Abstract
Neurochemicals are likely to play key roles in physiological/behavioral control in the copepod crustacean Calanus finmarchicus, the biomass dominant zooplankton for much of the North Atlantic Ocean. Previously, a de novo assembled transcriptome consisting of 206,041 unique sequences was used to characterize the peptidergic signaling systems of Calanus. Here, this assembly was mined for transcripts encoding enzymes involved in amine biosynthesis. Using known Drosophila melanogaster proteins as templates, transcripts encoding putative Calanus homologs of tryptophan-phenylalanine hydroxylase (dopamine, octopamine and serotonin biosynthesis), tyrosine hydroxylase (dopamine biosynthesis), DOPA decarboxylase (dopamine and serotonin biosynthesis), histidine decarboxylase (histamine biosynthesis), tyrosine decarboxylase (octopamine biosynthesis), tyramine β-hydroxylase (octopamine biosynthesis) and tryptophan hydroxylase (serotonin biosynthesis) were identified. Reverse BLAST and domain analyses show that the proteins deduced from these transcripts possess sequence homology to and the structural hallmarks of their respective enzyme families. Developmental profiling revealed a remarkably consistent pattern of expression for all transcripts, with the highest levels of expression typically seen in the early nauplius and early copepodite. These expression patterns suggest roles for amines during development, particularly in the metamorphic transitions from embryo to nauplius and from nauplius to copepodite. Taken collectively, the data presented here lay a strong foundation for future gene-based studies of aminergic signaling in this and other copepod species, in particular assessment of the roles they may play in developmental control.
Keywords: tryptophan-phenylalanine hydroxylase (TPH), tyrosine hydroxylase (TH), DOPA decarboxylase (DDC), histidine decarboxylase (HDC), tyrosine decarboxylase (TDC), tyramine β-hydroxylase (TβH), tryptophan hydroxylase (TRH), Illumina sequencing, functional genomics
1. Introduction
One major class of molecules used by nervous systems for chemical communication are amines. In crustaceans, four amines are generally recognized, dopamine, histamine, octopamine and serotonin (Christie, 2011), each of which is produced via the enzymatic processing of an amino acid substrate. While regulation of aminergic signaling systems can occur at many levels, modulation of the transcription, translation, and degradation of the enzymes involved in their biosynthesis is a major component in their control.
In arthropods, both dopamine and octopamine are produced from the amino acid phenylalanine in a three-step process (e.g. Coleman and Neckameyer, 2005; Monastirioti, 1999). The first step in the biosynthesis of both of these amines involves the enzymatic conversion of phenylalanine to tyrosine via the action of tryptophan-phenylalanine hydroxylase (TPH). To produce dopamine, tyrosine is subsequently converted to L-3,4-dihydroxyphenylalanine (L-DOPA) via the action of tyrosine hydroxylase (TH), and then to dopamine by DOPA decarboxylase (DDC). For the production of octopamine, tyrosine is converted by tyrosine decarboxylase (TDC) to tyramine, which in turn is converted to octopamine via the action of tyramine β-hydroxylase (TβH). To produce histamine, the amino acid histidine is decarboxylated via a reaction catalyzed by histidine decarboxylase (HDC) (e.g. Stuart, 1999). The amino acid tryptophan is the initial substrate for the production of serotonin. This amino acid is converted to 5-hydroxytryptophan via either TPH or tryptophan hydroxylase (TRH), which in turn is converted to serotonin by DDC (e.g. Coleman and Neckameyer, 2005; Monastirioti, 1999).
As part of an ongoing effort to identify and characterize the neurochemical signaling systems of the copepod crustacean Calanus finmarchicus (Christie et al., 2013a, 2013b), the biomass dominant zooplankton for much of the North Atlantic Ocean (Dale et al., 2001; Marshall and Orr, 1955; Meise and O’Reilly, 1996), we have mined a de novo assembled transcriptome for sequences encoding the enzymes responsible for the generation of dopamine, histamine, octopamine and serotonin. This study complements and augments an earlier report describing transcripts/proteins that contribute to peptidergic signaling in this ecologically important species (Christie et al., 2013a). As our data will show, C. finmarchicus transcripts putatively encoding TPH, TH, DDC, TDC, TβH, HDC, and TRH were identified. Reverse BLAST and domain analyses of the proteins deduced from the identified transcripts show that they possess sequence homology to and the structural hallmarks of their respective enzyme families. In addition, RNA-Seq profiling of the identified transcripts across C. finmarchicus development (embryo, early nauplius, late nauplius, early copepodite, late copepodite, and adult), revealed peaks in expression in the early nauplius and early copepodite stages, which suggests the amines may play roles in the metamorphic transitions between embryo/nauplius and nauplius/copepodite in this species. Collectively, the data presented in this study not only provide the first descriptions of amine biosynthetic enzymes in C. finmarchicus, but also lay a strong foundation for future gene-based studies of aminergic signaling in this species, including possible neurochemical roles for the amines in metamorphic control.
2. Materials and methods
2.1 De novo transcriptome assembly
A de novo transcriptome for C. finmarchicus was generated as described in detail in Christie et al. (2013a). In brief, multiplexed gene libraries were prepared from RNA extracted from six developmental stages of wild-caught or laboratory cultured C. finmarchicus: egg (which represents a mixture of embryonic stages; cultured), early nauplius (stages NI and NII; cultured), late nauplius (stages NV and NVI; cultured), early copepodite (stages CI and CII; cultured), late copepodite (stage CV; wild-caught) and adult female (wild-caught). All libraries were sequenced at the HudsonAlpha Institute for Biotechnology (Huntsville, AL, USA) in a single lane using an Illumina HiSeq 2000 instrument (Illumina Inc.). In total, 415,469,690 raw, 100 base pair (bp), paired-end reads were obtained. After quality filtering and trimming, the combined raw reads from all six developmental stage libraries were assembled de novo using Trinity 2012-03-17-IU_ZIH_TUNED software (Grabherr et al., 2011) on a node of the National Center for Genome Analysis Support’s (NCGAS; Indiana University, Bloomington, IN, USA) Mason Linux cluster. In total, 206,041 unique nucleotide sequences >300 bp in length were generated using Trinity.
2.2. Transcriptome mining
Searches of the transcriptome assembly produced by Trinity were conducted using the DeCypher Tera-BLASTP algorithm on the Mount Desert Island Biological Laboratory’s TimeLogic DeCypher server (MDIBL, Salisbury Cove, ME, USA; http://decypher.mdibl.org/decypher/algo-tera-blast/tera-tblastn_an.shtml) as described in several recent publications (Christie et al., 2013a, 2013b). For all searches, the DeCypher program database was set to the combined Trinity assembly and a known fruit fly Drosophila melanogaster protein was used as the query sequence. All hits were translated (Supplemental Figure 1) and checked manually for homology to the query protein. Table 1 provides the BLAST-generated E-value (the number of alignments expected by chance that have the same score as the alignment) for each hit that was identified as encoding a putative target transcript, as well as the length of identified transcripts; the length of the protein deduced from each target sequence is also provided in this table.
Table 1.
Putative Calanus finmarchicus amine biosynthetic enzyme-encoding transcripts and proteins identified via in silico transcriptome mining
| Query protein | Calanus transcript/protein identifications | |||||
|---|---|---|---|---|---|---|
| Transcripts | Proteins | |||||
| Trinity ID Number | Name | E-value | Length* | Name | Length† | |
| Tryptophan-phenylalanine hydroxylase1,3,4 | comp34563_c3_seq1 | calfi-tph | 2.9e-175 | 1531 | Calfi-TPH | 443 |
| Tyrosine hydroxylase1 | comp541083_c3_seq1 | calfi-th | 5.1e-139 | 1932 | Calfi-TH | 516 |
| DOPA decarboxylase1,4 | comp374436_c0_seq5 | calfi-ddc | 1.7e-168 | 1560 | Calfi-DDC | 487 |
| Histidine decarboxylase2 | comp175390_c0_seq2 | calfi-hdc | 2.3e-230 | 2659 | Calfi-HDC | 724 |
| Tyrosine decarboxylase3 | comp374436_c0_seq1 | calfi-tdc | 6.7e-154 | 2127 | Calfi-TDC | 617 |
| Tyramine β-hydroxylase3 | comp975856_c0_seq1 | calfi-tβh | 4.1e-110 | 2122 | Calfi-TβH | 672 |
| Tryptophan hydroxylase4 | comp441733_c0_seq2 | calfi-trh | 2.8e-133 | 1976 | Calfi-TRH | 541 |
Length in nucleotides
Length in amino acids
Dopamine biosynthesis
Histamine biosynthesis
Octopamine biosynthesis
Serotonin biosynthesis
2.3. Analyses of protein conservation and structure
Analyses of protein conservation and structure were conducted using a workflow described in Christie et al. (2013a, 2013b). To identify the proteins most similar to each of the C. finmarchicus amine biosynthetic enzymes identified in this study, the deduced Calanus sequence was used to query the annotated protein dataset present in FlyBase (version FB2013_01), as well as the non-redundant arthropod protein dataset (taxid:6656) curated in GenBank (excluding C. finmarchicus proteins, obvious partial proteins, synthetic constructs, and provisional/unannotated hypothetical protein sequences); for both searches the blastp algorithm was used (Altschul et al., 1997). The results of these searches, including BLAST scores (the similarity between the queried sequence and the found sequence) and E-values (see Section 2.2 for description), are summarized in Tables 2 and 3.
Table 2.
BLAST analyses* of putative Calanus finmarchicus amine biosynthetic enzymes vs. all annotated Drosophila melanogaster proteins in FlyBase
| Calanus query | Top Drosophila protein hit | |||
|---|---|---|---|---|
| FlyBase No. | Protein description | BLAST score | E-value | |
| Calfi-TPH | FBpp0306707 | Henna†, isoform C | 629 | 3e-180 |
| Calfi-TH | FBpp0076665 | Pale‡, isoform A | 482 | 5e-136 |
| Calfi-DDC | FBpp0288730 | DOPA decarboxylase, isoform D | 593 | 2e-169 |
| Calfi-HDC | FBpp0304256 | Histidine decarboxylase, isoform B | 833 | 0.0 |
| Calfi-TDC | FBpp0085475 | Tyrosine decarboxylase 2, isoform A | 734 | 0.0 |
| Calfi-TβH | FBpp0089042 | Tyramine β-hydroxylase, isoform B | 385 | 1e-106 |
| Calfi-TRH | FBpp0072534 | Tryptophan hydroxylase, isoform A | 498 | 1e-140 |
All searches conducted on or before June 1, 2013.
A synonym for tryptophan-phenylalanine hydroxylase.
A synonym for tyrosine hydroxylase.
Abbreviations: Calfi-TPH, Calanus finmarchicus tryptophan-phenylalanine hydroxylase; Calfi-TH, Calanus finmarchicus tyrosine hydroxylase; Calfi-DDC, Calanus finmarchicus DOPA decarboxylase; Calfi-HDC, Calanus finmarchicus histidine decarboxylase; Calfi-TDC, Calanus finmarchicus tyrosine decarboxylase; Calfi-TβH, Calanus finmarchicus tyramine β-hydroxylase; Calfi-TRH, Calanus finmarchicus tryptophan hydroxylase.
Table 3.
blastp analyses* of putative Calanus finmarchicus amine biosynthetic enzymes vs. all GenBank curated non-redundant arthropod proteins†
|
Calanus Query |
Top 10 blastp arthropod protein hits | ||||
|---|---|---|---|---|---|
| Accession No. |
Species | Protein description | BLAST score |
E- value |
|
| Calfi-TPH |
AAM28250 AAT78350 EFR29984 BAJ83477 EFZ21127 ADM33594 BAE66652 EHJ64587 AFL02790 EFN61629 |
Aedes aegypti Armigeres subalbatus Anopheles darlingi Gryllus bimaculatus Solenopsis invicta Bombyx mori Papilio xuthus Danaus plexippus Polyrhachis vicina Camponotus floridanus |
Phenylalanine hydroxylase Phenylalanine hydroxylase Hypothetical protein AND_000708 Phenylalanine-tryptophan hydroxylase Hypothetical protein SINV_04627 Phenylalanine hydroxylase Phenylalanine hydroxylase Phenylalanine hydroxylase Phenylalanine hydroxylase Protein henna |
646 645 643 643 642 642 640 639 637 632 |
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 |
| Calfi-TH |
ABQ95974 CAA53802 AAW47987 EEB18101 EFX75078 ACU77882 BAJ07593 BAJ07587 BAE43824 BAF32574 |
Tribolium castaneum Drosophila melanogaster Apis mellifera Pediculus humanus corporis Daphnia pulex Tenebrio molitor Papilio polytes Papilio machaon Papilio xuthus Mythimna separata |
Tyrosine hydroxylase Pale, isoform A Tyrosine hydroxylase Tyrosine 3-monooxygenase Hypothetical protein DAPPUDRAFT_323742 Tyrosine hydroxylase Tyrosine hydroxylase Tyrosine hydroxylase Tyrosine hydroxylase Tyrosine hydroxylase type 2, brain form |
518 515 514 514 513 512 495 494 493 489 |
1e-177 8e-177 1e-176 5e-176 5e-176 3e-175 4e-168 5e-168 2e-167 1e-166 |
| Calfi- DDC |
BAJ83478 BAJ07594 BAJ07588 BAE43825 BAB68545 AAR23825 BAB68549 EFA03481 ABU25222 AAK48988 |
Gryllus bimaculatus Papilio polytes Papilio machaon Papilio xuthus Mamestra brassicae Antheraea pernyi Mythimna separata Tribolium castaneum Tribolium castaneum Bombyx mori |
Aromatic L-amino-acid decarboxylase DOPA decarboxylase DOPA decarboxylase DOPA decarboxylase DOPA decarboxylase DOPA-decarboxylase DOPA decarboxylase Hypothetical protein TcasGA2_TC 13480 DOPA decarboxylase DOPA decarboxylase |
650 647 645 645 644 640 640 638 637 636 |
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 |
| Calfi- HDC |
EFX79676 EDS34715 EAA14857 EFN88410 EGI58389 EFZ20538 EDX06463 AGB93389 CAA49989 EDW32340 |
Daphnia pulex Culex quinquefasciatus Anopheles gambiae Harpegnathos saltator Acromyrmex echinatior Solenopsis invicta Drosophila simulans Drosophila melanogaster Drosophila melanogaster Drosophila persimilis |
Hypothetical protein D APPUDRAFT_304481 Aromatic amino acid decarboxylase AGAP009001-AP Histidine decarboxylase Histidine decarboxylase Hypothetical protein SINV_06729 GD10721 Histidine decarboxylase, isoform C Histidine decarboxylase GL11582 |
876 865 854 848 847 838 842 832 840 839 |
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 |
| Calfi-TDC |
EEB13798 EFA10348 EDS34186 EAT45251 EDW89072 AAM70812 EDV38125 EDV59802 EDW61660 EAL24715 |
Pediculus humanus corporis Tribolium castaneum Culex quinquefasciatus Aedes aegypti Drosophila yakuba Drosophila melanogaster Drosophila ananassae Drosophila erecta Drosophila virilis Drosophila pseudoobscura pseudoobscura |
Aromatic-L-amino-acid decarboxylase Hypothetical protein TcasGA2_TC012567 Aromatic-L-amino-acid decarboxylase Aromatic-L-amino-acid- decarboxylase GE24598 Tyrosine decarboxylase 2 GF13800 GG10812 GJ20190 GA15851 |
791 791 772 766 764 764 764 764 763 761 |
0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 0.0 |
| Calfi-TβH |
EEB12867 AFO63077 AFO63079 AFO63076 AFO63080 EFA00428 EFZ13313 EFN89747 EFX67831 EGI65668 |
Pediculus humanus corporis Periplaneta americana Periplaneta americana Periplaneta americana Periplaneta americana Tribolium castaneum Solenopsis invicta Harpegnathos saltator Daphnia pulex Acromyrmex echinatior |
Dopamine beta-hydroxylase precursor Tyramine beta hydroxylase Tyramine beta hydroxylase Tyramine beta hydroxylase Tyramine beta hydroxylase Similar to tyramine beta hydroxylase Hypothetical protein SINV_01889 Dopamine beta-hydroxylase Hypothetical protein DAPPUDRAFT_1839 Tyramine beta-hydroxylase |
536 526 526 526 525 520 512 489 489 487 |
0.0 8e-178 1e-177 1e-177 4e-177 3e-175 8e-172 8e-164 2e-163 9e-163 |
| Calfi-TRH |
EFN85713 BAJ83476 EFN68680 EFA01002 BAK19479 EHJ65579 EEB18903 EAA11742 EFX86644 EDX08760 |
Harpegnathos saltator Gryllus bimaculatus Camponotus floridanus Tribolium castaneum Gryllus bimaculatus Danaus plexippus Pediculus humanus corporis Anopheles gambiae Daphnia pulex Drosophila simulans |
Tryptophan 5-hydroxylase Tryptophan hydroxylase Tryptophan 5-hydroxylase Similar to AGAP006020-PA Tryptophan hydroxylase long variant Hypothetical protein KGM_15172 Tryptophan 5-hydroxylase, putative AGAP006020-PA Hypothetical protein DAPPUDRAFT_44381 GD13494 |
551 543 526 522 523 514 507 509 496 498 |
0.0 0.0 4e-180 3e-179 2e-178 2e-175 1e-173 1e-173 7e-170 3e-169 |
All searches conducted on or before June 1, 2013.
Excluding C. finmarchicus proteins, obvious partial proteins, obvious provisional proteins and synthetic constructs. Crustacean hits are shown in red font.
Abbreviations: Calfi-TPH, Calanus finmarchicus tryptophan-phenylalanine hydroxylase; Calfi-TH, Calanus finmarchicus tyrosine hydroxylase; Calfi-DDC, Calanus finmarchicus DOPA decarboxylase; Calfi-HDC, Calanus finmarchicus histidine decarboxylase; Calfi-TDC, Calanus finmarchicus tyrosine decarboxylase; Calfi-TβH, Calanus finmarchicus tyramine β-hydroxylase; Calfi-TRH, Calanus finmarchicus tryptophan hydroxylase.
To determine amino acid identity/similarity between proteins, the sequences in question were aligned using MAFFT version 7 (http://align.bmr.kyushu-u.ak.jp/mafft/online/server/; Katoh and Standley, 2013), and amino acid identity/similarity was subsequently determined using the alignment output. Percent identity was calculated as the number of identical amino acids (denoted by “*” in the MAFFT output) divided by the total number of amino acids in the longest sequence (x100). Percent similarity was calculated as the number of identical and similar amino acids (the latter denoted by the “:” and “.” symbols in the protein alignment) divided by the total number of amino acids in the longest sequence (x100). In the MAFFT output “:” indicates that one of the following strong groups is fully conserved: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY or FYW (Kazutaka Katoh, personal communication). The “.” symbol in MAFFT indicates full conservation of one of the following weaker groups: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM or HFY (Kazutaka Katoh, personal communication).
Protein structural motifs were analyzed using the online program Pfam version 27.0 (http://pfam.sanger.ac.uk/; Punta et al., 2012). A common highlighting scheme has been used to denote functional domains in Figures 1 and 2 and Supplemental Figure 2: ACT, green; biopterindependent aromatic amino acid hydroxylase, yellow; pyridoxal-dependent decarboxylase conserved, blue; DOMON, pink; copper type-II ascorbate-dependent monooxygenase N-terminal, red; copper type-II ascorbate-dependent monooxygenase C-terminal, dark red.
Figure 1.
Deduced amino acid sequences of Calanus finmarchicus amine biosynthetic enzymes and their functional domains as predicted by the online program Pfam. (A) Calanus finmarchicus tryptophan-phenylalanine hydroxylase. (B) Calanus finmarchicus tyrosine hydroxylase. (C) Calanus finmarchicus DOPA decarboxylase. (D) Calanus finmarchicus histidine decarboxylase. (E) Calanus finmarchicus tyrosine decarboxylase. (F) Calanus finmarchicus tyramine β-hydroxylase. (G) Calanus finmarchicus tryptophan hydroxylase. Highlighting code for functional domains identified by Pfam: ACT, green; biopterin-dependent aromatic amino acid hydroxylase, yellow; pyridoxal-dependent decarboxylase conserved, blue; DOMON, pink; copper type-II ascorbate-dependent monooxygenase N-terminal, red; copper type-II ascorbate-dependent monooxygenase C-terminal, dark red.
Figure 2.
Alignment of Drosophila melanogaster tryptophan-phenylalanine hydroxylase (Drome-TPH; Accession No. CAA66797) with a putative Calanus finmarchicus homolog (Calfi-TPH) deduced from the Trinity de novo transcriptome assembly. In the line immediately below each sequence grouping, “*” indicates amino acids that are identical in the two proteins, while “.” and “:” denote amino acids that are similar in structure between the two sequences. In this figure, ACT and biopterin-dependent aromatic amino acid hydroxylase domains identified by Pfam analyses are highlighted in green and yellow, respectively.
2.4. Vetting of deduced protein sequences using publicly accessible expressed sequence tags
In an attempt to confirm the amino acid sequences of the proteins deduced from the Trinity assembly, each of the putative Calanus proteins was used as a query to search the extant C. finmarchicus ESTs (∼11,000 in total; Lenz et al., 2012) curated at GenBank using the tblastn algorithm; see Christie et al. (2013a, 2013b) for details.
2.5. Developmental expression mapping
To compare the relative levels of transcript expression in embryos, early nauplii, late nauplii, early copepodites, late copepodites, and adult females, Illumina reads from each of these developmental stages were mapped against the sequence in question using Bowtie software (Johns Hopkins University, Baltimore, MD, USA; http://bowtie-bio.sourceforge.net/index.shtml; Langmead et al., 2009) as described in Christie et al. (2013a, 2013c). For five stages, four biological replicates (one generated from animals collected in 2011 and three from material collected in 2012), with two technical replicates for each biological replicate, were aligned against each reference sequence independently. For one stage grouping, early copepodites, we had three biological replicates (one from 2011 and two from 2012), which were also included in the analyses.
2.6. Statistical analyses
The mapped data were normalized by dividing the reads for each technical replicate that mapped to a given target transcript by the total number of mapped reads in the technical replicate (Christie et al., 2013a). A two-way ANOVA was used to test for differences in relative transcript expression between years and among stages. Since both variables showed significant differences, a Bonferroni test was used to identify which stages showed significant differences in expression between years. Differences in expression among stages were calculated for the data collected in 2012 only, since it included biological and technical replication. Here, we used a one-way ANOVA followed by a Bonferroni post hoc test to calculate p-values for all pair-wise comparisons; GraphPad Prism v.6 (GraphPad Software, Inc., La Jolla, CA, USA) was used for all statistical analyses.
3. Results
3.1. Discovery of aminergic biosynthetic enzymes using a de novo assembled transcriptome
3.1.1. Dopamine biosynthesis
As described in Section 1, dopamine is synthesized from phenylalanine via the actions of the enzymes TPH, TH and DDC. Using known Drosophila proteins as query sequences, transcripts encoding putative Calanus homologs of each enzyme were identified within our de novo assembled transcriptome; these sequences are described in turn below.
3.1.1.1. Tryptophan-phenylalanine hydroxylase (TPH)
Using D. melanogaster phenylalanine hydroxylase (Accession No. CAA66797; Ruiz-Vázquez et al., 1996) as the query sequence, a single, 1,531 bp C. finmarchicus transcript (comp34563_c3_seq1; named here calanus finmarchicus tryptophan-phenylalanine hydroxylase or calfi-tβh) was identified as encoding a putative TPH homolog (Table 1). Translation of calfi-tβh yielded a 443 amino acid, full-length TPH isoform (Calfi-TPH; Fig. 1A and Table 1), with the open reading frame (ORF) bounded on both sides by stop codons (Supplemental Fig. 1A).
Alignment of the D. melanogaster query protein and Calfi-TPH revealed the two proteins share 65.3% identity/87.6% similarity in amino acid composition (Fig. 2). Structural analysis of the Drosophila protein using the online program Pfam identified a single biopterin-dependent aromatic amino acid hydroxylase domain within the sequence (Fig. 2); this domain was also identified by Pfam within Calfi-TPH (Figs. 1 and 2), as was a single ACT domain (Figs. 1 and 2). The biopterin-dependent aromatic amino acid hydroxylase domains are identically positioned in the two proteins and are nearly identical in amino acid composition (Fig. 2). Interestingly, while not identifed as an ACT domain, the amino acids in the Drosophila protein are also nearly identical to those corresponding to this motif in Calfi-TPH (Fig. 2). These data support Calfi-TPH being a member of the tryptophan-phenylalanine hydroxylase family.
3.1.1.2. Tyrosine hydroxylase (TH)
Using D. melanogaster TH (Drome-TH; Accession No. CAA53802; Neckameyer and Quinn, 1989) as the query sequence, a single, 1,932 bp C. finmarchicus transcript (comp541083_c3_seq1; named here calanus finmarchicus tyrosine hydroxylase or calfi-th) was identified as encoding a putative TH homolog (Table 1). Translation of calfi-th yielded a 516 amino acid, full-length TH isoform (Calfi-TH; Fig. 1B and Table 1), with the ORF bounded on both sides by stop codons (Supplemental Fig. 1B).
Alignment of Drome-TH and Calfi-TH revealed the two proteins share 48.6% identity/82.6% similarity in amino acid composition (Supplemental Fig. 2A). Pfam analysis of the two proteins identified a single biopterin-dependent aromatic amino acid hydroxylase domain within each sequence (Fig. 1B and Supplemental Fig. 2A); the location of this domain was the same within the two proteins, and the amino acid composition of the motif is highly conserved between the two THs (Supplemental Fig. 2A). These data support Calfi-TH being a member of the tyrosine hydroxylase family.
3.1.1.3. DOPA decarboxylase (DDC)
Using an isoform of D. melanogaster DDC (Drome-DDC; Accession No. AAF53763; Adams et al., 2000) as the query sequence, a single, 1,560 bp C. finmarchicus transcript (comp374436_c0_seq5; named here calanus finmarchicus dopa decarboxylase or calfi-ddc) was identified as encoding a putative DDC homolog (Table 1). Translation of calfi-ddc yielded a 487 amino acid, putative full-length DDC isoform (Calfi-DDC; Fig. 1C and Table 1), with the ORF bounded on its 3’ but not 5’ end by a stop codon (Supplemental Fig. 1C).
The alignment of Drome-DDC and Calfi-DDC revealed the two proteins share 60.0% identity/84.6% similarity in amino acid composition (Supplemental Fig. 2B). Pfam analysis of the two proteins identified a single pyridoxal-dependent decarboxylase conserved domain within each sequence (Fig. 1C and Supplemental Fig. 2B), the location and amino acid sequence of which is highly conserved between the two DDCs (Supplemental Fig. 2B). These data support Calfi-DDC being a member of the DOPA decarboxylase family.
3.1.2. Histamine biosynthesis
3.1.2.1. Histidine decarboxylase (HDC)
As described in Section 1, histamine is produced from the amino acid histidine in a one-step reaction involving the enzyme HDC. Using an isoform of D. melanogaster HDC (Drome-HDC; Accession No. AAF58823; Adams et al., 2000) as the query sequence, a single, 2,659 bp C. finmarchicus transcript (comp175390_c0_seq2; named here calanus finmarchicus histidine decarboxylase or calfi-hdc) was identified as encoding a putative HDC homolog (Table 1). Translation of calfi-hdc yielded a 724 amino acid, full-length HDC isoform (Calfi-HDC; Fig. 1D and Table 1), with the ORF bounded on both sides by stop codons (Supplemental Fig. 1D).
Alignment of the Drome-HDC and Calfi-HDC revealed the two proteins share 49.9% identity/71.9% similarity in amino acid composition (Supplemental Fig. 2C). Pfam analysis of the two proteins identified a single pyridoxal-dependent decarboxylase conserved domain within each sequence (Fig. 1D and Supplemental Fig. 2C), the locations and the amino acid sequences of which are nearly identical in the two HDCs (Supplemental Fig. 2C). These data support Calfi-HDC being a member of the histidine decarboxylase family.
3.1.3. Octopamine biosynthesis
Octopamine, like dopamine, is synthesized from phenylalanine in a three-step process. As with dopamine, TPH is the first enzyme in the biosynthesis of octopamine, with TDC and TβH catalyzing the two remaining reactions. The identification of a Calanus TPH-encoding transcript is described in Section 3.1.1.1, while those encoding C. finmarchicus TDC and TβH are presented below.
3.1.3.1. Tyrosine decarboxylase (TDC)
Using D. melanogaster TDC (Drome-TDC; Accession No. AAM70810; Adams et al., 2000) as the query sequence, a single, 2,127 bp C. finmarchicus transcript (comp374436_c0_seq1; named here calanus finmarchicus tyrosine decarboxylase or calfi-tdc) was identified as encoding a putative TDC homolog (Table 1). Translation of calfi-tdc yielded a 617 amino acid, putative full-length TDC isoform (Calfi-TDC; Fig. 1E and Table 1), with the ORF bounded on its 3’ but not 5’ end by a stop codon (Supplemental Fig. 1E).
Alignment of the Drome-TDC and Calfi-TDC revealed the two proteins share 46.7% identity/74.6% similarity in amino acid composition (Supplemental Fig. 2D). Pfam analysis of the two proteins identified a single pyridoxal-dependent decarboxylase conserved domain within each sequence (Fig. 1E and Supplemental Fig. 2D); in both TDCs, the locations and amino acid sequences of this motif are nearly identical (Supplemental Fig. 2D). These data support Calfi-TDC being a member of the tyrosine decarboxylase family.
3.1.3.3. Tyramine β-hydroxylase (TβH)
Using D. melanogaster TβH (Drome-TβH; Accession No. AAO41640; Adams et al., 2000) as the query sequence, a single, 2,122 bp C. finmarchicus transcript (comp975856_c0_seq1; named here calanus finmarchicus tyramine β-hydroxylase or calfi-tβh) was identified as encoding a putative TβH homolog (Table 1). Translation of calfi-tβh yielded a 672 amino acid, putative full-length TβH isoform (Calfi-TβH; Fig. 1F and Table 1), with the ORF bounded on its 3’ but not 5’ end by a stop codon (Supplemental Fig. 1F).
Alignment of the Drome-TβH and Calfi-TβH revealed the two proteins share 36.0% identity/65.5% similarity in amino acid composition (Supplemental Fig. 2E). Pfam analysis of the two proteins identified one DOMON, one copper type-II ascorbate-dependent monooxygenase N-terminal, and one copper type-II ascorbate-dependent monooxygenase C-terminal domain within each sequence (Fig. 1F and Supplemental Fig. 2E). The location of each of these domains is similar in the two proteins (Supplemental Fig. 2E). Likewise, the amino acid composition of each of the domains is highly conserved between the two TβHs (Supplemental Fig. 2E). These data support Calfi-TβH being a member of the tyramine β-hydroxylase family.
3.1.4. Serotonin biosynthesis
As described in Section 1, serotonin is synthesized from tryptophan via the actions of TPH or TRH and DDC. Discovery of the Calanus transcript encoding the former protein is discussed in Section 3.1.1.1, while discovery of a C. finmarchicus DDC-encoding sequence is discussed in Section 3.1.1.3. The identification of a TRH-encoding transcript is provided below.
3.1.4.2. Tryptophan hydroxylase (TRH)
Using D. melanogaster TRH (Drome-TRH; Accession No. AAF47444; Adams et al., 2000) as the query sequence, a single, 1,976 bp C. finmarchicus transcript (comp441733_c0_seq2; named here calanus finmarchicus tryptophan hydroxylase or calfi-trh) was identified as encoding a putative TRH homolog (Table 1). Translation of calfi-trh yielded a 541 amino acid, full-length TRH isoform (Calfi-TRH; Fig. 1G and Table 1), with the ORF bounded on both sides by stop codons (Supplemental Fig. 1G).
Alignment of the Drome-TRH and Calfi-TRH revealed the two proteins share 44.7% identity/76.9% similarity in amino acid composition (Supplemental Fig. 2F). Pfam analysis of the two proteins identified a single biopterin-dependent aromatic amino acid hydroxylase domain within each sequence (Fig. 1G and Supplemental Fig. 2F), which is similarly positioned and highly conserved in amino acid composition between the two TRHs (Supplemental Fig. 2F). These data support Calfi-TRH being a member of the tryptophan hydroxylase family.
3.2. Reverse BLAST analyses using deduced protein sequences
To confirm the annotations attributed to the putative aminergic biosynthetic enzyme-encoding transcripts described in Section 3.1, each deduced protein was used to query the extant annotated proteins in FlyBase and the extant non-redundant arthropod proteins in GenBank for the most similar sequences. As can be seen in Table 2, for the searches of FlyBase, an isoform of the protein to which the Calanus sequence was annotated was identified as the most similar protein for each query. For example, the query of FlyBase using Calfi-TPH returned Henna (FlyBase No. FBpp0306707), a synonym for TPH, as the most similar protein. Likewise, the query using Calfi-TH returned Pale (FlyBase No. FBpp0076665), a synonym for TH, as most similar to this Calanus sequence. While not the case here, we have noted that for some targeted protein discoveries, a Drosophila member of an unrelated family is returned as the top FlyBase hit (e.g. (6-4)-photolyase rather than cryptochrome for C. finmarchicus protein initially identified using D. melanogaster cyptochrome as the query; Christie et al., 2013b), which immediately calls into question the functional attribution ascribed to the query sequence. Like the reciprocal BLASTs conducted against FlyBase, those directed against the non-redundant arthropod proteins curated at GenBank typically identified proteins from the same family attributed to the Calanus query as the most similar arthropod sequence (Table 3), again supporting our functional attributions, e.g. TβH from the cockroach Periplaneta americana (Accession No. AFO63077; Chatel, Murillo, Bourdin, Quinchard, Picard and Legros, unpublished direct submission) for Calfi-TβH and tryptophan 5-hydroxylase 1 from the ant Harpegnathos saltator (Accession No. EFN85713; Bonasio et al., 2010) for Calfi-TRH. Taken collectively, the reverse BLASTs conducted against FlyBase and GenBank support the functional attributions given to the transcripts/proteins described in Section 3.1.
3.3. Expressed sequence tag support for deduced protein sequences
The C. finmarchicus transcriptome used here for protein discovery was assembled from over 400,000,000 paired-end reads (100-bp long). As the software used to assemble the reads is not infallible, there may be errors in some output sequences. If such errors occur in protein coding regions or result in frame shifts upstream of ORFs, then there would be a high likelihood of spurious protein predictions from the misassemblies. Thus, to strengthen our confidence in the sequences of the proteins presented in Section 3.1, each protein was BLASTed against the C. finmarchicus ESTs curated in GenBank. The extant Calanus ESTs, while small in number relative to the sequences present in our Trinity assembly, are single-pass sequences (Lenz et al., 2012), and thus are not subject to assembly errors. Via this protocol, EST support for the deduced amino acid sequences of Calfi-TH, Calfi-TDC and Calfi-TRH was obtained (Table 4). However, no ESTs were identified that encoded any portion of Calfi-TPH, Calfi-DDC, Calfi-HDC, or Calfi-TβH (Table 4). An example of the EST vetting of one of the deduced Calanus amine biosynthetic enzymes, Calfi-TDC, is shown in Figure 3. As can be seen from this figure, the protein deduced from FK041551 is a 232 amino acid, partial protein that differs from an internal portion of the amino (N)-terminus of Calfi-TDC at just two residues (99.1% identity), both conservative amino acid substitutions (Fig. 3). While the agreement between EST sequences and those derived from the de novo assembled transcriptome increases our confidence in the predicted proteins, it also illustrates the need for deep sequencing in order to obtain the predicted complement of full-length protein sequences.
Table 4.
Expressed sequence tag (EST) support for deduced Calanus finmarchicus amine biosynthetic enzyme protein sequences
| Calanus query | Expressed sequence tags | ||||
|---|---|---|---|---|---|
| Accession No. | E-value | Protein length* |
Location on query |
% identity to query |
|
| Calfi-TPH | - | - | - | - | - |
| Calfi-TH | EH666558 | 8e-68 | 130 | N | 94.6 |
| Calfi-DDC | - | - | - | - | - |
| Calfi-HDC | - | - | - | - | - |
| Calfi-TDC | FK042551 | 3e-158 | 232 | I | 99.1 |
| Calfi-TβH | - | - | - | - | - |
| Calfi-TRH | EL585923 | 1e-141 | 200 | N | 98.0 |
Abbreviations: Calfi-TPH, Calanus finmarchicus tryptophan-phenylalanine hydroxylase; Calfi-TH, Calanus finmarchicus tyrosine hydroxylase; Calfi-DDC, Calanus finmarchicus DOPA decarboxylase; Calfi-HDC, Calanus finmarchicus histidine decarboxylase; Calfi-TDC, Calanus finmarchicus tyrosine decarboxylase; Calfi-TβH, Calanus finmarchicus tyramine p-hydroxylase; Calfi-TRH, Calanus finmarchicus tryptophan hydroxylase; N, amino-terminal partial protein; I, internal fragment; C, carboxy-terminal partial protein.
In amino acids.
Figure 3.
Confirmation of protein sequence of Calanus finmarchicus tyrosine decarboxylase (Calfi-TDC) using publicly accessible expressed sequence tags (ESTs). To increase our confidence in the deduced amino acid sequences of the amine biosynthetic enzymes reported in this study, each protein was used to query the extant C. finmarchicus ESTs curated at GenBank for identical sequences. As can be seen here, the BLAST searches using Calfi-TDC identified one EST, FK041551, as encoding partial protein homologs of the full-length sequence; the overlapping regions of the two proteins differ at just two positions (black highlighting).
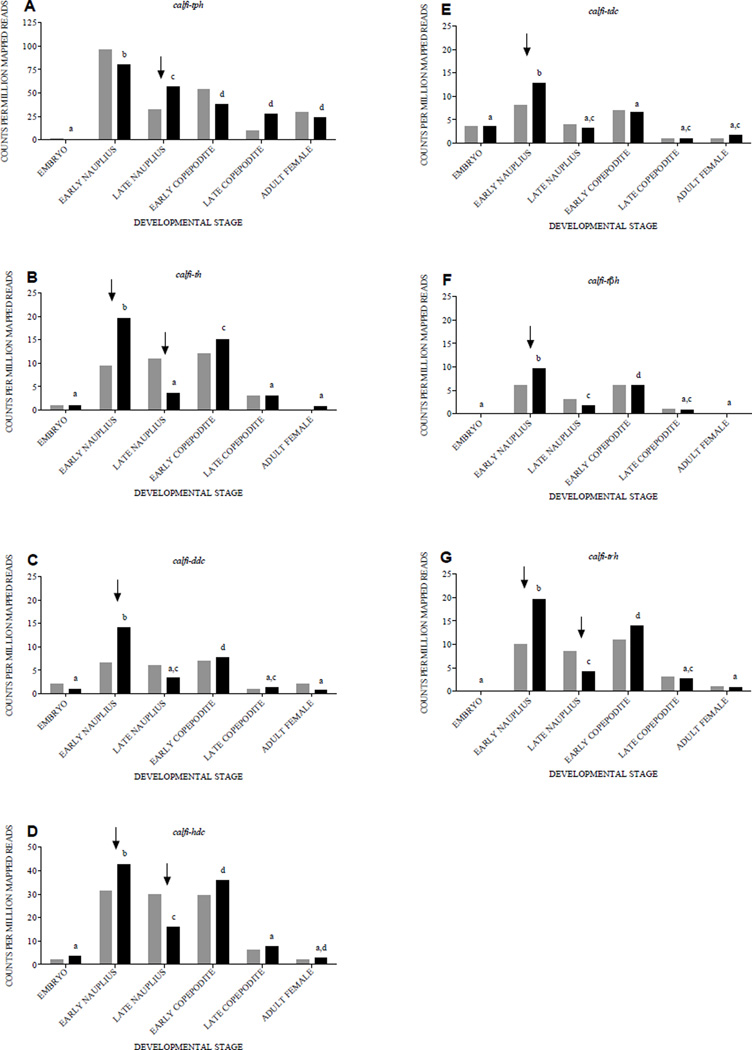
3.4. Expression mapping of aminergic signaling transcripts
Illumina RNA-Seq reads from six C. finmarchicus developmental stages (embryo, early nauplius, late nauplius, early copepodite, late copepodite, and adult female) were mapped against the transcripts encoding Calfi-TPH (Fig. 4A), Calfi-TH (Fig. 4B), Calfi-DDC (Fig. 4C), Calfi-HDC (Fig. 4D), Calfi-TDC (Fig. 4E), Calfi-TβH (Fig. 4F) and Calfi-TRH (Fig. 4G) to determine their patterns of developmental expression. Normalized data from samples collected in both 2011 (gray bars) and 2012 (black bars) are shown in Figure 4, and as can be seen from this figure, significant differences between years were noted for 10 of the 42 comparisons (arrows in Fig. 4). In all cases, the differences between years were in either the early or late nauplii, and with the exception of calfi-th, calfi-hdc and calfi-trh in the late nauplii, expression in 2012 was significantly higher than in 2011. The factor(s) responsible for the variation seen between these 2011 and 2012 samples remain unknown, but as both stages were laboratory cultured in eachyear, environmental variables (e.g. ocean temperature, salinity, etc) seem unlikely to be major contributors.
Figure 4.
Expression patterns of amine biosynthetic enzyme-encoding transcripts across six Calanus finmarchicus developmental stages in material collected in 2011 (grey bars) and 2012 (black bars) based on RNA-Seq data. Expression levels are presented as normalized counts per million reads that mapped to each target transcript sequence in embryo, early nauplius (stages NI-II), late nauplius (stages NV-NVI), early copepodite (stages CI-II), late copepodite (stage CV) and adult female (stage CVI). (A) calanus finmarchicus tryptophan-phenylalanine hydroxylase (calfi-tph). (B) calanus finmarchicus tyrosine hydroxylase (calfi-th). (C) calanus finmarchicus dopa decarboxylase (calfi-ddc). (D) calanus finmarchicus histidine decarboxylase (calfi-hdc). (E) calanus finmarchicus tyrosine decarboxylase (calfi-tdc). (F) calanus finmarchicus tyramine β-hydroxylase (calfi-tβh). (G) calanus finmarchicus tryptophan hydroxylase (calfi-trh). Significant differences in stage-specific expression between years are indicated by arrows. Significant differences among stages are indicated by small letters over the black bars (2012): the same letter indicates that two or more stages have similar expression levels (p-values given in Supplemental Fig. 3).
Comparisons among stages for the 2012 data (Fig. 4 and Supplemental Fig. 3), for which both biological and technical replication was included, show a common pattern of expression for all of the enzyme-encoding transcripts except calfi-tph (Fig. 4A). Specifically, calfi-th, calfi-ddc, calfi-hdc, calfi-tdc, calfi-tβh and calfi-trh all showed a low level of relative expression in the embryo, high relative expression in the early nauplius and early copepodite, and intermediate expression levels the late nauplius, late copepodite and adult (Fig. 4B–G). For calfi-tph, an increase in relative expression was noted between the embryo and early nauplius, with relative expression levels subsequently decreasing through the early copepodite stage and then being maintained at this lower level in the late copepodite and adult (Fig. 4A).
4. Discussion
4.1. In silico characterization of neurochemical signaling systems in C. finmarchicus
Large-scale shifts in the geographic range of C. finmarchicus, presumably due at least in part to rising ocean temperature (Helaouët et al., 2011; Pepin and Head, 2009; Pepin et al., 2011; Reygondeau and Beaugrand 2011; Speirs et al., 2006), have resulted in an increased focus on understanding how this species adapts to its environment physiologically. As part of this ongoing effort, we have generated a transcriptome for Calanus that consists of over 200,000 unique sequences (Christie et al., 2013a) and are currently using this resource as a platform for targeted gene/protein discovery (Christie et al., 2013a, 2013b). Among our ongoing projects is the identification and characterization of the neurochemical signaling systems of C. finmarchicus (Christie et al., 2013a), which are likely key components in the control systems that allow this species to adapt physiologically to environmental challenges.
In a recent publication, we described the peptidergic systems of C. finmarchicus (Christie et al., 2013a). In this study, evidence for over twenty neuropeptide families was presented (Christie et al., 2013a). Here, we have expanded this work by identifying the transcripts and deducing the proteins responsible for amine biosynthesis. Specifically, the amino acid sequences of known Drosophila amine biosynthetic enzymes were used to query our C. finmarchicus transcriptome for sequences encoding proteins implicated in the production of dopamine, histamine, octopamine and serotonin. Via this approach, putative Calanus transcripts encoding TPH (dopamine, octopamine and serotonin biosynthesis), TH (dopamine biosynthesis), DDC (dopamine and serotonin biosynthesis), HDC (histamine biosynthesis), TDC (octopamine biosynthesis), TβH (octopamine biosynthesis) and TRH (serotonin biosynthesis) were identified. Each transcript appears to encode a full-length enzyme, and reverse BLAST and domain analyses confirm that these proteins possess sequence homology to and structural hallmarks of their respective enzyme families. Collectively, the data described here are the first identifications of the gene/protein components of aminergic signaling systems in C. finmarchicus. Moreover, while a number of other crustacean amine biosynthetic enzymes have been identified, only those of the cladoceran Daphnia pulex were among the top arthropod BLAST hits returned when the Calanus sequences were used to query the extant GenBank database, and here for just four of the seven queries, with but two as top 5 hits. This finding mirrors that seen for the peptidergic signaling systems of Calanus (Christie et al., 2013a), and reinforces the hypothesis that C. finmarchicus may occupy a position in arthropod phylogeny that is at or near the point where the crustacean and insect lineages diverged (e.g. Andrew, 2011; Andrew et al., 2012; Strausfeld and Andrew, 2011). If this hypothesis continues to be supported as sequence data for the Crustacea are expanded (it is currently less specious than that for the hexapods), C. finmarchicus may be a key species for studying the evolution of neurochemical signaling within the Arthropoda.
4.2. Developmental acquisition of aminergic systems in C. finmarchicus and a possible role for them in metamorphic transitions
Currently, little work has focused on the developmental acquisition of aminergic systems in crustaceans, and the few studies that are extant have focused on nervous systems, primarily those of decapods, in particular members of the Astacidea. Immunohistochemical mapping of aminergic profiles in the nervous systems of lobsters (Homarus americanus and/or Homarus gammarus) and crayfish (the parthenogenetic variant of Procambarus fallax, i.e. Marmorkrebs) suggest each amine to be initially expressed during embryonic development, with serotonin expression likely occurring early in embryogenesis, and the others in mid- to late embryonic life (Beltz et al., 1990; Cournil et al., 1995; Kilman et al., 1999; Le Feuvre et al., 2001; Pulver et al., 2003; Richards et al., 2003; Rieger and Harzsch, 2008; Schneider et al., 1996). For histamine and serotonin, the embryonic expression patterns are reported to be largely stable through neural developmet (Beltz et al., 1990; Rieger and Harzsch, 2008), whereas acquisition of the full complement of dopaminergic and octopaminergic neurons appears to be a more protracted event (Cournil et al., 1995; Schneider et al., 1996). Regardless, the early appearance of amines in crustacean nervous systems relative to other neuroactive compounds, e.g. many peptides (e.g. Beltz et al., 1990), suggests roles for them in the developmental control of this and other (see Section 4.3) organ systems (e.g. Beltz et al., 2001; Benton and Beltz, 2001; Benton et al., 1997; Sullivan et al., 2000).
Our developmental profiling of the C. finmarchicus transcripts encoding the biosynthetic enzymes for dopamine, histamine, octopamine and serotonin suggests that the initial acquisition of all four amines occurs at least by early naupliar life, as the highest levels of expression for each of the enzyme-encoding transcripts in this species was observed at this time point. As in the decapods nervous systems, this developmental peak in expression occurs earlier than was seen for many of the peptide/peptide receptor-encoding transcripts in Calanus (Christie et al., 2013a), and is suggestive that the amines may play a regulatory role in development control in this species too.
An interesting phenomenon that was noted for each of the amine biosynthetic enzyme-encoding transcripts except calfi-tph was the presence of two peaks in expression, one in the early nauplius and the other in the early copepodite. For most of the sequences, the differences in expression seen at these stages were significant compared to the other developmental time points that were examined. The early nauplius and the early copepodite stages represent two major transitions in the life history of C. finmarchicus. The transition from the embryo to the nauplius represents the activation of the sensory motor system to allow for swimming and escape responses (Bradley et al., 2013), but not yet feeding (Mauchline, 1998). In copepods, the transition from nauplius to copepodite involves a major change in body plan, including changes in the nervous system (Wilson and Hartline, 2011). Thus, it is possible that some or all of the amines may play key neurochemical roles in controlling these events, potentially including neurogenesis, for which at least serotonin has been implicated in decapods (Benton et al., 2008; Sandeman et al., 2009; Zhang et al., 2011).
4.3. Other roles for biogenic amines in C. finmarchicus
The acquisition of aminergic systems in C. finmarchicus appears to occur early in development and suggests a possible role for amines in the development and/or reorganization of the nervous system. While the distributions of aminergic neurons have not yet been mapped across development in Calanus, those expressing dopamine, histamine and serotonin have been examined in adults (Hartline and Christie, 2010). For these three amines, the distributions of labeled profiles were consistent with them serving as locally-released neuromodulators (Hartline and Christie, 2010), i.e. present, and presumably released, within central neuropil. For serotonin, and possibly dopamine, putative endocrine-like structures were also labeled in the nervous system, suggesting neurohormonal release is also possible (Hartline and Christie, 2010).
The specific roles served by locally-released and/or circulating amines in Calanus are currently unknown. However, work done on other vertebrate and invertebrate species, including numerous decapod crustaceans, suggests that they will ultimately be found to be both numerous and diverse. For example, in other crustaceans, dopamine, histamine, octopamine and serotonin have all been shown to affect rhythmic behaviors (e.g. locomotion, heartbeat and foregut movements) by acting directly on neural circuits and/or at neuromuscular junctions (e.g. Battelle and Kravitz, 1978; Callaway and Stuart, 1999; Christie et al., 2004; Djokaj et al., 2001; Flamm and Harris-Warrick, 1986a, 1986b; Florey and Rathmayer, 1978; Fort et al., 2004; Glusman and Kravitz, 1982; Harzsch and Glötzner, 2002; Johnson and Harris-Warrick, 1990, 1997; Johnson et al., 1993, 1994, 1995; Jorge-Rivera et al., 1998; Kravitz et al., 1980; Kvarta et al., 2012; Kwiatkowski et al., 2013; McCoole et al., 2011; Mulloney and Hall, 1991; Peck et al., 2006; Pulver et al., 2003; Rieger and Harzsch, 2008). In addition, dopamine, octopamine and serotonin have all been shown to play roles in the control of crustacean osmoregulation (e.g. Lohrmann and Kamemoto, 1987; Morris and Ahern, 2003; Morris et al., 2000), with the latter two amines also key components in the control of aggression/social dominance (e.g. Edwards and Kravitz, 1997; Huber, 2005; Huber et al., 2001; Kravitz, 2000; Panksepp et al., 2003; Pedetta et al., 2010; Sosa and Baro, 2002), and dopamine implicated in immune function (e.g. Chang et al., 2007; Cheng et al., 2005; Li et al., 2005). Clearly much work will be needed to assess the roles of the amines in these and other actions in Calanus, but as additional studies are conducted, it will be interesting to see how their functions in C. finmarchicus correspond to those described for decapod crustaceans given differences in lifestyle (planktonic vs. benthic) and the evolutionary relationship between copepods and members of the Malacostraca.
4.4. Conclusions and future directions
Using a well-vetted bioinformatics workflow, we have mined for and characterized the transcripts and proteins that are likely responsible for the synthesis of dopamine, histamine, octopamine and serotonin in the copepod C. finmarchicus. These data complement and augment those previously described for the peptidergic systems of this species (Christie et al., 2013a), which is the biomass dominant zooplankton in the North Atlantic Ocean. Our ultimate goal is to characterize all of the neurochemical signaling systems of Calanus, which will allow for genebased studies of physiological adaptation, including the contributions of neurochemical signaling, in this ecologically critical species.
Supplementary Material
Highlights.
Calanus transcripts encoding amine biosynthetic enzymes were identified
Dopamine, histamine, octopamine and serotonin pathways proteins were deduced
Expression mapping showed all amines are produced early in development
Two peaks in expression were noted (i.e. early nauplius and early copepodite)
Expression peaks suggest hormonal roles for amines in metamorphic transitions
Acknowledgements
We would like to thank Roy McMorran (Mount Desert Island Biological Laboratory [MDIBL]), Benjamin King (MDIBL) and Daniel Hartline (University of Hawaii at Manoa) for sharing resources and helpful discussions concerning this study. Le-Shin Wu (National Center for Genome Analysis Support, Indiana University) is thanked for his assistance in producing the C. finmarchicus transcriptome used in this study. Karl Brunner and the crew of the Elizabeth T (Downeast Friendship Sloop Charters) are thanked for their help with plankton collections. Kazutaka Katoh (Osaka University) is thanked for his helpful discussion on issues concerning the interpretation of MAFFT output formatting. The authors acknowledge financial support from The National Science Foundation (NSF OCE 1040597), the National Institutes of Health National Institute of General Medical Sciences (NIH 2T34GM007684-32 to Patricia A. Couvillon and P.H.L.), and the Cades Foundation of Honolulu, Hawaii. V.R. thanks the David W. Towle Fellowship for supporting her 2012 summer stay at MDIBL. The authors also acknowledge that this research was based upon work supported by NSF ABI-1062432 to Indiana University, and any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of NSF, the National Center for Genome Analysis Support, or Indiana University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew DR. A new view of insect-crustacean relationships II. Inferences from expressed sequence tags and comparisons with neural cladistics. Arthropod Struct. Dev. 2011;40:289–302. doi: 10.1016/j.asd.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Andrew DR, Brown SM, Strausfeld NJ. The minute brain of the copepod Tigriopus californicus supports a complex ancestral ground pattern of the tetraconate cerebral nervous systems. J. Comp. Neurol. 2012;520:3446–3470. doi: 10.1002/cne.23099. [DOI] [PubMed] [Google Scholar]
- Battelle BA, Kravitz EA. Targets of octopamine action in the lobster: cyclic nucleotide changes and physiological effects in hemolymph, heart and exoskeletal muscle. J. Pharmacol. Exp. Ther. 1978;205:438–448. [PubMed] [Google Scholar]
- Beltz BS, Benton JL, Sullivan JM. Transient uptake of serotonin by newborn olfactory projection neurons. Proc. Natl. Acad. Sci. USA. 2001;98:12730–12735. doi: 10.1073/pnas.231471298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz BS, Pontes M, Helluy SM, Kravitz EA. Patterns of appearance of serotonin and proctolin immunoreactivities in the developing nervous system of the American lobster. J. Neurobiol. 1990;21:521–542. doi: 10.1002/neu.480210402. [DOI] [PubMed] [Google Scholar]
- Benton JL, Beltz BS. Effects of serotonin depletion on local interneurons in the developing olfactory pathway of lobsters. J. Neurobiol. 2001;46:193–205. doi: 10.1002/1097-4695(20010215)46:3<193::aid-neu1002>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Benton JL, Goergen EM, Rogan SC, Beltz BS. Hormonal and synaptic influences of serotonin on adult neurogenesis. Gen. Comp. Endocrinol. 2008;158:183–190. doi: 10.1016/j.ygcen.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton JL, Huber R, Ruchhoeft M, Helluy S, Beltz BS. Serotonin depletion by 5,7-dihydroxytryptamine alters deutocerebral development in the lobster, Homarus americanus . J. Neurobiol. 1997;33:357–373. doi: 10.1002/(sici)1097-4695(199710)33:4<357::aid-neu2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C, Zhang P, Huang Z, Berger SL, Reinberg D, Wang J, Liebig J. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator . Science. 2010;329:1068–1071. doi: 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley CJ, Strickler JR, Buskey EJ, Lenz PH. Swimming and escape behavior in two species of calanoid copepods from nauplius to adult. J. Plankton Res. 2013;35:49–65. [Google Scholar]
- Callaway JC, Stuart AE. The distribution of histamine and serotonin in the barnacle's nervous system. Microsc. Res. Tech. 1999;44:94–104. doi: 10.1002/(SICI)1097-0029(19990115/01)44:2/3<94::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Chang CC, Wu ZR, Kuo CM, Cheng W. Dopamine depresses immunity in the tiger shrimp Penaeus monodon . Fish Shellfish Immunol. 2007;23:24–33. doi: 10.1016/j.fsi.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Cheng W, Chieu HT, Tsai CH, Chen JC. Effects of dopamine on the immunity of white shrimp Litopenaeus vannamei . Fish Shellfish Immunol. 2005;19:375–385. doi: 10.1016/j.fsi.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Christie AE. Crustacean neuroendocrine systems and their signaling agents. Cell Tissue Res. 2011;345:41–67. doi: 10.1007/s00441-011-1183-9. [DOI] [PubMed] [Google Scholar]
- Christie AE, Fontanilla TM, Nesbit KT, Lenz PH. Prediction of a putative Calanus finmarchicus (Crustacea, Copepoda) circadian signaling system using a de novo assembled transcriptome. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2013b;8:165–193. doi: 10.1016/j.cbd.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Lona PB, McCoole MD, King BL, Bucklin A, Hartline DK, Lenz PH. In silico characterization of the insect diapause-associated protein couch potato (CPO) in Calanus finmarchicus (Crustacea: Copepoda) Comp. Biochem. Physiol. Part D Genomics Proteomics. 2013c;8:45–57. doi: 10.1016/j.cbd.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Wu LS, Ganote CL, Doak T, Lenz PH. Peptidergic signaling in Calanus finmarchicus (Crustacea, Copepoda): In silico identification of putative peptide hormones and their receptors using a de novo assembled transcriptome. Gen. Comp. Endocrinol. 2013a;187:117–135. doi: 10.1016/j.ygcen.2013.03.018. [DOI] [PubMed] [Google Scholar]
- Christie AE, Stein W, Quinlan JE, Beenhakker MP, Marder E, Nusbaum MP. Actions of a histaminergic/peptidergic projection neuron on rhythmic motor patterns in the stomatogastric nervous system of the crab Cancer borealis . J. Comp. Neurol. 2004;469:153–169. doi: 10.1002/cne.11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman CM, Neckameyer WS. Serotonin synthesis by two distinct enzymes in Drosophila melanogaster . Arch. Insect Biochem. Physiol. 2005;59:12–31. doi: 10.1002/arch.20050. [DOI] [PubMed] [Google Scholar]
- Cournil I, Casasnovas B, Helluy SM, Beltz BS. Dopamine in the lobster Homarus gammarus: II. Dopamine-immunoreactive neurons and development of the nervous system. J. Comp. Neurol. 1995;362:1–16. doi: 10.1002/cne.903620102. [DOI] [PubMed] [Google Scholar]
- Dale T, Kaartverdt S, Ellertsen B, Amundsen R. Large-scale oceanic distribution and population structure of Calanus finmarchicus, in relation to physical environment, food and predators. Mar. Biol. 2001;139:561–574. [Google Scholar]
- Djokaj S, Cooper RL, Rathmayer M. Presynaptic effects of octopamine, serotonin, and cocktails of the two modulators on neuromuscular transmission in crustaceans. J. Comp. Physiol. A. 2001;187:145–154. doi: 10.1007/s003590100187. [DOI] [PubMed] [Google Scholar]
- Edwards DH, Kravitz EA. Serotonin, social status and aggression. Curr. Opin. Neurobiol. 1997;7:812–819. doi: 10.1016/s0959-4388(97)80140-7. [DOI] [PubMed] [Google Scholar]
- Flamm RE, Harris-Warrick RM. Aminergic modulation in lobster stomatogastric ganglion. I. Effects on motor pattern and activity of neurons within the pyloric circuit. J. Neurophysiol. 1986a;55:847–865. doi: 10.1152/jn.1986.55.5.847. [DOI] [PubMed] [Google Scholar]
- Flamm RE, Harris-Warrick RM. Aminergic modulation in lobster stomatogastric ganglion. II. Target neurons of dopamine, octopamine, and serotonin within the pyloric circuit. J. Neurophysiol. 1986b;55:866–881. doi: 10.1152/jn.1986.55.5.866. [DOI] [PubMed] [Google Scholar]
- Florey E, Rathmayer M. The effects of octopamine and other amines on the heart and on neuromuscular transmission in decapod crustaceans: further evidence for a role as neurohormone. Comp. Biochem. Physiol. C. 1978;61C:229–237. doi: 10.1016/0306-4492(78)90136-3. [DOI] [PubMed] [Google Scholar]
- Fort TJ, Brezina V, Miller MW. Modulation of an integrated central pattern generator-effector system: dopaminergic regulation of cardiac activity in the blue crab Callinectes sapidus . J. Neurophysiol. 2004;92:3455–3470. doi: 10.1152/jn.00550.2004. [DOI] [PubMed] [Google Scholar]
- Glusman S, Kravitz EA. The action of serotonin on excitatory nerve terminals in lobster nerve-muscle preparations. J. Physiol. 1982;325:223–241. doi: 10.1113/jphysiol.1982.sp014147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartline DK, Christie AE. Immunohistochemical mapping of histamine, dopamine, and serotonin in the central nervous system of the copepod Calanus finmarchicus (Crustacea; Maxillopoda; Copepoda) Cell Tissue Res. 2010;341:49–71. doi: 10.1007/s00441-010-0974-8. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Glötzner J. An immunohistochemical study of structure and development of the nervous system in the brine shrimp Artemia salina Linnaeus, 1758 (Branchiopoda, Anostraca) with remarks on the evolution of the arthropod brain. Arthropod Struct. Dev. 2002;30:251–270. doi: 10.1016/s1467-8039(02)00012-9. [DOI] [PubMed] [Google Scholar]
- Helaouët P, Beaugrand G, Reid PC. Macrophysiology of Calanus finmarchicus in the North Atlantic Ocean. Prog. Oceanogr. 2011;91:217–228. [Google Scholar]
- Huber R. Amines and motivated behaviors: a simpler systems approach to complex behavioral phenomena. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2005;191:231–239. doi: 10.1007/s00359-004-0585-5. [DOI] [PubMed] [Google Scholar]
- Huber R, Panksepp JB, Yue Z, Delago A, Moore P. Dynamic interactions of behavior and amine neurochemistry in acquisition and maintenance of social rank in crayfish. Brain. Behav. Evol. 2001;57:271–282. doi: 10.1159/000047245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Harris-Warrick RM. Aminergic modulation of graded synaptic transmission in the lobster stomatogastric ganglion. J. Neurosci. 1990;10:2066–2076. doi: 10.1523/JNEUROSCI.10-07-02066.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BR, Harris-Warrick RM. Amine modulation of glutamate responses from pyloric motor neurons in lobster stomatogastric ganglion. J. Neurophysiol. 1997;78:3210–3221. doi: 10.1152/jn.1997.78.6.3210. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Peck JH, Harris-Warrick RM. Amine modulation of electrical coupling in the pyloric network of the lobster stomatogastric ganglion. J. Comp. Physiol. A. 1993;172:715–732. doi: 10.1007/BF00195397. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Peck JH, Harris-Warrick RM. Differential modulation of chemical and electrical components of mixed synapses in the lobster stomatogastric ganglion. J. Comp. Physiol. A. 1994;175:233–249. doi: 10.1007/BF00215119. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Peck JH, Harris-Warrick RM. Distributed amine modulation of graded chemical transmission in the pyloric network of the lobster stomatogastric ganglion. J. Neurophysiol. 1995;74:437–452. doi: 10.1152/jn.1995.74.1.437. [DOI] [PubMed] [Google Scholar]
- Jorge-Rivera JC, Sen K, Birmingham JT, Abbott LF, Marder E. Temporal dynamics of convergent modulation at a crustacean neuromuscular junction. J. Neurophysiol. 1998;80:2559–2570. doi: 10.1152/jn.1998.80.5.2559. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman V, Fénelon VS, Richards KS, Thirumalai V, Meyrand P, Marder E. Sequential developmental acquisition of cotransmitters in identified sensory neurons of the stomatogastric nervous system of the lobsters, Homarus americanus and Homarus gammarus . J. Comp. Neurol. 1999;408:318–334. doi: 10.1002/(sici)1096-9861(19990607)408:3<318::aid-cne2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Kravitz EA. Serotonin and aggression: insights gained from a lobster model system and speculations on the role of amine neurons in a complex behavior. J. Comp. Physiol. A. 2000;186:221–238. doi: 10.1007/s003590050423. [DOI] [PubMed] [Google Scholar]
- Kravitz EA, Glusman S, Harris-Warrick RM, Livingstone MS, Schwarz T, Goy MF. Amines and a peptide as neurohormones in lobsters: actions on neuromuscular preparations and preliminary behavioural studies. J. Exp. Biol. 1980;89:159–175. doi: 10.1242/jeb.89.1.159. [DOI] [PubMed] [Google Scholar]
- Kvarta MD, Harris-Warrick RM, Johnson BR. Neuromodulator-evoked synaptic metaplasticity within a central pattern generator network. J. Neurophysiol. 2012;108:2846–2856. doi: 10.1152/jn.00586.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski MA, Gabranski ER, Huber KE, Chapline MC, Christie AE, Dickinson PS. Coordination of distinct but interacting rhythmic motor programs by a modulatory projection neuron using different co-transmitters in different ganglia. J. Exp. Biol. 2013;216:1827–1836. doi: 10.1242/jeb.082503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Feuvre Y, Fenelon VS, Meyrand P. Ontogeny of modulatory inputs to motor networks: early established projection and progressive neurotransmitter acquisition. J. Neurosci. 2001;21:1313–1326. doi: 10.1523/JNEUROSCI.21-04-01313.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz PH, Unal E, Hassett RP, Smith CM, Bucklin A, Christie AE, Towle DW. Functional genomics resources for the North Atlantic copepod, Calanus finmarchicus: EST database and physiological microarray. Comp. Biochem. Physiol. Part D Genomics Proteomics. 2012;7:110–123. doi: 10.1016/j.cbd.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JT, Lee PP, Chen OC, Cheng W, Kuo CM. Dopamine depresses the immune ability and increases susceptibility to Lactococcus garvieae in the freshwater giant prawn, Macrobrachium rosenbergii . Fish Shellfish Immunol. 2005;19:269–280. doi: 10.1016/j.fsi.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Lohrmann DM, Kamemoto FI. The effect of dibutyryl cAMP on sodium uptake by isolated perfused gills of Callinectes sapidus . Gen. Comp. Endocrinol. 1987;65:300–305. doi: 10.1016/0016-6480(87)90177-8. [DOI] [PubMed] [Google Scholar]
- Marshall SM, Orr AP. On the biology of Calanus finmarchicus VIII. Food uptake, assimilation and excretion in adult and stage V Calanus . J. Mar. Biol. Ass. UK. 1955;34:495–529. [Google Scholar]
- Mauchline J. The biology of calanoid copepods. Adv. Mar. Biol. 1998;33:1–710. [Google Scholar]
- McCoole MD, Baer KN, Christie AE. Histaminergic signaling in the central nervous system of Daphnia and a role for it in the control of phototactic behavior. J. Exp. Biol. 2011;214:1773–1782. doi: 10.1242/jeb.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meise CJ, O’Reilly JE. Spatial and seasonal patterns in abundance and age-composition of Calanus finmarchicus in the Gulf of Maine and on Georges Bank: 1977–1987. Deep-Sea Res. II. 1996;43:1473–1501. [Google Scholar]
- Monastirioti M. Biogenic amine systems in the fruit fly Drosophila melanogaster . Microsc. Res. Tech. 1999;45:106–121. doi: 10.1002/(SICI)1097-0029(19990415)45:2<106::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Morris S, Ahern MD. Regulation of urine reprocessing in the maintenance of sodium and water balance in the terrestrial Christmas Island red crab Gecarcoidea natalis investigated under field conditions. J. Exp. Biol. 2003;206:2869–2881. doi: 10.1242/jeb.00499. [DOI] [PubMed] [Google Scholar]
- Morris S, Greenaway P, Adamczewska AM, Ahern MD. Adaptations to a terrestrial existence in the robber crab Birgus latro L. IX. Hormonal control of post-renal urine reprocessing and salt balance in the branchial chamber. J. Exp. Biol. 2000;203:389–396. doi: 10.1242/jeb.203.2.389. [DOI] [PubMed] [Google Scholar]
- Mulloney B, Hall WM. Neurons with histaminelike immunoreactivity in the segmental and stomatogastric nervous systems of the crayfish Pacifastacus leniusculus and the lobster Homarus americanus . Cell Tissue Res. 1991;266:197–207. doi: 10.1007/BF00678725. [DOI] [PubMed] [Google Scholar]
- Neckameyer WS, Quinn WG. Isolation and characterization of the gene for Drosophila tyrosine hydroxylase. Neuron. 1989;2:1167–1175. doi: 10.1016/0896-6273(89)90183-9. [DOI] [PubMed] [Google Scholar]
- Panksepp JB, Yue Z, Drerup C, Huber R. Amine neurochemistry and aggression in crayfish. Microsc. Res. Tech. 2003;60:360–368. doi: 10.1002/jemt.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck JH, Gaier E, Stevens E, Repicky S, Harris-Warrick RM. Amine modulation of Ih in a small neural network. J. Neurophysiol. 2006;96:2931–2940. doi: 10.1152/jn.00423.2005. [DOI] [PubMed] [Google Scholar]
- Pedetta S, Kaczer L, Maldonado H. Individual aggressiveness in the crab Chasmagnathus: Influence in fight outcome and modulation by serotonin and octopamine. Physiol. Behav. 2010;101:438–445. doi: 10.1016/j.physbeh.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Pepin P, Head EJH. Seasonal and depth-dependent variations in the size and lipid contents of stage 5 copepodites of Calanus finmarchicus in the waters of the Newfoundland Shelf and the Labrador Sea. Deep Sea Res. I. 2009;56:6989–1002. [Google Scholar]
- Pepin P, Parrish CC, Head EJH. Late autumn condition of Calanus finmarchicus in the northwestern Atlantic: evidence of size-dependent differential feeding. Mar. Ecol. Prog. Ser. 2011;423:155–166. [Google Scholar]
- Pulver SR, Thirumalai V, Richards KS, Marder E. Dopamine and histamine in the developing stomatogastric system of the lobster Homarus americanus . J. Comp. Neurol. 2003;462:400–414. doi: 10.1002/cne.10767. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reygondeau G, Beaugrand G. Future climate-driven shifts in distribution of Calanus finmarchicus . Global Change Biol. 2011;17:756–766. [Google Scholar]
- Richards KS, Simon DJ, Pulver SR, Beltz BS, Marder E. Serotonin in the developing stomatogastric system of the lobster, Homarus americanus . J. Neurobiol. 2003;54:380–392. doi: 10.1002/neu.10136. [DOI] [PubMed] [Google Scholar]
- Rieger V, Harzsch S. Embryonic development of the histaminergic system in the ventral nerve cord of the marbled crayfish (Marmorkrebs) Tissue Cell. 2008;40:113–126. doi: 10.1016/j.tice.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Ruiz-Vázquez P, Moulard M, Silva FJ. Structure of the phenylalanine hydroxylase gene in Drosophila melanogaster and evidence of alternative promoter usage. Biochem. Biophys. Res. Commun. 1996;225:238–242. doi: 10.1006/bbrc.1996.1160. [DOI] [PubMed] [Google Scholar]
- Sandeman DC, Benton JL, Beltz BS. An identified serotonergic neuron regulates adult neurogenesis in the crustacean brain. Dev Neurobiol. 2009;69:530–545. doi: 10.1002/dneu.20722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Budhiraja P, Walter I, Beltz BS, Peckol E, Kravitz EA. Developmental expression of the octopamine phenotype in lobsters, Homarus americanus . J. Comp. Neurol. 1996;371:3–14. doi: 10.1002/(SICI)1096-9861(19960715)371:1<3::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Sosa MA, Baro DJ. Amine effects on aggression in the giant tropical freshwater prawn Macrobrachium rosenbergii. In: Wiese K, editor. The Crustacean Nervous System. Berlin: Springer-Verlag; 2002. pp. 143–155. [Google Scholar]
- Speirs DC, Gurney WSC, Heath MR, Horbelt W, Wood SN, de Cuevas BA. Ocean-scale modeling the distribution, abundance, and seasonal dynamics of the copepod Calanus finmarchicus . Mar. Ecol. Prog. Ser. 2006;313:173–192. [Google Scholar]
- Strausfeld NJ, Andrew DR. A new view of insect-crustacean relationships I. Inferences from neural cladistics and comparative neuroanatomy. Arthropod Struct. Dev. 2011;40:276–288. doi: 10.1016/j.asd.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Stuart AE. From fruit flies to barnacles, histamine is the neurotransmitter of arthropod photoreceptors. Neuron. 1999;22:431–433. doi: 10.1016/s0896-6273(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Sullivan JM, Benton JL, Beltz BS. Serotonin depletion in vivo inhibits the branching of olfactory projection neurons in the lobster deutocerebrum. J. Neurosci. 2000;20:7716–7721. doi: 10.1523/JNEUROSCI.20-20-07716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CH, Hartline DK. Novel organization and development of copepod myelin. i. ontogeny. J. Comp. Neurol. 2011;519:3259–3280. doi: 10.1002/cne.22695. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Benton JL, Beltz BS. 5-HT receptors mediate lineage-dependent effects of serotonin on adult neurogenesis in Procambarus clarkii . Neural Dev. 2011;6:2. doi: 10.1186/1749-8104-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.