Abstract
Background
Porphyromonas gingivalis (Pg) is one of a constellation of oral organisms associated with human chronic periodontitis. While adaptive immunity to periodontal pathogen proteins has been investigated and is an important component of periodontal bone resorption, the effect of periodontal pathogen DNA in eliciting systemic and mucosal antibody and modulating immune responses has not been investigated.
Methods
Rowett rats were locally injected with whole genomic Pg DNA in alum. Escherichia coli (Ec) genomic DNA, Fusobacterium nucleatum (Fn) genomic DNA, and saline/alum injected rats served as controls. After various time points, serum IgG and salivary IgA antibody to Ec, Fn or Pg were detected by ELISA. Serum and salivary antibody reactions with Pg surface antigens were determined by western blot analyses and the specific antigen was identified by mass spectrometry. Effects of genomic DNA immunization on Pg bacterial colonization and experimental periodontal bone resorption were also evaluated.
Results
Sera from Pg DNA, Ec DNA and Fn DNA-injected rats did not react with Ec or Fn bacteria. Serum IgG antibody levels to Pg and Pg surface extracts were significantly higher in animals immunized with Pg DNA as compared to the control groups. Rats injected with Pg DNA demonstrated a strong serum IgG and salivary IgA antibody reaction solely to Pg fimbrillin (41kD), the major protein component of Pg fimbriae. In the Pg DNA-immunized group, the numbers of Pg bacteria in oral cavity and the extent of periodontal bone resorption were significantly reduced after Pg infection.
Conclusions
This study suggests that infected hosts may select specific genes from whole genomic DNA of the periodontal pathogen for transcription and presentation. The results indicate that the unique gene selected can initiate a host protective immune response to the parent bacterium.
Keywords: Immune response, Periodontal disease, Porphyromonas gingivalis, Bacterial genomic DNA, Bone resorption, Vaccine
INTRODUCTION
Porphyromonas gingivalis (Pg) is one of a constellation of oral microorganisms associated with human chronic periodontitis [1]. Among a variety of virulence determinants expressed, Pg fimbriae are important cell surface virulence factors involved in Pg colonization of the periodontal surface and pathogenicity [2]. Pg fimbriae are critical determinants for induction of periodontitis in rats and, when used as immunogens, can reduce periodontal destruction [3]. Studies have shown that Pg fimbriae have important immunomodulating properties and can stimulate the production of inflammatory cytokines in human monocytes and polymorphonuclear leukocytes [2, 4]. Studies indicated that Pg has evolved multiple levels of control of fimbrial gene expression to enhance its survival in hostile environments [5, 6]. Mutation of the fimA gene, encoding fimbrillin, the major subunit of the fimbriae, prevents Pg to adhere to host cells [7]. Furthermore, it was suggested that genes encoding the minor components of the fimbriae fimC, fimD and fimE, play critical roles in the adhesive activities of the mature FimA fimbriae in Pg [8]. Thus, Pg fimbriae represent important cell structures involved in mucosal pathogenesis and periodontitis by facilitating colonization and invasion of mucosal cells and induction of inflammatory responses [9].
Adaptive immunity can be an important component in response to periodontal pathogens [10-12]. Considerable efforts have been made to seek effective antigens that can elicit functional protection against periodontal infection and tissue destruction. Studies have shown that DNA immunization can induce host immune responses in both systemic and mucosal compartments [13-15].
Recent studies have used plasmid DNA encoding a protein for vaccination, which usually consists of a cytomegalovirus (CMV) promoter for efficient gene expression in mammalian cells, followed by a region encoding the desired protein antigen. Vaccines of DNA encoding a single component of Pg (including fimbriae, Arg-gingipain and Lys-gingipain) have been described [16-18].
Naked genomic DNA is also effective as a vaccine [19] and epitopes encoded in such DNA can be expressed in recipient cells and can induce antigen-specific immune responses [20-22]. However, the ability and the efficacy of such genomic DNA to elicit antibody and modulate immune response have not be explored. This entity could be of considerable clinical importance since it has been suggested that bacterial DNA liberated at the site of infection is likely to sustain the local inflammatory response [23] and host immune responses to bacterial DNA may contribute to immunity to bacteria[24].
In this study, we tested the hypothesis that host selects the gene from naked whole genomic DNA that encodes an antigen that will initiate a protective immune response. Therefore, we allowed the host to select antigens by using bacterial whole genomic DNA as an immunogenicity probe.
MATERIALS AND METHODS
Preparation of Whole Genomic DNA
Pg bacteria (strain 33277) were grown in trypticase soy broth (TSB) containing 1% yeast extract, 5μg/mL hemin and 2.5μg/mL menadione. Fn bacteria (strain 25586) were grown in mycoplasma broth, and Ec bacteria (strain DHI) were grown in LB broth. Whole genomic DNA was prepared by phenol-chloroform isoamyl alcohol extraction and ethanol precipitation to remove protein contents, followed by anion exchange chromatography (Qiagen) to remove LPS. The purity of each DNA preparation was checked by the limulus amebocyte lysate (LAL) test to quantitate LPS (Associates of Cape Cod, Inc, Falmouth, MA). Plasmid DNA containing full length or partial FimA gene (aa224-337), and FimA mutant Pg strain (DPG3) were a kindly gift from Dr. Ashu Sharma at The State University of New York, University at Buffalo.
Animals and Injection Protocol
All animals were inbred Rowett rats maintained under pathogen-free conditions in laminar flow cabinets. Experiments using these animals were approved by the Forsyth Institute’s Internal Animal Care and Use Committee (IACUC). Female Rowett rats (6-9 rats/group) were injected subcutaneously in the salivary gland vicinity with PBS buffer in alum (group I), alum/Ec DNA (group II), alum/Fn DNA (group III), or alum/Pg DNA (group IV). Four milligram of alum and 100μg DNA were injected to each animal. Animals were immunized first time on Day 0 and a second time on Week 15. Blood and saliva samples were taken at approximately 3-week intervals. Animals were infected with Pg for 4 consecutive days starting on Week 24 and the bacteria in oral cavity were recovered 16 weeks later. A detailed schedule of immunization, infection and bacterial recovery is shown in Figure 1.
Figure 1. A scheme of detailed schedule of immunization, infection and bacterial recovery.
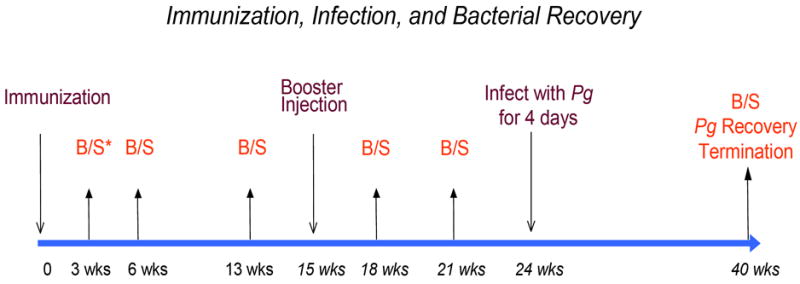
Female Rowett rats (6-9 rats/group) were injected subcutaneously in the salivary gland vicinity with PBS buffer in alum (group I) or with alum/DNA (group II, alum/Ec DNA; group III, alum/Fn DNA, group IV, alum/Pg DNA). Four milligram of alum and 100μg DNA were injected to each animal. Animals were immunized first time on Day 0 and a second time on Week 15. Blood samples were taken at approximately 3-week intervals. Animals were infected with Pg for 4 consecutive days starting on Week 24. The Pg bacteria in oral cavity were counted and periodontal bone resorption were measured 16 weeks later.
*B/S: Bleed and Salivation.
Determination of Antibody Levels by ELISA
Antibody levels in sera and in saliva were determined using isotype-specific reagents for IgG in serum and for IgA in saliva [25]. Antigens tested in ELISA included Pg whole bacteria (107/well), Pg membrane extracts (50μg/well) and purified Pg fimbriae [26] (2μg/well) (a kindly gift from Dr. Caroline Genco at Boston University Medical Center). Briefly, each of these antigens was coated onto 96-well ELISA plate. Rat serum (1:100-1:500) was applied to the plate and rabbit anti-rat IgG antibody was added followed by ALP-conjugated goat anti-rabbit IgG antibody. Rat saliva (1:4-1:20) was applied to the plate and mouse anti-rat IgA antibody was added followed by ALP-conjugated goat anti-mouse IgG antibody. Colorimetric reactions were developed with p-nitropenyl phosphate (pNPP) substrate. ELISA measurements were performed and expressed as ELISA units (EU) converted based on a reference curve provided by dilution of an antibody-containing hyperimmune rat serum collected from rats immunized with Pg [27].
All readings were recorded with a microplate reader (Bio-Tek Instruments) at 405nm.
T Cell Proliferation Assays
Cells from cervical lymph nodes were recovered and cultured in 96-well plates (1 × 104 cells/ well) in the presence or absence of Pg 33277 (1 × 107), Fimbriae deficient Pg DPG3 (1 × 107), or purified Pg fimbriae (2μg/well). [3H]-thymidine (0.5 μCi/well) was added for the last 16 h of a total of 4 days in culture. Samples were harvested onto glass fiber filters, and radioactivity (cpm) was measured in a scintillation spectrometer (Beckman Coulter, Fullerton, CA).
Determination of Unknown Antigen by Western Blot and Mass Spectrometry
Pg membrane extracts or purified fimbrial protein were resolved on 10% SDS-PAGE gel and transferred onto nitrocellulose membrane. The membrane was incubated with sera (1:200 dilution) or saliva (1:3.5 dilution) samples from each rat for 2hrs at room temperature. After washing, the membrane was incubated with goat anti-rat antibody (1:5000) conjugated with HRP (Biosource, Camarillo, CA) for 1hr. The color was then developed with HRP conjugate substrate kit (BioRad, Hercules, CA). A duplicate SDS-PAGE gel with the same sample used for western blot was fixed in a solution containing 40% methanol and 10% acetic acid for 20min. The gel was then stained with coomassie blue stain (Invitrogen, Carlsbad, CA) and the single band corresponding to the band detected in western blot was sliced out from the gel. After destaining in a solution containing 50% methanol and 5% acetic acid, the gel slice was digested and analyzed by a MALDI Orthogonal Time of Flight Mass Spectrometer (prOTOF™ 2000, PerkinElmer, Shelton, CT). The detected peptide sequences were searched against various databases including SwissProt, PIR, PRF, PDB to identify the protein.
Bacterial Growth & Quantitation
Oral swabs from each rat were grown on blood agar plates containing menadione and hemin for 3 days and DNA was extracted from each plate by proteinase K digestion. Equal amount of DNA from each sample was subjected to PCR amplification of Pg-specific DNA fragments using the following primers: Forward: 5’-GAGTTTGATYMTGGCTCAG-3’ and Reverse: 5’-TCAGTCGCAGTATGGCAA-3’. PCR reactions were then resolved in 1% agarose gel and separated by electrophoresis. The density of amplified PCR products was measured and the densitometry readings were converted to the number of Pg bacteria based on a standard curve determined from direct bacterial counts.
Measurement of Periodontal Bone Loss
After defleshing of mandibular and maxillary jaws, the distance from the cementoenamel junction (CEJ) to the alveolar crest of both buccal and lingual roots of all six molar teeth in each rat (n= 4-6) was measured using a microscope with a reticule eyepiece under 25x magnification. The sum of the recordings was used as a measure of the total bone loss.
Statistical Analysis
Results obtained from periodontal bone resorption and quantitation of Pg DNA are expressed as means ± standard deviations (SD). Data compiled from T cell proliferation assays, from ELISAs for serum IgG and salivary IgA are expressed as means ± standard errors (SE). A Student’s t test was used to evaluate significance and P values of ≤0.05 were considered statistically significant.
RESULTS
Ec or Fn DNA-immunized rats did not produce an antibody response to Ec, Fn or Pg bacteria, but Pg DNA-immunized rats responded robustly to Pg bacterial antigen(s)
Three and six weeks post immunization, antibody levels to Ec or Fn from DNA immunized rats (group II, III, and IV) did not differ from rats injected with alum only (group I), suggesting that immunization with Ec or Fn whole genomic DNA does not elicit a response associated with these bacteria (Fig. 2A-B). However, the Pg DNA immunized group (group IV) demonstrated significantly elevated antibody levels to Pg whole bacteria compared to the control groups (group I, II and III) at both three and six weeks post immunization (Fig. 2C).
Figure 2. Detection of serum IgG antibody to Ec, Fn or Pg bacteria and to Pg extracts by ELISA.
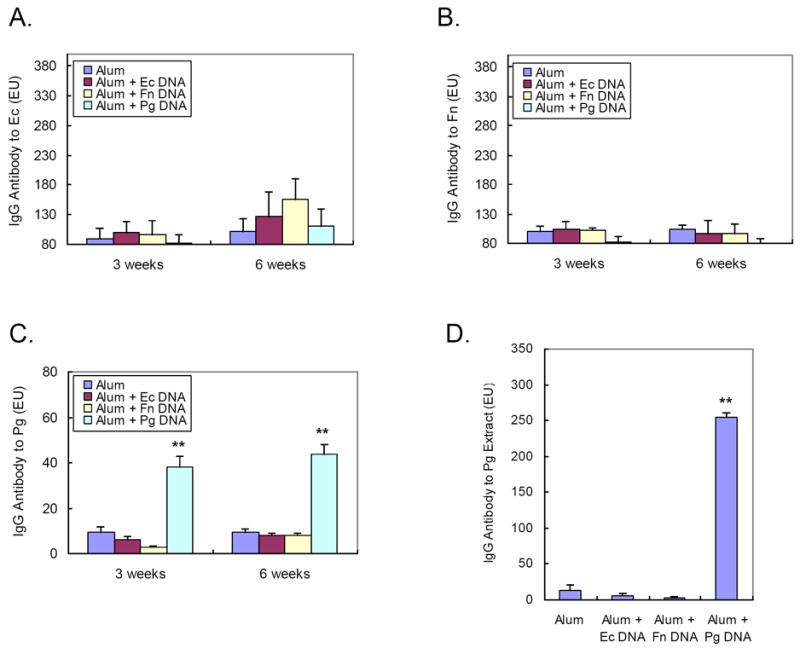
Three weeks and six weeks after immunization, rat sera were collected and reacted with ELISA plates coated with (A) Ec bacteria, (B) Fn bacteria or (C) Pg bacteria. (D) Rat sera from six weeks post immunization were also reacted with ELISA plates coated with Pg extracts as described in “Materials and Methods”. Each experiment was performed in duplicate and readings were recorded by a microplate reader at 405nm. (Mean ± SE, **p< 0.01)
Pg membrane extract contains antigen(s) encoded by naked Pg genomic DNA
We prepared the Pg extracts containing mostly Pg surface proteins and determined the serum IgG antibody response to these cell surface proteins by ELISA. Serum samples taken from rats of six weeks post-immunization were used. Sera from the Pg DNA immunized group (group IV) demonstrated significantly elevated antibody levels to component(s) in the Pg membrane extract compared to other groups (group I, II and III) (Fig. 2D).
Pg fimbriae are the sole component in the Pg extract that reacts with the IgG anti-Pg antibody
The potential antigen(s) to which the observed serum responses were directed were investigated by western blot analyses. When the Pg extract was reacted with the sera from Pg DNA-immunized rats, only a single antigen with molecular weight of ~41kD was detected in the Pg extract (Fig. 3A). The corresponding band from an identical SDS-PAGE gel was excised and subjected to Mass Spectrometry analysis (Harvard Medical School Mass Spectrometry Core). The composition determined was identical to that of Pg fimbrillin precursor (NCBI accession#: JN0915, strain BH18/10, also see Supplementary Data). The findings were further confirmed by a western blot analysis showing that serum IgG antibody from Pg DNA-immunized animals reacted with purified Pg fimbriae and not with other Pg components such as Lys-gingipain, Arg-Gingipain or hemin/hemoglobin utilization receptor (Fig. 3B).
Figure 3. Characterization of antigen responsible for the observed antibody responses by western blot.
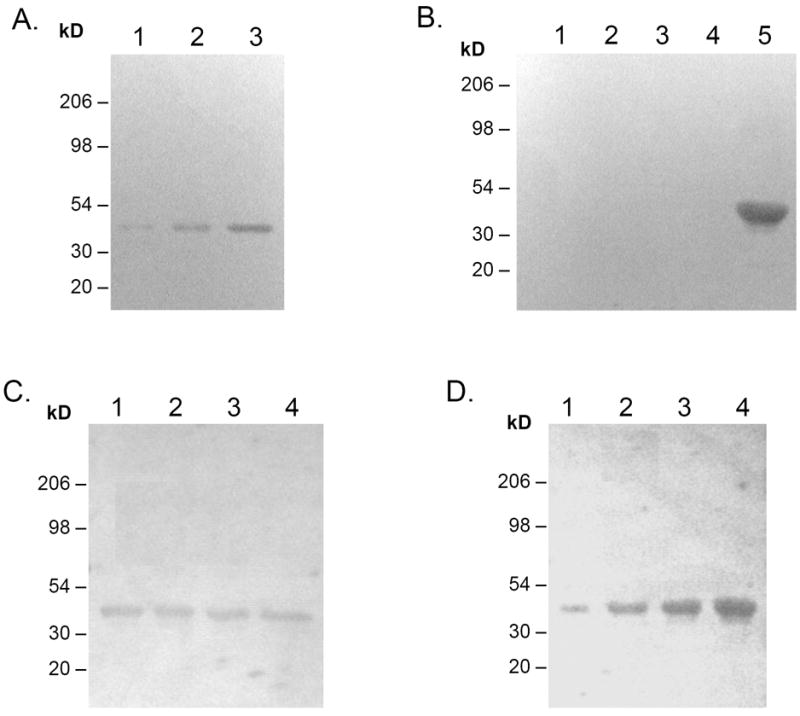
To determine antibody response to immunization, Pg extract or purified Pg fimbriae were blotted onto nitrocellulose as follows: (A) Various amount of Pg extracts was blotted onto nitrocellulose as follows: 1. 10μg, 2. 30μg, 3. 90μg. (B) Purified Pg components were blotted onto nitrocellulose as follows: 1. Lys-gingipain, 2. Gingipain R, 3. Gingipain R1, 4. Recombinant hemin/hemoglobin utilization receptor, 5. Purified Pg fimbriae (67kD + 41kD). (C) Lane 1-4, 100μg Pg extract each; (D) Lane 1-4, 1μg, 3μg, 6μg, 9μg of purified Pg fimbriae. The membrane was incubated with pooled serum (1:200) or pooled saliva (1:3.5) from rats immunized with Pg DNA (6 weeks post immunization), then with goat anti-rat IgG-HRP (1:10000). Color was developed with HRP substrate kit (BioRad).
Pg fimbriae are the sole component in the Pg extract that reacts with the salivary IgA antibody from Pg DNA-immunized rats
In order to assess the effect of Pg DNA immunization on mucosal immune response, salivary IgA antibody response was tested against Pg extract and purified Pg fimbriae. Western blot showed similar results as those observed in serum antibody response (Fig. 3C-D). Salivary IgA antibody from Pg DNA immunized rats reacted only with a single component in the Pg extract with molecular weight of 41kD (Fig. 3C), which was confirmed to be Pg fimbriae in a separate experiment (Fig. 3D).
Kinetics of serum IgG and salivary IgA response to Pg fimbriae (41 kD) in DNA-immunized rats
Throughout the tested period, serum IgG and salivary IgA antibody to Pg fimbriae in Pg DNA-immunized rats were significantly higher than in those injected with Ec DNA or alum alone (Fig. 4). Different patterns were observed for systemic and mucosal responses. The increase of serum IgG antibody to Pg fimbriae peaked at 13 weeks post immunization and gradually decreased despite a second booster injection at 15 weeks (Fig. 4A). The increase of salivary IgA antibody to Pg fimbriae quickly peaked at 6 weeks post immunization and began to decrease afterward, until after the second booster injection at 15 weeks, when the antibody level was increased again and to the highest level at 21 weeks (Fig. 4B). These differences may stem from the nature of the injection route for immunization and demonstrate a coordinate host immune responses to foreign antigens.
Figure 4. Kinetics of serum IgG and salivary IgA antibody response to Pg fimbriae (41kD) in DNA immunized rats.
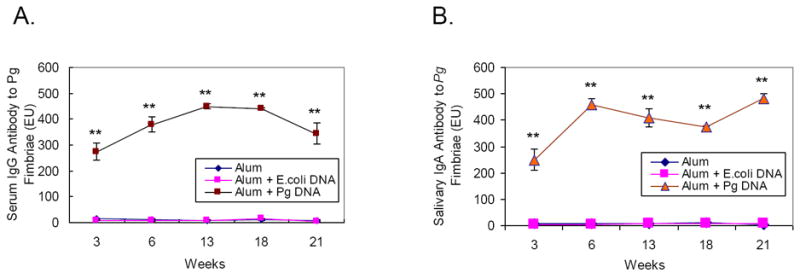
After immunization, rat sera (A) and saliva (B) were collected at different time points (3 weeks, 6 weeks, 13 weeks, 18 weeks, and 21 weeks) and reacted with ELISA plates coated with purified Pg fimbriae (41kD) as described in “Materials and Methods”. Each experiment was performed in duplicate and readings were recorded by a microplate reader at 405nm. (Mean ± SE, **p < 0.01)
Pg DNA immunization significantly decreased the number of Pg bacteria recoverable from the rat oral cavity and resulted in significantly reduced periodontal bone resorption of Pg infected rats
The number of Pg recovered from rats immunized with Pg DNA was significantly reduced compared to the control group, suggesting a protective effect of Pg DNA immunization against Pg infection (Fig. 5A). Interestingly, the number of Pg recovered from rats immunized with Fn DNA was also reduced compared to control group, although the extent of this reduction was not as great as those observed in Pg DNA immunized group. This suggests that non-antigen specific immune reactions could exist that lead to the reduction of bacterial infection in animals immunized with various Pg DNA. Furthermore, only rats immunized with Pg DNA demonstrated a significant reduction of total bone resorption as compared to control rats (without DNA injection) after infection with live Pg (Fig. 5B). As expected, rats devoid of Pg infection exhibited minimal level of bone resorption.
Figure 5. Recoveries of Pg bacteria and periodontal bone resorption after immunization with bacterial whole genomic DNA and infection with live Pg.
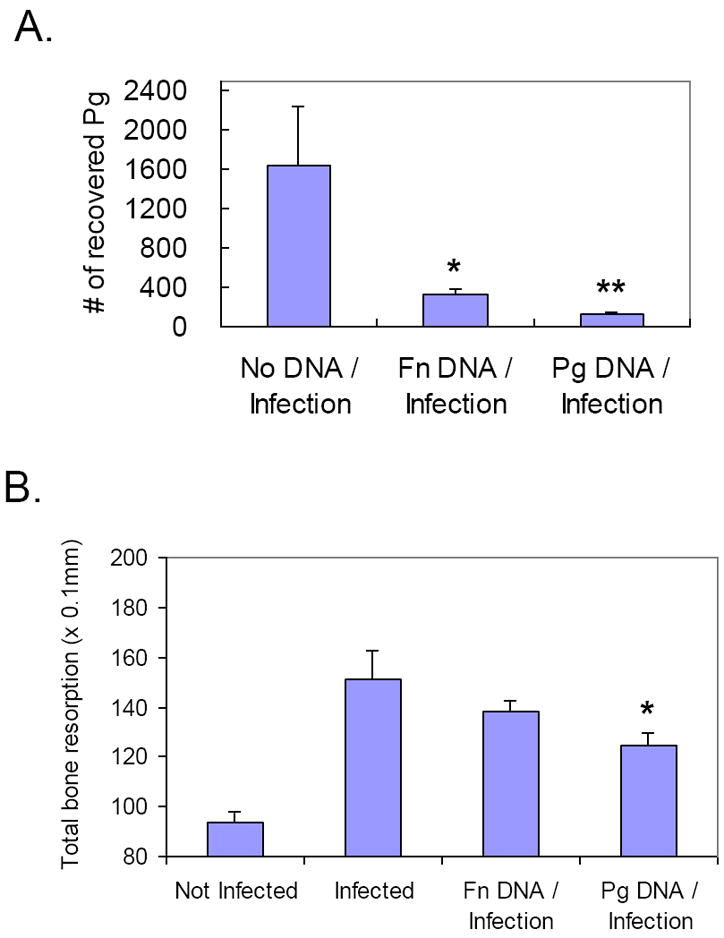
A. At the termination of the experiment (40 weeks), oral swabs from each rat were grown on blood agar plates for 3 days and were then subjected to DNA extraction. An equal amount of DNA from each sample was amplified by PCR using Pg-specific primers. Quantitative estimates of the numbers of Pg was performed by measuring the density of amplified PCR product and the densitometry readings were converted to the number of Pg bacteria based on a standard curve. B. At the termination of the experiment (40 weeks), rat heads were defleshed and processed to measure periodontal bone resorption. The distances from cemento-enamel junction (CEJ) to the alveolar crest (AC) of each root was measured and the sum of the recordings was used as a measure of the total bone loss. (Mean ± SD, n=5, *p< 0.05, **p < 0.01).
Immunization with Pg DNA fragment encoding fimbriae is not sufficient enough to elicit immune response
Three weeks and six weeks after immunization, rats immunized with DNase I-treated Pg DNA demonstrated little or no response to Pg fimbriae (Fig. 6A-B). However, rats immunized with proteinase K-treated Pg DNA demonstrated a level of serum IgG and salivary IgA response to Pg fimbriae similar to that of rats immunized with untreated Pg DNA (Fig. 6A-B). These results indicated that the observed host responses were initiated by Pg DNA rather than the trace amount of antigenic protein possibly contained in the Pg DNA preparation. Rats immunized with plasmid DNA containing full length or partial FimA gene (aa224-337) did not demonstrated any serum IgG and salivary IgA response to Pg fimbriae (Fig. 6A-B). This suggests that the gene encoding antigen fimbrillin is not sufficient enough to elicit immune response after its introduction into host tissues. Specific promoter element(s) in Pg genomic DNA are required for the expression of the protein antigen. Comparable results were obtained from T cell proliferation assay, showing that only rats immunized with intact Pg DNA (untreated or treated with proteinase K) demonstrated increased [3H]-thymidine incorporation of cultured lymphocytes (Fig. 6C). This indicates an involvement of T cells in the local draining lymph node, to which the antigen was presented by the primed APCs migrated from the DNA injection sites, in the adaptive immune response to DNA immunization.
Figure 6. Immunization with Pg DNA fragment encoding fimbriae is not sufficient enough to elicit immune response.
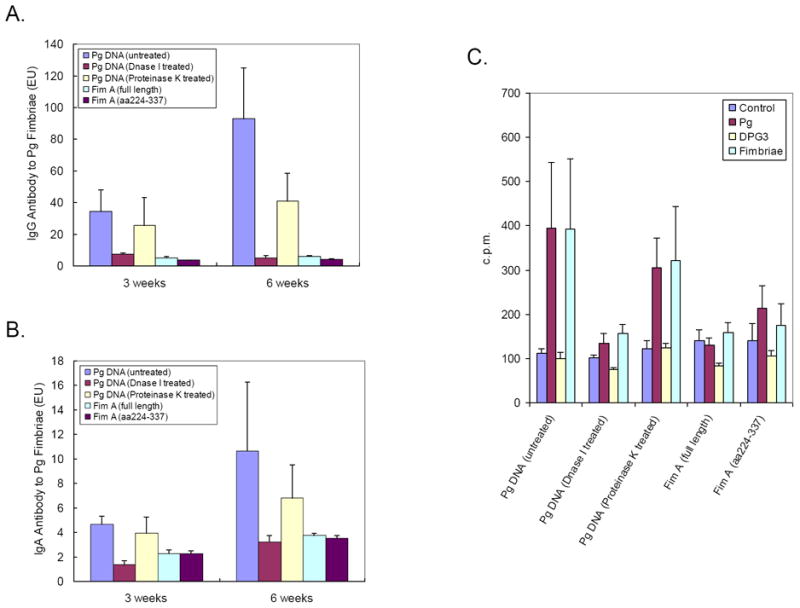
To further verify the immunogenicity of Pg whole genomic DNA, rats were divided into five groups (n=5 per group) and were immunized with 100 μg of untreated Pg DNA (as positive control), Pg DNA treated with 0.1U/μL DNase I at RT for 30 min (to digest all double-stranded DNA to oligonucleotides), Pg DNA treated with 1mg/mL proteinase K at 50°C for 2hrs (to remove trace amount of proteins), plasmid DNA containing full length FimA gene (encoding aa1-337), or plasmid DNA containing truncated FimA gene (encoding aa224-337), respectively. Both plasmid DNA were kindly provided by Dr. Ashu Sharma from University at Buffalo, Buffalo, NY. Three weeks and six weeks after immunization, rat sera and saliva were collected and reacted with ELISA plates coated with purified Pg fimbriae for the detection of serum IgG (A) and salivary IgA (B) as described in “Materials and Methods”. Each experiment was performed in duplicate and readings were recorded by a microplate reader at 405nm. (C) Six weeks after immunization, cells from rat cervical lymph nodes were cultured on 96-well plates in the presence or absence of Pg (107/well), fimA-deletion mutant DPG3 (107/well) [34], and purified Pg fimbriae (2μg/well). [3H]-thymidine (0.5 μCi/well) was added for the last 16 h of a total of 4 days in culture. Samples were harvested and radioactivity (cpm) was measured in a β-scintillation spectrometer. The data are Mean ± SE cpm values of an experiment performed in triplicate.
DISCUSSION
DNA vaccines represent a novel approach to the control of infectious disease. Both cellular and humoral immune responses are induced without the concerns associated with traditional vaccines (low cellular immune activation with non-viable vaccines and potential reversal to pathogenicity within the individual immunized with live vaccines). Most of the recent reports have used eukaryotic expression vectors carrying a virus (such as cytomegalovirus, CMV) promoter to induce immune responses to some identified antigens [16-18, 28].
It has long been considered that bacterial DNA is not inert but is sensed through unmethylated DNA motifs (CpG) and activates immune cells such as macrophages and dendritic cells [29-32] involved in the innate immunity. Our preliminary data have demonstrated that Pg DNA appears to modulate pro-inflammatory cytokines (IL-1β, IL-8) and the anti-inflammatory cytokine (IL-10) production in cultured THP-1 cells or primary monocytes (data not shown). In this study, we have indicated a novel mechanism by which bacterial genomic DNA could be sensed and processed by the host to present an antigen that triggers the systemic and mucosal adaptive immune system. This is probably achieved by the recognition of unique genetic features and the subsequent transcription of specific genes in bacterial genomic DNA, leading to an adaptive immune response towards major immunogenic antigens. Previous studies have shown that epitopes encoded in tumor-derived genomic DNA can be expressed in recipient cells after transduction and can induce tumor-specific T cell formation and immune responses [20, 21]. More importantly, it has been demonstrated that protein expression of transferred genes can be detected after direct injection of genes into mouse skeletal muscle in vivo without any special delivery system [33]. Our results demonstrated that antibody reactivity with Pg could be found in the Pg extract and was subsequently found to be directed to Pg fimbriae but not to other common Pg antigen tested. These results substantiated the notion that the host can select an antigen from the whole genomic DNA that is encoded and transcribed. Pg whole genomic DNA induced host immune response to Pg fimbriae alone suggested that possibly a unique promoter element in the Pg genome is needed for the elicitation of such immune responses. The reduction system using plasmid FimA gene substantiate that the antibody response elicited by the genomic DNA cannot be achieved by FimA gene alone but by multiple components involving other features in DNA sequences or structures. Involvement of the fimA promoter could be one of such features contributing to the development of host immune response. It is also possible that other antigens are expressed from the Pg genomic DNA but are not as immunogenic as FimA and thus do not elicit an antibody response. Furthermore, it is conceivable that CpG motifs within genomic DNA act as a ‘danger signal’ for the immune cells and triggers a robust response to DNA-encoded products. This may also explain why plasmid DNA, which lacks CpG motifs, was not very immunogenic (Fig. 6A-B). Further investigations are warranted to elucidate the full mechanism leading to the genomic DNA-elicited antibody responses to Pg.
Our results indicated that when rats immunized with Pg DNA were infected with live Pg bacteria, they demonstrated strong serum IgG and salivary IgA antibody responses (Fig. 4). Colonization with Pg and periodontal bone loss were significantly reduced (Fig. 5). This suggests that immunization of rats with Pg whole genomic DNA elicited a potent systemic and mucosal immune response resulting in reduction of Pg bacteria and the subsequent protection from periodontal bone resorption. Interestingly, the mucosal response was brought to a higher level when animals received a booster injection of Pg DNA, as demonstrated by a further increased salivary IgA antibody titer (Fig. 4B). The protective effect of such immunization against Pg bacteria colonization and periodontal bone resorption can be observed 40 weeks post immunization (Fig. 5). These results indicated that local immune responses (such as IgA antibody production) are inducible and maintained for a sustained period (over 10 months) following the repeated Pg whole genomic DNA immunization. This longevity is probably due to the formation of abundant memory cells that elicit sustained adaptive immunity.
Interestingly, our results demonstrated that Fn DNA immunization led to significantly reduced Pg colonization (Fig. 5A) and some reduction in periodontal bone loss (though not significant, Fig. 5B), even though no cross-reactive antibodies were observed (Fig. 2). The Fn DNA elicited protection against Pg colonization could be due to the CpG motifs in genomic DNA sequence that trigger TLR9-mediated protective immune responses, which is universal to all bacterial genomic DNAs. Therefore, the observed Pg DNA-elicited protection against Pg-induced bone loss might not derived solely from FimA- mediated antibody responses but also be partially contributed by CpG-induced TLR9-mediated protective reactions.
To our knowledge, this is the first demonstration of the host selection from whole genomic DNA of a gene encoding an important antigen that confers virulence. But more importantly it can elicit a protective response. These are very intriguing findings that open a lot of new questions and avenues for investigations in the future. Whether humans will likewise induce a protective response remains to be determined but this is a good proof-of-principle study for vaccine development against periodontitis. This also represents a novel host defense strategy which may have ramifications in other disease situations. Evidence for the bacterial genomic DNA elicited-antibody response is convincing, questions still remain to be answered and warranted for further studies. How bacterial DNA interacts with host machinery so that the major pathogenic antigen can be selected and presented to elicit protective immune response? Why only the antigen from a specific bacterial genomic DNA (Pg) was selected for encoding and presentation by APCs for the subsequent activation of adaptive immunity is totally unclear. The signaling pathway(s) involved in the process of direct or indirect induction of protective immune responses are also unknown. While working on these unanswered questions, it could also be a rewarding task to examine the potential application of whole genomic DNA vaccination as a novel approach to protect susceptible individuals against pathogenic colonization and inflammatory-mediated bone resorption.
Supplementary Material
Highlights.
Rats immunized with P. gingivalis (Pg) genomic DNA had elevated serum IgG to Pg.
A strong serum IgG and salivary IgA response to a major component of Pg fimbriae.
The numbers of Pg in oral cavity were significantly reduced after Pg infection.
The extent of periodontal bone resorption was significantly reduced after Pg infection.
Host may select specific genes from whole genomic DNA of the periodontal pathogen.
Acknowledgments
The work described above was supported by NIH Grants DE-03420, DE-04733, DE-014551 and DE-015254 from National Institute of Dental and Craniofacial Research (NIDCR).
List of Abbreviations
- Pg
Porphyromonas gingivalis
- Ec
Escherichia coli
- Fn
Fusobacterium nucleatum
- LPS
lipopolysaccharide
- ELISA
Enzyme-linked immunosorbent assay
- MS
Mass spectrometry
- APCs
antigen presenting cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ezzo PJ, Cutler CW. Microorganisms as risk indicators for periodontal disease. Periodontol 2000. 2003;32:24–35. doi: 10.1046/j.0906-6713.2003.03203.x. [DOI] [PubMed] [Google Scholar]
- 2.Ogawa T, Uchida H, Hamada S. Porphyromonas gingivalis fimbriae and their synthetic peptides induce proinflammatory cytokines in human peripheral blood monocyte cultures. FEMS Microbiol Lett. 1994;116:237–42. doi: 10.1111/j.1574-6968.1994.tb06707.x. [DOI] [PubMed] [Google Scholar]
- 3.Evans RT, Klausen B, Sojar HT, Bedi GS, Sfintescu C, Ramamurthy NS, et al. Immunization with Porphyromonas (Bacteroides) gingivalis fimbriae protects against periodontal destruction. Infect Immun. 1992;60:2926–35. doi: 10.1128/iai.60.7.2926-2935.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa T, Asai Y, Hashimoto M, Uchida H. Bacterial fimbriae activate human peripheral blood monocytes utilizing TLR2, CD14 and CD11a/CD18 as cellular receptors. Eur J Immunol. 2002;32:2543–50. doi: 10.1002/1521-4141(200209)32:9<2543::AID-IMMU2543>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Xie H, Chung WO, Park Y, Lamont RJ. Regulation of the Porphyromonas gingivalis fimA (Fimbrillin) gene. Infect Immun. 2000;68:6574–9. doi: 10.1128/iai.68.12.6574-6579.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie H, Kozlova N, Lamont RJ. Porphyromonas gingivalis genes involved in fimA regulation. Infect Immun. 2004;72:651–8. doi: 10.1128/IAI.72.2.651-658.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamada N, Watanabe K, Sasakawa C, Yoshikawa M, Yoshimura F, Umemoto T. Construction and characterization of a fimA mutant of Porphyromonas gingivalis. Infect Immun. 1994;62:1696–704. doi: 10.1128/iai.62.5.1696-1704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiyama S, Murakami Y, Nagata H, Shizukuishi S, Kawagishi I, Yoshimura F. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology. 2007;153:1916–25. doi: 10.1099/mic.0.2006/005561-0. [DOI] [PubMed] [Google Scholar]
- 9.Jotwani R, Cutler CW. Fimbriated Porphyromonas gingivalis is more efficient than fimbria-deficient P. gingivalis in entering human dendritic cells in vitro and induces an inflammatory Th1 effector response. Infect Immun. 2004;72:1725–32. doi: 10.1128/IAI.72.3.1725-1732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katz J, Michalek SM. Effect of immune T cells derived from mucosal or systemic tissue on host responses to Porphyromonas gingivalis. Oral Microbiol Immunol. 1998;13:73–80. doi: 10.1111/j.1399-302x.1998.tb00716.x. [DOI] [PubMed] [Google Scholar]
- 11.Yanagita M, Hiroi T, Kitagaki N, Hamada S, Ito HO, Shimauchi H, et al. Nasopharyngeal-associated lymphoreticular tissue (NALT) immunity: fimbriae-specific Th1 and Th2 cell-regulated IgA responses for the inhibition of bacterial attachment to epithelial cells and subsequent inflammatory cytokine production. J Immunol. 1999;162:3559–65. [PubMed] [Google Scholar]
- 12.Michalek SM, Katz J, Childers NK, Martin M, Balkovetz DF. Microbial/host interactions: mechanisms involved in host responses to microbial antigens. Immunol Res. 2002;26:223–34. doi: 10.1385/IR:26:1-3:223. [DOI] [PubMed] [Google Scholar]
- 13.Eo SK, Gierynska M, Kamar AA, Rouse BT. Prime-boost immunization with DNA vaccine: mucosal route of administration changes the rules. J Immunol. 2001;166:5473–9. doi: 10.4049/jimmunol.166.9.5473. [DOI] [PubMed] [Google Scholar]
- 14.Hamajima K, Hoshino Y, Xin KQ, Hayashi F, Tadokoro K, Okuda K. Systemic and mucosal immune responses in mice after rectal and vaginal immunization with HIV-DNA vaccine. Clin Immunol. 2002;102:12–8. doi: 10.1006/clim.2001.5141. [DOI] [PubMed] [Google Scholar]
- 15.He XW, Wang F, Jiang L, Li J, Liu SK, Xiao ZY, et al. Induction of mucosal and systemic immune response by single-dose oral immunization with biodegradable microparticles containing DNA encoding HBsAg. J Gen Virol. 2005;86:601–10. doi: 10.1099/vir.0.80575-0. [DOI] [PubMed] [Google Scholar]
- 16.Kawabata S, Terao Y, Fujiwara T, Nakagawa I, Hamada S. Targeted salivary gland immunization with plasmid DNA elicits specific salivary immunoglobulin A and G antibodies and serum immunoglobulin G antibodies in mice. Infect Immun. 1999;67:5863–8. doi: 10.1128/iai.67.11.5863-5868.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonezawa H, Ishihara K, Okuda K. Arg-gingipain a DNA vaccine induces protective immunity against infection by Porphyromonas gingivalis in a murine model. Infect Immun. 2001;69:2858–64. doi: 10.1128/IAI.69.5.2858-2864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuboniwa M, Amano A, Shizukuishi S, Nakagawa I, Hamada S. Specific antibodies to Porphyromonas gingivalis Lys-gingipain by DNA vaccination inhibit bacterial binding to hemoglobin and protect mice from infection. Infect Immun. 2001;69:2972–9. doi: 10.1128/IAI.69.5.2972-2979.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pardoll DM, Beckerleg AM. Exposing the immunology of naked DNA vaccines. Immunity. 1995;3:165–9. doi: 10.1016/1074-7613(95)90085-3. [DOI] [PubMed] [Google Scholar]
- 20.Whiteside TL, Gambotto A, Albers A, Stanson J, Cohen EP. Human tumor-derived genomic DNA transduced into a recipient cell induces tumor-specific immune responses ex vivo. Proc Natl Acad Sci U S A. 2002;99:9415–20. doi: 10.1073/pnas.142302399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura M, Kikuchi T, Kufe DW, Ohno T. Antitumor effects of fusions composed of dendritic cells and fibroblasts transfected with genomic DNA from tumor cells. Cancer Immunol Immunother. 2004;53:690–6. doi: 10.1007/s00262-004-0511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O-Sullivan I, Ng LK, Martinez DM, Kim TS, Chopra A, Cohen EP. Immunity to squamous carcinoma in mice immunized with dendritic cells transfected with genomic DNA from squamous carcinoma cells. Cancer Gene Ther. 2005;12:825–34. doi: 10.1038/sj.cgt.7700847. [DOI] [PubMed] [Google Scholar]
- 23.Heeg K, Sparwasser T, Lipford GB, Hacker H, Zimmermann S, Wagner H. Bacterial DNA as an evolutionary conserved ligand signalling danger of infection to immune cells. Eur J Clin Microbiol Infect Dis. 1998;17:464–9. doi: 10.1007/BF01691128. [DOI] [PubMed] [Google Scholar]
- 24.Elkins KL, Rhinehart-Jones TR, Stibitz S, Conover JS, Klinman DM. Bacterial DNA containing CpG motifs stimulates lymphocyte-dependent protection of mice against lethal infection with intracellular bacteria. J Immunol. 1999;162:2291–8. [PubMed] [Google Scholar]
- 25.Eastcott JW, Holmberg CJ, Dewhirst FE, Esch TR, Smith DJ, Taubman MA. Oligonucleotide containing CpG motifs enhances immune response to mucosally or systemically administered tetanus toxoid. Vaccine. 2001;19:1636–42. doi: 10.1016/s0264-410x(00)00422-9. [DOI] [PubMed] [Google Scholar]
- 26.Lee JY, Sojar HT, Amano A, Genco RJ. Purification of major fimbrial proteins of Porphyromonas gingivalis. Protein Expr Purif. 1995;6:496–500. doi: 10.1006/prep.1995.1066. [DOI] [PubMed] [Google Scholar]
- 27.Kawai T, Matsuyama T, Hosokawa Y, Makihira S, Seki M, Karimbux NY, et al. B and T lymphocytes are the primary sources of RANKL in the bone resorptive lesion of periodontal disease. The American journal of pathology. 2006;169:987–98. doi: 10.2353/ajpath.2006.060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonezawa H, Kato T, Kuramitsu HK, Okuda K, Ishihara K. Immunization by Arg-gingipain A DNA vaccine protects mice against an invasive Porphyromonas gingivalis infection through regulation of interferon-gamma production. Oral Microbiol Immunol. 2005;20:259–66. doi: 10.1111/j.1399-302X.2005.00220.x. [DOI] [PubMed] [Google Scholar]
- 29.Stacey KJ, Sweet MJ, Hume DA. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157:2116–22. [PubMed] [Google Scholar]
- 30.Sparwasser T, Miethke T, Lipford G, Erdmann A, Hacker H, Heeg K, et al. Macrophages sense pathogens via DNA motifs: induction of tumor necrosis factor-alpha-mediated shock. Eur J Immunol. 1997;27:1671–9. doi: 10.1002/eji.1830270712. [DOI] [PubMed] [Google Scholar]
- 31.Lipford GB, Sparwasser T, Bauer M, Zimmermann S, Koch ES, Heeg K, et al. Immunostimulatory DNA: sequence-dependent production of potentially harmful or useful cytokines. Eur J Immunol. 1997;27:3420–6. doi: 10.1002/eji.1830271242. [DOI] [PubMed] [Google Scholar]
- 32.Sparwasser T, Miethke T, Lipford G, Borschert K, Hacker H, Heeg K, et al. Bacterial DNA causes septic shock. Nature. 1997;386:336–7. doi: 10.1038/386336a0. [DOI] [PubMed] [Google Scholar]
- 33.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, et al. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–8. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 34.Malek R, Fisher JG, Caleca A, Stinson M, van Oss CJ, Lee JY, et al. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. Journal of bacteriology. 1994;176:1052–9. doi: 10.1128/jb.176.4.1052-1059.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


