Abstract
Previously, we described a novel pH-responsive RNA element in Escherichia coli that resides in the 5′ untranslated region of the alx gene and controls its translation in a pH-dependent manner. Under normal growth conditions, this RNA region forms a translationally inactive structure, but when transcribed under alkaline conditions, it forms an active structure producing the Alx protein. We identified two distinct transcriptional pause sites and proposed that pausing at these sites interfered with the formation of the inactive structure while facilitating folding of the active one. Alkali increases the longevity of pausing at these sites, thereby promoting folding of the translationally active form of alx RNA. We show here that mutations that modify the extent and/or position of pausing, although silent with regard to structure stability per se, greatly influence the dynamics of folding and thereby translation. Our data illustrate the mechanistic design of alx regulation, relying on precise temporal and spatial characteristics. We propose that this unique design provides an opportunity for environmental signals such as pH to introduce structural changes in the RNA and thereby modulate expression.
INTRODUCTION
RNA conformation plays a major role in processes such as transcription termination, RNA processing and translation initiation. Hence, the folding of RNA is an important determinant in regulation of gene expression in many cellular systems, in both prokaryotes and eukaryotes. Furthermore, the structure of RNAs may change in response to environmental signals. Thus, understanding the mechanisms by which RNAs assemble to form active units is of considerable interest.
An increasing body of evidence suggests that much of the regulation of gene expression that is based on RNA folding is accomplished during the elongation phase of transcription and involves pausing of RNA polymerase (RNAP) (1). Pausing, a temporal inhibition of the transcription elongation complex, plays an essential role in coupling transcription and translation in bacteria (2). Pausing also allows interaction with, or recruitment of regulators (3,4) and ligands and acts as the first step leading to transcription termination (5–8). Transcriptional pausing has also been shown to assist in the correct folding of certain Escherichia coli non-coding RNAs, including signal recognition particle (SRP), RNase P and transfer-messenger RNA (tmRNA) (9). Importantly, pausing plays a crucial role in gene expression regulation by riboswitches, coordinating RNA folding with ligand binding (10–13).
Recently, we have described the first example of a pH-responsive RNA regulator that operates in the absence of a ligand, undergoing a conformational change depending on RNAP pausing. This pH-responsive RNA element (PRE) precedes the alx gene, encoding a putative transporter, and controls its translation in a pH-dependent manner. In cells grown under neutral conditions, the PRE-alx transcript forms a translationally inactive structure, whereas growth under high pH results in the formation of an active structure and production of Alx (14). The pH-dependent folding of PRE-alx RNA into an active structure requires de novo synthesis, during which RNAP, by pausing at two sites, plays an active role in controlling which structure forms. The location of the pause sites led us to suggest that both correlate with the formation of the alx translatable structure. By pausing at the first site (PC in Figure 1), RNAP sequesters part of the sequence that is required to form hairpin C and thereby allows formation of an alternative hairpin (hairpin S in Figure 1). Once formed, this hairpin precludes annealing of the former one. Similarly, by pausing at the second site (PD), the elongation complex prevents the formation of hairpin D and thereby indirectly promotes annealing of complementary sequences in hairpins C and D, forming a new hairpin (hairpin C/D). As a consequence, the alternative translationally active structure forms. In this study, we show that mutations that change the precise location of pausing or mutations leading to increased pausing have a profound effect on the folding path of PRE-alx.
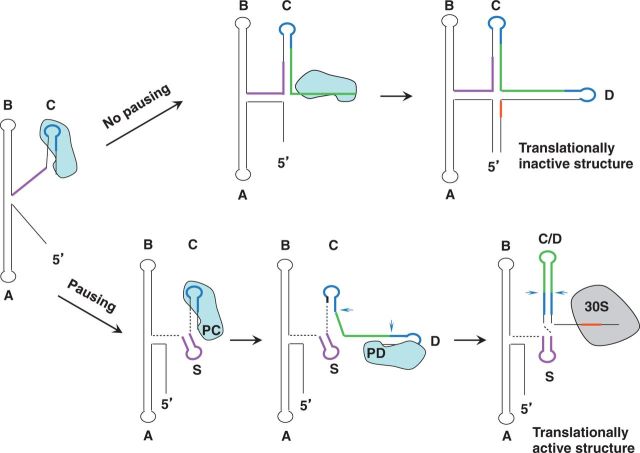
Figure 1.
A model for PRE-alx regulation by transcriptional pausing [adapted from (14)]. In the absence of transcriptional pausing, PRE-alx forms a translationally inactive structure, whereas when folding is affected by RNAP pausing, the newly synthesized RNA adopts an active structure in which the ribosome-binding site (in red) is exposed for 30 S ribosomal subunit binding. Formation of the active structure involves pausing of RNAP at two sites located at hairpin C (PC) and hairpin D (PD). On pausing of RNAP (shaded in light blue) at PC, the sequence required to complete stem C is sequestered within the transcription elongation complex and thus formation of hairpin C is prevented. Instead, hairpin S (indicated by a purple line) forms. Similarly, pausing at PD inhibits the formation of hairpin D, allowing base pairing between the complementary sequences in hairpins C and D (in blue) and formation of the apical part of hairpin C/D (in green).
MATERIALS AND METHODS
Bacterial growth conditions
Escherichia coli MC4100 cells were grown at 37°C (200 rpm) in Luria-Bertani broth (LB) medium (pH 6.8) or in potassium modified LB medium (LBK; pH 8.4) (14). Ampicillin (100 µg/ml) and kanamycin (40 µg/ml) were added where appropriate.
Plasmid construction
The construction of plasmids carrying Palx-PRE-alx'-'lacZ (pSA60; translation fusion), PT7-PRE-alx' in pGEM4 (pSA66), Palx-PRE-alx' in pGEM4 (pSA64) and Palx-PRE-alx' in pZE12 (pSA65) has been described previously (14,15).
Site-directed mutagenesis
Mutants A132G and C135U were generated by PCR using plasmid carrying Palx-PRE-alx' (pSA64) as template and two tail-to-tail divergent primers (5′ phosphorylated) of which one carried the desired mutation. The PCR product was subjected to blunt-end ligation and then used as template to amplify the desired fragment for sub-cloning. To construct Palx-PRE-alx' deleted for bases 12–91 of PRE (pSA64 Δ12–91), divergent primers spaced by the site of the deletion were used instead of tail to tail. Mutant A137G was constructed in two steps using overlapping divergent primers, both carrying the desired mutation. The first two PCR reactions contained each, one of the primers, the corresponding fragment end primer used to generate the wild-type fragment and plasmid carrying Palx-PRE-alx'-'lacZ (pSA60) as template. The products from these two reactions were used as template in a second PCR reaction containing the fragment end primers used to generate the wild-type fragment. The resulting fragment was then sub-cloned.
β-galactosidase assays
Overnight cultures were diluted 1:100 in fresh LB or LBK medium and grown to OD600 of 0.4. β-galactosidase activity was assayed as described (16).
In vitro structure probing of RNA generated by T7 RNAP
RNA was synthesized from pSA66 (wild-type and mutants) and probed as described previously (14).
Native gels
RNA synthesized from pSA66 (wild-type and mutants) was incubated for 30 min at 22°C and then analyzed on 5% non-denaturing polyacrylamide gels as described previously (14).
Cotranscriptional in vitro structure probing
RNA was synthesized using E. coli RNAP (1 U; Epicentre) at pH 7.2 in 50 µl reactions containing 20 mM Tris–HCl, 150 mM KCl, 10 mM MgCl2, 5 mM dithiothreitol (DTT), NTPs (250 µM each), 20 units RNase inhibitor (Takara Bio) and 2.5 µg of plasmid DNA carrying Palx-PRE-alx' (pSA64; wild-type and mutants). The mixture was preincubated for 5 min at 37°C before the addition of enzyme. After 5 min of synthesis at 37°C, 1.7 µl of dimethyl sulfate (DMS) (diluted 1:10 in ethanol) was added. The reactions were stopped 5 min after the addition of DMS by phenol/chloroform extraction followed by ethanol precipitation in the presence of 0.3 M sodium acetate and 1 µl of Quick-Precip (Edge BioSystems). To detect the modified sites, the treated RNA was annealed with 0.6 pmol end-labeled primer (70°C for 10 min, followed by incubation for 5–10 min on ice) and then subjected to primer extension (at 43°C for 60 min) in 15 µl reactions containing 0.5% (v/v) Tween 20, 50 mM Tris–HCl (pH 8.3), 40 mM KCl, 5 mM MgCl2, 10 mM DTT, deoxynucleotide triphosphate (dNTPs) (1 mM each) and 25 units Expand reverse transcriptase (Roche). The extension products were separated on 6% polyacrylamide, 7.8 M urea gels, alongside sequencing reactions.
Single round transcription
Single round transcription reactions were performed at pH 7.2 as described in (17) with slight modifications. To allow formation of halted elongation complexes and incorporation of label, 1.25 µg of plasmid DNA carrying Palx-PRE-alx' (pSA64; wild-type and mutants) was incubated at 37°C for 10 min in the presence of 10 µCi of α-32 P-ATP, 20 mM Tris–HCl, 150 mM KCl, 10 mM MgCl2, 2.77 mM DTT, 1 µM ATP, 2.5 µM GTP, 20 µg/ml BSA, 150 µM GpC dinucleotide (Sigma), glycerol to 3.33% (v/v) (including glycerol of the enzyme storage buffer) and 0.5 unit E. coli RNAP in a 45-µl volume. Thereafter, aliquots of 9 µl were transferred to pre-warmed tubes, and transcription was resumed by adding 1 µl of pre-warmed solution containing NTPs (2.5 mM each), 20 mM Tris–HCl, 150 mM KCl, 10 mM MgCl2 and 150 µg/ml rifampicin (to block re-initiation). The reactions were stopped by freezing in liquid nitrogen. Single round transcription reactions with PCR generated templates were performed similarly, with the following modifications—reactions were conducted at 30°C (Supplementary Figure S2) or 25°C (Figure 4b), the incubation time for formation of halted elongation complexes was 15 min (Supplementary Figure S2) or 20 min (Figure 4b) and 225 ng of template and 1 unit of enzyme were used. Samples were analyzed on 6% polyacrylamide and 7.8 M urea gels. The position of PC in reactions using PCR generated templates was verified by comparison with reactions using plasmid templates (Supplementary Figure S2). Band intensities were measured using the Quantity One program (Bio-Rad).
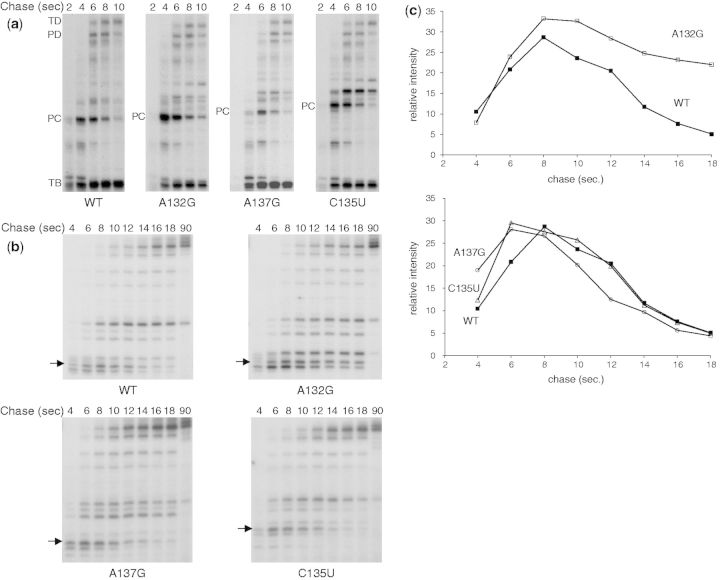
Figure 4.
The mutations modulate pausing position and longevity. (a) Synchronized single round in vitro transcription of full-length PRE wild-type and mutant RNAs carried out using E. coli RNAP. After formation of initiation complexes, transcription was allowed to proceed for the indicated times (in seconds) by adding all four unlabeled NTPs in excess and rifampicin. TB and TD indicate RNA products generated by the rho-independent transcription terminators at hairpins B and D, respectively. PC and PD indicate pause sites at hairpins C and D, respectively. (b) Single round transcription reactions of truncated (Δ12–91) PRE using PCR generated templates were performed at 25°C as described in ‘Materials and Methods’ section. The last lane in each run indicates runoff transcripts obtained with PCR generated templates (10 sec. (a) and 90 sec (b)). Arrows indicate the position of pause C as verified by comparison with reactions using plasmid templates (Supplementary Figure S2). (c) Pause intensities of wild-type and mutants relative to total transcription are plotted as a function of time (chase).
In vivo DMS structure probing
The structure of PRE-alx' RNA (wild-type and mutants) was probed in vivo in LB medium as described previously for pSA65 (14). Detection of modified sites was performed with 50 µg of total RNA for each sample.
RESULTS
Mutations designed to modify pausing at PRE affect alx translation
The nature of the sequence located upstream of a pause site greatly influences RNAP transcriptional elongation (18–20). To learn about the relationship between pausing and RNA folding, we introduced single point mutations (A132G, C135U and A137G) upstream of the first pause site, which were expected to affect pausing efficiency and/or position. The mutations designed were assumed to have a minimal effect, or none at all, on the stability of either the active or the inactive structure (Figure 2). The mutations A132G, C135U and A137G were first analyzed for their effect on expression using Palx-PRE-alx'-'lacZ translation fusions. These assays showed that the mutations substantially increased the reporter expression level compared with wild-type. The response of these mutants to high pH was more moderate than that of wild-type (Table 1).
Figure 2.
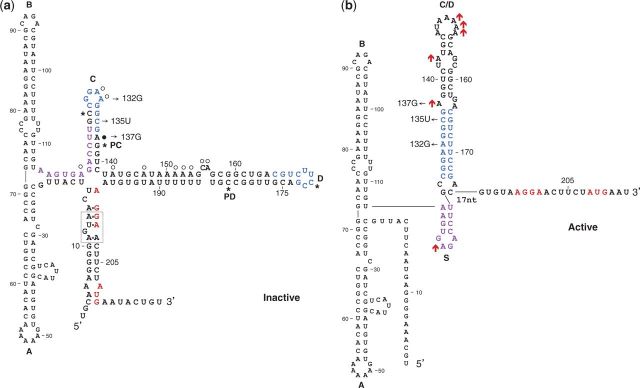
Translationally inactive (a) and active (b) structures. The Shine–Dalgarno and the initiation codon of alx are in red. Complementary sequences in hairpins C and D are in blue. Thin arrows indicate specific nucleotide changes generated by site-directed mutagenesis. To form the translationally active structure, the proximal part of hairpin C folds into the small hairpin S (nucleotides marked in purple), whereas basepairing between loops C and D forms stem C/D. A loop E motif in the inactive structure is boxed (dashed line). PC and PD denote the pause sites at hairpins C and D, respectively. Asterisks in (a) delineate the regions typically sequestered within the paused transcription elongation complex. The cotranscriptional structure probing data obtained on synthesis with E. coli RNAP (Figure 5) is presented on the structures. (a) Data obtained with wild-type RNA. Circles indicate strong (filled circles) and weak (open circles) DMS modification sites. (b) Data obtained with the high-level expression mutant A132G. Red upward arrows indicate increased modification intensity compared with wild-type. The two structures, the inactive and the active one, were previously shown to form under neutral and high pH conditions, respectively.
Table 1.
Mutations modifying pause C affect alx expression
| Translation fusiona | β-galactosidase units |
Positionb | |
|---|---|---|---|
| pH 6.8 | pH 8.4 | ||
| Wild-type | 68 ± 17 | 531 ± 182 | 138 |
| A132G | 182 ± 27 | 1243 ± 163 | 138 |
| C135U | 424 ± 5 | 1708 ± 225 | 141 |
| A137G | 178 ± 30 | 885 ± 111 | 140 |
aTranslation fusion plasmids carry the alx promoter followed by PRE and the first 53 nucleotides of alx (Palx-PRE-alx'-'lacZ). Results (in Miller units) are displayed as mean ± standard deviation.
bThe numbers indicate the estimated location of pause C.
Structural analysis of the mutants
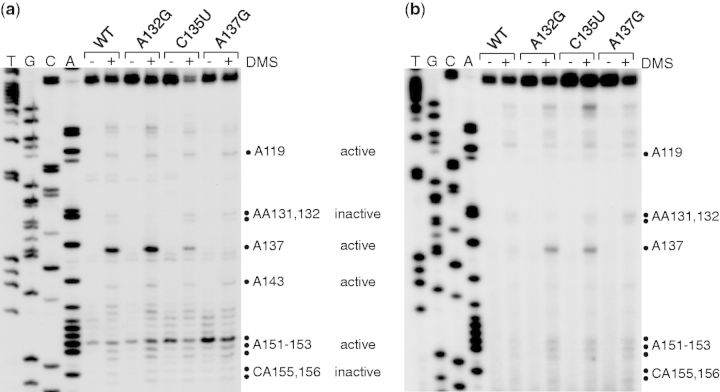
The mutations could influence alx translation by affecting the dynamics of RNA folding via pausing or by changing the stability of one of the structures, shifting the equilibrium towards the alternative one. To rule out an effect on structure stability that could lead to changes in the ratio between the two forms, we probed the structure of RNAs synthesized in vitro using T7 RNAP. As opposed to E. coli RNAP, T7 RNAP exhibits much less pausing during elongation. In accordance with this feature, we showed previously that T7 RNAP could not promote formation of the active structure of PRE. The structures were probed under conditions of neutral pH, using DMS that methylates unpaired adenosine and cytidine residues at N1 and N3 positions, respectively, and by subjecting the RNAs to partial cleavage by RNase T1 that is specific for single-stranded guanosine residues. The modified nucleotides and the cleavage sites were mapped by primer extension (Figure 3a, b and d). As a control, we probed the structure of mutant G134A that was previously shown to maintain the active structure. In G134A RNA, residues A149–A153, located in loop C/D and A137 of the interior loop of this stem are much more accessible to DMS modification than in wild-type RNA. Conversely, in wild-type RNA that is predominantly found in the inactive form (14), the modification rates of residues C155, A156, located in the bulge of stem D and A131, A132 located in loop C of the inactive structure, are higher than in G134A mutant, where these nucleotides base pair forming hairpin C/D. Thus, residues A131, A132 and C155, A156 are indicative of the inactive form, whereas residues A149–A153 and A137 are indicative of the active form. Comparing the intensities of these modifications in wild-type and G134A RNAs with those of the mutants indicates that the modification pattern of the high-level expression pause site mutants resembles the pattern displayed by wild-type RNA, and the ratio of active to inactive forms is similar to that of the wild-type. Notably, in A137G RNA, the position A137 is no longer modified because of the mutation. Also, in C135U RNA, the position A137 can no longer be modified, possibly because of stabilization of stacking due to formation of pairs of UG/GU in tandem (21). In A132G RNA, the position A132 is no longer modified because of the A to G substitution, and A131 seems to be less accessible to DMS. However, the modification pattern of residues C155, A156 and A149–153 in these mutants resembles the pattern displayed by wild-type RNA, contrasting with the pattern displayed by the high-level expression mutant G134A. Furthermore, probing of the ribosome binding site by RNase T1 shows that the accessibility of this region in the pause site mutants is similar to that displayed by the wild-type, in contrast to the high-level expression mutant G134A, which forms almost exclusively the active structure (Figure 3b and d). In addition, to visualize the different conformers, we analyzed the mutant RNAs on non-denaturing polyacrylamide gels. Previously, we observed that the high-level expression mutant G134A RNA is found almost exclusively in one structure, whereas the majority of wild-type is found in the other conformation (14). The native gel shows that the distribution pattern of the mutant RNAs is similar to wild-type (Figure 3c). Taken together, the data show that when synthesized with limited pausing, the ratio between the active and inactive structures in the mutant RNAs is similar to that observed in the wild-type. These results indicate that the mutations influence translation of alx not because they affect structure stability, shifting the equilibrium in favor of one of the structures, but possibly because they modify pausing and thereby the dynamics of folding.
Figure 3.
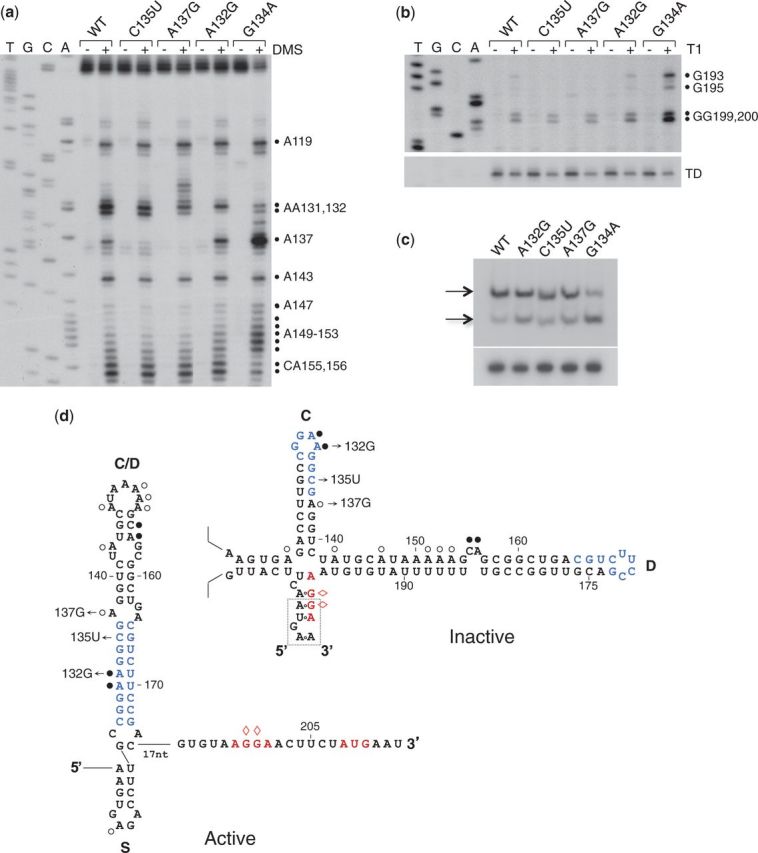
Structure determination of RNAs synthesized in vitro by T7 RNAP. RNA synthesized from PT7-PRE-alx' wild-type and mutants was treated with DMS (a) or RNase T1 (b). Reverse transcription of untreated (−) and DMS or T1 treated (+) RNA samples. The right panel (b; cleavage by T1) shows the region of the ribosome-binding site. In the lower part of this panel, a shorter exposure of a reverse transcription stop typical to the terminator at stem D (TD) is shown as a loading control. The numbers to the right indicate sequence positions relative to the transcription start site. (c) RNA as above was separated on non-denaturing polyacrylamide gels and transferred to nylon membranes by electroblotting (upper panel). To avoid detection of the short terminated transcripts, the membranes were probed with end-labeled primer complementary to sequences of alx (512). Arrows indicate the two conformations observed. To verify the integrity of this RNA, the samples were analyzed on denaturing 6% polyacrylamide, 7.8 M urea gel (lower part of the panel). (d) The relevant parts of PRE inactive and active conformations are shown with DMS modification data obtained with wild-type RNA. Circles indicate strong (filled circles) and weak (open circles) DMS modification sites. Red diamonds indicate T1 cleavage sites. The modification and the cleavage patterns of the pause site mutants resemble the patterns displayed by wild-type RNA. G134A mutant that forms the translationally active structure was used as a control.
The mutations modify the extent and/or position of pausing
To characterize the effect of the mutations on pausing, we monitored pausing in these mutants by synchronized single-round in vitro transcription (Figure 4a). These assays suggest that the position of pause C shifts forward in mutants A137G and C135U. For better resolution, we further characterized the effect of the mutations, using templates deleted for the majority of the upstream sequence including hairpins A and B (Δ12–91). As single point mutations can perturb separation that is based on RNA length, we estimated the position of pausing by using suitable RNA size markers, each carrying the corresponding point mutation. This mapping shows that mutations A137G and C135U shift the location of pause C two to three nucleotides forward, relative to wild-type, whereas in mutant A132G, the natural position of pause C is maintained (Supplementary Figure S1). The data showing that shifting the site forward is accompanied by a rise in alx translation endorse the mechanistic perception we previously suggested for alx regulation. Given that typically 12 ± 1 nucleotides of the nascent RNA are sequestered within the transcription elongation complex (22), RNAP, when paused at position G138, sequesters the sequence required to form the stem of hairpin C, from position G138 to C127 (Figure 2a). Once the formation of stem C is excluded, the free transcript preceding position C127 is capable of forming hairpin S (A114–U125). Shifting the site forward from position 138 and farther increases the chances of hairpin S to form and thereby stimulates the formation of the translatable structure.
Given that the high-level expression mutant A132G retains the natural position of pause C, we examined the effect of this mutation on the extent of pausing using linear templates. Single round transcription using linear templates enabled estimation of pause intensity relative to the total transcription as represented by the runoff. Mutant A132G exhibits increased pause intensity as well as extended pause duration (Figure 4b and c). As for pause duration, the decrease in the intensity of pausing in mutant A132G over a 10 s interval is significantly different from that of wild-type (33 and 83%, respectively). These results suggest that an increase in the rate of pausing by RNAP accompanied by a substantially slower pause escape increase the proportion of transcripts in which the formation of stem C is inhibited, thereby allowing the formation of the translationally active structure. Pause extent measurements of mutants A137G and C135U show that these mutants exhibit pause characteristics similar to wild-type (Figure 4b and c), suggesting that in these mutants the location of pausing plays a major role.
Changes in transcriptional pausing influence RNA folding
The differences in RNA folding caused by the changes in transcriptional pausing were investigated in assays of cotranscriptional structure probing. In these assays, the structures of the different mutant RNAs were determined using DMS during synthesis with E. coli RNAP (Figure 5a; summarized in Figure 2). To learn about changes in the proportion between the structures, active and inactive, we calculated the ratio of intensities of DMS modification of nucleotides that are indicative of the active or the inactive structures. Because in C135U and A137G mutants, the position A137 is no longer modified and in A132G RNA, position A131 seems to be less accessible to DMS, we measured the modification ratio of residues A153/C155 (indicative of the active and inactive structure, respectively). The calculated ratios in A132G, C135U and A137G RNAs are 2.8, 3.4 and 4.1, respectively, whereas wild-type RNA exhibits a ratio of 1.8. On the basis of these measurements, the active to inactive ratios of these high-level expression mutants are higher than that of wild-type RNA. Our results show that shifting the pause site farther from hairpin S or increasing the extent of pausing leads to an increase in the formation of the alternative, translationally active structure, correlating with the rise in expression observed in vivo.
Figure 5.
In vitro cotranscriptional and in vivo structure probing. (a) RNA was transcribed in vitro by E. coli RNAP for 5 min before the addition of DMS. (b) Cells containing plasmids carrying the locus Palx-PRE-alx' were grown in LB and treated with DMS for 5 min before RNA extraction. Reverse transcription of DMS modified (+) and unmodified (−) RNA. Modifications indicative of the active structure are marked as ‘active’, whereas modifications indicative of the inactive structure are marked as ‘inactive’. A summary of cotranscriptional structure probing data is displayed in Figure 2.
In vivo structure probing
To correlate the effect of the mutations on RNA folding as detected in vitro with RNA folding occurring in vivo, we compared the in vivo RNA structure of the mutants with wild-type. Notably, position A137 in mutant C135U is modified in vivo in contrast to the in vitro assays. The in vivo probing shows that the modification ratios of A137/A131 and A153/C155 increase (Figure 5b), thus demonstrating that the ratio of active to inactive structures in the high-level expression mutants shifts towards the translationally active structure. Taken together, the results demonstrate that transcription that involves pausing governs the process of PRE RNA folding, providing the mechanistic basis for PRE-alx regulation.
DISCUSSION
In recent years, a large number of riboregulators have been discovered and characterized, of which, a subset that includes riboswitches affects expression in cis, by adopting different conformations in response to cellular and/or environmental signals (23,24). With respect to mechanisms, riboswitches can be assigned to two categories: thermodynamically driven and kinetically driven switches. The action of riboswitches with a thermodynamic regime is mainly governed by a thermodynamic equilibrium between ligand bound and unbound conformations. The conformation of such riboswitches changes post-transcriptionally on ligand binding or ligand dissociation. For example, in the add riboswitch of Vibrio vulnificus, the ribosome binding site of the downstream gene is sequestered in the absence of adenine. Following adenine binding, a structural rearrangement occurs that exposes the site to ribosomes, thereby leading to add translation. The conformational change conferred by adenine binding, as well as the rise in translation initiation, have been shown to occur in vitro with full-length add riboswitch transcripts, indicating that this adenine riboswitch is a thermodynamically driven switch (12).
In riboswitches with a kinetic regime, RNA folding and ligand binding occur cotranscriptionally. Cotranscriptional folding allows formation of structures that are often different from those displayed when a full transcript folds as a whole. These structures are in many cases not optimally stable. The process of cotranscriptional folding is influenced by two main determinants, transcription progression and transcription pausing. As transcription is directional and basepairing is fast compared with transcription rate, upstream sequences can basepair before downstream sequences have been transcribed or emerged from the polymerase exit channel (25,26). Transcriptional pausing is a temporary inhibition of transcript elongation. By halting the transcribing enzyme at strategic positions, it can affect regulatory interactions. In some cases, transcriptional pausing may prevent formation of competing structures, thus allowing the formation of complex, or less stable, structures (10,27). In other cases, pausing enables binding of ligand by extending the time frame available for interactions (11). One such example is the ribD riboswitch of Bacillus subtilis. In this kinetically driven riboswitch, an antiterminator stem that prevents premature termination forms in the absence of flavin mononucleotide (FMN). Conversely, cotranscriptional binding of FMN to ribD RNA results in formation of a transcription terminator. Two pause sites located between the aptamer and the expression platform delay the regulatory decision, extending the time frame for FMN binding (11). Another example of a kinetically driven switch is pbuE of B. subtilis, which depends on cotranscriptional binding of adenine to function (12,28). Interestingly, pbuE of B. subtilis and add of V. vulnificus both respond to adenine, yet their mechanistic properties differ. Whereas pbuE is kinetically driven, add is a thermodynamically driven genetic switch. Unlike the aforementioned examples of kinetically driven riboswitches that regulate transcription termination, the E. coli btuB riboswitch controls translation initiation in response to coenzyme B12 binding. Here, adequately positioned pause sites serve to isolate the aptamer region from the rest of the molecule so as to allow it to form a complex tertiary structure before downstream competing sequences become available (10).
The pH-responsive RNA element of alx is a riboregulator displaying kinetic properties. Its action depends on transcriptional pausing and cotranscriptional folding. The location of the pause sites led us to suggest that the paused enzyme sequesters sequences required for the formation of the stable, translationally inactive structure, thus allowing formation of a less stable, however translationally active structure. Here, we show that changing the position of the first pause site influences the regulatory result dramatically. Shifting the pause site farther from stem S increases the likelihood of hairpin S assembly, a requisite for the formation of the active structure. Consequently, the probability of the active structure to form is increased. Mutations A137G and C135U shift the position of PC from 138 to 140 and 141, respectively. These mutations increase alx expression by 2–6-fold. Because no other changes in pause characteristics can be detected in these mutants, the increase in alx expression is most probably due to the new location of pausing.
Mutant A132G pauses at the same position as wild-type. However, it exhibits a dramatic effect on the rate of decay; pause intensity in this mutant decreases slowly compared with wild-type. A132G mutant also shows a higher proportion of pausing. Both characteristics can explain the rise in alx translation. An increase in the number of transcription complexes paused at PC, as well as an extension in the time these complexes spend at that site, can both lead to an increase in the formation of stem S, and thus the active structure.
Mutations like A132G that extends the duration of pausing at PC and A137G and C135U that shift the position of PC, increase the probability of formation of stem S and thus the translationally active structure. The physiological and molecular characteristics of these mutations substantiate the model that describes the role of pH in this system, whereby high pH extends pausing at PC, thus leading to activation of alx translation. It is interesting to note that the response of these mutants to high pH is more moderate than that of wild-type, suggesting that mutations affecting pause behavior are less susceptible to the pH effects on pausing, either because of their direct effect on pause characteristics or due to an effect on the pH sensing mechanism.
Taken together, the mutational data illustrate the mechanistic design of alx regulation that relies on precise temporal and spatial characteristics, demonstrating the importance of both the precise location and the extent of pausing to RNA folding and control of gene expression. We propose that the mechanistic design of the alx regulatory RNA element is based on a delicate balance that provides an opportunity for environmental signals such as pH to introduce structural changes in the RNA and thereby modulate expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [29].
FUNDING
This work was supported by the Israel Science Foundation founded by The Israel Academy of Sciences and Humanities [911/09]; by Deutsche Forschungsgemeinschaft research grant [VO 875/5-1]; and by Israel Centers of Research Excellence (ICORE), Chromatin and RNA [1796/12]. Funding for open access charge: The Israel Science Foundation founded by The Israel Academy of Sciences and Humanities [911/09] and Deutsche Forschungsgemeinschaft research grant [VO 875/5-1].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Robert Landick for his advice throughout the study.
REFERENCES
- 1.Pan T, Sosnick T. RNA folding during transcription. Annu. Rev. Biophys. Biomol. Struct. 2006;35:161–175. doi: 10.1146/annurev.biophys.35.040405.102053. [DOI] [PubMed] [Google Scholar]
- 2.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 3.Artsimovitch I, Landick R. The transcriptional regulator RfaH stimulates RNA chain synthesis after recruitment to elongation complexes by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 4.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc. Natl Acad. Sci. USA. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morgan WD, Bear DG, von Hippel PH. Rho-dependent termination of transcription. II. Kinetics of mRNA elongation during transcription from the bacteriophage lambda PR promoter. J. Biol. Chem. 1983;258:9565–9574. [PubMed] [Google Scholar]
- 6.Lau LF, Roberts JW, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J. Biol. Chem. 1983;258:9391–9397. [PubMed] [Google Scholar]
- 7.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 8.Santangelo TJ, Artsimovitch I. Termination and antitermination: RNA polymerase runs a stop sign. Nat. Rev. Microbiol. 2011;9:319–329. doi: 10.1038/nrmicro2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong TN, Sosnick TR, Pan T. Folding of noncoding RNAs during transcription facilitated by pausing-induced nonnative structures. Proc. Natl Acad. Sci. USA. 2007;104:17995–18000. doi: 10.1073/pnas.0705038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perdrizet GA, II, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl Acad. Sci. USA. 2012;109:3323–3328. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 12.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, Masse E, Lafontaine DA. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TN, Pan T. RNA folding during transcription: protocols and studies. Methods Enzymol. 2009;468:167–193. doi: 10.1016/S0076-6879(09)68009-5. [DOI] [PubMed] [Google Scholar]
- 14.Nechooshtan G, Elgrably-Weiss M, Sheaffer A, Westhof E, Altuvia S. A pH-responsive riboregulator. Genes Dev. 2009;23:2650–2662. doi: 10.1101/gad.552209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 16.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. [DOI] [PubMed] [Google Scholar]
- 18.Bochkareva A, Yuzenkova Y, Tadigotla VR, Zenkin N. Factor-independent transcription pausing caused by recognition of the RNA-DNA hybrid sequence. EMBO J. 2012;31:630–639. doi: 10.1038/emboj.2011.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sidorenkov I, Komissarova N, Kashlev M. Crucial role of the RNA: DNA hybrid in the processivity of transcription. Mol. Cell. 1998;2:55–64. doi: 10.1016/s1097-2765(00)80113-6. [DOI] [PubMed] [Google Scholar]
- 20.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masquida B, Westhof E. On the wobble GoU and related pairs. RNA. 2000;6:9–15. doi: 10.1017/s1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Monforte JA, Kahn JD, Hearst JE. RNA folding during transcription by Escherichia coli RNA polymerase analyzed by RNA self-cleavage. Biochemistry. 1990;29:7882–7890. doi: 10.1021/bi00486a015. [DOI] [PubMed] [Google Scholar]
- 23.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 24.Bastet L, Dube A, Masse E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol. Microbiol. 2011;80:1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- 25.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 27.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc. Natl Acad. Sci. USA. 1999;96:9545–9550. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frieda KL, Block SM. Direct observation of cotranscriptional folding in an adenine riboswitch. Science. 2012;338:397–400. doi: 10.1126/science.1225722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.