1 suppressor mutations potentially restore NIMA kinase function by repairing the structure of this α-helix. These results also suggest that the region of NIMA just downstream of the catalytic domain plays an important role in NIMA kinase functions, supporting the previously published finding that the C-terminus of NIMA acts in a dominant-negative manner and is likely important for targeting NIMA to its substrates (Lu and Means 1994; De Souza et al. 2004).
The first genetic screen to identify nimA1 suppressors identified two proteins, SonA and SonB, both of which are components of the nuclear pore complex and are known to physically interact in other systems with single alleles of each being identified (Bailer et al. 1998; Wu et al. 1998; Pritchard et al. 1999; De Souza et al. 2003). We identified two additional alleles of sonA that suppress nimA1 temperature sensitivity, sonA2 and sonA3. All three sonA alleles contain mutations within the WD40 repeat-like domain. It is possible that all three of these mutations change the structure of SonA, resulting in weakening of protein–protein interactions with other NPC proteins, including SonB, which could subsequently help lead to mitotic NPC disassembly in the absence of full NIMA activity. These alleles could also suppress nimA1 by modifying non-NPC-related functions of SonA because SonA orthologs have been shown to have important cell cycle functions in addition to their NPC and RNA export roles, including those related to the spindle assembly checkpoint and transcription (Brown et al. 1995; Babu et al. 2003; Blower et al. 2005; Jeganathan et al. 2005; Wong et al. 2006; Grill et al. 2012; Rajani et al. 2012).
We re-isolated the sonB1 allele but did not isolate any new alleles of SonB, indicating that the specific amino acid change in sonB1 is unique in its ability to suppress nimA1 and cause temperature-dependent DNA damage and cold sensitivity. SonB and its orthologs contain a GLEBS motif. This motif is necessary and sufficient for binding SonA orthologs in both S. cerevisiae and Schizosaccharomyces pombe (Bailer et al. 1998). The structure of the human SonA ortholog Rae1 in complex with the GLEBS motif of Nup98 was recently determined (Ren et al. 2010), and the sonB1 mutation occurs within this interacting region (De Souza et al. 2003). The GLEBS domain is highly conserved from yeast to humans, and two amino acids essential for the interaction with Rae1 are two tandem glutamate residues (Ren et al. 2010). The sonB1 mutation (K193E) (De Souza et al. 2003) occurs in a disordered region of the solved structure between two β-strands upstream of the two tandem glutamate residues (Ren et al. 2010), suggesting that this disordered region may be important for stabilizing the interaction of Rae1 with the GLEBS motif. The K193E substitution changes the charge of the amino acid, which could destabilize this interaction.
DNA damage sensitivity and temperature
Another interesting result from our studies was the identification of a multi-copy-number suppressor of DNA damage sensitivity caused by either sonA1 or sonC1. The copy-number suppressor is orthologous to S. cerevisiae Mag1, a DNA glycosylase repair protein that protects DNA from alkylating agents (Chen et al. 1989). Mag1 has been linked in other fungi to the DNA damage response (Prakash and Prakash 1977; Chen et al. 1989; Alseth et al. 2005), and deletion of MAG1 in S. cerevisiae causes sensitivity to DNA-damaging agents (Chen et al. 1990). Consistent with it playing a role in the DNA damage response in A. nidulans, deletion of mag1 results in DNA damage sensitivity that is, however, surprisingly temperature-sensitive. It was expected that deletion of mag1 would result in DNA damage sensitivity because lack of Mag1 3-methyl-adenine DNA glycosylase activity would hinder the initiation of the base excision repair (BER) pathway, leaving damaged bases unrepaired. However, what was not expected was that lack of Mag1 would cause high temperature-dependent sensitivity to DNA damage. One possible reason for this is that the chemical modifications to DNA caused by DEO or MMS could lead to more drastic DNA damage when the temperature is raised. Higher temperatures cause increased rates of DNA depurination (Kunkel 1984; Alberts et al. 2002), which is repaired by the BER pathway (Croteau and Bohr 1997). Hyperthermia has also been found to generate double-strand breaks, particularly during G1 and G2 in human cells in culture (Velichko et al. 2012). DNA damage in cells exposed to DNA-damaging agents at higher temperatures therefore may not be adequately repaired in the absence of Mag1, leading to a type of induced synthetic lethality (Krawczyk et al. 2011). Increased temperatures can also affect other DNA damage response mechanisms, and hyperthermia has been shown to sensitize cells to radiation (Ben-Hur et al. 1974; Dewey et al. 1977) and is being investigated as a way for sensitizing cancer cells to various therapies (Habash et al. 2011; Krawczyk et al. 2011). Given the role of high temperature in the DNA damage sensitivity displayed by An-mag1-deleted strains, investigating the roles of Mag1-like proteins in hyperthermia-sensitizing studies might be informative.
SonC contains a unique putative DNA-binding domain
We cloned the gene responsible for the nimA1 suppression and DNA damage sensitivity from one of our mutants and identified the locus designated systematically as AN1232, which we named sonC. SonC and its orthologs are unique among all Zn2Cys6 domain-containing proteins in that there are an unusually large number of amino acids (12) present between the last two cysteine residues (Wortman et al. 2009). Because SonC contains a zinc finger domain, it has been annotated as a potential transcription factor (AN1232) (Wortman et al. 2009; Arnaud et al. 2012). A previous study identified a Forkhead-like transcription factor, MCNB, as a multi-copy-number suppressor of nimA1 (Ukil et al. 2008) related to the S. pombe transcription factor Sep1. Overexpression of MCNB leads to increased nimA1 protein levels, which is likely the mechanism by which it is able to suppress the temperature sensitivity caused by nimA1 (Ukil et al. 2008). The sonC1 mutation does not result in increased nimA1 expression (data not shown), so if it is a transcription factor, it must suppress nimA1 temperature sensitivity by modifying the transcriptional pattern of other genes. A. nidulans contains >300 Zn2Cys6 predicted binuclear cluster domain-containing encoding genes (Pel et al. 2007; Wortman et al. 2009), and the vast majority of these have not been studied. It is therefore possible that some of these proteins, including SonC, are not transcription factors. Instead, the Cys(6) binuclear cluster domain could target these proteins to DNA for other functions, including those related to genome organization or chromosome segregation. One example of this is S. cerevisiae Cep3, a component of the Cbf3 complex, which connects the Cbf3 complex to centromeres and is essential for chromosome segregation (Lechner 1994). Our cell biological analysis indicates that SonC is a nuclear protein that likely associates with DNA either directly or indirectly because a significant percentage of the protein associates with condensing chromatin during mitosis.
Mitotic nuclear SonC localizes in reverse to histone H1 along a distinct region of DNA that contains the NOR
SonC-GFP localizes to the main chromatin mass defined by histone H1-mCherry and additionally along a distinct region of DNA that forms a projection away from the main chromatin mass. This projection is most evident during prophase as chromatin is being condensed prior to anaphase. The SonC projection likely corresponds to the NOR as both RNA polymerase Pol I (Figure 7E) and the nucleolar marker Bop1 (Figure 7D) locate on the SonC projection. Histone H1 is also located along the SonC projection during interphase but, as chromatin condensation proceeds to anaphase, H1 histones intriguingly become largely evicted from the middle region of the projection while remaining at its distal region. This indicates that dramatic chromatin modifications occur at this chromosome region during its mitotic condensation. The eviction of histone H1 may therefore be necessary to allow access of other DNA-binding proteins to this region of the genome so that it can be transcriptionally silenced and compacted for mitotic segregation. During budding yeast mitosis, condensation of the NOR involves loading of condensin onto the rDNA to promote its condensation from an extended loop structure to enable its segregation during anaphase (Freeman et al. 2000; Lavoie et al. 2000, 2002, 2004; Bhalla et al. 2002). However, unlike S. cerevisiae in which the chromosome arm housing the NOR (the right arm of chromosome XII) is the longest chromosome arm and requires higher levels of condensation for its mitotic segregation (Petes 1979; Lavoie et al. 2004; Torres-Rosell et al. 2004), the chromosome arm of A. nidulans retaining the NOR is not its longest. The longest chromosome arm of A. nidulans is the right arm of chromosome VIII, which by sequence (Galagan et al. 2005) and CHEF analysis (Brody and Carbon 1989) is >4 Mbp long whereas the NOR-containing left arm of ChV is only ∼2 Mbp in length. The removal of histone H1 from the NOR region during A. nidulans mitosis is therefore unlikely to involve a higher level of NOR condensation for segregation. We instead suggest that, because A. nidulans mitotic nucleoli undergo disassembly (unlike mitotic yeast nucleoli), H1 removal from the NOR might be involved in releasing the nucleolus from the NOR. In this regard, it is interesting that a recent proteomic study identified an extensive network of nucleolar proteins that interact with human histone H1 (Kalashnikova et al. 2013). Of further interest, it has also been shown that histone H1 of Trypanosoma cruzi is concentrated in the nucleolus and disperses upon mitotic H1 phosphorylation (Gutiyama et al. 2008). These studies provide some further support for the idea that mitotic H1 phosphorylation might promote histone H1 release from the NOR region to aid release of the nucleolus from its chromatin tether.
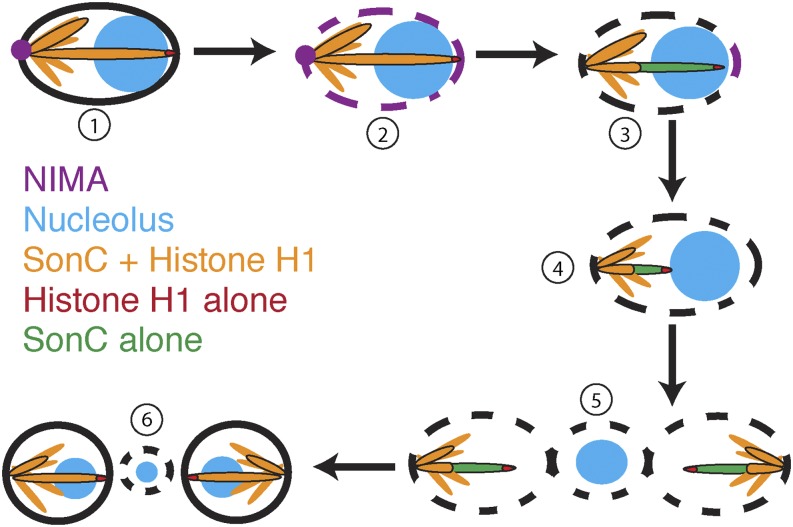
Although histone H1 is removed from the NOR during prophase, it still localizes distinctly to a focus at the distal end of the nuclear chromatin projection, and this domain resides outside of the nucleolus. This pattern is consistent with this region representing the telomere proximal region of chromosome V. Intriguingly, SonC is excluded from this H1 focus. In practical terms, the identification of SonC and its location to the NOR during mitosis at a stage when histone H1 is removed from the NOR has allowed us to more directly monitor NOR mitotic behavior, which we were unable to do previously using H1 as a general chromatin marker. However, at this time it is not clear why SonC is globally excluded from the telomere proximal region of chromosome V yet present along the NOR. One possibility is that we can observe this exclusion from part of the right arm of chromosome V because this arm is physically stretched across the nucleus from its centromere region anchored at the SPBs into and through the nucleolus. It therefore remains formally possible that SonC is similarly excluded from the telomere regions of other chromosome arms, but because these arms are not physically separated from the main chromatin mass, it is difficult to distinguish this exclusion. Why SonC is excluded globally from the end region of at least one chromosome arm remains an interesting unanswered question. Equally interesting questions remain regarding how mutation of sonC is able to suppress the nimA1 mutation and allow some mitotic progression without normal NIMA function and also cause temperature-dependent DNA damage sensitivity. Because SonC is excluded in a selective manner from large-scale chromatin domains evident during mitosis, we speculate that the spatial regulation of mitotic events by NIMA involves SonC (Figure 9). During mitotic initiation, NIMA locates first to SPBs to promote their separation, allowing spindle formation and also local NPC disassembly (Shen and Osmani 2013). NIMA then transitions around the nuclear periphery, locating finally to a cluster of NPCs adjacent to the nucleolus, which we here show likely associates with the telomere region of the chromosome arm containing the NOR (Figure 8 and Figure 9). It is likely that sequential triggering of events around the nuclear periphery starting at SPBs and ending near the nuclear periphery adjacent to the nucleolus helps to orient and to order mitotic events. For example, the release of the nucleolus from the NOR and from the nuclear periphery might be coordinated with spindle formation and chromosome condensation, and the sequential progression of NIMA to different mitotic-specific locations could provide a mechanism by which such coordination might be achieved. In further support of this, recent partial NIMA inactivation studies have shown that NIMA is required sequentially for successful completion of stage-specific mitotic events (Govindaraghavan et al. 2013). Because SonC locates to chromosome domains that have specific locations within the 3D structure of the nucleus, and because a specific sonC mutation can suppress partial NIMA function, it is tempting to speculate that NIMA and SonC interact, perhaps to help orchestrate chromatin states during mitosis and the DNA damage response.
Figure 9.
Model of mitotic events coordinated by NIMA and SonC. NIMA first localizes to the spindle pole body (1) and then spreads around the nuclear periphery, leading to the partial disassembly of NPCs, until it locates to an NPC cluster adjacent to the nucleolus (2). The left arm of chromosome V becomes detached from the nuclear periphery, and histone H1 is evicted from the NOR (3). As chromatin condenses, the NOR is pulled out of the nucleolus (4). Once the NOR is pulled out of the nucleolus, the nucleolus is separated from the daughter nuclei and from the cytoplasm by the nuclear envelope (5). As nuclei enter G1, NPCs are reassembled, and the nucleolus is disassembled and then reassembled into two daughter nuclei (6).
Future identification of the other proteins isolated from our genetic screen should provide additional insights into how SonC and NIMA are involved in the DNA damage response and in the regulation of mitosis.
Supplementary Material
Acknowledgments
We thank Kelly Tatchell (Louisiana State University Health Sciences Center) for critically reading the manuscript and members of the Osmani lab for assistance and for insightful discussions. This work was supported by a National Institutes of Health (NIH) National Research Service Award (T32CA106196) and an American Heart Association postdoctoral fellowship (to J.R.L.) and a grant from the NIH (GM042564) (to S.A.O.).
Footnotes
Communicating editor: O. Cohen-Fix
Literature Cited
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., et al. , 2002. Molecular Biology of the Cell, Ed. 4 Garland Science, New York. [Google Scholar]
- Alseth I., Osman F., Korvald H., Tsaneva I., Whitby M. C., et al. , 2005. Biochemical characterization and DNA repair pathway interactions of Mag1-mediated base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 33: 1123–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud M. B., Cerqueira G. C., Inglis D. O., Skrzypek M. S., Binkley J., et al. , 2012. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40: D653–D659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu J. R., Jeganathan K. B., Baker D. J., Wu X., Kang-Decker N., et al. , 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer S. M., Siniossoglou S., Podtelejnikov A., Hellwig A., Mann M., et al. , 1998. Nup116p and nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor gle2p. EMBO J. 17: 1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur E., Elkind M. M., Bronk B. V., 1974. Thermally enhanced radioresponse of cultured Chinese hamster cells: inhibition of repair of sublethal damage and enhancement of lethal damage. Radiat. Res. 58: 38–51. [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A. W., 2002. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell 13: 632–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M. D., Nachury M., Heald R., Weis K., 2005. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell 121: 223–234. [DOI] [PubMed] [Google Scholar]
- Boisvert F. M., Van Koningsbruggen S., Navascues J., Lamond A. I., 2007. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8: 574–585. [DOI] [PubMed] [Google Scholar]
- Borrow J., Shearman A. M., Stanton V. P., Jr, Becher R., Collins T., et al. , 1996. The t(7;11)(p15;p15) translocation in acute myeloid leukaemia fuses the genes for nucleoporin NUP98 and class I homeoprotein HOXA9. Nat. Genet. 12: 159–167. [DOI] [PubMed] [Google Scholar]
- Brody H., Carbon J., 1989. Electrophoretic karyotype of Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 86: 6260–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody H., Griffith J., Cuticchia A. J., Arnold J., Timberlake W. E., 1991. Chromosome-specific recombinant DNA libraries from the fungus Aspergillus nidulans. Nucleic Acids Res. 19: 3105–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. A., Bharathi A., Ghosh A., Whalen W., Fitzgerald E., et al. , 1995. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J. Biol. Chem. 270: 7411–7419. [DOI] [PubMed] [Google Scholar]
- Chen J., Derfler B., Maskati A., Samson L., 1989. Cloning a eukaryotic DNA glycosylase repair gene by the suppression of a DNA repair defect in Escherichia coli. Proc. Natl. Acad. Sci. USA 86: 7961–7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Derfler B., Samson L., 1990. Saccharomyces cerevisiae 3-methyladenine DNA glycosylase has homology to the AlkA glycosylase of E. coli and is induced in response to DNA alkylation damage. EMBO J. 9: 4569–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen P. L., Chen C. F., Jiang X., Riley D. J., 2008. Never-in-mitosis related kinase 1 functions in DNA damage response and checkpoint control. Cell Cycle 7: 3194–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck, A. J., and M. Farman, 2008 Aspergillus nidulans linkage map and genome sequence: closing gaps and adding telomeres in The Aspergilli Genomics: Medical Aspects, Biotechnology, and Research Methods, edited by G. H. Goldman and S. A. Osmani. Taylor and Francis Group, Boca Raton, FL. [Google Scholar]
- Cole C., Barber J. D., Barton G. J., 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36: W197–W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau D. L., Bohr V. A., 1997. Repair of oxidative damage to nuclear and mitochondrial DNA in mammalian cells. J. Biol. Chem. 272: 25409–25412. [DOI] [PubMed] [Google Scholar]
- David H., Ozcelik I. S., Hofmann G., Nielsen J., 2008. Analysis of Aspergillus nidulans metabolism at the genome-scale. BMC Genomics 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani S. A., 2007. Mitosis, not just open or closed. Eukaryot. Cell 6: 1521–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani S. A., 2009. Double duty for nuclear proteins: the price of more open forms of mitosis. Trends Genet. 25: 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Wu L. P., Spotts J. L., Osmani S. A., 2000. Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102: 293–302. [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Horn K. P., Masker K., Osmani S. A., 2003. The SONB(NUP98) nucleoporin interacts with the NIMA kinase in Aspergillus nidulans. Genetics 165: 1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Osmani A. H., Hashmi S. B., Osmani S. A., 2004. Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14: 1973–1984. [DOI] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Horn K. P., Osmani S. A., 2006. A point mutation in the Aspergillus nidulans sonBNup98 nuclear pore complex gene causes conditional DNA damage sensitivity. Genetics 174: 1881–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Nayak T., Oakley B., Osmani S. A., 2009. Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol. Biol. Cell 20: 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P., Hashmi S. B., Yang X., Osmani S. A., 2011. Regulated inactivation of the spindle assembly checkpoint without functional mitotic spindles. EMBO J. 30: 2648–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey W. C., Hopwood L. E., Sapareto S. A., Gerweck L. E., 1977. Cellular responses to combinations of hyperthermia and radiation. Radiology 123: 463–474. [DOI] [PubMed] [Google Scholar]
- Dimario P. J., 2004. Cell and molecular biology of nucleolar assembly and disassembly. Int. Rev. Cytol. 239: 99–178. [DOI] [PubMed] [Google Scholar]
- Freeman L., Aragon-Alcaide L., Strunnikov A., 2000. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell Biol. 149: 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry A. M., O’Regan L., Sabir S. R., Bayliss R., 2012. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125: 4423–4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funasaka T., Nakano H., Wu Y., Hashizume C., Gu L., et al. , 2011. RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle 10: 1456–1467. [DOI] [PubMed] [Google Scholar]
- Galagan J. E., Calvo S. E., Cuomo C., Ma L. J., Wortman J. R., et al. , 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Gough S. M., Slape C. I., Aplan P. D., 2011. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood 118: 6247–6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraghavan M., Lad A. A., Osmani S. A., 2013. The NIMA kinase is required to execute stage specific mitotic functions after initiation of mitosis. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24186954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill B., Chen L., Tulgren E. D., Baker S. T., Bienvenut W., et al. , 2012. RAE-1, a novel PHR binding protein, is required for axon termination and synapse formation in Caenorhabditis elegans. J. Neurosci. 32: 2628–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiyama L. M., Da Cunha J. P., Schenkman S., 2008. Histone H1 of Trypanosoma cruzi is concentrated in the nucleolus region and disperses upon phosphorylation during progression to mitosis. Eukaryot. Cell 7: 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habash R. W., Krewski D., Bansal R., Alhafid H. T., 2011. Principles, applications, risks and benefits of therapeutic hyperthermia. Front. Biosci. (Elite Ed.) 3: 1169–1181. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D., 2011. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus 2: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeganathan K. B., Malureanu L., Van Deursen J. M., 2005. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438: 1036–1039. [DOI] [PubMed] [Google Scholar]
- Kalashnikova A. A., Winkler D. D., Mcbryant S. J., Henderson R. K., Herman J. A., et al. , 2013. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus. Nucleic Acids Res. 41: 4026–4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk P. M., Eppink B., Essers J., Stap J., Rodermond H., et al. , 2011. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc. Natl. Acad. Sci. USA 108: 9851–9856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Thorsteinsdottir U., Mayotte N., Nakamura T., Sauvageau G., 2001. NUP98-HOXA9 expression in hemopoietic stem cells induces chronic and acute myeloid leukemias in mice. EMBO J. 20: 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., 1984. Mutational specificity of depurination. Proc. Natl. Acad. Sci. USA 81: 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., et al. , 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144: 539–550. [DOI] [PubMed] [Google Scholar]
- Lavoie B. D., Hogan E., Koshland D., 2002. In vivo dissection of the chromosome condensation machinery: reversibility of condensation distinguishes contributions of condensin and cohesin. J. Cell Biol. 156: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Hogan E., Koshland D., 2004. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 18: 76–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B. D., Tuffo K. M., Oh S., Koshland D., Holm C., 2000. Mitotic chromosome condensation requires Brn1p, the yeast homologue of Barren. Mol. Biol. Cell 11: 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., 1994. A zinc finger protein, essential for chromosome segregation, constitutes a putative DNA binding subunit of the Saccharomyces cerevisiae kinetochore complex, Cbf3. EMBO J. 13: 5203–5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., et al. , 2004. Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166: 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L., De Souza C. P., Osmani A. H., Osmani S. A., 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84–120 complex. Mol. Biol. Cell 20: 616–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. L., Osmani A. H., Ukil L., Son S., Markossian S., et al. , 2010. Single-step affinity purification for fungal proteomics. Eukaryot. Cell 9: 831–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Ho C. K., Ouyang J., Zou L., 2013. Nek1 kinase associates with ATR-ATRIP and primes ATR for efficient DNA damage signaling. Proc. Natl. Acad. Sci. USA 110: 2175–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K. P., Hunter T., 1995. Evidence for a NIMA-like mitotic pathway in vertebrate cells. Cell 81: 413–424. [DOI] [PubMed] [Google Scholar]
- Lu K. P., Means A. R., 1994. Expression of the noncatalytic domain of the NIMA kinase causes a G2 arrest in Aspergillus nidulans. EMBO J. 13: 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M., 2011. Physiological relevance of cell cycle kinases. Physiol. Rev. 91: 973–1007. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Sambrook J., Fritsch E. F., 1989. Molecular Cloning: A Laboratory Manual. Cold Srping Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Melixetian M., Klein D. K., Sorensen C. S., Helin K., 2009. NEK11 regulates CDC25A degradation and the IR-induced G2/M checkpoint. Nat. Cell Biol. 11: 1247–1253. [DOI] [PubMed] [Google Scholar]
- Mi J., Guo C., Brautigan D. L., Larner J. M., 2007. Protein phosphatase-1alpha regulates centrosome splitting through Nek2. Cancer Res. 67: 1082–1089. [DOI] [PubMed] [Google Scholar]
- Miles T. D., Jakovljevic J., Horsey E. W., Harnpicharnchai P., Tang L., et al. , 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 25: 10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz L. S., Stambolic V., 2011. Nek10 mediates G2/M cell cycle arrest and MEK autoactivation in response to UV irradiation. Mol. Cell. Biol. 31: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz L., Dutt P., Haider N., Stambolic V., 2011. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. A., Chung K. Y., Plasilova M., Schuringa J. J., Shieh J. H., et al. , 2007. NUP98 dysregulation in myeloid leukemogenesis. Ann. N. Y. Acad. Sci. 1106: 114–142. [DOI] [PubMed] [Google Scholar]
- Morris N. R., 1975. Mitotic mutants of Aspergillus nidulans. Genet. Res. 26: 237–254. [DOI] [PubMed] [Google Scholar]
- Morris N. R., James S. W., O’Connell M. J., 1992. Mitotic regulation in Aspergillus nidulans. Ciba Found. Symp. 170: 115–123; discussion 123–129. [DOI] [PubMed] [Google Scholar]
- Moura D. J., Castilhos B., Immich B. F., Canedo A. D., Henriques J. A., et al. , 2010. Kin3 protein, a NIMA-related kinase of Saccharomyces cerevisiae, is involved in DNA adduct damage response. Cell Cycle 9: 2220–2229. [DOI] [PubMed] [Google Scholar]
- Moy T. I., Boettner D., Rhodes J. C., Silver P. A., Askew D. S., 2002. Identification of a role for Saccharomyces cerevisiae Cgr1p in pre-rRNA processing and 60S ribosome subunit synthesis. Microbiology 148: 1081–1090. [DOI] [PubMed] [Google Scholar]
- Nassirpour R., Shao L., Flanagan P., Abrams T., Jallal B., et al. , 2010. Nek6 mediates human cancer cell transformation and is a potential cancer therapeutic target. Mol. Cancer Res. 8: 717–728. [DOI] [PubMed] [Google Scholar]
- Nayak T., Szewczyk E., Oakley C. E., Osmani A., Ukil L., et al. , 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak T., Edgerton-Morgan H., Horio T., Xiong Y., De Souza C. P., et al. , 2010. Gamma-tubulin regulates the anaphase-promoting complex/cyclosome during interphase. J. Cell Biol. 190: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K., Fukazawa H., Murakami Y., Uehara Y., 2002. Nek11, a new member of the NIMA family of kinases, involved in DNA replication and genotoxic stress responses. J. Biol. Chem. 277: 39655–39665. [DOI] [PubMed] [Google Scholar]
- Noguchi K., Fukazawa H., Murakami Y., Uehara Y., 2004. Nucleolar Nek11 is a novel target of Nek2A in G1/S-arrested cells. J. Biol. Chem. 279: 32716–32727. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R., 1980. Nuclear movement is beta–tubulin-dependent in Aspergillus nidulans. Cell 19: 255–262. [DOI] [PubMed] [Google Scholar]
- Oakley B. R., Morris N. R., 1983. A mutation in Aspergillus nidulans that blocks the transition from interphase to prophase. J. Cell Biol. 96: 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. J., Norbury C., Nurse P., 1994. Premature chromatin condensation upon accumulation of NIMA. EMBO J. 13: 4926–4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell M. J., Krien M. J., Hunter T., 2003. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13: 221–228. [DOI] [PubMed] [Google Scholar]
- Ong T. M., Serres F. J., 1975. Mutation induction by difunctional alkylating agents in Neurospora crassa. Genetics 80: 475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherov N., May G., 2000. Conidial germination in Aspergillus nidulans requires RAS signaling and protein synthesis. Genetics 155: 647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., McGuire S. L., O’Donnell K. L., Pu R. T., Osmani S. A., 1991. Role of the cell-cycle-regulated NIMA protein kinase during G2 and mitosis: evidence for two pathways of mitotic regulation. Cold Spring Harb. Symp. Quant. Biol. 56: 549–555. [DOI] [PubMed] [Google Scholar]
- Osmani A. H., Davies J., Liu H. L., Nile A., Osmani S. A., 2006a Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol. Biol. Cell 17: 4946–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani A. H., Oakley B. R., Osmani S. A., 2006b Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 1: 2517–2526. [DOI] [PubMed] [Google Scholar]
- Osmani S. A., May G. S., Morris N. R., 1987. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J. Cell Biol. 104: 1495–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani S. A., Pu R. T., Morris N. R., 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53: 237–244. [DOI] [PubMed] [Google Scholar]
- Park H. S., Yu J. H., 2012. Multi-copy genetic screen in Aspergillus nidulans. Methods Mol. Biol. 944: 183–190. [DOI] [PubMed] [Google Scholar]
- Pel H. J., De Winde J. H., Archer D. B., Dyer P. S., Hofmann G., et al. , 2007. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 25: 221–231. [DOI] [PubMed] [Google Scholar]
- Pelegrini A. L., Moura D. J., Brenner B. L., Ledur P. F., Maques G. P., et al. , 2010. Nek1 silencing slows down DNA repair and blocks DNA damage-induced cell cycle arrest. Mutagenesis 25: 447–454. [DOI] [PubMed] [Google Scholar]
- Pestov D. G., Stockelman M. G., Strezoska Z., Lau L. F., 2001. ERB1, the yeast homolog of mammalian Bop1, is an essential gene required for maturation of the 25S and 5.8S ribosomal RNAs. Nucleic Acids Res. 29: 3621–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes T. D., 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA 76: 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polci R., Peng A., Chen P. L., Riley D. J., Chen Y., 2004. NIMA-related protein kinase 1 is involved early in the ionizing radiation-induced DNA damage response. Cancer Res. 64: 8800–8803. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G., Roper J. A., Hemmons L. M., Macdonald K. D., Bufton A. W., 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5: 141–238. [DOI] [PubMed] [Google Scholar]
- Prakash L., Prakash S., 1977. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics 86: 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C. E., Fornerod M., Kasper L. H., Van Deursen J. M., 1999. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J. Cell Biol. 145: 237–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu R. T., Osmani S. A., 1995. Mitotic destruction of the cell cycle regulated NIMA protein kinase of Aspergillus nidulans is required for mitotic exit. EMBO J. 14: 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu R. T., Xu G., Wu L., Vierula J., O’Donnell K., et al. , 1995. Isolation of a functional homolog of the cell cycle-specific NIMA protein kinase of Aspergillus nidulans and functional analysis of conserved residues. J. Biol. Chem. 270: 18110–18116. [DOI] [PubMed] [Google Scholar]
- Quarmby L. M., Mahjoub M. R., 2005. Caught Nek-ing: cilia and centrioles. J. Cell Sci. 118: 5161–5169. [DOI] [PubMed] [Google Scholar]
- Rajani K. R., Pettit Kneller E. L., McKenzie M. O., Horita D. A., Chou J. W., et al. , 2012. Complexes of vesicular stomatitis virus matrix protein with host Rae1 and Nup98 involved in inhibition of host transcription. PLoS Pathog. 8: e1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Seo H. S., Blobel G., Hoelz A., 2010. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl. Acad. Sci. USA 107: 10406–10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K. F., Osmani S. A., 2013. Regulation of mitosis by the NIMA kinase involves TINA and its newly discovered partner An-WDR8 at spindle pole bodies. Available at: http://www.ncbi.nlm.nih.gov/pubmed/24152731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen C. S., Melixetian M., Klein D. K., Helin K., 2010. NEK11: linking CHK1 and CDC25A in DNA damage checkpoint signaling. Cell Cycle 9: 450–455. [DOI] [PubMed] [Google Scholar]
- Strezoska Z., Pestov D. G., Lau L. F., 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S RRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 20: 5516–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Boettner D., Huebner N., Xu F., Rhodes J. C., et al. , 2001. Molecular cloning of cgrA, the gene encoding the Aspergillus nidulans ortholog of Saccharomyces cerevisiae CGR1. Curr. Microbiol. 42: 403–407. [DOI] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C. E., Edgerton H., Xiong Y., et al. , 2006. Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1: 3111–3120. [DOI] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Carmo-Fonseca M., Hurt E. C., 1991. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 10: 573–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Jarmuz A., Aragon L., 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network. Cell Cycle 3: 496–502. [PubMed] [Google Scholar]
- Tsunoda N., Kokuryo T., Oda K., Senga T., Yokoyama Y., et al. , 2009. Nek2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 100: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukil L., Varadaraj A., Govindaraghavan M., Liu H. L., Osmani S. A., 2008. Copy number suppressors of the Aspergillus nidulans nimA1 mitotic kinase display distinctive and highly dynamic cell cycle-regulated locations. Eukaryot. Cell 7: 2087–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukil L., De Souza C. P., Liu H. L., Osmani S. A., 2009. Nucleolar separation from chromosomes during Aspergillus nidulans mitosis can occur without spindle forces. Mol. Biol. Cell 20: 2132–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velichko A. K., Petrova N. V., Kantidze O. L., Razin S. V., 2012. Dual effect of heat shock on DNA replication and genome integrity. Mol. Biol. Cell 23: 3450–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R. W., Blobel G., Coutavas E., 2006. Rae1 interaction with NuMA is required for bipolar spindle formation. Proc. Natl. Acad. Sci. USA 103: 19783–19787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortman J. R., Gilsenan J. M., Joardar V., Deegan J., Clutterbuck J., et al. , 2009. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 46(Suppl. 1): S2–S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Osmani S. A., Mirabito P. M., 1998. A role for NIMA in the nuclear localization of cyclin B in Aspergillus nidulans. J. Cell Biol. 141: 1575–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ukil L., Osmani A., Nahm F., Davies J., et al. , 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3: 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X. S., Xu G., Pu R. T., Fincher R. R., McGuire S. L., et al. , 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14: 986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X. S., Fincher R. R., Tang A., O’Donnell K., Osmani S. A., 1996. Two S-phase checkpoint systems, one involving the function of both BIME and Tyr15 phosphorylation of p34cdc2, inhibit NIMA and prevent premature mitosis. EMBO J. 15: 3599–3610. [PMC free article] [PubMed] [Google Scholar]
- Ye X. S., Fincher R. R., Tang A., Osmani A. H., Osmani S. A., 1998. Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA. Mol. Biol. Cell 9: 3019–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Chen H. W., Mu R. L., Zhang W. K., Zhao M. Y., et al. , 2011. NIMA-related kinase NEK6 affects plant growth and stress response in Arabidopsis. Plant J. 68: 830–843. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.