Abstract
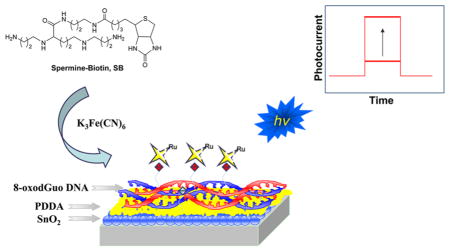
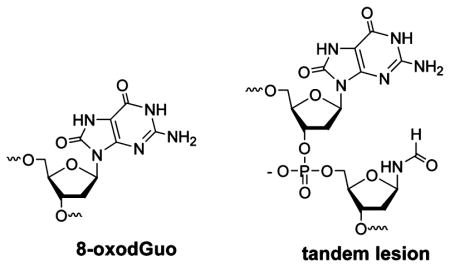
Exposure of DNA to oxidative stress conditions results in the generation of 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo). 8-OxodGuo is genotoxic if left unrepaired. We quantified 8-oxodGuo lesions in double-stranded DNA films by using a photoelectrochemical DNA sensor in conjunction with a specific covalent labeling method. A lesion-containing DNA film was assembled on a SnO2 nanoparticle modified indium tin oxide electrode through layer-by-layer electrostatic adsorption. The lesions were covalently labeled with a biotin conjugated spermine derivative, and ruthenium tris(bipyridine) labeled streptavidin was introduced as the signal reporter molecule. Photocurrent increased with the number of lesions in the strand and decreased as the film was diluted with intact DNA. Quantification of 8-oxodGuo was achieved with an estimated detection limit of ~1 lesion in 650 bases or 1.6 fmol of 8-oxodGuo on the electrode. Incubation of the film with a DNA base excision repair enzyme, E. coli formamidopyrimidine–DNA glycosylase (Fpg), resulted in complete loss of the signal, indicating efficient excision of the isolated lesions in the nucleotide. Oxidatively generated DNA damage to a double-stranded calf thymus DNA film by the Fenton reaction was then assessed. One 8-oxodGuo lesion in 520 bases was detected in DNA exposed to 50 μM Fe2+/200 μM H2O2. Treatment with Fpg reduced the photocurrent by 50%, indicating only partial excision of 8-oxodGuo. This suggests that tandem lesions, which are resistant to Fpg excision, are generated by the Fenton reaction. Unlike repair enzyme dependent methods, the sensor recognizes 8-oxodGuo in tandem lesions and can avoid underestimating DNA damage.
The 8-Oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo) molecule is one of the major products of oxidatively damaged DNA.1 It has been commonly employed as a molecular biomarker in the studies of DNA oxidation, screening of carcinogenic chemicals, and health effects of carcinogenic chemicals.2–4 Methods for the selective, accurate, and rapid quantification of 8-oxodGuo in DNA are highly desirable. We developed a photoelectrochemical DNA sensor and used it to quantify 8-oxodGuo in double-stranded calf thymus DNA after oxidative damage by the Fenton reaction and after DNA repair. The sensor also recognizes 8-oxodGuo in tandem lesions, and unlike methods that utilize repair enzymes, it can avoid underestimating DNA damage.5,6
Oxidative stress refers to the imbalance between the generation of reactive oxygen species (ROS) and the antioxidant defenses in a living organism and has been implicated in age-related disorders such as cancer and neurological and heart diseases.7–9 ROS arising from exposure to environmental pollutants, normal metabolic process, and inflammation leads to the oxidation of many biomolecules including proteins and DNA. Oxidation of DNA nucleobases is of great concern because of their high mutagenic potential. Generation of 8-oxodGuo by the oxidation of 2′-deoxyguanosine (dG) in DNA is one of the most frequently produced oxidative lesions. 8-OxodGuo mispairs with 2′-deoxyadenosine (dA) and induces G→T transversion mutations during DNA replication. The lesion is therefore mutagenic if left unrepaired.10,11 Consequently, the 8-oxodGuo level has been proposed as a molecular biomarker in the studies of the effects of oxidative stress on DNA and related human diseases.12,13 The importance of 8-oxodGuo as a biomarker has provided the impetus for developing methods for its selective quantification.
Living organisms have DNA repair mechanisms to protect against genomic instability. Base excision repair (BER) is a very important repair system for oxidatively damaged DNA. For example, formamidopyrimidine-DNA glycosylase (Fpg) is responsible for repairing 8-oxodGuo in bacterial species.14 In the repair process, Fpg first releases the modified base from duplex DNA and then cleaves the 3′- and 5′-phosphodiester bonds of the resulting apurinic site (AP site) by its class I AP lyase activity to produce a single nucleotide gap. Successful repair of oxidative DNA lesions depends on efficient DNA repair. In the past, studies focused on isolated 8-oxodGuo lesions, which are removed very efficiently by DNA glycosylases.15 More recently, considerable attention has been paid to the repair of clustered lesions that are composed of two or more lesions within one or two helical turns. Clustered lesions are considered to be more toxic compared to individual DNA lesions in part because their repair is often less efficient.16–18 Tandem lesions consist of two modified bases adjacent to each other and located on the same strand. Tandem lesions are a subset of clustered lesions and are produced in DNA exposed to ionizing radiation or Fenton-type reactions. 19–22 Under oxidative conditions, 8-oxodGuo is formed by either the addition of a hydroxyl radical to the C8 position of the purine moiety or hydration of the guanine radical cation.23 One mechanism proposed for tandem lesions containing 8- oxodGuo involves addition of pyrimidine peroxyl radicals to C8 of an adjacent purine base.19 According to a study in which DNA was subjected to γ-irradiation, tandem lesions were proposed to account for ~50% of the total 8-oxodGuo.24 It was also found that much of the 8-oxodGuo involved in the tandem lesions was refractory to excision by DNA glycosylases.16,24–26 This indicates that methods for detecting 8-oxodGuo that rely on repair enzymes may underestimate the formation of these lesions. As a result, developing methods that are independent of the local DNA environment are required for detecting the lesion.
HPLC/GC-MS and HPLC-EC are the most common techniques for quantifying 8-oxodGuo. These methods require degradation of the DNA sample and complicated extraction and/or derivatization steps before detection.27 To develop alternative methods, Sasaki designed fluorescent probes that interacted specifically with 8-oxodGuo by forming multiple hydrogen bonds with the lesion.28–31 In addition, Rusling developed voltammetric and electrochemiluminescence sensors based on the electrochemical oxidation of 8-oxodGuo for the rapid detection of oxidized DNA bases.32,33 Furthermore, 8- oxodGuo detection has been achieved using nanopores and DNA aptamers.34,35 We recently demonstrated that 8-oxodGuo was selectively tagged with a spermine–biotin conjugate (SB) under mild oxidation conditions.36,37 When used in conjunction with an ELISA type assay, SB provided a facile, inexpensive method for quantifying 8-oxodGuo, but the process is time-consuming. Photoelectrochemistry-based detection has many inherent advantages. Because of the separation of excitation source (light) and detection signal (photocurrent), photoelectrochemical detection has very low background. In addition, the required instrumentation only requires inexpensive low-powered light sources (e.g., light-emitting diodes) and simple electronic circuits.38,39 In the current work, we employed SB as a specific label for 8-oxodGuo in conjunction with a photoelectrochemical signal reporter and used this method to quantify the lesion in double-stranded DNA films after damage by the Fenton reaction as well as after repair by Fpg. The photoelectrochemical DNA sensor we developed provides a simple, rapid, and relatively sensitive technique for the quantification of 8-oxodGuo. As DNA repair can be investigated readily, the sensor can also be employed as a useful tool for the investigation of DNA damage and repair mechanism.
EXPERIMENTAL SECTION
Chemicals and Materials
Oligonucleotides containing different number of 8-oxodGuo were purchased from Integrated DNA Technologies (Coralville, IA). The complementary oligonucleotide was from Sangon Corp. (Shanghai, P. R. China). E. coli formamidopyrimidine-DNA glycosylase (Fpg) and 1 × REC Buffer 10 (10 mM HEPES-KOH, 100 mM KCl, 10 mM EDTA, 0.1 mg/mL BSA, pH 7.4) were obtained from Trevigen (Gaithersburg, MD). Ruthenium bis(2,2′-bipyridine)- (4-methyl-4′-carboxyl-2,2′-bipyridine) NHS ester (Ru–NHS) was purchased from Fluka (Milwaukee, WI). Poly-(diallydimethyl ammonium chloride) (PDDA), bovine serum albumin (BSA), Rose Bengal sodium salt, and double-stranded calf thymus DNA (CT-DNA) were obtained from Sigma (St. Louis, MO). Tin(IV) oxide nanoparticle (15 nm particle size, 15% colloidal dispersion in water) and hydrogen peroxide were purchased from Alfa Aesar (Ward Hill, MA). Streptavidin was from Promega Corp. (Madison, WI). Solutions and buffers were prepared in water from a Millipore Milli-Q (Biocel) water purification system. Indium tin oxide (ITO) conductive glass was obtained from Weiguang Corp. (Shenzhen, Guangdong Province, P. R. China) and cut into 2.5 cm × 0.5 cm sized electrodes.
Oligonucleotide hybridization was performed in 2 × SSC buffer (0.3 M NaCl, 30 mM sodium citrate, pH 7.0) on a Biometra T-Gradient thermocycler (Goettingen, Germany) and then slowly cooled to room temperature. The concentration of the hybridized double-stranded DNA was determined by absorbance at 260 nm.
Streptavidin was labeled with Ru–NHS by following the published procedure.38 The labeling ratio was found to be 1.5:1 Ru–streptavidin. SB was synthesized according to the published literature.36
Sensor Preparation
SnO2 nanoparticle modified ITO electrodes were prepared as previously described.38,39 DNA films were assembled on SnO2 nanoparticle-modified electrodes by layer-by-layer electrostatic adsorption, with polycationic PDDA as the first layer and polyanionic nucleic acid as the second layer. The film was deposited on the bottom of an electrode over a 0.5 cm × 0.5 cm area. The concentration of PDDA was 2.0 mg/mL (10 μL) and that of 8-oxodGuo containing oligonucleotide was 62.5 μg/mL (10 μL contains 40 pmol).
To generate 8-oxodGuo lesions in CT-DNA, the DNA film was shaken (200 rpm/min speed) in a Fe2+/H2O2 solution (Fenton reagent) at 37 °C for 30 min. The oxygen-enriched Fenton reagents were prepared by the Fenton reagent solution with O2 for 30 min. After the Fenton reaction, the electrodes were removed and rinsed with water. DNA damage induced by Rose Bengal was prepared by exposure of the electrodes to 10 μM Rose Bengal solution for 30 min under irradiation of visible light, which was provided by a commercial 500 W xenon lamp (Trusttech, Inc., Beijing, P. R. China) at a distance of 16 cm.
Biotinylation of 8-oxodGuo lesions in DNA films was carried out by coating the film with SB (0.5 mM, 10 μL) for 30 min at 25 °C, followed by addition of an aqueous solution of K3Fe(CN)6 (6 mM, 2 μL), after which the DNA film was incubated for an additional 30 min before the electrodes were rinsed. Biotinylation was verified by carrying out the reaction in solution and running the product on a polyacrylamide nondenaturing gel (20% concentration, 5% cross-link). After the DNA film was biotinylated, it was reacted with 50 μg/mL Ru-labeled streptavidin for 1 h at 25 °C to attach the photoelectrochemical signal reporter to the film.
To verify the cleavage of 8-oxodGuo lesions by Fpg, synthesized DNA containing one 8-oxodGuo was incubated with different concentrations of Fpg solution and then the cleavage products were resolved by PAGE (Figure S1 of the Supporting Information). Repair of the lesions in the double-stranded DNA immobilized on the electrodes was conducted by coating the DNA films with Fpg solution (10 μL) for 1 h at 37 °C.
Photoelectrochemical Detection
Photocurrent was measured on a CHI 800 electrochemical analyzer (CH Instruments, Austin, TX, USA) by placing the electrode in a 30 mM oxalate buffer (pH 5.8), using a Pt flag as the counter electrode, a Ag/AgCl (3 M KCl) reference electrode, and a bias voltage of +0.5 V. The excitation light source was a 473 nm blue light emitting diode (Lamp Inc., Shenzhen, P. R. China) with an illumination area of 0.2 cm2.
RESULTS AND DISCUSSION
A critical component of this photoelectrochemical sensor is the ability of the SB conjugate to trap oxidized 8-oxodGuo. 8- OxodGuo possesses an oxidation potential of ~0.75 V (vs NHE), which is lower than all other nucleotides including dG (1.29 V vs NHE).40 When K3Fe(CN)6 was used as a mild oxidant, 8-oxodGuo was selectively oxidized and subsequently trapped by SB in ~80% yield.36 In our experiment, the biotin tagged DNA migrated more slowly in PAGE than the untagged sample (Figure S2, lanes 4 and 6, of the Supporting Information). This is consistent with the previous report,36 and proves that DNA biotinylation was successful.

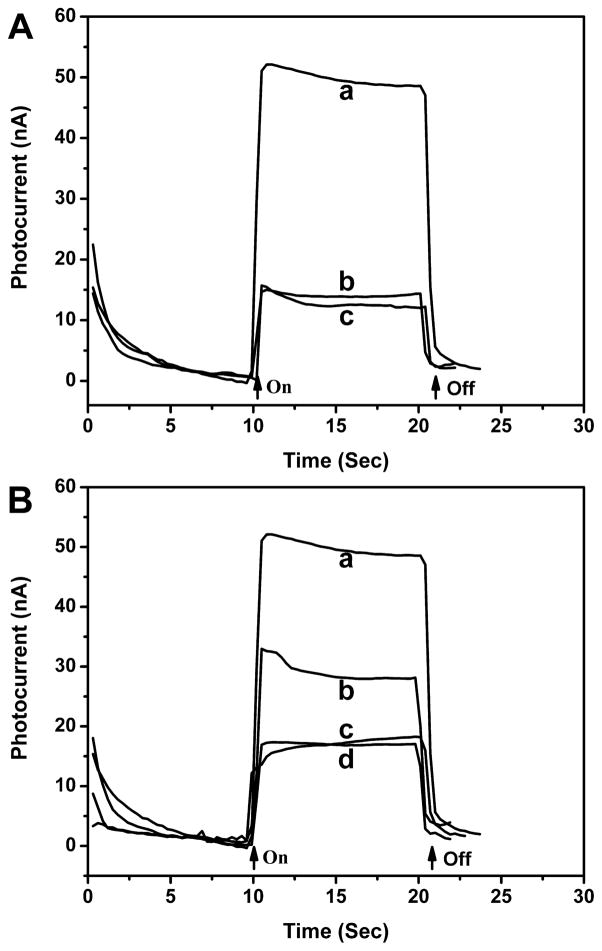
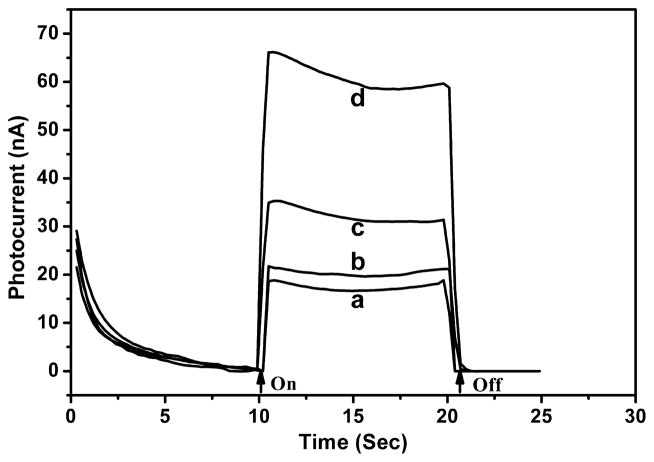
To establish the photoelectrochemical sensor detection method, Duplex 3, a double-stranded oligonucleotide containing four 8-oxodGuo lesions, was assembled on an electrode, tagged with SB, and reacted with Ru–streptavidin. The anodic photocurrent measured in an oxalate electrolyte was ~50 nA (Figure 1A), more than 3 fold higher than that of the same DNA film that was not treated with SB. The latter was close to the background photocurrent of the DNA film electrode itself (without reacting with SB and Ru–-streptavidin), indicating that nonspecific adsorption of the labeled protein directly on the electrode was negligible. Therefore, the photocurrent enhancement was attributed to the covalent linkage of SB with 8-oxodGuo lesion sites and the subsequent binding of Ru–streptavidin. The photocurrent signal was found to be dependent on the amount of 8-oxodGuo in the oligonucleotide. In Figure 1B, the photocurrent response of three different oligonucleotides, Duplex 1 (no lesion), Duplex 2 (1 lesion), and Duplex 3 (4 lesions) is displayed. The photocurrent was measured to be 17, 30, and 50 nA, respectively. Because Duplex 1 produced a photocurrent response similar to the film not treated with SB tag, we concluded that reaction of SB with dG and other nucleotides was negligible under the experimental conditions. Furthermore, the proportionality between photo-current signal and 8-oxodGuo level in the oligonucleotide indicated that quantitative analysis was feasible.
Figure 1.
(A) Photocurrent response of Ru–streptavidin labeled Duplex 3 film on SnO2 electrode measured in an oxalate buffer: (a) oligonucleotide with biotinylation, (b) oligonucleotide without biotinylation, and (c) oligonucleotide without biotinylation and Ru– streptavidin labeling. (B) Photocurrent response of Ru–Streptavidin labeled (a) Duplex 3 film, (b) Duplex 2 film, (c) Duplex 1 film, and (d) Duplex 1 film without biotinylation. Arrows indicate light on and off.
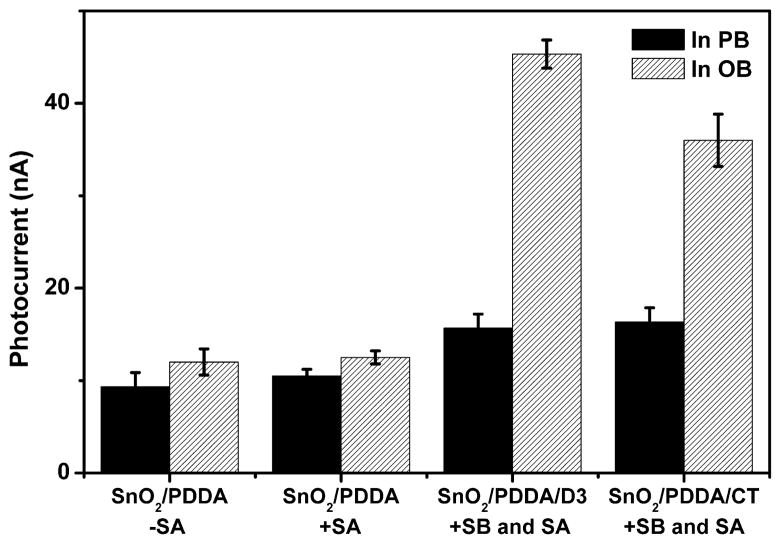
As proposed and verified in our previous work,38,39 photocurrent production is assisted by either guanine bases in the DNA sequence or oxalate in the measuring buffer as electron donor. Contribution from guanine oxidation renders the signal dependent on DNA composition and thus complicates the detection of 8-oxodGuo. To distinguish the photocurrent source, the SnO2/PDDA/Duplex 3 electrodes after Ru–streptavidin labeling were measured in a phosphate buffer (without oxalate). As shown in Figure 2, the photocurrent is about 15 nA, which is the sum of background photocurrent (~10 nA) and photocurrent from guanine oxidation (~5 nA). In comparison, the photo-current measured in the oxalate buffer is 45 nA. Therefore, the contribution from dG oxidation to the overall photocurrent is quite low (about 1/ 7th).
Figure 2.
Photocurrent response of different films in PB buffer and oxalate buffer. PB: 20 mM phosphate buffer, pH 7.4. OB: 30 mM oxalate buffer, pH 5.8. –SA: without the coating of Ru–streptavidin. +SA: with the coating of Ru–streptavidin. SnO2/PDDA/D3: electrodes with the assembly of Duplex 3. SnO2/PDDA/CT: electrodes with the assembly of CT-DNA after the exposure to 50 μM Fe2+/200 μM H2O2 for 30 min.
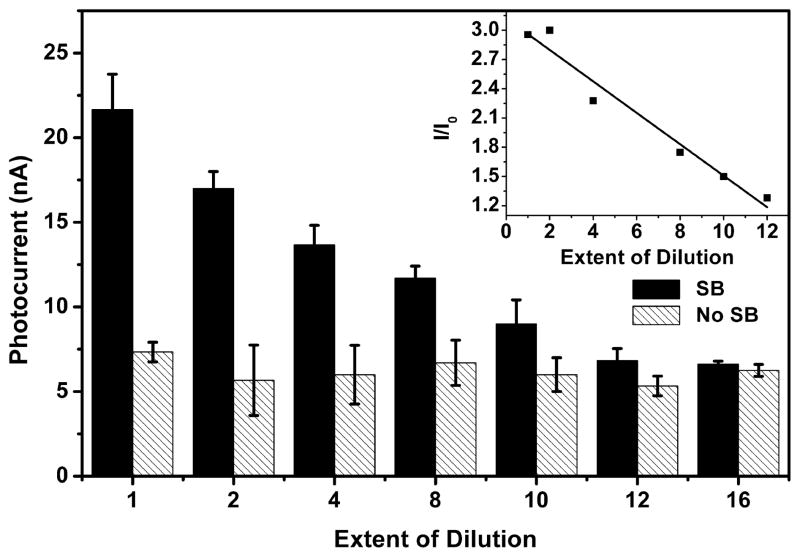
The linear response range and lower detection limit of the photoelectrochemical sensor were determined by preparing DNA films from mixtures of solutions containing different ratios of Duplex 1 and Duplex 2, while maintaining the total DNA concentration constant. Because the total DNA concentration in solution was fixed, the amount of DNA adsorbed on the electrode surface was assumed to remain constant. Increasing the fraction of Duplex 1 in the mixture in effect diluted the surface coverage of 8-oxodGuo. The photocurrent was proportional to the content of Duplex 2 in the mixture (Figure 3). To offset the batch-to-batch variation in electrode preparation, the photocurrent of the sample electrode was divided by that of the control electrode (electrode without biotinylation) from the same batch. The plot of this relative photocurrent as a function of the extent of dilution is reasonably linear in the range examined (Figure 3 inset). The lower limit of detection was calculated to be approximately 1 Duplex 2 in 13 total duplexes (3SD), which corresponds to 1 8- oxodGuo in 650 bases. In our previous work, the amount of DNA immobilized on the electrode surface was measured using a quartz crystal microbalance.41 On the basis of the DNA concentration and electrode size used in the current work, the amount of DNA on the electrode was estimated to be less than 1 pmol, which means that less than 1.6 fmol 8-oxodGuo on the electrode was detectable. Compared with the voltammetric and electrochemiluminescence methods based on 8-oxodGuo electrochemical oxidation, our method is in principle more accurate. This is because other oxidatively damaged DNA bases such as 5-hydroxy-2′-deoxyuridine and 5-hydroxy-2′-deoxycytidine have lower oxidation potentials than 8-oxodGuo and therefore can be electrochemically oxidized together with 8-oxodGuo, which might lead to an overestimate of the 8-oxodGuo level.42 However, a previous study showed that these lesions were not tagged by SB after Fe(CN)63− oxidation, probably because in the Fe(CN)63− oxidation product there is no functional group to react with SB.36
Figure 3.
Photocurrent intensity of Ru–streptavidin labeled Duplex 2 films with different extent of dilution. Filled column: DNA films with biotinylation. Shaded column: DNA film without biotinylation. Inset is a plot of the relative photocurrent intensity as a function of the extent of dilution. Error bars represent the standard deviation of three replicate electrodes.
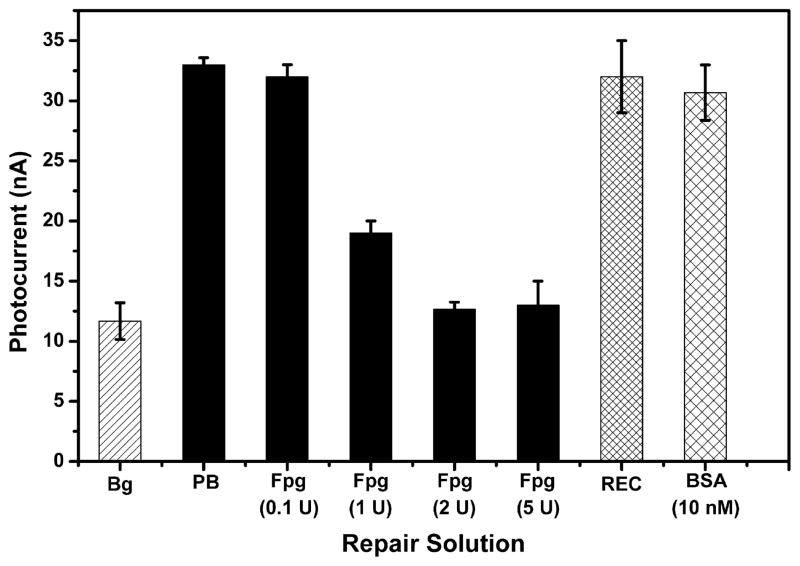
In addition to detecting 8-oxodGuo, the photoelectrochemical sensor can be employed to study DNA repair processes. For example, a Duplex 2 film was incubated with Fpg repair protein for 1 h before the biotinylation procedure. With increasing Fpg dose, the photocurrent response of the film decreased (Figure 4). When the protein dose was 2 U or higher, the photocurrent was reduced to background level. To eliminate the possibility of other interferences, the same film was exposed to either REC buffer in which Fpg was dissolved or 10 nM BSA (a nonrepairing protein with its concentration equal to 5 U Fpg) instead of the repair protein. Neither REC buffer nor BSA had any effect on the photocurrent intensity. Consequently, the reduced photocurrent can be attributed to the excision of 8-oxodGuo lesions in the DNA film by Fpg. More than 90% of the lesions were excised by 2 U Fpg in 1 h, leading to the complete loss of signal. Fpg was highly efficient in repairing isolated 8-oxodGuo lesions in the oligonucleotides.
Figure 4.
Photocurrent intensity of Ru–streptavidin labeled Duplex 2 films after incubation with different solutions for 1 h. Bg: Duplex 2 without biotinylation and incubated in PB buffer. PB: Duplex 2 with biotinylation and incubated in 20 mM phosphate buffer, pH 7.4. Others: Duplex 2 with biotinylation and incubated in Fpg, REC (Fpg buffer) or BSA. Error bars represent the standard deviation of three replicate electrodes.
After the photoelectrochemical detection method was established, it was applied to the quantification of 8-oxodGuo in chemically induced oxidatively damaged DNA. The Fenton reaction is a frequently employed system, in which Fe2+ and H2O2 react to produce highly reactive hydroxyl radicals that cause oxidatively damaged DNA, including 8-oxodGuo.43–45 CT–DNA films were assembled on electrodes and then exposed to a solution of Fe2+ and H2O2. The amount of 8-oxodGuo was then determined by photoelectrochemical measurement. The photocurrent increased progressively with the concentration of Fenton reagents (Figure 5). Lesions were detected in CT–DNA films exposed to as low as 50 μM Fe2+ and 200 μMH2O2. The amount of 8-oxodGuo was determined by comparing the photocurrent signal with the calibration curve in Figure 3 inset. On the basis of the comparison, it is estimated that 50 μM Fe2+ and 200 μM H2O2 produced 1 8-oxodGuo in 520 bases of double-stranded CT–DNA.
Figure 5.
Photocurrent response of Ru–streptavidin labeled CT–DNA films after exposure to (a) 20 mM phosphate buffer, pH 7.4, (b) 10 μM Fe2+/40 μM H2O2, (c) 50 μM Fe2+/200 μM H2O2, and (d) 100 μM Fe2+/400 μM H2O2 for 30 min at 37 °C.
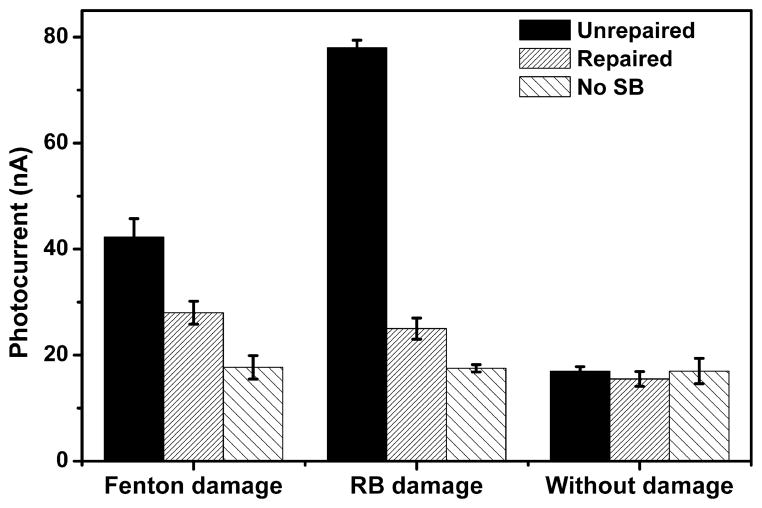
Repair of the CT–DNA film after its exposure to the Fenton reagents was also investigated. Under aerobic conditions, a Fenton reaction produced 42 nA photocurrent on the CT–DNA film, which decreased to 28 nA upon treatment with 2 U Fpg protein (Figure 6). The decreased signal is still significantly higher than the control electrode (17 nA), indicating that some of the 8-oxodGuo was resistant to Fpg excision. This result is in contrast to the Duplex 2 film, on which the photocurrent was reduced to background after Fpg repair (Figure 4). In a recent report, repair efficiency of 8-oxodGuo lesions in CT–DNA induced by γ-radiation was investigated24 and was found to be around 50%. The low efficiency was attributed to the formation of 8-oxodGuo tandem lesions through addition of a transiently generated peroxyl radial onto the C8 of an adjacent purine base, which unlike the isolated lesions in Duplex 2, are refractory to Fpg repair. The similarly low repair efficiency observed in the current work suggests that tandem lesions are also produced in Fenton reaction induced oxidatively damaged DNA. Previously, a tandem lesion involving 8-oxodGuo and thymidine glycol was detected in CT–DNA solution exposed to Cu(II)/ascobate/ H2O2.25 As a comparison, Fpg repair of oxidatively damaged DNA induced by Rose Bengal was also investigated. Rose Bengal is a type II photosensitizer that damages DNA by the formation of singlet oxygen (1O2) under visible light. As there is no hydroxyl radical formation, tandem lesions are not produced.46 After exposing the DNA-coated electrode to xenon lamp irradiation in the presence of 10 μM Rose Bengal, a 78 nA photocurrent was measured. After incubation with Fpg, the photocurrent decreased to 26 nA (Figure 6). The repair efficiency is about 87%, which is much higher than that induced by Fenton reaction (about 50%). The results are in accordance with the finding in the published literature that 8-oxodGuo produced in the reaction of Rose Bengal with dGuo is sensitive to Fpg.46,47
Figure 6.
Photocurrent intensity of Ru–streptavidin labeled CT– DNA films after exposure to 50 μM Fe2+/200 μM H2O2 (Fenton damage) or 10 μM Rose Bengal under irradiation (RB damage) for DNA damage and then incubation in 2 U Fpg solution for DNA repair. The Fenton reagents were aerated with O2 before use. Error bars represent the standard deviation of three replicate electrodes.
CONCLUSIONS
Quantitative and selective photoelectrochemical detection of 8- oxodGuo in double-stranded DNA films was achieved by employing a spermine–biotin conjugate (SB) to covalently label the lesion. The new sensor was capable of detecting about 1 lesion in 650 bases or as little as 1.6 fmol 8-oxodGuo on the electrode. Using this sensor, oxidative damage of double-stranded CT–DNA films by the Fenton reaction and DNA repair by Fpg protein were investigated. The amount of 8- oxodGuo produced by the Fenton reaction was dependent on the concentration of the reagents and was determined to be 1 in 520 bases with 50 μM Fe2+/200 μM H2O2. Although repair of the isolated 8-oxodGuo in oligonucleotides by Fpg protein was complete, the efficiency was only 50% for the lesions produced by the Fenton reaction. This result supports the hypothesis that tandem lesions are formed in ROS induced oxidatively damaged DNA.24 It also suggests the formation of such toxic lesions might be a general phenomenon that occurs in oxidatively damaged DNA. The photoelectrochemical DNA sensor we developed provides a simple, rapid, and low-cost method for the study DNA damage and repair.
Supplementary Material
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (2012AA062804), National Natural Science Foundation of China (20825519, 20890112, 20921063), and National Science Foundation, USA (M.M.G., CHE-0956466).
Footnotes
Notes
The authors declare no competing financial interest.
PAGE electrophoresis images of Duplex 2 before and after Fpg repair and the three nucleotide duplexes before and after biotinylation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Beckman KB, Ames BN. J Biol Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 2.Wu LL, Chiou CC, Chang PY, Wu JT. Clin Chim Acta. 2004;339:1–9. doi: 10.1016/j.cccn.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Kawanishi S, Hiraku Y, Murata M, Oikawa S. Free Radicals Biol Med. 2002;32:822–832. doi: 10.1016/s0891-5849(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 4.Singh N, Manshian B, Jenkins GJS, Griffiths SM, Williams PM, Maffeis TGG, Wright CJ, Doak SH. Biomaterials. 2009;30:3891–3914. doi: 10.1016/j.biomaterials.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Azqueta A, Shaposhnikov S, Collins AR. Mutat Res, Genet Toxicol Environ Mutagen. 2009;674:101–108. [Google Scholar]
- 6.Collins A, Cadet J, Epe B, Gedik C. Carcinogenesis. 1997;18:1833–1836. doi: 10.1093/carcin/18.9.1833. [DOI] [PubMed] [Google Scholar]
- 7.Dalle-Donne I, Rossi R, Colombo R, Giustarini D, Milzani A. Clin Chem. 2006;52:601–623. doi: 10.1373/clinchem.2005.061408. [DOI] [PubMed] [Google Scholar]
- 8.Loft S, Poulsen HE. J Mol Med. 1996;74:297–312. doi: 10.1007/BF00207507. [DOI] [PubMed] [Google Scholar]
- 9.Jenner P, Olanow CW. Neurology. 1996;47:S161–S170. doi: 10.1212/wnl.47.6_suppl_3.161s. [DOI] [PubMed] [Google Scholar]
- 10.Wood ML, Esteve A, Morningstar ML, Kuziemko GM, Essigmann JM. Nucleic Acids Res. 1992;20:6023–6032. doi: 10.1093/nar/20.22.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neeley WL, Essigmann JM. Chem Res Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 12.Roszkowski K, Jozwicki W, Blaszczyk P, Mucha-Malecka A, Siomek A. Med Sci Monit. 2011;17:CR329–CR333. doi: 10.12659/MSM.881805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malins DC, Hellstrom KE, Anderson KM, Johnson PM, Vinson MA. Proc Natl Acad Sci USA. 2002;99:5937–5941. doi: 10.1073/pnas.082111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson DM, Sofinowski TM, McNeill DR. Front Biosci. 2003;8:963–981. doi: 10.2741/1109. [DOI] [PubMed] [Google Scholar]
- 15.David SS, O’Shea VL, Kundu S. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.David-Cordonnier MH, Boiteux S, O’Neill P. Nucleic Acids Res. 2001;29:1107–1113. doi: 10.1093/nar/29.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eot-Houllier G, Gonera M, Gasparutto D, Giustranti C, Sage E. Nucleic Acids Res. 2007;35:3355–3366. doi: 10.1093/nar/gkm190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H, Cao H, Wang Y. Chem Res Toxicol. 2006;19:614–621. doi: 10.1021/tx060025x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Box HC, Budzinski EE, Dawidzik JB, Gobey JS, Freund HG. Free Radicals Biol Med. 1997;23:1021–1030. doi: 10.1016/s0891-5849(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 20.Bourdat AG, Douki T, Frelon S, Gasparutto D, Cadet J. J Am Chem Soc. 2000;122:4549–4556. [Google Scholar]
- 21.Douki T, Riviere J, Cadet J. Chem Res Toxicol. 2002;15:445–454. doi: 10.1021/tx0155909. [DOI] [PubMed] [Google Scholar]
- 22.Carter KN, Greenberg MM. J Am Chem Soc. 2003;125:13376–13378. doi: 10.1021/ja036629u. [DOI] [PubMed] [Google Scholar]
- 23.Cadet J. Mutat Res, Fundam Mol Mech Mutagen. 2003;531:5–23. [Google Scholar]
- 24.Bergeron F, Auvre F, Radicella JP, Ravanat JL. Proc Natl Acad Sci USA. 2010;107:5528–5533. doi: 10.1073/pnas.1000193107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang Y, Wang Y, Wang Y. Chem Res Toxicol. 2009;22:574–583. doi: 10.1021/tx8003449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budworth H, Dianova II, Podust VN, Dianov GL. J Biol Chem. 2002;277:21300–21305. doi: 10.1074/jbc.M201918200. [DOI] [PubMed] [Google Scholar]
- 27.Collins AR, Cadet J, Moller L, Poulsen HE, Vina J. Arch Biochem Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi Y, Kawaguchi R, Sasaki S. J Am Chem Soc. 2011;133:7272–7275. doi: 10.1021/ja200327u. [DOI] [PubMed] [Google Scholar]
- 29.Nakagawa O, Ono S, Li Z, Tsujimoto A, Sasaki S. Angew Chem, Int Edit. 2008;47:8983–8983. [Google Scholar]
- 30.Nasr T, Li Z, Nakagawa O, Taniguchi Y, Ono S, Sasaki S. Bioorg Med Chem Lett. 2009;19:727–730. doi: 10.1016/j.bmcl.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 31.Koga Y, Fuchi Y, Nakagawa O, Sasaki S. Tetrahedron. 2011;67:6746–6752. [Google Scholar]
- 32.Dennany L, Forster RJ, White B, Smyth M, Rusling JF. J Am Chem Soc. 2004;126:8835–8841. doi: 10.1021/ja048615+. [DOI] [PubMed] [Google Scholar]
- 33.Mugweru A, Wang BQ, Rusling J. Anal Chem. 2004;76:5557–5563. doi: 10.1021/ac049375j. [DOI] [PubMed] [Google Scholar]
- 34.Schibel AEP, An N, Jin Q, Fleming AM, Burrows CJ, White HS. J Am Chem Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy J, Chirania P, Ganguly S, Huang HD. Bioorg Med Chem Lett. 2012;22:863–867. doi: 10.1016/j.bmcl.2011.12.046. [DOI] [PubMed] [Google Scholar]
- 36.Xue L, Greenberg MM. J Am Chem Soc. 2007;129:7010–7011. doi: 10.1021/ja072174n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. J Am Chem Soc. 1999;121:9423–9428. [Google Scholar]
- 38.Liang MM, Liu SL, Wei MY, Guo LH. Anal Chem. 2006;78:621–623. doi: 10.1021/ac051926y. [DOI] [PubMed] [Google Scholar]
- 39.Liang MM, Guo LH. Environ Sci Technol. 2007;41:658–664. doi: 10.1021/es0617688. [DOI] [PubMed] [Google Scholar]
- 40.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 41.Wei MY, Guo LH, Chen H. Microchim Acta. 2006;155:409–414. [Google Scholar]
- 42.Rivière J, Bergeron F, Tremblay S, Gasparutto D, Cadet J, Wagner JR. J Am Chem Soc. 2004;126:6548–6549. doi: 10.1021/ja049438f. [DOI] [PubMed] [Google Scholar]
- 43.Aruoma OI, Halliwell B, Dizdaroglu M. J Biol Chem. 1989;264:13024–13028. [PubMed] [Google Scholar]
- 44.Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. J Biol Chem. 1989;264:20509–20512. [PubMed] [Google Scholar]
- 45.Aruoma OI, Halliwell B, Gajewski E, Dizdaroglu M. Biochem J. 1991;273:601–604. doi: 10.1042/bj2730601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cadet J, Berger M, Douki T, Ravanat J. Rev Physiol Biochem Pharmacol. 1997;31:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 47.Pouget JP, Douki T, Richard MJ, Cadet J. Chem Res Toxicol. 2000;13:541–549. doi: 10.1021/tx000020e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.