Background: p24 family proteins function as cargo receptors for ER-Golgi transport.
Results: Genetic and physical interactions among eight known and one newly identified S. cerevisiae p24 proteins were determined.
Conclusion: Yeast p24 proteins function in several functionally redundant complexes, including one containing a novel p24 isoform induced under respiratory conditions.
Significance: This is the first comprehensive study of the yeast p24 complexes.
Keywords: Endoplasmic Reticulum (ER), Golgi, Membrane Trafficking, Protein Assembly, Protein Sorting, Yeast
Abstract
p24 family proteins are evolutionarily conserved transmembrane proteins involved in the early secretory pathway. Saccharomyces cerevisiae has 8 known p24 proteins that are classified into four subfamilies (p24α, -β, -γ, and -δ). Emp24 and Erv25 are the sole members of p24β and -δ, respectively, and deletion of either destabilizes the remaining p24 proteins, resulting in p24 null phenotype (p24Δ). We studied genetic and physical interactions of p24α (Erp1, -5, and -6) and γ (Erp2, -3, and -4). Deletion of the major p24α (Erp1) partially inhibited p24 activity as reported previously. A second mutation in either Erp5 or Erp6 aggravated the erp1Δ phenotype, and the triple mutation gave a full p24Δ phenotype. Similar genetic interactions were observed among the major p24γ (Erp2) and the other two γ members. All the p24α/γ isoforms interacted with both p24β and -δ. Interaction between p24β and -δ was isoform-selective, and five major α/γ pairs were detected. These results suggest that the yeast p24 proteins form functionally redundant αβγδ complexes. We also identified Rrt6 as a novel p24δ isoform. Rrt6 shows only limited sequence identity (∼15%) to known p24 proteins but was found to have structural properties characteristic of p24. Rrt6 was induced when cells were grown on glycerol and form an additional αβγδ complex with Erp3, Erp5, and Emp24. This complex was mainly localized to the Golgi, whereas the p24 complex containing Erv25, instead of Rrt6 but otherwise with the same isoform composition, was found mostly in the ER.
Introduction
Membrane traffic between the endoplasmic reticulum (ER)3 and the Golgi apparatus is bidirectional and mediated by two coat protein complexes, COPI and COPII (1). Anterograde transport of newly synthesized proteins and lipids from the ER depends on COPII. COPII consists of the inner coat complex Sec23/Sec24 and the outer coat complex Sec13/Sec31, whose assembly is regulated by the small GTPase Sar1 (2). Retrograde transport of ER-resident and ER-Golgi recycling proteins back to the ER is mediated by COPI, the heptameric coatomer complex (α, β, β′, γ, δ, ϵ, ζ), which is assembled by the small GTPase Arf1 (3). COPI is also implicated in intra-Golgi retrograde transport of resident proteins and in forward transport of cargo protein into and through the Golgi (4–9).
Selective incorporation of cargo proteins into the transport vesicles basically relies on their interaction with the coat proteins. The coat complexes have multiple cargo binding sites and capture a variety of cargo proteins into transport vesicles (10–13). However, some cargo proteins require additional sorting machinery for efficient transport. For example, certain soluble cargo proteins are incorporated into transport vesicles in a manner dependent on cargo receptors. Cargo receptors are transmembrane proteins that can interact with both cargo and coat proteins and bridge their interaction during the formation of the transport vesicles (14). A well documented example is the KDEL receptor, which is responsible for the Golgi-to-ER retrieval of soluble ER-resident proteins bearing a KDEL retention signal (15, 16). ERGIC-53 and its paralogs have a luminal carbohydrate recognition domain and are thought to facilitate transport of newly synthesized glycoproteins (17, 18). Cargo receptors for transmembrane proteins have also been reported (19–23).
p24 family proteins are evolutionarily conserved transmembrane proteins of ∼24 kDa that have been characterized as cargo receptors for mammalian and yeast glycosylphosphatidylinositol-anchored proteins (24–27), Drosophila Wingless protein (28, 29), Xenopus pro-opiomelanocortin (30), and others (31, 32). p24 proteins are single transmembrane proteins with a short C-terminal cytoplasmic tail of 10–20 residues. The cytoplasmic tail can interact with both COPI and COPII subunits, thus enabling the protein to continuously cycles between the ER and the Golgi. The luminal portion of the proteins consists of two domains. The membrane proximal domain has a helical structure and is implicated in the p24 oligomer formation. The N-terminal domain of ∼100 residues is referred to as GOLD domain (33), which is predicted to form a β-sandwich structure and to participate in the cargo recognition (27).
In the present study we focused on the formation of the p24 complexes in Saccharomyces cerevisiae. Most eukaryotes have 4–10 p24 proteins that are classified into four subfamilies (p24α, -β, -γ, and -δ) (34). Knockdown or overexpression of a single p24 isoform often leads to mislocalization and degradation of other isoforms (27, 28, 35–38). These observations together with the results of physical interaction studies led to the conclusion that p24 proteins form heteromeric complexes, most likely dimers and tetramers. Although how p24 proteins recognize their cargoes remains unknown, it is conceivable that p24 complexes with different isoform compositions have different cargo specificity. Assuming that the cargo recognition site is located at the interface of two or more p24 proteins, cargo binding would depend on and could even be regulated by the complex formation. Interestingly, evidence suggests that the complex formation in mammals and plants is reversible and is coupled to the ER-Golgi transport (39, 40). The reversible assembly of the p24 complex has been discussed mainly in terms of the localization of individual p24 isoforms, because the oligomeric state of the cytosolic tail has been shown to influence the interaction with the coat subunits and other transport factors. However, an intriguing possibility is that the each repeated cycle of complex assembly drives the cycle of cargo capture and release. In this way the p24 complex formation can be directly coupled to their function. However, limited information is available on the subunit composition of individual complexes and their relative abundance.
We examined genetic and physical interactions of S. cerevisiae p24 proteins. The yeast p24 family consists of one each of p24β and -δ, which has shown to be essential for the cellular p24 activity (24, 35), and three each of p24β and -δ. We show that the yeast p24 proteins function in several different αβγδ heteromeric complexes with mostly overlapping functions. We also identified a novel p24δ isoform, Rrt6. Rrt6 is induced under respiratory conditions to form another αβγδ complex, which has unique features not found in the other p24 complexes, although the physiological role of the Rrt6-containing complex remains to be determined.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
Yeast strains used in this study (Table 1) are of BY4741 background (41). Strains expressing p24 proteins that are N-terminally tagged with the myc epitope (EQKLISEEDL) in triplicate (3myc) were constructed by two-step pop-in pop-out methods as described in Rothstein (42). The tag was inserted at one (Erp1 to Erp6) or three (Rrt6) amino acid carboxyl sites to the potential signal peptide cleavage site. Gene disruption by KanMX4, natNT2, or HIS3 cassette was done as in Janke et al. (43). Yeast cells were grown in YPD medium (1% yeast extract (Difco), 2% bacto peptone (Difco), and 2% glucose) or in MCD medium (0.67% yeast nitrogen base (Difco), 0.5% casamino acids (Difco), and 2% glucose). MCD medium was supplemented with nutrients when required as described in Burke et al. (44). YPGlycerol and MCGlycerol are same as YPD and MCD, respectively, except that they contain 3% glycerol instead of glucose. BY4513 (rrt6Δ) was crossed to the following mutants to see possible genetic interactions: COPI/COPII mutants, sec13-1, sec21-1, and sec27-1; ER/Golgi transport mutants, gcs1Δ, glo3Δ, gsg1Δ, erv14Δ, erv29Δ, and svp26Δ; ER-associated degradation pathway mutants, ire1Δ and hac1Δ.
TABLE 1.
Strains used in This study
| Strains | Genotype | References |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Invitrogen |
| BY7153 | erp1::KanMX4 in BY4741 | Invitrogen |
| BY401 | erp2::KanMX4 in BY4741 | Invitrogen |
| BY3714 | erp3::KanMX4 in BY4741 | Invitrogen |
| BY1792 | erp4::KanMX4 in BY4741 | Invitrogen |
| BY1938 | erp5::KanMX4 in BY4741 | Invitrogen |
| BY4370 | erp6::KanMX4 in BY4741 | Invitrogen |
| BY4567 | emp24::KanMX4 in BY4741 | Invitrogen |
| BY562 | erv25::KanMX4 in BY4741 | Invitrogen |
| BY4513 | rrt6::KanMX4 in BY4741 | Invitrogen |
| CNY246 | erp1::natNT2 erp5::KanMX4 in BY4741 | This study |
| CNY247 | erp1::natNT2 erp6::KanMX4 in BY4741 | This study |
| CNY258 | erp5:: KanMX4 erp6::HIS3 in BY4741 | This study |
| CNY256 | erp1::natNT2 erp5::HIS3 erp6::KanMX4 in BY4741 | This study |
| CNY253 | erp2:: KanMX4 erp3::natNT2 in BY4741 | This study |
| CNY242 | erp2:: KanMX4 erp4::natNT2 in BY4741 | This study |
| CNY248 | erp3:: natNT2 erp4:: KanMX4 in BY4741 | This study |
| CNY250 | erp2::KanMX4 erp3::HIS3 erp4::natNT2 in BY4741 | This study |
| CNY249 | erp1::KanMX4 erp2::natNT2 erp4::HIS3 in BY4741 | This study |
| CNY201 | 3myc-ERP1 in BY4741 | This study |
| CNY202 | 3myc-ERP2 in BY4741 | This study |
| CNY203 | 3myc-ERP3 in BY4741 | This study |
| CNY204 | 3myc-ERP4 in BY4741 | This study |
| CNY205 | 3myc-ERP5 in BY4741 | This study |
| CNY206 | 3myc-ERP6 in BY4741 | This study |
| CNY207 | 3myc-RRT6 in BY4741 | This study |
Plasmids
Plasmids used in this study (Table 2) were constructed on centromere-based low-copy vectors. RRT6 (YGL146c) was amplified with 900-base 5′ and 600-base 3′ flanking regions by polymerase chain reaction (PCR) and cloned into pRS316 (45). A BglII restriction site was introduced after the 54th codon of the RRT6 open reading frame, and the 3myc tag was inserted in triplicate (pCNY611). Mutant versions of pCNY611 were constructed by overlap extension PCR (46).
TABLE 2.
Plasmids used in this study
| Plasmids | Description | References |
|---|---|---|
| pRS313 | S. cerevisiae low-copy vector (HIS3) | (45) |
| pRS316 | S. cerevisiae low-copy vector (URA3) | (45) |
| pCNY611 | pRS316 3myc-RRT6 | This study |
| pCNY608 | pRS316 3myc-rrt6–293YL294-AA | This study |
| pCNY601 | pRS316 3myc-rrt6–295II296-AA | This study |
| pCNY602 | pRS316 3myc-rrt6–297KI298-AA | This study |
| pCNY603 | pRS316 3myc-rrt6–299KS300-AA | This study |
| pCNY604 | pRS316 3myc-rrt6–301NP302-AA | This study |
| pCNY605 | pRS316 3myc-rrt6–303SS304-AA | This study |
| pCNY606 | pRS316 3myc-rrt6–305HI306-AA | This study |
| pCNY609 | pRS316 3myc-rrt6–307KKK309-AAA | This study |
| pCNY607 | pRS316 3myc-rrt6–310GL311-AA | This study |
| pCNY611 | pRS316 3myc-rrt6–297KIK299-AIA | This study |
| pCNY571 | pRS316 3myc-rrt6-N96D | This study |
| pCNY572 | pRS316 3myc-rrt6-C105S | This study |
| pCNY575 | pRS316 3myc-rrt6-C183S | This study |
| pCNY576 | pRS316 3myc-rrt6-C189S | This study |
| pCNY033 | pRS316 3HA-RRT6 | This study |
| pCNY033–2 | pRS316 3HA-RRT6 (tag inserted after the 5th codon) | This study |
| pCNY033–3 | pRS316 3HA-RRT6 (tag inserted after the 31st codon) | This study |
| pYS283 | pRS316 PERP2-ERP3 | This study |
| pYS285 | pRS316 PERP1-ERP5 | This study |
| pYS289 | pRS316 PERP2-ERP3 PERP1-ERP5 | This study |
| pYS296 | pRS313 PERP2-ERP3 PERP1-ERP5 | This study |
| pYS74 | pRS316 PERV25-ERV25 signal peptide-3myc-RRT6 | This study |
Antibodies
Antibodies were raised against synthetic peptides corresponding to residues 194–203 of Emp24, 70–87 of Erv25, 294–311 of Rrt6, 463–480 of Och1, 55–69 of Pma1, and 171–188 of Rer1. Antigen peptides were synthesized with an additional cysteine residue at the N terminus, conjugated to keyhole limpet hemocyanin, and used to inject into rabbits. Anti-Vma22 antiserum was raised against His6-tagged Vma22 expressed in Escherichia coli. His6-Vma22 was purified with a nickel-nitrilotriacetic acid agarose column (Qiagen) under denaturing conditions according to the manufacturer's instructions. Antisera were affinity-purified over antigen-immobilized columns. Affinity-purified rabbit antibodies against COPI (47), Sec12 (48), Sec23 (18), and Kex2 (18) were as described. Mouse monoclonal antibodies against Vph1 and Por1 were purchased from Life Technologies. Mouse anti-HA (12CA5) and anti-myc (9B11) antibodies were from Roche Diagnostics and Cell Signaling Technology, respectively. Anti-myc tag-agarose conjugate (4A6) was purchased from Merck Millipore. Rabbit anti-myc antibodies were from Sigma.
p24 Subtype Isolation and Mass Spectrometry
Logarithmically growing cells were harvested and converted to spheroplasts by incubation at 30 °C for 30 min in a buffer containing 20 mm sodium phosphate, pH 7.2, 1 m sorbitol, 1 mm sodium azide, and 0.15 mg/ml zymolyase 100T (Nakalai). Spheroplasts (∼1 × 1010 cells) were lysed in ∼30 ml of HBS buffer (10 mm HEPES, 0.15 m NaCl, pH 7.4) containing 1 mm MgCl2 and protease inhibitor mixture (Sigma). Unbroken cells and cell debris were removed by centrifugation at 1000 × g for 5 min. The cleared lysate was centrifuged at 13,000 × g for 10 min to obtain S13 (supernatant) and P13 (pellet) fractions. S13 fraction was further centrifuged at 100,000 × g for 60 min to yield S100 (supernatant) and P100 (pellet) fractions. P13 and P100 fractions were each suspended in ∼3 ml of HBS buffer containing 1% (w/v) Triton X-100, and proteins were solubilized by incubation at 4 °C for 30 min. Insoluble materials were removed by centrifugation (13,000 × g 10 min), and 5 μg of anti-myc monoclonal antibody conjugated to agarose beads (clone 4A6, Millipore) was added to the solubilized fractions. After overnight incubation at 4 °C, beads were collected and washed 5 times with the same buffer, and bound proteins were eluted into modified Laemmli sample buffer (50 mm Tris, 1% (w/v) SDS, 2% (v/v) 2-mercaptoethanol, 1 mm EDTA, pH 6.8). For protein identification, eluted proteins were separated by SDS-PAGE and stained with Coomassie Brilliant Blue, and gel regions containing the proteins of interest were excised. Proteins in the excised gel blocks were destained, reduced, alkylated at the cysteine residues, and digested with modified trypsin. Trypsin-digested peptides were extracted from the gel blocks and subjected to mass spectrometric analysis. Liquid chromatography-tandem mass spectrometry analysis was performed on Paradigm MS2 HPLC (Michrom BioResources) and LTQ linear ion trap mass spectrometer with Nanospray ion source housing (Thermo Fisher Scientific). Peptides were separated through L-column2 C18 (0.1 × 150 mm, 3 μm particle size, CERI, Tokyo) using a linear gradient (5–65% acetonitrile in 0.1% formic acid, 2%/min) at a flow rate of 1 μl/min. Mass spectrometric data were searched against NCBI database using MASCOT software (Matrix Science).
Membrane Fractionation
Membrane association of Rrt6 was determined as in Sato and Nakano (18). Spheroplasts were lysed in phosphate buffer saline, and cleared lysate was adjusted to either 0.5 m NaCl, 0.1 m Na2CO3, pH 11, or 1% (w/v) Triton X-100. After incubation on ice for 30 min, the mixtures were centrifuged at 100,000 × g for 1 h, and the extraction of Rrt6 was assayed by Western blot analysis.
Sucrose density gradient fractionation was done as described in Sato and Nakano (18). Briefly, spheroplasts (∼3 × 108 cells) were lysed in 1 ml of a buffer containing 10 mm HEPES, pH 7.4, 1 mm MgCl2, and 12.5% (w/v) sucrose. Cleared lysate was layered on the top of a 9-ml sucrose gradient (20–60%, 5% steps) in 10 mm HEPES, pH 7.4, and 1 mm MgCl2, and the gradient was centrifuged for 2.5 h at 200,000 × g in a swing-bucket rotor (Hitachi P40). Fractions of 0.7 ml were collected from the top and assayed for Rrt6 and organelle markers by Western blot analysis.
Limited Protease Digestion of Rrt6
Spheroplasts were lysed in HBS buffer, and cleared lysate was centrifuged at 100,000 × g for 1 h. The pellet fraction (crude membrane fraction) was suspended in TBS (20 mm Tris, 0.15 m NaCl, pH 8.0) with or without 0.1% (w/v) Triton X-100 at a protein concentration of ∼0.3 mg/ml. Aliquots (100 μl) were incubated with trypsin (5 units/μg, 0–100 μg/ml) at 37 °C for 30 min. Proteins were precipitated by trichloroacetic acid and were subjected to Western blot analysis to determine the residual level of Rrt6.
Endoglycosidase Treatment
The crude membrane fraction was suspended in a buffer (∼0.3 mg protein/ml) containing 0.5% (w/v) SDS and 40 mm dithiothreitol, and proteins were solubilized under denaturing conditions by heating at 70 °C for 10 min. Aliquots (∼80 μl) were buffered with 50 mm sodium citrate, pH 5.5, and incubated with or without 2000 units of recombinant Streptomyces plicatus endoglycosidase H (EndoHf, New England Biolabs) at 37 °C for 2 h. Removal of the sugar chains from Rrt6 was examined by Western blot analysis.
COPI/COPII Binding to p24 C-tail Peptides
In vitro COPI binding assay of p24 cytoplasmic tail peptides was done as in Belden and Barlowe (49). Synthetic peptides used were Erv25C (CLKNYFKTKHII), Erv25YF-AA (CLKNAAKTKHII), Rrt6C (CTYLIIKIKSNPSSHIKKKGL), and Rrt6-KQ (CTYLIIKIKSNPSSHIQQQGL). These peptides (0.4 μmoles) were conjugated to 1 ml of iodoacetyl-activated agarose beads (Sulfolink, Thermo) according to the supplier's protocol. The amount of peptides coupled to the beads was quantified by amino acid analysis of the acid hydrolysates of the beads. Coupling efficiency was ∼50%. Spheroplasts (∼1.5 × 108 cells) were lysed in 1 ml of HBS containing 0.5% (w/v) Triton X-100 and incubated with ∼15 μl of peptide-conjugated beads (adjusted to 3 nmol of tail peptide) at 4 °C for 1 h. Beads were collected and washed four times with the lysis buffer, and bound proteins were analyzed by Western blot analysis.
RESULTS
S. cerevisiae has eight known p24 proteins (Table 3) (35). Emp24 and Erv25 are the sole yeast counterparts of the mammalian p24β and p24δ, respectively, and their mutants show the p24 null phenotype (p24Δ) (49, 50). Erp1 and Erp2 are similar to p24α and p24γ, respectively, and are assembled with Emp24 and Erv25 to form a major p24 complex in this organism (35). However, deletion of Erp1 or Erp2 gives only a partial p24Δ phenotype. The residual p24 activity in these mutants can be attributed to the presence of additional p24α (Erp5 and Erp6) and p24γ (Erp3 and Erp4) isoforms, but the functionality of these isoforms remains elusive. Therefore, we decided to examine the phenotype of single, double, and triple mutants of the p24α and p24γ subfamilies.
TABLE 3.
p24 family members in S. cerevisiae and mammals
| p24 subtype | S. cerevisiae | Mammals (34)a | Other names |
|---|---|---|---|
| p24α | Erp1 | TMED11 (p24α1)b | gp25L |
| Erp5 | TMED9 (p24α2) | p25, GMP25, gp25L2, p24d | |
| Erp6 | TMED4 (p24α3) | GMP25iso, ERS25 | |
| p24β | Emp24 | TMED2 (p24β) | p24, p24a |
| p24γ | Erp2 | TMED1 (p24γ1) | tp24 |
| Erp3 | TMED5 (p24γ2) | p28 | |
| Erp4 | TMED7 (p24γ3) | gp27, p27 | |
| TMED3 (p24γ4) | p26, p24b | ||
| TMED6 (p24γ5) | |||
| p24δ | Erv25 | TMED10 (p24δ) | p23, p24c, TMP21 |
| Rrt6 |
p24β and -δ Isoforms Are Essential Components of Active p24 Complexes
S. cerevisiae p24Δ phenotype includes slow ER export of glycosylphosphatidylinositol-anchored proteins, anomalous accumulation of the secreted form of invertase (50), and genetic interactions with COPI/COPII mutations (51). The mutants also show activation of unfolded protein response, resulting in increased expression and secretion of an ER-resident chaperone Kar2/BiP (52). Although how p24Δ mutation triggers the stress response remains unknown, the Kar2 secretion phenotype is highly responsive to the p24 dysfunction and thus is a good indicator of cellular p24 activity. Therefore, we first examined the Kar2 secretion of p24α/γ mutant cells.
As reported previously, erp1Δ and erp2Δ mutant cells secreted Kar2 at a level intermediate (50∼60% of p24Δ) between those of the wild-type and p24Δ cells (Fig. 1A, 1Δ and 2Δ) (35). The phenotype was less prominent in the other four mutants. Kar2 secretion was weak (∼15%) in erp4Δ and almost undetectable (∼10%) in erp3Δ, erp5Δ, and erp6Δ (Fig. 1A, 3Δ to 6Δ). However, these four mutations showed clear synergy when combined with erp1Δ or erp2Δ. p24α double mutants erp1Δ erp5Δ and erp1Δ erp6Δ secreted an increased amount of Kar2 than the erp1Δ mutant, and the triple mutant (p24αΔ) secreted even more to a level comparable with those of emp24Δ and erv25Δ (Fig. 1A). Similarly, both erp3Δ and erp4Δ aggravated the phenotype of erp2Δ, and the triple deletion (p24γΔ) gave a full p24Δ phenotype.
FIGURE 1.
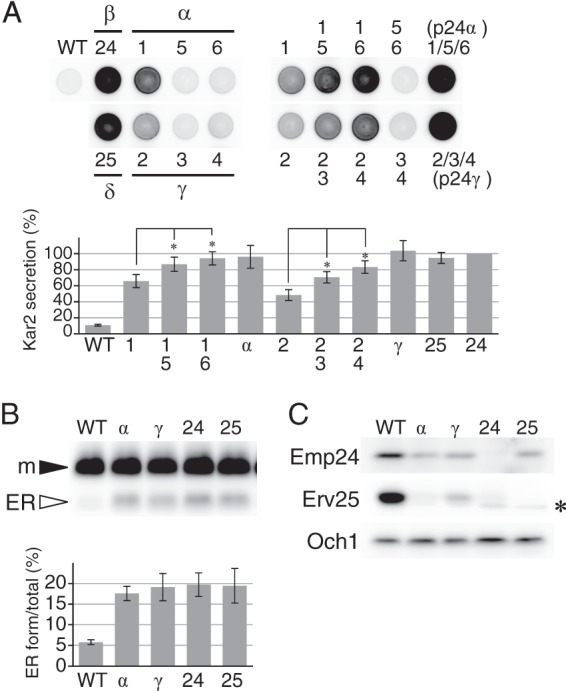
Phenotype of single, double, and triple deletion mutants of p24α and p24γ subfamilies. A, Kar2 secretion of p24 mutant cells. Mid-log cultures were harvested and suspended in fresh medium, and aliquots (∼1.5 × 105 cells) were spotted on a YPD agar plate. The plate was overlaid with a nitrocellulose filter and incubated for 5 h at 30 °C. Kar2 protein secreted from the cells and bound to the filter was detected by Western blot analysis and quantified by densitometry. The graph represents the relative amount of secreted Kar2. The amount of Kar2 from emp24Δ cells was set as the standard (100%). Error bars represent S.E. (n = 4). Asterisks indicate p < 0.05. Strains used were: the parental wild-type cells (WT); single mutants of p24β (emp24Δ, 24Δ) and p24δ (erv25Δ, 25Δ); single (Δ1, Δ5 and Δ6), double (Δ1Δ5, Δ1Δ6, and Δ5Δ6), and triple (Δα) mutants of p24α (Erp1, -5, and -6); single (Δ2, Δ3, and Δ4), double (Δ2Δ3, Δ2Δ4, and Δ3Δ4), and triple (Δγ) mutants of p24γ (Erp2, -3, and -4). B, accumulation of the ER form of Gas1 in p24 mutant cells. Mature (m) and ER-precursor forms (ER) of Gas1 in whole-cell extracts (∼1 × 106 cells) were detected by Western blog analysis. Strains used are the wild-type (WT) and p24 mutants lacking the three p24α (αΔ), three p24γ (γΔ), p24β (24Δ), or p25δ (25Δ). The intensities of the bands were quantified by densitometry, and the ratio of ER form to the total Gas1 was calculated and shown in the graph. Error bars represent S.E. (n = 4). C, decreased steady-state levels of p24β (Emp24) and p24δ (Erv25) in p24 mutant cells. Steady-state levels of Emp24 and Erv25 in whole-cell extracts (∼2 × 106 cells) were determined by densitometry of immunoblots. The strains used are the same as in B. Och1, a Golgi-resident mannosyltransferase, was used as a control. The asterisk represents an unrelated cross-reacting protein.
Western blot analysis of whole-cell extracts showed that p24αΔ and p24γΔ mutants are also defective in the transport of a glycosylphosphatidylinositol-anchored protein, Gas1. The triple mutants accumulated increased amounts of the ER-precursor form of Gas1 (Fig. 1B, αΔ and γΔ), just like emp24Δ and erv25Δ (24Δ and 25Δ), whereas erp1Δ or erp2Δ alone showed a weaker phenotype (data not shown) (35, 53). Stability of p24 proteins depends on their complex formation, and erp1Δ and erp2Δ are reported to decrease the steady-state levels of Emp24 and Erv25 (35). Again, p24αΔ and p24γΔ showed severer phenotype, and in these mutant cells steady-state levels of Emp24 and Erp25 were decreased as observed in p24Δ mutant cells (Fig. 1C). These results indicate that all the p24β and -δ isoforms are functional and that the p24α and p24γ subfamilies are also essential for the cellular p24 activity.
p24β and -δ Isoforms Interact in an Isoform-selective Manner
Marzioch et al. (35) identified a major S. cerevisiae p24 complex consisting of Emp24, Erv25, Erp1, and Erp2. Their results also indicated the presence of additional p24 complexes, and our data suggested such complexes would contain p24β and -δ isoforms in several different combinations. Therefore, we examined physical interactions between p24α and -γ. For immunoprecipitation experiments, we constructed a series of strains in which one of the p24 proteins is tagged at the N terminus with three copies of myc epitope (3myc) (Fig. 2A). In these strains, the tag-coding DNA fragment was inserted at the chromosomal locus of the respective genes so that only the tagged version of the protein is expressed under their own promoter. Kar2 secretion assay confirmed that the tagged proteins are functional, although 3myc-Erp1 appeared to be partially defective (Fig. 2B).
FIGURE 2.
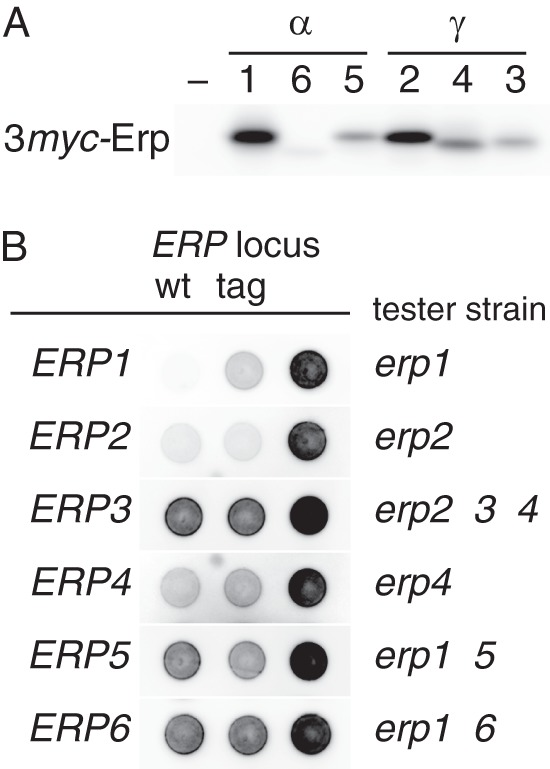
Epitope tagging of p24α and p24γ proteins. A, detection of 3myc-tagged p24α (Erp1, -5, and -6) and p24γ (Erp2, -3, and -4). Whole-cell extracts (∼2 × 106 cells) of wild-type (–) and 3myc-tagged p24 recombinant strains (1–6, the numbers correspond to the ERP gene numbers) were analyzed as in Fig. 1C. B, complementation of p24α/γ mutations by the 3myc-tagged versions of the respective genes. Tester strains (also as negative controls; Δ, right spots) used are indicated at the right side of each row. Wild-type (positive controls; wt, left spots) or 3myc-tagged (tag, center spots) ERP genes of interest were introduced to the tester strains on an integration vector. Kar2 secretion was assayed as in Fig. 1A. Blots for Erp1, Erp2, and Erp4 were overexposed for maximum clarity.
The tagged p24 proteins were solubilized with 1% (w/v) Triton X-100 from ER-enriched P13 membrane fractions (13,000 × g pellet fraction) and precipitated with anti-myc-agarose beads. Fig. 3A shows SDS-PAGE patterns of proteins co-immunoprecipitated with the 3myc-tagged p24γ isoforms. The identity of the proteins shown in the figure was inferred from the results of Western blot and mass spectrometric analyses (Table 4). The three p24γ isoforms efficiently coprecipitated Emp24 and Erv25 to significant levels that suggest formation of stable complexes. Similar levels of interaction were observed with p24α isoforms but in an isoform-selective manner. Specifically, 3myc-Erp2 and 3myc-Erp4 interacted with Erp1 and Erp6 but not with Erp5, whereas 3myc-Erp3 preferentially interacted with Erp5. These results were confirmed by the mass spectrometric analysis of the 3myc-p24α immunoprecipitates (Table 4). In this experiment, Erp4 was not efficiently coprecipitated with 3myc-Erp1 (Table 4). Although the exact reasons are unknown, this might represent preferential binding of Erp1 to Erp2 rather than to Erp4. Alternatively, the introduction of the 3myc tag might somehow inhibit normal interaction with Erp4, resulting in the partial inhibition of this construct (Fig. 2B).
FIGURE 3.
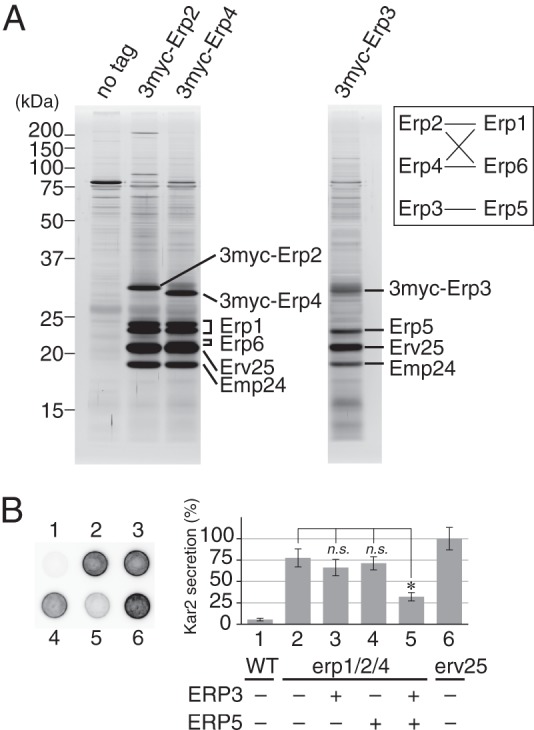
Physical interactions among p24 proteins. A, coprecipitation of p24 proteins with p24γ isoforms. ER-enriched P13 fractions were prepared as described under “Experimental Procedures” from cells expressing 3myc-tagged p24γ (Erp2, Erp3, or Erp4). The tagged p24γ proteins in the P13 fraction were solubilized with 1% (w/v) Triton X-100 and immunoprecipitated with anti-myc-agarose beads. Immunoprecipitated proteins were separated by SDS-PAGE and detected by silver staining. The protein identity shown in the figure was inferred from the result of the mass spectrometry (Table 4) and Western blot analysis. Erp1 was separated as a doublet for unknown reasons. The identities of these two bands were individually confirmed by mass spectrometric analysis. We could not clearly identified the band of Erp6 protein in the silver-stained gel, but because the gel fragment containing Erv25 gave mass signals of Erp6, we speculate that the band overlapping with that of Erv25 and slightly less mobile represents Erp6. The band of 3myc-Erp3 was also not clearly visible, most likely because it overlapped with those of proteins of unknown identity. The inset shows a schematic representation of the observed interactions (solid lines) between p24α and -γ isoforms. B, partial restoration of erp1Δ erp2Δ erp4Δ mutation by simultaneous overexpression of ERP3 and ERP5. Kar2 secretion was assayed as in Fig. 1A. The graph represents the relative amount of secreted Kar2, in which the average amount of Kar2 from erv25Δ cells was set to 100%. Column (spot) 1, wild-type cells carrying the empty vector. Column 2, erp1Δerp2Δerp4Δ cells carrying the empty vector. Columns 3–5, erp1Δerp2Δerp4Δ cells overexpressing ERP3 (column 3), ERP5 (column 4), or both (column 5). Column 6, erv25Δ cells carrying the empty vector. Error bars represent S.E. (n = 4). The asterisk indicates p < 0.05; n.s., not significant.
TABLE 4.
Mass spectrometry of p24 immunoprecipitates
3 myc-p24 immunoprecipitates were resolved by SDS-PAGE. Gel regions expected to contain proteins of 15–37 kDa were excised and subjected to mass spectrometry. Total number of mass matched, the number of sequences uniquely assigned to the proteins of interest, sequence coverage, and emPAI are shown. emPAI (exponentially modified protein abundance index), which offers approximate and relative quantification of the proteins in a mixture, was calculated on two parameters, the number of experimentally observed peptides as in Ishihama et al. (79). The emPAI values for p24α and -β (Emp24) were calculated to be higher than those for p24γ and -δ (Erv25). We speculate this reflects ionization efficiency of these proteins rather than their stoichiometry in the complex.
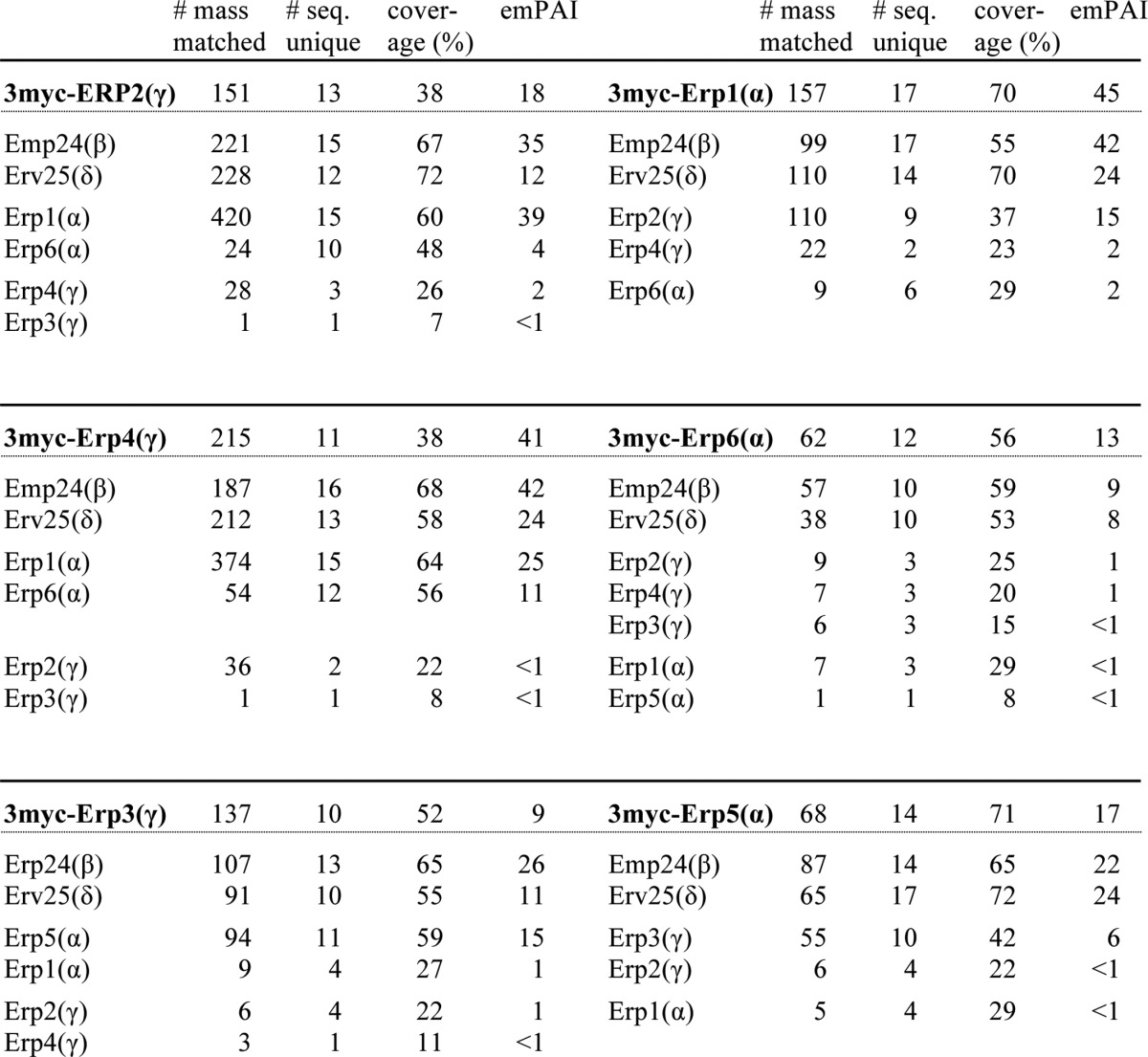
The mass spectrometric analysis identified five major p24α/γ interaction pairs, Erp1/Erp2, Erp1/Erp4, Erp6/Erp2, Erp6/Erp4, and Erp5/Erp3 (Fig. 3A and Table 4). The pairing pattern implies that Erp1 and Erp6 are interchangeable concerning the complex formation and so are Erp2 and Erp4, whereas Erp5 and Erp3 are independent of the other four (Fig. 3A). This is consistent with the result of a previous cross complementation experiment. Marzioch et al. (35) reported that, when overexpressed, Erp6 restores the loss of Erp1, and Erp4 restores the loss of Erp2, whereas Erp5 and Erp3 did not show such complementing activity. The apparent lack of complementation by Erp5 and Erp3 can be explained by their lower affinity for Erp2 and Erp1, respectively. However, the phenotype of the double and triple mutants suggests that Erp3 and Erp5 are also functionally redundant with their subfamily members (Fig. 1). We thought that simultaneous overexpression of Erp3 and Erp5 could complement the absence of the other p24α/γ isoforms and tested this hypothesis. We constructed plasmids designed to overexpress ERP3 by the ERP2 promoter, ERP5 by the ERP1 promoter, or both. These plasmids were introduced into a strain lacking the major p24α/γ isoforms (erp1Δ erp2Δ erp4Δ) and tested for complementation activity. As expected, Kar2 secretion of the parental strain was restored when both Erp3 and Erp5 were overexpressed (Fig. 3B, 5), whereas overexpression of Erp3 or Erp5 alone was not effective (3 and 4). These results suggest that in S. cerevisiae p24 proteins form several αβγδ heteromeric complexes with at least partially overlapping functions.
Unexpectedly, the mass spectrometry data also indicated physical interactions between two different members within the p24α subfamily and within p24γ (Table 4). This result is interesting because it might suggest the existence of previously unidentified p24 complexes. However, the signals were weak, and their significance should be confirmed by further studies.
Identification of Rrt6 as a p24-binding Protein
As seen in Fig. 3A, the p24 immunoprecipitates contained various proteins other than the p24 isoforms. Most of them appeared nonspecific because their recovery was not consistent among different preparations, but we found a protein that was constantly recovered in the 3myc-Erp3/Erp5 binding fractions (Fig. 4A). Mass spectrometric analysis identified this protein as Rrt6. Rrt6 was originally reported in screening for mutants with increased levels of ribosomal DNA transcription (54). Rrt6 has no reported function, but Pfam, a sequence alignment database (55), annotates this protein as a putative member of p24 family proteins. Gene chip studies suggested, and we confirmed that the protein is repressed under fermentative conditions and induced under respiratory conditions (data not shown) (56). We, therefore, examined whether rrt6Δ shows the p24Δ phenotype under respiratory growth conditions but found no discernible phenotype or genetic interactions that can be ascribed to the loss of p24 or to any other dysfunction in membrane traffic (see “Experimental Procedures” for details on the crossed strains). Therefore, the function of Rrt6 still remains unknown. However, because the aim of this study was to investigate all the p24 family members for their ability to form p24 complexes, we decided to see whether Rrt6 is a bona fide member of this protein family and whether the protein also forms a complex with other members.
FIGURE 4.
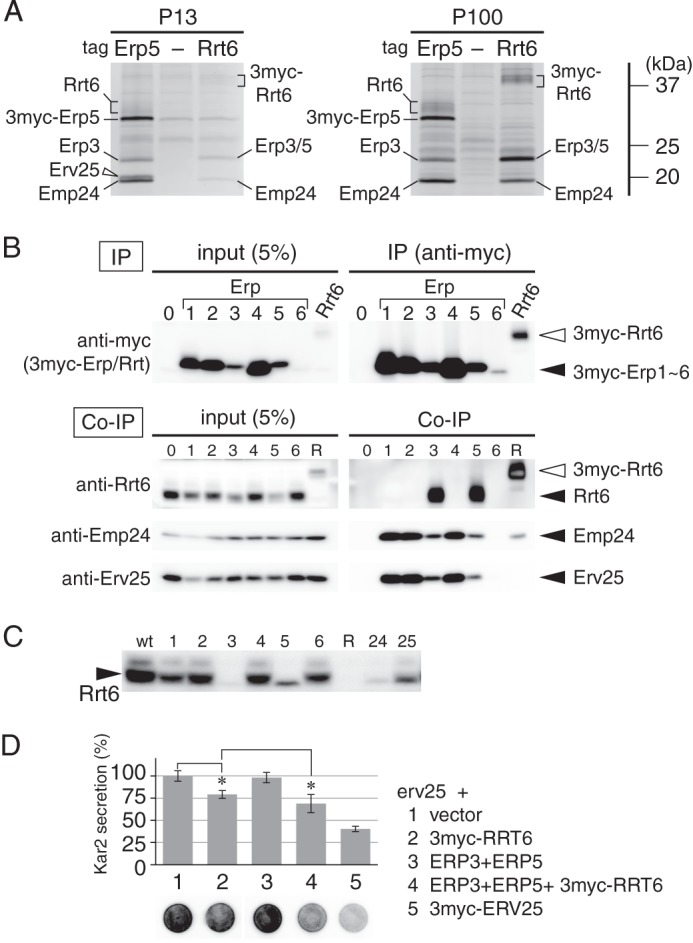
Identification of Rrt6 as a p24-binding protein. A, coprecipitation of Rrt6 with an Erp3/5-containing p24 complex. The ER-enriched P13 (P13) and the Golgi-containing P100 (P100) fractions were isolated from cells expressing either 3myc-Erp5 (Erp5) or 3myc-Rrt6 (Rrt6). From these fractions (P13–3myc-Erp5, P13–3myc-Rrt6, P100–3myc-Erp5, and P100–3myc-Rrt6), the tagged proteins were immunoprecipitated as in Fig. 3A, and proteins in the precipitated fractions were resolved by SDS-PAGE and detected by silver staining. Parental wild-type cells were processed in the same way as control (P13-(−), P100-(−)). Positions of Rrt6 and p24 proteins are shown. B, coprecipitation of Rrt6 with Erp3 and Erp5. Cells expressing one of the 3myc-tagged p24α/γ (lanes 1–6, numbers correspond to the ERP gene numbers), 3myc-tagged Rrt6 (lane R), and control wild-type cells (lane 0) were grown in glycerol medium, and tagged proteins were immunoprecipitated as in Fig. 3A. Proteins in the solubilized fractions (input) and immunoprecipitated fractions (IP and Co-IP) were resolved by SDS-PAGE, and the myc-tagged proteins (anti-myc), Rrt6 (anti-Rrt6), Emp24 (anti-Emp24), and Erv25 (anti-Erv25) were detected by Western blot analysis. Despite that 3myc-Erp6 is functional and has shown to be assembled with Emp24 and Erv25 (Fig. 2A and Table 4), these p24 isoforms were not detected in the 3myc-Erp6 immunoprecipitates in this experiment. We speculate that this is due to the low expression level of 3myc-Erp6. C, steady-state levels of Rrt6 in wild-type (wt), erp1Δ to erp6Δ (Δ1 to Δ6), emp24Δ (Δ24), and erv25Δ (Δ25) cells. Steady-state levels of Rrt6 were determined as in Fig. 1C. D, partial restoration of erv25Δ by overexpression of RRT6. Kar2 secretion was assayed as in Fig. 1A. The graph represents the relative amount of secreted Kar2. Column 1 (spot 1), erv25Δ cells carrying the empty vector. Columns 2–4, erv25Δ cells overexpressing 3myc-RRT6 (column 2), ERP3 and ERP5 (column 3), or 3myc-RRT6, ERP3, and ERP5 (column 4). Column 5, erv25Δ cells expressing 3myc-ERV25. 3myc-Rrt6 used in this experiment has ERV25 promoter and Erv25 signal peptide. Error bars represent S.E. (n = 6). Asterisks indicate p < 0.05.
Fig. 4A shows coprecipitation of Rrt6 by 3myc-Erp5 with other p24 proteins. To optimize the expression of Rrt6, we used glycerol medium and the following experiments. 3myc-Erp5 was mainly found in the ER-enriched P13 fraction in glucose-grown cells (data not shown). In glycerol-grown cells, the protein was equally distributed between P13 and the Golgi-containing P100 (100,000 × g pellet) fractions (Fig. 4A, P13-Erp5 and P100-Erp5). 3myc-Erp5 in P13 fraction coprecipitated Erp3, Emp24, and Erv25 as in glucose-grown cells. 3myc-Erp5 in P100 fraction also coprecipitated Erp3 and Emp24 but not Erv25. Instead, Rrt6, a broadly migrating protein of ∼32 kDa, was found in this fraction (Fig. 4A, P100-Erp5). The interaction between Rrt6 and the p24 proteins was confirmed by reverse immunoprecipitation in which 3myc-Rrt6 was precipitated. 3myc-Rrt6 was mainly found in the P100 fraction and coprecipitated Erp3, Erp5, and Emp24 as expected (Fig. 4A, P100-Rrt6).
The isoform-selective interaction of Rrt6 was further confirmed by another immunoprecipitation experiment. Among the six 3myc-tagged p24α/γ isoforms, only 3myc-Erp3 and 3myc-Erp5 coprecipitated Rrt6 (Fig. 4B, Co-IP lanes 3 and 5). Consistently, the steady-state level of Rrt6 was significantly decreased in erp3Δ and erp5Δ cells (Fig. 4C, 3Δ and 5Δ). Deletion of another binding partner Emp24 (p24β) also destabilized Rrt6 (Fig. 4C, 24Δ). The loss of Erv25 (p24δ) apparently affected the cellular level of Rrt6 as well, but we think this was an indirect effect due to the destabilization of Emp24 in erv25Δ mutant. These results indicate that Rrt6 interacts with Erp3 (p24γ), Erp5 (p24α), and Emp24 (p24β) but not with Erv25 (p24δ) and, therefore, suggest that Rrt6 is a p24δ isoform. Accordingly, we examined genetic interaction between rrt6Δ and erv25Δ, but erv25Δ alone was sufficient to give a full p24Δ phenotype even when cells were grown on glycerol, and we did not detect any additive effect by introduction of rrt6Δ. However, when overexpressed under the ERV25 promoter, 3myc-Rrt6 could partially restore the Kar2 secretion phenotype of erv25Δ cells grown in glucose medium (Fig. 4D, 2). In addition, the complementing activity was facilitated by simultaneous overexpression of Erp3 and Erp5 (Fig. 4D, 4). The functional complementation further suggests that Rrt6 is a novel p24δ isoform.
Rrt6 Has Structural Properties Characteristic of Known p24 Proteins
Pfam database annotates Rrt6 as a member of the p24 family as described earlier; however, Rrt6 is larger (with a calculated mass of ∼34,000) and shows only limited sequence similarity (∼15% identity) to known p24 proteins. We wanted to confirm that Rrt6 is in fact structurally related to the known p24 proteins. Fig. 5A shows schematic representations of Emp24, Erv25, and Rrt6. p24 proteins generally show low amino acid sequence conservation. For example, Emp24 and Erv25 have only 18% sequence identity. However, predicted secondary structures of p24 proteins are very similar (34, 57). The N-terminal domain of the mature p24 protein is predicted to have high β-sheet content (Fig. 5A) (33) and has two invariably conserved cysteine residues that are thought to be important for the structural integrity of this domain. The C-terminal half of the protein is predicted to form a long helical structure extending across the transmembrane domain to the C-terminal cytosolic tail, which is implicated in the complex formation (58–60). The cytosolic tail contains one or more of the known COPI/COPII binding motifs including the di-phenylalanine motif, the di-lysine motif, and the C-terminal valine motif (49, 60, 61). A secondary structure prediction of Rrt6 suggests that these characteristics are well conserved in this protein. In Rrt6, the β-rich domain follows a nonhomologous insertion of ∼50 amino acids at the N terminus. The domain has three cysteine residues, and two of them (Cys-108 and Cys-189) were found to be important for the stability of this protein (data not shown). The C-terminal half of Rrt6 is also predicted to have high α-helix content, and the cytosolic tail contains a functional di-lysine motif as described later.
FIGURE 5.
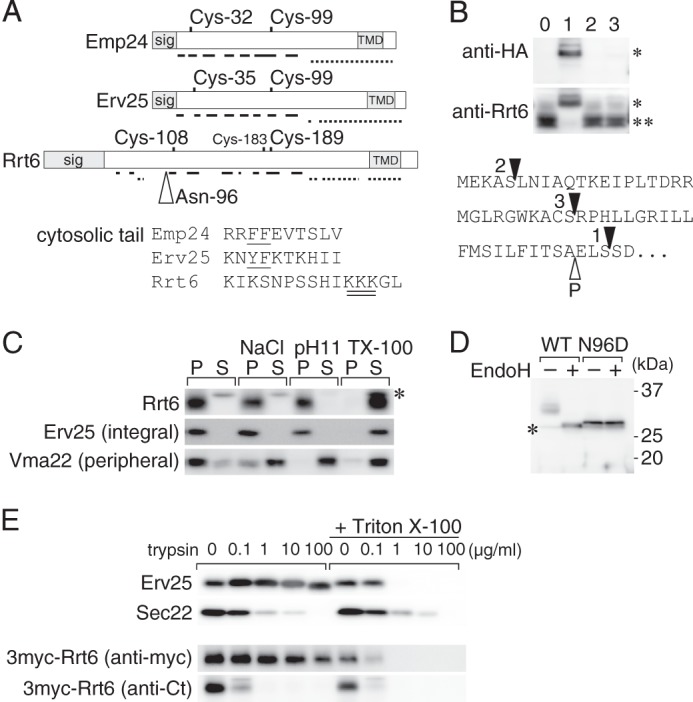
Rrt6 is structurally related to the known p24 proteins. A, schematic representations of Emp24, Erv25, and Rrt6. Location of the signal peptide (sig), the two invariably conserved cysteine residues, and the transmembrane domain (TMD) are shown. Solid and broken lines represent regions predicted to form β-strands and α-helices, respectively (57). Rrt6 has three cysteine residues (Cys-108, -183, and -189) in the luminal domain. Of these, Cys-108 and -189 appeared to correspond to the conserved cysteine pair (see text). The open triangle indicates the N-glycosylation site (Asp-96) of Rrt6. An alignment of the cytosolic tail sequences of the three proteins is also shown. In the alignment, amino acids were expressed in the single-letter codes. The COPI/COPII binding di-phenylalanine motifs of Emp24 and Erv25 are underlined, and the di-lysine motif of Rrt6 is double underlined. B, N-terminal epitope tagging of Rrt6. RRT6 gene was cloned into a low-copy plasmid, tagged with a 3×HA epitope at three different positions, and expressed in rrt6Δ cells. The bottom shows the predicted N-terminal sequence of Rrt6 with the tag insertion sites indicated by closed triangles. The open triangle indicates the predicted signal cleavage site. The tagged (*) and untagged (**) Rrt6 were detected by Western blot analysis using anti-HA or anti-Rrt6 antibodies. Lane 0, untagged Rrt6, Lanes 1–3, 3HA-Rrt6 variants. The lane numbers correspond to those in the bottom sequence. C, extraction of Rrt6 from the membrane. Whole-cell extract was adjusted to either 0.5 m NaCl (NaCl), 0.1 m sodium carbonate, pH 11, or 1% (w/v) Triton X-100 (TX-100), incubated at 4 °C for 30 min, and centrifuged at 100,000 × g for 1 h. The amounts of Rrt6, Erv25, and Vma22 in the pellet (P) and the supernatant (S) fractions were determined by Western blot analysis. Asterisk indicates an unrelated cross-reacting protein. D, N-glycosylation of Rrt6. Crude membrane fractions prepared from rrt6Δ cells expressing plasmid-borne RRT6 or rrt6-N96D were treated with recombinant endoglycosidase H (EndoH) as described under “Experimental Procedures.” Wild-type and the N96D mutant Rrt6 were detected by Western blot analysis. The asterisk indicates an unrelated cross-reacting protein. E, limited trypsin digestion of Rrt6 in the membrane. A crude membrane fraction prepared from the cells expressing 3myc-Rrt6 was treated with trypsin at the concentrations indicated in the figure. The reaction was done at 37 °C for 30 min in the presence or absence of 0.1% (w/v) Triton X-100. Residual amounts of 3myc-Rrt6, Erv25, and Sec22 were determined by Western blot analysis. Anti-Erv25 antibodies recognize the luminal domain of the protein. 3myc-Rrt6 was probed with anti-myc and anti-Rrt6 C-tail (anti-Ct) antibodies.
We next examined the topology of Rrt6. Signal peptide prediction by SignalP algorithm (62) suggests that the most probable cleavage site is located between alanine 50 and glutamic acid 51. Although we have not succeeded in determining the N-terminal sequence of the mature Rrt6, epitope-tagging experiments suggested that the N-terminal hydrophobic region is cleaved as predicted. Introduction of a triplicate hemagglutinin epitope tag (3HA) just behind the putative cleavage site resulted in expression of a recombinant Rrt6 with the expected size and reactivity to both anti-Rrt6 and anti-HA antibodies (Fig. 5B, lane 1). In contrast, the other two Rrt6 recombinants, in which the tag was inserted upstream of the cleavage site, yielded proteins that co-migrated with the non-tagged protein and lost reactivity to anti-HA antibodies, suggesting that the tag-containing N-terminal segments were cleaved off from the mature proteins (Fig. 5B, lanes 2 and 3).
To confirm the integral membrane association of Rrt6, we conducted a protein extraction experiment (Fig. 5C). Total membrane faction was suspended in phosphate-buffered saline (PBS) or PBS containing either 1 m NaCl, 50 mm Na2CO3, pH 11, or 1% (w/v) Triton X-100, and solubilized proteins were separated by centrifugation. Membrane attachment of the peripheral membrane marker Vma22 (63) was sensitive to high salt and alkaline conditions as well as to the detergent treatment. In contrast, Rrt6 and the integral membrane marker Erv25 were extracted from the membrane only when treated with Triton X-100. This result indicates that Rrt6 is an integral membrane protein.
The orientation of the N- and C-terminal domains was investigated by determining the N-glycosylation site of Rrt6 and by limited protease digestion. The calculated mass of mature Rrt6 is ∼29 kDa, whereas the protein migrated as a diffuse band of 32∼35 kDa in SDS-PAGE gels (Figs. 4A and 5D). The apparent size difference and the diffuse migration pattern are predicted to be due to N-glycosylation of Rrt6 because endoglycosidase H converted the protein to migrate as a more focused band of ∼30 kDa (Fig. 5D). Rrt6 has two potential N-glycosylation sites, one (Asn-96) in the N-terminal domain and the other (Asn-301) in the C-terminal tail. We found that replacement of Asn-96 by aspartic acid reproduces the mobility shift by the glycosidase treatment (Fig. 5D, N96D). These results indicate that Rrt6 is glycosylated at Asn-96 (Fig. 5A) and, hence, that the N-terminal domain is located in the lumen.
Limited protease digestion gave a consistent result (Fig. 5E). Total membrane fractions prepared from cells expressing 3myc-Rrt6 were treated with various concentration of trypsin. Two single transmembrane proteins, Erv25 and Sec22, were used as controls. Erv25 is a p24 protein with a larger luminal domain and short cytosolic tail, whereas Sec22 is a tail-anchored protein with the bulk of the protein exposed to the cytosol (64). As shown in Fig. 5E, Erv25 was more resistant to the trypsin digestion than Sec22 but showed an increased sensitivity upon membrane permeabilization by Triton X-100. In contrast, digestion of Sec22 was not stimulated by the addition of the detergent. Their digestion patterns indicate that the membranes were in the right-side-out orientation. 3myc-Rrt6, when probed with anti-myc antibodies, showed a digestion pattern similar to that of Erv25 that indicates that the N terminus of Rrt6 is in the lumen. Susceptibility of Rrt6 C-terminal tail antigen (Fig. 5E, anti-Ct) confirmed that the C terminus is exposed to the cytosol. From these results, we concluded that Rrt6 is a type-I membrane protein like other p24 family members.
Cellular Localization of Rrt6
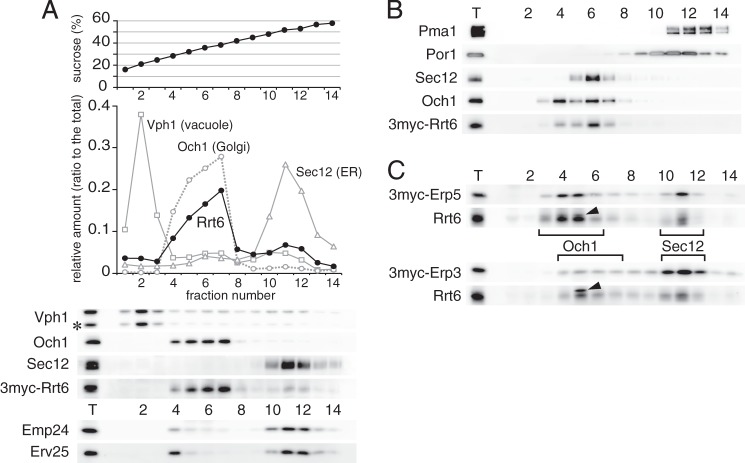
As shown in Fig. 4A, Rrt6 was mainly found in the P100 fraction, whereas the other p24δ isoform Erv25 was mostly in the P13 fraction. We examined detailed cellular localization of Rrt6 by sucrose density gradient fractionation. Logarithmically growing cells expressing 3myc-Rrt6 were spheroplasted and lysed, and membranes were separated through a 20–60% sucrose gradient. Fig. 6A shows the distribution of 3myc-Rrt6 and the organelle markers including Sec12 (ER), Och1 (Golgi), and Vph1 (vacuole) (65–67). A major peak of 3myc-Rrt6 (fractions 4–7) overlapped with the Golgi marker Och1 (Fig. 6A). Another peak of 3myc-Rrt6 (fractions 10–12) coincided with that of the ER-marker Sec12. The heavier Rrt6 peak fractions also contained markers of the plasma membrane (Pma1) and mitochondria (Por1). However, when fractionation was done in the presence of 1 mm EDTA (19), 3myc-Rrt6 and Sec12 in these fractions were shifted to the lighter fractions and well separated from Pma1 and Por1 (Fig. 6B). We concluded that Rrt6 is localized mainly to the Golgi and to a lesser extent to the ER. We next examined the localization of the components of the Rrt6-containing p24 complex. Sucrose density gradient fractionation confirmed that 3myc-Erp3 and 3myc-Erp5 were also partly localized to the Golgi (Fig. 6C, Och1), supporting our interpretation that the Rrt6 complex found in P100 fraction (Fig. 4A) is localized to the Golgi. However, the Golgi localization of 3myc-Erp3 appeared to be weaker than that of 3myc-Erp5 (Fig. 6C). This is unexpected because the two isoforms were expressed at similar levels and were mostly present in the same complexes (Fig. 2A and Table 4). Because the localization of Rrt6 was also shifted more to the ER in 3myc-Erp3 cells, we speculate that the addition of the tag on Erp3 might partially inhibit the traffic of the Rrt6-containing complex to the Golgi. Under the same fractionation conditions, Emp24, the invariant component of the p24 complexes, was mostly found in the ER as reported previously (Fig. 6A) (50, 68). Emp24 is most abundant of the p24 isoforms, whereas Rrt6, Erp3, and Erp5 are minor in abundance (Figs. 2A and 4B). Therefore, although Emp24 was apparently scarce in the Golgi fractions, we consider that the amount of Emp24 in these fractions would actually be comparable to those of Rrt6, Erp3, and Erp5. Erv25, the constitutively expressed p24δ isoform, showed a similar distribution pattern to that of Emp24 but appeared to be more concentrated into the fraction 4, as if the protein has another peak here (Fig. 6A). We speculate that this fraction might contain Erv25 (and possibly also Emp24) present in specialized microdomains of the ER or the Golgi, but further studies are required to reach a conclusion.
FIGURE 6.
Cellular localization of Rrt6. A, sucrose density gradient fractionation of Rrt6. Cells expressing 3myc-Rrt6 were converted to spheroplasts and lysed, and cleared lysate was layered on a 25–60% (w/v) sucrose density gradient. After centrifugation at 200,000 × g for 2.5 h, fractions were collected from the top and assayed for the amounts of 3myc-Rrt6 and organelle markers. The asterisk indicates a major degradation product of Vph1. B, sucrose density gradient in the presence of 1 mm EDTA. Fractionation was done as in A, except that the lysis buffer and the gradient contained 1 mm EDTA. C, sucrose density gradient fractionation of Erp3 and Erp5. Membranes of cells expressing 3myc-Erp3 or 3myc-Erp5 were fractionated as in A. Closed triangles indicate an unrelated cross-reacting protein. Brackets show the distribution of the Golgi marker Och1 and the ER marker Sec12.
The cellular localization of p24 proteins mostly depends on the interaction with COPI/COPII components and other transport factors at the cytosolic tail. We searched Rrt6 C-terminal tail for putative transport signals by site-directed mutagenesis (Fig. 7A) and found that it has a functional di-lysine motif (KKKGL). When the lysine residues were replaced with alanines, the mutant Rrt6 was mislocalized to the vacuole, as indicated by the Pep4 (vacuolar protease)-dependent degradation (Fig. 7A). Several other mutations also destabilized Rrt6, but fractionation experiments suggested that they do not have clear localization defects (Fig. 7, A and B). Mutations of the last two amino acids (GL) modestly increased the amount of Rrt6 in the ER-enriched P13 fraction. So these residues might function as an ER export signal like the C-terminal valine motif in Emp24 (60, 61).
FIGURE 7.
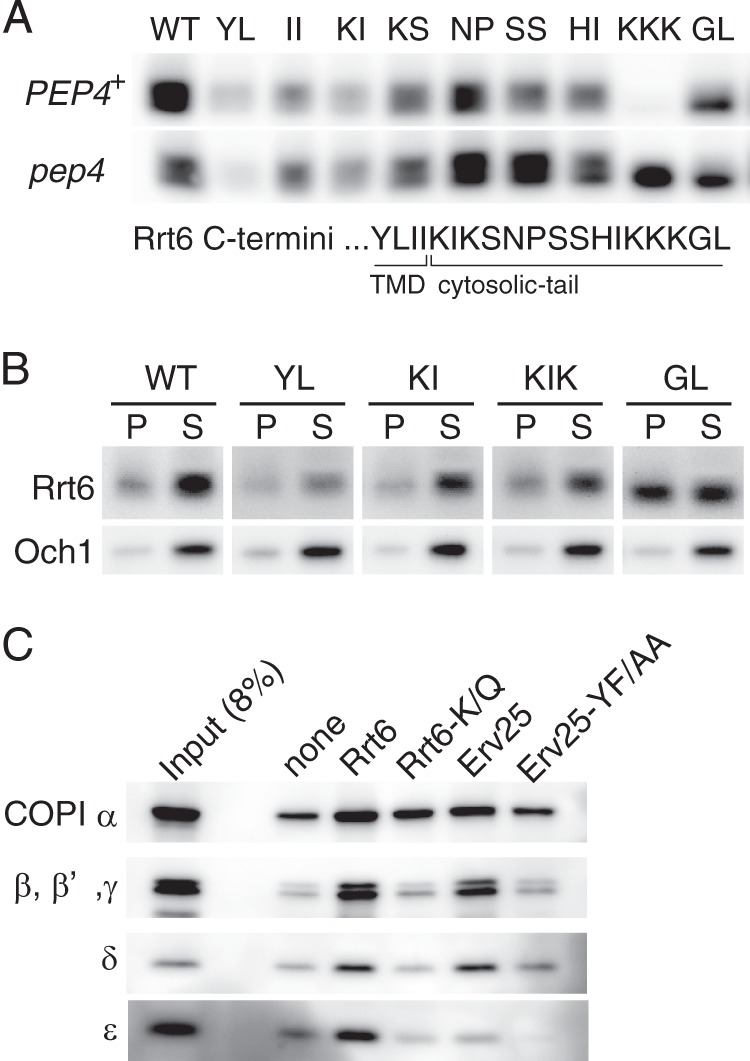
COPI-dependent steady-state localization of Rrt6. A, mutational analysis of Rrt6 cytosolic tail. Wild-type (WT) and mutant 3myc-RRT6 were cloned into a centromere-based low-copy plasmid and introduced into rrt6Δ and rrt6Δ pep4Δ cells. In the mutant constructs, two alanine residues replaced two consecutive residues shown in the figure. The di-lysine motif (−5 to −3) was disrupted by triple alanine substitutions for the lysine residues. TMD, transmembrane domain. B, fractionation of mutant Rrt6 proteins. Cellular localizations of the three mutants (YL, KI, GL) shown in A were determined by cell fractionation. A KIKS-to-AIAS mutant (KIK) was also examined. Cell lysates were centrifuged at 13,000 × g for 10 min to yield P13 (pellet (P)) and S13 (supernatant (S)) fractions. Under the conditions, the ER membrane was sedimented into P13, whereas the Golgi membrane mostly remained in the S13 fraction. C, in vitro binding of synthetic C-tail peptides and COPI. Synthetic peptides corresponding to the cytosolic tails of Erv25 and Rrt6 were conjugated to agarose beads. Rrt6-K/Q is a mutant version of Rrt6 peptide in which the C-terminal three lysine residues (−5 to −3) are substituted by glutamine residues. Erv25-YF/AA has alanine substitutions at the COPI-binding di-phenylalanine (YF) motif. The peptide-conjugated beads (corresponding to 3 nmol of peptide) were incubated with whole-cell extracts (∼1.5 × 108 cells) at 4 °C for 1 h, and COPI binding was assayed by Western blot analysis. Beads without peptide (none) were used as control.
Vacuolar degradation of the di-lysine motif mutant suggests that the COPI-dependent retrograde transport is required for the localization of Rrt6. We examined COPI binding activity of the Rrt6 cytosolic tail by in vitro binding experiments. Wild-type and mutant cytoplasmic tail peptides were synthesized, conjugated to agarose beads, and incubated with whole-cell extracts. Bound proteins were separated by SDS-PAGE, and COPI was detected by Western blotting analysis. As controls, cytoplasmic tail peptides of Erv25 were used. Under the conditions that reproduced the COPI binding to the Erv25 di-phenylalanine motif (YF in Erv25) (49), the Rrt6 tail peptide interacted with COPI in a manner dependent on the di-lysine motif (Fig. 7C). These results indicate that the steady-state localization of Rrt6 is dynamically maintained by a mechanism involving retrograde transport by COPI.
DISCUSSION
S. cerevisiae expresses eight p24 proteins when grown in standard medium containing glucose. They are classified into four subfamilies, p24α (Erp1, -5, and -6), β (Emp24), γ (Erp2, -3, and -4), and δ (Erv25). Marzioch et al. (35) identified a p24 complex consisting of Emp24, Erv25, Erp1, and Erp2. This complex accounts for a major part of the cellular p24 activity; however, we thought that a more comprehensive description of the p24 complexes would help to interpret previous data and to design future studies. We examined genetic and physical interactions of p24α and -γ isoforms, which we thought specify the structural variation of p24 complexes, and identified several p24 complexes with redundant functions. We also identified Rrt6 as a novel p24δ isoform. Rrt6 is unique in that it is the only S. cerevisiae p24 isoform that is inducibly expressed and receives N-linked glycosylation, although its physiological role remains unknown.
S. cerevisiae p24 Proteins Function in αβγδ Heteromeric Complexes
A major conclusion of this study is that p24α and -γ are essential for the p24 activity. Previous studies demonstrated that p24β (Emp24) and p24δ (Erv25) are the essential components of the S. cerevisiae p24 complex (35). We show that the triple mutant of either p24α or p24γ is phenotypically indistinguishable from the p24β/δ mutants (Fig. 1). This implies that active p24 complexes contain at least one each of p24α and -γ isoform as well as the invariant p24α and -γ. On the other hand, the results of coprecipitation experiments suggest that the majority of p24 complexes contain only one each of p24α and -γ. Specifically, each p24α/γ isoform coprecipitated only substoichiometric amounts of the other two isoforms of its own subfamily, whereas those of the other subfamilies were efficiently coprecipitated (Table 4). A simple interpretation of these results would be that active p24 complexes have a composition of αβγδ, although the copy number of each subunit remains to be determined.
The interaction between p24α and -γ was selective, yielding five major α/γ interaction pairs (Fig. 3). Therefore, S. cerevisiae would have 5 major p24 complexes consisting of the constitutive p24β/δ (Emp24/Erv25) and variable p24α/γ (Erp1/Erp2, Erp1/Erp4, Erp6/Erp2, Erp6/Erp4, or Erp5/Erp3). We currently prefer a model in which all these and other putative minor αβγδ complexes are active and functionally redundant. This is an oversimplification but is still well consistent with the previously reported genetic interactions (35, 53). The apparent compatibility between Erp1 and Erp6 (or between Erp2 and Erp4) in the complex formation gives a reasonable account for the high-copy suppression of erp1Δ by ERP6 (erp2Δ by ERP4). The selective interaction between Erp5 and Erp3 can explain why only simultaneous, but not individual, overexpression of the two isoforms could restore erp1Δ, erp2Δ, or both (Fig. 3B). The model can also explain the synergetic interaction between erp1Δ and erp2Δ (53).
Although the current data are not strictly quantitative, relative abundance of the p24α/γ isoforms (Fig. 2A) appeared to mirror the severity of their mutant phenotype. Erp1 was found to be the dominant p24α isoform, and its deletion gave the most severe phenotype among the p24α/γ mutations. Erp2 and Erp4 shared the majority of p24γ isoforms, and deletion of either alone resulted in weaker but still detectable phenotype, whereas a single deletion of the other less abundant p24α/γ isoforms showed almost no visible defects.
p24 Complexes Other Than αβγδ
Phylogenetic studies indicate that p24 proteins have evolved differently in different species (34, 69, 70). This appears to be reflected in the diversity in the regulation of complex formation. Marzioch et al. (35) reported that the majority of the yeast p24 proteins are assembled into heterodimers and tetramers. Early studies suggested that the mammalian p24 proteins also form heteromeric complexes, most likely tetramers. However, Jenne et al. (39) reported that p24 proteins exist mostly in monomers and dimers in HeLa cells. Although the conclusion is still somewhat controversial, available data suggest that p24 proteins in mammals are in dynamic equilibrium among monomers, dimers, and tetramers (27, 36, 37). Martens and co-workers (71–73) proposed another model in which p24 proteins in Xenopus melanotrope cells can function as monomers, and each has an individual role in prohormone processing.
We consider that in yeast the major αβγδ complexes constitute most of the active p24 complexes as described above, but other type of complexes could have significant functional contributions in certain conditions. Our results are generally well consistent with those of the earlier studies, but there is a major discrepancy in the phenotype of double and triple mutants of p24α and p24γ. Marzioch et al. (35) reported that the triple mutant of either p24α (p24αΔ) or p24γ (p24γΔ) shows the same phenotype as erp1Δ or erp2Δ, respectively, and the second and third mutations do not have an additive effect. They found that p24α and -γ could form a dimer even in the absence of p24α, although at a much lower level than the αβγδ complex(es) in wild-type cells. Based on these findings, they proposed that the p24β/δ dimer remains active and are responsible for the residual p24 activity in the mutants devoid of p24α or p24γ. In contrast, p24α and -γ were less stable in our p24αΔ and p24δΔ strains (Fig. 1C). We speculate that the strain background and/or culture conditions modulate the stability of p24β/δ or any other partially active subcomplexes, resulting in the different phenotypes observed in the two studies. These subcomplexes, although their molecular compositions and relative abundance in wild-type cells are unknown yet, could represent putative intermediates of biosynthetic assembly or regulated disassembly of the αβγδ complexes. Future studies will be directed toward the dynamic aspects of the αβγδ complexes and its potential coupling with the ER-Golgi transport.
Functional Diversity of p24 Complexes
We identified a novel p24δ isoform, Rrt6. rrt6Δ showed no apparent growth defects, and the physiological roles of this protein remain unknown. It is also unknown how the loss of Rrt6 confers yeast cells the rrt (regulator of ribosomal DNA transcription) phenotype. However, we think that the protein would serve as a good model to study functional diversity of p24 complexes. In Xenopus melanotrope cells, six p24 isoforms are expressed, and four of them (p24α3, -β1, -γ3, and -δ2) are strongly up-regulated during background color adaptation, which is associated with increased pro-opiomelanocortin synthesis and secretion (74). Strating et al. (34) demonstrated that 2 of the 11 p24 proteins in mouse show tissue-specific expression, and both are implicated in the insulin secretion (31). These studies suggest that the isoform composition determines, at least in part, the cargo specificity of the p24 complexes. However, it is still elusive whether a single cell actually uses different p24 complexes for different cargo proteins. The Rrt6-containing p24 complex is unique in that it was specifically expressed under respiratory conditions, whereas the other complexes were constitutively expressed. The Rrt6 complex was mainly localized to the Golgi, whereas the others were mostly to the ER (Fig. 6). We, therefore, speculate that the Rrt6 complex may have its specific cargo proteins. If so, it would be interesting to compare the cargo specificity of Rrt6- and Erv25-containing complexes that otherwise have the same isoform composition. In this context it is intriguing that, in yeast, Rrt6 is the only p24 protein that receives N-glycosylation (Fig. 5D), which could modulate cargo specificity.
We also identified several constitutively expressed p24 complexes (Fig. 3). Although our results indicate that they have redundant functions, this does not exclude the possibility that they also have their specific cargo proteins. Li et al. (75) identified erp2Δ by screening for resistance to exogenously expressed ricin A-chain. When the toxin is expressed in yeast and targeted to the ER lumen, it is translocated back to the cytosol and exhibits cytotoxicity. The authors proposed that the toxin translocation is mediated by the ER-associated quality control mechanism that depends on the ER-Golgi transport of the target proteins and that Erp2 is involved in the toxin transport (76, 77).
Unexpectedly, erp2Δ is the only p24 mutant that shows the toxin-resistant phenotype. The major Erp2-containing complex also contains Erp1, Emp24, and Erv25, but the deletion of any one of the latter three does not show the toxin resistance. The authors explain their lack of the resistant phenotype as follows; in yeast, p24Δ (emp24Δ) was originally identified as an extragenic suppressor of sec13 mutation (51). Based on this and other observations (53), p24 proteins have been proposed to have an additional function as negative regulators of the COPII-dependent protein export from the ER. According to this model, the loss of p24 activity would increase the bulk flow of protein traffic out of the ER. Therefore, in mutants that show a stronger p24Δ phenotype than erp2Δ, such as erp1Δ, emp24Δ, and erv25Δ, increased bulk flow transport could compensate for the reduced toxin transport by the loss of Erp2-containing p24 complex. Interestingly, Shi et al. (78) identified erp2Δ, also as a sole p24Δ mutant, among mutants defective in cell surface expression of the Can1 arginine permease. Immunoprecipitation of p24 complexes and mass spectrometric analysis, as shown in Fig. 3, would be applicable to examine whether Erp2 does specifically interact with these putative cargo proteins and to screen for novel cargo candidates.
Cellular Localization of Rrt6
We show that the Rrt6-containing p24 complex is mainly localized to the Golgi, and when Erv25 replaces Rrt6, the resulting complex stayed mainly in the ER (Fig. 4A). This implies that the p24δ can determine the cellular localization of the p24 complexes. Then how is the steady-state localization of Rrt6 regulated? We found that the COPI binding di-lysine motif in the C terminus is essential for the ER-Golgi localization of Rrt6, which indicates that the steady-state localization of this protein is maintained by the COPI-dependent retrograde transport. Mutation of the two C-terminal residues inhibited the Golgi localization of Rrt6. These residues are most likely involved in the anterograde transport of this protein; however, there would be additional molecular mechanisms ensuring the Golgi localization of Rrt6. We are preparing for more comprehensive mutational studies to answer the question.
Acknowledgments
We thank Rainer Duden (Univ. Lübeck) for providing anti-coatomer antibodies. We are grateful to the Support Unit for Bio-material Analysis and the Support Unit for Animal Resources Development in RIKEN BSI Research Resources Center for help with amino acid analysis (T. Morishita), mass spectrometry (K. Otsuki, A. Abe, and M. Usui), peptide synthesis (R. Itoh, J. Ishikawa, and S. Kurata), and antibody production (M. Kumai and S. Kashiwagi). We thank Y. Sugisawa, Y. Zenke, R. Nakazawa, and Y. Ichikawa for technical assistance and K. Kurokawa and Y. Suda for helpful discussions.
This work was supported by a grant-in-aid for Specially Promoted Research, grants-in-aid for Scientific Research, and the Targeted Proteins Research Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Bioarchitect, the Extreme Photonics, and the Cellular Systems Biology Projects of RIKEN.
- ER
- endoplasmic reticulum
- COPI
- coat protein complex I
- COPII
- coat protein complex II.
REFERENCES
- 1. Brandizzi F., Barlowe C. (2013) Organization of the ER-Golgi interface for membrane traffic control. Nat. Rev. Mol. Cell Biol. 14, 382–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sato K., Nakano A. (2007) Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 581, 2076–2082 [DOI] [PubMed] [Google Scholar]
- 3. Popoff V., Adolf F., Brügger B., Wieland F. (2011) COPI budding within the Golgi stack. Cold Spring Harb. Perspect. Biol. 3, a005231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Glick B. S., Nakano A. (2009) Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 25, 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Losev E., Reinke C. A., Jellen J., Strongin D. E., Bevis B. J., Glick B. S. (2006) Golgi maturation visualized in living yeast. Nature 441, 1002–1006 [DOI] [PubMed] [Google Scholar]
- 6. Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. (2006) Live imaging of yeast Golgi cisternal maturation. Nature 441, 1007–1010 [DOI] [PubMed] [Google Scholar]
- 7. Rizzo R., Parashuraman S., Mirabelli P., Puri C., Lucocq J., Luini A. (2013) The dynamics of engineered resident proteins in the mammalian Golgi complex relies on cisternal maturation. J. Cell Biol. 201, 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lavieu G., Zheng H., Rothman J. E. (2013) Stapled Golgi cisternae remain in place as cargo passes through the stack. eLife 2, e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang J.-S., Valente C., Pollshchuk R. S., Turacchio G., Layre E., Moody D. B., Leslie C. C., Gelb M. H., Brown W. J., Corda D., Luini A., Hsu V. W. (2011) COPI acts in both vesicular and tubular transport. Nat. Cell Biol. 13, 996–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. (2003) Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 [DOI] [PubMed] [Google Scholar]
- 11. Michelsen K., Yuan H., Schwappach B. (2005) Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6, 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato K., Nakano A. (2005) Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat. Struct. Mol. Biol. 12, 167–174 [DOI] [PubMed] [Google Scholar]
- 13. Ma W., Goldberg J. (2013) Rules for the recognition of dilysine retrieval motifs by coatomer. EMBO J. 32, 926–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otte S., Barlowe C. (2004) Sorting signals can direct receptor-mediated export of soluble proteins into COPII vesicles. Nat. Cell Biol. 6, 1189–1194 [DOI] [PubMed] [Google Scholar]
- 15. Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. (1990) ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 16. Lewis M. J., Pelham H. R. (1990) A human homologue of the yeast HDEL receptor. Nature 348, 162–163 [DOI] [PubMed] [Google Scholar]
- 17. Zheng C., Page R. C., Das V., Nix J. C., Wigren E., Misra S., Zhang B. (2013) Structural characterization of carbohydrate binding by LMAN1 protein provides new insight into the endoplasmic reticulum export of factors V (FV) and VIII (FVIII). J. Biol. Chem. 288, 20499–20509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sato K., Nakano A. (2002) Emp47p and its close homolog Emp46p have a tyrosine-containing endoplasmic reticulum exit signal and function in glycoprotein secretion in Saccharomyces cerevisiae. Mol. Biol. Cell 13, 2518–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Powers J., Barlowe C. (1998) Transport of Axl2p depends on Erv14p, an ER-vesicle protein related to the Drosophila cornichon gene product. J. Cell Biol. 142, 1209–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sato K., Sato M., Nakano A. (2003) Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol. Biol. Cell 14, 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bue C. A., Barlowe C. (2009) Molecular dissection of Erv26p identifies separable cargo binding and coat protein sorting activities. J. Biol. Chem. 284, 24049–24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noda Y., Yoda K. (2010) Svp26 facilitates endoplasmic reticulum to Golgi transport of a set of mannosyltransferases in Saccharomyces cerevisiae. J. Biol. Chem. 285, 15420–15429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herzig Y., Sharpe H. J., Elbaz Y., Munro S., Schuldiner M. (2012) A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol. 10, e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Muñiz M., Nuoffer C., Hauri H. P., Riezman H. (2000) The Emp24 complex recruits a specific cargo molecule into endoplasmic reticulum-derived vesicles. J. Cell Biol. 148, 925–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castillon G. A., Aguilera-Romero A., Manzano-Lopez J., Epstein S., Kajiwara K., Funato K., Watanabe R., Riezman H., Muñiz M. (2011) The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell 22, 2924–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bonnon C., Wendeler M. W., Paccaud J.-P., Hauri H.-P. (2010) Selective export of human GPI-anchored proteins from the endoplasmic reticulum. J. Cell Sci. 123, 1705–1715 [DOI] [PubMed] [Google Scholar]
- 27. Fujita M., Watanabe R., Jaensch N., Romanova-Michaelides M., Satoh T., Kato M., Riezman H., Yamaguchi Y., Maeda Y., Kinoshita T. (2011) Sorting of GPI-anchored proteins into ER exit sites by p24 proteins is dependent on remodeled GPI. J. Cell Biol. 194, 61–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buechling T., Chaudhary V., Spirohn K., Weiss M., Boutros M. (2011) p24 proteins are required for secretion of Wnt ligands. EMBO Rep. 12, 1265–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Port F., Hausmann G., Basler K. (2011) A genome-wide RNA interference screen uncovers two p24 proteins as regulators of Wingless secretion. EMBO Rep. 12, 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bouw G., Van Huizen R., Jansen E. J., Martens G. J. (2004) A cell-specific transgenic approach in Xenopus reveals the importance of a functional p24 system for a secretory cell. Mol. Biol. Cell 15, 1244–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang L., Volchuk A. (2010) p24 family type 1 transmembrane proteins are required for insulin biosynthesis and secretion in pancreatic β-cells. FEBS Lett. 584, 2298–2304 [DOI] [PubMed] [Google Scholar]
- 32. Luo W., Wang Y., Reiser G. (2011) Proteinase-activated receptors, nucleotide P2Y receptors, and μ-opioid receptor-1B are under the control of the type I transmembrane proteins p23 and p24A in post-Golgi trafficking. J. Neurochem. 117, 71–81 [DOI] [PubMed] [Google Scholar]
- 33. Anantharaman V., Aravind L. (2002) The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 3, research0023.1–research0023.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strating J. R., van Bakel N. H., Leunissen J. A., Martens G. J. (2009) A comprehensive overview of the vertebrate p24 family. Identification of a novel tissue-specifically expressed member. Mol. Biol. Evol. 26, 1707–1714 [DOI] [PubMed] [Google Scholar]
- 35. Marzioch M., Henthorn D. C., Herrmann J. M., Wilson R., Thomas D. Y., Bergeron J. J., Solari R. C., Rowley A. (1999) Erp1p and Erp2p, partners for Emp24p and Erv25p in a yeast p24 complex. Mol. Biol. Cell 10, 1923–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dominguez M., Dejgaard K., Füllekrug J., Dahan S., Fazel A., Paccaud J. P., Thomas D. Y., Bergeron J. J., Nilsson T. (1998) gp25L/emp24/p24 protein family members of the cis-Golgi network bind both COP I and II coatomer. J. Cell Biol. 140, 751–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Füllekrug J., Suganuma T., Tang B. L., Hong W., Storrie B., Nilsson T. (1999) Localization and recycling of gp27 (hp24γ3). Complex formation with other p24 family members. Mol. Biol. Cell 10, 1939–1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Denzel A., Otto F., Girod A., Pepperkok R., Watson R., Rosewell I., Bergeron J. J., Solari R. C., Owen M. J. (2000) The p24 family member p23 is required for early embryonic development. Curr. Biol. 10, 55–58 [DOI] [PubMed] [Google Scholar]
- 39. Jenne N., Frey K., Brugger B., Wieland F. T. (2002) Oligomeric state and stoichiometry of p24 proteins in the early secretory pathway. J. Biol. Chem. 277, 46504–46511 [DOI] [PubMed] [Google Scholar]
- 40. Montesinos J. C., Sturm S., Langhans M., Hillmer S., Marcote M. J., Robinson D. G., Aniento F. (2012) Coupled transport of Arabidopsis p24 proteins at the ER-Golgi interface. J. Exp. Bot. 63, 4243–4261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brachmann C. B., Davies A., Cost G. J., Caputo E., Li J., Hieter P., Boeke J. D. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C. A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 [DOI] [PubMed] [Google Scholar]
- 42. Rothstein R. (1991) Targeting, disruption, replacement, and allele rescue. Integrative DNA transformation in yeast. Methods Enzymol. 194, 281–301 [DOI] [PubMed] [Google Scholar]
- 43. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes. New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 44. Burke D., Dawson D. S., Stearns T., Laboratory C. S. H. (2000) Methods in Yeast Genetics 2000: A Cold Spring Harbor Laboratory Course Manual, pp. 171–181, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Higuchi R., Krummel B., Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments. Study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sato K., Sato M., Nakano A. (2001) Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 152, 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nishikawa S., Nakano A. (1993) Identification of a gene required for membrane protein retention in the early secretory pathway. Proc. Natl. Acad. Sci. U.S.A. 90, 8179–8183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Belden W. J., Barlowe C. (2001) Distinct roles for the cytoplasmic tail sequences of Emp24p and Erv25p in transport between the endoplasmic reticulum and Golgi complex. J. Biol. Chem. 276, 43040–43048 [DOI] [PubMed] [Google Scholar]
- 50. Schimmöller F., Singer-Krüger B., Schröder S., Krüger U., Barlowe C., Riezman H. (1995) The absence of Emp24p, a component of ER-derived COPII-coated vesicles, causes a defect in transport of selected proteins to the Golgi. EMBO J. 14, 1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Elrod-Erickson M. J., Kaiser C. A. (1996) Genes that control the fidelity of endoplasmic reticulum to Golgi transport identified as suppressors of vesicle budding mutations. Mol. Biol. Cell 7, 1043–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Belden W. J., Barlowe C. (2001) Deletion of yeast p24 genes activates the unfolded protein response. Mol. Biol. Cell 12, 957–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Čopič A., Latham C. F., Horlbeck M. A., D'Arcangelo J. G., Miller E. A. (2012) ER cargo properties specify a requirement for COPII coat rigidity mediated by Sec13p. Science 335, 1359–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hontz R. D., Niederer R. O., Johnson J. M., Smith J. S. (2009) Genetic identification of factors that modulate ribosomal DNA transcription in Saccharomyces cerevisiae. Genetics 182, 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Roberts G. G., 3rd, Hudson A. P. (2009) Rsf1p is required for an efficient metabolic shift from fermentative to glycerol-based respiratory growth in S. cerevisiae. Yeast 26, 95–110 [DOI] [PubMed] [Google Scholar]
- 57. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 58. Ciufo L. F., Boyd A. (2000) Identification of a lumenal sequence specifying the assembly of Emp24p into p24 complexes in the yeast secretory pathway. J. Biol. Chem. 275, 8382–8388 [DOI] [PubMed] [Google Scholar]
- 59. Weidler M., Reinhard C., Friedrich G., Wieland F. T., Rösch P. (2000) Structure of the cytoplasmic domain of p23 in solution. Implications for the formation of COPI vesicles. Biochem. Biophys. Res. Commun. 271, 401–408 [DOI] [PubMed] [Google Scholar]
- 60. Barr F. A., Preisinger C., Kopajtich R., Körner R. (2001) Golgi matrix proteins interact with p24 cargo receptors and aid their efficient retention in the Golgi apparatus. J. Cell Biol. 155, 885–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nakamura N., Yamazaki S., Sato K., Nakano A., Sakaguchi M., Mihara K. (1998) Identification of potential regulatory elements for the transport of Emp24p. Mol. Biol. Cell 9, 3493–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0. Discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 63. Graham L. A., Hill K. J., Stevens T. H. (1998) Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 142, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sacher M., Stone S., Ferro-Novick S. (1997) The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J. Biol. Chem. 272, 17134–17138 [DOI] [PubMed] [Google Scholar]
- 65. Nakano A., Brada D., Schekman R. (1988) A membrane glycoprotein, Sec12p, required for protein transport from the endoplasmic reticulum to the Golgi apparatus in yeast. J. Cell Biol. 107, 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Harris S. L., Waters M. G. (1996) Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J. Cell Biol. 132, 985–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Finnigan G. C., Hanson-Smith V., Houser B. D., Park H. J., Stevens T. H. (2011) The reconstructed ancestral subunit a functions as both V-ATPase isoforms Vph1p and Stv1p in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 3176–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Belden W. J., Barlowe C. (1996) Erv25p, a component of COPII-coated vesicles, forms a complex with Emp24p that is required for efficient endoplasmic reticulum to Golgi transport. J. Biol. Chem. 271, 26939–26946 [DOI] [PubMed] [Google Scholar]
- 69. Chen S., Zhang Y. E., Long M. (2010) New genes in Drosophila quickly become essential. Science 330, 1682–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kellis M., Birren B. W., Lander E. S. (2004) Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428, 617–624 [DOI] [PubMed] [Google Scholar]
- 71. Strating J. R., Bouw G., Hafmans T. G., Martens G. J. (2007) Disparate effects of p24α and p24δ on secretory protein transport and processing. PLoS ONE 2, e704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Strating J. R., Hafmans T. G., Martens G. J. (2009) Functional diversity among p24 subfamily members. Biol. Cell 101, 207–219 [DOI] [PubMed] [Google Scholar]
- 73. Strating J. R., Martens G. J. (2009) The p24 family and selective transport processes at the ER-Golgi interface. Biol. Cell 101, 495–509 [DOI] [PubMed] [Google Scholar]
- 74. Rötter J., Kuiper R. P., Bouw G., Martens G. J. (2002) Cell-type-specific and selectively induced expression of members of the p24 family of putative cargo receptors. J. Cell Sci. 115, 1049–1058 [DOI] [PubMed] [Google Scholar]
- 75. Li S., Spooner R. A., Allen S. C., Guise C. P., Ladds G., Schnöder T., Schmitt M. J., Lord J. M., Roberts L. M. (2010) Folding-competent and folding-defective forms of ricin A chain have different fates after retrotranslocation from the endoplasmic reticulum. Mol. Biol. Cell 21, 2543–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Haynes C. M., Caldwell S., Cooper A. A. (2002) An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158, 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vashist S., Ng D. T. (2004) Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J. Cell Biol. 165, 41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shi Y., Stefan C. J., Rue S. M., Teis D., Emr S. D. (2011) Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol. Biol. Cell 22, 4093–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]