Abstract
Bacteria use type IV secretion systems for two fundamental objectives related to pathogenesis — genetic exchange and the delivery of effector molecules to eukaryotic target cells. Whereas gene acquisition is an important adaptive mechanism that enables pathogens to cope with a changing environment during invasion of the host, interactions between effector and host molecules can suppress defence mechanisms, facilitate intracellular growth and even induce the synthesis of nutrients that are beneficial to bacterial colonization. Rapid progress has been made towards defining the structures and functions of type IV secretion machines, identifying the effector molecules, and elucidating the mechanisms by which the translocated effectors subvert eukaryotic cellular processes during infection.
The year 2003 marks the fiftieth anniversary of the first description of a type iv secretion (T4S) system: the conjugation apparatus of the F plasmid1. This is a dynamic bacterial surface organelle, the activities of which are now known to include the contact-dependent delivery of DNA to bacterial recipients and the assembly and retraction of a conjugal pilus2. In the past decade, reports describing systems that are ancestrally related to the F-transfer system and other conjugation machines have emerged. Instead of mediating DNA transfer between bacteria, these systems deliver DNA or protein substrates, known as effectors, to eukaryotic target cells during infection3–5. More recently, several new T4S systems have been described that are also ancestrally related to the conjugation machines, but these systems mediate the exchange of DNA with the extracellular milieu6–8. Collectively, this diversity of function in the face of a common ancestry makes the T4S machines attractive subjects for comparative studies that explore the dynamics of organelle assembly and action. Additionally, from a medical perspective, it is of enormous interest to develop a detailed understanding of how the inter-kingdom transfer of type IV effector molecules contributes to pathogenesis. This review will summarize the recent advances in our knowledge of T4S, with an emphasis on machine structure and function, and the activities of effectors after translocation into the eukaryotic host.
The T4S family
This fascinatingly versatile translocation family can be classified into three subfamilies, each of which contributes in unique ways to pathogenesis (Fig. 1; Table 1). The largest subfamily, the conjugation systems, are found in most species of Gram-negative and Gram-positive bacteria. These systems mediate DNA transfer both within and between phylogenetically diverse species, and some systems even deliver DNA to fungi, plants and human cells2,9–13. Conjugation is an important contributor to genome plasticity, and therefore bacterial fitness under changing environmental conditions, as encountered during infection of the human host. Moreover, conjugation is problematic in nosocomial settings because the dissemination of conjugative plasmids and other mobile elements — often reservoirs of antibiotic resistance genes — can lead to an explosive emergence of multiple drug resistance among populations of clinically significant pathogens14. Recent work has also shown that plasmid-encoded conjugative pili of Gram-negative bacteria, or surface glycoproteins of Gram-positive bacteria, contribute to biofilm formation and colonization of various human tissues15,16.
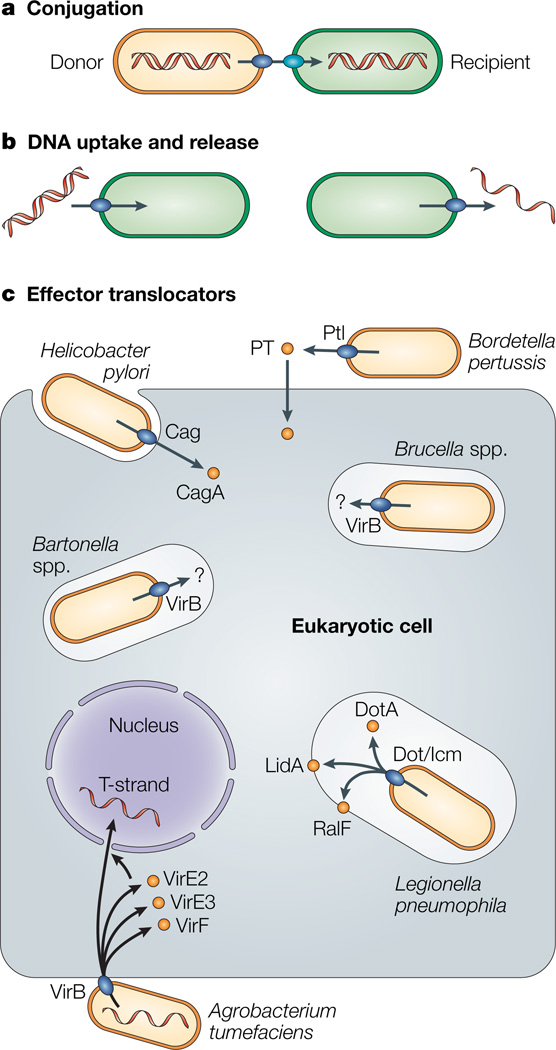
Figure 1. Schematic representation of the different type-IV-dependent mechanisms.
The three subfamilies of type IV secretion (T4S) systems are shown. Conjugation machines deliver DNA to recipient bacteria and other cell types by cell-to-cell contact. DNA-uptake and -release systems exchange DNA with the extracellular milieu independently of contact with target cells. Effector translocators deliver DNA or protein substrates to eukaryotic cells during infection. The effector translocators contribute in markedly different ways to the infection processes of the bacterial pathogens shown. PT, pertussis toxin.
Table 1.
Type IV secretion (T4S) systems and disease manifestations
| Bacterial species | T4S system | Target-cell or host alterations | References |
|---|---|---|---|
| Conjugation | |||
| Escherichia coli F plasmid (IncF) | Tra | Genetic exchange | 2 |
| Escherichia coli RP4 (IncP) | Trb | Genetic exchange | 28 |
| Escherichia coli R388 (IncW) | Trw | Genetic exchange | 28 |
| Shigella Collb-P9 (IncI) | Tra | Genetic exchange | 28 |
| *Agrobacterium tumefaciens | VirB | Crown gall/genetic exchange | 11 |
| *Legionella pneumophila | Dot/Icm | Genetic exchange | 5 |
| DNA uptake and release | |||
| Campylobacter jejuni | Cjp/VirB | DNA uptake | 6 |
| Helicobacter pylori | ComB | DNA uptake | 18 |
| Neisseria gonorrhoeae | Tra | DNA release | 8 |
| Effector translocation | |||
| Agrobacterium tumefaciens | VirB | Crown gall | 11 |
| Helicobacter pylori | Cag | Gastritis, peptic ulcer | 4 |
| Bordetella pertussis | Ptl | Whooping cough | 45 |
| Legionella pneumophila | Dot/Icm | Legionnaire’s pneumonia | 5 |
| Brucella spp. | VirB | Brucellosis | 105 |
| Bartonella spp. | VirB, Trw | Cat-scratch, angiomatosis | 26,72 |
| ‡Actinobacillus | MagB | Periodontitis | 106 |
| ‡Ehrlichia spp. | VirB | Ehrlichiosis | 107 |
| ‡Wolbachia spp. | VirB | Host sexual alterations | 108 |
| ‡Rickettsia spp. | – | Epidemic typhus, Mediterranean spotted fever | 109 |
| ‡Xylella fastidiosa | VirB | Leaf scorch disease, citrus variegated chlorosis | 110 |
| ‡Coxiella burnetii | Dot/Icm | Q fever | 111 |
These systems have been shown to function both as conjugation machines, transferring DNA to bacterial recipients, and as effector translocators, delivering effector molecules to eukaryotic target cells during infection.
These systems are presumed to be functional T4S systems on the basis of sequence similarities with conjugation systems.
The second subfamily, the so-called ‘DNA uptake and release’ systems, function independently of contact with a target cell (Fig. 1; Table 1). This recently discovered subfamily presently comprises two competence (DNA-uptake) systems — the Campylobacter jejuni Cjp/VirB system and the Helicobacter pylori ComB system6,7,17,18 — and one DNA-release system, an F-plasmid Tra-like system that is carried on the gonococcal genetic island (GGI) of Neisseria gonorrhoeae8,19. As with the conjugation machines, these systems promote genetic exchange and therefore also represent potential mechanisms for the transfer of survival traits during infection20.
The third subfamily, here designated the ‘effector translocator’ systems, is indispensable in the infection processes of several prominent pathogens of plants and mammals (Fig. 1; Table 1; Box 1). In general, these machines can be viewed as ‘injectisomes’, reminiscent of the type III secretion (T3S) machines21, because they deliver their substrates through direct contact with the eukaryotic target cell. The list of pathogens that are dependent on effector translocators for disease progression includes the phytopathogen Agrobacterium tumefaciens and several pathogens of mammals, such as H. pylori, Legionella pneumophila, and Brucella and Bartonella species. Bordetella pertussis also uses an effector translocator, but this system functions as a true exporter to deliver its toxin substrate to the extracellular milieu. Related systems of several additional pathogens are also implicated in the trafficking of substrates to eukaryotic cells, so the list of T4S effector translocators continues to grow (Table 1).
Box 1 | Some pathogens that use type IV effector translocators during infection*.
Agrobacterium tumefaciens
A phytopathogen that is responsible for crown gall disease, which manifests as an uncontrolled proliferation of plant tissue. The disease affects many agriculturally important dicotyledenous plant species. A. tumefaciens is also economically valuable as a widely used gene-delivery system for the construction of transgenic plants.
Bartonella henselae
The causative agent of cat-scratch disease, a relatively benign disease that is transmitted to humans by blood-sucking arthropods or by direct contact with domestic cats. The clinical manifestations are broad and include intermittent fever, cerebral arteriosis and lethargy. In North America, the incidence is ~5–8/100,000.
Bordetella pertussis
Responsible for a respiratory disease known as ‘whooping cough’ or pertussis, and transmitted by aerosol droplets. Although a vaccine exists, the incidence worldwide is ~40–60 million cases and ~300,000 deaths annually.
Brucella spp
The causative agents of brucellosis, or Malta fever, these organisms are transmitted to humans through direct contact with infected animals, carcasses or milk products. A febrile disease with effects on the musculoskeletal system accompanied by irregular fevers. The annual incidence is ~80/100,000 in countries of southern Europe, the Mediterranean and the Middle East.
Helicobacter pylori
The causative agent of chronic gastric disorders, and is important in the development of peptic ulcer and gastric cancers. H. pylori is also able to persist in the human stomach without inciting disease. Approximately 50% of the world population is infected with this bacterium.
Legionella pneumophila
Responsible for pneumonia known as ‘legionnaire’s disease’. Humans are infected through contact with contaminated water or aerosol sources from ventilation, air conditioning or shower systems. In developed countries, the annual incidence is 4/100,000, with a mortality of ~20%; the incidence of disease is appreciably higher in developing countries.
* Information on clinical manifestations and disease incidence is from the World Health Organization (WHO) (see further information in Online links).
T4S machine assembly and architecture
Since the discovery of conjugation systems, a vast body of literature has accumulated describing the factors governing the expression of transfer (tra) genes, the assembly and overall architectures of the conjugation machines, the contributions of various subunits to substrate transfer, and, most recently, high-resolution structures of some of the machine subunits and associated extracellular appendages. Researchers are continuing to build on this foundation of knowledge, with ongoing mechanistic studies that are focused particularly on the VirB/D4 transfer system of A. tumefaciens and the conjugation systems of the F, RP4 and R388 plasmids. In the next sections, we will summarize the present understanding of how conjugation systems are arranged and how they function in a dynamic sense, using the A. tumefaciens VirB/D4 system22 as a framework for discussion (Fig. 2; Table 2). Additionally, owing to their tremendous clinical significance, there is a strong interest in defining the architectures and modes of action of the effector translocator systems. Already, these studies have identified several fascinating variations on some of the mechanistic themes established for the conjugation systems. Where available, we will include this information to highlight the structural and functional diversity of this translocation family. Finally, although it is beyond the scope of this review, another rich area of investigation concerns the question of how the various T4S gene sets are regulated in non-pathogenic and pathogenic settings. We refer the reader to several excellent recent studies exploring this topic23–27.
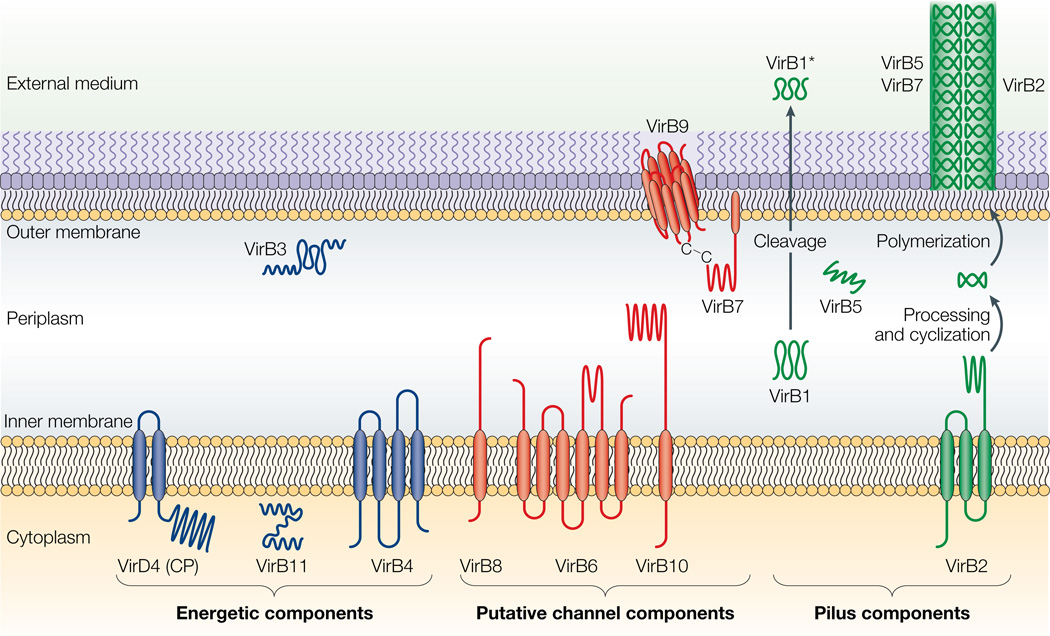
Figure 2. Topologies of the VirB/D4 subunits of the A. tumefaciens type IV secretion (T4S) system.
The coupling protein (CP) VirD4 and the mating-pore-formation components (VirB1–VirB11) are represented according to their proposed functions: energetic (blue), channel (red) or pilus (green) components. Several proteins are post-translationally modified in the periplasm. Signal sequences of VirB1, VirB2, VirB5, VirB7 and VirB9 are cleaved by signal peptidases. VirB1 is processed to form VirB1*, which is exported across the outer membrane. VirB2 undergoes a novel head-to-tail cyclization reaction, and polymerizes as the T-pilus. VirB7 is modified as a lipoprotein that associates with the T-pilus and also forms an intermolecular disulphide crosslink with VirB9, a possible secretin113. The VirB and VirD4 proteins are postulated to assemble as a supramolecular structure composed of a transenvelope channel and an extracellular pilus.
Table 2.
VirB/D4 subunit contacts, postulated functions and related components of effector translocator systems
| VirB proteins | Localization | Protein–protein contacts* | Proposed functions | Homologues‡ | |
|---|---|---|---|---|---|
| Dihybrid screen |
Biochemical assay |
||||
| Coupling protein | |||||
| VirD4 | IM | D4, E2 | D4, E2 | Recruitment of DNA and protein substrates to Mpf complex; translocase |
HP0524 (H.p.), DotL (L.p.), VirD4 (B.t.) |
| Channel subunits | |||||
| VirB6 | IM | B7, B9 | Assembly factor; channel subunit | PtlD (B.p.), VirB6 (Bart./Bruc.), TrwI1–5 (B.t.) | |
| VirB8 | IM | B1, B8, B9, B10 |
B8, B9, B10 | Assembly factor; bridge between subcomplexes; IM & OM VirB channel subunit |
HP0530 (H.p.), PtlE(B.p.), VirB8 (Bart./Bruc.), TrwG (B.t.) |
| VirB10 | IM | B8, B9, B10 | B8, B9, B10 | Bridge between IM & OM subcomplexes; channel subunit |
HP0527 (H.p.), PtlF (B.p.), DotB (L.p.), VirB10 (Bart./ Bruc.), TrwG (B.t.) |
| VirB3 | OM | PtlB (B.p.), VirB3 (Bart./Bruc.), TrwM (B.t.) | |||
| VirB7 | OM lipoprotein |
B7, B9 | B2, B6, B7, B9, 10 |
Stabilizes B9 by disulphide crosslink; B7–B9 dimer stabilizes other VirB proteins |
HP0532 (H.p.), PtlI(B.p.), VirB7 (Bart./Bruc.), TrwH1–5 (B.t.) |
| VirB9 | OM | B1, B7, B8, B9, B10, B11 |
B6, B7, B9, B10 |
OM pore? | HP0528 (H.p.), PtlF (B.p.), VirB9 (Bart./Bruc.), TrwF (B.t.) |
| Energetics | |||||
| VirB4 | IM | B4, B8, B10, B11 |
B4 | ATPase; homomultimer; energy for substrate export & pilus biogenesis |
CagE(H.p.), PtlC (B.p.), VirB4 (Bart./Bruc.), TrwF (B.t.) |
| VirB11 | IM | B4, B9 | B11 | ATPase; homomultimer; energy for substrate export & pilus biogenesis |
HP0525 (H.p.), PtlH (B.p.), DotB (L.p.) |
| Periplasmic factors | |||||
| VirB1 | Periplasm | B1, B4, B8, B9, B10, B1 |
Peptidoglycan hydrolase; channel assembly |
PtlE (B.p.), VirB1 (Bruc.), TrwN (B.t.) | |
| AcvB & VirJ | Periplasm | B1 to B11, D4, E2, D2 |
Possible periplasmic chaperones | ||
| T pilus | |||||
| VirB2 | IM, exocellular | B2, B5, B7 | Cyclized pilin subunit | PtlA (B.p.), VirB2 (Bart./ Bruc.), TrwL1-7 (B.t.) | |
| VirB5 | Periplasm, exocellular |
B2 | Pilus subunit, chaperone? | VirB5 (Bart./ Bruc.), TrwJ1-5 (B.t.) | |
| VirB7 | OM, exocellular |
B2 | Pilus assembly? | HP0532 (H.p.), Ptll(B.p.), VirB7 (Bart./Bruc.), TrwH1–5 (B.t.) |
|
| Effector translocator proteins associated with surface structures | |||||
| HP0532 (B7) (H.p.), HP0527 (B10) (H.p.) HP0529 (B9) (H.p.), DotO (L.p.), DotH (L.p.) |
|||||
Evidence for subunit–subunit contacts on the basis of results of yeast or bacterial dihybrid screens or biochemical assays. References are presented in the text.
Bacterial species are in parentheses: (Bart.) Bartonella henselae and B. tribocorum; (B.p.) B. pertussis; (Bruc.) Brucella spp.; (B.t.) Bartonella tribocorum; (H.p.) H. pylori; (L.p.) L. pneumophila. A comprehensive and continuously updated list of type IV secretion systems and gene–protein relationships can be found at Peter J. Christie’s laboratory website (see Online links). The Vir8 homologue HP0530 (H.p.) is reviewed in ref. 112. IM, inner membrane; OM, outer membrane; Mpf; mating pore formation.
The conjugation systems of Gram-negative bacteria can be viewed as assemblages of three distinct substructures: the coupling protein (CP) homomultimer; a transenvelope protein complex; and the conjugative pilus (transfer- or T-pilus)28. The latter two substructures are assembled from the mating-pore-formation (Mpf) proteins — for example, VirB1–VirB11 of the A. tumefaciens VirB/D4 T4S system. The CP, trans-envelope complex and the T-pilus act coordinately, probably as a single, supramolecular organelle, to mediate the various stages of translocation. These stages include the recruitment of cognate DNA and protein substrates to the transfer machine, the transfer of substrates across the cell envelope and the delivery of substrates to target cells.
The hexameric coupling protein: a substrate-recruitment factor and possible translocase
For conjugal DNA transfer, a set of processing proteins, known as the DNA transfer and replication (Dtr) proteins, act at the origin of transfer (oriT) sequence(s) of mobile DNA elements and process the DNA into a single-stranded substrate (T-strand). One processing protein, the relaxase, generates a strand-specific nick at oriT and remains covalently bound to the 5′ end of the T-strand. So, the translocation-competent form of the DNA substrate corresponds to a T-strand relaxase nucleoprotein complex11,22,28. The coupling proteins are so-called because they recruit DNA substrates, through specific interactions with relaxases and other Dtr proteins, to the Mpf structure. The biochemical and genetic data supporting this DNA-substrate-recruitment activity are summarized elsewhere29; here, we will highlight recent structural findings for the Escherichia coli TrwB CP of plasmid R388, which provide important insights into how this family of proteins function.
The CPs are composed of an amino-terminal-proximal region, which includes two transmembrane helices and a small periplasmic domain, and a large carboxy-terminal region that resides in the cytoplasm (see the topology of the VirD4 CP; Fig. 2). An X-ray crystal structure of the cytoplasmic domain of TrwB (TrwBΔN70) showed that six equivalent protomers form a spherical particle of 110 Å in diameter and 90 Å in height30,31. The view that is parallel to the membrane shows a ring-like structure with a central channel of 20-Å diameter, which is restricted to 8 Å at the entrance of the channel facing the cytoplasm. This channel is proposed to traverse the structure and connects the cytoplasm with the periplasm.Originally, the amino-terminal transmembrane domain was modelled in the crystal structure, but recent electron-microscopy and image-averaging studies have shown a discernible appendix, thereby validating the predicted presence of a transmembrane stem32.
This overall structure bears a striking resemblance to the F1-ATPase α3β3 heterohexamer, whereas the structure of the soluble domain closely resembles DNA ring helicases and other proteins that translocate along single- or double-stranded DNA31. Indeed, the CPs share sequence similarity with two known DNA translocases, FtsK and SpoIIIE31,33, and, consistent with a possible DNA-translocase activity, CPs bind ATP and single-stranded DNA32,34. Intriguingly, TrwBΔN70 purifies as both hexamers and monomers, indicating that the full-length CP might undergo dynamic monomer–hexamer transitions in vivo32. This dynamic action, mediated by nucleotide and/or substrate binding, might be important in substrate translocation29.
Although it is clear that the CP coordinates with the Mpf complex to drive DNA transfer, until recently it was not known whether the CP physically interacts with the Mpf structure. Now, two studies have reported that CPs form stable interactions with homologues of the A. tumefaciens VirB10 protein35,36. The VirB10-type proteins are bitopic inner-membrane proteins that are probably responsible for bridging inner- and outer-membrane Mpf protein subassemblies (Fig. 2). Interestingly, one study showed that a CP of one T4S machine interacts not only with the VirB10 protein of the cognate T4S system, but also with several other VirB10 homologues. On the basis of these findings, it can be proposed that the CP recruits DNA substrates to the T4S machine through contacts with the DNA-processing proteins. Then, through the VirB10 contact, the CP coordinates passage of the T-strand through the Mpf protein channel36.
CPs are ubiquitous components of conjugation machines, but they are also common components of effector translocators, raising the question of whether the CP also participates in effector protein recruitment. Cytological evidence favouring this role was recently presented. In A. tumefaciens, the VirD4 CP localizes at the cell poles37 or it can be artificially localized at the mid-cell38,39, so marking distinct cellular positions for monitoring the recruitment of substrates tagged with green fluorescent protein (GFP). Indeed, VirD4 was shown to recruit GFP fused to the VirE2 effector protein, and further cytological and biochemical studies established that the interaction occurs independently of any requirement for Mpf proteins, the T-strand or even VirE1, a secretion chaperone for VirE2 (ref. 39). Of further interest, VirD4 was shown to interact with the carboxy-terminal region of VirE2, adding to evidence that secretion signals are localized at the carboxyl termini of VirE2 and other protein substrates of the VirB/D4 T4S system39–42.
Further supporting a general substrate-recruitment function for CPs, the H. pylori Cag and L. pneumophila Dot/Icm systems require cognate CPs to translocate effectors43,44 (discussed in further detail below). However, we also note that some effector translocator systems seem to have evolved alternative mechanisms for the recruitment of secretion substrates. The Bordetella pertussis Ptl system provides an example of a T4S machine that operates independently of a CP. In this system, sub-units of the effector molecule, pertussis toxin (PT), are secreted across the inner membrane by the general secretory pathway (GSP), thereby bypassing the requirement for a CP45. Furthermore, the H. pylori Cag T4S system induces the secretion of interleukin-8 (IL-8) without a requirement for its CP43. This could result from the translocation of unidentified effector proteins by a CP-independent mechanism, although as noted below, CP-independent induction of IL-8 secretion might alternatively result from receptor engagement by the T4S apparatus. Finally, the Brucella spp. VirB T4S system, and a recently described Bartonella tribocorum trw pathogenicity (Trw-PAI) system, also contribute to virulence independently of a CP. These systems might use the Mpf structure itself, the GSP or another mechanism for the recruitment and translocation of effector molecules across the inner membrane26,46.
The transenvelope Mpf structure
Once DNA and protein substrates are recruited to the T4S apparatus, they are delivered across one or both membranes by the Mpf structure. For the A. tumefaciens VirB/D4 T4S system, the subcellular locations and topologies of the VirB Mpf proteins have been defined based on computer predictions and a combination of subcellular fractionation and analyses of reporter-protein fusion studies (Fig. 2; Table 2). In general, the VirB proteins can be divided into three classes according to known or postulated functions22,28. First, the putative channel components include the inner-membrane proteins VirB6,VirB8 and VirB10, and the outer-membrane proteins VirB3,VirB7 and VirB9. Second, two ATPases, VirB4 and VirB11, which are localized at the cytoplasmic face of the inner membrane, probably provide energy to drive substrate transfer and, possibly, biogenesis of the transfer channel and the pilus. Finally, the pilin subunit, VirB2, assembles as the T-pilus in association with VirB5 and the VirB7 lipoprotein47–50.VirB1 is a transglycosylase that has been implicated in machine biogenesis, and VirB1*, a truncated VirB1 derivative, is delivered across the outer membrane for an unspecified function51.
Early studies of the VirB/D4 T4S machine showed that certain VirB proteins exert stabilizing effects on other VirB subunits52. Further investigations exploring these stabilizing effects, as well as recent cell biology studies, have led to the development of a biogenesis pathway for this transfer apparatus22,53.A crucial intermediate in this pathway is a ‘core’ structure that is composed of VirB4, VirB7–VirB10 and, probably, VirB6. The existence of this structure is now supported by data from dihybrid screens and complementary biochemical assays54–59 (Table 2). Additionally, some of the interactions required for assembly of the putative core are conserved in the B. pertussis Ptl system. For example, it has been postulated that the A. tumefaciens VirB8 protein functions in part by recruiting the VirB1 transglycosylase for localized lysis of the peptidoglycan at the site of machine assembly57. B. pertussis PtlE is the VirB8 homologue, but PtlE itself shows transglycosylase activity, apparently eliminating the requirement for a VirB1 homologue in the Ptl system60. Additionally, PtlI and PtlF are homologues ofVirB7 and VirB9, respectively, and, as determined for the VirB counterparts, these proteins form intermolecular disulphide bridges that are required for stabilization of the Ptl proteins61. Finally, for the E. coli F plasmid T4S system, there is evidence for assembly of a transenvelope core that is composed of the TraV (VirB7-like lipoprotein), TraK (VirB9-like) and TraB (VirB10-like) subunits62.
We suggest these core structures might correspond to ancestral protein organelles, to which function-specifying subunits or protein subassemblies were added to evolve the present-day T4S family. Indeed, two observations support the notion that the core structure itself might function as a translocation channel in vivo. First, it has been shown that the VirB proteins that are produced by agrobacterial recipient cells greatly stimulate the acquisition of plasmid DNA during matings with donor cells. Recently, the proteins required for this effect were shown to correspond largely to the core subunits63. This core structure might therefore assemble as a conduit to facilitate DNA uptake across the recipient cell envelope. Second, it is noteworthy that the DNA-uptake systems of C. jejuni and H. pylori are composed of homologues of the VirB core proteins7,17. It is intriguing to speculate that these competence systems might have evolved by the simple addition of a surface-localized DNA receptor to the ancestral core.
The Mpf ATPases
The VirB4 and VirB11 ATPases are postulated to mediate VirB/D4 T4S machine assembly or function through dynamic, ATP-driven conformational changes. Homologues of both ATPases are widely conserved among the T4S system family members and, intriguingly, VirB11-like ATPases constitute a protein superfamily that extends to the transport machines of many Gram-negative and Gram-positive bacteria and several species of the Archaea64. A crystal structure has been determined for H. pylori HP0525, a homologue of the VirB11 ATPase65. Like the TrwB CP, HP0525 is homohexameric, but the protomers assemble as a double-stacked ring that is formed by self-association of the amino- and carboxy-terminal domains. The structure of the HP0525 apoprotein bound to ADP has an external diameter of 100 Å and an internal lumen with a diameter of 50 Å.As is also observed for TrwB, the entrance of the channel is narrow, approximately 10 Å in diameter. On the basis of this crystal structure, it was postulated that the VirB11 family of ATPases function as hexameric pores, the closure and opening of which is regulated by the concerted binding and release of ATP, respectively. Recently, the predicted dynamic nature of these ATPases was supported by studies showing that nucleotide binding ‘locks’ the hexamer into a symmetric and compact structure, whereas in the absence of nucleotide, the amino-terminal domains show a collection of rigid-body conformations66. So, the structural findings validate earlier models in which the VirB11 family of ATPases direct the assembly of the T4S machine and/or drive the passage of substrates across the envelope through dynamic conformational changes that are mediated by ATP binding and release.
Information about the VirB4 family of ATPases is emerging67,68. For A. tumefaciens VirB4, enzymatic activity is required for substrate export, and this ATPase also seems to function as a homomultimer in vivo. VirB4 possesses two domains that embed into, or protrude across, the cytoplasmic membrane, possibly forming contacts with machine subunits across the inner membrane68. Recently, evidence was presented for an interaction between VirB4 and VirB11 (ref. 57). An intriguing area for further study is how these two ATPases functionally interact.
Extracellular filaments
Conjugation systems elaborate several morphologically distinct pili. These pili can be long and flexible, for example the F-plasmid pilus2, or short and rigid, for example, the RP4-plasmid pilus48. These pili have long been considered essential for substrate transfer, minimally by promoting mating-pair formation. For the A. tumefaciens T4S system, the T-pilus morphologically resembles the RP4 pilus and, as noted above, is composed of VirB2 pilin and the associated VirB5 protein and VirB7 lipoprotein48–50. Early work showed the importance of the T-pilus for the delivery of DNA and protein substrates into plant cells. Intriguingly however, recent studies have identified mutations in the VirB11 and VirB6 subunits that ‘uncouple’ pilus biogenesis from substrate transfer. Some mutations do not affect assembly of a wild-type pilus, but block substrate transfer, whereas, conversely, others prevent pilus formation without affecting substrate transfer59,69. So, the formation of a conjugative pilus extending from the cell surface is not, in fact, an obligatory feature of conjugation machines of Gram-negative bacteria.
Although morphological variations exist, all conjugative pili are thought to be composed of a single pilin subunit that forms a helical filament of ~8–16 nm in diameter. Intriguingly, recent studies have shown that several effector translocator systems elaborate completely new types of extracellular appendages — for example, by the Cag T4S system of H. pylori. In one study, a rigid needle-like structure that is covered by a sheath was shown to protrude from the poles of H. pylori cells70. The sheathed structures differ morphologically from the conjugative pili and they are much larger — the needle structure is ~40 nm in diameter and the cross-section of the sheathed structure is ~70 nm. Of considerable further interest, the sheath is composed, at least in part, of HP0527, a protein that has sequence similarity to A. tumefaciens VirB10, whereas at the base of the organelle there is a cluster of the lipoprotein HP0532, a protein that is related to A. tumefaciens VirB7. In fact, HP0527 is a large protein with five domains, of which only one is homologous to VirB10. One domain — the middle repeat region (MRR) — consists of 74 contiguous segments of six different consensus sequences of variable lengths between 5 and 14 residues. Intragenic frame-shifts result in derivatives of HP0527 of differing sizes and antigenicities, and so it is postulated that HP0527 covers the T4S filament to protect it from a deleterious host antibody response during infection70.
A second study also identified HP0532 (VirB7-like) as a component of a T4S filament extending from the H. pylori cell surface71. Furthermore, evidence was presented for the association of HP0528, a VirB9 homologue, with the filament. As expected from work on the conjugation machines, the H. pylori CP (HP0524) was not required for formation of the filament. Interestingly, however, HP0525 (a VirB11 ATPase homologue) was found to be dispensable for HP0532 (a VirB7 homologue) surface localization and association with extracellular filaments. These observations prompted a suggestion that HP0525 contributes to morphogenesis or function of the Cag T4S system in ways that are distinct from those of other VirB11 ATPases in mediating assembly of conjugative pili71. Although this proposal needs further study, it is nevertheless evident that the H. pylori Cag system elaborates filaments that differ strikingly in composition and morphology from the T-pili of conjugation systems. However, a feature that might be common to T4S appendages is the association of a lipoprotein at the base, or along the length, of the filament.
Recent studies of B. tribocorum have identified two T4S systems that are both required for pathogenesis26,72. One of these systems is highly similar to the transfer system of the IncW plasmid R388, with the exception that it lacks a CP homologue. In addition, of particular interest with respect to pilus assembly, the B. tribocorum Trw-PAI system carries multiple tandem duplications of trwL, which codes for a homologue of VirB2 pilin, and of the trwJIH genes, which code for homologues of the VirB5 and VirB7 pilus-associated proteins, as well as a VirB6 homologue26. In view of the discovery that the sheath surrounding the H. pylori needle is composed of an antigenically variable HP0527 protein, it is intriguing to speculate that B. tribocorum combines its repertoire of pilus proteins to achieve the same goal as H. pylori — production of antigenically variable surface appendages to aid the infection process.
Finally, the L. pneumophila Dot/Icm system also elaborates a surface structure that is morphologically distinct from the conjugative pili73. This is a diffuse web of fibrous material around the cell surface that is composed of the DotO and DotH proteins. During the infection cycle, this material is produced transiently, shortly before the bacteria burst from the macrophage and spread to other host cells. These observations prompted a proposal that the DotO/H fibres facilitate lysis out of the macrophage or infection of neighbouring cells73.
Possible T4S translocation routes
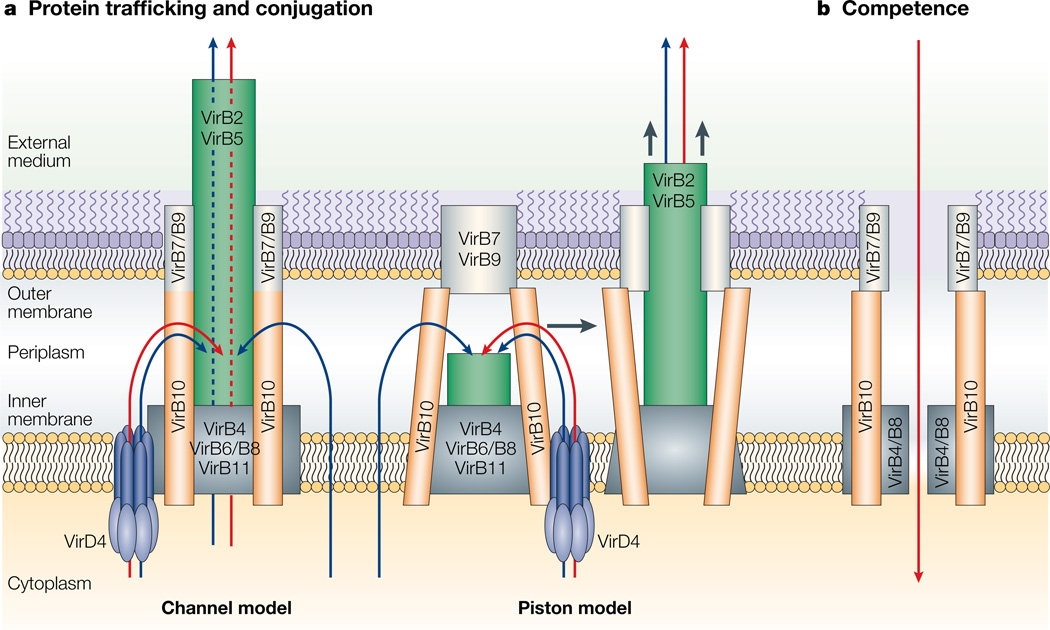
The CP must coordinate its activities with the Mpf structure to direct the passage of substrates across the cell envelope. Although the molecular details of the translocation route are at present unknown, we envision two working models (Fig. 3). According to the ‘channel’ model, the CP (for example, VirD4) recruits and then delivers DNA and protein substrates to an Mpf channel. Substrates might access the channel at the cytoplasmic face of the inner membrane, where the role of the CP is restricted to that of a recruitment factor, or in the periplasm, where the CP functions as an inner-membrane translocase. On engagement with the Mpf channel, substrates are then delivered through the lumen of a pilus-like structure extending, minimally, through the cell envelope. Alternatively, the ‘piston’ model is more dynamic as substrates are first translocated across the inner membrane by the CP, then, in the periplasm, they are delivered to the tip of a pilus-like structure. By a piston-like action, the rudimentary pilus extends and pushes the substrates across the outer membrane. On substrate export, the pilus retracts for a second cycle of translocation. It is noteworthy that the F-plasmid T4S machine elaborates a retractile pilus2, although this dynamic activity has yet to be shown for other T4S surface filaments. Also of interest, the B. pertussis Ptl system requires PtlA, a VirB2 pilin homologue, for PT export, yet so far no extracellular pilus has been detected. Additionally, recent work has established that the S1 subunit of PT associates with the outer membrane and so might nucleate PT assembly at the outer membrane. These observations indicate that the Ptl system extrudes PT across the outer membrane by a piston-like mechanism74. At this time, however, both the channel and piston models can accommodate these and other recent experimental findings52,75,76.
Figure 3. Models of type IV secretion (T4S) system-mediated substrate translocation.
The Agrobacterium tumefaciens VirB/D4 system is presented as a model (a), with the possible architecture in accordance with the results of topological (Fig. 2) and interaction (Table 2) studies. Two working models are depicted – the ‘channel’ model in which the pilus acts as a channel for passage of the substrate across the cell envelope, and the ‘piston’ model in which the pilus acts as a piston motor, pushing the substrates to the medium or into the eukaryotic cell. Possible translocation routes are represented (blue arrows represent protein substrates; red arrows represent DNA substrates). DNA and protein substrates might be translocated through the same or different pathways, using the coupling protein (CP; for example, VirD4), the mating-pore-formation (Mpf) complex, the general secretion pathway (GSP) or another pathway for secretion across the inner membrane. The Mpf structure is used to deliver substrates across the outer membrane. The competence model (b) is shown as a comparison.
Consequences of effector translocation
Conjugation, competence or other gene-transfer mechanisms often endow a bacterium with the capacity to survive in changing environments through the acquisition of adaptive traits. Conversely, both the T3S77 and T4S effector translocators seem to have evolved for the opposite purpose: to render the harsh environment of the eukaryotic host a more habitable place. This is achieved through subversion of a myriad of host cellular processes, as illustrated below for the four systems for which effector molecules have been identified so far (Fig. 4; Table 3).
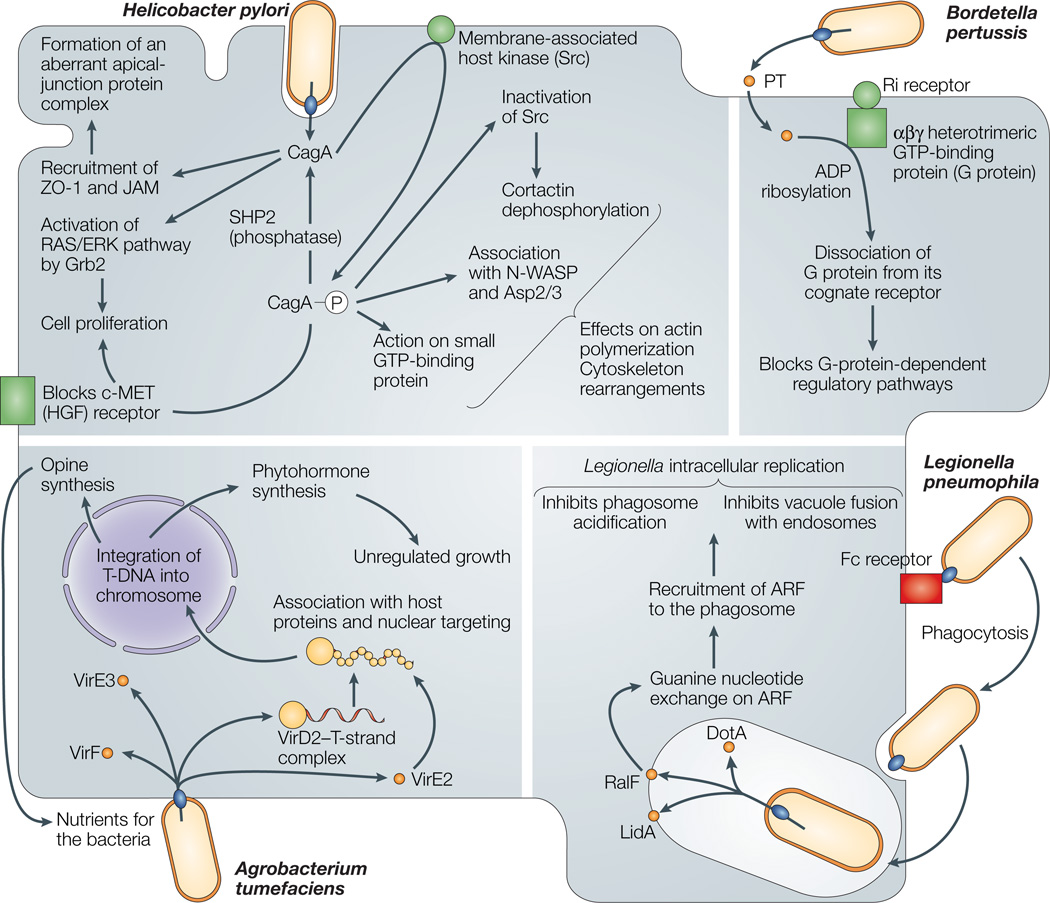
Figure 4. Schematic representation of the cellular consequences of type IV secretion (T4S) system effector translocation.
T4S effector translocation alters various eukaryotic cellular processes, as illustrated for the four systems in which effector molecules have been identified so far. Agrobacterium tumefaciens delivery of T-DNA and effector proteins induces synthesis of opine food substrates and also induces tumour production through modulation of phytohormone levels. Helicobacter pylori CagA modulates various pathways associated with eukaryotic-cell differentiation, proliferation and motility. Bordetella pertussis pertussis toxin (PT) interferes with G-protein-dependent signalling pathways, and Legionella pneumophila RalF recruits the ARF (ADP ribosylation factor) family of guanosine triphosphatases to the phagosome to promote intracellular survival.
Table 3.
Type IV secretion substrates and host interacting partner proteins*
| Bacteria | Exported substrates | Host-cell interacting partners |
|---|---|---|
|
Agrobacterium tumefaciens |
VirD2 | AtKAPα, CypA, PP2C, RocA, Roc4 |
| VirE2 | VIP1, VIP2 | |
| VirE3 | ? | |
| VirF | ASK (Skp1-like), VIP1 | |
| Helicobacter pylori | CagA | c-Src, SHP2, c-MET Grb2, ARP2/3, Csk, Rho GTPases, (Rac1,Cdc42), N-WASP, ZO-1, JAM |
| Bordetella pertussis | Pertussis toxin | αβγ heterotrimeric G proteins |
| Legionella pneumophila | RalF | ADP ribosylation factor (ARF) |
| LidA | ? | |
| DotA | ? | |
| Brucella spp. | ? | ? |
| Bartonella spp. | ? | ? |
| Rickettsia spp. | RalF? | ? |
Some protein–protein contacts might be mediated by mutual interactions with partner proteins.
A. tumefaciens T-DNA and effector protein transfer
A. tumefaciens translocates T-DNA and effector proteins to induce plant cells to synthesize opine food substrates and to induce proliferation of the transformed plant cells. The outcome of infection is a plant tumour, known as a crown gall, which for the bacterium represents a good ecological niche as it acts as a food-producing factory (Fig. 4). The A. tumefaciens T4S system is both a conjugation system and an effector translocator. It delivers oncogenic T-DNA as a single-stranded T-strand that is covalently bound at its 5′ end to the VirD2 relaxase11, and it independently translocates three protein effectors, VirE2 (ref. 78), VirE3 (ref. 42) and VirF (ref. 40). Whereas VirE2 interacts with the T-strand VirD2 particle to form the so-called T-complex, VirE3 and VirF participate in largely unspecified ways to promote infection of certain plant species. VirD2 and VirE2 carry nuclear-localization sequences (NLS) that are thought to mediate interactions with plant cellular factors for nuclear targeting and import, and T-DNA integration into the host genome.
Specific interactions between these two bacterial proteins and several eukaryotic factors have been identified (Table 3). For example, VirD2 binds three members of the Arabidopsis cyclophilin chaperone family; these interactions might maintain the proper conformation of VirD2 in the host-cell cytoplasm or nucleus during T-complex transit79. VirD2 also interacts with a serine/threonine phosphatase (PP2C) and, correspondingly, there is evidence for VirD2 phosphorylation in plant tissues. In fact, A. tumefaciens shows enhanced virulence following infection of an Arabidopsis strain bearing a PP2C gene (abi1) mutation, and so it is proposed that PP2C acts to suppress virulence by dephosphorylation of VirD2 (ref. 80). VirD2 also interacts with a member of the Arabidopsis karyopherin-α family, AtKAPα; these proteins mediate the nuclear import of NLS-containing proteins, indicating their involvement in nuclear import of VirD2 and, hence, the T-DNA81.
VirE2 interacts with two Arabidopsis proteins termed VIP1 and VIP2. These proteins localize to plant nuclei and so also probably facilitate delivery of the T-complex to its site of integration81. Similarly, the Ran GTPase is implicated in T-complex targeting to the nucleus81. Mutational studies of Arabidopsis, as well as a surrogate yeast host, are uncovering many additional cellular factors that contribute to successful T-strand delivery and integration into the plant genome80–83. Given the large numbers of cellular factors identified so far, it is likely that the T-DNA and the reported effector proteins represent only a subset of the molecules translocated by the VirB/D4 T4S system during infection.
H. pylori CagA transfer
When H. pylori cells carrying the cag pathogenicity island attach to cultured AGS cells, a human gastric adenocarcinoma epithelial cell line, they induce the ‘hummingbird phenotype’, the features of which include cytoskeletal rearrangements, cell elongation and increased cell motility84 (Fig. 4). For induction of this phenotype, H. pylori uses the Cag T4S system to translocate the ~145-kDa CagA protein into the eukaryotic cell. As discussed above, mechanistic studies of the Cag T4S system are advancing rapidly — HP0525 now represents a structural paradigm for the VirB11 family of ATPases65 and new extracellular appendages have been visualized70,71. In parallel, studies by several laboratories are generating extensive information about the cellular consequences of CagA translocation. On transfer, CagA localizes on the inner surface of the plasma membrane where it interacts with and is phosphorylated by the Src family of protein tyrosine kinases, such as c-Src84–91. The CagA phosphorylation sites have been mapped to the so-called EPIYA motifs that share homology with c-Src consensus phosphorylation sites and are present in variable numbers in the carboxy-terminal half of the protein89,91.
Both non-phosphorylated and phosphorylated forms of CagA alter the activities of a large number of cellular factors that are associated with distinct signalling pathways (Fig. 4; Table 3). For example, on phosphorylation, CagAP-Tyr inactivates c-Src kinase activity, which results in dephosphorylation of another c-Src substrate, cortactin91. Dephosphorylated cortactin relocalizes in the cell and is thought to have enhanced ACTIN cross-linking activity, causing actin rearrangements associated with the hummingbird phenotype. Additionally, CagA interacts with Src homology-2 (SH2) domain-containing proteins. A CagA interaction with the tyrosine phosphatase SHP2 stimulates SHP2 phosphatase activity, which, in turn, might be responsible for the observed dephosphorylation of several cellular proteins92. Intriguingly, CagA also interacts with the hepatocyte growth factor/scatter factor (HGF/SF) receptor, c-MET, which is involved in invasive growth of tumour cells. This interaction leads to deregulation of the c-MET signal-transduction pathway and induction of the motogenic response93. CagA interactions with additional eukaryotic factors are implicated (Table 3), although in several cases further studies are needed to determine whether the suspected contacts are mediated by a mutual partner protein.
Non-phosphorylated CagA binds directly to Grb2 by its EPIYA motifs, and this interaction is thought to activate the stress-kinase pathway, resulting in the scatter phenotype94. Moreover, a recent study further showed that non-phosphorylated CagA associates with the epithelial tight-junction scaffolding protein, ZO-1, and the transmembrane protein junctional adhesion molecule (JAM)95 (Fig. 4). These interactions induce the formation of an aberrant APICAL JUNCTION protein complex, which results in the loss of cell polarity, proliferation and differentiation. The finding that CagA recruitment of ZO-1 and JAM occurs independently of CagA phosphorylation led to a proposal that CagA might mediate its effects on host cells through at least two functional domains, one that interacts with SH2-domain-containing proteins and another that interacts with components of the apical junction complex95.
Finally, although the interactions of CagA with several eukaryotic factors exert complex effects on signalling networks that lead to H. pylori-mediated cancer onset and tumour progression, the broad range of cellular consequences accompanying CagA translocation tell only part of the story. There is also compelling evidence that the Cag T4S machine, through receptor-dependent activation96 or translocation of an unidentified effector(s)43, operates independently of CagA and of HP0524 (VirD4) to elicit stress-response pathways that result in induction of IL-8 secretion.
B. pertussis PT export
As with A. tumefaciens and H. pylori, B. pertussis uses a type IV system (Ptl) to deliver its cargo (PT) to the mammalian cell surface. However, the Ptl system is unique among the known effector translocators in that its sole substrate is the multimeric PT and it functions exclusively to deliver PT across the outer membrane by a cell-contact-independent mechanism45,74 (Fig. 4; Table 3). Although it is known that PT assembles in the periplasm, possibly at the outer membrane through interactions with the outer-membrane-associated S1 subunit74, little else is known about the signals mediating export of the holotoxin.
However, PT itself is by far the best characterized of the T4S substrates and a crystal structure is available97. This is a hexameric A/B toxin of ~105 kDa that consists of an enzymatically active A component (S1 sub-unit) and a pentameric B component. The A component is delivered to the mammalian cell membrane by binding of the B pentamer to surface glycoproteins, and it is internalized by receptor-mediated endocytosis. Once in the cytosol, the A component ADP-ribosylates Gα isoforms of the Gi subfamily of heterotrimeric G proteins in the presence of βγ subunits. The consequence of ADP-ribosylation is the uncoupling of G proteins from their receptor, with alteration of all signals that are transduced by them. This can elicit different cellular consequences in different tissues, but common effects are increased insulin secretion and sensitization to histamine98.
L. pneumophila Dot/Icm substrates
In contrast to the pathogens described above, L. pneumophila and Brucella and Bartonella species are facultative intracellular pathogens, the infection cycles of which depend on type IV secretion after internalization into the host cell (Fig 1). The L. pneumophila infection cycle involves host-cell entry by phagocytosis, creation of a specialized vacuole for replication, replication and macrophage lysis, and infection of neighbouring cells99,100 (Fig. 4). Intriguingly, the Dot/Icm T4S system functions as a bona fide conjugation system that is closely related in ancestry to the ColIb-P9 plasmid-transfer system5,101,102. However, during infection L. pneumophila uses this transfer system to inject effector proteins into the phagosome, both to control biogenesis of the replicative vacuole and to modulate the activity of host factors involved in vesicle traffic.
So far, three Dot/Icm secretion substrates have been identified (Fig. 4; Table 3). Intriguingly, one is DotA, a protein that was originally reported to assemble as a polytopic protein in the inner membrane of L. pneumophila cells103. No known effector function has been identified for DotA, although an observation that purified DotA forms oligomeric ring structures prompted speculation that it might form a pore in the eukaryotic membrane for the passage of other T4S effectors103. Second, a protein termed LidA is translocated to the phagosome membrane44. In the bacterium, LidA regulates Dot/Icm assembly and is therefore thought to be one of the first translocated substrates. After delivery, LidA is postulated to function in vesicle recruitment during the biogenesis of the replication vacuole44.
Third, L. pneumophila use the Dot/Icm complex to export RalF, a protein with a Sec7 homology domain that in eukaryotes mediates guanine nucleotide exchange. RalF is translocated through the phagosomal membrane where it recruits and activates ARF1, a member of the adp ribosylation factor family of guanosine triphosphatases (GTPases) to the phagosomal membrane. Although the function of ARF1 during infection is not yet defined, RalF recruitment and activation of ARF1 is postulated to enhance the efficiency of replicative vacuole formation104. As with the A. tumefaciens and H. pylori T4S systems, there are recent indications from several laboratories that the L. pneumophila Dot/Icm system might translocate additional effectors during its infection cycle.
Concluding remarks and future directions
The functional versatility of the T4S systems is unparalleled among the known bacterial translocation machines, as evidenced by the fact that these systems export both DNA and protein substrates by cell-contact-dependent and cell-contact-independent mechanisms, and also import DNA from the extracellular milieu. Furthermore, these systems translocate DNA and protein substrates to phylogenetically diverse taxa, including many bacterial species and, astoundingly, many different eukaryotic cell types. The cellular consequences of transfer are numerous, ranging from the introduction of heritable alterations in genomic structure to the transient alteration of a myriad of eukaryotic physiological processes and signalling pathways. In spite of this diversity of function, however, it is clear that the T4S systems have all evolved for a single purpose: to promote bacterial survival and propagation in the face of a changing environment.
Investigations of T4S machines are advancing extremely rapidly, making this arguably one of the most exciting areas of study in the field of bacterial pathogenesis. In the very near future, the mechanistic studies, using a combination of classical genetics and biochemical, molecular, and state-of-the-art cell-biological and structural technologies, promise to offer unprecedented insights into a 50-year old question — how do bacteria conjugally transfer DNA to bacterial recipients? The answer will be exciting, but as we now know, in the past 50 years many more questions have surfaced about these machines that also await investigation (Box 2). Studies of many different T4S machines are crucial to the advancement of our knowledge, because already many mechanistic variations on the conjugation themes have been identified. Besides, discoveries of the new type IV effector translocator systems have opened up an entirely new field of study that is aimed at understanding how bacteria suppress, mimic, subvert or otherwise ‘hijack’ eukaryotic cellular processes to promote their own selfish means. Identifying the effectors and uncovering the spectrum of cellular consequences they exact on their targets will supply new information about pathogenic strategies and, equally importantly, contribute to a broader understanding of complex signalling networks in eukaryotic cells.
Box 2 | Questions that remain to be answered.
What are the overall architectures of T4S machines?
Is the coupling protein a DNA and/or protein translocase? Do alternative/redundant pathways exist for substrate trafficking across the inner membrane? How are substrates delivered across the outer membrane of Gram-negative bacteria?
How do the type IV ATPases direct machine assembly, pilus biogenesis and substrate transfer?
What are the functions of the extracellular pili and other type-IV-dependent surface structures?
What are the identities and cellular activities of other T4S effector proteins?
Acknowledgements
We dedicate this review to Brian Wilkins in loving memory. We apologize for any omissions in citation of primary reports owing to space limitations. We thank members of the laboratory for helpful comments and critical appraisals of this manuscript. We also gratefully acknowledge the National Institutes of Health for supporting our studies of the Agrobacterium VirB/D4 T4S system.
Glossary
- TYPE IV SECRETION (T4S) APPARATUS OR SYSTEM
A bacterial organelle that is ancestrally related to a conjugation machine that translocates DNA or protein substrates across the cell envelope, often for purposes associated with pathogenesis. Other bacterial translocation systems include the type II secretion (T2S) machines that deliver protein substrates across the outer membrane and the type III secretion (T3S) machines that translocate effectors in one step across the cell envelope through a structure that is ancestrally related to the bacterial flagellum. Like the T4S systems, the T3S systems elaborate syringe- or pilus-like surface organelles and deliver effector proteins to plant and mammalian cells during infection.
- CONJUGATION
A mechanism for transfer of a DNA substrate from a bacterial donor cell to a recipient cell by direct cell-to-cell contact.
- PILUS
A filamentous organelle that extends from the surface of the bacterial cell. Composed of pilin subunits, these structures mediate attachment to target cells or inert matter. They might also participate directly in the delivery of secretion substrates to target cells.
- COMPETENCE
The ability of a bacterial cell to import exogenous DNA and stably incorporate it into the bacterial genome.
- PERIPLASM
An aqueous compartment between the inner and outer membranes of Gram-negative bacteria.
- CHAPERONE
A protein or protein complex that participates in folding or unfolding of protein substrates. Secretion chaperones prevent their substrates from aggregating or interacting prematurely with other substrates or cellular factors. They might also mediate the delivery of substrates to a secretory apparatus.
- SECRETION SIGNAL
A motif that confers recognition of a protein that is destined for export by a cognate secretory apparatus.
- GENERAL SECRETORY PATHWAY (GSP)
The main pathway used for delivery of protein substrates into or across the bacterial inner membrane.
- INTERLEUKIN-8 (IL-8)
A peptide that is produced by epithelial cells and is an indicator of infection. IL-8 secretion is induced by pathogens preceding clinical complications.
- TRANSGLYCOSYLASE
A protein, the enzymatic activity of which degrades peptidoglycan. These proteins participate in assembly of supramolecular transenvelope structures by ‘punching’ holes in the peptidoglycan.
- PEPTIDOGLYCAN
A shape-determining polymer that is present within the periplasm of Gram-negative bacteria.
- SECRETIN
A protein that forms oligomeric pores to allow the passage of macromolecular substrates across the outer membrane.
- KARYOPHERIN
These proteins have a central role in nuclear import processes, mediating substrate recognition and release at the nuclear-pore complex by GTP-hydrolysis-dependent reactions.
- CORTACTIN
Cortactin is an actin-binding protein, regulated by the membrane-associated c-Src kinase. Cortactin transduces signals from the cell surface to the cytoskeleton.
- ACTIN
A eukaryotic protein that polymerizes to form microfilaments. Microfilaments have a dual role, acting as a passive structural complex that maintains cell shape and anchors cytoskeletal proteins, and an active function for the transport of vesicles and organelles that can result in cell movement.
- APICAL JUNCTION
Apical junctions consist of protein complexes that join the actin cytoskeleton to the apical pole of epithelial cells. Adherent junctions have pivotal roles in cell organization by mediating cell adhesion and signalling.
- ADP RIBOSYLATION FACTOR (ARF)
The ADP-ribosylation factor family of small GTP-binding proteins is involved in the regulation of membrane traffic (vesicular transport) and in the control of the actin cytoskeleton.
Footnotes
Online links
DATABASES
The following terms in this article are linked online to:
Entrez: http://www.ncbi.nlm.nih.gov/Entrez/
HP0527 | HP0528 | HP0532
Protein Data Bank: http://www.rcsb.org/pdb/
HP0525 | TrwB
SwissProt: http://www.ca.expasy.org/sprot/
ARF1 | JAM | VirB1 | VirB11 | VirD4
FURTHER INFORMATION
Peter J. Christie’s laboratory: http://mmg.uth.tmc.edu/webpages/faculty/pchristie.html
World Health Organization: http://who.int/en/
Access to this interactive links box is free online.
References
- 1.Cavelli-Sforza L, Lederberg J, Lederberg E. An infective factor controlling sex compatibility. B. coli. J. Gen. Microbiol. 1953;8:89–103. doi: 10.1099/00221287-8-1-89. [DOI] [PubMed] [Google Scholar]
- 2.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 3.Winans SC, Burns DL, Christie PJ. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Censini S, et al. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl Acad. Sci. USA. 1997;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila . Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. Reports the discovery of a type IV secretion system that is evolutionarily related to the IncI plasmids and functions dually in conjugative transfer and effector translocation
- 6.Bacon DJ, et al. Involvement of a plasmid in virulence of Campylobacter jejuni 81–176. Infect. Immun. 2000;68:4384–4390. doi: 10.1128/iai.68.8.4384-4390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 2001;41:379–391. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 8. Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. References 6–8 report the discoveries of new type IV translocation systems, herein termed the ‘DNA uptake and release’ subfamily
- 9.Bundock P, den Dulk Ras A, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae . EMBO J. 1995;14:3206–3214. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bates S, Cashmore AM, Wilkins BM. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae . J. Bacteriol. 1998;180:6538–6543. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu J, et al. The bases of crown gall tumorigenesis. J. Bacteriol. 2000;182:3885–3895. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waters VL. Conjugation between bacterial and mammalian cells. Nature Genet. 2001;29:375–376. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 13.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de la Cruz I, Davies J. Horizontal gene transfer and the origin of species: lessons from bacteria. Trends Microbiol. 2000;8:128–133. doi: 10.1016/s0966-842x(00)01703-0. [DOI] [PubMed] [Google Scholar]
- 15.Ghigo JM. Natural conjugative plasmids induce bacterial biofilm development. Nature. 2001;412:442–445. doi: 10.1038/35086581. [DOI] [PubMed] [Google Scholar]
- 16.Schlievert PM, et al. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 1998;66:218–223. doi: 10.1128/iai.66.1.218-223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bacon DJ, et al. DNA sequence and mutational analyses of the pVir plasmid of Campylobacter jejuni 81–176. Infect. Immun. 2002;70:6242–6250. doi: 10.1128/IAI.70.11.6242-6250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofreuter D, Odenbreit S, Puls J, Schwan D, Haas R. Genetic competence in Helicobacter pylori: mechanisms and biological implications. Res. Microbiol. 2000;151:487–491. doi: 10.1016/s0923-2508(00)00164-9. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton HL, Schwartz KJ, Dillard JP. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 2001;183:4718–4726. doi: 10.1128/JB.183.16.4718-4726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen I, Dubnau D. DNA transport during transformation. Front. Biosci. 2003;8:S544–S556. doi: 10.2741/1047. [DOI] [PubMed] [Google Scholar]
- 21.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl Acad. Sci. USA. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christie PJ. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilbi H, Segal G, Shuman HA. Icm/Dot-dependent upregulation of phagocytosis by Legionella pneumophila . Mol. Microbiol. 2001;42:603–617. doi: 10.1046/j.1365-2958.2001.02645.x. [DOI] [PubMed] [Google Scholar]
- 24.Schmiederer M, Arcenas R, Widen R, Valkov N, Anderson B. Intracellular induction of the Bartonella henselae virB operon by human endothelial cells. Infect. Immun. 2001;69:6495–6502. doi: 10.1128/IAI.69.10.6495-6502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boschiroli ML, et al. The Brucella suis virB operon is induced intracellularly in macrophages. Proc. Natl Acad. Sci. USA. 2002;99:1544–1549. doi: 10.1073/pnas.032514299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seubert A, Hiestand R, de la Cruz F, Dehio C. A bacterial conjugation machinery recruited for pathogenesis. Mol. Microbiol. 2003;49:1253–1266. doi: 10.1046/j.1365-2958.2003.03650.x. [DOI] [PubMed] [Google Scholar]
- 27.Rouot B, et al. Production of the type IV secretion system differs among Brucella species as revealed with VirB5- and VirB8-specific antisera. Infect. Immun. 2003;71:1075–1082. doi: 10.1128/IAI.71.3.1075-1082.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baron C, O’Callaghan D, Lanka E. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 2002;43:1359–1365. doi: 10.1046/j.1365-2958.2002.02816.x. [DOI] [PubMed] [Google Scholar]
- 29.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 30.Gomis-Ruth FX, Coll M. Structure of TrwB, a gatekeeper in bacterial conjugation. Int. J. Biochem. Cell Biol. 2001;33:839–843. doi: 10.1016/s1357-2725(01)00060-7. [DOI] [PubMed] [Google Scholar]
- 31. Gomis-Ruth FX, et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–641. doi: 10.1038/35054586. Reports the first structure of a coupling protein, a hexameric F1-ATPase-like multimer that is highly indicative of a translocase function
- 32.Hormaeche I, et al. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J. Biol. Chem. 2002;277:46456–46462. doi: 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 33.Errington J, Bath J, Wu LJ. DNA transport in bacteria. Nature Rev. Mol. Cell Biol. 2001;2:538–545. doi: 10.1038/35080005. [DOI] [PubMed] [Google Scholar]
- 34.Schroder G, et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 2002;184:2767–2779. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol. Microbiol. 2003;49:105–116. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- 36.Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl Acad. Sci. USA. 2003;100:10465–10470. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 2002;43:1523–1532. doi: 10.1046/j.1365-2958.2002.02829.x. [DOI] [PubMed] [Google Scholar]
- 38.Ding Z, et al. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a type IV secretion system. J. Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at the cell poles of Agrobacterium tumefaciens . Mol. Microbiol. 2003;49:1699–1713. doi: 10.1046/j.1365-2958.2003.03669.x. Reports the first direct evidence for recruitment of a protein effector to a coupling protein, supporting a proposal that the coupling proteins function as general recruitment factors for type IV secretion substrates
- 40. Vergunst AC, et al. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–982. doi: 10.1126/science.290.5493.979. This elegant study supplied the first incontrovertible evidence for protein trafficking by the A. tumefaciens VirB/D4 type IV secretion system
- 41.Simone M, McCullen CA, Stahl LE, Binns AN. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 2001;41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 42.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jacome E, Hooykaas PJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucl. Acids Res. 2003;31:860–868. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Selbach M, Moese S, Meyer TF, Backert S. Functional analysis of the Helicobacter pylori cag pathogenicity island reveals both VirD4-CagA-dependent and VirD4-CagA-independent mechanisms. Infect. Immun. 2002;70:665–667. doi: 10.1128/iai.70.2.665-671.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol. Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. Describes the discovery of a novel effector, LidA, that is required for elaboration of the L. pneumophila Dot/Icm complex in the bacterium and, on translocation, functions in vesicle recruitment during biogenesis of the replication vacuole
- 45.Burns DL. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 2003;6:1–6. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 46.O’Callaghan D, et al. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis . Mol. Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 47.Lai EM, Kado CI. The T-pilus of Agrobacterium tumefaciens . Trends Microbiol. 2000;8:361–369. doi: 10.1016/s0966-842x(00)01802-3. [DOI] [PubMed] [Google Scholar]
- 48. Eisenbrandt R, et al. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 1999;274:22548–22555. doi: 10.1074/jbc.274.32.22548. Provides evidence that the pilin subunits of the A. tumefaciens VirB/D4 and the plasmid RP4 Tra systems undergo head-to-tail cyclization during maturation, a completely new reaction in bacteria
- 49.Schmidt-Eisenlohr H, et al. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens . J. Bacteriol. 1999;181:7485–7492. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 2001;183:3642–3651. doi: 10.1128/JB.183.12.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baron C, Llosa M, Zhou S, Zambryski PC. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J. Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 1994;176:3646–3660. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar RB, Xie YH, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol. Microbiol. 2000;36:608–617. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- 54.Das A, Anderson LB, Xie YH. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J. Bacteriol. 1997;179:3404–3409. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beaupré CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 2000;182:758–763. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ward DV, Draper O, Zupan JR, Zambryski PC. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl Acad. Sci. USA. 2002;99:11493–11500. doi: 10.1073/pnas.172390299. Reports a comprehensive linkage map of the VirB proteins by yeast dihybrid screens
- 58.Krall L, et al. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens . Proc. Natl Acad. Sci. USA. 2002;99:11405–11410. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 2003;185:2867–2878. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rambow-Larsen AA, Weiss AA. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J. Bacteriol. 2002;184:2863–2869. doi: 10.1128/JB.184.11.2863-2869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farizo KM, Cafarella TG, Burns DL. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 1996;271:31643–31649. doi: 10.1074/jbc.271.49.31643. [DOI] [PubMed] [Google Scholar]
- 62.Harris RL, Hombs V, Silverman PM. Evidence that F-plasmid proteins TraV, TraK and TraB assemble into an envelope-spanning structure in Escherichia coli . Mol. Microbiol. 2001;42:757–766. doi: 10.1046/j.1365-2958.2001.02667.x. [DOI] [PubMed] [Google Scholar]
- 63.Liu Z, Binns AN. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J. Bacteriol. 2003;185:3259–3269. doi: 10.1128/JB.185.11.3259-3269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: Identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl Acad. Sci. USA. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yeo H-J, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol. Cell. 2000;6:1461–1472. doi: 10.1016/s1097-2765(00)00142-8. Reports the first structure of a member of the VirB11 superfamily of traffic ATPases
- 66.Savvides SN, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–1980. doi: 10.1093/emboj/cdg223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rabel C, Grahn AM, Lurz R, Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J. Bacteriol. 2003;185:1045–1058. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dang TA, Zhou X-R, Graf B, Christie PJ. Dimerization of the Agrobacterium tumefaciens VirB4 ATPase and the effect of ATP-binding cassette mutations on assembly and function of the T-DNA transporter. Mol. Microbiol. 1999;32:1239–1251. doi: 10.1046/j.1365-2958.1999.01436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sagulenko Y, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J. Bacteriol. 2001;183:5813–5825. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol. Microbiol. 2003;49:219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 71. Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell. Microbiol. 2003;5:395–404. doi: 10.1046/j.1462-5822.2003.00286.x. References 70 and 71 report that the Cag type IV secretion system elaborates new sheathed surface structures that contribute to H. pylori pathogenesis
- 72.Schulein R, Dehio C. The VirB/VirD4 type IV secretion system of Bartonella is essential for establishing intraerythrocytic infection. Mol. Microbiol. 2002;46:1053–1067. doi: 10.1046/j.1365-2958.2002.03208.x. [DOI] [PubMed] [Google Scholar]
- 73.Watarai M, Andrews HL, Isberg R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol. Microbiol. 2001;39:313–329. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 74.Farizo KM, Fiddner S, Cheung AM, Burns D. L Membrane localization of the S1 subunit of pertussis toxin in Bordetella pertussis and implications for pertussis toxin secretion. Infect. Immun. 2002;70:1193–1201. doi: 10.1128/IAI.70.3.1193-1201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grahn AM, Haase J, Bamford DH, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J. Bacteriol. 2000;182:1564–1574. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol. Microbiol. 2002;45:1325–1335. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 77.Galan JE, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 78.Ward DV, Zambryski PC. The six functions of Agrobacterium VirE2. Proc. Natl Acad. Sci. USA. 2001;98:385–386. doi: 10.1073/pnas.98.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng W, et al. Agrobacterium VirD2 protein interacts with plant host cyclophilins. Proc. Natl Acad. Sci. USA. 1998;95:7040–7045. doi: 10.1073/pnas.95.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gelvin SB. Agrobacterium-mediated plant transformation: the biology behind the ‘gene-jockeying’ tool. Microbiol. Mol. Biol. Rev. 2003;67:16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tzfira T, Citovsky V. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium . Trends Cell Biol. 2002;12:121–129. doi: 10.1016/s0962-8924(01)02229-2. [DOI] [PubMed] [Google Scholar]
- 82.Zhu Y, et al. Identification of Arabidopsis rat mutants. Plant Physiol. 2003;132:494–505. doi: 10.1104/pp.103.020420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roberts RL, et al. Purine synthesis and increased Agrobacterium tumefaciens transformation of yeast and plants. Proc. Natl Acad. Sci. USA. 2003;100:6634–6639. doi: 10.1073/pnas.1132022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Segal E, Cha J, Lo J, Falkow S, Tompkins L. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori . Proc. Natl Acad. Sci. USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hatakeyama M. Helicobacter pylori CagA — a potential bacterial oncoprotein that functionally mimics the mammalian Gab family of adaptor proteins. Microbes Infect. 2003;5:143–150. doi: 10.1016/s1286-4579(02)00085-0. [DOI] [PubMed] [Google Scholar]
- 86.Asahi M, et al. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 2000;191:593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl Acad. Sci. USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Odenbreit S, et al. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- 89.Stein M, et al. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- 90. Backert S, et al. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2000;2:155–164. doi: 10.1046/j.1462-5822.2000.00043.x. References 84, 86, 87, 88 and 90 report the first evidence for CagA translocation and subsequent phosphorylation in human cells
- 91.Selbach M, Moese S, Hauck CR, Meyer TF, Backert S. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo . J. Biol. Chem. 2002;277:6775–6778. doi: 10.1074/jbc.C100754200. [DOI] [PubMed] [Google Scholar]
- 92.Higashi H, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 93.Churin Y, et al. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J. Cell Biol. 2003;161:249–255. doi: 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mimuro H, et al. Grb2 is a key mediator of Helicobacter pylori CagA protein activities. Mol. Cell. 2002;10:745–755. doi: 10.1016/s1097-2765(02)00681-0. [DOI] [PubMed] [Google Scholar]
- 95. Amieva MR, et al. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–1434. doi: 10.1126/science.1081919. Shows that non-phosphorylated CagA induces formation of an aberrant apical-junction protein complex, resulting in loss of cell polarity, proliferation and differentiation
- 96.Fischer W, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 2001;42:1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 97.Stein PE, et al. The crystal structure of pertussis toxin. Structure. 1994;2:45–57. doi: 10.1016/s0969-2126(00)00007-1. [DOI] [PubMed] [Google Scholar]
- 98.Albert PR, Robillard L. G-protein specificity: traffic direction required. Cell Signal. 2002;14:407–418. doi: 10.1016/s0898-6568(01)00259-5. [DOI] [PubMed] [Google Scholar]
- 99.Vogel JP, Isberg RR. Cell biology of Legionella pneumophila . Curr. Opin. Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- 100.Swanson MS, Sturgill-Koszycki S. Exploitation of macrophages as a replication niche by Legionella pneumophila . Trends Microbiol. 2000;8:47–49. doi: 10.1016/s0966-842x(99)01674-1. [DOI] [PubMed] [Google Scholar]
- 101.Segal G, Purcell M, Schuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc. Natl Acad. Sci. USA. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Segal G, Shuman HA. Intracellular multiplication and human macrophage killing by Legionella pneumophila are inhibited by conjugal components of IncQ plasmid RSF1010. Mol. Microbiol. 1998;30:197–208. doi: 10.1046/j.1365-2958.1998.01054.x. [DOI] [PubMed] [Google Scholar]
- 103.Nagai H, Roy CR. The DotA protein from Legionella pneumophila is secreted by a novel process that requires the Dot/Icm transporter. EMBO J. 2001;20:5962–5970. doi: 10.1093/emboj/20.21.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–682. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 105.Boschiroli ML, et al. Type IV secretion and Brucella virulence. Vet. Microbiol. 2002;90:341–348. doi: 10.1016/s0378-1135(02)00219-5. [DOI] [PubMed] [Google Scholar]
- 106.Novak KF, Dougherty B, Pelaez M. italic> Actinobacillus actinomycetemcomitans harbours type IV secretion system genes on a plasmid and in the chromosome. Microbiology. 2001;147:3027–3035. doi: 10.1099/00221287-147-11-3027. [DOI] [PubMed] [Google Scholar]
- 107.Ohashi N, Zhi N, Lin Q, Rikihisa Y. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 2002;70:2128–2138. doi: 10.1128/IAI.70.4.2128-2138.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Masui S, Sasaki T, Ishikawa H. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J. Bacteriol. 2000;182:6529–6531. doi: 10.1128/jb.182.22.6529-6531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Andersson SG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 110.Bhattacharyya A, et al. Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl Acad. Sci. USA. 2002;99:12403–12408. doi: 10.1073/pnas.132393999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zusman T, Yerushalmi G, Segal G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila . Infect. Immun. 2003;71:3714–3723. doi: 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Burghdorf R, Forster C, Haas R, Fischer W. Topological analysis of a putative VirB8 homologue essential for the cag type IV secretion system in Helicobacter pylori . Int. J. Med. Microbiol. 2003;29:213–217. doi: 10.1078/1438-4221-00260. [DOI] [PubMed] [Google Scholar]
- 113.Cao TB, Saier MH., Jr Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: distribution, structural constraints, and evolutionary conclusions. Microbiology. 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]