Abstract
The incidence of food allergy in developed countries is rising at a rate that cannot be attributed to genetic variation alone. In this review we discuss the environmental factors that may contribute to the increasing prevalence of potentially fatal anaphylactic responses to food. Decreased exposure to enteric infections due to advances in vaccination and sanitation, along with the adoption of high-fat (Western) diets, antibiotic use, Caesarian birth, and formula feeding of infants, have all been implicated in altering the enteric microbiome away from its ancestral state. This collection of resident commensal microbes performs many important physiological functions and plays a central role in the development of the immune system. We hypothesize that alterations in the microbiome interfere with immune system maturation, resulting in impairment of IgA production, reduced abundance of regulatory T cells, and Th2-skewing of baseline immune responses which drive aberrant responses to innocuous (food) antigens.
Keywords: Microbiome, hygiene hypothesis, dysbiosis, atopy, bacteria
Introduction
The incidence of food allergy is on the rise. In the US, the Centers for Disease Control and Prevention documented an 18% increase in the prevalence of reported food allergy in children within a span of just ten years [1]. As reviewed elsewhere in this volume, genetic variation associated with allergic responses to food accounts for only a small fraction of the overall disease risk and cannot explain a dramatic increase in disease prevalence over such a short time period [2–5]. The focus has, therefore, shifted toward gene-by-environment interactions. Several hypotheses have been offered for how the environment may be interacting with the immune system to promote allergic disease; of these, the role of the commensal microbiota has recently come to the fore. This chapter will review the epidemiological and experimental evidence suggesting that various environmentally driven alterations in the microbiome are contributing to the increasing prevalence of atopy. We will then examine some of the complex host-microbiome interactions that influence the development of the immune system and establish a framework for understanding how these changes can promote allergic responses to food. Finally, potential therapeutic applications will be considered.
The hygiene hypothesis: influence of environmental microbial exposure on disease
The idea that microbes and microbial exposure influence the development of allergic disease was first proposed about twenty years ago, when it was noted that children from larger families had a lower incidence of allergic rhinitis [6]. Strachan suggested that, in large families, younger children were more frequently exposed to infections due to the presence of older siblings. This observation led to the “hygiene hypothesis”, which postulated that societal efforts to reduce exposure to infectious disease early in life, including widespread compulsory vaccination and improvements in sanitation, were also depriving the immune system of immunoregulatory stimulation necessary for protection against allergic disease. Further evidence for the hygiene hypothesis came from a study on the effects of an anthroposophic lifestyle on the incidence of atopy in children [7]. The study focused on two anthroposophic schools in Sweden at which vaccination and the use of antibiotics was greatly restricted and exposure to childhood infections correspondingly increased. Children at these schools were found to have reduced allergen-specific IgE in their serum and fewer reports of atopic symptoms, supporting Strachan’s contention that exposure to infection may protect against allergic disease. The diet associated with an anthroposophic lifestyle was also considerably different from that of the rest of Sweden, containing a large amount of fermented vegetables. As a possible consequence of this diet, these children were found to have an increased proportion of lactobacilli in their intestinal microbial communities. Since lactobacilli have been shown to be more prevalent in populations with a lower incidence of atopic disease, this shift may be indicative of a mechanism of protection in these children [8]. The hygiene hypothesis received additional support from studies which demonstrated that the increasing prevalence of allergic disease is mostly restricted to industrialized nations where the use of antibiotics and Western standards of sanitation are more prevalent than they are in developing nations. In fact, certain developing nations have almost no cases of food allergy [9]. Comparisons between genetically similar populations in two different countries, such as East and West Germany after reunification or populations in Finnish versus Russian Karelia, have shown that children living in more developed areas have an increased likelihood of asthma and atopy [10,11]. Even within developing nations, the prevalence of allergic disease is increased in affluent urban areas compared to rural communities [12]. Epidemiological studies of children who live on farms also lend support to the hygiene hypothesis. These studies showed that children who grew up on farms and were exposed to livestock during the first year of life were considerably less likely to develop allergies than their urban counterparts [13–16]. In keeping with the idea that increased exposure to bacteria protects against allergic sensitization, bacterial ligands such as endotoxin (lipopolysaccharide, LPS, from the cell wall of Gram negative bacteria) were found at much higher levels on the mattresses and floors of farming homes than non-farming or urban homes and their presence was negatively correlated with allergic sensitization [17,18]. The inverse association between farm life and incidence of atopy seems to hold true only for affluent countries, however, supporting the contention that multiple environmental influences control allergic susceptibility [19].
Both the innate and adaptive arms of the immune system are influenced by exposure to microbes, and each may play a role in protection against allergy. Prenatal (or neonatal) exposure to stables or barns has been correlated with an increased expression of innate immune receptors (TLR2, TLR4 and CD14) on peripheral blood mononuclear cells as well as decreased atopic sensitization in school-age children [20,21]. Alterations in innate signaling may provide protection against allergy by regulating dendritic cell function and isotype-switching by B cells, primarily by promoting a Th1 response (with increased IFN-γ and IL-12). Evidence for adaptive immune alteration comes from studies examining the concentrations of various antibody isotypes in the serum of children exposed to farm animals [22]; these children had reduced levels of circulating Th2-dependent antibody isotypes, including IgE, IgG1, and IgG4. Increased proportions of regulatory T cells (Tregs) in farm children may also play a role in disease protection [23]. These data, along with the longitudinal association studies, demonstrate an association of early life farm exposure with decreased allergic immune responses, although the precise mechanism(s) behind these correlations remain unclear. In an effort to gain insight into the mechanism involved and to provide direct experimental evidence for the relationship between farm living and reduced atopy, murine models that mimic these environmental exposures have been developed. Individual microbial components from the farm environment were shown to provide protection against allergic airway disease in BALB/c mice primed by OVA-alum and sensitized and challenged with aerosolized OVA [24]. Treatment with dust extract from homes on farms, or with components from specific bacterial strains found at particularly high concentrations in barns, led to reduced eosinophilia and airway hyperresponsiveness in sensitized mice [24,25]. Thus, specific stimuli in the farm environment may be uniquely suited to interact with the immune system and provide signals that drive both innate and adaptive immune responses away from an allergic Th2 response.
In keeping with a role for animal-associated microbes in protection against allergic disease, studies in urban populations have demonstrated that early life exposure to pets may also protect against the development of asthma and allergy. Some evidence suggests that exposure at an early age induces tolerance to pet allergens and prevents the generation of an allergic or anaphylactic response [26,27]. Neonatal exposure to household pets (particularly dogs) correlated with increased production of IL-10 (and decreased IL-13) by circulating monocytes and protection from atopic dermatitis [28,29]. Beyond increasing exposure to common pet allergens, however, early life exposure to pets has also been shown to alter the bacterial composition of house dust. A comparison of the bacterial populations present in dust collected from homes with dogs, cats, or no pets showed that keeping pets increased microbial abundance and diversity in these samples, particularly when animals were allowed to move between indoor and outdoor environments [30]. Lynch and colleagues hypothesized that pet ownership in early life protects against allergy by exposing infants to a greater number and diversity of microbes and tested this hypothesis by conducting a longitudinal study of children who were or were not exposed to pets during the first year of their life. Using serum IgE levels as a measure of the likelihood of developing atopy, they showed that exposure to pets reduced serum IgE in young children [31]. However, other cohort studies have found that microbial products, such as endotoxin, are unchanged by the presence of pets in the home [28,32–34] and that exposure to pets in early life does not alter the incidence of allergic disease in later childhood [35]. While this variability in outcomes does not invalidate this hypothesis, it does speak to the complexity inherent in examining how environmental factors impact host microbial interactions to regulate susceptibility to allergic disease.
The hygiene hypothesis revised: the commensal microbiome and regulation of allergic disease
Given the wealth of data relating changes in the environment to allergic disease, it seems clear that environmental stimuli are at the heart of the question. In its original conception, the hygiene hypothesis suggested that reduced exposure to infectious disease was the major environmental factor responsible for the increasing prevalence of atopy. The revised “counter-regulatory” hypothesis suggests that a balance of microbial signals during the development and education of the immune system is required to prevent both Th1 and Th2 driven inflammatory diseases; environmental changes that disrupt this balance are responsible for the increased incidence of both allergic and autoimmune diseases [36]. The commensal microbiome has moved to center stage as it has become increasingly clear that it integrates signals from the environment and the host to impact immune homeostasis and health.
The human intestine is colonized by a complex ecosystem of 1014 bacteria that comprise an estimated 1500 different species [37]. These bacteria, collectively called the microbiome, have maintained a symbiotic relationship with their hosts over millions of years of co-evolution [38]. Given the extensive evidence that these bugs have far-reaching effects on both the systemic and mucosal immune systems, the earlier hypotheses have been further modified to encompass the unique role of the microbiota in the regulation of allergic disease [39,40]. Early epidemiological studies supported the idea that the microbiota plays a central role in the regulation of allergic disease [41–44]. A study comparing the fecal microbiota of allergic and non-allergic children in two different countries revealed similar patterns of colonization among allergic children including reduced lactobacilli and increased coliforms [42]. Other evidence suggested that atopic children have reduced populations of Gram positive bacteria prior to weaning [45]. Ongoing research is focused on exploring the precise nature of the cross talk between the immune system and communities of bacteria in the gut. Lifestyle changes in developed nations may be contributing to perturbations in a long-established mutualistic relationship between gut bacteria and the host and these perturbations may be driving the observed increases in immune-mediated diseases in the human population [38]. Although more than fifty bacterial phyla have been identified, most of the bacteria that comprise the human microbiota represent only six phyla [46]. The gut microbiome of mammals is dominated by just two phyla: the Bacteroidetes (Gram negative) and the Firmicutes (Gram positive) [47]. Mammalian species share broad similarities in the composition of their microbiome but also exhibit considerable individual variation. In humans, the composition of each individual’s microbiome has been shown to change rapidly from birth through early childhood, but become fairly stable in adulthood [48]. Among their many contributions to host physiology, commensal bacteria extract energy and nutrients from food and produce short chain fatty acids (SCFAs) from fibers that cannot be digested by the host [49]. Thus, when the appropriate balance of these bacteria is disrupted, there can be considerable detriment to the host, including incomplete or improper stimulation of the immune system, which will be discussed in detail below.
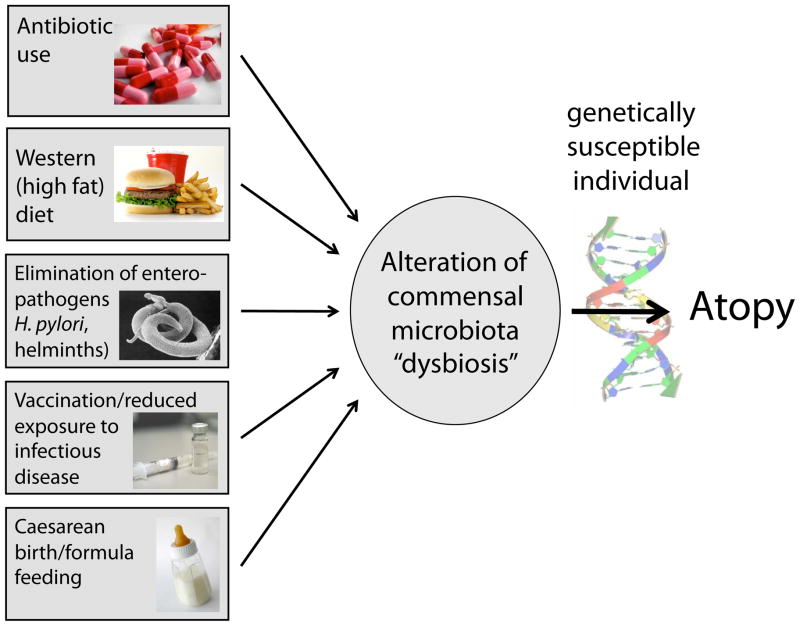
The commensal bacteria that populate mucosal surfaces exist at the interface between the outside world and the rest of the body and are subjected to a plethora of environmental stimuli. Some of these stimuli can disrupt the delicate homeostatic balance that the host must maintain with its microbiota. Indeed, compelling data supports a role for antibiotic use [50], diet [51,52], elimination of parasitic infections [53], Caesarean birth [54] and formula feeding [55] in the regulation of the composition and diversity of the microbiota. Each of these stimuli induces alterations to the microbiota (or “dysbiosis”). In a genetically susceptible individual, these shifts in the microbiome away from homeostasis may promote the development of atopy (Figure 1). Evidence for a role for environment-induced dysbiosis in atopic disease is rapidly emerging and may provide new insights into the mechanisms driving the increasing prevalence of atopy as well as novel therapeutic targets to prevent or treat food allergy and other allergic diseases.
Figure 1. Why is the prevalence of food allergy increasing?
Gene-by-environment interactions are likely to be at the heart of the increasing prevalence of allergic disease in Western countries. Environmental factors that may influence allergic susceptibility include antibiotic use, high-fat diet, elimination of enteric pathogens, reduced exposure to infection, and Caesarean birth and formula feeding. All of these stimuli can alter the commensal microbiome of the host and lead to dysbiosis, which, in a genetically susceptible individual may drive the development of atopic disease.
Environmental stimuli that drive dysbiosis
Antibiotics
A major environmental factor implicated in the rapidly increasing prevalence of allergic disease is the use (and abuse) of antibiotics, particularly in infancy and childhood. Antibiotics have broad-spectrum activities on bacterial function (such as inhibiting ribosome function or cell wall assembly) and can act on both the beneficial members of the microbiome and those causing the infection for which they were prescribed. Thus, when a course of antibiotics is administered for the treatment of an infection, it can also alter the commensal microbiome. Work from multiple laboratories has demonstrated the profound impact of antibiotic administration on the composition and diversity of the intestinal microbiota, particularly with powerful “last resort” antibiotics like vancomycin [52,56–58]. Unfortunately, as antibiotic use has become more prevalent, antibiotic-resistant bacteria have emerged and spread, compounding the problem by necessitating the more frequent use of more potent and longer-lasting antibiotic regimens. The impact of antibiotic-induced alterations of the microbiome on conditions such as allergy and atopy is only beginning to be understood [59].
A full appreciation for how antibiotics influence the commensal microbiome is dependent upon the detailed categorization of the bacteria that populate various body habitats. The assembly of this bacterial catalog has been facilitated, in part, by the Human Microbiome Project [60]. Most of the bacterial inhabitants of the gut are strict anaerobes and only about 10% are culturable by standard microbiological techniques. Culture independent methods of analysis are transforming our understanding of the composition and diversity of the intestinal microbiota. Much of the work published to date relies on the use of the 16S rRNA gene, which is highly conserved among bacterial species in some regions and highly variable in others. “Universal” primers target conserved regions of this gene and allow for amplification and sequencing of species-specific hypervariable regions for bacterial identification. New high-throughput sequencing methods allow for metagenomic and transcriptomic studies. Instead of sequencing just the 16S rRNA gene for identification of species present, new technologies allow sequencing of all genes present in a population and, alternatively, all transcribed mRNAs. This information will continue to lead to great insights into the structure and function of the microbiome.
16S rRNA gene based analysis of cecal samples obtained from antibiotic treated mice showed that oral administration of a cocktail of multiple drugs resulted in a reduction in both Bacteroidetes and Firmicutes while Proteobacteria, normally a minor phylum in the murine and human intestinal microbiome, expanded significantly and enduringly [56]. When a short course of a single antibiotic was used and mice were allowed to recover, the bacterial communities detected were both dissimilar to the original populations and significantly less diverse. Mucosa associated bacterial populations may be among the most vulnerable to depletion by oral antibiotics [57]. Depletion of intestinal bacterial populations by antibiotic treatment often induces phenotypic changes in intestinal anatomy similar to those seen in germ free (GF) mice (discussed below) which include cecal enlargement, intestinal hyperplasia and altered villus length and width [57,61]. Antibiotic-induced shifts in microbial composition also affect host gene transcription, including alterations in antimicrobial peptide secretion by intestinal epithelial cells and cytokine production by lamina propria (LP) T cell populations [57,61]. This indicates that intestinal microbial communities influence both hematopoietic and non-hematopoietic cells. Studies in humans have found similarly dramatic changes in microbiome composition after antibiotic use [50,62–65]. Pyrosequence analyses of fecal samples from antibiotic treated subjects showed reductions in species richness that were only partially restored after cessation of antibiotic treatment. Dethlefsen and colleagues examined gut bacterial communities in stool samples collected before and after treatment with a short course of oral ciprofloxacin, administered at a dose typical for treatment of urinary tract infection and other common bacterial illnesses [50]. They found that oral ciprofloxacin treatment led to a reduced abundance of about one third of the bacterial taxa present in the gut, as well as an overall reduction in community diversity (both richness and evenness) [50]. The magnitude of the response of different subjects to antibiotic treatment varied, as did the composition of the gut microbiome of each individual. Multiple rounds of antibiotic treatment and recovery resulted in long-lasting changes in the composition of the gut microbiome [62]. When analyzed two months after ciprofloxacin treatment, community composition had stabilized but was different than before treatment. This work suggests that the effects of antibiotics are long lasting and can contribute to prolonged dysbiosis even after the course of antibiotics has ended.
The influences of antibiotic treatment on gut microbial communities may be most detrimental early in life when the majority of immune system education occurs. Disruptions to key commensal bacterial populations during this window may ablate signals important for healthy immune system development with consequences that last throughout the life of the host and impact the later development of allergy or other diseases. Several clinical studies demonstrate an association between antibiotic administration during the first few months of life and the incidence of asthma and other allergic disorders [66–68]. A study of children from anthroposophic schools in New Zealand, many of whom had never been exposed to antibiotics, showed that children aged 5 to 10 who had received antibiotics during their first year of life were significantly more likely to have a history of asthma and wheezing [69]. Interestingly, repeated courses of antibiotics during the first year of life were more strongly associated with developing atopic disease later in childhood than single or no courses of treatment. A similar study of children in East Germany found that the risk of developing asthma increased with earlier exposure to antibiotics [70]. A meta-analysis of data from 1966 to 2006 concluded that, based on data collected in both prospective and retrospective studies, antibiotic use during the first year of life is a risk factor for developing asthma between ages 1 and 18 [71]. Prenatal antibiotic exposure has also been correlated with increased allergic disease during childhood [72]. Although larger, carefully controlled cohorts need to be studied, dysbiosis of gut bacterial communities induced by exposure to oral antibiotics, particularly in early life, is likely to be one environmental factor that influences allergic disease susceptibility.
Murine model studies have begun to provide insight into the mechanisms by which antibiotic-induced alterations in the commensal microbiota regulate allergic disease [73–77]. Early work from our laboratory showed that mice unable to signal via TLR4, the receptor for bacterial LPS, are highly susceptible to an allergic response to food. [73]. We hypothesized that the commensal microbiota was the source of the TLR4 ligand and demonstrated that neonatal administration of a cocktail of broad-spectrum antibiotics induced an allergic response in TLR4 sufficient mice similar to that seen in TLR4 mutant mice [73]. Subsequent work showed that antibiotic-treated, food allergic mice exhibit impairments in two compartments critical for the maintenance of mucosal homeostasis [74]. Both the proportion of CD4+Foxp3+ Tregs in the colonic LP (LP) and the concentration of fecal IgA was significantly reduced in antibiotic treated mice and correlated with elevated levels of food allergen specific IgE and IgG1 [74]. Transfer of a conventional microbiota to antibiotic treated mice restored the Treg and IgA compartments and blocked the allergic response. Atarashi et al showed that vancomycin treatment depletes Clostridium species indigenous to the proximal colon and specifically reduces the proportions of CD4+Foxp3+Tregs in that tissue, but not at other peripheral sites [75]. Administration of a mixture of clostridia strains to two week old SPF mice increased the proportion of Foxp3+Tregs in the colonic LP and reduced the OVA specific IgE response induced by intraperitioneal immunization with OVA plus alum [75]. Other work has emphasized the age dependence of the effects of antibiotic treatment on the composition of the microbiota. Antibiotic-treated neonatal, but not adult, mice exhibited enhanced susceptibility to allergic airway disease. [76]. Antibiotic treatment also influences other cellular compartments; another report showed that spontaneously elevated levels of serum IgE correlate with increased numbers of circulating basophils in both antibiotic-treated and GF mice [77]. Challenge of antibiotic-treated mice sensitized with house dust mite antigen resulted in increased basophil mediated Th2 responses and exacerbated airway inflammation [77]. Taken together these studies support a role for early life exposure to antibiotics in promoting dysbiosis and increasing susceptibility to allergic disease.
Diet
Diet strongly influences the composition of the microbiome. The modern Western diet, low in fiber and high in fat, sugar, and processed foods, is markedly different from the diet of our Neolithic predecessors with which our microbiome co-evolved. The significance of this change was demonstrated in a simple but elegant study which compared the composition of the intestinal microbiota of children in rural Africa to an age-matched cohort in urban Europe [52]. The African children ate a plant-based diet high in fiber and low in fat and similar to the type of diet with which our ancestors co-evolved. The European children, by contrast, ate a Western type diet, which was high in animal fat and sugar and low in plant polysaccharides. Analysis of the species present in the feces of these children showed a substantial, diet-induced shift in the Bacteroidetes: Firmicutes ratio; the high plant fiber diet favored the growth of Bacteroidetes while animal fat favored the growth of Firmicutes. Moreover, the diet of the European children promoted a microbiota that had fewer bacteria that could produce certain SCFAs, fiber-derived metabolites essential for healthy gut function. The availability of SCFAs has been implicated as an important player in modulating mucosal homeostasis. One SCFA, butyrate, is a major energy source for colonocytes. Butyrate is associated with the maintenance of a healthy epithelial barrier through, for example, the assembly and organization of tight junctions [78–80]. Furthermore, SCFAs are known to regulate inflammation through the G protein-coupled receptor GPR43, which is expressed primarily by innate immune cells as well as inflammatory cells such as neutrophils, eosinophils and activated macrophages [78]. One study found that mice deficient in GPR43 displayed severe inflammatory responses in models of colitis, arthritis and asthma [81]. A diet-altered microbiome may therefore lead to inflammatory disease through the loss of bacterial taxa that can maintain high levels of SCFAs in the gut. Indeed, GF mice have very low levels of SCFAs [82] and, like Gpr43−/− mice, exhibit increased responses in inflammatory models [81]. These studies support the therapeutic potential of a high-fiber diet that drives the selective expansion of bacteria that produce high levels of SCFAs. In this respect, a few studies have looked at the effect of including foods high in fermentable dietary fiber, such as broccoli, in mouse diets and have found a beneficial effect on the colonic mucosal surface [83,84]. So far, direct administration of SCFAs has already been shown to have clinical benefits in the treatment of colitis, though further study is required to elucidate its direct effect for other inflammatory diseases [85,86]. A reduced proportion of SCFAs has also been documented in fecal samples of allergic children, suggesting this therapy may also treat atopic diseases [44]. Therefore, the evidence suggests that diet is a selecting factor for certain functional members of the microbiome which then influence the metabolic potential and downstream products that can be derived from the host diet.
As further evidence of the complex interaction between diet, the microbiome, and disease, diet-induced changes in the microbiome have been linked not only to dysbiosis but also to obesity. Work from Gordon and colleagues demonstrated that both genetically driven [87] and diet-induced obesity [51] in mice lead to alterations in the microbiota. Ob/ob mice, which lack the leptin receptor and eat 42% more than their lean, leptin sufficient, littermates, have more Firmicutes and significantly fewer Bacteroidetes. This phenotype was recapitulated in a model of obesity using genetically wild type mice. When compared to mice raised on conventional mouse chow (which is low in fat and high in polysaccharides), mice fed a high fat (Western-like) diet had a significantly less diverse microbiota and decreased abundance of Bacteroidetes. Most interestingly, when the microbiome of mice with diet-induced obesity was transferred to lean GF mice, the GF mice gained significantly more weight than those colonized with the microbiome of chow-fed mice, indicating that the changes in the microbiome caused by diet can actually potentiate weight gain and eventual obesity. When obese mice were placed on diets with reduced carbohydrates or reduced fat, their weight gain was reduced while Bacteroidetes were partially increased. This suggests that changes in the microbiota driven by diet are reversible but can have a dramatic impact on the health of the host. Other work demonstrated that the phylum-level shifts in microbial composition noted when mice were switched to a high fat diet were independent of obesity, suggesting that it was not the obese state, but the high fat diet itself, that drove the changes observed [88]. Recent work from Chang and colleagues added an additional wrinkle to our understanding of the relationship between diet, the microbiome and disease by demonstrating that not all dietary fat is the same. Mice switched to a diet high in milk-derived fat (as opposed to the lard-derived fat examined in other studies) had expanded populations of Bacteroidetes (not Firmicutes), as well as the outgrowth of a rare Proteobacterial species, the sulphite-reducing pathobiont Bilophila wadsworthia [89]. Novel mechanistic insight was provided by the observation that milk fats, due to their hydrophobicity, promote hepatic taurine conjugation (TC) of bile acids. These compounds are a rich source of sulphur, which B. wadsworthia uses as its terminal electron acceptor during cellular respiration. Thus, in the presence of milk fats, populations of B. wadsworthia thrive and promote a pathogenic pro-inflammatory Th1 immune response and colitis in genetically susceptible Il10−/− mice. Ongoing studies are elucidating how diet-induced shifts in the composition of the intestinal microbiome are linked to human disease. Comparatively little is known about the influence of diet on allergic disease, in particular. There is evidence for an association between the intake of dietary fat, especially omega-3 and omega-6 fatty acids, and decreased susceptibility to allergy [90–93], but these studies have not investigated whether the connection between dietary fat and allergy is mediated by the microbiome.
Although, as discussed above, the composition of the intestinal microbiota is sensitive to dietary alterations, some work suggests that, in humans, the intestinal microbiome can be grouped into three stable “enterotypes” characterized by clusters of particular microbial taxa [94, 95]. These enterotypes are strongly influenced by long-term dietary patterns [95]. Rapid changes in microbial composition induced by short-term dietary modifications are not sufficient to induce a shift in an individual’s enterotype cluster [95]. Whether this seemingly simplistic grouping of a very complex microbial ecosystem will hold up in larger studies and inform our understanding of disease will be a topic for future work.
Enteropathogens
Although pathogens are, by definition, not part of the commensal microbiota, they can have profound effects on the homeostasis of the host. One group of pathogens, gastrointestinal helminths, is particularly interesting when considering non-host regulation of allergic disease. Gastrointestinal helminth infection elicits a Th2-biased response from the host that is characterized by high levels of circulating parasite-induced IgE and IgG1. Much of this response is induced at the site of initial infection in the gut and its associated lymphoid tissue. To create a niche that allows for survival for long periods of time in the host, gastrointestinal helminths elicit high levels of immunoregulatory mediators such IL-10 and TGFβ [96]. The immunosuppressive capacity of helminths may also be linked to their ability to alter the composition of the host microbiome. Murine models of helminth infection have demonstrated a site-specific effect of infection on the microbiota of the host; the ileum of Heligmosomoides polygyrus-infected mice contains more Clostridiaceae and Lactobacillaceae family members than that of uninfected mice [53]. This shift in is not necessarily detrimental but may fundamentally change the homeostasis between the host and the microbiota, particularly because helminth infections are often chronic.
As recently as fifty years ago, helminth infection was endemic in much of the world. Epidemiological data suggests that, as helminth infection was being eradicated from the populations of developed nations, allergic diseases began to rise, suggesting that these infections provide protection against allergic disease [97]. This is somewhat surprising since helminths and allergens are unique in their ability to induce a Th2 biased immune response leading one to expect an exacerbation of allergic disease. Children in helminth-infected populations were shown to have high levels of allergen-specific IgE in the absence of symptomatic disease or positive reactions in response to skin testing. In other words, they exhibited a Th2 response without atopy [97, 98]. One study found that children who received anti-helminthics had greater responses to skin-prick tests but remained negative for clinical disease, raising the possibility that helminth infection induces long-lasting residual protection from allergic effector responses [99]. Some researchers hypothesize that very high levels of parasite-induced IgE and IgG protect against allergy by blocking the binding of allergen-specific antibodies to mast cells [100]. This suggests that the mechanism of protection is passive, though other evidence suggests that there is cytokine and cell-mediated suppression of the allergic response during a helminth infection [101].
Work in animal models of allergic disease validates the numerous associations found between helminth infection and reduced allergy in humans. Several studies have demonstrated a reduction in allergic airway inflammation and anaphylaxis in animals infected with helminths including Heligmosomoides polygyrus and Schistosoma mansoni [102–105]. Sensitization and challenge of helminth-infected mice resulted in reduced airway constriction, fewer cellular infiltrates in the lungs (particularly eosinophils), and reduced Th2 cytokine secretion. The protection was found to be either IL-10 or TGFβ-dependent and mediated by either CD4+CD25+ Tregs or B cells, depending on the source of both the worm and the allergen [101,102,104,106]. Other studies found that a similar protective effect could be elicited by individual worm antigens or worm products [105,107]. Murine model work from our laboratory demonstrated that gastrointestinal helminth infection protects against allergic responses to food by promoting global immunosuppression [108]. Intragastric sensitization of H. polygyrus infected mice with peanut plus cholera toxin resulted in reduced antigen specific IgE and IgG1, abrogated anaphylactic responses, and reduced Th2 cytokine production upon in vitro restimulation with peanut antigen when compared to sensitized, uninfected counterparts. Protection from allergy was shown to be at least partially IL-10 dependent. Taken together with the other studies cited above, this work indicates that helminth infection can stimulate a cytokine response that is immunosuppressive and protective against anaphylactic responses to food.
Given the evidence in support of helminth mediated suppression of allergic disease in both epidemiological studies and murine models, it comes as no surprise that helminth infection has been targeted as a potential therapy for allergic diseases. Two trials have looked at whether deliberate infection with helminths can treat preexisting allergy. The first, using the hookworm Necator americanus, demonstrated that infection with a small number of larvae marginally improved airway hyperreactivity in patients with allergic asthma analyzed 16 weeks post infection [109]. The other study examined the effect of Trichuris suis on allergic rhinitis and reached a Phase 2 clinical trial. These authors found that infection with eggs of this whipworm, which normally infects pigs, did not provide statistically significant improvement in allergic symptoms but patients that received treatment took fewer antihistamine pills [110]. Given that there have been successful studies using Trichuris to treat inflammatory bowel disease (reviewed in [111]), improvements in the dosing or treatment protocol may demonstrate efficacy of T. suis for the treatment of allergic disease.
Another well-known pathogen that is associated with protection from allergic disease is Helicobacter pylori. Although H. pylori is more commonly associated with increased risk of stomach ulcers and cancer, considerable evidence suggests that colonization with H. pylori early in life has beneficial effects, including protection from infection and asthma [112–115]. Colonization of humans with this gastric commensal was, at one time, nearly ubiquitous. Increased use of antibiotics has decreased the proportion of colonized individuals to about 6% in developed nations [59]. Clinical studies of human patient cohorts demonstrated an inverse correlation between colonization status and childhood asthma [113]. Further examination of adult cohorts of asthmatic or control patients revealed a similar correlation [115]. Certain strains of H. pylori appear to have greater protective effects. CagA, a protein encoded in the cag pathogenicity island, is injected into host cells through a type IV secretion system and interferes with signaling pathways. Although invasive interaction between bacteria and host is associated with increased gastric inflammation, adults colonized with CagA+ H. pylori had the lowest incidence of asthma [115]. Experimental evidence for a role for H. pylori in protection against allergic disease comes from mouse models of asthma [116]. Colonization of young mice with H. pylori, followed by sensitization with OVA-alum and challenge with aerosolized OVA, resulted in significantly reduced airway hyperresponsiveness and cellular infiltration in the lung tissue and bronchoalveolar fluid compared to uninfected controls. Resolution of H. pylori infection with antibiotic treatment eliminated the protective effects. On a cellular level, H. pylori-infected mice had fewer mature, fully activated pulmonary dendritic cells (DCs) and increased numbers of highly suppressive Tregs in their lungs [116]. Thus, Helicobacter colonization early in life seems to have a beneficial effect against allergic disease despite the risk of gastric disease later in life. Ultimately, H. pylori might be harnessed as an anti-asthma therapy through early-life colonization followed by narrow-spectrum antibiotic treatment to eliminate it later in life [59].
Given that the original idea behind the hygiene hypothesis related atopy to exposure to infection, and that more recent data suggests a role for the microbiome, it is interesting to consider how infection and the microbiome may be linked. Might certain infections influence the composition of the microbiome and, thus, allergic susceptibility? In support of this idea, there is evidence that infectious inflammation drastically alters the intestinal microbiome. Murine models have demonstrated that infection with Citrobacter rodentium and Salmonella typhimurium induces transient changes in the composition of the microbiome and in the abundance of certain bacterial families [117, 118]. Infection with either bacterium results in a transient reduction in the total bacterial load, which rebounds upon clearance of the infection. Each infection causes distinct changes in the microbiome. Citrobacter infection leads to an outgrowth of Firmicutes, specifically those in the order Clostridiales, while Salmonella infection depletes the Eubacterium rectale/Clostridium coccoides group. When both infections are cleared, however, the microbial communities of previously infected mice return to their original state, with minimal remaining evidence of the pathogen. These transient alterations in the microbiome may play an important role in the education of the immune system and the establishment of a homeostatic balance within the microbial communities of the gut. Moreover, acute gastrointestinal infections may act as a trigger for the initiation of the allergic cascade.
Mode of birth and formula feeding
While antibiotic use, diet, and enteropathogen infection can modulate the microbiome throughout the life of the host, early life events may be of particular importance for the initial establishment of a healthy microbiome. Initial colonization occurs immediately after birth and bacteria are therefore present in the intestines during the earliest periods of post-natal immune system development and education [119]. The neonatal intestinal microbiome is dominated by lactobacilli and other facultative anaerobes typically present in the vagina [119,120]; these founder bacteria seem to function to make the intestine anaerobic and promote later waves of colonization with strict anaerobes such as Bacteroides and clostridia [121]. The unique body habitats of the adult human harbor different bacterial communities [122] and recent evidence suggests that mode of birth influences an infant’s initial bacterial colonization. Infants born by Caesarean section are colonized with a microbiome more closely resembling that of the skin than of the vagina [54]. The changes in composition of the enteric microbiome attributed to Caesarean birth have been shown to persist for many months [119]. Although the composition of the microbiome is highly dynamic throughout childhood, mode of delivery and early colonization events have lasting effects that have been linked to dysbiosis and disease including asthma and food allergy later in life [123–126]. A study of allergic children showed reduced bifidobacteria colonization while healthy children had increased lactobacilli colonization during the first few months of life [41]. Caesarean birth can also delay colonization, further exposing these neonates to infection with pathogens [127]. Clinical studies of children born via Caesarean section provide evidence that they have stronger, non-specific humoral responses at a young age [123] and are more likely to develop asthma [128] and celiac disease [129].
Another factor that has been hypothesized to provide protection against the development of dysbiosis and allergy early in life is breast-feeding [55,130]. Breast milk, especially colostrum, transfers passive immunity in the form of IgM, IgG and IgA from mother to infant. IgA is of particular importance in the gut because of its role in immune exclusion of dietary antigen and in the maintenance of commensal homeostasis. Studies in mice have shown that IgA has a profound effect on controlling both the composition and abundance of commensal bacteria [131]. Breast milk also contains biologically active molecules including soluble CD14 (a receptor for bacterial LPS), tolerogenic cytokines, and fatty acids including the omega-3 and omega-6 fatty acids associated with protection against atopy as part of a solid diet [132]. Thus, additional protection provided by breast milk during the first months of life can influence the development of allergic disease [132], perhaps by reducing the outgrowth of harmful bacteria and promoting the establishment of a healthy microbiome. Clinical studies of the effect of breastfeeding on the incidence of allergic disease report mixed effects; some reports suggest a protective role against developing atopy while others do not [134–136]. Those that cite a protective effect suggest that the important window is the first four months of life [135]. Analysis of the fecal microbiome of breast-fed versus formula-fed infants showed distinct clustering patterns, confirming that different modes of feeding profoundly influence the microbiome [55]. Interestingly, bacterial diversity was enriched in formula-fed infants, with other cohort studies showing that this increased diversity was associated with a higher incidence of allergic disease [137]. The effects of formula feeding seen in this study could be reversed by supplementation with bifidobacteria, indicating that therapeutic intervention with probiotics to prevent dysbiosis may be possible and efficacious. Another group demonstrated that feeding human milk lysozyme to neonatal pigs altered the composition of the microbiome and promoted the growth of protective bacterial populations such as bifidobacteria and lactobacilli [138]. This demonstrates a potential mechanism by which breastfeeding can directly alter the microbiome to the benefit of the host immune system and protect against allergic disease.
The association of mode of delivery and diet with alteration of the microbiome and increased autoimmune and allergic disease suggests that a suboptimal microbiome--one depleted of “beneficial” bacterial taxa--can alter the profile of host homeostasis, thereby skewing away from protective immune responses and increasing aberrant responses to innocuous allergens.
Food allergy as a failure of oral tolerance
The most common way to conceptualize food allergies is a loss of oral tolerance. Oral tolerance is the process by which the immune system promotes mucosal and systemic non-responsiveness to orally administered antigens [139]. When this process breaks down, an aberrant immune response is mounted, typically in the form of food-specific IgE and IgG1 (mouse and human) or IgG4 (human), increased infiltration of mast cells into the intestinal mucosa, and, frequently, anaphylaxis upon reexposure to the same antigen [140].
The mechanisms regulating the induction of oral tolerance may be more complicated than originally appreciated. Food antigen is taken up by antigen presenting cells (APCs) in the intestine and presented to naïve T cells in the draining mesenteric lymph node (MLN), which are then educated not to respond to this antigen upon reexposure [141]. Under normal conditions, food antigens are acquired in an environment that favors anti-inflammatory responses and some T cells reactive to food antigens undergo anergy or deletion by a mechanism similar to central tolerance [142,143]. The peripheral induction of food antigen specific Tregs is thought, however, to be the primary mechanism by which tolerance to dietary antigens is induced and maintained [141,143]. Some animal model studies suggested that high doses of oral antigen induce anergy or deletion, while frequent low-dose antigen exposure favors the induction of Tregs [143]. Mechanistic hypotheses based on antigen dose may, however, be of limited physiological relevance [141]. Recent work supports a two-step model of oral tolerance which focuses on the peripheral induction of food antigen specific Tregs [144,145]. In the first step, antigen may be taken up through a variety of pathways, including transcytosis through M cells (specialized epithelial cells present in Peyer’s patches and other organized mucosal lymphoid structures), transcytosis through enterocytes, and diffusion between epithelial cells [146]. Once antigen reaches the LP, it is phagocytosed by a subset of migratory DC characterized by their expression of CD11c and CD103 [147,148]. This DC population processes the antigen and traffics to the MLN to present it to naïve T cells. The environment of the MLN promotes and supports the induction of Tregs that become highly effective producers of IL-10 and help to suppress effector responses by other cells that may escape tolerization [145,149]. Importantly, secretion of retinoic acid and TGF-β by CD103+ DC upregulates key gut homing molecules such as α4β7 and CCR9, allowing antigen specific Tregs to traffic back to the small intestinal LP and expand. Expansion of food antigen specific Foxp3+Tregs locally in the LP, after initial induction in the MLN, is postulated to be the second required step in this model [144,145,147]. This expanded LP Treg population contributes to the anti-inflammatory environment in which APCs see antigen, further limiting the chance of generating deleterious effector responses to food antigens. While it is not completely known what drives the expansion of Tregs in the LP, there is evidence that it is dependent on a resident CX3CR1+ macrophage population [140]. Since these CX3CR1+ macrophages do not migrate, any antigen they take up may be passed to the migratory CD103+ DCs or may promote the expansion of newly arrived but already-educated Foxp3+Tregs in the LP [146,148]. CX3CR1+ macrophages may also promote Treg survival and expansion through the production of IL-10; myeloid cells in the LP have been shown to be an important source of IL-10 for sustained Foxp3 expression in induced Tregs [151]. In the absence of this CX3CR1+ macrophage population, there are significantly fewer Tregs in the LP and oral tolerance is impaired [143]; there is also an increase in bacterial translocation and the production of inflammatory cytokines in the intestinal mucosa [152]. Thus, both dendritic cells and macrophages in the LP are involved in promoting oral tolerance and regulating homeostasis in the gut. Other cells have also been implicated in oral tolerance, albeit with less clearly defined roles and without strict necessity. APCs in the liver been have suggested to play a role in promoting the systemic non-responsiveness that is characteristic of, and necessary for, successful oral tolerance [146,153]. Plasmacytoid DCs, in particular, have been implicated in inducing anergy in antigen-specific T cells after oral antigen administration [154,155]. Of note, the CX3CR1+ DC subset is related to plasmacytoid DCs based on gene-expression profiling [156].
Keeping in mind the two step model of oral tolerance discussed above, we can consider how signals from the microbiome can influence each of the steps in this model by altering the availability of allergen, the proportion and composition of T cell subsets, the phenotype of innate cells, or the microenvironment in which these interactions take place. Experimental evidence discussed below will examine how bacterial subpopulations can interact with the immune system and influence host homeostasis and tolerance to dietary antigen.
Regulation of lymphocyte homeostasis by the microbiome
Under homeostatic conditions, there is a balance between effector and regulatory cells that insures a productive response to infection while avoiding excessive or misdirected responses to non-infectious stimuli such as food antigens. When various environmental factors induce dysbiosis, the balance between effector and regulatory cells can be lost [37,38]. The profound impact of the commensal microbiota on the development and maturation of the immune system is readily apparent in GF mice. In the absence of colonization with commensal bacteria, secondary and tertiary lymphoid organs remain underdeveloped [157, 158], various T cell subsets are absent or underrepresented [159–161], serum antibody titers are skewed [162], and intestinal architecture is disrupted; the cecum, in particular, is dramatically enlarged [163]. Colonization of GF mice with a complex microbiome reverses many of these phenotypes. Certain commensal bacteria have been shown to drive the development and/or differentiation of particular cell types, especially Tregs versus Th17 cells [159–161, 164, 165]. There is also a body of literature demonstrating that, in the gut, the microbiome plays a role in the induction of secretory IgA, an important neutralizing defense mechanism at mucosal surfaces [166].
Gnotobiotic (“known flora”) mice have been employed to examine the effects of individual bacterial species or groups of species on the development of the immune system [157]. Gnotobiotic models have shown that colonization of GF mice with certain members of the conventional microbiome can drive the development of specific T cell subsets. For example, segmented filamentous bacteria (SFB) induce the development of proinflammatory Th17 cells [160] that exacerbate intestinal inflammation [167] and drive systemic autoimmune arthritis in a genetically susceptible mouse model [168]. Other work showed that colonization with Bacteroides fragilis drives both Th1 and Th2 cell development [161] while a cocktail of heat-resistant bacteria was found to recapitulate the balance of all T helper subsets (Th1, Th2, Th17, and Treg) in formerly GF mice [159]. A recent study also demonstrated that Altered Schaedler Flora (ASF), a cocktail of eight bacteria from both the Bacteroidetes and Firmicutes phyla that has historically been considered to be mutualistic, can specifically induce Tregs in the colonic LP without inducing proinflammatory T cells [164]. Atarashi et al showed that Clostridium species indigenous to the proximal colon (members of which are present in the ASF consortium) specifically induce Tregs at this site [75]. Taken together, these studies show that particular commensal bacterial populations can alter the profile of inflammatory versus regulatory T cells at mucosal sites, particularly in the intestine, while also exerting systemic effects.
The simplified microbiomes of gnotobiotic mice may not, however, fully recapitulate the complex ecosystem of an SPF microbiome. Targeted antibiotic treatment is another method that has been used experimentally to examine the contributions of specific bacterial populations to immune system regulation. Recent work has shown that alteration of the SPF microbiome with antibiotics can reduce the size of the Treg compartment or favor the expansion/recruitment of effector cells. Atarashi et al. found that in both GF and vancomycin-treated mice, there is a significant reduction in the CD4+Foxp3+ Treg compartment of the colonic LP, suggesting that a vancomycin-sensitive (Gram positive) bacterial population is required for the induction of these colonic Tregs [75]. In order to narrow down the identity of these bacteria, the spore-forming fraction of the SPF microbiome was extracted with chloroform and used to colonize GF free mice. Upon colonization with these chloroform-extracted species, the Treg compartment of the colonic LP was restored to the levels seen in unmanipulated SPF mice. The essential chloroform-extracted microbes were identified as a mixture of Clostridium species. Administration of a clostridia cocktail to SPF mice further increased their colonic Treg compartment and provided protection against both immunization with OVA plus alum (a Th2-biased response) and Dextran sodium sulfate (DSS)-induced colitis (a Th1-biased response). The influence of clostridia on the immune system has been well documented [169]. In addition to inducing Foxp3+ Tregs, Clostridium species drive the recruitment of intraepithelial lymphocytes and promote the production of IgA by LP B cells, both of which may improve immune defenses at the barrier [170,171]. Clostridiales have been shown to reside in close proximity to the mucosa, among the folds of the proximal colon, which may provide insight into their potent immunostimulatory ability [172]. Whether this stimulation is protective or pathogenic, however, depends on the host context. Although Atarashi et al demonstrated protection from allergic sensitization, other studies implicate a flagellin protein (CBir1) from Lachnospiraceae, a specific family of clostridia, in the induction of colitis and demonstrate an association between inflammatory bowel disease and an antigen-specific T cell response to this protein [173,174]. This suggests that the same bacterial families may result in differential responses depending on the genetics of the host or the environmental context (ie. Th1 vs. Th2 inflammation). It is also noteworthy that Clostridium spp. are not the only bacteria shown to have this immunoregulatory effect; Bacteroides fragilis (specifically its polysaccharide A) and certain probiotic mixes of lactobacilli can also induce Foxp3+ Tregs [165, 175–177]. A unique role for extrathymically generated, Foxp3+Tregs at mucosal surfaces was further emphasized by Josefowicz et al [178]. They created a knockout mouse lacking CNS-1, a TGFβ responsive element required for the development of peripherally-induced, but not thymically-derived, Foxp3+Tregs. CNS-1 deficient mice exhibit spontaneous Th2-biased inflammation in both the gut and the lungs, suggesting that peripherally induced Foxp3+Tregs play a non-redundant role in the prevention of allergic, Th2-biased inflammation at these sites.
Other work demonstrated that CD4+Foxp3+ Tregs induced in the presence of commensal bacteria bear T cell receptors specific for the bacteria themselves [179]. Lathrop et al. screened the endogenous colonic TCR repertoire of mice with a fixed TCRβ chain but a fully rearranged TCRα chain; these mice generate a reduced, but polyclonal, repertoire of T cells. An examination of Foxp3+Tregs, effector/memory T cells, and naïve T cells in these mice showed that each compartment had distinct TCR usage. Further analysis of the TCRs from the Treg compartment indicated that hybridomas expressing certain TCRs from the colon were responsive to bacterial stimuli in vitro. These hybridomas were only activated by colonic contents from mice housed in the same facility, suggesting that activation was due to TCR-mediated recognition of specific bacterial epitopes rather than non-specific activation by bacterial PAMPS. Finally, a TCR clone was identified that could be activated by members of the genus Parabacteroides but not by members of the closely related genus Bacteroides, confirming the specifity of these TCRs. Furthermore, these Tregs converted to effector T cells under conditions that promote development of colitis. This study established that some induced Tregs are specific for members of the commensal microbiome and that these Tregs are important for the maintenance of homeostasis.
Recent work has demonstrated that invariant NKT cells (iNKTs) are also educated by the commensal microbiota. Olszak et al demonstrated that an accumulation of iNKT cells in the lungs of GF mice was associated with an exacerbated response to sensitization in an OVA-induced model of allergic asthma [180]. Colonization of neonatal, but not adult, GF mice with feces from an adult SPF mouse led to reduced numbers of mucosal iNKT cells and ameliorated allergic disease, demonstrating that early colonization events in the intestine have a profound impact on effector cell subsets other than T cells, and have effects beyond the site of colonization.
Food allergy and the microbiome: a working model
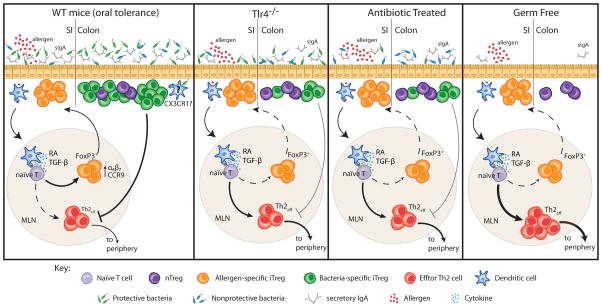
How does all of this data relate to the increasing prevalence of food allergy? We propose that the cumulative effect of various environmental changes that have occurred over the past two decades have altered the composition of the commensal microbiome in a manner that impairs the induction of tolerance to dietary antigen. We suggest that both food antigen-specific Tregs and those induced by the commensal microbiome are required for proper maintenance of oral tolerance and the prevention of food allergy. In support of this model, data from our laboratory has demonstrated that there is a significant reduction in Tregs in the colonic LP of allergy susceptible Tlr4−/−, antibiotic treated and GF mice. Antibiotic treatment of neonatal mice leads to reduced proportions of Tregs in the colonic LP, impaired production of intestinal IgA, and elevated peanut specific IgE and IGg1 in response to sensitization [73,74]. When a conventional microbiome is reintroduced to antibiotic treated mice, the Treg and IgA compartments are restored and the allergic response is blocked, formally demonstrating a role for the microbiota in protection against the allergic response induced by sensitization of antibiotic treated mice. TLR4-deficient mice, which cannot sense LPS and are thus unable to sense a large component of their microbiome, also have a reduced colonic Treg/IgA compartment. TLR4 deficient mice have enhanced allergic responses to food when compared to their littermate controls. GF mice, which have no bacteria-induced colonic Tregs, are highly susceptible to allergic sensitization. Systematic colonization of GF mice showed that only bacterial consortia that induce the production of secretory IgA and the expansion of Tregs in the colonic LP are protected from an allergic response to food upon sensitization with peanut plus cholera toxin. Our data therefore supports a model in which the maintenance of tolerance to dietary antigen is reliant on bacteria-induced Tregs and IgA (Figure 2).
Figure 2. Both allergen-specific and bacterially induced Tregs are required to prevent allergic responses to food.
In a healthy individual, dietary antigen is carried to the MLN by migratory CD103+ DC and presented to naïve T cells to induce antigen specific Foxp3+Tregs. Secretion of retinoic acid (RA) and TGF-β by these CD103+DC upregulates the expression of the homing molecules α4β7 and CCR9 which allows these antigen specific Foxp3+ Tregs to migrate back to the small intestinal LP and expand for the induction of oral tolerance. New work from our laboratory shows that tolerance to dietary antigen also relies on bacteria-induced Tregs and IgA. Microbiota-induced Tregs and IgA are reduced in the intestines of Tlr4−/− and antibiotic treated mice (and absent in germ free mice). Impairment of this mucosal Treg/IgA compartment correlates with the enhanced production of food allergen specific IgE and IgG1 in response to sensitization. Reintroduction of a conventional microbiota to antibiotic-treated mice restores the Treg and IgA compartments and blocks the allergic response. Colonization of germ free mice with a microbiota containing Clostridium species induces both Foxp3+ Tregs and intestinal IgA and attenuates allergic sensitization.
Future directions: potential new therapeutic targets
The evidence presented here linking the immune system, allergy, and the composition of the microbiome provides new insights into the causes of allergic disease and suggests potential new therapeutic targets. Probiotics, defined as live microbial supplements that have beneficial, non-pathogenic effects on health, have been the focus of much attention for their potential--but often disputed--therapeutic effects. The probiotic theory, or the hypothesis that probiotics can be consumed to repopulate the microbiome with the correct balance of beneficial microbes, was first introduced by Metchnikoff as early as 1907 in an attempt to account for an unexplained association between the health and long lives of Bulgarian peasants and their daily intake of fermented milk products. Metchnikoff surmised that the daily consumption of beneficial bacteria within fermented milk was maintaining a healthy microbiome that improved the length and quality of life of this population [reviewed in 181]. Subsequent research has focused on lactic acid bacteria, especially Lactobacillus species, due to their common use in the manufacture of cheese, yogurt, and other fermented food products [182]. Interestingly, archeological evidence suggests that the technique of using lactic acid bacteria to ferment foods has been used for thousands of years, and may have been invented as long as 1.5 million years BCE. This would suggest that our intestinal tract has evolved to support a daily intake of lactic acid bacteria, and would thus benefit from the use of probiotics that could sustain this evolutionarily stable composition [183]. Probiotics have been shown to have positive effects against several diseases, including irritable bowel syndrome and inflammatory bowel disease [184,185] and as a treatment for chronic infections such as those caused by Helicobacter pylori and Clostridium difficile [186]. Here, however, we will focus on research that has demonstrated efficacy in allergy.
The effect of probiotics on atopic eczema has been widely investigated. In one report, pregnant women were treated with capsules containing Lactobacillus rhamnosus from 35 weeks of gestation until their infants were six months old (if the mother breastfed). Infants began receiving the capsules between 2 and 16 days after birth and continued until two years of age. The study found that by age two, there was a 50% reduction in eczema prevalence in the L. rhamnosus treated group [187]. This effect was sustained at age 4 [188]. Other studies have not demonstrated sustained protection from eczema. Kalliomaki et al found that a similar probiotic (Lactobacillus rhamnosus GG) led to a high relative risk reduction of 49% by age 2 but risk reduction fell to 43% by age 4 and by 36% by age 7 [189]. Another study administered a mixture of four Lactobacillus strains with prebiotic oligosaccharides and found a reduced risk for eczema by age 2 but no extended effect by age 5 [190]. The disagreement between these results may be due to differences in methodology between studies such as the duration and timing of administration, characterization of allergic responses, or probiotic formulations. Expanded populations of lactobacilli are detected in the gut after administration, indicating that these bacteria are able to survive passage through the stomach and successfully colonize the gut [187,191,192]. The expanded Lactobacillus population dwindles in size upon cessation of probiotic intake, however. Some evidence suggests that transiently expanded populations of lactobacilli ameliorate intestinal inflammation by replacing pathobionts in the gut [191]; other studies did not replicate these findings [192,193].
The murine model studies cited above suggest that the predominant focus on Lactobacillus-based probiotic formulations, to date, may provide some explanation for their limited therapeutic efficacy. Other bacterial populations, like the Clostridiales, may be more effective at inducing Tregs and secretory IgA that protect against allergic disease. The translation of recent mouse model work to human disease holds great promise for the development of novel therapeutic approaches to reshape the microbiota to prevent or treat food allergy.
Conclusions
The experimental evidence presented in this review strongly supports a role for the commensal microbiota in regulating susceptibility to food allergy. The commensal microbiome exists in a dynamic relationship with its host and has profound effects on the development and maturation of the immune system. When environmental influences alter the microbiome and disrupt homeostasis, a state of dysbiosis can result. Experimental and clinical evidence suggests that early life events are particularly important for the establishment of a “healthy” microbiome. The mechanisms by which the microbiome provides signals to the host immune system to establish immunoregulatory pathways critical to protect against allergic disease are only beginning to be understood. Fortunately, because the microbiome is so attuned to the external environment, many opportunities for therapeutic intervention exist. It will be the task of current and future immunologists and clinicians to further elucidate the complex interactions of environment, the microbiome, and host responses to food and to use this knowledge for the treatment of food allergic disease.
Acknowledgments
This work was supported by the Food Allergy Initiative. We thank Joanna Wroblewska for assistance with preparation of the figures and review of the manuscript.
Abbreviations
- TLR
Toll-like receptor
- Treg
Regulatory T cells
- SCFA
short chain fatty acids
- LP
Lamina propria
- GF
germ free
- APC
antigen presenting cell
- DC
dendritic cell
- MLN
mesenteric lymph node
- OVA
chicken egg ovalbumin
References
- 1.Branum AM, Lukacs SL. Food allergy among U.S. children: trends in prevalence and hospitalizations. National Center for Health Statistics Data Brief; 2008. [PubMed] [Google Scholar]
- 2.Hong X, Tsai H-J, Wang X. Genetics of food allergy. Current Opinion in Pediatrics. 2009;21(6):770–776. doi: 10.1097/MOP.0b013e32833252dc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vercelli D. Genetic polymorphism in allergy and asthma. Curr Opin Immunol. 2003;15(6):609–613. doi: 10.1016/j.coi.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Vercelli D. Advances in asthma and allergy genetics in 2007. J Allergy Clin Immunol. 2008;122(2):267–271. doi: 10.1016/j.jaci.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 5.Vercelli D. Genetics, epigenetics, and the environment: switching, buffering, releasing. J Allergy Clin Immunol. 2004;113(3):381–386. doi: 10.1016/j.jaci.2004.01.752. [DOI] [PubMed] [Google Scholar]
- 6.Strachan DP. Hay fever, hygiene, and household size. Brit Med J. 1989;299(6710):1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alm JS, et al. Atopy in children of families with an anthroposophic lifestyle. Lancet. 1999;353(9163):1485–1488. doi: 10.1016/S0140-6736(98)09344-1. [DOI] [PubMed] [Google Scholar]
- 8.Sepp E, et al. Intestinal microflora of Estonian and Swedish infants. Acta Pædiatrica. 1997;86(9):956–961. doi: 10.1111/j.1651-2227.1997.tb15178.x. [DOI] [PubMed] [Google Scholar]
- 9.Graham-Rowe D. Lifestyle: When allergies go west. Nature. 2011;479(7374):S2–S4. [Google Scholar]
- 10.Mutius EV, et al. Prevalence of asthma and allergic disorders among children in united Germany: a descriptive comparison. Brit Med J. 1992;305(6866):1395–1399. doi: 10.1136/bmj.305.6866.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Hertzen L, et al. Growing disparities in atopy between the Finns and the Russians: A comparison of 2 generations. J of Allergy Clin Immunol. 2006;117(1):151–157. doi: 10.1016/j.jaci.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 12.Addo-Yobo EOD, et al. Exercise-Induced Bronchospasm and Atopy in Ghana: Two Surveys Ten Years Apart. PLoS Med. 2007;4(2):e70. doi: 10.1371/journal.pmed.0040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riedler J, et al. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358(9288):1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 14.von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol. 2010;10(12):861–868. doi: 10.1038/nri2871. [DOI] [PubMed] [Google Scholar]
- 15.Braun-Fahrländer C, et al. Prevalence of hay fever and allergic sensitization in farmer’s children and their peers living in the same rural community. SCARPOL team. Swiss Study on Childhood Allergy and Respiratory Symptoms with Respect to Air Pollution. Clin Exp Allergy. 1999;29(1):28–34. doi: 10.1046/j.1365-2222.1999.00479.x. [DOI] [PubMed] [Google Scholar]
- 16.Riedler J, et al. Austrian children living on a farm have less hay fever, asthma and allergic sensitization. Clin Exp Allergy. 2000;30(2):194–200. doi: 10.1046/j.1365-2222.2000.00799.x. [DOI] [PubMed] [Google Scholar]
- 17.Braun-Fahrlander C, et al. Environmental exposure to endotoxin and its relation to asthma in school-age children. N Engl J Med. 2002;347(12):869–877. doi: 10.1056/NEJMoa020057. [DOI] [PubMed] [Google Scholar]
- 18.Waser M, et al. Exposure to pets, and the association with hay fever, asthma, and atopic sensitization in rural children. Allergy. 2005;60(2):177–184. doi: 10.1111/j.1398-9995.2004.00645.x. [DOI] [PubMed] [Google Scholar]
- 19.Brunekreef B, et al. Early life exposure to farm animals and symptoms of asthma, rhinoconjunctivitis and eczema: an ISAAC Phase Three Study. Int J Epidemiol. 2012;41(3):753–761. doi: 10.1093/ije/dyr216. [DOI] [PubMed] [Google Scholar]
- 20.Lauener RP, et al. Expression of CD14 and Toll-like receptor 2 in farmers’ and nonfarmers’ children. Lancet. 2002;360 (9331):465–466. doi: 10.1016/S0140-6736(02)09641-1. [DOI] [PubMed] [Google Scholar]
- 21.Ege MJ, et al. Prenatal farm exposure is related to the expression of receptors of the innate immunity and to atopic sensitization in school-age children. J Allergy Clin Immunol. 2006;117(4):817–823. doi: 10.1016/j.jaci.2005.12.1307. [DOI] [PubMed] [Google Scholar]
- 22.Stern DA, Riedler J, Nowak D, Braun-Fahrlander C, Swoboda I, Balic N, Chen KW, Vrtala S, Gronlund H, van Hage M, et al. Exposure to a farming environment has allergen-specific protective effects on TH2-dependent isotype switching in response to common inhalants. J Allergy Clin Immunol. 2007;119(2):351–358. doi: 10.1016/j.jaci.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Schaub B, et al. Maternal farm exposure modulates neonatal immune mechanisms through regulatory T cells. J Allergy Clin Immunol. 2009;123(4):774–782. e775. doi: 10.1016/j.jaci.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 24.Peters M, et al. Inhalation of stable dust extract prevents allergen induced airway inflammation and hyperresponsiveness. Thorax. 2006;61(2):134–139. doi: 10.1136/thx.2005.049403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Debarry J, et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J Allergy Clin Immunol. 2007;119(6):1514–1521. doi: 10.1016/j.jaci.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 26.Platts-Mills T, et al. Sensitisation, asthma, and a modified Th2 response in children exposed to cat allergen: a population-based cross-sectional study. Lancet. 2001;357(9258):752–756. doi: 10.1016/S0140-6736(00)04168-4. [DOI] [PubMed] [Google Scholar]
- 27.Smallwood J, Ownby D. Exposure to Dog Allergens and Subsequent Allergic Sensitization: An Updated Review. Curr Allergy Asthma Rep. 2012 doi: 10.1007/s11882-012-0277-0. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 28.Gern JE, et al. Effects of dog ownership and genotype on immune development and atopy in infancy. J Allergy Clin Immunol. 2004;113(2):307–314. doi: 10.1016/j.jaci.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 29.Ownby DR, Johnson CC, Peterson EL. Exposure to dogs and cats in the first year of life and risk of allergic sensitization at 6 to 7 years of age. JAMA. 2002;288(8):963–972. doi: 10.1001/jama.288.8.963. [DOI] [PubMed] [Google Scholar]
- 30.Fujimura KE, et al. Man’s best friend? The effect of pet ownership on house dust microbial communities. J Allergy Clin Immunol. 2010;126(2):410–412. doi: 10.1016/j.jaci.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havstad S, et al. Effect of prenatal indoor pet exposure on the trajectory of total IgE levels in early childhood. J Allergy Clin Immunol. 2011;128(4):880–885. doi: 10.1016/j.jaci.2011.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130(1):44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 33.Litonjua AA, et al. A longitudinal analysis of wheezing in young children: The independent effects of early life exposure to house dust endotoxin, allergens, and pets. J Allergy Clin Immunol. 2002;110(5):736–742. doi: 10.1067/mai.2002.128948. [DOI] [PubMed] [Google Scholar]
- 34.Bufford JD, et al. Effects of dog ownership in early childhood on immune development and atopic diseases. Clin Exp Allergy. 2008;38(10):1635–1643. doi: 10.1111/j.1365-2222.2008.03018.x. [DOI] [PubMed] [Google Scholar]
- 35.Wegienka G, et al. Indoor pet exposure and the outcomes of total IgE and sensitization at age 18 years. J Allergy Clin Immunol. 2010;126(2):274–279. doi: 10.1016/j.jaci.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wills-Karp M, Santeliz J, Karp CL. The germless theory of allergic disease: revisiting the hygiene hypothesis. Nat Rev Immunol. 2001;1(1):69–75. doi: 10.1038/35095579. [DOI] [PubMed] [Google Scholar]
- 37.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330(6012):1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol. 2010;3(5):450–460. doi: 10.1038/mi.2010.20. [DOI] [PubMed] [Google Scholar]
- 39.Noverr MC, Huffnagle GB. The ‘microflora hypothesis’ of allergic diseases. Clin Exp Allergy. 2005;35(12):1511–1520. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 40.Prioult G, Nagler-Anderson C. Mucosal immunity and allergic responses: lack of regulation and/or lack of microbial stimulation? Immunol Rev. 2005;206:204–218. doi: 10.1111/j.0105-2896.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 41.Björkstén B, et al. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108(4):516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 42.Björkstén B, et al. The intestinal microflora in allergic Estonian and Swedish 2-year-old children. Clin Exp Allergy. 1999;29(3):342–346. doi: 10.1046/j.1365-2222.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 43.Kalliomaki M, et al. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107(1):129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 44.Böttcher MF, et al. Microflora-associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy. 2000;30(11):1591–1596. doi: 10.1046/j.1365-2222.2000.00982.x. [DOI] [PubMed] [Google Scholar]
- 45.Kirjavainen PV, et al. Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol Med Microbiol. 2001;32(1):1–7. doi: 10.1111/j.1574-695X.2001.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 46.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ley RE, et al. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6(10):776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dominguez-Bello MG, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenteroloy. 2011;140(6):1713–1719. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587(Pt 17):4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dethlefsen L, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6(11):e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turnbaugh PJ, et al. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Filippo C, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walk ST, et al. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis. 2010;16(11):1841–1849. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hill DA, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3(2):148–158. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120(12):4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476(7361):393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 60.Turnbaugh PJ, et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reikvam DH, et al. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One. 2011;6(3):e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Pro Natl Acad Sci USA. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1(1):56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 64.De La Cochetière MF, et al. Resilience of the Dominant Human Fecal Microbiota upon Short-Course Antibiotic Challenge. J Clin Microbiol. 2005;43(11):5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakobsson HE, et al. Short-Term Antibiotic Treatment Has Differing Long-Term Impacts on the Human Throat and Gut Microbiome. PLoS One. 2010;5(3):e9836. doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson CC, et al. Antibiotic exposure in early infancy and risk for childhood atopy. J Allergy Clin Immunol. 2005;115(6):1218–1224. doi: 10.1016/j.jaci.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 67.Droste JHJ, et al. Does the use of antibiotics in early childhood increase the risk of asthma and allergic disease. Clin Exp Allergy. 2000;30(11):1547–1553. doi: 10.1046/j.1365-2222.2000.00939.x. [DOI] [PubMed] [Google Scholar]
- 68.Kozyrskyj AL, Ernst P, Becker AB. Increased Risk of Childhood Asthma From Antibiotic Use in Early Life. Chest. 2007;131(6):1753–1759. doi: 10.1378/chest.06-3008. [DOI] [PubMed] [Google Scholar]
- 69.Wickens K, et al. Antibiotic use in early childhood and the development of asthma. Clin Exp Allergy. 1999;29(6):766–771. doi: 10.1046/j.1365-2222.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 70.Wjst MHB, et al. Early antibiotic treatment and later asthma. Eur J Med Res. 2001;6(6):263–271. [PubMed] [Google Scholar]
- 71.Marra F, et al. Does antibiotic exposure during infancy lead to development of asthma?: a systematic review and metaanalysis. Chest. 2006;129(3):610–618. doi: 10.1378/chest.129.3.610. [DOI] [PubMed] [Google Scholar]
- 72.Dom S, et al. Pre- and post-natal exposure to antibiotics and the development of eczema, recurrent wheezing and atopic sensitization in children up to the age of 4 years. Clin Exp Allergy. 2010;40(9):1378–1387. doi: 10.1111/j.1365-2222.2010.03538.x. [DOI] [PubMed] [Google Scholar]
- 73.Bashir ME, et al. Toll-like receptor 4 signaling by intestinal microbes influences susceptibility to food allergy. J Immunol. 2004;172(11):6978–6987. doi: 10.4049/jimmunol.172.11.6978. [DOI] [PubMed] [Google Scholar]
- 74.Stefka AT, et al. Commensal bacteria-induced regulatory Tcells prevent systemic allergic responses to food. 2012 Submitted for publication. [Google Scholar]
- 75.Atarashi K, et al. Induction of Colonic Regulatory T Cells by Indigenous Clostridium Species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell SL, et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep. 2012;13(5):440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hill DA, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18(4):538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Maslowski KM, Mackay CR. Diet, gut microbiota and immune responses. Nat Immunol. 2011;12(1):5–9. doi: 10.1038/ni0111-5. [DOI] [PubMed] [Google Scholar]
- 79.Peng L, et al. Butyrate Enhances the Intestinal Barrier by Facilitating Tight Junction Assembly via Activation of AMP-Activated Protein Kinase in Caco-2 Cell Monolayers. J Nutr. 2009;139(9):1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lewis K, et al. Enhanced translocation of bacteria across metabolically stressed epithelia is reduced by butyrate. Inflamm Bowel Dis. 2010;16(7):1138–1148. doi: 10.1002/ibd.21177. [DOI] [PubMed] [Google Scholar]
- 81.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Høverstad T, Midtvedt T. Short-Chain Fatty Acids in Germfree Mice and Rats. J Nutr. 1986;116(9):1772–1776. doi: 10.1093/jn/116.9.1772. [DOI] [PubMed] [Google Scholar]
- 83.Paturi G, et al. Cecal and Colonic Responses in Rats Fed 5 or 30% Corn Oil Diets Containing Either 7.5% broccoli dietary fiber or microcrystalline cellulose. J Agric Food Chem. 2010;58(10):6510–6515. doi: 10.1021/jf100296m. [DOI] [PubMed] [Google Scholar]
- 84.Paturi G, et al. Influence of dietary blueberry and broccoli on cecal microbiota activity and colon morphology in mdr1a−/− mice, a model of inflammatory bowel diseases. Nutrition. 2012;28(3):324–330. doi: 10.1016/j.nut.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 85.Harig JM, et al. Treatment of Diversion Colitis with Short-Chain-Fatty Acid Irrigation. N Engl J Med. 1989;320(1):23–28. doi: 10.1056/NEJM198901053200105. [DOI] [PubMed] [Google Scholar]
- 86.Vernia P, et al. Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment Pharmacol Ther. 1995;9(3):309–313. doi: 10.1111/j.1365-2036.1995.tb00386.x. [DOI] [PubMed] [Google Scholar]
- 87.Ley RE, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hildebrandt MA, et al. High Fat Diet Determines the Composition of the Murine Gut Microbiome Independently of Obesity. Gastroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Devkota S, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kull I, et al. Fish consumption during the first year of life and development of allergic diseases during childhood. Allergy. 2006;61(8):1009–1015. doi: 10.1111/j.1398-9995.2006.01115.x. [DOI] [PubMed] [Google Scholar]
- 91.Dunstan JA, et al. Fish oil supplementation in pregnancy modifies neonatal allergen-specific immune responses and clinical outcomes in infants at high risk of atopy: A randomized, controlled trial. J Allergy Clin Immunol. 2003;112(6):1178–1184. doi: 10.1016/j.jaci.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 92.Manley BJ, et al. High-Dose Docosahexaenoic Acid Supplementation of Preterm Infants: Respiratory and Allergy Outcomes. Pediatrics. 2011;128(1):e71–e77. doi: 10.1542/peds.2010-2405. [DOI] [PubMed] [Google Scholar]
- 93.Kankaanpaa P, et al. Polyunsaturated fatty acids in maternal diet, breast milk, and serum lipid fatty acids of infants in relation to atopy. Allergy. 2001;56(7):633–638. doi: 10.1034/j.1398-9995.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- 94.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu GD, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hewitson JP, Grainger JR, Maizels RM. Helminth immunoregulation: the role of parasite secreted proteins in modulating host immunity. Mol Biochem Parasitol. 2009;167(1):1–11. doi: 10.1016/j.molbiopara.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van den Biggelaar AH, van Ree R, Rodrigues LC, et al. Decreased atopy in children infected with Schistosoma haematobium: a role for parasite-induced IL-10. Lancet. 2000;356(9243):1723–1727. doi: 10.1016/S0140-6736(00)03206-2. [DOI] [PubMed] [Google Scholar]
- 98.Yazdanbakhsh M, van den Biggelaar A, Maizels RM. Th2 responses without atopy: immunoregulation in chronic helminth infections and reduced allergic disease. Trends Immunol. 2001;22(7):372–377. doi: 10.1016/s1471-4906(01)01958-5. [DOI] [PubMed] [Google Scholar]
- 99.Flohr C, et al. Reduced helminth burden increases allergen skin sensitization but not clinical allergy:a randomized, double-blind, placebo-controlled trial in Vietnam. Clin Exp Allergy. 2010;40(1):131–142. doi: 10.1111/j.1365-2222.2009.03346.x. [DOI] [PubMed] [Google Scholar]
- 100.Lynch NR, et al. Effect of anthelmintic treatment on the allergic reactivity of children in a tropical slum. J Allergy Clin Immunol. 1993;92(3):404–411. doi: 10.1016/0091-6749(93)90119-z. [DOI] [PubMed] [Google Scholar]
- 101.Wilson MS, et al. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J Exp Med. 2005;202(9):1199–1212. doi: 10.1084/jem.20042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mangan NE, et al. Helminth infection protects mice from anaphylaxis via IL-10-producing B cells. J Immunol. 2004;173(10):6346–6356. doi: 10.4049/jimmunol.173.10.6346. [DOI] [PubMed] [Google Scholar]
- 103.Mangan NE, et al. Helminth-modified pulmonary immune response protects mice from allergen-induced airway hyperresponsiveness. J Immunol. 2006;176(1):138–147. doi: 10.4049/jimmunol.176.1.138. [DOI] [PubMed] [Google Scholar]
- 104.Kitagaki K, et al. Intestinal helminths protect in a murine model of asthma. J Immunol. 2006;177(3):1628–1635. doi: 10.4049/jimmunol.177.3.1628. [DOI] [PubMed] [Google Scholar]
- 105.Cardoso LS, et al. Schistosoma mansoni antigens modulate the allergic response in a murine model of ovalbumin-induced airway inflammation. Clin Exp Immunol. 2010;160(2):266–274. doi: 10.1111/j.1365-2249.2009.04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wilson MS, et al. Helminth-induced CD19+CD23hi B cells modulate experimental allergic and autoimmune inflammation. Eur J Immunol. 2010;40(6):1682–1696. doi: 10.1002/eji.200939721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McSorley HJ, et al. Suppression of type 2 immunity and allergic airway inflammation by secreted products of the helminth Heligmosomoides polygyrus. Eur J Immunol. 2012 doi: 10.1002/eji.201142161. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bashir ME, et al. An enteric helminth infection protects against an allergic response to dietary antigen. J Immunol. 2002;169(6):3284–3292. doi: 10.4049/jimmunol.169.6.3284. [DOI] [PubMed] [Google Scholar]
- 109.Feary JR, et al. Experimental hookworm infection: a randomized placebo-controlled trial in asthma. Clin Exp Allergy. 2010;40(2):299–306. doi: 10.1111/j.1365-2222.2009.03433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bager P, et al. Trichuris suis ova therapy for allergic rhinitis: A randomized, double-blind, placebo-controlled clinical trial. J Allergy Clin Immunol. 2010;125(1):123–130. e123. doi: 10.1016/j.jaci.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 111.Jouvin M-H, Kinet J-P. Trichuris suis ova: Testing a helminth-based therapy as an extension of the hygiene hypothesis. J Allergy Clin Immunol. 2012;130(1):3–10. doi: 10.1016/j.jaci.2012.05.028. [DOI] [PubMed] [Google Scholar]
- 112.Perry S, et al. Infection with Helicobacter pylori Is Associated with Protection against Tuberculosis. PLoS One. 2010;5(1):e8804. doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen Y, Blaser MJ. Helicobacter pylori colonization is inversely associated with childhood asthma. J Infect Dis. 2008;198(4):553–560. doi: 10.1086/590158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167(8):821–827. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 115.Reibman J, et al. Asthma is inversely associated with Helicobacter pylori status in an urban population. PLoS ONE. 2008;3(12):e4060. doi: 10.1371/journal.pone.0004060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arnold IC, et al. Helicobacter pylori infection prevents allergic asthma in mouse models through the induction of regulatory T cells. J Clin Invest. 2011;121(8):3088–3093. doi: 10.1172/JCI45041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(3):204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 118.Barman M, et al. Enteric Salmonellosis Disrupts the Microbial Ecology of the Murine Gastrointestinal Tract. Infect Immun. 2008;76(3):907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Grölund M-M, et al. Fecal Microflora in Healthy Infants Born by Different Methods of Delivery: Permanent Changes in Intestinal Flora After Cesarean Delivery. J Pediatr Gastroenterol Nutr. 1999;28(1):19–25. doi: 10.1097/00005176-199901000-00007. [DOI] [PubMed] [Google Scholar]
- 120.Savage DC, Dubos R, Schaedler RW. The gastrointestinal epithelium and its autochthonous bacterial flora. J Exp Med. 1968;127(1):67–76. doi: 10.1084/jem.127.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Martin R, et al. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1(4):367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- 122.Costello EK, et al. Bacterial Community Variation in Human Body Habitats Across Space and Time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huurre A, et al. Mode of delivery - effects on gut microbiota and humoral immunity. Neonatology. 2008;93(4):236–240. doi: 10.1159/000111102. [DOI] [PubMed] [Google Scholar]
- 124.van Nimwegen FA, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128(5):948–955. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 125.Björkstén B. Genetic and environmental risk factors for the development of food allergy. Curr Opin Allergy Clin Immunol. 2005;5(3):249–253. doi: 10.1097/01.all.0000168790.82206.17. [DOI] [PubMed] [Google Scholar]
- 126.Salminen S, et al. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut. 2004;53(9):1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Eggesbø M, et al. Is delivery by cesarean section a risk factor for food allergy? J Allergy Clin Immunol. 2003;112(2):420–426. doi: 10.1067/mai.2003.1610. [DOI] [PubMed] [Google Scholar]
- 128.Kero J, et al. Mode of delivery and asthma -- is there a connection? Pediatr Rev. 2002;52(1):6–11. doi: 10.1203/00006450-200207000-00004. [DOI] [PubMed] [Google Scholar]
- 129.Decker E, Hornef M, Stockinger S. Cesarean delivery is associated with celiac disease but not inflammatory bowel disease in children. Gut Microbes. 2011;2(2):91–98. doi: 10.4161/gmic.2.2.15414. [DOI] [PubMed] [Google Scholar]
- 130.Kramer MS. Breastfeeding and Allergy: The Evidence. Ann Nutr Metab. 2011;59(Suppl 1):20–26. doi: 10.1159/000334148. [DOI] [PubMed] [Google Scholar]
- 131.Fagarasan S, Honjo T. Intestinal IgA synthesis: regulation of front-line body defences. Nat Rev Immunol. 2003;3(1):63–72. doi: 10.1038/nri982. [DOI] [PubMed] [Google Scholar]
- 132.Greer FR, et al. Effects of Early Nutritional Interventions on the Development of Atopic Disease in Infants and Children: The Role of Maternal Dietary Restriction, Breastfeeding, Timing of Introduction of Complementary Foods, and Hydrolyzed Formulas. Pediatrics. 2008;121(1):183–191. doi: 10.1542/peds.2007-3022. [DOI] [PubMed] [Google Scholar]
- 133.Iyengar SR, Walker WA. Immune Factors In Breastmilk And The Development of Atopic Disease. Journal of Pediatric Gastroenterology and Nutrition. 2012 doi: 10.1097/MPG.0b013e3182617a9d. Publish Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 134.von Berg A, et al. The effect of hydrolyzed cow’s milk formula for allergy prevention in the first year of life: The German Infant Nutritional Intervention Study, a randomized double-blind trial. J Allergy Clin Immunol. 2003;111(3):533–540. doi: 10.1067/mai.2003.101. [DOI] [PubMed] [Google Scholar]
- 135.Laubereau B, et al. Effect of breast-feeding on the development of atopic dermatitis during the first 3 years of life—results from the GINI-birth cohort study. J Pediatr. 2004;144(5):602–607. doi: 10.1016/j.jpeds.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 136.Yang YW, Tsai CL, Lu CY. Exclusive breastfeeding and incident atopic dermatitis in childhood: a systematic review and meta-analysis of prospective cohort studies. Br J Dermatol. 2009;161(2):373–383. doi: 10.1111/j.1365-2133.2009.09049.x. [DOI] [PubMed] [Google Scholar]
- 137.Hascoët J-M, et al. Effect of Formula Composition on the Development of Infant Gut Microbiota. J Pediatr Gastroenterol Nutr. 2011;52(6):756–762. doi: 10.1097/MPG.0b013e3182105850. [DOI] [PubMed] [Google Scholar]
- 138.Maga EA, et al. Consumption of lysozyme-rich milk can alter microbial fecal populations. Appl Environ Microbiol. 2012 doi: 10.1128/AEM.00956-12. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Smith DW, Nagler-Anderson C. Preventing Intolerance: The Induction of Nonresponsiveness to Dietary and Microbial Antigens in the Intestinal Mucosa. J Immunol. 2005;174(7):3851–3857. doi: 10.4049/jimmunol.174.7.3851. [DOI] [PubMed] [Google Scholar]
- 140.Vickery BP, et al. Mechanisms of immune tolerance relevant to food allergy. J Allergy Clin Immunol. 2011;127(3):576–584. doi: 10.1016/j.jaci.2010.12.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim JS, Sampson HA. Food allergy: a glimpse into the inner workings of gut immunology. Curr Opin Gastroenterol. 2012;28(2):99–103. doi: 10.1097/MOG.0b013e32834e7b60. [DOI] [PubMed] [Google Scholar]
- 142.Wang J, Sampson HA. Food allergy. J Clin Invest. 2011;121(3):827–835. doi: 10.1172/JCI45434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Weiner HL, et al. Oral tolerance. Immunol Rev. 2011;241(1):241–259. doi: 10.1111/j.1600-065X.2011.01017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hadis U, et al. Intestinal Tolerance Requires Gut Homing and Expansion of FoxP3+ Regulatory T Cells in the Lamina Propria. Immunity. 2011;34(2):237–246. doi: 10.1016/j.immuni.2011.01.016. [DOI] [PubMed] [Google Scholar]
- 145.Cassani B, et al. Gut-Tropic T Cells That Express Integrin a4β7 and CCR9 Are Required for Induction of Oral Immune Tolerance in Mice. Gastroenterology. 2011;141(6):2109–2118. doi: 10.1053/j.gastro.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pabst O, Mowat AM. Oral tolerance to food protein. Mucosal Immunol. 2012;5(3):232–239. doi: 10.1038/mi.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jaensson E, et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med. 2008;205(9):2139–2149. doi: 10.1084/jem.20080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schulz O, et al. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J Exp Med. 2009;206(13):3101–3114. doi: 10.1084/jem.20091925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun C-M, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204(8):1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rubtsov YP, et al. Regulatory T Cell-Derived Interleukin-10 Limits Inflammation at Environmental Interfaces. Immunity. 2008;28(4):546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 151.Murai M, et al. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10(11):1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Medina-Contreras O, et al. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest. 2011;121(12):4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Crispe IN. The Liver as a Lymphoid Organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 154.Goubier A, et al. Plasmacytoid Dendritic Cells Mediate Oral Tolerance. Immunity. 2008;29(3):464–475. doi: 10.1016/j.immuni.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dubois B, et al. Sequential Role of Plasmacytoid Dendritic Cells and Regulatory T Cells in Oral Tolerance. Gastroenterology. 2009;137(3):1019–1028. doi: 10.1053/j.gastro.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 156.Bar-On L, et al. CX3CR1+ CD8alpha+ dendritic cells are a steady-state population related to plasmacytoid dendritic cells. Pro Natl Acad Sci USA. 2010;107(33):14745–14750. doi: 10.1073/pnas.1001562107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol. 2007;19(2):59–69. doi: 10.1016/j.smim.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 158.Hamada H, et al. Identification of multiple isolated lymphoid follicles on the antimesenteric wall of the mouse small intestine. J Immunol. 2002;168(1):57–64. doi: 10.4049/jimmunol.168.1.57. [DOI] [PubMed] [Google Scholar]
- 159.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–688. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 160.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mazmanian SK, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122(1):107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 162.McCoy KD, et al. Natural IgE production in the absence of MHC Class II cognate help. Immunity. 2006;24(3):329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 163.Itoh K, Mitsuoka T. Characterization of clostridia isolated from faeces of limited flora mice and their effect on caecal size when associated with germ-free mice. Lab Anim. 1985;19(2):111–118. doi: 10.1258/002367785780942589. [DOI] [PubMed] [Google Scholar]
- 164.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 165.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107(27):12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fagarasan S, Honjo T. Regulation of IgA synthesis at mucosal surfaces. Curr Opin Immunol. 2004;16(3):277–283. doi: 10.1016/j.coi.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 167.Feng T, et al. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J Immunol. 2011;186(11):6313–6318. doi: 10.4049/jimmunol.1001454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu H-J, et al. Gut-Residing Segmented Filamentous Bacteria Drive Autoimmune Arthritis via T Helper 17 Cells. Immunity. 2010;32(6):815–827. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nagano Y, Itoh K, Honda K. The induction of Treg cells by gut-indigenous Clostridium. Curr Opin Immunol. 2012 doi: 10.1016/j.coi.2012.05.007. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 170.Umesaki Y, et al. Differential Roles of Segmented Filamentous Bacteria and Clostridia in Development of the Intestinal Immune System. Infect Immunol. 1999;67(7):3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Cong Y, et al. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106(46):19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nava GM, Friedrichsen HJ, Stappenbeck TS. Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 2011;5(4):627–638. doi: 10.1038/ismej.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Duck LW, et al. Isolation of flagellated bacteria implicated in Crohn’s disease. Inflamm Bowel Dis. 2007;13(10):1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 174.Shen C, et al. Enhanced CBir1-specific innate and adaptive immune responses in Crohn’s disease. Inflamm Bowel Dis. 2008;14(12):1641–1651. doi: 10.1002/ibd.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Di Giacinto C, et al. Probiotics Ameliorate Recurrent Th1-Mediated Murine Colitis by Inducing IL-10 and IL-10-Dependent TGF-β-Bearing Regulatory Cells. J Immunol. 2005;174(6):3237–3246. doi: 10.4049/jimmunol.174.6.3237. [DOI] [PubMed] [Google Scholar]
- 177.Karimi K, et al. Lactobacillus reuteri–induced Regulatory T cells Protect against an Allergic Airway Response in Mice. Am J Respir Crit Care Med. 2009;179(3):186–193. doi: 10.1164/rccm.200806-951OC. [DOI] [PubMed] [Google Scholar]
- 178.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lathrop SK, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature. 2011;478(7368):250–254. doi: 10.1038/nature10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Olszak T. Microbial Exposure During Early Life Has Persistent Effects on Natural Killer T Cell Function. Science. 2012;336(6080):489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Noverr MC, Huffnagle GB. Does the microbiota regulate immune responses outside the gut? Trends Microbiol. 2004;12(12):562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 182.Willyard C. Microbiome: Gut reaction. Nature. 2011;479(7374):S5–S7. [Google Scholar]
- 183.Molin G. Probiotics in foods not containing milk or milk constituents, with special reference to Lactobacillus plantarum 299v. Am J Clin Nutr. 2001;73(2 Suppl):380S–385S. doi: 10.1093/ajcn/73.2.380s. [DOI] [PubMed] [Google Scholar]
- 184.Clarke G, et al. Review article: probiotics for the treatment of irritable bowel syndrome – focus on lactic acid bacteria. Aliment Pharmacol Ther. 2012;35(4):403–413. doi: 10.1111/j.1365-2036.2011.04965.x. [DOI] [PubMed] [Google Scholar]
- 185.Jonkers D, et al. Probiotics in the Management of Inflammatory Bowel Disease: A Systematic Review of Intervention Studies in Adult Patients. Drugs. 2012;72(6):803–823. doi: 10.2165/11632710-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 186.Heineman J, et al. Fighting Fire with Fire: Is it Time to Use Probiotics to Manage Pathogenic Bacterial Diseases? Curr Gastroenterol Rep. 2012;14(4):343–348. doi: 10.1007/s11894-012-0274-4. [DOI] [PubMed] [Google Scholar]
- 187.Wickens K, et al. A differential effect of 2 probiotics in the prevention of eczema and atopy: A double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2008;122(4):788–794. doi: 10.1016/j.jaci.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 188.Wickens K, et al. A protective effect of Lactobacillus rhamnosus HN001 against eczema in the first 2 years of life persists to age 4 years. Clin Exp Allergy. 2012;42(7):1071–1079. doi: 10.1111/j.1365-2222.2012.03975.x. [DOI] [PubMed] [Google Scholar]
- 189.Kalliomaki N, et al. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet. 2001;357(9262):1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 190.Kukkonen K, et al. Probiotics and prebiotic galacto-oligosaccharides in the prevention of allergic diseases: a randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol. 2007;119(1):192–198. doi: 10.1016/j.jaci.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 191.Johansson ML, et al. Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol. 1993;59(1):15–20. doi: 10.1128/aem.59.1.15-20.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Goossens D. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther. 2003;18(5):495–505. doi: 10.1046/j.1365-2036.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 193.Tannock GW, et al. Analysis of the Fecal Microflora of Human Subjects Consuming a Probiotic Product Containing Lactobacillus rhamnosusDR20. Appl Environ Microbiol. 2000;66(6):2578–2588. doi: 10.1128/aem.66.6.2578-2588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]