Abstract
Acute myeloid leukemia subtype M4 with eosinophilia is associated with a chromosome 16 inversion that creates a fusion gene CBFB-MYH11. We have previously shown that CBFB-MYH11 is necessary but not sufficient for leukemogenesis. Here, we report the identification of genes that specifically cooperate with CBFB-MYH11 in leukemogenesis. Neonatal injection of Cbfb-MYH11 knock-in chimeric mice with retrovirus 4070A led to the development of acute myeloid leukemia in 2-5 months. Each leukemia sample contained one or a few viral insertions, suggesting that alteration of one gene could be sufficient to synergize with Cbfb-MYH11. The chromosomal position of 67 independent retroviral insertion sites (RISs) was determined, and 90% of the RISs mapped within 10 kb of a flanking gene. In total, 54 candidate genes were identified; six of them were common insertion sites (CISs). CIS genes included members of a zinc finger transcription factors family, Plag1 and Plagl2, with eight and two independent insertions, respectively. CIS genes also included Runx2, Myb, H2T24, and D6Mm5e. Comparison of the remaining 48 genes with single insertion sites with known leukemia-associated RISs indicated that 18 coincide with known RISs. To our knowledge, this retroviral genetic screen is the first to identify genes that cooperate with a fusion gene important for human myeloid leukemia.
Acute myeloid leukemia (AML) arises from uncontrolled clonal expansion of hematopoietic progenitor cells. Approximately half of AML cases present specific and recurrent chromosomal translocations that affect transcription factors with critical functions in hematopoietic differentiation. The heterodimeric transcription factor, core binding factor (CBF), is the main target of translocations in AML (1). These translocations alter the RUNX1 and CBFB genes, which encode the α- and β-subunits of CBF respectively, and create fusion proteins with dominant effects in leukemogenesis. For example, practically all cases of AML subtype M4 with eosinophilia express the CBFB-MYH11 fusion gene [encoding the CBFβ-smooth muscle myosin heavy chain (SMMHC) fusion protein] as a result of the chromosome 16 break-and-join inversion, inv(16)(p13q22), or its equivalent, t(16;16)(p13; q22) (2).
Previous studies using a Cbfb+/Cbfb-MYH11 knock-in mouse model (henceforth called Cbfb+/MYH11) have convincingly shown that Cbfβ-SMMHC is a dominant repressor of CBF function. Cbfβ-SMMHC blocks embryonic definitive hematopoiesis as well as the adult hematopoietic stem cell differentiation to the myeloid and lymphoid lineages (3). This phenotype is similar to that resulting from homozygous disruption of either the α (Runx1-/-)- or β (Cbfb-/-)-subunits in the mouse (4-8). Furthermore, mutagenesis studies using Cbfb+/MYH11 knock-in mice indicated that whereas Cbfβ-SMMHC participates in leukemogenesis, additional (cooperating) genetic events are required (9).
Retroviral insertional mutagenesis (RIM) provides an efficient approach to alter gene expression and identify candidate cancer genes (10). Retroviral insertions near a gene can up-regulate its expression by promoter activation, up- or down-regulate its expression at any distance or orientation by enhancer interference, generate truncated transcripts when inserted within the coding sequence, or stabilize mRNAs by excluding destabilizing sequences when inserted in the 3′ UTR (10). An additional advantage of RIM is that the proviral sequence serves as a tag to identify flanking genes. The power of RIM is best exemplified by recent reports identifying >2,000 tags associated with hematologic malignancies in the mouse (11-13). Interestingly, in mice carrying oncogenes associated with lymphomagenesis, including CD2-myc (14), E2A-PBX1 (15), Pim1 and Pim2 (16), Cdkn1b (encoding p27Kip1) (17), and Cdkn2a (18), RIM studies have allowed the identification of genes that synergize with defined genetic alterations in cancer development.
This article reports for the first time, to our knowledge, the use of RIM in mice carrying a human myeloid leukemia translocation-associated fusion gene, Cbfb-MYH11, and identifies cooperating genes in AML development. The leukemic cells are predominantly myeloblasts and monoblasts, are mono- or oligoclonal, and are transplantable. Genetic evidence is reported suggesting that misexpression of known (i.e., Myb and Runx2) and other (i.e., Plag1 and Plagl2) leukemia-associated transcription factors may specifically synergize with Cbfb-MYH11 in AML.
Materials and Methods
Retroviral Infection. Molecularly cloned amphotropic virus 4070A (19, 20) was propagated in NIH 3T3 cells in DMEM containing 10% FCS. The titer of the infectious virus in stocks from the medium of infected cells was determined in an assay by using PG-4 cat cell line (21). Briefly, PG-4 cells were infected with serial dilutions of virus in the presence of 8 μg/ml polybrene. After an overnight incubation, the virus-containing medium was removed and the cells were maintained in fresh medium for 5-6 days. After fixation in methanol and staining with carbol fuchsin and methylene blue, plaques were counted. Two- to 6-day-old mice were inoculated i.p. with 105 infectious particles in 0.05 ml.
Phenotype Analysis. Mice were under daily observation for early signs of leukemia. These signs included limited motility, pale paws, and dehydration. At the first signs of illness, peripheral blood was analyzed for cell number and the presence of immature cells. Peripheral blood was collected from the retroorbital sinus for smears, and fluorescence-activated cell sorting analysis was performed by using cell-surface markers Gr-1, CD11b, B220, CD3, Ter119, and CD117.
Leukemic cells were harvested from bone marrow or spleen of affected mice in RPMI medium 1640 (Life Technologies) and 10% FBS, and single cell suspension aliquots of 1 × 106 cells were injected intravenously in 4- to 6-week-old F1/129SvEv C57BL6 recipients.
Inverse PCR (iPCR). One microgram of spleen DNA from leukemic mice was digested for 12-16 h with EcoRI or BamHI in a 40-μl final volume. DNA fragments were ligated in a 600-μl volume by using 4,000 units of T4-Ligase (M0202M, New England Biolabs) at 16°C for 12 h, and were resuspended in 20 μl of water after precipitation. Two sequential PCRs were performed by using nested primers specific for 4070A retrovirus. The PCR mix included 2 μl of template DNA, 400 μM dNTP, 0.4 μM of each primer, and 2.5 units of Taq enzyme (Expand Long Template PCR, Roche Applied Sciences, Indianapolis), in a 50-μl final volume. Primers for the first PCR included LTR3 (CCGAACTTCCCTATTCTCAGTTC) and gagF1 (GATTCTGGTAGGAGACAGAGA) for BamHI 5′ LTR amplification, LTR1 (CTCTTGCTGTTGCATCGGACT) and 5env1(GTCAGGGAGAACATGGTAATAGG) for EcoRI 3′ LTR amplification, and LTR1 and env4R (AGATTGAGCACCGTGGTGGAC) for BamHI 3′LTR amplification. The first PCR was performed with a denaturation step (5 min at 94°C), followed by 30 cycles of amplification (each cycle included 30 sec of denaturing at 94°C, 1 min annealing at 60°C, and 10 min extension at 68°C), and a final extension step (5 min at 68°C). In most cases, 1 μl of the first PCR was used as a template for the second PCR. In the case an amplicon was identified after the first PCR when analyzed by electrophoretic separation, dilutions to 1:10 or 1:100 were used as template for the second PCR. The second PCR was performed by using nested primers LTR4 (GCGTTACTTAAGCTAGCTTGC) and gagF5 (GACAGGGACAGAGATGATGGA) for BamHI 5′ LTR amplification, LTR2 (CTCCTCAGAGTGATTGACTAC) and 5env2 (TACAGTCTCAGTCCCCACGAT) for EcoRI 3′ LTR, and LTR2 and env5 (AGTCAATCCAGTGCTGCAAGC) for BamHI 3′ LTR amplification. Amplicons from the second PCR were analyzed by electrophoretic separation in a 1% agarose gel, were cloned into XL- or TA-TOPO vectors (Invitrogen), and were sequenced using M13 vector primers.
Southern Blot Analysis. Southern blot analysis was performed following standard protocols. Briefly, spleen DNA from leukemic and control mice was cut with EcoRI, was separated in a 0.8% agarose gel, and was transferred onto a nylon membrane (Hybond-XL, Amersham Biosciences, Buckinghamshire, U.K.). The viral probe included a 1.2-kb EcoRI-ClaI fragment from the 4070A envelope sequence. Genomic probes were generated from clones isolated by iPCR, and included 100 μg/ml denatured salmon sperm DNA in the hybridization solutions. Probes were hybridized overnight at 45°C in formamide hybridization solution (Hybrisol-I, Serologicals, Norcross, GA).
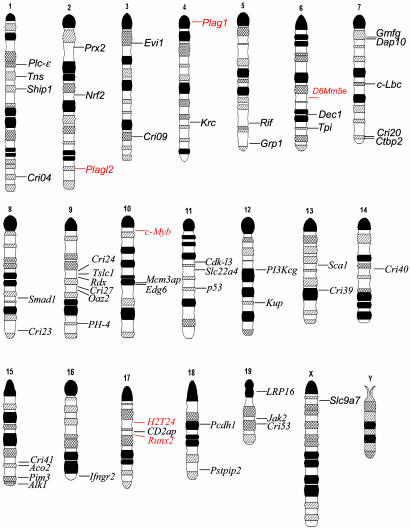
Database Analysis and URL Information. Genomic sequence tags after the end of the proviral LTR sequence TCA were analyzed by using a combination of public (genome.ucsc.edu. and www.ensembl.org, February 2003 freeze) and private (Celera Discovery System, www.celera.com) databases. Each insertion site was assigned with a Cbfb-MYH11 retrovirus insertion (Cri) number, and the name of the proximal gene. Uncharacterized genes include ESTs, published expressed genes of unknown function, and predicted genes. These genes (and their human homologues) were annotated as they appear in public databases. In the case more than one candidate gene is located near a Cri, the gene symbol and accession number of both flanking genes are denoted. Of note is that this assignment is relative to the quality of the mouse genome sequence and gene annotation available at the time of study, which may need to be adjusted after new available information is discovered. Sequence tags were deposited into GenBank with accession nos. CL001593-CL001659, as shown in Table 2, which is published as supporting information on the PNAS web site. These tags can also be found in the Retroviral Tagged Cancer Gene Database (http://genome2.ncifcrf.gov/RTCGD). The chromosomal position of flanking genes was obtained from the Ensembl Mouse Genome Server (www.ensembl.org/Mus_musculus/blastview) and was adapted to the Standard Mouse ideogram (Fig. 4), as accepted by the International Mouse Nomenclature Committee. The reference Standard Ideogram/Anomaly Breakpoints of the Mouse can be accessed at www.mgu.har.mrc.ac.uk/anomaly/anomaly-intro.html.
Fig. 4.
Ideogram representation of chromosomal location of genes flanking RISs. CISs are red and SISs are black. Uncharacterized genes are identified by their Cri number.
Results
Cbfb+/MYH11 Knock-in Chimeras Efficiently Develop AML After Infection with Amphotropic Retrovirus 4070A. Amphotropic murine leukemia virus (MLV) strain 4070A (known henceforth as 4070A) is a weak inducer of hematologic malignancies in the mouse. It can induce follicular lymphoma and myeloid leukemia in NIH/Swiss mice with a mean latency of 278 days and a tumor incidence of 64% (22). Also, ≈50% of DBA/2N mice treated with 4070A develop myeloid leukemia only after induction of chronic inflammatory response by pretreatment with pristane (23).
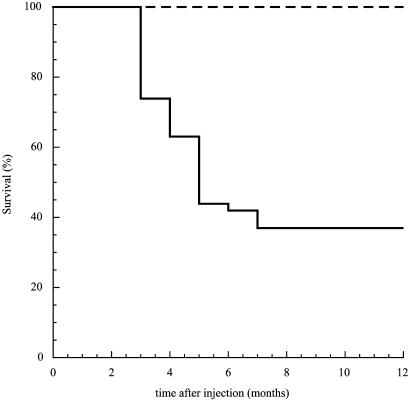
As Cbfb+/MYH11 knock-in chimeras developed AML efficiently after N-ethyl-N-nitrosourea treatment, we first asked whether these mice could also develop AML after 4070A mutagenesis. Forty-three Cbfb+/MYH11 knock-in chimeras and 40 control mice were injected with retrovirus 4070A during their first week of life. Sixty three percent (27 of 43) of the infected Cbfb+/MYH11 knock-in mice developed AML 2-5 months after viral injection and reached terminal disease 3-6 weeks later (Fig. 1). The 37% disease-free chimeras probably escaped leukemia due to low contribution of Cbfb+/MYH11 embryonic stem cells to the hematopoietic compartment or to an inefficient viral injection at birth. The 20 129Sv/Ev and 20 C57BL/6 wild-type controls did not develop leukemia 1 year after injection (experimental endpoint), indicating that 4070A-induced AML depended on CbfbMYH11 expression.
Fig. 1.
Induction of AML in Cbfb-MYH11 knock-in chimeras by RIM. Kaplan-Mayer survival plot of 40 wild-type mice (dashed line) and 43 Cbfb+/MYH11 chimeras (solid line) after neonatal injection with MuLV-4070A retrovirus. Test and control mice were followed up to 1 year after treatment.
The clinical presentation and histological findings of the 4070A-induced leukemias in the Cbfb+/MYH11 chimeras were similar to those induced by N-ethyl-N-nitrosourea. Peripheral blood analysis showed a progressive anemia (hematocrit between 18% and 35%) and the presence of immature cells in the circulation (Fig. 2A), which were predominantly myeloblasts and monocytic blasts by morphology (Fig. 2B). Infrequent immature erythroid, granulocytic, and apoptotic cells could also be detected in some samples. Fluorescence-activated cell sorting analysis consistently indicated that most of the leukemic cells in peripheral blood were CD117+ (59.46 ± 14.07%, n = 5 and 1.9 ± 0.26%, n = 3 for the leukemic and control mice, respectively) and negative for lineage markers (CD3, CD45, CD11b, and Gr-1). In addition, some samples contained CD117+/ter119+ cells (5.2 to 24.1% of the total cell populations, n = 5), as detected previously in N-ethyl-N-nitrosourea studies (9). Spectral karyotyping analysis was performed on leukemic cells from two mice with 10 cells each being analyzed, which failed to identify chromosomal rearrangements, suggesting that AML arose with no evidence of genome instability (data not shown). Twenty of the 27 leukemic mice were killed at terminal stage for further analysis. Pathology analysis consistently revealed splenomegaly (a 10- to 35-fold increase in size), with massive infiltration of leukemic cells and disruption of splenic architecture (Fig. 2C). In addition, hepatomegaly was frequently observed with focal infiltration of leukemic cells around blood vessels (Fig. 2D). Thymus and lymph nodes were not affected. These results indicate that Cbfb+/MYH11chimeras readily develop AML after acquisition of additional genetic changes, and that the leukemic phenotype is independent of the mutagen used.
Fig. 2.
Histological features of leukemic cells. (A) Wright-Giemsa staining of peripheral blood (×40 magnification) from an affected mouse showing a high count of white blood cells (dark blue) and a decreased number of red blood cells. (B) Wright-Giemsa staining of leukemic cells (×100 magnification) showing the presence of predominantly myeloblasts and monoblasts in peripheral blood. Hematoxylin/eosin stained histology sections at ×60 magnification of enlarged spleen (C) and infiltrated liver (D; at ×40 magnification) show focal infiltration of leukemic cells.
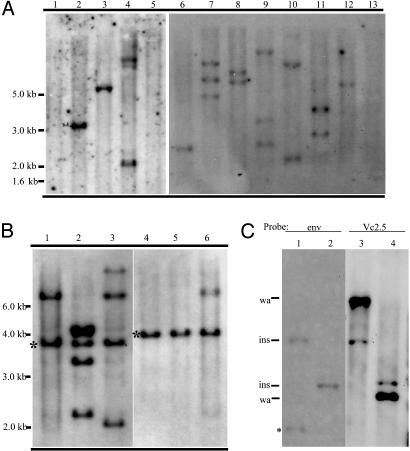
Clonal Expansion of RIM Leukemic Samples. The number of proviral insertions present in the leukemic samples was estimated by Southern blot analysis (Fig. 3A) of spleen DNA from moribund mice. The 4070A-envelope-specific probe identified one to three proviral integrations, with an average of 1.7 insertions per tumor. In tumor VC4 (Fig. 3A, lane 5), no viral integrations were identified. One insertion was later identified in this tumor by iPCR, using primers specific for the 5′ end of the provirus. Sequence analysis of the viral integration site isolated by PCR revealed a truncated provirus 4070A lacking the envelope and 3′ LTR sequences (data not shown).
Fig. 3.
Southern blot analysis of retroviral insertions in spleen DNA of Cbfb+/MYH11 chimeras with AML. (A) A representative EcoRI-digested DNA panel of 11 AML samples (lanes 2-12) and two wild-type controls (lanes 1 and 13) was hybridized with the 4070A envelop probe. (B)An EcoRI-digested DNA panel of primary leukemia samples (lanes 1 and 4) and of secondary leukemia samples after transplantation (lanes 2, 3, 5, and 6) hybridized with the 4070A envelope probe. Asterisks indicate bands present in both the primary (donor) and transplanted leukemic samples. (C) Cri10 insertions map back to the expected DNA fragment. DNA from leukemia sample VC2 digested with BamHI (lanes 1 and 3) or EcoRI (lanes 2 and 4). Viral probe envelope (env, lanes 1 and 2) and Cri10 clone Vc2.5 genomic probe (vc2.5, lanes 3 and 4) were used to sequentially hybridize to the same blot. Bands shown include wild-type allele (wa), inserted allele (ins), and an internal viral fragment (asterisks). Note that the band representing the inserted allele is expected to be fainter than the wild-type allele, because the spleen sample used includes a large number of wild-type cells, in addition to the leukemic cells carrying one inserted allele and one wild-type allele.
To verify the functional significance of the insertions during leukemogenesis, bone marrow and spleen cell suspensions from affected mice were transplanted into isogenic wild-type mice. Recipient mice developed AML within 8 weeks of transplantation with a phenotype similar to the original retrovirus-infected chimeras. The clonality of the leukemia was assessed by analysis of the pattern of proviral integration in primary and transplanted tumors (Fig. 3B). One band (representing one viral insertion) from primary tumors (see asterisks in lanes 1 and 4) was always present in all transplanted samples (lanes 2 and 3 and lanes 5 and 6). As 4070A is replication-competent, additional bands were found in the transplanted samples. These results indicate that a discrete number of genetic alterations (one to three), caused by 4070A proviral insertion, can synergize with Cbfb-MYH11 to generate leukemia in the mouse. Furthermore, these leukemic cells expanded in a clonal fashion.
Cloning of Retroviral Insertion Sites (RISs). To identify candidate genes that cooperate with Cbfb-MYH11 in AML, iPCR was carried out on spleen DNA from 20 tumors. The iPCR primers matching the 4070A envelope region did not generate any background (false-positive amplicons) because they do not match with endogenous sequences in the mouse genome. In addition, the efficiency of cloning 4070A insertions was increased by independently using two restriction enzymes (EcoRI and BamHI), and using primers for both ends of the provirus. A total of 67 retroviral insertion tags were identified (Table 2), rendering an average of 3.3 tags per tumor. This tag per tumor ratio is higher than that detected by Southern blot hybridization described above, and this difference is probably due to integration of broken copies of the viral genome that were only detectable by iPCR and/or the higher sensitivity of PCR. Nonetheless, 3.3 tags per tumor is still a relatively low ratio, which may be related to Cbfb-MYH11 function and/or neonatal injection of the retrovirus.
Flanking genes were then defined by their genomic proximity to a Cbfb-MYH11-associated RIS (Cri). In case a Cri mapped near two genes (11 cases), both gene names were included in Table 2. Ninety percent (60 of 67) of the insertions mapped within 10 kb of a candidate gene (Table 3, which is published as supporting information on the PNAS web site). In total, the chromosomal position of the tags in the mouse genome unambiguously defined 54 Cris. Six Cris were identified as common insertion sites (CISs), because insertions in these six sites occurred in more than one leukemia sample: eight for Cri10, three for Cri48, and two each for Cri07, Cri14, Cri30, and Cri46.
The definition used in this study for CIS is as described (11). For each of these CISs, the proviral insertions were all contained within a 30-kb segment of DNA.
The chromosome position of the 54 Cris with the associated candidate genes is shown as a mouse standard ideogram in Fig. 4. From these data, 10 Cris mapped within 10 kb upstream of the transcription initiation site and 16 mapped in the 5′ UTR (Table 3), suggesting that promoter activation was a frequent event. In addition, one Cri disrupted an exon and 21 others are located in introns of the protein coding region, which probably generated truncated proteins or altered expression of the full length transcript. Only seven Cris mapped >10 kb away from the flanking genes.
Candidate Genes That Synergize with Cbfb-MYH11 in Leukemogenesis.Next, we analyzed the genes flanking the CIS (Table 3). Cri10 represents eight independent insertions near the transcription start site (introns 1 and 2) of the transcription factor gene Plag1, and the inserted viral genomes were in the same orientation as the Plag1 gene. These insertions were also detected by Southern blot analysis using genomic DNA probes (Fig. 3C), confirming that Plag1 insertions were predominant in the related tumors and probably participated in the transformation process. PLAG1 has been identified at the breakpoints of a t(3;8)(p21;q12) translocation, which is the most common recurrent chromosomal abnormality in human pleomorphic adenomas of the salivary gland, lipomas, and lipoblastomas (24-26). The translocation results in a promoter swapping between PLAG1 and CTNNB1 that led to the activation of PLAG1 expression. The data reported here is evidence of Plag1 involvement in leukemia, and the likely consequence of retroviral insertions near the transcriptional start site of the Plag1 gene is also activation of expression. Plagl2, a Plag1-related gene, has recently been proposed also to be a protooncogene (27), Two insertions (Cri07) were identified near the transcriptional start site of Plagl2. Therefore, it is predicted that a promoter activation event for Plagl2 could play a role in Cbfb-MYH11-dependent leukemogenesis.
Two insertions (Cri30) mapped at a common site of MLV insertions in mouse chromosome 10, called Mml-3 locus (28), 80 and 83 kb upstream of the Myb transcription factor and 60 and 57 kb upstream of Hbs1 helicase. This region is a hot spot for MLV insertions (see Table 1 and references therein), but its actual effect on Myb or Hbs1 expression is poorly understood. Recent data, however, suggest that viral insertions at Mml loci may alter Myb expression (29).
Table 1. Cbfb-MYH11-associated CISs.
| Cri | No. of tags | Gene | Ref(s). |
|---|---|---|---|
| Cbfb-MYH11-specific CISs | |||
| Cri07 | 2 | Plagl2 | |
| Cri10 | 8 | Plag1 | |
| Cri14 | 2 | D6Mm5e | |
| Cri46 | 2 | H2T24 | |
| Previously Known CISs | |||
| Cri30 | 2 | Myb | 12, 13, 14, 16-18, 23 |
| Cri48 | 3 | Runx2 | 14 |
| SISs matching known CISs | |||
| Cri08 | 1 | Evi1 | 12, 13 |
| Cri12 | 1 | Rif | 12 |
| Cri18 | 1 | Dap10 | 12, 18 |
| Cri19 | 1 | Lbc | 13, 18 |
| Cri43 | 1 | Pim3 | 16 |
| Cri45 | 1 | Ifngr2 | 12 |
| Cri53 | 1 | MGC30540 | 12, 13 |
The runt-related RUNX2 protein forms a heterodimeric complex with CBFβ, and plays a critical role in osteoblast and chondrocyte differentiation (30). Sustained expression of Runx2 alters T cell activation (31, 32), whereas MLV insertions upstream of Runx2 induce lymphomagenesis in cooperation with Myc (14, 33, 34). In our study, however, three insertions (Cri48) mapped within Runx2 intron 5. Such an event would predict expression of a truncated Runx2 including the runt domain (excluding the transactivation and repression domains) with a putative dominant-negative function. Alternatively, a yet unidentified regulatory sequence in intron 5 could up- or down-regulate expression of the full-length Runx2 transcript.
Two insertions (Cri14) were identified in introns 3 and 6 of the D6Mm5e gene in mouse chromosome 6. The function of this gene is unknown, and analysis of its amino acid sequence does not predict any known conserved domains. Alternatively, these insertions could affect expression of Dok1, a gene located 18 and 45 kb from these insertions toward the telomere. P62-Dok (encoded by Dok1) belongs to the Dok family of adaptor proteins that interacts with Ras-GAP, Csk, and Nck, and participates in a variety of cellular processes, such as migration (35) and activation of T cells (36), and the pathogenesis of cancer (37).
Two insertions (Cri46) mapped within the MHC Class 1 T gene cluster, 3 kb upstream and 7 kb downstream of the cell-surface glycoprotein gene H2T24, a histocompatibility 2 T region locus 24 antigen-related gene. The guanine nucleotide-binding protein gene Gna-rs1, which maps 25 and 40 kb from these insertions, could also be affected in these tumors.
In addition to the six CISs identified, six single insertion sites (SISs) have previously been identified as CIS with MLV insertions (Table 1), suggesting a common role in cell transformation. We identified SIS genes associated with regulation of IFN receptor signaling (receptor Ifngr2 and kinase Jak2), TGFβ pathway (type I receptor Acvrl-1 and the transcription factors Dec1 and Smad1), and Ras/PI3K signaling pathway (adaptor Dap10, PI3Kcg, and Ship1). Finally, genes associated with cytoskeleton organization and cell adhesion that may have key transforming functions in AML were also targeted. Cytoskeleton-associated genes included structural genes (radixin and tensin genes), as well as regulatory genes (phosphatase gene Pstpip2, the Rho GDP-GTP exchange factor gene Lbc, and the Rho GTPase gene Rif). Genes associated with cell adhesion included the protocadherin gene Pcdh1, homeoprotein gene Prx2, and the tumor suppressor gene Tslc1.
Discussion
Fusion genes generated by chromosome translocations/inversions play important roles in leukemogenesis. Previously, we demonstrated that one of the fusion genes, CBFB-MYH11, generated by chromosome 16 inversion in human AML subtype M4 with eosinophilia, is necessary but not sufficient for leukemia development in our knock-in mouse model (9). In more recent years, it has been demonstrated that many other fusion genes are not sufficient to induce leukemia (38-40). In this manuscript, we identified specific candidate genes that can cooperate with Cbfb-MYH11 for leukemogenesis in our knock-in mouse model. AML was efficiently induced by injecting retrovirus 4070A in newborn Cbfb+/MYH11 chimeras with a latency of 2-5 months. RISs were detected by Southern blot hybridization and were identified easily and efficiently by iPCR. The insertion sites were mapped to the genome and flanking genes were identified. We showed that as few as one additional alteration (a second genetic “hit”) could efficiently accelerate AML in the mouse. This finding was verified by the fact that one insertion per leukemia was retained after secondary transplantation, and the fact that no additional mutations were identified at cytogenetic level. However, we cannot rule out the presence of subtle mutations, such as point mutations, that may also play a role in the leukemogenic process. Because a similar type of myeloid leukemia was induced in this insertional mutagenesis model and in our previous N-ethyl-N-nitrosourea mutagenesis model (9), it seems that Cbfb-MYH11 expression is an important factor in determining the morphology of the leukemic cells, whereas the secondary alterations are associated with proliferative or antiapoptotic events.
Of particular interest is the identification of CISs specific for Cbfb-MYH11. This study identified, for the first time, the role of the C2H2 zinc finger transcription factors Plag1 and Plagl2 in AML. Both proteins recognize the same DNA-binding element and regulate IGFII expression (27, 41). PLAGL2 also regulates expression of the proapoptotic gene Nip3 in response to hypoxia or iron deficiency (42). However, little is known about the mechanism of action of these proteins in cancer formation.
It is intriguing that Myb and Runx2 were identified as CISs in Cbfb-MYH11-induced AML. CBFβ/RUNX1 heterodimer regulation of normal proliferation and differentiation of myeloid and lymphoid lineages depend on the combined functions of Myb and CBF proteins (43, 44). Therefore, because CBFβ-SMMHC inhibits CBF function, it is surprising that Myb or Runx2 may play a synergistic effect in transformation. We speculate that Cbfb-MYH11 expression may have an incomplete effect on CBF function that is sufficient to affect differentiation, although the residual CBF function may rescue the cells from transformation. Additional alterations on CBF regulation of target genes, due to Runx2 or Myb misexpression, may transform the cells to AML. Alternatively, these genes may have CBFindependent functions that cooperate with CBFβ-SMMHC in AML.
Many SIS genes in this study encode members of signaling pathways linked to proliferation, apoptosis, and cancer. These pathways include Ras/PI3K, TGFβ IFN signaling, and cytoskeleton organization. Whereas the SIS matching previously identified sites may represent genes with broad functions associated with transformation, the majority of SISs are candidates and may be specifically associated with AML subtype M4 with eosinophilia.
The efficiency of our approach is highlighted by the absence of leukemia in MLV-mutagenized 129Sv/Ev wild-type mice and in untreated Cbfb-MYH11 mice. In addition, each leukemia case harbored very few insertions (sometimes only one), which was probably attributed to the transformation predisposition provided by Cbfb-MYH11. The fact that the 4070A envelope sequence does not share homology with endogenous retroviruses contributes to the easy identification of the insertion sites by iPCR with relatively low backgrounds. As a result, unlike previous studies that show a background incidence of lymphoma/leukemia and numerous insertions per sample (16-18), most of the genes identified in this study are likely to play a role in Cbfb-MYH11-associated AML. Finally, the use of a systematic mutagenesis approach, such as RIM in transgenic mice expressing leukemia-associated fusion genes, will provide a wealth of genetic information for the study of human leukemia, including new directions for improved therapies in the future.
Supplementary Material
Acknowledgments
We thank Carolyn Wilson for advice on quantitation of replication competent retrovirus. L.H.C. is a Leukemia and Lymphoma Society Special Fellow. This work was supported in part by National Institutes of Health Grant RO1-CA096983 (to L.H.C.) and American Cancer Society Grant IRG 93-033.
Abbreviations: AML, acute myeloid leukemia; RIS, retroviral insertion site; CIS, common insertion site; SIS, single insertion site; SMMHC, smooth muscle myosin heavy chain; CBF, core binding factor; RIM, retroviral insertional mutagenesis; iPCR, inverse PCR; MLV, murine leukemia virus; Cri, Cbfb-MYH11 retrovirus insertion.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. CL001593-CL001659).
References
- 1.Look, A. T. (1997) Science 278, 1059-1064. [DOI] [PubMed] [Google Scholar]
- 2.Liu, P., Tarle, S. A., Hajra, A., Claxton, D. F., Marlton, P., Freedman, M., Siciliano, M. J. & Collins, F. S. (1993) Science 261, 1041-1044. [DOI] [PubMed] [Google Scholar]
- 3.Castilla, L. H., Wijmenga, C., Wang, Q., Stacy, T., Speck, N. A., Eckhaus, M., Marin-Padilla, M., Collins, F. S., Wynshaw-Boris, A. & Liu, P. P. (1996) Cell 87, 687-696. [DOI] [PubMed] [Google Scholar]
- 4.Okuda T., van Duersen, J., Hiebert, S. W., Grosveld, G. & Downing, J. R. (1996) Cell 84, 321-330. [DOI] [PubMed] [Google Scholar]
- 5.Wang, Q., Stacy, T., Miller, J. D., Lewis, A. F., Gu, T. L., Huang, X., Bushweller, J. H., Bories, J. C., Alt, F. W., Ryan, G., et al. (1996) Cell 87, 697-708. [DOI] [PubMed] [Google Scholar]
- 6.Wang, Q., Stacy, T., Binder, M., Marin-Padilla, M., Sharpe, A. H. & Speck, N. A. (1996) Proc. Natl. Acad. Sci. USA 93, 3444-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sasaki, K., Yagi, H., Bronson, R. T., Tominaga, K., Matsunashi, T., Deguchi, K., Tani, Y., Kishimoto, T. & Komori, T. (1996) Proc. Natl. Acad. Sci. USA 93, 12359-12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niki, M., Okada, H., Takano, H., Kuno, J., Tani, K., Hibino, H., Asano, S., Ito, Y., Satake, M. & Noda, T. (1997) Proc. Natl. Acad. Sci. USA 94, 5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castilla, L. H., Garrett, L., Adya, N., Orlic, D., Dutra, A., Anderson, S., Owens, J., Eckhaus, M., Bodine, D. & Liu, P. P. (1999) Nat. Genet. 23, 144-146. [DOI] [PubMed] [Google Scholar]
- 10.Jonkers, J. & Berns, A. (1996) Biochim. Biophys. Acta 1287, 29-57. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki, T., Shen, H., Akagi, K., Morse, H. C., Malley, J. D., Naiman, D. Q., Jenkins, N. A. & Copeland, N. G. (2002) Nat. Genet. 32, 166-174. [DOI] [PubMed] [Google Scholar]
- 12.Li, J., Shen, H., Himmel, K. L., Dupuy, A. J., Largaespada, D. A., Nakamura, T., Shaughnessy, J. D., Jr., Jenkins, N. A. & Copeland, N. G. (1999) Nat. Genet. 23, 348-353. [DOI] [PubMed] [Google Scholar]
- 13.Joosten, M., Vankan-Berkhoudt, Y., Tas, M., Lunghi, M., Jenniskens, Y., Parganas, E., Valk, P. J., Lowenberg, B., van den Akker, E. & Delwel, R. (2002) Oncogene 21, 7247-7255. [DOI] [PubMed] [Google Scholar]
- 14.Stewart, M., Terry, A., Hu, M., O'Hara, M., Blyth, K., Baxter, E., Cameron, E., Onions, D. E. & Neil, J. C. (1997) Proc. Natl. Acad. Sci. USA 94, 8646-8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feldman, B. J., Hampton, T. & Cleary, M. L. (2000) Blood 96, 1906-1913. [PubMed] [Google Scholar]
- 16.Mikkers, H., Allen, J., Knipscheer, P., Romeijn, L., Hart, A., Vink, E., Berns, A. & Romeyn, L. (2002) Nat. Genet. 32, 153-159. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, H. C., Martins, C. P., Bronkhorst, Y., Randel, E., Berns, A., Fero, M. & Clurman, B. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11293-11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund, A., Turner, G., Trubetskoy, A., Verhoeven, E., Wientjens, E., Hulsman, D., Russell, R., DePinho, R., Lenz, J. & van Lohuizen, M. (2002) Nat. Genet. 32, 160-165. [DOI] [PubMed] [Google Scholar]
- 19.Chattopadhyay, S. K., Oliff, A. I., Linemeyer, D. L., Lander, M. R. & Lowy, D. R. (1981) J. Virol. 39, 777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oliff, A., McKinney, M. D. & Agranovsky, O. (1985) J. Virol. 54, 864-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassin, R. H., Ruscetti, S., Ali, I., Haapala, D. K. & Rein, A. (1982) Virology 123, 139-151. [DOI] [PubMed] [Google Scholar]
- 22.Ott, D. E., Keller, J., Sill, K. & Rein, A. (1992) J. Virol. 66, 6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolff, L., Koller, R. & Davidson, W. (1991) J. Virol. 65, 3607-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kas, K., Voz, M. L., Roijer, E., Astrom, A. K., Meyen, E., Stenman, G. & Van de Ven, W. J. (1997) Nat. Genet. 15, 170-174. [DOI] [PubMed] [Google Scholar]
- 25.Foa, C., Mainguene, C., Dupre, F., Coindre, J. M., Huguet, C., Kober, C. & Pedeutour, F. (2002) Cancer Genet. Cytogenet. 133, 156-159. [DOI] [PubMed] [Google Scholar]
- 26.Hibbard, M. K., Kozakewich, H. P., Dal Cin, P., Sciot, R., Tan, X., Xiao, S. & Fletcher, J. A. (2000) Cancer Res. 60, 4869-4872. [PubMed] [Google Scholar]
- 27.Hensen, K., Van Valckenborgh, I. C., Kas, K., Van de Ven, W. J. & Voz, M. L. (2002) Cancer Res. 62, 1510-1517. [PubMed] [Google Scholar]
- 28.Haviernik, P., Festin, S. M., Opavsky, R., Koller, R. P., Barr, N. I., Neil, J. C. & Wolff, L. (2002) J. Gen. Virol. 83, 819-827. [DOI] [PubMed] [Google Scholar]
- 29.Hanlon, L., Barr, N. I., Blyth, K., Stewart, M., Haviernik, P., Wolff, L., Weston, K., Cameron, E. R. & Neil, J. C. (2003) J. Virol. 77, 1059-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., et al. (1997) Cell 89, 765-771. [DOI] [PubMed] [Google Scholar]
- 31.Satake, M., Nomura, S., Yamaguchi-Iwai, Y., Takahama, Y., Hashimoto, Y., Niki, M., Kitamura, Y. & Ito, Y. (1995) Mol. Cell. Biol. 15, 1662-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogawa, E., Maruyama, M., Kagoshima, H., Inuzuka, M., Lu, J., Satake, M., Shigesada, K. & Ito, Y. (1993) Proc. Natl. Acad. Sci. USA 90, 6859-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blyth, K., Terry, A., Mackay, N., Vaillant, F., Bell, M., Cameron, E. R., Neil, J. C. & Stewart, M. (2001) Oncogene 20, 295-302. [DOI] [PubMed] [Google Scholar]
- 34.Vaillant, F., Blyth, K., Terry, A., Bell, M., Cameron, E. R., Neil, J. & Stewart, M. (1999) Oncogene 18, 7124-7134. [DOI] [PubMed] [Google Scholar]
- 35.Sattler, M., Verma, S., Pride, Y. B., Salgia, R., Rohrschneider, L. R. & Griffin, J. D. (2001) J. Biol. Chem. 276, 2451-2458. [DOI] [PubMed] [Google Scholar]
- 36.Martelli, M. P., Boomer, J., Bu, M. & Bierer, B. E. (2001) J. Biol. Chem. 276, 45654-45661. [DOI] [PubMed] [Google Scholar]
- 37.Murakami, H., Yamamura, Y., Shimono, Y., Kawai, K., Kurokawa, K. & Takahashi, M. (2002) J. Biol. Chem. 277, 32781-32790. [DOI] [PubMed] [Google Scholar]
- 38.Pandolfi, P. P. (2001) Oncogene 20, 5726-5735. [DOI] [PubMed] [Google Scholar]
- 39.Higuchi, M., O'Brien, D., Kumaravelu, P., Lenny, N., Yeoh, E. J. & Downing, J. R. (2002) Cancer Cell 1, 63-74. [DOI] [PubMed] [Google Scholar]
- 40.Yuan, Y. Z., Zhou, L. M., Miyamoto, T., Iwasaki, H., Harakawa, N., Hetherington, C. J., Burel, S. A., Lagasse, E., Weissman, I. L., Akashi, K. & Zhang, D. E. (2001) Proc. Natl. Acad. Sci. USA 98, 10398-10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Voz, M. L., Agten, N. S., Van de Ven, W. J. & Kas, K. (2000) Cancer Res. 60, 106-113. [PubMed] [Google Scholar]
- 42.Mizutani, A., Furukawa, T., Adachi, Y., Ikehara, S. & Taketani, S. (2002) J. Biol. Chem. 277, 15851-15858. [DOI] [PubMed] [Google Scholar]
- 43.Britos-Bray, M. & Friedman, A. D. (1997) Mol. Cell. Biol. 17, 5127-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hernandez-Munain, C. & Krangel, M. S. (1994) Mol. Cell. Biol. 14, 473-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.