Abstract
Background. The development of active tuberculosis disease has been shown to be multifactorial. Interactions between host and bacterial genotype may influence disease outcome, with some studies indicating the adaptation of M. tuberculosis strains to specific human populations. Here we investigate the role of the human leukocyte antigen (HLA) class I genes in this biological process.
Methods. Three hundred patients with tuberculosis from South Africa were typed for their HLA class I alleles by direct sequencing. Mycobacterium tuberculosis genotype classification was done by IS6110 restriction fragment length polymorphism genotyping and spoligotyping.
Results. We showed that Beijing strain occurred more frequently in individuals with multiple disease episodes (P < .001) with the HLA-B27 allele lowering the odds of having an additional episode (odds ratio, 0.21; P = .006). Associations were also identified for specific HLA types and disease caused by the Beijing, LAM, LCC, and Quebec strains. HLA types were also associated with disease caused by strains from the Euro-American or East Asian lineages, and the frequencies of these alleles in their sympatric human populations identified potential coevolutionary events between host and pathogen.
Conclusions. This is the first report of the association of human HLA types and M. tuberculosis strain genotype, highlighting that both host and pathogen genetics need to be taken into consideration when studying tuberculosis disease development.
Keywords: Mycobacterium tuberculosis, tuberculosis, human leukocyte antigens, host–pathogen, coadaptation, susceptibility
The human leukocyte antigen (HLA) class I molecules are involved in various biological processes and play an essential role in immunity [1]. These molecules act as multisite receptors; including peptides for antigen presentation, αβ T-cell receptors, and CD8 for cytotoxic T-cell response stimulation. The HLAs represent an unsurpassed example of polymorphism with thousands of SNPs identified to date [2]. This extreme diversity is believed to have occurred by selection events, allowing for the identification of peptide antigens from various pathogens and stimulation of an effective immune response by upregulation of the Th1 pathway [3]. HLA data for various populations demonstrate significant differences in allele frequencies between different geographical populations, with some HLA alleles completely absent from certain populations [4, 5]. The HLA genes were the first to be associated with susceptibility to tuberculosis, and many different HLA alleles have been associated in different populations [6].
The present-day population structure of several pathogens, including Mycobacterium leprae and Mycobacterium tuberculosis, can be attributed to ancient human migrations [7, 8]. Such long-standing host–pathogen interactions could lead to adaptive genetic changes in both the host and pathogen populations [9]. Evidence of this can be seen from studies that have shown that M. tuberculosis lineages have adapted to specific human populations [9–11] and the selection of strains from a distinct sublineage by a human population in a defined geographical setting [12]. In cosmopolitan settings, the association between particular M. tuberculosis complex (MTBC) lineages and their human hosts have remained even though some degree of intermingling of the different human populations has occurred [9–11].
Mycobacterium tuberculosis strains of the Beijing lineage are probably the most well characterized and are associated with increased virulence and transmission [13]. They are the most dominant strain lineage globally and have been reported in many Asian countries, and are emerging as the dominant strain in several other countries, including South Africa [14], Argentina, Cuba, Malawi, Vietnam, countries of the former Soviet Union, and parts of Western Europe [15]. Different M. tuberculosis strains induce different patterns of host immune response [16] as well as resulting in different disease phenotypes [17]. Beijing strains are also thought to be able to evade the protective effect of the BCG vaccine [18] and have evolved properties that allow them to cause disease more frequently than non-Beijing strains [19]. This could be due to their ability to modulate the host immunity toward a Th2 instead of a Th1 response or their inhibition of TNF-α release as demonstrated in activated THP-1 macrophages [20].
Within the Cape Town metropolis in South Africa, disease caused by the Beijing strains was found to be rising exponentially over a decade whereas disease caused by non-Beijing strains has remained constant [14]. Furthermore, Hanekom et al showed that the Beijing sublineage 7 strains were able to transmit and cause disease more frequently than strains from sublineages 2–6 in urban and rural populations of the Western Cape [12]. It appears that this is due to an evolutionary selection in local populations for this sublineage instead of a founder effect.
In this study, we investigate the relationship between HLA class I molecules and disease by specific M. tuberculosis lineages, specifically those lineages occurring in the Western Cape, South Africa. We show that HLA class I variants are associated with several strains in our study cohort and that taking host and pathogen genetic factors into consideration is necessary for further understanding of the tuberculosis disease burden.
MATERIALS AND METHODS
Ethics Statement
Human blood and sputum samples were obtained with written informed consent for DNA extraction or culture and with approval from the Health Research Ethics Committee of Stellenbosch University, South Africa (project registration numbers 95/072 and 2003/022/N).
Study Participants and Genotyping
Participants for this study were recruited from suburbs in Cape Town, South Africa, where the incidence of tuberculosis was high (1005 per 100 000 in 2007) [21] and the prevalence of human immunodeficiency virus (HIV) was low [22]. These individuals belong to the South African Coloured (SAC) population, a highly admixed population [23]. Three hundred patients with bacteriologically confirmed pulmonary tuberculosis were included in this study (mean age, 34.8 ± 12.6 years, 53% males). All participants were HIV negative and unrelated.
The HLA class I genes were genotyped by direct sequencing. In brief, locus-specific primers flanking exons 2 and 3 were used to amplify the HLA-A, -B and -C loci. The purified polymerase chain reaction products were then sequenced using exon-specific primers and a BigDye Terminator version 3.1 cycle sequencing kit (Applied Biosystems) on the ABI-3130XL DNA analyzer. Sequence traces were analyzed using the Assign SBT 3.5.1 software (Conexio Genomics). HLA class I supertype classification was done as described by Sidney et al for HLA-A and HLA-B [24]. HLA-C alleles were defined based on their KIR2DL1 and KIR2DL2 binding [25] as previously done by Balamurugan et al [26]. Individuals who had alleles (4-digit) that could not be classified into a supertype were identified as “undefined.” HLA class I allele frequency data for white and East Asian populations were obtained from the online Allele Frequency Net Database (AFND; www.allelefrequencies.net) [27] (see Supplementary Data for lists of countries).
Bacterial Isolates and Genotyping
Sputum samples were collected for culture at diagnosis from all new and retreatment tuberculosis patients who attended primary healthcare clinics and who were resident in an epidemiological field site in Cape Town, South Africa, during the period January 1993 to December 2004.
Mycobacterium tuberculosis isolates were classified by culturing the sputum on mycobacteria growth indicator tube and/or Löwenstein-Jensen media, and isolates were classified by IS6110 restriction fragment length polymorphism genotyping and spoligotyping using internationally standardized protocols [14]. Strains were identified according to distinct IS6110 banding patterns using Gelcompar II (Applied Maths) and were subsequently grouped into evolutionary clades that were classified based on their spoligotype signatures. Strains having <6 IS6110 bands (low-copy clade) comprise a single lineage as defined by IS6110 and were therefore regarded as a single clade. Sublineages of the Beijing clade were identified as previously described [12]. Mycobacterium tuberculosis lineages were inferred as members of the East Asian or Euro-American MTBC lineages [9].
Statistical Analysis
Logistic regression models were used to analyze the likelihood of tuberculosis cases having a specific strain, compared to having any other strain, because they enable us to adjust for other variables by including them in the models as covariates. All results were corrected for age and sex in this way. All P values, odds ratios (ORs), and their confidence intervals (CIs) were derived from these models. Genetic association with the susceptibility to different strains was tested using each of the following as predictors in the models: genotypes, additive allelic effect, and additive haplotypes. The haplotypes were inferred, with their probabilities of being correct, and haplotypes were used as predictors in logistic regression models, with their probabilities as weights as previously described [28]. We tested for Hardy-Weinberg equilibrium (HWE) using the exact test [29].
A result or effect was described as significant if P < .05. Bonferroni correction for multiple testing was not used, as this method is considered to be overly conservative when several genetic associations are tested in the same group of individuals [30], resulting in the potential rejection of important findings. Bonferroni correction might also be inappropriate in a situation such as this where there is a priori evidence that the genes are associated with tuberculosis [31], whereas Bayesian methods for correction rely on knowledge of prior probability of involvement, which is currently unknown for most genetic variants [32]. Current methods for multiple testing are only applicable if all tests are independent, which is not the case in this study, and therefore no appropriate method is available. All analyses were done in R (freely available from www.r-project.org) using functions from base R and R packages genetics and haplo.stats.
RESULTS
Host Genotype and Multiplicity of Infections
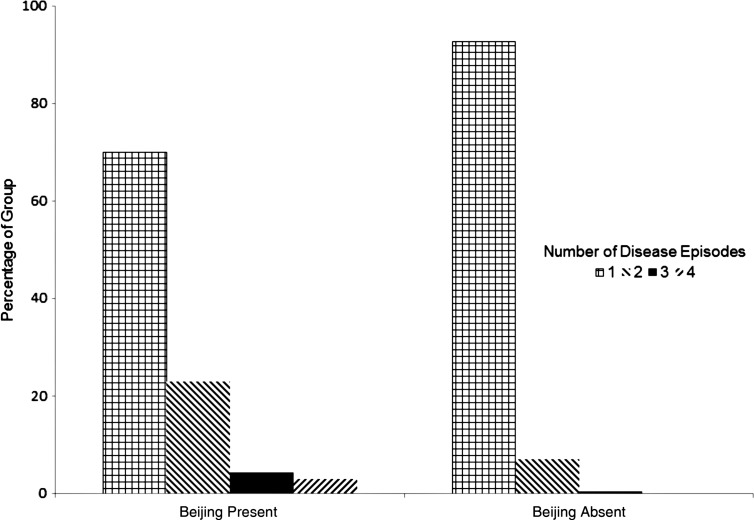
MTBC lineage, strain, and sublineage frequencies in this study cohort are listed in Table 1. Most of the tuberculosis patients (90%) had only 1 episode of disease with infection caused by 1 strain, but 27 (9%) and 3 individuals (1%) had disease episodes caused by 2 and 3 different strains, respectively. Of the 30 individuals with >1 disease episode, 19 (63%) had 1 infection with the Beijing strain, and other episodes with another strain, whereas 37% of those with a non-Beijing strain had an additional episode with another strain. Figure 1 shows that of the 70 individuals with a Beijing strain, 19 (27%) had >1 episode with different strains. Of the 19 individuals who had ≥2 infections of which 1 was Beijing, Beijing was the first infection in 6 cases, and a subsequent infection in 13 cases. Having the HLA-B*27 allele was found to be significantly associated (OR, 0.21 [95% CI, .03–.68], P = .006) with having fewer strains.
Table 1.
Mycobacterium tuberculosis Strain Frequencies in the South African Coloured Population of the Western Cape
| MTBC Lineage | Frequency | Strain | Frequency | Sublineage | Frequency |
|---|---|---|---|---|---|
| Euro-American | 0.79 | ||||
| LAM | 0.32 | ||||
| LCC | 0.19 | ||||
| Quebec | 0.11 | ||||
| Haarlem | 0.10 | ||||
| Haarlem-like | 0.02 | ||||
| East Asian | 0.21 | ||||
| CAS1 | 0.02 | ||||
| Beijing | 0.23 | ||||
| Beijing | Sublineage 2–6 | 0.26 | |||
| Beijing | Sublineage 7 | 0.74 | |||
| Other | 0.11 |
Abbreviation: MTBC, M. tuberculosis complex.
Figure 1.
Presence of Beijing strain in individuals according to number of infections. Individuals with a Beijing strain of infection were more likely to have subsequent infections (P < .001). Of the 19 individuals who had ≥2 disease episodes, 1 of which was Beijing, Beijing was the first infection in 6 individuals, and a subsequent infection in 13 individuals.
Relationship Between Host and Bacterial Genotype in Tuberculosis Disease
All genotype distributions were in HWE. The allele, genotype, and haplotype distributions for HLA-A, -B, and -C were significantly associated with the genotype of the M. tuberculosis strain causing disease in the host. The Beijing strain was significantly associated with each class of variation of the HLA class I genes, with the A*01, B*08, and C2 alleles increasing the odds of having disease with a Beijing strain (Table 2) with ORs ranging between 1.58 and 2.32, while conversely, each B*27 and C1 allele lowered the odds of having disease with a Beijing strain (ORs, 0.35 and 0.60, respectively). Disease with a Beijing strain was also influenced by HLA-B and HLA-C genotypes, as well as 3 class I haplotypes (Table 3). However, due to the small sample sizes and the resulting large CIs, these results are imprecise.
Table 2.
Significant Associations Only, of HLA Class I Alleles and Mycobacterium tuberculosis Lineages in the South African Coloured Population of the Western Cape
| HLA Allele | Frequency (No.) | Strain | P Valuea | OR (95% CI)b |
|---|---|---|---|---|
| A*01 | 0.33 (167) | Beijing | .031 | 1.58 (1.04–2.40) |
| A*03 | 0.24 (120) | LAM | .022 | 1.65 (1.08–2.54) |
| B*07 | 0.25 (123) | LCC | .019 | 0.49 (.25–.89) |
| B*08 | 0.06 (31) | Beijing | .045 | 2.32 (1.02–5.13) |
| B*27 | 0.17 (84) | Beijing | .002 | 0.35 (.16–.68) |
| B*44 | 0.23 (114) | LCC | .007 | 2.07 (1.22–3.52) |
| B*58 | 0.17 (86) | Quebec | .001 | 2.69 (1.27–5.75) |
| C1 | 0.45 (243) | Beijing | .011 | 0.60 (.40–.89) |
| C2 | 0.32 (171) | Beijing | <.001 | 2.03 (1.35–3.08) |
Abbreviations: CI, confidence interval; HLA, human leukocyte antigen; OR, odds ratio.
a P value adjusted for age and sex. P values in bold are statistically significant.
b Odds of having a specific lineage, vs any other lineage, for each extra HLA allele carried.
Table 3.
Significant Associations Only, of HLA Class I Genotypes and Haplotypes With Mycobacterium tuberculosis Lineages in the South African Coloured Population of the Western Cape
| Lineage | HLA Factor | Frequency (No.) | P Valuea | OR (95% CI)b |
|---|---|---|---|---|
| Genotype | ||||
| LAM | A*01/A*01c | 0.13 (33) | .036 | 1 |
| A*01/A*02 | 0.11 (27) | 3.89 (1.37–11.04) | ||
| A*03/Undefined | 0.03 (8) | 6.33 (1.19–33.67) | ||
| Beijing | B*07/B*07c | 0.06 (15) | .001 | 1 |
| B*07/B*08 | 0.04 (10) | 19.6 | ||
| B*07/B*44 | 0.09 (23) | 10.4 | ||
| B*08/B*62 | 0.01 (3) | 25.4 | ||
| Beijing | C1/C1c | 0.23 (62) | <.001 | 1 |
| C1/C2 | 0.24 (65) | 3.61 (1.39–9.33) | ||
| C2/C2 | 0.12 (32) | 4.39 (1.49–12.97) | ||
| C2/Undefined | 0.16 (42) | 4.46 (1.62–12.29) | ||
| Haplotype | ||||
| Beijing | A*01-B*58-C1c | 0.07 (30) | <.001 | 1 |
| A*01-B*08-C2 | 0.05 (20) | 7.8 (1.2–50.0) | ||
| A*02-B*07-C2 | 0.05 (24) | 8.3 (1.5–45.6) | ||
| A*01-B*44-Undefined | 0.03 (17) | 7.6 (1.2 50.30) |
Abbreviations: CI, confidence interval; HLA, human leukocyte antigen; OR, odds ratio.
a P value for genotype and haplotype models, adjusted for age and sex. P values in bold are statistically significant.
b Odds of having a specific lineage and genotype or haplotype, vs any other lineage, compared to the reference genotype/haplotype (OR = 1). The 95% CI could not be calculated for HLA-B genotypes due to their very low frequencies in patients whose infections were not Beijing.
c Reference genotypes/haplotype—the most common (having the highest frequency, so assumed to be the wild type) homozygous genotype and haplotype in the study population.
Tuberculosis caused by LAM genotype strains was found to be significantly associated with the A*03 allele (Table 2), where each additional allele increased the risk with an OR of 1.65. Two HLA-A genotypes were also associated with a LAM infection (Table 3). For the LCC strain (Table 2), each B*44 allele increased the risk of disease (OR, 2.07) whereas the presence of the B*07 allele lowered the chances of disease (OR, 0.49). The odds of disease with a Quebec strain (Table 2) was increased by the presence of the B*58 allele (OR, 2.69).
As Beijing sublineage 7 is the most frequent sublineage in the Western Cape but not the rest of South Africa, we tested whether this could be attributed to the HLAs in the human host. We identified 2 significant associations: where the A*30:02 allele occurred only in individuals with tuberculosis due to a Beijing sublineage 7 strain (P = .02) and is thus a potential risk factor, and with A*02:02 having a protective role against disease with Beijing sublineage 7 strain (OR, 0.04 [95% CI, .0–.51], P = .012). However, it should be noted that these Beijing sublineage 7 results are preliminary due to the small number of individuals (53 individuals with a sublineage 7 infection and 19 individuals with a sublineage 2–6 infection) that could be included in this analysis.
Relationship Between M. tuberculosis Phylogenetic Lineages and HLA Class I Allele Frequencies in Specific Geographical Populations
Table 4 contains a summary of associations between MTBC lineages and HLA types in the SAC population, as well as the bacterial “footprint” of these MTBC lineages in various regions globally. Mycobacterium tuberculosis strains in our study group were separated into members of Euro-American or East Asian, the 2 MTBC lineages most prevalent in the Western Cape. HLA class I allele frequencies of the ancestral populations were derived from AFND. In our SAC sample set there were 199 individuals with an Euro-American MTBC strain only, 57 with an East Asian MTBC strain only, 18 with strains from both MTBCs, and 26 with neither. Several significant associations were identified, with the following alleles associated with both MTBC member strains: A*23:01, B*14:01, B*14:02, and C*16:01. The A*23:01 and C*16:01 alleles were found to be less prevalent in those individuals with disease caused by a Euro-American MTBC strain, while increasing the risk of having disease caused by an East Asian MTBC strain. However, the HLA allele frequencies in the white and East Asian human populations do not correlate with this, as these alleles were found to be more prevalent in white populations than in East Asian populations. The opposite effect was seen for the B*14:01 and B*14:02 alleles, with all individuals carrying these alleles having a Euro-American MTBC strain. In this instance, the HLA population data for B*14:02 was in line with this finding as the allele occurs more frequently in white populations than in East Asian populations.
Table 4.
Significant Associations Only, Between Mycobacterium tuberculosis Phylogenetic Strains and HLA Class I Alleles in Geographic Populations
| MTBC Phylogenetic Lineages |
Allele Frequency per Population |
||||||
|---|---|---|---|---|---|---|---|
| Euro-American | East Asian | SACa | Whiteb | East Asianb | |||
| Allele | P Valuec | OR (95% CI)d | P Valuec | OR (95% CI)d | |||
| A*23:01 | .026 | 0.43 (.20–.90) | .043 | 2.24 (1.03–4.84) | 0.065 | 0.023 | 0.008 |
| A*74:01 | .016 | All are EuroAm | .338 | 0.39 (.02–2.31) | 0.016 | 0.003 | 0.003 |
| B*07:05 | .051 | 0.23 (.05–1.01) | .020 | 5.66 (1.32–29.02) | 0.016 | 0.003 | 0.009 |
| B*14:01 | .018 | All are EuroAm | .019 | None are East Asian | 0.018 | 0.005 | 0.008 |
| B*14:02 | .024 | All are EuroAm | .028 | None are East Asian | 0.016 | 0.019 | 0.007 |
| B*35:01 | .129 | 3.93 (.72–75.44) | .010 | None are East Asian | 0.022 | 0.057 | 0.039 |
| B*58:02 | .001 | 4.64 (1.73–16.48) | .134 | 0.53 (.20–1.21) | 0.087 | 0.003 | 0.002 |
| C*08:01 | .021 | 0.20 (.04–.78) | .054 | 3.83 (.97–16.10) | 0.017 | 0.002 | 0.107 |
| C*16:01 | .028 | 0.35 (.14–.89) | .002 | 4.48 (1.78–11.69) | 0.039 | 0.021 | 0.005 |
Abbreviations: CI, confidence interval; EuroAm, European American; HLA, human leukocyte antigen; OR, odds ratio; SAC, South African Coloured.
a Frequency in the SAC population.
b List of countries included and their respective numbers can be found in the Supplementary Data. Frequency data from Allele Frequency Net Database (www.allelefrequencies.net).
c P value adjusted for age and sex. P Values in bold are statistically significant.
d Odds of having a specific Mycobacterium tuberculosis complex (MTBC) phylogenetic lineage, vs the other MTBC phylogenetic lineage.
Individuals with disease caused by Euro-American MTBC strains were less likely to have the C*08:01 allele and more likely to have the A*74:01 and B*58:02 alleles. At the population level, alleles A*74:01 and B*58:02 occurred at the same frequency in both human populations, whereas allele C*08:01 was found at an extremely low frequency in the white population and at a very high frequency in the East Asian population, providing an inconsistent correlation between risk in the population of specific strains and frequency of HLA alleles.
Statistically significant associations with disease caused by East Asian MTBC strains were seen for the B*07:05 and B*35:01 alleles, with the former increasing the risk of having this strain and the latter reducing the chance (to zero in this study). These findings largely concur with the population data where the B*07:05 allele is found more frequently in East Asian populations than in white populations and the B*35:01 allele occurs more frequently in white populations than in East Asian populations.
DISCUSSION
We report for the first time a number of associations between human HLA class I types and specific M. tuberculosis strains. The role of the coevolution of host and pathogen in disease development has been difficult to study in humans, with most of the proof of concept to date provided by studies of pathogen [9–12] and animal models [33]. We postulated a natural experiment in coevolution taking place in the Cape Town area, which has experienced a multiplicity of human visitors and their mycobacterial strains over the past 350 years. The resident population is extremely diverse [23], with inputs from Khoisan, Bantu, European, and Asian people and could therefore be assumed to have HLA types from all these ancestral populations. The M. tuberculosis strains present can be expected to have experienced intense competition and the incidence of tuberculosis is one of the highest in the world (1005 per 100 000 in 2007 [21]), thereby enabling us to investigate correlations between bacterial strain and HLA type in adequate numbers of patients. In this study we identified associations between HLA class I gene variants with certain strain genotypes, excluding Haarlem, Haarlem-like, and CAS1 strains, which occurred at very low frequencies within our study cohort. The strongest associations were identified for disease with Beijing genotype strains, which was found to be associated with several alleles, genotypes, and haplotypes of the HLA class I genes in the SAC population. Specific allelic associations were also identified for the LAM, LCC, and Quebec genotype strains. We showed that the Beijing genotype strains occurred more frequently in individuals with multiple disease episodes (P < .001) compared to infections by non–Beijing genotype strains.
The B*27 supertype reduced the odds of having multiple disease episodes, as well as having a Beijing strain. This supertype allele is found frequently in individuals who are able to control their HIV infections without any antiretroviral treatment and slow disease progression [34]. This is thought to be due to an increased CD8 T-cell response in individuals with this allele and induction of the apoptotic pathway through the increased presence of cytotoxic proteins [35]. The B*27 supertype has not previously been shown to be associated with susceptibility to tuberculosis [6].
Even though CD4+ T cells (HLA class II restricted) represent the predominant immune response mechanism against M. tuberculosis infection [36], there is growing evidence that suggests an important role for CD8+ T cells (HLA class I restricted) in protection against M. tuberculosis infection [37, 38]. Studies in animals and humans have shown that M. tuberculosis is capable of stimulating MHC class I restricted CD8+ T cells and the involvement of several different pathways for class I presentation of mycobacterial antigens via cross-presentation [39], where HLA class I recognition of mycobacterial antigens includes ESAT-6 (HLA-B*52), 19 kDa lipoprotein (HLA-A*02:01), and Ag85B (HLA-A*02:01) [37, 40, 41]. CD8+ T cells also have direct microbicidal activity and kill M. tuberculosis through the expression of granulysin and perforin [42]. HLA class I alleles have been associated with leprosy susceptibility [43], providing further support for the role of class I genes in immunity against mycobacterial infections. It is, however, possible that the strong LD between genes within the MHC complex [44] could mean that the associations found here reflect the involvement of the class II genes, which remain to be genotyped in this population.
To date, variants in the TLR2 [45], IRGM [46], and SLC11A1 [47] genes have shown a correlation between human and bacterial genotype. Variants in SLC11A1 and TLR2 were found to be associated with an increased risk of having tuberculosis with a Beijing strain in Asian populations, whereas in Ghana, the IRGM polymorphism was found to protect against disease caused by the Euro-American lineage. The phenotype of tuberculosis disease may be affected by the bacterial strain, as strains of the Euro-American lineage appear to be less likely to cause extrapulmonary disease [45], whereas strains of the Beijing and S genotypes were associated with an increased risk of extrathoracic disease [48]. In Vietnam, the relapse rate was significantly increased in tuberculosis cases caused by Beijing strains, and this probably contributes to the successful spread of this strain family [49]. It is therefore evident that the outcome of exposure to M. tuberculosis depends on both the human and bacterial genotypes, and Alter et al [43] speculated that genetic heterogeneity in common infectious diseases could be at least partially explained by the pathogen strain differences, and patient strain types should therefore be incorporated into the analysis to overcome genetic heterogeneity.
Both MTBC lineages and HLA allele frequencies are found in specific geographical settings; for example, lineages of an East Asian origin occur more frequently in human populations from the same region [9]. HLA allele frequencies are hugely dissimilar between different ethnic groups, with certain alleles completely absent in some populations [4, 5]. We therefore investigated the frequencies of HLA class I alleles associated with the Euro-American and East Asian M. tuberculosis lineages, in their sympatric populations. We postulated that an allele more frequent in individuals with a Euro-American strain would also occur more frequently in white populations, whereas an allele that lowered the risk of having a Euro-American strain infection would occur at an extremely low frequency or be absent in white populations. The same rationale would apply to East Asian M. tuberculosis strains and human populations from East Asia. However, although results fitting the postulate were found in several cases, there was no fit in an equivalent number. This could be explained by the use of allele frequency averages across a number of populations listed in the databases. The A*23:01 allele for example, occurs between allele frequencies of 0.075 in the Beijing Han population (AFND), and 0.004 in the Shijiazhuang Tianjian Han, highlighting the enormous discrepancies between allele frequencies in populations of the same geographical region. Second, HLA genes are involved in several biological processes [1] and some could thus be under balancing selective pressures [3], which could have led to the discrepant findings. In spite of the limitations of this broad categorization of populations, we did find several cases where the predominant MTBC lineage and the HLA class I allele frequency fit the hypothesis of the coevolution of M. tuberculosis strains with the HLA class I genes. We now show that specific strains are associated with HLA types of the host, thus providing a molecular genetic explanation for the previous observation by Gagneux et al, who correlated M. tuberculosis strain lineages with geography [9].
The evolutionary forces on HLA have been extremely complex [3], including many bacterial and viral infections. We could thus be seeing the remaining association due to coevolution with M. tuberculosis and/or other diseases with similar clinical pathologies. Hershberg et al has postulated an “out-of-and-back-to-Africa” migration of MTBC that coincided with the out-of-Africa human migration pattern and the subsequent global human exploration quests [7]. Considering this hypothesis, the bottleneck events that accompanied the out-of-Africa migration, and the expansion of disease-causing variants within the last 5000 years [50], it is quite likely that coevolution between MTBC and their human hosts could have occurred.
In summary, this study highlights the role of HLA class I molecules in infection with M. tuberculosis strains and emphasizes the importance of considering both host and pathogen genotype in understanding tuberculosis disease development and vaccine efficacy. Host–pathogen coevolution has significant biomedical and epidemiological implications and by identifying the genes involved in this interaction, the adaptation mechanisms of host and pathogen can be understood, as well as the limitations they impose upon each other. It is also likely that the complexity of HLA types within any given population, and the possible balancing effects of increased susceptibility to tuberculosis vs other pathogens or conditions, will prevent any simple correlations being seen between the predominant HLA type in a population and the strain of M. tuberculosis in that area.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank all study participants for their cooperation.
Financial support. This work was supported by the Wellcome Trust (053844/Z/98/Z to P. D. vH. and E. G. H.); Frederick National Laboratory for Cancer Research (contract number HHSN261200800001E); the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research; the Harry Crossley Foundation; the National Research Foundation (DAAD-NRF); and the Columbia University-Southern African Fogarty AIDS International Training and Research Program through the Fogarty International Center, National Institutes of Health (grant number 2 D43 TW000231 16 to M. S.).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kostyu DD, Hannick LI, Traweek JL, Ghanayem M, Heilpern D, Dawson DV. HLA class I polymorphism: structure and function and still questions. Hum Immunol. 1997;57:1–18. doi: 10.1016/s0198-8859(97)00175-4. [DOI] [PubMed] [Google Scholar]
- 2.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62:1009–30. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 3.Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc Biol Sci. 2010;277:979–88. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lombard Z, Brune AE, Hoal EG, et al. HLA class II disease associations in southern Africa. Tissue Antigens. 2006;67:97–110. doi: 10.1111/j.1399-0039.2006.00530.x. [DOI] [PubMed] [Google Scholar]
- 5.Abi-Rached L, Jobin MJ, Kulkarni S, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kettaneh A, Seng L, Tiev KP, Tolédano C, Fabre B, Cabane J. Human leukocyte antigens and susceptibility to tuberculosis: a meta-analysis of case-control studies. Int J Tuberc Lung Dis. 2006;10:717–25. [PubMed] [Google Scholar]
- 7.Hershberg R, Lipatov M, Small PM, et al. High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol. 2008;6:e311. doi: 10.1371/journal.pbio.0060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monot M, Honoré N, Garnier T, et al. On the origin of leprosy. Science. 2005;308:1040–2. doi: 10.1126/science/1109759. [DOI] [PubMed] [Google Scholar]
- 9.Gagneux S, Deriemer K, Van T, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:2869–73. doi: 10.1073/pnas.0511240103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci U S A. 2004;101:4871–6. doi: 10.1073/pnas.0305627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed MB, Pichler VK, McIntosh F, et al. Major Mycobacterium tuberculosis lineages associate with patient country of origin. J Clin Microbiol. 2009;47:1119–28. doi: 10.1128/JCM.02142-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanekom M, van der Spuy GD, Streicher E, et al. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. J Clin Microbiol. 2007;45:1483–90. doi: 10.1128/JCM.02191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanekom M, Gey van Pittius NC, McEvoy C, Victor TC, Van Helden PD, Warren RM. Mycobacterium tuberculosis Beijing genotype: a template for success. Tuberculosis (Edinb) 2011;91:510–23. doi: 10.1016/j.tube.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Van der Spuy GD, Kremer K, Ndabambi SL, et al. Changing Mycobacterium tuberculosis population highlights clade-specific pathogenic characteristics. Tuberculosis (Edinb) 2009;89:120–5. doi: 10.1016/j.tube.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 15.European Concerted Action on New Generation Genetic Markers and Techniques for the Epidemiology and Control of Tuberculosis. Beijing/W genotype Mycobacterium tuberculosis and drug resistance. Emerg Infect Dis. 2006;12:736–43. doi: 10.3201/eid1205.050400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manca C, Reed MB, Freeman S, et al. Differential monocyte activation underlies strain-specific Mycobacterium tuberculosis pathogenesis. Infect Immun. 2004;72:5511–4. doi: 10.1128/IAI.72.9.5511-5514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez B, Aguilar D, Orozco H, et al. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin Exp Immunol. 2003;133:30–7. doi: 10.1046/j.1365-2249.2003.02171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremer K, van-der-Werf MJ, Au BKY, et al. Vaccine-induced immunity circumvented by typical Mycobacterium tuberculosis Beijing strains. Emerg Infect Dis. 2009;15:335–9. doi: 10.3201/eid1502.080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sreevatsan S, Pan X, Stockbauer KE, et al. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci U S A. 1997;94:9869–74. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theus S, Eisenach K, Fomukong N, Silver RF, Cave MD. Beijing family Mycobacterium tuberculosis strains differ in their intracellular growth in THP-1 macrophages. Int J Tuberc Lung Dis. 2007;11:1087–93. [PubMed] [Google Scholar]
- 21.Health Systems Trust. Incidence of TB in the provinces of South Africa. 2009. Available at: http://www.hst.org.za/healthstats/16/data . Accessed 17 May 2011.
- 22.Kritzinger FE, den BS, Verver S, et al. No decrease in annual risk of tuberculosis infection in endemic area in Cape Town, South Africa. Trop Med Int Health. 2009;14:136–42. doi: 10.1111/j.1365-3156.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 23.De Wit E, Delport W, Rugamika CE, et al. Genome-wide analysis of the structure of the South African Coloured population in the Western Cape. Hum Genet. 2010;128:145–53. doi: 10.1007/s00439-010-0836-1. [DOI] [PubMed] [Google Scholar]
- 24.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90:12000–4. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balamurugan A, Sharma SK, Mehra NK. Human leukocyte antigen class I supertypes influence susceptibility and severity of tuberculosis. J Infect Dis. 2004;189:805–11. doi: 10.1086/381689. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Galarza FF, Christmas S, Middleton D, Jones AR. Allele frequency net: a database and online repository for immune gene frequencies in worldwide populations. Nucleic Acids Res. 2011;39:D913–9. doi: 10.1093/nar/gkq1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–34. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wigginton JE, Cutler DJ, Abecasis GR. A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet. 2005;76:887–93. doi: 10.1086/429864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–9. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;316:1236–8. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell H, Rudan I. Interpretation of genetic association studies in complex disease. Pharmacogenomics J. 2002;2:349–60. doi: 10.1038/sj.tpj.6500132. [DOI] [PubMed] [Google Scholar]
- 33.Di Pietrantonio T, Correa JA, Orlova M, Behr MA, Schurr E. Joint effects of host genetic background and mycobacterial pathogen on susceptibility to infection. Infect Immun. 2011;79:2372–8. doi: 10.1128/IAI.00985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Migueles SA, Connors M. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA. 2010;304:194–201. doi: 10.1001/jama.2010.925. [DOI] [PubMed] [Google Scholar]
- 35.Migueles SA, Weeks KA, Nou E, et al. Defective human immunodeficiency virus-specific CD8+ T-cell polyfunctionality, proliferation, and cytotoxicity are not restored by antiretroviral therapy. J Virol. 2009;83:11876–89. doi: 10.1128/JVI.01153-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaufmann SH. How can immunology contribute to the control of tuberculosis? Nat Rev Immunol. 2001;1:20–30. doi: 10.1038/35095558. [DOI] [PubMed] [Google Scholar]
- 37.Geluk A, van Meijgaarden KE, Franken KL, et al. Identification of major epitopes of Mycobacterium tuberculosis AG85B that are recognized by HLA-A*0201-restricted CD8+ T cells in HLA-transgenic mice and humans. J Immunol 1950. 2000;165:6463–71. doi: 10.4049/jimmunol.165.11.6463. [DOI] [PubMed] [Google Scholar]
- 38.Lazarevic V, Flynn J. CD8+ T cells in tuberculosis. Am J Respir Crit Care Med. 2002;166:1116–21. doi: 10.1164/rccm.2204027. [DOI] [PubMed] [Google Scholar]
- 39.Sigal LJ, Crotty S, Andino R, Rock KL. Cytotoxic T-cell immunity to virus-infected non-haematopoietic cells requires presentation of exogenous antigen. Nature. 1999;398:77–80. doi: 10.1038/18038. [DOI] [PubMed] [Google Scholar]
- 40.Lalvani A, Brookes R, Wilkinson RJ, et al. Human cytolytic and interferon gamma-secreting CD8+ T lymphocytes specific for Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 1998;95:270–5. doi: 10.1073/pnas.95.1.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mohagheghpour N, Gammon D, Kawamura LM, van Vollenhoven A, Benike CJ, Engleman EG. CTL response to Mycobacterium tuberculosis: identification of an immunogenic epitope in the 19-kDa lipoprotein. J Immunol 1950. 1998;161:2400–6. [PubMed] [Google Scholar]
- 42.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 43.Alter A, Huong NT, Singh M, et al. Human leukocyte antigen class I region single-nucleotide polymorphisms are associated with leprosy susceptibility in Vietnam and India. J Infect Dis. 2011;203:1274–81. doi: 10.1093/infdis/jir024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traherne JA. Human MHC architecture and evolution: implications for disease association studies. Int J Immunogenet. 2008;35:179–92. doi: 10.1111/j.1744-313X.2008.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caws M, Thwaites G, Dunstan S, et al. The influence of host and bacterial genotype on the development of disseminated disease with Mycobacterium tuberculosis. PLoS Pathog. 2008;4:e1000034. doi: 10.1371/journal.ppat.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Intemann CD, Thye T, Niemann S, et al. Autophagy gene variant IRGM -261 T contributes to protection from tuberculosis caused by Mycobacterium tuberculosis but not by M. africanum strains. PLoS Pathog. 2009;5:e1000577. doi: 10.1371/journal.ppat.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Crevel R, Parwati I, Sahiratmadja E, et al. Infection with Mycobacterium tuberculosis Beijing genotype strains is associated with polymorphisms in SLC11A1/NRAMP1 in Indonesian patients with tuberculosis. J Infect Dis. 2009;200:1671–4. doi: 10.1086/648477. [DOI] [PubMed] [Google Scholar]
- 48.Hesseling AC, Marais BJ, Kirchner HL, et al. Mycobacterial genotype is associated with disease phenotype in children. Int J Tuberc Lung Dis. 2010;14:1252–8. [PubMed] [Google Scholar]
- 49.Huyen MNT, Buu TN, Tiemersma E, et al. Tuberculosis relapse in Vietnam is significantly associated with Mycobacterium tuberculosis Beijing genotype infections. J Infect Dis. 2013;207:1516–24. doi: 10.1093/infdis/jit048. [DOI] [PubMed] [Google Scholar]
- 50.Fu W, O'Connor TD, Jun G, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–20. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.