Abstract
Patterns of DNA polymorphisms are a primary tool for dissecting signatures of selection; however, the underlying selective forces are poorly understood for most genes. A classical example of diversifying selection is the complementary sex-determining locus that is found in the very large insect order Hymenoptera (bees, wasps, ants, and sawflies). The gene responsible for sex determination, the complementary sex determiner (csd), has been most recently identified in the honey bee. Females are heterozygous at this locus. Males result when there is only one functional allele present, as a result of either homozygosity (fertilized eggs) or, more commonly, hemizygosity (unfertilized eggs). The homozygotes, diploid males, do not reproduce and have zero fitness, which implies positive selection in favor of rare alleles. Large differences in csd cDNA sequences within and between four populations were found that fall into two major groups, types I and II. Type I consists of several allelic lineages that were maintained over an extended period, an indication of balancing selection. Diversifying selection has operated on several confined parts of the protein, as shown by an excess of nonsynonymous differences. Elevated sequence differences indicate another selected part near a repeat region. These findings have general implications about the understanding of both the function of the multiallelic mechanism and the adaptive processes on the level of nucleotide sequences. Moreover, the first csd sequence data are a notable basis for the avoidance of diploid males in bee selection programs by allele-assisted breeding.
Complementary sex determination is a genetic mechanism that controls sexual development in many, probably most, haplodiploid hymenopteran insects (bees, wasps, ants, and sawflies) (1, 2), ≈200,000 species. In many hymenopteran species, the complementary mechanism is provided by a single locus. Males develop from unfertilized eggs and are hemizygous for the sex-determining locus, whereas females are derived from fertilized eggs and are heterozygous. Diploid males arise when fertilized eggs are homozygous at the csd locus. Virtually all successfully reproducing males in natural populations, however, are haploid because diploid males produce diploid sperm, resulting in sterile offspring (3–6), or, in the case of honey bees, larval diploid males are eaten by worker bees (7, 8). The failure of diploid males to produce fertile offspring implies a strong advantage of heterozygotes at the sex locus and leads to the expectation of a high number of distinct alleles at selection–mutation–drift equilibrium in populations (9, 10). For the honey bee, Apis mellifera, the estimated number of alleles segregating in populations ranges from 11 to 19 (7, 11, 12).
The population dynamics of sex-determining alleles resembles those of the self-incompatibility (S) locus in fungi and plants (10, 13–15). Typically, these self-incompatibility systems are controlled by a single genetic locus having multiple allelic versions or specificities. However, the complementary sex-determining system differs from the S locus systems in the way that homozygotes can form and in that genotypic state governs two pathways of male and female development.
The fact that allelic composition governs sexual fate has long been of interest to biologists (16), not only because of its differences from sex chromosomal-based sex-determining systems (1, 17, 18) but also because of its major impact on population genetics (9, 10). The sex-determining alleles provide an excellent example for the maintenance of genetic variation by natural selection under a well characterized mode of selection, in which homozygotes have zero fitness. In addition to the general biological aspects, the occurrence of diploid males in the economically important honey bee has considerable consequences for applied bee management and for bee selection programs (6, 19).
Positional cloning and functional analyses have identified in the honey bee a single complementary sex determiner (csd) gene that governs sexual regulation by means of its allelic composition (18). The transcript encodes an SR-type protein with a putative protein-binding and -splicing function. The isolation of the gene and the first allelic cDNA sequences data from csd transcripts provides us with the unique power to dissect the evolutionary forces and historical processes proposed to act on this gene. The evolutionary past can be explored by constructing a csd genealogy, providing insight on the historical process of increase and maintenance of sex alleles, that can be compared to theoretical predictions. The pattern of synonymous and nonsynonymous differences among csd alleles can reveal selective signatures in the gene. A significant excess of nonsynonymous over synonymous differences would indicate that diversifying selection acted on this gene by favoring amino acid changes. In addition, the distribution of synonymous and nonsynonymous differences across the gene may suggest regions of the gene with more or less intense selective constraints. These analyses may enable the identification of the regions of the gene that determine the functional differentiation between sequences and will help to dissect its underlying molecular process and evolution.
Materials and Methods
Samples. Embryos (n = ≈200–300, 0–30 h after laying) were collected from two to three colonies of A. mellifera from four geographical locations: Davis (CA), Berlin, Stellenbosch (South Africa), and Ribeirão Preto (Brazil). Colony samples of hundreds of eggs represent population samples from at least 10 different genetic sources. Because of multiple matings of the queen, these eggs have 10–19 sources of genotypes derived from as many different fathers (20).
Molecular Methods. csd alleles were identified based on nucleotide differences in the cDNA sequences. Therefore, total RNA from egg samples was isolated by using the TRIzol protocol (GIBCO), and mRNAs were obtained by using Dynabeads Oligo(dT)25 (Dynal Biotech, Hamburg, Germany). First- and second-strand cDNA was synthesized by following the instructions of the RevertAid H Minus kit and the protocol of the supplier (Fermentas, St. Leon-Rot, Germany). csd sequences encompassing the ORF were amplified and cloned. Conserved sets of primers were designed based on sequences of 19 different 5′ and 3′ RACE fragments that were derived from different geographical origins (Davis, Berlin, and Ribeirão Preto) by following the protocol of the FirstChoice RLM-RACE kit (Ambion, Austin, TX). Primers were as follows: fw_csd, 5′-CTTGTTCGGTAT TWTCATAAA-3′; rev1_csd, 5′-TSAAA TRTCATCTCATWTTTC-3′; rev2_csd, 5′-ARTTRTCCAATYTCGATA TAT-3′; conscsd_for, 5′-GGTGATTTATACATTTGCAGGT-3′; cons2csd_for, 5′-TCATAAAAATGAAACGAAATATATC-3′; cons3csd_for, 5′-TCATAAAA ATGA A ACGGA ATA CAAC-3′; and conscsd_rev, 5′-RTCATCTCATWTTTCATTAT TCAAT-3′. Primer combinations were as follows: fw_csd/rev1_csd, fw_csd/rev2_csd, conscsd_for/conscsd_rev, cons2csd_for/conscsd_rev, and cons3csd_for/conscsd_rev. PCRs were performed under standard conditions (21) including proofreading Pwo DNA Polymerase (PEQLAB, Erlangen, Germany). PCR conditions were an initial 94°C step for 160 s, followed by 35 cycles of 94°C for 30 s, 48°C for 40 s, 72°C for 140 s, and then by a 72°C terminating step for 15 min. PCR fragments were cloned into pGEM-T vector (Promega). Cloned fragments (n = 60–100) from each geographical origin were screened for fragment polymorphism by using two restriction enzymes, ApoI and MseI (Fermentas). Cloned fragments were amplified by using designated primer combinations, restricted, and resolved in high-resolution gels (21). Fragments were subjected to doublestrand sequencing (MWG Biotech, Ebersberg, Germany). Single-sequence reads were assembled by using staden package version 4.6 (22).
Analysis of Sequence Data. To analyze sequence differences between alleles, the ORFs of 30 cDNA sequences were aligned by using the clustalx program (23). The sequences were manually edited by using the bioedit program (24) so that the frame of coding was conserved. Mean diversity estimates within sequences, SE, and genealogy analyses with 1,000 bootstrap replications were performed with the mega version 2.1 program (25), by using the Poisson-corrected distance and the complete-deletion-of-gaps option. To test whether the length of terminal branches in the genealogy exceeds that of the internal ones, the branches were compared by using the interior branch length test. All following analyses were performed on 15 sequences that represent the allelic lineages (sequences used are shown in Figs. 2 and 3). Synonymous mean pairwise diversity per synonymous site (Ks) and nonsynonymous mean pairwise diversity per nonsynonymous site (Ka) were calculated with the complete-deletion-of-gaps option and a sliding window of 50 bases (step size of 10 bases), implemented in the k-estimator version 6.0 program (26). Comparison of Ks and Ka across exons was calculated by using dnasp version 3.5.3 (27). To test whether evolution of allelic lineages differs across the genealogy, synonymous differences per site (bS) and nonsynonymous differences per site (bN) for each tree branch in the genealogy were estimated by using the method of ref. 28 in the bn–bs program (29). The tree for this calculation was obtained by using the dnaml program included in the phylip version 3.57C package (30). Heterogeneity in the distribution of nucleotide differences across csd type I alleles was tested by using the ratio of polymorphism (those that differed within type I) to fixed differences (those that were fixed within type I but differed within type II). Significant heterogeneity in this ratio was tested based on 10,000 replicate Monte Carlo simulations using different recombination parameters (r = 0, 1, 2, 4, 8, 16, 32, and 64) as implemented in the dna slider program (31).
Fig. 2.
Amino acid alignment of the hypervariable region with a variable number of (N)1–4/Y repeats that were excluded in the evolutionary sequence analysis. Several gaps must be introduced to maximize homology. Flanking amino acids shown in bold are conserved. Amino acid sequence among replicate variants (as shown in Fig. 1) is the same, except for an amino acid difference for replicate D2-38 (position 390, Y → H). The numbers above the alignment represent the position in the overall sequence alignment, including gaps.
Fig. 3.
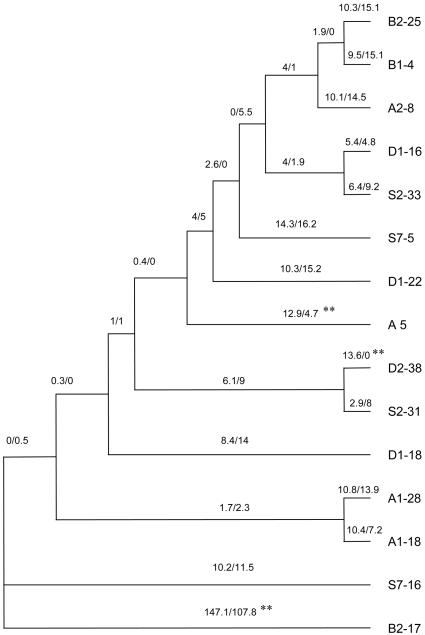
Evolutionary tree of csd type I/II alleles. The numbers of nonsynonymous differences (bN) and synonymous differences (bS) per site and for each branch in the genealogy are presented above each branch as bN × 1,000/bS × 1,000 with a transition/transversion ratio of R = 2. The statistical significance was tested with a one-tailed Z test (P < 0.05) and is indicated by asterisks. The tree is unrooted, and its topology differs from that in Fig. 1 because of the omission of replicate variants.
Results
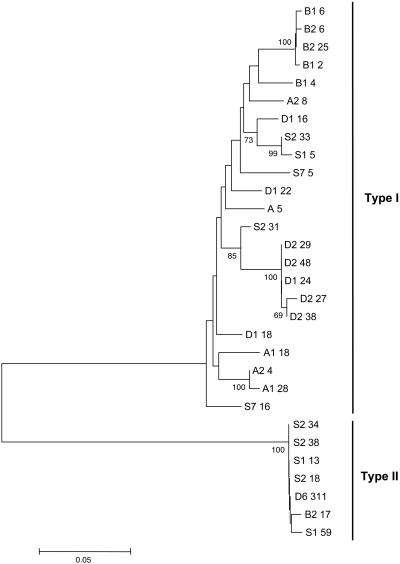
Genealogy and Diversification of csd Alleles. Differences in csd cDNA fragments were identified by restriction site polymorphism, and these clones were sequenced. Only unambiguous sites were included in the subsequent analysis; thus, gaps in the alignment were completely deleted. Three striking results are illustrated by the csd genealogy of 30 deduced amino acid sequences (Fig. 1). First, the genealogy reveals two major branches that suggest an extraordinary degree of divergence between two subset classes of sequences, types I and II. The notion of high divergence is supported by the amino acid mean diversity between types I and II (0.307 ± 0.03), which greatly exceeds the mean diversity within type I (0.056 ± 0.007) and within type II (0.003 ± 0.002) (one-tailed Z test, P < 0.001 for both comparisons). Second, type I sequences from different populations tended to differ more than sequences from the same source population, which were sometimes extremely similar and probably represent variants of the same functional specificity. Four of the Berlin, two of the Stellenbosch, two of the Ribeirão Preto, and five of the Davis sequences were such “replicates.” Nevertheless, these populations included highly divergent type I sequences (one sequence different in the Berlin sample, four in the Stellenbosch sample, four in the Ribeirão Preto sample, and three in the Davis sample). Given the small samples of colonies and alleles sequenced, it is clear that these sequences are polymorphic within each of the four populations studied. In addition, three of the four geographical regions yielded at least one type II, each of which was nearly identical. The finding of various allelic replicates and lineages that are derived from distinct genotypes within the small colony sample is consistent with the multiple-mating behavior of the queen (20). Third, the terminal branches among allelic lineages from type I are very long compared with the internal branches (ordinary leastsquares test of branch length, Mann–Whitney U test, P < 0.001). The finding of longer terminal branches still held when analyses were performed on the synonymous sites that are presumed to be under no selection (Mann–Whitney U test, P < 0.001), demonstrating that alleles within type I evolve or are maintained discontinuously over time. Very few differences are observed within allelic lineages that cluster at the end of long branches, which most likely represent replicate variants of the same allelic lineage. The assumption that bushes at the tips of branches are replicates of the same allelic lineage is supported by the finding that the average genetic distances between allelic lineages greatly exceed the average genetic distances within replicates (one-tailed Z test, P < 0.001). Moreover, the amino acid sequence in the most diverse region with repeats that appear a different number of times in different sequences is the same within replicates but differs extensively between allelic lineages (Fig. 2) except for one difference that resulted in an amino acid replacement in replicate D2-38. It is unknown which of these sequences encode functional specificities. The differences among the 15 allelic lineages are within the order of magnitude of those for which functional analyses were carried out (18), suggesting that the identified lineages generate functionally distinct proteins. However, further functional studies are needed to test for their specificities.
Fig. 1.
csd genealogy based on neighbor-joining analysis of genetic differences obtained from the deduced amino acid sequence, excluding ambiguous sites in which gaps have to be introduced in the alignment. The sequences fall into two major branches, types I and II. Fifteen allelic lineages were found; other sequences represent replicate variants of the same lineage, as indicated by bushes at the end of some tips (see text for further details). A genealogy of nearly identical topology was obtained when a hypervariable repeat region (rich in NY) was included. Numbers to the left of allele and replicate numbers indicate bootstrap percentages (of 1,000 resamplings) in support of each note. Bootstrap values <90% were not included. The scale bar indicates amino acid differences per site. Geographical origin is indicated by letter: D, Davis; B, Berlin; S, Stellenbosch; and A, Ribeirão Preto.
Positive Selection Operates on Parts of the Protein. To detect signatures of selection, Ks and Ka values were compared among allelic lineages. Ka values did not exceed Ks values for the full-length coding sequence for all comparisons, only for those involving type I sequences (with each other or against type II sequences) (Z test, P > 0.1; Table 1). When differences were estimated for each branch (28) in the csd genealogy (Fig. 3), a great excess of nonsynonymous differences was found in the branch that separates types I and II, which suggests that positive selection has operated on a common ancestor sequence of types I and II. In addition, an overrepresentation of nonsynonymous changes was found in two terminal branches in the genealogy (alleles D2-38 and A-5) but not in other terminal or internal branches in the type I genealogy. Although no test was carried out for multiple testing in the genealogy, a possible explanation is that positive selection has operated on some of these type I sequences.
Table 1. Ks and Ka values.
| Coding region
|
Exon 2+3
|
Exon 4+5
|
Exon 6-9
|
|||||
|---|---|---|---|---|---|---|---|---|
| Ks | Ka | Ks | Ka | Ks | Ka | Ks | Ka | |
| Types I + II | 0.058 ± 0.004 | 0.041 ± 0.004 | 0.043 ± 0.006 | 0.028 ± 0.004 | 0.027 ± 0.003 | 0.032 ± 0.004 | 0.085 ± 0.004* | 0.059 ± 0.004* |
| Type I | 0.041 ± 0.001 | 0.025 ± 0.0006 | 0.020 ± 0.0015 | 0.0128 ± 0.0008 | 0.0137 ± 0.0015 | 0.0164 ± 0.0011† | 0.0727 ± 0.0023* | 0.0428 ± 0.0013* |
| Type I vs. II | 0.17 ± 0.003 | 0.145 ± 0.001 | 0.1915 ± 0.0026 | 0.1298 ± 0.0011 | 0.1137 ± 0.0014 | 0.1398 ± 0.0028†† | 0.1645 ± 0.0058 | 0.1648 ± 0.0025 |
Ks and Ka values were calculated within and between types I and II for the entire coding region and the groups of exons. Ka values exceeded Ks values in exon 4+5 when types I and II were compared (††, P < 0.001) but failed slightly for the comparison within type I (†, P < 0.1). The test for heterogeneity showed that Ka and Ks values in exon 6-9 were higher than the corresponding values of other groups of exons when all or type I lineages were included (*, P < 0.001).
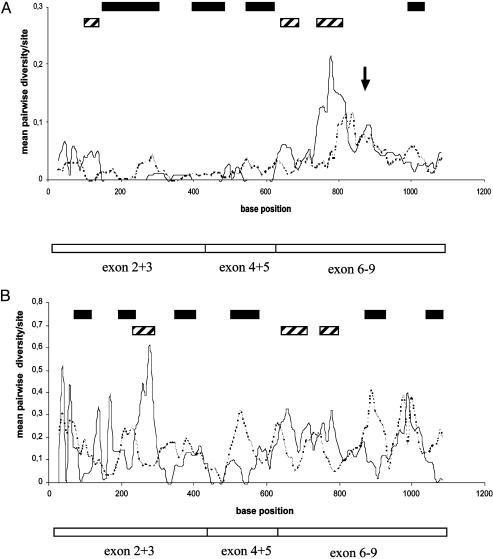
To further investigate whether confined parts of the protein show signatures of selection, the coding sequences were split into groups of exons that physically cluster in the genomic sequence (18) and were tested for nonsynonymous and synonymous differences. Ka values exceeded Ks values in exon 4 + 5 between types I and II (P < 0.001) but failed slightly to be significant for comparison within type I (P < 0.1). This finding led us to conclude that positive selection may have operated on more confined parts of the protein. Thus, Ks and Ka plots for sites with windows of 50 bases sliding along the csd sequence alignment were constructed. Within type I pairs, Ka values exceeded Ks values in consecutive windows (filled boxes in Fig. 4A; one-tailed Z test, P < 0.05) ranging from 145 to 295, 395 to 615, and 985 to 1,025 bases (base positions were assigned to the middle of the sliding window). Although it is unclear in this approach whether the differences from one region of the sequence to another are significant, the large overall proportion of windows showing Ka > Ks (46 of 107, 43%) and the considerable number of consecutive windows in which Ka values exceeded Ks values strongly suggest that in confined parts of the protein diversifying selection has operated. Consecutive windows were found in which Ks values exceeded Ka values (bases 635–675 and 735–805; striped boxes in Fig. 4B), indicating possible regions in which selective constraints have operated on the protein. Similar to the type I plot, the plot between types I and II sequences showed several consecutive Ka > Ks windows in the first half of the coding region (Fig. 4B). Unique to the type I/II plot is a region from base 865 to 925 in which Ka values were elevated. A possible explanation is that these differences were selectively favored after types I and II diverged from a common ancestor sequence but then were not further selected in the subsequent diversification process of type I sequences. The region of 635–785 bases showed elevated Ks values (Fig. 4B), supporting the finding of the type I plot. The alternative hypothesis that the observed pattern in the type I/II plot results from differences within type I sequences can be excluded based on the finding that Ks and Ka values between types I and II greatly exceed those within type I (one-tailed Z test, P < 0.001).
Fig. 4.
Mean pairwise diversity plots of Ks and Ka values, which were constructed for windows of 50 bases sliding along the csd sequence alignment. Only sequences from separate allelic lineages were used in the analysis, which did not include the replicate variants. (A) Average of these statistics for all pairwise comparisons of type I lineages. The arrow indicates the position of the hypervariable repeat region (see Fig. 2) not included in this analysis. (B) The comparison between types I and II. Filled boxes indicate base positions at the middle of the window (step size of 10 bases) in which nonsynonymous differences significantly exceed synonymous differences (one-tailed Z test, P < 0.05). Striped boxes indicate base positions in the sequence in which the reverse significant condition is found (Ks > Ka, P < 0.05).
Ka and Ks Values Vary Across Type I. The type I plot gives the appearance of high heterogeneity of Ks and Ka values across the gene, which is illustrated by the elevated values in the C-terminal part (Fig. 4A). To test for heterogeneity, Ka and Ks values were compared across the groups of exons (Table 1). Ka and Ks values of exon 6–9 exceeded diversity values of exon 2 + 3 and exon 4 + 5 only when type I differences were compared (P < 0.001). No heterogeneity was found for other exons and type combinations (Table 1). Further support for heterogeneity across csd sequences of type I lineages comes from the analysis of the ratio of polymorphisms within type I alleles to the fixed differences (fixed within type I alleles but different within type II) (31). The ratios deviate from the expected uniform distribution arrived at by using different recombination parameters and various test statistics (the Kolmogorov–Smirnov test, maximum sliding and average sliding G test, and Goss and Lewontin's modified variance test all show P < 0.05). These results indicate that evolutionary forces and constraints have operated differently across type I lineages. In addition to the elevated differences, the region contains the hypervariable region (Fig. 2; for position in the sequence, see the arrow in Fig. 4A) with a variable number of repeats that were, because of unambiguous sites, excluded in the former analysis. The region consists of (N)1–4/Y (asparagine/tyrosine) repeats that appear 5–25 times in different lineages of type I. Several amino acid positions flanking this hypervariable region are highly conserved (shown in bold in Fig. 2), suggesting strong functional constraints on these amino acids flanking the repeat region.
Discussion
The finding of huge differences in the molecular analyses of csd sequences is consistent with predictions of the single-locus population genetic model of balancing selection of sex-determining alleles (9, 10). Heterozygotes at the locus develop into females, but homozygous males are eaten by worker bees shortly after they hatch from the egg. Reproductive males are hemizygous at the locus. Input of new functional alleles by mutation is initially favored, because they participate in fewer diploid males than commoner alleles do; hence, many alleles can be maintained in a population. The gain of new alleles by mutation will eventually be balanced by the loss of alleles by genetic drift. A huge mutation rate alone without selection cannot explain the observed pattern of Ka values exceeding Ks values. New allelic specificities, i.e., new functional alleles, must result from nonsynonymous substitutions, and we have found an excess of these. The ability to specify identical and nonidentical alleles may increase as a function of differences.
In addition to increased nonsynonymous differences, the extended residence time of csd alleles is a strong indication of balancing selection. The number of synonymous differences between lineages greatly exceeds that of replicate sequences of types I and II (Fig. 1). The replicates are an internal measure of neutral variation and thus give a clear demonstration of the extended residence time of type I allelic lineages, a signature of balancing selection. Theoretical results (32), findings for self-incompatible (S locus) (33, 34) fungal mating types (35, 36), and MHC genes in vertebrates (37, 38) show that residence time under balancing selection exceeds that of neutral alleles, which appears to be a general property of these loci. The highly divergent sequences of csd allelic lineages are thus a consequence of the extreme age of these alleles.
Taking the less divergent exons 2 + 3 and 3 + 4 as a reference, it appears that exons 6–9 of type I are evolving differently from other parts of the gene and thus show considerably elevated synonymous and nonsynonymous differences. Positive selection (Ka > Ks) can explain the elevated nonsynonymous differences but fails to explain the enormous increase of the synonymous differences, particularly near bases 700–900 (Fig. 4A). The cause of this pattern may be differences in mutation rate or relaxed selective constraints operating on this part of the protein. Under this hypothesis, however, we should find patterns of heterogeneity also in the evolutionary old type II, which we failed to detect. This result allows us to conclude that across type I alleles both the mutation rate and the selective constraints should be quite similar. One possible explanation for the observed pattern is positive selection and hitchhiking. Tightly linked nondeleterious (mostly synonymous) differences have hitchhiked, along with a strongly selective favored part. They have thus gone more often to fixation than differences in a less favored region. The hypervariable repeat region (amino acids 335–397; Fig. 2) is a candidate for a selectively favored part of the protein. This region is unique to type I sequences, which is consistent with the observation that heterogeneity is not found in type II. We cannot rule out, however, that there may be other parts in the designated region with a strong selective advantage. Consistent with a hitchhiking model is the finding that the extent of polymorphism is related to proximity to the hypervariable region. The diversity steadily increases near the hypervariable region (Ka and Ks values near the arrow in Fig. 4A), an effect predicted by theory (39, 40) and found in the MHC complex (41) for neutral genetically linked loci. In our study, we propose the same effect for sequences within our gene.
Up to 19 alleles have been proposed to exist in natural populations (12), and we have identified 15 lineages in four populations that show huge differences in their sequence. A challenging problem is to determine how the allelic specificities are encoded in the amino acid sequences, transforming the compositional information into the binary switch of male and female development (18). csd combines two essential functions for its activity: one function that determines the specificity of alleles in various combinations and another that governs the onset of female development irrespective of the various heterozygous combinations (18). We have found signatures of diversifying selection in confined parts of the first half of the protein, encoding candidate regions that determine the specificity of alleles. No domain and function have been proposed for these parts of the protein. We have deduced that regions of selective constraints in parts of the RS domain are candidates for regions in which purifying selection has operated. In these regions, purifying selection may have operated more strongly than in other parts of the protein, because the overall diversity was significantly higher than that of the rest of the gene (test of heterogeneity). It has been proposed that the RS domain is involved in activating the female pathway by alternative splicing (18). In agreement with this finding, it seems reasonable that parts of the RS domain are functionally conserved among alleles.
The strong signatures of hitchhiking near the repeat region suggest functional significance for the allelic specificity of type I sequences. The variable number of (N)1–4/Y repeats lies between two protein-binding domains, the RS domain and the P-rich region (18). An obvious mechanistic interpretation is that the variable number of repeats modifies the protein-binding of type I polypeptides. A possible explanation for the low diversity found within type II is the lack of this repeat, which has the potential function of establishing specificities and, thus, diversity within type I lineages. Further analysis of type I and II sequences from related species will further insight on the processes of diversification of these types.
Population studies of csd sequences will be a basis for the establishment of tools for bee breeders to test bee lines before mating to ensure that fertilized eggs will not result in diploid males. The occurrence of diploid males due to inbreeding is a major problem in bee selection programs.
Regions with signatures of selection are prime candidate parts of the protein to further our knowledge of the molecular multiallelic mechanism. Further molecular and functional studies may lead to a combined understanding of the molecular function and its evolution. The existence of multiple versions of specificity thus provides an elegant system to study adaptive processes directly on the level of nucleotide sequences.
Acknowledgments
We thank R. E. Page, D. Charlesworth, J. Evans, and R. Crozier for critical comments on the manuscript; K. Hartfelder, M. Allsopp, and J. Pflugfelder for providing eggs; R. E. Page and M. K. Fondrk for providing bee facilities in Davis; and J. H. McDonald for the ratio heterogeneity test. This research was funded by Deutsche Forschungsgemeinschaft Grant 2194/3 and Kultusminister of Sachsen-Anhalt Grant 3266A/0020L (to M.B.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ks, synonymous mean pairwise diversity per synonymous site; Ka, nonsynonymous mean pairwise diversity per nonsynonymous site.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY569694–AY569721, AY350617, and AY350618).
References
- 1.Bull, J. J. (1983) Evolution of Sex Determining Mechanisms (Benjamin Cummings, Menlo Park, CA).
- 2.Cook, J. M. (1993) Heredity 71, 421–435. [Google Scholar]
- 3.Woyke, J. & Skowronek, W. (1967) Proc. XXI Internatl. Apic. Congr., Maryland (Apimondia, Bucharest), pp. 470–471.
- 4.Awadalla, P. (2003) Nat. Rev. Genet 4, 50–60. [DOI] [PubMed] [Google Scholar]
- 5.Duchateau, M. J. & Marien, J. (1995) Insectes Soc. 42, 255–266. [Google Scholar]
- 6.Rinderer, T. E. (1986) Bee Genetics and Breeding (Academic, Orlando, FL).
- 7.Mackensen, O. (1955) J. Hered. 46, 72–74. [Google Scholar]
- 8.Woyke, J. (1963) J. Apic. Res. 2, 19–24. [Google Scholar]
- 9.Kimura, M. & Crow, J. F. (1964) Genetics 49, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama, S. & Nei, M. (1979) Genetics 91, 609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laidlaw, H. H., Gomes, F. P. & Kerr, W. E. (1956) Genetics 41, 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams, J., Rothman, E. D., Kerr, W. E. & Paulino, Z. L. (1977) Genetics 86, 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright, S. (1939) Genetics 24, 538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth, D. (2002) Curr. Biol 12, R424–R426. [DOI] [PubMed] [Google Scholar]
- 15.Casselton, L. A. (2002) Heredity 88, 142–147. [DOI] [PubMed] [Google Scholar]
- 16.Page, R. E., Jr., Gadau, J. & Beye, M. (2002) Genetics 160, 375–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whiting, P. W. (1939) Genetics 24, 110–111. [Google Scholar]
- 18.Beye, M., Hasselmann, M., Fondrk, M. K., Page, R. E., Jr., & Omholt, S. W. (2003) Cell 114, 419–429. [DOI] [PubMed] [Google Scholar]
- 19.Page, R. E., Jr., & Laidlaw, H. H. (1982) J. Apic. Res. 21, 30–37. [Google Scholar]
- 20.Palmer, K. A. & Oldroyd, B. P. (2000) Apidologie 31, 235–248. [Google Scholar]
- 21.Hasselmann, M., Fondrk, M. K., Page, R. E., Jr., & Beye, M. (2001) Insect Mol. Biol. 10, 605–608. [DOI] [PubMed] [Google Scholar]
- 22.Staden, R., Beal, K. F. & Bonfield, J. K. (2000) Methods Mol. Biol. 132, 115–130. [DOI] [PubMed] [Google Scholar]
- 23.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleid Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, T. A. (1999) Nucl. Acids Symp. Ser. 41, 95–98. [Google Scholar]
- 25.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244–1245. [DOI] [PubMed] [Google Scholar]
- 26.Comeron, J. M. (1999) Bioinformatics 15, 763–764. [DOI] [PubMed] [Google Scholar]
- 27.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- 28.Rzhetsky, A. & Nei, M. (1993) Mol. Biol. Evol. 10, 1073–1095. [DOI] [PubMed] [Google Scholar]
- 29.Zhang, J., Rosenberg, H. F. & Nei, M. (1998) Proc. Natl. Acad. Sci. USA 95, 3708–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein, J. (1995) phylip, The Phylogeny Inference Package (Dept. of Genetics, University of Washington, Seattle), Version 3.57c.
- 31.McDonald, J. H. (1998) Mol. Biol. Evol. 15, 377–384. [DOI] [PubMed] [Google Scholar]
- 32.Takahata, N. (1990) Proc. Natl. Acad. Sci. USA 87, 2419–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vekemans, X. & Slatkin, M. (1994) Genetics 137, 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark, A. G., ed. (1993) in Mechanisms of Molecular Evolution, eds. Takahata, N. and Clark, A. G. (Japan Sci. Soc. Press, Tokyo/Sinauer Associates, Sunderland, MA), pp. 79–107.
- 35.Wu, J., Saupe, S. J. & Glass, N. L. (1998) Proc. Natl. Acad. Sci. USA 95, 12398–12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.May, G., Shaw, F., Badrane, H. & Vekemans, X. (1999) Proc. Natl. Acad. Sci. USA 96, 9172–9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahata, N. & Clark, A. G., eds. (1993) in Mechanisms of Molecular Evolution (Japan Sci. Soc. Press, Tokyo/Sinauer Associates, Sunderland, MA), pp. 1–21.
- 38.Salamon, H., Klitz, W., Easteal, S., Gao, X., Erlich, H., Fernandez-Vina, M., Trachtenberg, E., McWeeney, S., Nelson, M. & Thomson, G. (1999) Genetics 152, 393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takahata, N. & Satta, Y. (1998) Genetica (The Hague) 102/103, 157–169. [PubMed] [Google Scholar]
- 40.Slatkin, M. (2000) Genetics 154, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'hUigin, C., Satta, Y., Hausmann, A., Dawkins, R. L. & Klein, J. (2000) Genetics 156, 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]