Abstract
Poly(ADP-ribose) polymerase 1 (PARP-1) activity is detected in both neuronal and nonneuronal cells in the CNS, and excessive PARP-1 activity is known to be detrimental to tissue because of the cellular energy loss. Accordingly, PARP-1-deficient (PARP-1-/-) mice have been shown to be resistant to cerebral ischemia and several forms of inflammation. Recently, PARP-1 in glial cells has been shown to mediate the expression of proinflammatory genes in response to inflammatory stimuli by, in part, enhancing cognate DNA-binding capacities of transcription factors such as NF-κB and activator protein 1. Here, we demonstrate an additional mechanism whereby a significant reduction of proinflammatory gene expression such as IL-1β, tumor necrosis factor α, and inducible nitricoxide synthase in PARP-1-/- glial cells is linked to defective inflammatory stimuli-induced p38MAPK-mediated phosphorylation of ATF-2 and cAMP-response element-binding protein and phosphorylation of NF-κB p65. Importantly, an inflammatory stimuli-induced p38MAPK activation is impaired in PARP-1-/- glial cells in a signaling pathway- and cell/tissue type-specific manner. These findings indicate that PARP-1 is an essential host factor among factors that actively mediate excessive production of proinflammatory molecules in glial cells, which may in turn contribute to the initiation of neuronal injuries.
Poly(ADP-ribose) polymerase 1 (PARP-1, EC 2.4.2.30) is a multifunctional nuclear enzyme (1). DNA strand breaks activate PARP-1, which adds branched chains of up to 200 ADP-ribose units covalently to various nuclear proteins such as histone proteins and especially PARP-1 itself. PARP-1 is involved in chromatin remodeling, DNA repair, replication, transcription, and the maintenance of genomic stability by, in part, poly(ADP-ribosyl)ation (1). With moderate amounts of DNA damage, PARP-1 is thought to participate in the DNA repair process (2, 3). However, with excessive activation of abundant PARP-1, its substrate NAD+ is depleted, and in efforts to resynthesize NAD+, ATP is also depleted such that cells may die from energy loss (4-7). A role for excessive PARP-1 activation in cell death is indicated by the protection against cell death observed after treatment with PARP inhibitors (5, 8) and the pronounced protection against neuronal ischemia (9, 10), myocardial ischemia (11), acute lung inflammation (12), acute septic peritonitis (13), zymogen-induced multiple organ failure (14), and diabetic pancreatic damage (8, 15, 16) in PARP-1-deficient (PARP-1-/-) mice.
Glial cells are nonneuronal cells, and they have been considered to be merely supportive elements in the CNS. However, activated glial cells, mainly composed of microglia and astrocytes, can mediate neuronal inflammation and toxicity by releasing substances such as excitotoxins, reactive oxygen/nitrogen species, cytokines, and chemokines in response to inflammation, infection, and injury (17, 18). Activation of microglia and the expression of inflammatory mediators are rapidly induced by means of phosphorylation of p38MAPK (19). The p38MAPK signal-transduction pathway seems to mediate microglial activation and neuronal injury, occurring in most chronic neurodegenerative diseases such as Alzheimer's disease, multiple sclerosis, and HIV-associated dementia (17, 19, 20). Microglia from amyloid precursor protein transgenic mice also show p38MAPK activation (21). In addition, activation of p38MAPK is detected in acute brain injuries such as stroke and brain trauma (22). Permanent occlusion of the middle cerebral artery and gerbil transient forebrain ischemia are shown to induce p38MAPK activation in microglia cells located in periinfarct areas and adjacent to dying CA1 neurons, respectfully (23). Chemokine and cytokine promote p38MAPK activity, which in turn phosphorylates and activates transcription factors such as ATF-2 and MEF-2C (24, 25).
Stimuli-induced phosphorylation of transcription factors is essential for their transcriptional competence. Activated p38MAPK rapidly translocates from the cytoplasm to the nucleus and phosphorylates its substrates such as transcription factors, i.e., cAMP-response element-binding protein (CREB), myocyte enhancer factor 2C, and activating transcription factor 2 (ATF-2), or other kinases, such as mitogen- and stress-activated protein kinase (17, 24-26). In addition, activation of NF-κB involves posttranslational modification of NF-κB/Rel proteins, particularly p65. Several studies have shown that IL-1β and tumor necrosis factor α (TNF-α) induce phosphorylation and activation of the p65 NF-κB subunit by pathways that are distinct from those that lead to IκB degradation and NF-κB nuclear translocation (27, 28). Together, a signal-induced phosphorylation of transcription factors such as ATF-2, CREB, and NF-κB p65 is essential for their transcriptional competence, which in turn provides an additional mechanism for the tightly controlled gene expression in response to diverse signals.
In our previous reports, PARP-1-/- glial cells showed significantly reduced proinflammatory genes, including IL-1β, IL-6, TNF-α, and inducible NO synthase (iNOS) with lack of cognate DNA-binding capacities of transcription factors, such as NF-κB and activator protein 1, in response to lipopolysaccharide (LPS) (29). The expression of proinflammatory genes requires coordinated activation of numerous transcription factors. Therefore, the present study investigates whether reduced activation of transcription factors leads to the lack of proinflammatory gene expression in PARP-1-/- glial cells in response to inflammatory stimuli. The present results show that the stimuli-induced phosphorylation of NF-κB p65, ATF-2, and CREB is absent in PARP-1-/- glial cells, which in turn result in a significant reduction of proinflammatory gene expression in PARP-1-/- glial cells. Importantly, we found a defective nuclear signaling of p38MAPK in PARP-1-/- glial cells in response to inflammatory stimuli.
Materials and Methods
Reagents and Materials. Reagents and materials were as described (29).
Isolation and Culture of Mouse Glial Cells and Embryonic Fibroblasts. The PARP-1-/- mice (30) were provided kindly by Z. Q. Wang (International Agency for Research on Cancer, Lyon, France). Primary cultures of glial cells were generated from 1- to 3-day-old mouse neopallium from wild type (PARP-1+/+, C57BL/6 × 129/Sv) and PARP-1-/- (C57BL/6 × 129/Sv), as described in ref. 29 and elsewhere (31). The resulting culture was determined by staining with glial fibrillary acidic protein for astrocytes and with isolectin B4 from Griffonia simplicifolia for microglia.
PARP Activity Assay, ELISA, and Electrophoretic Mobility-Shift Assay. These assays were performed as described (29).
Metabolic Labeling and Immunoprecipitation. For 32P metabolic labeling, cells were grown in phosphate-free medium with 2% serum for 3 h. [32P]H3PO4 were added 1 h before harvest. Nuclear extracts were prepared by the methods described in electrophoretic mobility-shift assay, and subsequent nuclear extracts in cold radioimmunoprecipitation assay buffer were subjected to immunoprecipitation with antibodies to NF-κB p65 (Santa Cruz Biotechnology). The precipitated proteins were separated on 4-12% SDS/PAGE gel, transferred to nitrocellulose membranes (NEN), and visualized by autoradiography.
Results
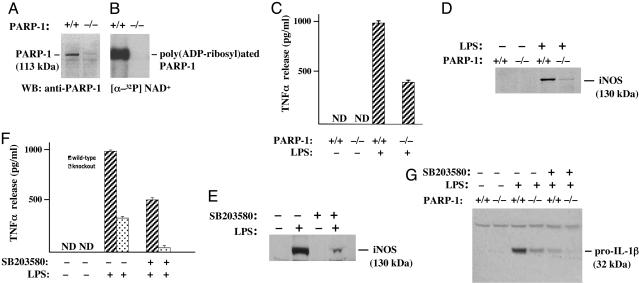
Proinflammatory Genes Regulated by PARP-1 in Glial Cells. In primary glial cultures from wild-type and PARP-1-/- mice, the expression of PARP-1 protein was abolished in PARP-1-/- preparations (Fig. 1A). PARP-1 activity, measured as poly(ADP-ribosyl)ated PARP-1, was detected only in wild-type glial cells. As expected with the abolished expression of PARP-1 protein, PARP-1-/- glial cells showed no PARP-1 activity (Fig. 1B). Immunocytochemical characterization of the glial cell cultures in this study consistently revealed 70-80% astrocytes, and the rest was mainly microglia, as determined by using specific markers glial fibrillary acidic protein and G. simplicifolia isolectin-B4, respectively. The number of glial fibrillary acidic protein-stained astrocytes and G. simplicifolia isolectin-B4-stained microglia was similar in PARP-1-/- and wild-type glial culture.
Fig. 1.
TNF-α release and pro-IL-1β and iNOS expression are reduced significantly in PARP-1-/- primary glial cells. (A) Expression of PARP-1 in wild-type and PARP-1-/- glial cells. The PARP-1-/- lane confirmed the absence of PARP-1 protein. (B) PARP-1 catalytic activity in wild-type and PARP-1-/- glial cells. Automodification of PARP-1 by [32P] poly(ADP-ribose) was abolished in PARP-1-/- glial cells. Glial cells were labeled with [32P] NAD+ for 20 min at 4°C. Subsequently, total protein (70 μg) from labeled glial cells was separated on 4-12% SDS/PAGE gel and visualized by autoradiography. (C) TNF-α release in primary glial culture. Treatment of wild-type glial cells with LPS markedly stimulated TNF-α release at 24 h. Contrarily, TNF-α release was significantly reduced in PARP-1-/- compared with wild-type glial cells. Released TNF-α was quantified by ELISA. Values are means ± SEM from triplicate samples. (D) Expression of iNOS in primary glial cells. Treatment of wild-type glial cells with LPS induced iNOS expression, but this LPS-stimulated expression of iNOS was notably abrogated in PARP-1-/- cells. (E) p38MAPK-dependent iNOS expression. The p38MAPK inhibitor SB203580 reduced expression of LPS-induced iNOS significantly. (F) p38MAPK-dependent TNF-α release. The p38MAPK inhibitor SB203580 reduced TNF-α release significantly in both wild-type (hatched bars) and PARP-1-/- (dotted bars) in response to LPS. (G) p38MAPK-dependent pro-IL-1β expression in primary glial cells. LPS-induced expression of pro-IL-1β was virtually abolished in PARP-1-/- cells, and the p38MAPK inhibitor SB203580 attenuated significantly pro-IL-1β expression in wild type and to a nondetectable level in PARP-1-/- cells. Cells were pretreated with SB203580 (20 μM) for 1 h before adding 1 μg/ml LPS. Expression of PARP-1, iNOS, and pro-IL-1β was visualized by separating total proteins from wild-type and PARP-1-/- glial cells on 4-12% SDS/PAGE gel and immunoblotting with antibodies to PARP-1, iNOS, or pro-IL-1β, respectively.
In studies (29), we have shown that LPS-stimulated TNF-α release is attenuated significantly in PARP-1-/- glial cells as compared with wild-type cells at 3 and 6 h after the treatment. To determine whether the attenuation of TNF-α release in PARP-1-/- glial cells was not simply a delay process, glial cells from both wild-type and PARP-1-/- mice were treated with LPS (Escherichia coli serotype, 055-B5) for 24 h. Treatment of wild-type glial cells markedly induced TNF-α release (Fig. 1C), whereas LPS-stimulated TNF-α release was reduced >50% in PARP-1-/- glial cells. Therefore, LPS-stimulated TNF-α release was reduced significantly, and not merely delayed, in PARP-1-/- glial cells.
Proinflammatory Genes Regulated by p38MAPK in Glial Cells. We have also demonstrated (29) that a p38MAPK inhibitor, SB203580, reduces LPS-induced iNOS expression dramatically in RAW264.7 macrophages. Accordingly, we further tested the role of p38MAPK on the proinflammatory gene expression in glial cells. Treatment of glial cells with LPS markedly induced the expression of iNOS, whereas the induction of iNOS expression was almost absent in PARP-1-/- glial cells (Fig. 1D). Moreover, pretreatment of wild-type glial cultures with SB203580 reduced LPS-induced iNOS expression efficiently, suggesting that the p38MAPK pathway also mediates LPS-induced iNOS expression in glial cells (Fig. 1E). We also examined LPS-induced TNF-α release and pro-IL-1β (32 kDa) expression. Pretreatment of wild-type glial cultures with SB203580 reduced dramatically LPS-induced TNF-α release and pro-IL-1β expression, whereas LPS-induced TNF-α release and pro-IL-1β expression was reduced further to an almost undetectable level in PARP-1-/- glial cells (Fig. 1 F and G).
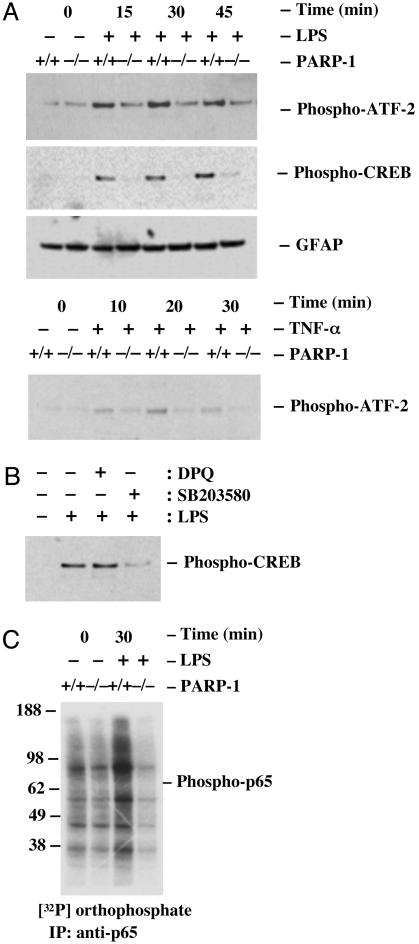
Lack of Stimuli-Induced ATF-2, CREB, and NF-κB p65 Phosphorylation in PARP-1-/- Glial Cells. Transcriptional regulation of iNOS, IL-1β, and TNF-α is complex and requires coordinated activation of a number of transcription factors including NF-κB, activator protein 1, C/EBP, Oct-1, CREB, and ATF-2 (32-34). The transcriptional activity of ATF-2 is controlled by phosphorylation. Cellular stress, such as inflammatory cytokines, activates ATF-2 by phosphorylation and stimulates the transcriptional activity of ATF-2 (35, 36). Mutation of this phosphorylation site results in the loss of stress-induced transcription by ATF-2 (35, 36). In glial cells, the phosphorylation-induced activation of ATF-2 is shown to enhance iNOS promoter activity, whereas a phosphorylation-defective form of ATF-2 is shown to have a suppressive effect on the iNOS promoter activity (26). To further investigate whether the impaired proinflammatory gene expression in PARP-1-/- glial cells is linked to a defective activation of ATF-2, we have assessed the phosphorylation of ATF-2 (Thr-71) in LPS-treated glial cells. Both wild-type and PARP-1-/- glial cells showed the basal phosphorylation of ATF-2 (Fig. 2A). However, treatment of wild-type glial cells with LPS rapidly induced the phosphorylation of ATF-2 at 15, 30, and 45 min, whereas the phosphorylation of ATF-2 was not induced from the basal phosphorylation of ATF-2 in PARP-1-/- glial cells (Fig. 2 A). Given that both LPS and TNF-α activate ATF-2 through Toll-like receptor 4 in microglia and TNFR1 in astrocytes and microglia, we further tested whether the lack of ATF-2 activation in PARP-1-/- glial cells was a stimulus- and receptor-specific manner. Reminiscent of LPS, treatment of wild-type glial cells with TNF-α rapidly induced the phosphorylation of ATF-2, whereas the phosphorylation of ATF-2 was not induced in PARP-1-/- glial cells (Fig. 2 A).
Fig. 2.
Activation of ATF-2, CREB, and NF-κB p65 is defective in PARP-1-/- primary glial cells. (A) LPS- or TNF-α-induced phosphorylation of ATF-2 and CREB. Treatment of wild-type glial cells with LPS rapidly induced the phosphorylation of ATF-2 or CREB within 15 min, whereas the phosphorylation of ATF-2 or CREB was not induced from the basal level or barely detectable at 45 min in PARP-1-/- glial cells. (B) p38MAPK-dependent phosphorylation of CREB. Pretreatment of wild-type cells with SB203580, a specific p38MAPK inhibitor, reduced dramatically LPS-induced phosphorylation of CREB, but DPQ, a specific PARP inhibitor, did not affect LPS-induced phosphorylation of CREB. Glial cells were pretreated with either 20 μM DPQ or SB203580 for 30 min and then stimulated with LPS for 30 min. (C) LPS-induced phosphorylation of NF-κB p65. Treatment of wild-type glial cells with LPS rapidly induced p65 phosphorylation at 30 min, whereas p65 phosphorylation was not induced from the basal level in PARP-1-/- glial cells. The phosphorylation of ATF-2 and CREB was determined by immunoblotting with antibodies specific to phospho-ATF-2 (Thr-71) and phospho-CREB (Ser-133). In addition, the phosphorylation of NF-κB p65 was determined by labeling glial cells with [32P] orthophosphate for 3 h and stimulating cells with LPS for 30 min. Subsequent nuclear extracts were analyzed by immunoprecipitation with antibodies to p65 and visualized by autoradiography. Data are representative of four independent experiments. GFAP, glial fibrillary acidic protein.
Cellular stress also activates a CREB to mediate the gene expression including iNOS and IL-1β (26, 33, 37). Activated p38MAPK is shown to phosphorylate a transcription factor CREB through mitogen- and stress-activated protein kinase (38). Treatment of wild-type glial cells with LPS markedly augmented phosphorylation of CREB in a time-dependent manner within 15 min, whereas PARP-1-/- glial cells had no augmented phosphorylation of CREB and was only slightly detectable at 45 min, as measured by phosphospecific antibodies to CREB (Ser-133) (Fig. 2 A). Treatment of TNF-α also failed to induce the phosphorylation of CREB as similar to LPS treatment in PARP-1-/- glial cells (data not shown). In addition, pretreatment of wild-type glial cultures with SB203580 reduced dramatically LPS-induced phosphorylation of CREB (Fig. 2B), thus further supporting that it is activated p38MAPK that leads to CREB phosphorylation. In contrast, pretreatment of wild-type glial cells with 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1-(2H)isoquinolinone (DPQ), a pharmacological PARP inhibitor (Fig. 2B), did not affect LPS-induced phosphorylation of CREB, indicating that the absence of augmented phosphorylation of CREB in response to LPS is not linked to the enzymatic activity of PARP-1.
A signal-induced phosphorylation of NF-κB is crucial for transcriptional competence, which in turn provides an additional mechanism to tightly regulate the NF-κB-dependent gene expression in response to diverse signals (27, 28, 39, 40). Glial cells labeled with [32P]orthophosphate were stimulated with LPS for 30 min, and the phosphorylation of p65 in the nucleus was analyzed by immunoprecipitation with antibodies to p65. The results demonstrated that p65 was phosphorylated under non-induced conditions, and treatment of wild-type glial cells with LPS induced further phosphorylation of p65, whereas the phosphorylation of p65 was not induced in PARP-1-/- glial cells (Fig. 2C).
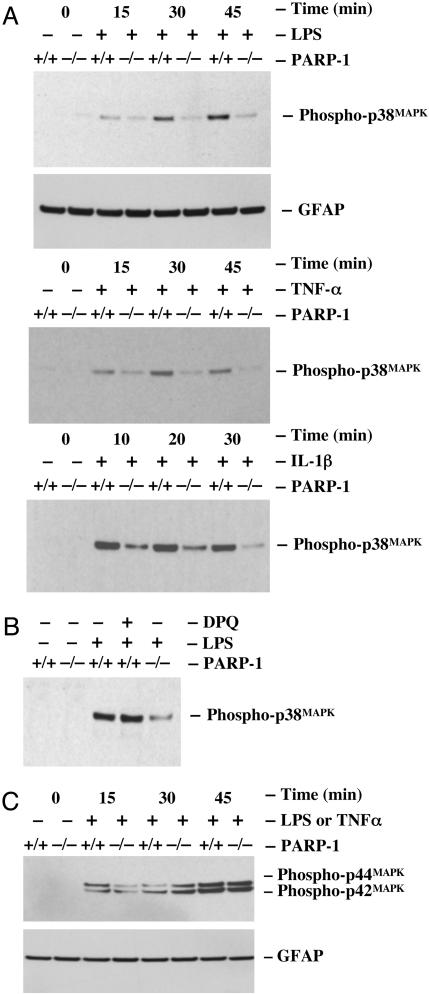
A Pathway-Specific Reduction of p38MAPK Activation in PARP-1-/- Glial Cells. LPS, TNF-α, or IL-1β activates p38MAPK in glial cells. Activated p38MAPK mediates a nuclear signaling through induced phosphorylation of CREB and ATF-2, resulting in an alteration of the proinflammatory gene expression. To further investigate whether defective nuclear signaling pathways were responsible for the lack of activation of CREB and ATF-2 in PARP-1-/- glial cells in response to inflammatory stimuli, we examined p38MAPK activation. Treatment of wild-type glial cells with LPS markedly activated p38MAPK, measured by phosphospecific antibodies to p38MAPK (Thr-180/Tyr-182), in a time-dependent manner. In contrast, p38MAPK was only slightly activated in PARP-1-/- glial cells, demonstrating that a defective p38MAPK nuclear signaling pathway is responsible for the lack of stimuli-induced CREB and ATF-2 activation and expression of certain inflammatory genes in response to inflammatory stimuli (Fig. 3A). To further test whether the lack of p38MAPK activation in PARP-1-/- glial cells depends on specific inflammatory stimuli, we examined the effects of TNF-α and IL-1β on p38MAPK activation in glial cells. Reminiscent of LPS, we found that both TNF-α- and IL-1β-induced p38MAPK activation was defective also in PARP-1-/- glial cells compared with wild-type glial cells (Fig. 3A). Previously, we found that DPQ, a specific PARP inhibitor, reduced LPS-induced iNOS expression in RAW264.7 macrophages. Accordingly, we examined whether this PARP activity might also influence the activation of p38MAPK. Pretreatment of wild-type glial cells with DPQ did not affect LPS-induced phosphorylation of p38MAPK (Fig. 3B), indicating that the reduced phosphorylation of p38MAPK in PARP-1-/- glial cells is not linked to the enzymatic activity of PARP-1.
Fig. 3.
Activation of the p38MAPK pathway is defective specifically in PARP-1-/- primary glial cells. (A) LPS-, TNF-α-, or IL-1β-induced phosphorylation of p38MAPK. Treatment of wild-type glial cells with LPS (1 μg/ml), TNF-α (1,000 units/ml), or IL-1β (1 ng/ml) rapidly induced the phosphorylation of p38MAPK within 10 or 15 min, whereas the phosphorylation of p38MAPK was significantly attenuated throughout the time course in PARP-1-/- glial cells. (B) PARP activity-independent activation of p38MAPK. DPQ had no effect on LPS-induced phosphorylation of p38MAPK even though PARP-1-/- glial cells showed dramatically reduced phosphorylation of p38MAPK.(C) LPS-induced phosphorylation of p42/44MAPK. Treatment of both wild-type and PARP-1-/- glial cells with LPS or TNF-α (data not shown) rapidly induced the phosphorylation of p42/44MAPK within 15 min. The phosphorylation of p38MAPK and p42/44MAPK was determined by immunoblotting with antibodies specific to phospho-p38MAPK (Thr-180/Tyr-182) and phospho-p42/44MAPK (Thr-202/Tyr-204). Wild-type glial cells were pretreated with a PARP inhibitor for 1 h before adding LPS. Data are representative of three independent experiments.
Given that a p42/44MAPK nuclear signaling mediates cell growth and differentiation but also mediates inflammatory responses to stress stimuli in glia, we assessed the activation of p42/44MAPK by phosphospecific antibodies to p44/42MAPK (Thr-202/Tyr-204) in response to inflammatory stimuli. In contrast to p38MAPK activity, treatment of wild-type and PARP-1-/- glial cells with LPS or TNF-α induced the phosphorylation of p44/42MAPK in a time-dependent manner (Fig. 3C), indicating p38MAPK regulates predominantly induced phosphorylation of ATF-2 and CREB in glial cells. Thus, the lack of a p38MAPK nuclear signaling in PARP-1-/- glial cells in response to inflammatory stimuli is pathway-specific signaling.
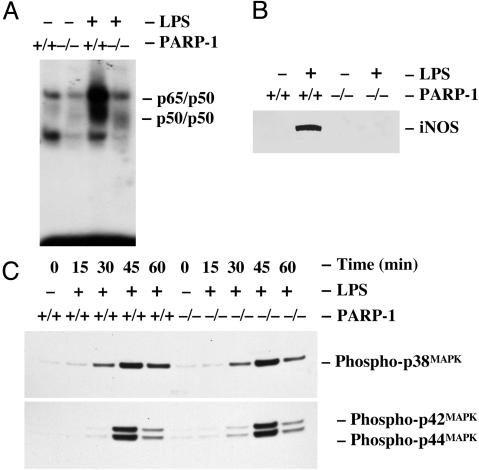
A Tissue Type-Specific Reduction of p38MAPK Activation in PARP-1-/- Glial Cells. Consistent with our previous data (29), treatment of wild-type peritoneal macrophages with LPS markedly augmented the binding activity of NF-κB to its cognate DNA sequences, whereas the cognate DNA binding activity of NF-κB was not induced in PARP-1-/- peritoneal macrophages (Fig. 4A). Reminiscent to NF-κB cognate DNA binding activity, treatment of wild-type peritoneal macrophages with LPS induced the iNOS expression, whereas the iNOS expression was not induced in PARP-1-/- peritoneal macrophages (Fig. 4B). Given that the expression of LPS-induced iNOS expression is dramatically abrogated in both primary glial cells and peritoneal macrophages from PARP-1-/- mice, we wondered whether inflammatory stimuli-induced intracellular signaling activities are also linked to lack of iNOS expression in peritoneal macrophages from PARP-1-/- mice. Accordingly, we examined LPS-induced p38MAPK and p42/44MAPK activation in primary peritoneal macrophages. In contrast to PARP-1-/- primary glial cells, treatment of both wild-type and PARP-1-/- peritoneal macrophages with LPS markedly induced p38MAPK and p42/44MAPK activity in a time-dependent manner (Fig. 4C). There was no difference in the level of p38MAPK and p42/44MAPK activation between wild-type and PARP-1-/- macrophages. Thus, the lack of a p38MAPK nuclear signaling in PARP-1-/- glial cells in response to inflammatory stimuli seems to be cell/tissue type-specific.
Fig. 4.
Activation of the p38MAPK pathway is normal in PARP-1-/- primary peritoneal macrophages. (A) Defective activation of NF-κB in PARP-1-/- primary peritoneal macrophages. Treatment of wild-type peritoneal macrophages with LPS rapidly induced cognate DNA binding activity of NF-κB. In contrast, LPS-induced cognate DNA binding activity of NF-κB was absent in PARP-1-/- macrophages. Primary peritoneal macrophages were treated with LPS for 30 min, and the extracted nuclear fractions were analyzed for the cognate DNA binding activity of NF-κB heterodimers (p65/p50) by electrophoretic mobility-shift assay. (B) Expression of iNOS in primary peritoneal macrophages. Treatment of wild-type peritoneal macrophages with LPS markedly induced iNOS expression. However, LPS-stimulated expression of iNOS was dramatically attenuated in PARP-1-/- glial cells. (C) LPS-induced phosphorylation of p38MAPK and p42/44MAPK in primary peritoneal macrophages. Treatment of wild-type and PARP-1-/- peritoneal macrophages with LPS rapidly induced the phosphorylation of p38MAPK and p42/44MAPK. The phosphorylation of p38MAPK and p42/44MAPK was determined by immunoblotting with antibodies specific to phospho-p38MAPK (Thr-180/Tyr-182) and phospho-p42/44MAPK (Thr-202/Tyr-204). Expression of iNOS was visualized by separating total proteins from wild-type and PARP-1-/- peritoneal macrophages on 4-12% SDS/PAGE gel and immunoblotting with antibodies to iNOS.
Discussion
In response to inflammation, infection, and injury, activated microglia and astrocytes can mediate neuronal inflammation and toxicity by releasing soluble factors. Rapid p38MAPK activation occurs in activated microglia and mediates the expression of inflammatory mediators, which may precipitate neurotoxicity. Our previous findings (29) that a significant reduction of proinflammatory cytokines and iNOS expression in PARP-1-/- glial cells in response to LPS are now shown to be a result of lacking not only cognate DNA-binding capacities of transcription factors but also lacking p38MAPK-mediated ATF-2 and CREB phosphorylation and NF-κB p65 phosphorylation.
LPS is a potent molecule for the activation of microglia in the CNS and peripheral immune cells (41). Receptors for LPS, TNF-α, and IL-1β are expressed in the brain and share downstream signaling pathways, including NF-κB and mitogen-activated protein kinase (42, 43). In glial culture, LPS initially triggers a robust inflammatory signaling cascade through a Toll-like receptor 4 expressed in microglia (44, 45) and induces the synthesis of proinflammatory cytokines, such as IL-1β and TNF-α, which in turn act as autocrine and paracrine factors to up-regulate more IL-1β and TNF-α and other inflammatory cytokine production (46). Our present studies clearly indicate that treatment of wild-type glial cells with LPS, TNF-α, or IL-1β induced activation of p38MAPK. Consequently, activated p38MAPK leads to phosphorylation of ATF-2 and CREB, which mediated TNF-α release and IL-1β and iNOS expression because a specific pharmacological inhibitor of p38MAPK, SB 203580, effectively reduced phosphorylation of ATF-2, CREB, and TNF-α release, as well as IL-1β and iNOS expression in wild-type glial cells treated with LPS. Interestingly, this activation of p38MAPK was also dramatically reduced in PARP-1-/- glial cells, causing a significant reduction of ATF-2 and CREB activation in PARP-1-/- glial cells. Activation of transcription factors by phosphorylation is essential for their transcriptional competence. Reminiscent to this reduced nuclear signaling of p38MAPK-dependent transcription activation, PARP-1-/- glial cells showed a significant reduction of TNF-α release and IL-1β and iNOS expression. Importantly, the lack of p38MAPK activation in PARP-1-/- glial cells was not due to defective receptors but, more likely, to the lack of a specific p38MAPK-signaling cascade step(s) common to LPS, TNF-α, and IL-1β in a pathway-specific and cell/tissue type-specific manner. Treatment of PARP-1-/- glial cells with LPS (or TNF-α) induced activation of p42/44MAPK as similar to wild-type cells; in addition, treatment of PARP-1-/- peritoneal macrophages with LPS induced activation of p38MAPK the same as with wild-type cells.
PARP-1 has been reported to participate in activation of NF-κB by interacting directly with NF-κB (47, 48) or poly(ADP-ribosyl)ating NF-κB (49, 50). Interaction of PARP-1 with NF-κB p65 may enhance its transcriptional competence by still unknown mechanism. However, poly(ADP-ribosyl)ation of transcription factors may hinder their interaction with cognate DNA sites or with their partners because of negatively charged poly(ADP-ribose) polymers. Interestingly, PARP-1-/-, not wild-type, glial cells lack cognate DNA-binding capacity of NF-κB and NF-κB-dependent gene transcription in response to inflammatory stimuli. Recently, Drosophila PARP with its activity has shown to be localized in the area of highly transcribed region, suggesting that PARP may regulate gene-specific transcription (51). In addition, PARP is shown to mediate stress-induced Drosophila heat shock protein (hsp70) (51). In our present studies, we provided an additional mechanism that severely impaired NF-κB-dependent gene expression in PARP-1-/- glial cells was, in part, a result of lacking induced phosphorylation of NF-κB p65, thus abrogating transcription competence of NF-κB p65. It is currently unknown which kinase phosphorylates NF-κB p65 in glial cells treated with LPS. Recently, p38MAPK-activated mitogen- and stress-activated protein kinase was shown to phosphorylate NF-κB p65 (52), CREB (53), and histone H3 (54), suggesting that p38MAPK-activated mitogen- and stress-activated protein kinase may phosphorylate NF-κB p65 in glial cells in response to LPS.
Previously, we and others showed that pharmacological inhibitors of PARP enzymatic activity, DPQ or PJ34, fail to reduce cognate DNA binding activity of NF-κB (29, 55); in addition, DPQ also fails to reduce expression of pro-IL-1β (29), which argues against a direct role for the PARP enzymatic activity on these transcription events. Conversely, DPQ reduces the expression of iNOS (29), and another PARP inhibitor, 6(5H)-phenanthridionone, is most recently shown to reduce cognate DNA binding activity of NF-κB and iNOS, TNF-α, and IL-1β expression/release in glial culture and organotypic hippocampal slices in response to LPS/IFN-γ or β-amyloid protein (56), implicating that PARP enzymatic activity may have a direct role in some, at least, proinflammatory gene expression. These variable effects of PARP enzymatic inhibitors on inflammatory gene expression may reflect the difference in efficacy of PARP inhibitors and/or PARP-1 enzymatic activity-independent mechanism(s) of the transcription regulation.
Our finding demonstrates that PARP-1 is an important contributory host factor among a number of factors to mediate proinflammatory cytokines and iNOS expression in the CNS. Therefore, PARP-1 can directly precipitate neuropathogenesis by mediating soluble factors released from glial cells and by activating its enzymatic activity, consequently depleting cellular ATP level in neuronal cells in various neurodegenerative diseases. Accordingly, understanding the precise molecular mechanism of PARP-1 on regulating inflammatory gene transcription in glial cells will facilitate the design of therapeutic intervention of inflammatory-related neurodegenerative disease.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CREB, cAMP-response element-binding protein; DPQ, 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1-(2H)-isoquinolinone; iNOS, inducible NO synthase; LPS, lipopolysaccharide; PARP-1, poly(ADP-ribose) polymerase 1; PARP-1-/-, PARP-1-deficient; TNF-α, tumor necrosis factor α; ATF, activating transcription factor.
References
- 1.D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G. G. (1999) Biochem. J. 342, 249-268. [PMC free article] [PubMed] [Google Scholar]
- 2.de Murcia, G. & Menissier de Murcia, J. (1994) Trends Biochem. Sci. 19, 172-176. [DOI] [PubMed] [Google Scholar]
- 3.Althaus, F. R., Kleczkowska, H. E., Malanga, M., Muntener, C. R., Pleschke, J. M., Ebner, M. & Auer, B. (1999) Mol. Cell. Biochem. 193, 5-11. [PubMed] [Google Scholar]
- 4.Berger, N. A. (1985) Radiat. Res. 101, 4-15. [PubMed] [Google Scholar]
- 5.Szabo, C. (1998) Eur. J. Pharmacol. 350, 1-19. [DOI] [PubMed] [Google Scholar]
- 6.Oleinick, N. L. & Evans, H. H. (1985) Radiat. Res. 101, 29-46. [PubMed] [Google Scholar]
- 7.Ha, H. C. & Snyder, S. H. (1999) Proc. Natl. Acad. Sci. USA 96, 13978-13982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pieper, A. A., Brat, D. J., Krug, D. K., Watkins, C. C., Gupta, A., Blackshaw, S., Verma, A., Wang, Z. Q. & Snyder, S. H. (1999) Proc. Natl. Acad. Sci. USA 96, 3059-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliasson, M. J., Sampei, K., Mandir, A. S., Hurn, P. D., Traystman, R. J., Bao, J., Pieper, A., Wang, Z. Q., Dawson, T. M., Snyder, S. H. & Dawson, V. L. (1997) Nat. Med. 3, 1089-1095. [DOI] [PubMed] [Google Scholar]
- 10.Endres, M., Wang, Z. Q., Namura, S., Waeber, C. & Moskowitz, M. A. (1997) J. Cereb. Blood Flow Metab. 17, 1143-1151. [DOI] [PubMed] [Google Scholar]
- 11.Zingarelli, B., Salzman, A. L. & Szabo, C. (1998) Circ. Res. 83, 85-94. [DOI] [PubMed] [Google Scholar]
- 12.Liaudet, L., Pacher, P., Mabley, J. G., Virag, L., Soriano, F. G., Hasko, G. & Szabo, C. (2002) Am. J. Respir. Crit. Care Med. 165, 372-377. [DOI] [PubMed] [Google Scholar]
- 13.Soriano, F. G., Liaudet, L., Szabo, E., Virag, L., Mabley, J. G., Pacher, P. & Szabo, C. (2002) Shock 17, 286-292. [DOI] [PubMed] [Google Scholar]
- 14.Szabo, C., Lim, L. H., Cuzzocrea, S., Getting, S. J., Zingarelli, B., Flower, R. J., Salzman, A. L. & Perretti, M. (1997) J. Exp. Med. 186, 1041-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masutani, M., Suzuki, H., Kamada, N., Watanabe, M., Ueda, O., Nozaki, T., Jishage, K., Watanabe, T., Sugimoto, T., Nakagama, H., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 2301-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burkart, V., Wang, Z. Q., Radons, J., Heller, B., Herceg, Z., Stingl, L., Wagner, E. F. & Kolb, H. (1999) Nat. Med. 5, 314-319. [DOI] [PubMed] [Google Scholar]
- 17.Kaul, M., Garden, G. A. & Lipton, S. A. (2001) Nature 410, 988-994. [DOI] [PubMed] [Google Scholar]
- 18.Kreutzberg, G. W. (1996) Trends Neurosci. 19, 312-318. [DOI] [PubMed] [Google Scholar]
- 19.Koistinaho, M. & Koistinaho, J. (2002) Glia 40, 175-183. [DOI] [PubMed] [Google Scholar]
- 20.Bhat, N. R., Zhang, P., Lee, J. C. & Hogan, E. L. (1998) J. Neurosci. 18, 1633-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koistinaho, M., Kettunen, M. I., Goldsteins, G., Keinanen, R., Salminen, A., Ort, M., Bures, J., Liu, D., Kauppinen, R. A., Higgins, L. S. & Koistinaho, J. (2002) Proc. Natl. Acad. Sci. USA 99, 1610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barone, F. C. & Feuerstein, G. Z. (1999) J. Cereb. Blood Flow Metab. 19, 819-834. [DOI] [PubMed] [Google Scholar]
- 23.Walton, K. M., DiRocco, R., Bartlett, B. A., Koury, E., Marcy, V. R., Jarvis, B., Schaefer, E. M. & Bhat, R. V. (1998) J. Neurochem. 70, 1764-1767. [DOI] [PubMed] [Google Scholar]
- 24.Jiang, Y., Chen, C., Li, Z., Guo, W., Gegner, J. A., Lin, S. & Han, J. (1996) J. Biol. Chem. 271, 17920-17926. [DOI] [PubMed] [Google Scholar]
- 25.Han, J., Jiang, Y., Li, Z., Kravchenko, V. V. & Ulevitch, R. J. (1997) Nature 386, 296-299. [DOI] [PubMed] [Google Scholar]
- 26.Bhat, N. R., Feinstein, D. L., Shen, Q. & Bhat, A. N. (2002) J. Biol. Chem. 277, 29584-29592. [DOI] [PubMed] [Google Scholar]
- 27.Wang, D., Westerheide, S. D., Hanson, J. L. & Baldwin, A. S., Jr. (2000) J. Biol. Chem. 275, 32592-32597. [DOI] [PubMed] [Google Scholar]
- 28.Madrid, L. V., Mayo, M. W., Reuther, J. Y. & Baldwin, A. S., Jr. (2001) J. Biol. Chem. 276, 18934-18940. [DOI] [PubMed] [Google Scholar]
- 29.Ha, H. C., Hester, L. D. & Snyder, S. H. (2002) Proc. Natl. Acad. Sci. USA 99, 3270-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, Z. Q., Auer, B., Stingl, L., Berghammer, H., Haidacher, D., Schweiger, M. & Wagner, E. F. (1995) Genes Dev. 9, 509-520. [DOI] [PubMed] [Google Scholar]
- 31.Vartanian, T., Li, Y., Zhao, M. & Stefansson, K. (1995) Mol. Med. 1, 732-743. [PMC free article] [PubMed] [Google Scholar]
- 32.Lowenstein, C. J., Alley, E. W., Raval, P., Snowman, A. M., Snyder, S. H., Russell, S. W. & Murphy, W. J. (1993) Proc. Natl. Acad. Sci. USA 90, 9730-9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stylianou, E. & Saklatvala, J. (1998) Int. J. Biochem. Cell Biol. 30, 1075-1079. [DOI] [PubMed] [Google Scholar]
- 34.Jongeneel, C. V. (1995) Immunobiology 193, 210-216. [DOI] [PubMed] [Google Scholar]
- 35.Livingstone, C., Patel, G. & Jones, N. (1995) EMBO J. 14, 1785-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta, S., Campbell, D., Derijard, B. & Davis, R. J. (1995) Science 267, 389-393. [DOI] [PubMed] [Google Scholar]
- 37.Gavrilyuk, V., Horvath, P., Weinberg, G. & Feinstein, D. L. (2001) J. Neurochem. 78, 129-140. [DOI] [PubMed] [Google Scholar]
- 38.Shaywitz, A. J. & Greenberg, M. E. (1999) Annu. Rev. Biochem. 68, 821-861. [DOI] [PubMed] [Google Scholar]
- 39.Bird, T. A., Schooley, K., Dower, S. K., Hagen, H. & Virca, G. D. (1997) J. Biol. Chem. 272, 32606-32612. [DOI] [PubMed] [Google Scholar]
- 40.Madrid, L. V., Wang, C. Y., Guttridge, D. C., Schottelius, A. J., Baldwin, A. S., Jr., & Mayo, M. W. (2000) Mol. Cell. Biol. 20, 1626-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, B. & Hong, J. S. (2003) J. Pharmacol. Exp. Ther. 304, 1-7. [DOI] [PubMed] [Google Scholar]
- 42.Aloisi, F. (2001) Glia 36, 165-179. [DOI] [PubMed] [Google Scholar]
- 43.Allan, S. M. & Rothwell, N. J. (2001) Nat. Rev. Neurosci. 2, 734-744. [DOI] [PubMed] [Google Scholar]
- 44.Lehnardt, S., Massillon, L., Follett, P., Jensen, F. E., Ratan, R., Rosenberg, P. A., Volpe, J. J. & Vartanian, T. (2003) Proc. Natl. Acad. Sci. USA 100, 8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bsibsi, M., Ravid, R., Gveric, D. & van Noort, J. M. (2002) J. Neuropathol. Exp. Neurol. 61, 1013-1021. [DOI] [PubMed] [Google Scholar]
- 46.Hanisch, U. K. (2002) Glia 40, 140-155. [DOI] [PubMed] [Google Scholar]
- 47.Hassa, P. O. & Hottiger, M. O. (1999) Biol. Chem. 380, 953-959. [DOI] [PubMed] [Google Scholar]
- 48.Ullrich, O., Diestel, A., Eyupoglu, I. Y. & Nitsch, R. (2001) Nat. Cell Biol. 3, 1035-1042. [DOI] [PubMed] [Google Scholar]
- 49.Kameoka, M., Ota, K., Tetsuka, T., Tanaka, Y., Itaya, A., Okamoto, T. & Yoshihara, K. (2000) Biochem. J. 346, 641-649. [PMC free article] [PubMed] [Google Scholar]
- 50.Chang, W. J. & Alvarez-Gonzalez, R. (2001) J. Biol. Chem. 276, 47664-47670. [DOI] [PubMed] [Google Scholar]
- 51.Tulin, A. & Spradling, A. (2003) Science 299, 560-562. [DOI] [PubMed] [Google Scholar]
- 52.Vermeulen, L., De Wilde, G., Van Damme, P., Vanden Berghe, W. & Haegeman, G. (2003) EMBO J. 22, 1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deak, M., Clifton, A. D., Lucocq, L. M. & Alessi, D. R. (1998) EMBO J. 17, 4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomson, S., Clayton, A. L., Hazzalin, C. A., Rose, S., Barratt, M. J. & Mahadevan, L. C. (1999) EMBO J. 18, 4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soriano, F. G., Virag, L. & Szabo, C. (2001) J. Mol. Med. 79, 437-448. [DOI] [PubMed] [Google Scholar]
- 56.Chiarugi, A. & Moskowitz, M. A. (2003) J. Neurochem. 85, 306-317. [DOI] [PubMed] [Google Scholar]