Abstract
In the early 1950’s, ‘host-controlled variation in bacterial viruses’ was reported as a non-hereditary phenomenon: one cycle of viral growth on certain bacterial hosts affected the ability of progeny virus to grow on other hosts by either restricting or enlarging their host range. Unlike mutation, this change was reversible, and one cycle of growth in the previous host returned the virus to its original form. These simple observations heralded the discovery of the endonuclease and methyltransferase activities of what are now termed Type I, II, III and IV DNA restriction-modification systems. The Type II restriction enzymes (e.g. EcoRI) gave rise to recombinant DNA technology that has transformed molecular biology and medicine. This review traces the discovery of restriction enzymes and their continuing impact on molecular biology and medicine.
INTRODUCTION
Restriction endonucleases (REases) such as EcoRI are familiar to virtually everyone who has worked with DNA. Currently, >19 000 putative REases are listed on REBASE (http://rebase.neb.com) (1). REases are classified into four main types, Type I, II, III and IV, with subdivisions for convenience; almost all require a divalent metal cofactor such as Mg2+ for activity (Table 1 and Figure 1). Type II REases represent the largest group of characterized enzymes owing to their usefulness as tools for recombinant DNA technology, and they have been studied extensively. Over 300 Type II REases, with >200 different sequence-specificities, are commercially available. Far fewer Type I, III and IV enzymes have been characterized, but putative examples are being identified daily through bioinformatic analysis of sequenced genomes (Table 1).
Table 1.
Characterization and organization of the genes and subunits of the four Types of restriction enzymes
| Type | Type I | Type II | Type III | Type IV |
|---|---|---|---|---|
| Features |
|
|
|
|
| Example | e.g. EcoKI | e.g. EcoRI | e.g. EcoP1I | No ‘typical’ example |
| Genes | hsdR, hsdM, hsdS | e.g. ecorIR, ecorIM | e.g. ecoP1IM, ecoP1IR | e.g. mcrA, mcrBC, mrr |
| Subunits | ∼135, ∼62 and ∼52 kDa | ∼31 and ∼38 kDa for EcoRI | ∼106 and ∼75 kDa for EcoP1I | Unrelated proteins |
| Proteins | REase: 2R + 2M + S | Orthodox REase: 2R | REase: 1 or 2 R + 2M | Varies |
| MTase: 2M + S (±2R) | Orthodox MTase: M | MTase: 2M (±2R) | ||
| REBASE |
|
|
|
|
Type I and II are currently divided in 5 and 11 different subclasses, respectively. Few enzymes have been well-characterized, but based on the current avalanche of sequence information many putative genes belonging to all Types and subtypes are being identified and listed on the restriction enzyme website (http://rebase.neb.com). The modification-dependent Type IV enzymes are highly diverse and only a few have been characterized in any detail. In each case, an example is given of one of the best-characterized enzymes within the different Types I, II and III. Note that Type II enzymes range from simple (shown here for EcoRI) to more complex systems (see Table 2 for the diversity of Type II subtypes).
REBASE count is as of 16 September 2013 (http://rebase.neb.com/cgi-bin/statlist).
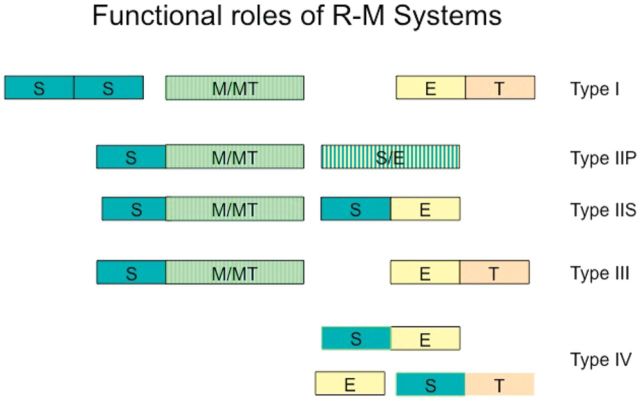
Figure 1.
Schematic representation of the functional roles filled in different ways in R-M systems. The functions served include the following: S, DNA sequence specificity; MT, methyltransferase catalytic activity; M, SAM binding; E, endonuclease; T, translocation. Boxes outline functions that are filled by distinct protein domains. Different colours indicate different functions, while different boxes represent distinct domains. Domains in a single polypeptide are abutted, and those in separate polypeptides spaced apart. The order of domains in a polypeptide may vary—e.g. not all Type IIS enzymes have the cleavage domain at the C-terminus. In some cases, functions are integrated with each other, e.g. S and E functions of Type IIP (striped box); in other cases, separate domains carry them out, e.g. Type IIS. See Table 2 for the complexity and diversity of Type II subtypes. The large families of Type I and Type II systems are currently subdivided in 5 and 11 different groups, respectively. The Type I families are distinguished by homology; the Type II groups are distinguished by catalytic properties rather than sequence homology. Type IV enzymes were initially identified as hydroxymethylation-dependent restriction enzymes and currently comprise a highly diverse family. Two examples are shown with and without a translocation domain.
Here we present a non-specialists perspective on important events in the discovery and understanding of REases. Studies of these enzymes have generated a wealth of information regarding DNA–protein interactions and catalysis, protein family relationships, control of restriction activity and plasticity of protein domains, as well as providing essential tools for molecular biology research. Discussion of the equally fascinating DNA-methyltransferase (MTase) enzymes that almost always accompany REases in vivo is beyond the scope of this review, but we note that base flipping, first discovered in the HhaI MTase (2), is not confined to these enzymes alone, but appears to be a common phenomenon that is also used by certain REases (3) and in other nucleic acid-binding enzymes (4–7).
Most research interest has focused on Type I and II enzymes for historical and practical reasons, so this history is weighted to their treatment. The molecular, genetic and enzymological properties of these have been extensively reviewed [see e.g. (8–12)], and separate reviews of the Type I, III and IV systems appear elsewhere in this journal.
THE FIRST HIGHLIGHTS
Discovery of ‘host-controlled variation’
Many important scientific developments in the first half of the 20th century laid the groundwork for to the discovery of restriction and modification (R-M). These included the discoveries of radiation and to the ability to incorporate isotopes in living cells; the molecular building blocks of DNA, RNA and protein; ‘filterable agents’ (viruses); the isolation of Escherichia coli and other bacteria, and of their viruses [called (bacterio)phage] and plasmids. Enabling technical advances included development of electron microscopy, ultracentrifugation, chromatography, electrophoresis and radiographic crystallography. Key was the emerging field of microbial genetics, which flourished owing to the discovery of lysogeny, conjugation, transduction, recombination and mutation.
Preliminary descriptions of the phenomenon of R-M were published by Luria and Human (1952) (13), Anderson and Felix (1952) (14) and Bertani and Weigle (1953) (15). These reports of ‘host-controlled variation in bacterial viruses’ were reviewed by Luria (1953) (16). Host-controlled variation referred to the observation that the efficiency with which phage infected new bacterial hosts depended on the host on which they previously grew. Phage that propagated efficiently on one bacterial strain could lose that ability if grown for even a single cycle on a different strain. The loss was not due to mutation, and one cycle of growth on the previous strain returned the virus to its original state once more. R-M systems of all types initially were investigated in this same way, by comparing the ‘efficiency of plating’ (eop; number of plaques on the test host divided by the number of plaques on a permissive host) on alternate bacterial hosts (17–21). Eop values would range from ∼10−1 to 10−5, thus indicating that R-M systems were effective barriers to the uptake of DNA; see (16,22–26) for early reviews.
DNA modification
A decade after these initial reports, Werner Arber and Daisy Dussoix, using phage lambda as experimental system, showed that it was the phage DNA that carried the host-range imprint (17). Different specificities could be imprinted concomitantly both by the bacterium itself (by what were later recognized to be the Type I EcoKI and EcoBI systems) and by phage P1 in its latent prophage state (the Type III EcoP1I system). Gunther Stent suggested that DNA methylation might be the basis for the modification imprint, thus prompting Arber to show that methionine was required in the growth medium to produce the imprint on the DNA (27). This important finding coincided with the discovery of RNA-methyltransferase and MTase activities in bacteria that catalysed the formation of m5C and m6A (28). Arber’s interest in the biochemical mechanisms of R-M was driven in part by insight that R-M enzymes would prove useful for analysing DNA molecules and DNA–protein interactions. He concluded a landmark 1965 review of host-controlled modification with the following words: ‘Looking toward future developments … it is to be hoped that the enzymes involved in production and control of host specificity will be isolated and characterized. Such studies, paralleled with investigations of the genes controlling R-M and of their expression, should eventually permit an explanation of the high degree of strain specificity, for example “by a mechanism of recognition of certain base sequences”. If this last idea should be correct one may further speculate that a restriction enzyme might “provide a tool for the sequence-specific cleavage of DNA” ’ (22) (our double quotes).
Sequence-specific DNA-cleavage
As chance would have it, the R-M systems studied by Arber, Type I and Type III (25), do not provide simple enzymes for the sequence-specific cleavage of DNA (see further below). However, REases with the desired sequence-specific cleavage were soon isolated, and these set the stage for the advances in gene analysis and manipulation, collectively called ‘recombinant DNA technology’, that quickly followed. The first of these new enzymes, HindII, was discovered in Hamilton (‘Ham’) Smith’s laboratory at Johns Hopkins Medical School in 1970 (29). This was subsequently termed a Type II REase as its properties were distinct from the Type I REases (25). Purified from Haemophilus influenzae serotype d, HindII (originally called endonuclease R) was found to act as a homodimer and to cleave DNA at the symmetric (though degenerate) sequence GTY’RAC (Y = C or T; R = A or G;’ indicates the cut site) (29,30). Subsequently, what was thought to be pure HindII was found to be a mixture of HindII and a second REase made by the same bacterium, HindIII. HindIII cleaved DNA at a different symmetric sequence, A’AGCTT (31,32); [see (33) for a thought-provoking discussion]. The existence of HindIII came to light during experiments to characterize the MTase activities of H. influenzae. These experiments showed that the HindII and HindIII MTases acted at the same DNA sequences as those cleaved by the REases. They modified these sequences rather than cleaving them, producing GTYRm6AC and m6AAGCTT, respectively (34–36).
The universe of enzymes in this Type II category expanded rapidly. As Smith’s work proceeded on the east coast of the USA, REases with similar behaviour but different specificity were discovered in the laboratory of Herb Boyer at the University of California, San Francisco, on the west coast. Here, PhD student Robert Yoshimori (37) benefited from the experience of Daisy Dussoix, who had moved from Werner Arber’s lab to UCSF. Yoshimori investigated restriction systems present on plasmids in clinical E. coli isolates, and purified what became known as EcoRI and EcoRII (37,38). The EcoRI REase was found to cleave G’AATTC (39,40) and the corresponding M.EcoRI MTase to modify the inner adenines in this sequence, producing GAm6ATTC (41). The EcoRII REase was found to cleave ‘CCWGG (W = A or T), and the M.EcoRII MTase to modify the inner cytosines, producing Cm5CWGG (42,43).
Staggered cuts and the advent of genetic engineering
In contrast to the Type I and Type III enzymes studied in the 1960’s, EcoRI and HindIII cleave DNA within their recognition sites and, most importantly, produce staggered cuts. Since the recognition sites are symmetric, this means that every fragment is flanked by the same single-stranded extension, allowing any fragment to anneal (via the extensions) to any other fragment, thus setting the stage for recombining DNA fragments and ‘cloning’. These findings were presented at the 1972 EMBO Workshop on Restriction, organized by Werner Arber (see Supplement S1 for details of the program and attendees). Figure 2 shows a photograph of participants at this Workshop, recalled by Noreen Murray as the most exciting meeting in the history of the REases, with discussions on the impact of this vital new information on ‘sticky ends’ and the implications for novel DNA manipulation. The recently described DNA ligase (44) would allow the joining of DNA fragments with the same sticky ends. EcoRI and HindIII spurred the development of recombinant DNA work through the availability of both purified enzymes and of replicatable carriers known as vectors. Both phage lambda (45) and various plasmids (46,47) were developed into vectors into which DNA fragments generated by EcoRI and HindIII could be ligated.
Figure 2.
Photograph of the participants at the EMBO Workshop on restriction in Leuenberg (Basel), Switzerland, 26–30 September 1972, organized by Werner Arber, who took the picture (Archive Noreen Murray). Supplement S1 contains a list with names of the attendees and the program of this meeting, and puts names to faces as far as the attendees could be identified (from the archives of Noreen Murray and Werner Arber).
Fittingly, in 1978, Werner Arber was awarded the Nobel Prize together with Dan Nathans and Ham Smith in recognition for their pioneering work on R-M (www.nobelprize.org).
Emerging genetic and enzymatic complexity
While the 1972 review by Matt Meselson et al. (26) mentions only the recognition sequence of HindII, the pace soon quickened. The discovery of new restriction enzymes skyrocketed, as laborious in vivo phage-plating assays were replaced by rapid in vitro DNA-cleavage assays of cell extracts. Elucidation of differences in recognition and cutting led to the classification of additional distinct classes, or types, of restriction enzymes (25,48), which with extensions and subdivisions has stood the test of time: Type I (exemplified by EcoKI, EcoBI, EcoR124, the ‘classical’ enzymes); Type II (EcoRI, HindIII, EcoRV, the ‘orthodox’ enzymes); and Type III (EcoP1I and EcoP15I), Table 1 and Figure 1. Type IV (modification-dependent REases Mcr and Mrr) was added later (49). Sequencing and biochemistry have since led to subdivisions within the Type I (see below) and Type II systems (Table 2) [see (49,50) and http://rebase.neb.com for nomenclature and details].
Table 2.
Nomenclature of Type II restriction enzymes
| Subtype | Features of restriction enzymesa | Examples |
|---|---|---|
| Type IIP | Palindromic recognition sequence; recognized by both homodimeric and monomeric enzymes; cleavage occurs symmetrically, usually within the recognition sequence | Prototypes EcoRI & EcoRV |
| Type IIA | Asymmetric recognition sequence | FokI |
| Type IIB | Cleavage on both sides of the recognition sequence | BcgI |
| Type IIC | Single, combination R-M polypeptide | HaeIV |
| Type IIE | Two sequences required for cleavage, one serving as allosteric effector | EcoRII, Sau3AI |
| Type IIF | Two sequences required for cleavage, concerted reaction by homotetramer | SfiI |
| Type IIG | Requires AdoMet cofactor for both R-M | Eco57I |
| Type IIH | Separate M and S subunits; MTase organization similar to Type I systems | BcgI |
| Type IIM | Require methylated recognition sequence; Type IIP or Type IIA | DpnI |
| Type IIS | Asymmetric recognition sequence; cleavage at fixed positions usually outside recognition sequence | FokI |
| Type IIT | Heterodimeric restriction enzyme. | Bpu10I, BslI |
| Putatives | All subtypes | |
| Control | Control proteins of Type II restriction enzymes | C.BamHI, C.PvuII |
The characteristics of the orthodox Type IIP enzymes originally distinguished this group of enzymes from the Type I and III R-M systems. Type IIP is the largest group, owing to its valuable role in molecular science and its commercial value, but the current classification and growing number of R-M systems (putatively) identified, makes it clear that Type II enzymes are highly diverse and the boundaries with the other types are beginning to blur; see also Figure 3 and text for details.
aThese classifications reflect enzyme properties and activities, and not their evolutionary relationships. The classifications are not exclusive, and one enzyme can often belong several classes. Thus BcgI, for example, is Type IIA, B, C, G and H (see text for details).
The recombinant DNA scare
In their 1975 review (51), Nathans and Smith discuss methods for DNA cleavage and separation of the resulting fragments on gels, as well as the use of REases in other applications, e.g. the physical mapping of chromosomes, taking Simian Virus SV40 (SV40) as an example. The debate on the safety of recombinant DNA technology started soon after the 1972 EMBO Workshop and reports on the transfer of eukaryotic DNA into E. coli [documented by (52)]. The debate was extremely heated, but by 1990 many of the fears had abated as the anticipated dangers did not materialize and the advantages of DNA cloning, and the ability to produce large quantities of pharmaceutically important proteins such as insulin, hormones and vaccines became clear.
FURTHER HIGHLIGHTS IN THE STUDY OF TYPE I R-M SYSTEMS
Type I families are defined by complementation and display sequence conservation
Type I REases were originally identified in E. coli and other enteric organisms as barriers to DNA entry. They turned out to be oligomeric proteins encoded by the three host specificity determinant (hsd) genes: a restriction (R), modification (M) and recognition (S for specificity) gene, respectively (Table 1 and Figure 1). Before the development of DNA sequencing, genetic complementation tests defined the hsdR, hsdM and hsdS genes (53,54). DNA hybridization studies, and probing with antibodies directed at EcoKI, established that EcoKI and EcoBI were more closely related to each other than to EcoAI, the Type I system in E. coli 15T− [reviewed in (8)]. This approach based on biological interaction led to the division of these systems into families: the Type IA (EcoKI, EcoBI, EcoDI and Salmonella typhimurium StySPI); Type IB (EcoAI, EcoEI and Citrobacter freundii CfrAI); Type IC (EcoR124, EcoDXXI, EcoprrI) (8); and later, Type ID (StyBLI and Klebsiella pneumoniae KpnAI) (9,55,56) and Type IE (KpnBI) (57); see reviews for further details (8–10,58,59). Other organisms will have their own families, for example, Staphylococcus aureus has at least two families [(60) and unpublished DTFD results].
Preparation, cofactor requirements and structures
Landmark studies on purified enzymes in the wake of the 1962 Arber and Dussoix articles (17,61) date to 1968. Stu Linn and Werner Arber in Switzerland and Matt Meselson and Bob Yuan in the USA, respectively, purified EcoBI and EcoKI. They used restriction of phages fd and lambda as their assay for detecting the enzymes during purification, a laborious process (62,63). This was no simple matter; Bob Yuan recalls that ‘the fall flew by in deep frustration’ until he and Matt discovered that the enzyme needed S-adenosylmethionine (SAM) for activity in addition to Mg2+ and ATP (See Supplement S2 for his personal story). The same cofactor requirement was also found for EcoBI (62,64), reviewed in (25,26). Twenty-five years later, we have come to appreciate that SAM, like ATP, is a widely used cofactor in many metabolic reactions (65,66).
A long-awaited breakthrough did not happen until much later: The structures of the subunits and assembled Type I R-M enzymes. Two structures of S subunits appeared in 2005 and culminated in 2012 with the structure of two complete R-M enzymes containing two R subunits, two M subunits and one S subunit (67–69).
Type I enzymes cut away from the target site
In 1972, Horiuchi and Zinder showed that the DNA recognition site of EcoBI is not the cleavage site (70). They cut 3H-labelled double-strand RF DNA of phage f1 (a relative of phage M13) with EcoBI, denatured and renatured the DNA and then treated with EcoBI a second time. This resulted in a heterogeneous distribution of small DNA fragments on alkaline sucrose gradients leading to the conclusion that EcoBI cuts at a variable distance from its target site. This feature is now known to be common to all Type I restriction enzymes. It was later shown by Studier that EcoKI would preferentially cleave DNA approximately half-way between consecutive target sites (71). This feature is also common to all Type I restriction enzymes although the distribution of cleavage locations can be broad.
DNA translocation to reach the cutting sites uses molecular motors
Translocation was first observed in electron microscope (EM) studies that showed DNA looping by EcoBI and EcoKI. These were interpreted as reaction intermediates, formed by the enzymes translocating along the DNA while remaining attached to their recognition sites (72,73). In the case of EcoKI, studies with full-length phage lambda DNA and relaxed or supercoiled circular DNA showed that EcoKI translocates the DNA past itself, concomitant with a large conformational change of the enzyme, creating large bidirectional loops clearly visible in the EM. Recent studies confirm that on DNA binding, the enzyme strongly contracts from an open to a compact form (58,69,74). In contrast, EcoBI appeared to form loops in only one direction. Later studies with EcoBI did show supercoiled structures like EcoKI; however, translocation was still unidirectional, and without any apparent strand selectivity in the cleavage reaction (75,76). The translocation process would explain the cleavage observed half-way between consecutive target sites on a DNA molecule as two translocating enzyme molecules would collide roughly half-way between target sites.
The R-subunit of Type I enzymes belongs to the SNF2 helicase/translocase superfamily of proteins. These appear to be the result of an ancient fusion between nuclease and ATP-dependent RecA-like (AAA+ or ‘motor’) domains, a linkage found in many enzymes involved in DNA repair, replication, recombination and chromosome remodelling (77–88). As such, Type I enzymes could prove useful for understanding the action of SNF2 enzymes in higher organisms, including the coordinated steps of DNA scanning, recognition, binding and alteration of the helical structure, that allow other domains or subunits to move and touch the DNA. All of these steps are required to prevent indiscriminate nuclease activity (69).
The key to the functionality of the Type I REase and other SNF2 proteins is their enormous flexibility, allowing large conformational changes. First noticed for EcoKI by Yuan et al (73), and more recently for RecB and EcoR124 (69,89), large protein motions may be a general feature of SNF2 proteins. In line with such large-scale domain movement, mutational analysis of EcoR124 showed long-range effects, e.g. nuclease mutants affect the distant helicase domain leading to a reduced translocation and ATP usage rate, a decrease in the off rate, slower restart and turnover. In other words, the nuclease and motor domain together are ‘more than the sum of their parts’ (89,90).
Plasticity of type I DNA sequence recognition: hybrid specificities and phase variation
Type I enzymes recognize bipartite DNA sequences [e.g. AAC(N6)GTGC for EcoKI]. The S subunit has a duplicated organization: two ∼150 aa variable regions alternate with smaller conserved regions, which are highly similar within each of the five families. Each variable region recognizes one part of the bipartite target sequence.
A key event in understanding the significance and mechanism of variation of sequence specificity was the discovery of a brand-new specificity resulting from a genetic cross (91,92). As a result of crossing-over in the conserved central region between the two variable regions, hybrid specificities were found. This change in specificity was found to occur in vivo and in vitro and was first noted for Salmonella species (91–94). An extensive treatment of this topic is found in the accompanying Type I review.
A variety of genetic processes promote variation in S subunits proteins of Type I and Type III enzymes. In Lactococcus lactis, entering plasmids may bring hsdS genes with them (95). The segmented organization of hsdS, with two DNA recognition domains, lends itself to variation by DNA rearrangement. Site-specific recombination leads to expression of S proteins with alternative recognition domains in Mycoplasma pulmonis, thus generating combinatorial variations of recognition sequence (96). Such plasticity of restriction specificity is also inferred in Bacteroides fragilis (97) and other species.
FURTHER HIGHLIGHTS IN THE STUDY OF TYPE II R-M SYSTEMS
Subdivisions of type II enzymes
Type II REases are defined rather broadly as enzymes that cleave DNA at a fixed position with respect to their recognition sequence, and produce distinct DNA-fragment banding patterns during gel electrophoresis. These REases are extremely varied and occur in many structural forms. The Type II classification was used originally to define the simplest kind of REases, exemplified by HindII and EcoRI, that recognize symmetric DNA sequences and require Mg2+ ions for cleavage activity (25). Enzymes of this sort generally act as homodimers and cleave DNA within their recognition sequences. In vivo, they function in conjunction with a separate modification MTase that acts independently as a monomer (Table 1). The first distinction made among Type II enzymes concerned REases such as HphI and FokI that recognize asymmetric sequences and cleave a short distance away, to one side. These were designated Type IIS (98).
As the number of REases producing distinct fragments grew, it became clear that many unrelated proteins were included in the category (99). Rather than dividing these into further Types based on their phylogenies, it was agreed that ‘Type II’ should become a utilitarian classification that reflected enzymatic behaviour rather than evolutionary relatedness, and for convenience, a number of Type II groups, corresponding to particular enzymatic behaviours, were defined (49) (Table 2). Each of these groups, A, B, C, E, F, G, H, M, P, S and T, should be thought of, not as an exclusive subdivision, but rather as an icon that signifies some specific property. Enzymes may exhibit more than one salient property and thus belong to more than one group. HindIII and EcoRI remain simple; they are members of just the one, Type IIP, group (‘P’ for Palindromic). BcgI, in contrast, is complicated since it recognizes an asymmetric DNA sequence (= Type IIA); cleaves on both sides of that sequence (= Type IIB); and comprises a fused endonuclease-methyltransferase subunit (= Type IIC) plus a Type I-like DNA-specificity subunit (= Type IIH). BcgI, thus, is a member of multiple groups (100–102). DpnI (Gm6A’TC) is a Type IIM REase, which cleaves its recognition sequence only when the sequence is methylated (103). DpnI is also a member of the Type IIP group since its recognition sequence is palindromic sequence and cleavage is internal and symmetric (Figure 3 and http://rebase.neb.com).
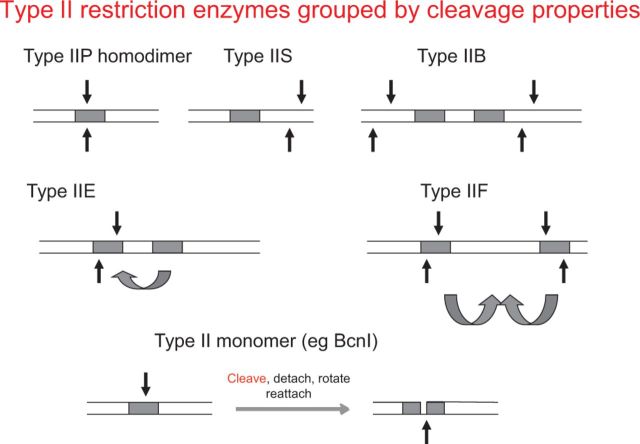
Figure 3.
Type II restriction enzymes grouped by cleavage properties. ‘Orthodox’IIP enzymes (e.g. EcoRI, EcoRV) cut at the recognition site. Type IIS cut away from the site (e.g. FokI, BfiI). Type IIB require two recognition sites and cut on the outside (e.g. BplI). Type IIE require two recognition sites, and one of the two sites acts as allosteric effector (e.g. EcoRII). Type IIF require two sites and cut at both sites as a tetramer after bringing the two regions together by looping the DNA (e.g. SfiI). Enzymes such as BcnI act as a monomer, in contrast to most Type II REases that act as dimers. See Table 2 and text for further details.
Type IIP (‘orthodox’) REases such as EcoRI and HindIII were crucial to the development of recombinant DNA technology. Certain ‘unorthodox’ enzymes have also been widely used. Sau3AI (’GATC) is a monomeric Type IIE REase that dimerizes on the DNA, inducing DNA loops. Two recognition sites must be bound for activity; one is cleaved while the other acts as allosteric effector (104). EcoRII is somewhat similar, and many REases are now known to cleave only as dimers of dimers bound to two separate sites.
Predicting enzyme families: sequences, structures and bioinformatics
Early amino acid sequences of Type II enzymes (e.g. EcoRI, EcoRV, PvuII and BamHI) showed them to be almost completely unrelated (105–109). When crystal structures appeared (110–114), commonalities began to emerge. The motif PD-(D/E)XK was identified as a common feature (115,116). This motif also appeared in other nucleases, e.g. lambda exonuclease (117) and the Tn7 transposase protein TnsA (118). This motif is the catalytic core of a Mg2+-dependent nuclease.
REBASE, the Restriction Enzyme Database set up by Rich Roberts to keep track of RM specificities and indicate how to acquire the enzymes, made possible the next phase of understanding. First on paper (119,120), then via the nascent Internet by File Transfer Protocol and finally on the World Wide Web (1,121) this resource makes available a focused organized data set allowing computational analysis of sequences and structures as well as access to individual topics of interest [e.g. (99,122–129)].
Recently, Sau3A (104) and several other REases proved able to cut DNA/RNA hybrids (130). The rarity of this property (6 of 223 surveyed) suggests that any biological roles for this ability will be specialized, but the property could be used to study the ubiquitous small RNA molecules that regulate expression in all domains of life (131).
FURTHER HIGHLIGHTS IN THE STUDY OF TYPE III R-M SYSTEMS
Type III enzymes have properties that are intermediate between Types I and II (Table 1 and Figure 1). In general, Type III enzymes recognize asymmetric sequences, cleave 25–27 nucleotides away from their recognition site and use ATP and SAM as cofactors, although they do not have an absolute requirement for the latter. Particularly interesting topics include control of the phage-borne R.EcoP1I REase activity following infection and how newly replicated DNA can be protected when only one strand of the recognition sequence is protected by methylation (132–139).
An early result showed that two copies of the target site were required for DNA cleavage but that these sites had to be in a head-to-head orientation (135,140). A head-to-tail orientation prevented cleavage. How this communication between the two target sites was achieved when ATP hydrolysis was insufficient for DNA translocation like the Type I enzymes (59) has provoked much discussion (141). It appears that DNA looping may have a role in bringing the sites together (142,143), but recent single-molecule analyses (144,145) show strong evidence for enzyme diffusion along the DNA triggered by an ATP-dependent conformational change as a novel mechanism for bringing two copies of the enzyme together to give cleavage, see also (83,146). The long-awaited atomic structure of a Type III R-M enzyme should resolve many of the complexities of these enzymes [AK Aggarwal, personal communication (147,148)].
FURTHER HIGHLIGHTS IN THE STUDY OF TYPE IV RESTRICTION SYSTEMS
Modification-dependent restriction was first observed with populations of phage T4 that contained hydroxymethylcytosine (hm5C)-substituted DNA (13), reviewed in (149,150). This original discovery relied on the fortuitous use of Shigella dysenteria SH as permissive host: it lacks both of the E. coli K-12 hm5C-targeted endonucleases and also the donor for the protective modification, glucosylation. This allowed glucoseless phage to be propagated in Shigella, while picking apart the E. coli K-12 set of restricting and modifying genes.
Key advances in the early years lay in determining the nature of the modifications in T-even phage DNA and the genes that enable them. hm5C is incorporated into the DNA during synthesis, and then glucose residues are added in different configurations. The host provides the glucosyl donor (151,152), while the phage provides the glucosyltransferase enzymes (153–155). With these genetic tools in hand, the host genes mediating the phage restriction activity were identified (156). These were named rglA and rglB (restricts glucoseless phage) because they mediate restriction of hmC-containing phage that lack the further glucose modification.
In the 1980s, the focus switched to other modifications, particularly m5C, with efforts to clone Type II MTases and eukaryotic DNA into E. coli (157–159). The m5C-specific functions mcrA and mcrB were mapped (160) and were shown to be identical to the rglA and rglB genes (161). A third modification–dependent enzyme was found to recognize m6A as well as m5C (162). Using the genetic tools described above, glucose-specific activity was identified (163,164). Most recently, a newly described DNA modification (165) has provided new targets (166) for Type IV enzymes: phosphorothioate linkages in the phosphodiester backbone.
The utility of all these discoveries was, at first, the ability to avoid them (167–169): these restriction systems were found to underlie difficulties encountered in the introduction of foreign MTases into E. coli (157,158,170). On the positive side, use of Type IV restriction in vivo also allowed enrichment of clone libraries for active eukaryotic genomic sequence, since much transcriptionally silenced DNA is heavily methylated [e.g. (171)].
Type IV enzymes have aroused considerable interest in recent years following the rediscovery of hm5C in the DNA of higher eukaryotes (172–175). This finding could portend the discovery of further, as yet unknown or neglected, DNA modifications. The ability of Type IV enzymes to distinguish between C, m5C, hm5C and other molecular variations of cytosine implicates these enzymes as useful tools for studies of epigenetic phenomena; the commercially available enzyme McrBC has been used for the study of such modification patterns (176,177).
Much history may remain to be written. The accompanying review focuses on structural and enzymatic properties of the systems that are known, and sketches some of the evolutionary pressures faced by restriction systems as they compete with each other and with invading replicons.
CONTROL OF RESTRICTION
Double-stranded cleavage of cellular DNA is extremely deleterious to the host cell, even when it can be repaired. Early in the study of restriction systems, the ease of moving systems among strains with differing systems by conjugation or transduction was noted. This suggested that regulation must be present to enable exchange of activities. More recently, the sporadic distribution of R-M systems in genomes of closely related strains strongly suggests that acquisition of a new system is a relatively frequent event in nature as well. Thus, coordination of expression or activity of the R-M activities is a key research topic. Transcriptional or translational control of Type I systems has not been documented, despite efforts to find it (8,178). However post-translational control is exerted at several levels and is described in the accompanying review on Type I R-M systems. The control of Type II R-M systems recapitulates the mechanisms for other regulatory systems and is described here.
Transcriptional control of Type II enzyme expression
In contrast to Type I enzymes, transcriptional control has been found for Type II enzymes. Most of the Type II systems that have been examined have the problem of integrating control of the modification and restriction activities separately, since they are embodied in separate proteins. Once again, the introduction of these genes into a naïve host is of special interest.
Control of restriction of Type II enzymes: the case of EcoRI
Expression of the MTase gene and methylation of the host DNA before synthesis of the REase is the obvious solution and the so-called ‘Hungarian trick’ was the basis for the cloning of many of the first restriction enzymes (179). The lab of Ichizo Kobayashi investigated the regulation of the EcoRI gene, ecoRIR (180–182). This gene is upstream of the modification gene, ecoRIM. The M gene has its own promoters embedded in ecoRIR and no transcription terminator between the genes, so ecoRIM can be transcribed with and without ecoRIR. Using primer extension to locate the start sites and gene fusions to assess expression, two adjacent promoters for ecoRIM as well as two reverse promoters were found within ecoRIR. These convergent promoters negatively affect each other [as in lambda (183)]. Transcription from the reverse promoter is terminated by the forward promoters and generates a small antisense RNA. The presence of the antisense RNA gene in trans reduced lethality mediated by cleavage of under-methylated chromosomes after loss of the EcoRI plasmid (post-segregational killing) (182,184).
Dual transcription control by C proteins
The Blumenthal laboratory provided the first evidence for temporal control in the plasmid-based PvuII system of Proteus vulgaris (108,185). A similar open reading frame with similar function was also found contemporaneously in the BamHI system (186,187). In the PvuII system, the MTase is expressed without delay from an independent promoter and protects the host DNA. The REase gene is in an operon with that for an autogenous activator/repressor protein, C.PvuII. Low basal expression from the pvuIIC promoter leads to accumulation of the activator, thereby boosting transcription of the C and REase genes (108,185) (Figure 4).
Figure 4.
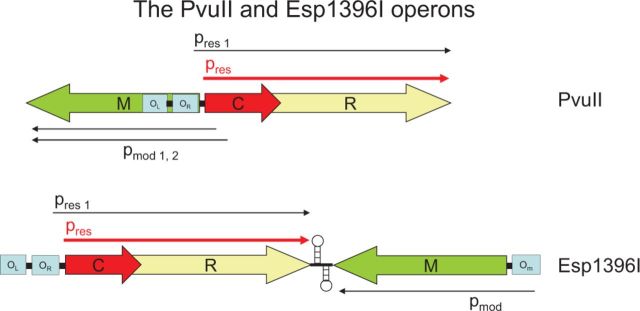
Intricate control of restriction in the operons of the Type II R-M systems of PvuII and Esp1396I by controlling C proteins. A small C gene upstream of, and partially overlapping with, R is coexpressed from pres1, located within the M gene, at low level with R after entry of the self-transmissible PvuII plasmid into a new host, while M is expressed at normal levels from its own two promoters pmod1 and pmod2 located within the C gene. A similar C protein operates in Esp1396I, but in this case the genes are convergently transcribed with transcription terminator structures in between, and M is expressed from a promoter under negative control of operator OR, when engaged by C protein in a manner similar to that of the PvuII system. Briefly, the C protein binds to two palindromic sequences (C boxes) defining operator sites OR and OL upstream of the C and R genes. After initial low-level expression of C.PvuII protein from the weak promoter pres1, positive feedback by high-affinity binding of a C protein dimer to the distal OL site later stimulates expression from the second promoter pres, resulting in a leaderless transcript and more C and R protein. The proximal site OR is a much weaker binding site, but C protein bound at OL enhances the affinity of OR for C protein, and at high levels of C protein, the protein-OR complex downregulates expression of C and R. In this way, C protein is both an activator and negative regulator of its own transcription. In addition, it is a negative regulator of M, which makes sense as overmethylation of DNA may also be harmful to the cell (see text for further details). C.Esp1396I controls OR, OL and OM in a similar manner as described above. In this way, C proteins keep both R-M under control, and have been tentatively identified in >300 R-M systems (Table 2).
The C protein binds to palindromic DNA sequences (C boxes) defining two sites upstream of its gene: OL, associated with activation, and OR, associated with repression. The C protein activates expression of its own gene as well as that of the REase (188). The regulation is similar to gene control in phage lambda: differential binding affinities for the promoters in turn depend on differential DNA sequence and dual symmetry recognition. C proteins belong to the helix-turn-helix family of transcriptional regulators that include the cI and cro repressor proteins of lambdoid phages.
In the wake of PvuII and BamHI, other R-M systems were discovered that were controlled by C proteins, including BglII (189), Eco72I (190), EcoRV (191), Esp396I (192) and SmaI (193). Currently, Rebase lists 19 documented C proteins, as well as 432 putatives based on sequence data (16 September 2013, http://rebase.neb.com). The organization of the genes in the system and regulatory details differ from system to system (108,185,194). There is no published evidence addressing the question of whether R-M systems as a whole evolve in concert with the C proteins. An interesting system would be one homologous to a C-regulated system but without the C gene.
Structure of C proteins
The first structures of C-proteins appeared in 2005: C.AhdI from Geoff Kneale’s laboratory (195), and C.BclI from a consortium of workers (196). The structures were solved without bound DNA, and while they confirmed the close relationship between C-proteins and helix-turn-helix DNA-binding proteins in general, they did not reveal details of the interactions between C-proteins and their C-box binding sites in DNA (195,197–204). That came 4 years later with the crystal structure of C.Esp1396I bound to DNA (205). This structure, coupled with experimental investigations, revealed the mechanics of the genetic switch and the nature of the sequence-specific and non-specific interactions with the promoters controlling the C/R and M genes (205–208). C.Esp1396I bound as a tetramer, with two dimers bound adjacently on the 35-bp operator sequence OL + OR (206). This cooperative binding of dimers to the DNA operator controls the switch from activation to repression of the C and R genes.
Biological consequences of transcriptional regulation
The existence of C proteins explains why it was difficult to introduce some R-M genes in E. coli. For instance, the BamHI system of Bacillus amyloliquefaciens could only be maintained in E. coli when the REase and MTase were present on one plasmid with an additional copy of the MTase on a second plasmid (209). Further analysis suggested that in Bacillus subtilis, a host more closely related to the original expression of R-M was even more stringently regulated (109). C.BamHI enhanced activity of the REase 100-fold in E. coli, but at least 1000-fold in B. subtilis. In E. coli, the C protein repressed expression of the MTase 15-fold. The B. subtilis vegetative RNA polymerase is known be more stringent in its promoter sequence requirements than that of E. coli (210), possibly accounting for the difference in behaviour in the two species.
Crosstalk among the C genes of similar specificity can allow exclusion with R-M systems of different sequence specificities because of the premature activation of the R gene. The pvuIIC and bamHIC genes define one incompatibility group of exclusion, whereas ecoRVC defines another (211). Entry of a second R-M system thus becomes lethal, a phenomenon called ‘apoptotic mutual exclusion’ (211).
THE IMPACT OF RESTRICTION ENZYMES
The technical ingenuity applied to the use of restriction enzymes warrants a separate detailed Survey and Summary or indeed an entire book. For instance, their use led to the production of insulin from recombinant bacteria and yeast by Genentech, thus greatly increasing the supply for diabetics and the production of a recombinant vaccine for Hepatitis B by Biogen to treat the hundreds of millions of people at risk of infection by this virus. More recently they have been redesigned to create artifical nucleases, the Zinc-finger nucleases and the TAL-effector nucleases, which have potential for gene targeting and gene therapy (212). Here, we limit ourselves to a few other examples with significant scientific or public impact.
Genetic engineering
Type II enzymes yielded many practical benefits, as E. coli K12, its genes and its vectors became the workhorses of molecular biology in the 1970s for cloning, generation of libraries, DNA sequencing, detection and overproduction of enzymes, hormones, etc [e.g. (45,213–224)]. The applications of Type II enzymes continued to expand, especially after the arrival of synthetic DNA, in vitro packaging of DNA in phage particles and improved bacterial hosts and vectors for overexpression and stabilization of proteins [see e.g. (225–232)].
A historical perspective on the above topics is beyond the scope of this review. However, a couple of vignettes illustrate how use of REases enabled the research community to leverage a store of understanding to create tools for new advances.
The lacZ gene, which had EcoRI sites suitable for early vectors, and its encoded enzyme, beta-galactosidase, had a long history of investigation. Its utility in the creation of cloning vectors relied on identification of domains within the encoded protein, namely a large catalytic domain and a small multimerization domain. This was discovered by the Muller-Hill group in 1974 (233,234). The 25 N-terminal residues of the small domain can be replaced by peptides of any size and origin without destroying the ability of the multimerization domain to interact with the catalytic domain (233,234). As a result, vectors with short stretches of DNA carrying multiple restriction sites could be created (235–237). Cloning into these sites interrupted the translation of the small domain, destroying its ability to interact with the separately expressed large one. This made possible rapid screening of bacterial colonies on an agar plate for those lacking the activity of LacZ using a colour assay.
In addition, vectors carrying the intact gene but with multiple cloning sites allowed EcoRI-based DNA constructs for transcriptional and translational fusions to the lacZ gene (238–250). The majority (90%) of such LacZ-fusion proteins are stable, allowing purification of chimeric antigens, as well as detection of positive clones with colour assays (238,251). Mutagenesis studies in the laboratory of Jeffrey Miller used the lacZ gene in phage f1, allowing the rapid detection of spontaneous or induced base substitutions and frameshifts (252–254). This resulted in e.g. LacZ-transgenic mice for studies on DNA damage in different organs and tissues in mammalian cells (255,256).
DNA fingerprinting
Restriction enzymes are tools for monitoring Restriction Fragment Length Polymorphisms, allowing the location of mutations, generation of human linkage maps, identification of disease genes (such as sickle cell trait or Huntington disease), and last, but not least, the DNA fingerprinting technique developed by Alec Jeffreys (257–267). DNA fingerprinting (268) allows the solution of paternity cases, the identification of criminals and their victims and the exoneration of the falsely accused. The use of REases in this system enabled the creation of suitable procedures for such identification, although PCR has largely displaced REases in this application.
REases have also proven useful for identifying pathogenic bacterial strains, most recently of S. aureus sp with antibiotic-resistance and virulence factors mediated by mobile genetic elements, e.g. the methicillin-resistant S. aureus (MRSA) bacteria (269). Such strains pose a great threat to humans and animals (270).
FINAL THOUGHTS
In 1977, Werner Arber proposed that REases might have additional functions in the cell (271), and this is an idea to keep in mind given that much of the study of restriction enzymes has been aimed at creating tools rather than a basic study of their behaviour in their natural hosts.
For example, the actions of translocating enzymes such as the Type I and IV enzymes at a replication fork or other variant structure are one such possibility (272,273). This activity may seem of arcane interest, but a broader understanding especially of the translocating enzymes could further understanding of genome stabilization activities in all domains of life. Applications to genome manipulation or medicine could emerge. Action at aberrant structures is a major topic of interest in medicine (274).
Lastly, it is interesting to speculate on the condition of molecular biology and all of its associated sciences at the present day if the simple experiment of spreading bacteria and phage on agar plates to follow the restriction-modification phenomenon (13–15) had not been pursued. It is clear that a multi-billion dollar biotechnology industry would not have been spawned, medical diagnostics and the treatment of many diseases would have been severely retarded, genomics and genome sequencing projects would have been difficult if not impossible and their support of bioinformatics and evolutionary studies would also not have been possible, thus greatly diminishing our current appreciation of the spectacular diversity of life on earth.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the many colleagues who generously provided information for this review. Attendees of the 1972 EMBO Workshop helped identify the people in Figure 2. Special thanks to Werner Arber, Joe Bertani and Rich Roberts for historical material, reprints from their archives and helpful emails over the past decade.
REFERENCES
- 1.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roberts RJ, Cheng X. Base flipping. Annu. Rev. Biochem. 1998;67:181–198. doi: 10.1146/annurev.biochem.67.1.181. [DOI] [PubMed] [Google Scholar]
- 3.Horton JR, Liebert K, Bekes M, Jeltsch A, Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J. Mol. Biol. 2006;358:559–570. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng X, Blumenthal RM. Cytosines do it, thymines do it, even pseudouridines do it—base flipping by an enzyme that acts on RNA. Structure. 2002;10:127–129. doi: 10.1016/s0969-2126(02)00710-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang W. Surviving the sun: repair and bypass of DNA UV lesions. Protein Sci. 2011;20:1781–1789. doi: 10.1002/pro.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks SC, Adhikary S, Rubinson EH, Eichman BF. Recent advances in the structural mechanisms of DNA glycosylases. Biochim. Biophys. Acta. 2013;1834:247–271. doi: 10.1016/j.bbapap.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tubbs JL, Pegg AE, Tainer JA. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 2007;6:1100–1115. doi: 10.1016/j.dnarep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol. Mol. Biol. Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray NE. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology. 2002;148:3–20. doi: 10.1099/00221287-148-1-3. [DOI] [PubMed] [Google Scholar]
- 10.Loenen WA. Tracking EcoKI and DNA fifty years on: a golden story full of surprises. Nucleic Acids Res. 2003;31:7059–7069. doi: 10.1093/nar/gkg944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luria SE, Human ML. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 1952;64:557–569. doi: 10.1128/jb.64.4.557-569.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson ES, Felix A. Variation in Vi-phage II of Salmonella typhi. Nature. 1952;170:492–494. doi: 10.1038/170492b0. [DOI] [PubMed] [Google Scholar]
- 15.Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J. Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luria SE. Host-induced modifications of viruses. Cold Spring Harb. Symp. Quant. Biol. 1953;18:237–244. doi: 10.1101/sqb.1953.018.01.034. [DOI] [PubMed] [Google Scholar]
- 17.Arber W, Dussoix D. Host specificity of DNA produced by Escherichia coli. I. Host controlled modification of bacteriophage lambda. J. Mol. Biol. 1962;5:18–36. doi: 10.1016/s0022-2836(62)80058-8. [DOI] [PubMed] [Google Scholar]
- 18.Arber W, Hattman S, Dussoix D. On the host-controlled modification of bacteriophage lambda. Virology. 1963;21:30–35. doi: 10.1016/0042-6822(63)90300-3. [DOI] [PubMed] [Google Scholar]
- 19.Glover W, Schell J, Symonds N, Stacey KA. The control of host-induced modification by phage P1. Genet. Res. 1963;4:480–482. doi: 10.1016/0006-291x(63)90193-1. [DOI] [PubMed] [Google Scholar]
- 20.Lederberg S. Genetics of host-controlled restriction and modification of deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 1966;91:1029–1036. doi: 10.1128/jb.91.3.1029-1036.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin NC, Dove WF. Genetic evidence for restriction targets in the DNA of phages lambda and phi 80. Genet. Res. 1969;14:151–157. doi: 10.1017/s0016672300001981. [DOI] [PubMed] [Google Scholar]
- 22.Arber W. Host-controlled modification of bacteriophage. Annu. Rev. Microbiol. 1965;19:365–378. doi: 10.1146/annurev.mi.19.100165.002053. [DOI] [PubMed] [Google Scholar]
- 23.Arber W, Linn S. DNA modification and restriction. Annu. Rev. Biochem. 1969;38:467–500. doi: 10.1146/annurev.bi.38.070169.002343. [DOI] [PubMed] [Google Scholar]
- 24.Arber W. Host-Controlled Variation. In: Hershey AD, editor. Bacteriophage Lambda. New York: Cold Spring Harbor Laboratory; 1971. pp. 83–96. [Google Scholar]
- 25.Boyer HW. DNA restriction and modification mechanisms in bacteria. Annu. Rev. Microbiol. 1971;25:153–176. doi: 10.1146/annurev.mi.25.100171.001101. [DOI] [PubMed] [Google Scholar]
- 26.Meselson M, Yuan R, Heywood J. Restriction and modification of DNA. Annu. Rev. Biochem. 1972;41:447–466. doi: 10.1146/annurev.bi.41.070172.002311. [DOI] [PubMed] [Google Scholar]
- 27.Arber W. Host specificit of DNA produced by Escherichia coli V The role of methionine in the production of host specificity. J. Mol. Biol. 1965;11:247–256. doi: 10.1016/s0022-2836(65)80055-9. [DOI] [PubMed] [Google Scholar]
- 28.Gold M, Hurwitz J, Anders M. The enzymatic methylation of RNA and DNA, II. On the species specificity of the methylation enzymes. Proc. Natl Acad. Sci. USA. 1963;50:164–169. doi: 10.1073/pnas.50.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelly TJ, Jr, Smith HO. A restriction enzyme from Hemophilus influenzae. II. J. Mol. Biol. 1970;51:393–409. doi: 10.1016/0022-2836(70)90150-6. [DOI] [PubMed] [Google Scholar]
- 30.Smith HO, Wilcox KW. A restriction enzyme from Hemophilus influenzae. I. Purification and general properties. J. Mol. Biol. 1970;51:379–391. doi: 10.1016/0022-2836(70)90149-x. [DOI] [PubMed] [Google Scholar]
- 31.Landy A, Ruedisueli E, Robinson L, Foeller C, Ross W. Digestion of deoxyribonucleic acids from bacteriophage T7, lambda, and phi 80h with site-specific nucleases from Hemophilus influenzae strain Rc and strain Rd. Biochemistry. 1974;13:2134–2142. doi: 10.1021/bi00707a022. [DOI] [PubMed] [Google Scholar]
- 32.Old R, Murray K, Boizes G. Recognition sequence of restriction endonuclease III from Hemophilus influenzae. J. Mol. Biol. 1975;92:331–339. doi: 10.1016/0022-2836(75)90232-6. [DOI] [PubMed] [Google Scholar]
- 33.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics? Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 34.Roy PH, Smith HO. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J. Mol. Biol. 1973;81:427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- 35.Roy PH, Smith HO. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J. Mol. Biol. 1973;81:445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- 36.Roszczyk E, Goodgal S. Methylase activities from Haemophilus influenzae that protect Haemophilus parainfluenzae transforming deoxyribonucleic acid from inactivation by Haemophilus influenzae endonuclease R. J. Bacteriol. 1975;123:287–293. doi: 10.1128/jb.123.1.287-293.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yoshimori,R. (1971) A genetic and biochemical analysis of the restriction and modification of DNA by resistance transfer factors. 146 p. Ph.D Thesis University of California, San Francisco. Thesis/Dissertation.
- 38.Yoshimori R, Roulland-Dussoix D, Boyer HW. R factor-controlled restriction and modification of deoxyribonucleic acid: restriction mutants. J. Bacteriol. 1972;112:1275–1279. doi: 10.1128/jb.112.3.1275-1279.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hedgpeth J, Goodman HM, Boyer HW. DNA nucleotide sequence restricted by the RI endonuclease. Proc. Natl Acad. Sci. USA. 1972;69:3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mertz JE, Davis RW. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc. Natl Acad. Sci. USA. 1972;69:3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dugaiczyk A, Hedgpeth J, Boyer HW, Goodman HM. Physical identity of the SV40 deoxyribonucleic acid sequence recognized by the Eco RI restriction endonuclease and modification methylase. Biochemistry. 1974;13:503–512. doi: 10.1021/bi00700a016. [DOI] [PubMed] [Google Scholar]
- 42.Boyer HW, Chow LT, Dugaiczyk A, Hedgpeth J, Goodman HM. DNA substrate site for the EcoRII restriction endonuclease and modification methylase. Nat. New Biol. 1973;244:40–43. doi: 10.1038/newbio244040a0. [DOI] [PubMed] [Google Scholar]
- 43.Bigger CH, Murray K, Murray NE. Recognition sequence of a restriction enzyme. Nat. New Biol. 1973;244:7–10. doi: 10.1038/newbio244007a0. [DOI] [PubMed] [Google Scholar]
- 44.Lehman IR. DNA ligase: structure, mechanism, and function. Science. 1974;186:790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- 45.Murray NE, Murray K. Manipulation of restriction targets in phage lambda to form receptor chromosomes for DNA fragments. Nature. 1974;251:476–481. doi: 10.1038/251476a0. [DOI] [PubMed] [Google Scholar]
- 46.Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl Acad. Sci. USA. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hershfield V, Boyer HW, Yanofsky C, Lovett MA, Helinski DR. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc. Natl Acad. Sci. USA. 1974;71:3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith HO, Nathans D. Letter: a suggested nomenclature for bacterial host modification and restriction systems and their enzymes. J. Mol. Biol. 1973;81:419–423. doi: 10.1016/0022-2836(73)90152-6. [DOI] [PubMed] [Google Scholar]
- 49.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Nathans D, Smith HO. Restriction endonucleases in the analysis and restructuring of DNA molecules. Annu. Rev. Biochem. 1975;44:273–293. doi: 10.1146/annurev.bi.44.070175.001421. [DOI] [PubMed] [Google Scholar]
- 52.Watson JD, Tooze J. The DNA Story: Documentary History of Gene Cloning. San Francisco: W.H. Freeman and Company; 1981. [Google Scholar]
- 53.Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 54.Hubacek J, Glover SW. Complementation analysis of temperature-sensitive host specificity mutations in Escherichia coli. J. Mol. Biol. 1970;50:111–127. doi: 10.1016/0022-2836(70)90108-7. [DOI] [PubMed] [Google Scholar]
- 55.Titheradge AJ, King J, Ryu J, Murray NE. Families of restriction enzymes: an analysis prompted by molecular and genetic data for type ID restriction and modification systems. Nucleic Acids Res. 2001;29:4195–4205. doi: 10.1093/nar/29.20.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kasarjian JK, Hidaka M, Horiuchi T, Iida M, Ryu J. The recognition and modification sites for the bacterial type I restriction systems KpnAI, StySEAI, StySENI and StySGI. Nucleic Acids Res. 2004;32:e82. doi: 10.1093/nar/gnh079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chin V, Valinluck V, Magaki S, Ryu J. KpnBI is the prototype of a new family (IE) of bacterial type I restriction-modification system. Nucleic Acids Res. 2004;32:e138. doi: 10.1093/nar/gnh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dryden DT, Murray NE, Rao DN. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 2001;29:3728–3741. doi: 10.1093/nar/29.18.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bourniquel AA, Bickle TA. Complex restriction enzymes: NTP-driven molecular motors. Biochimie. 2002;84:1047–1059. doi: 10.1016/s0300-9084(02)00020-2. [DOI] [PubMed] [Google Scholar]
- 60.Roberts GA, Houston PJ, White JH, Chen K, Stephanou AS, Cooper LP, Dryden DT, Lindsay JA. Impact of target site distribution for Type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res. 2013;41:7472–7484. doi: 10.1093/nar/gkt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dussoix D, Arber W. Host specificity of DNA produced by Escherichia coli. II. Control over acceptance of DNA from infecting phage lambda. J. Mol. Biol. 1962;5:37–49. doi: 10.1016/s0022-2836(62)80059-x. [DOI] [PubMed] [Google Scholar]
- 62.Linn S, Arber W. Host specificity of DNA produced by Escherichia coli, X. In vitro restriction of phage fd replicative form. Proc. Natl Acad. Sci. USA. 1968;59:1300–1306. doi: 10.1073/pnas.59.4.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meselson M, Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217:1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- 64.Roulland-Dussoix D, Boyer HW. The Escherichia coli B restriction endonuclease. Biochim. Biophys. Acta. 1969;195:219–229. doi: 10.1016/0005-2787(69)90618-2. [DOI] [PubMed] [Google Scholar]
- 65.Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- 66.Loenen WAM. S-adenosylmethionine: simple agent of methylation and secret to aging and metabolism? In: Tollefsbol TO, editor. Epigenetics of Aging. Springer; 2010. pp. 107–131. [Google Scholar]
- 67.Kim JS, DeGiovanni A, Jancarik J, Adams PD, Yokota H, Kim R, Kim SH. Crystal structure of DNA sequence specificity subunit of a type I restriction-modification enzyme and its functional implications. Proc. Natl Acad. Sci. USA. 2005;102:3248–3253. doi: 10.1073/pnas.0409851102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Calisto BM, Pich OQ, Pinol J, Fita I, Querol E, Carpena X. Crystal structure of a putative type I restriction-modification S subunit from Mycoplasma genitalium. J. Mol. Biol. 2005;351:749–762. doi: 10.1016/j.jmb.2005.06.050. [DOI] [PubMed] [Google Scholar]
- 69.Kennaway CK, Taylor JE, Song CF, Potrzebowski W, Nicholson W, White JH, Swiderska A, Obarska-Kosinska A, Callow P, Cooper LP, et al. Structure and operation of the DNA-translocating type I DNA restriction enzymes. Genes Dev. 2012;26:92–104. doi: 10.1101/gad.179085.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Horiuchi K, Zinder ND. Cleavage of bacteriophage fl DNA by the restriction enzyme of Escherichia coli B. Proc. Natl Acad. Sci. USA. 1972;69:3220–3224. doi: 10.1073/pnas.69.11.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Studier FW, Bandyopadhyay PK. Model for how type I restriction enzymes select cleavage sites in DNA. Proc. Natl Acad. Sci. USA. 1988;85:4677–4681. doi: 10.1073/pnas.85.13.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosamond J, Endlich B, Linn S. Electron microscopic studies of the mechanism of action of the restriction endonuclease of Escherichia coli B. J. Mol. Biol. 1979;129:619–635. doi: 10.1016/0022-2836(79)90472-8. [DOI] [PubMed] [Google Scholar]
- 73.Yuan R, Hamilton DL, Burckhardt J. DNA translocation by the restriction enzyme from E. coli K. Cell. 1980;20:237–244. doi: 10.1016/0092-8674(80)90251-2. [DOI] [PubMed] [Google Scholar]
- 74.Kennaway CK, Obarska-Kosinska A, White JH, Tuszynska I, Cooper LP, Bujnicki JM, Trinick J, Dryden DT. The structure of M.EcoKI Type I DNA methyltransferase with a DNA mimic antirestriction protein. Nucleic Acids Res. 2009;37:762–770. doi: 10.1093/nar/gkn988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. II. Further studies of the structure of DNA intermediates and products. J. Biol. Chem. 1985;260:5729–5738. [PubMed] [Google Scholar]
- 76.Endlich B, Linn S. The DNA restriction endonuclease of Escherichia coli B. I. Studies of the DNA translocation and the ATPase activities. J. Biol. Chem. 1985;260:5720–5728. [PubMed] [Google Scholar]
- 77.Davies GP, Martin I, Sturrock SS, Cronshaw A, Murray NE, Dryden DT. On the structure and operation of type I DNA restriction enzymes. J. Mol. Biol. 1999;290:565–579. doi: 10.1006/jmbi.1999.2908. [DOI] [PubMed] [Google Scholar]
- 78.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gorbalenya AE, Koonin EV. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Lett. 1991;291:277–281. doi: 10.1016/0014-5793(91)81301-n. [DOI] [PubMed] [Google Scholar]
- 80.Gorbalenya AE, Koonin EV, Donchenko AP, Blinov VM. A conserved NTP–motif in putative helicases. Nature. 1988;333:22. doi: 10.1038/333022a0. [DOI] [PubMed] [Google Scholar]
- 81.Hall MC, Matson SW. Helicase motifs: the engine that powers DNA unwinding. Mol. Microbiol. 1999;34:867–877. doi: 10.1046/j.1365-2958.1999.01659.x. [DOI] [PubMed] [Google Scholar]
- 82.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 83.Szczelkun MD, Friedhoff P, Seidel R. Maintaining a sense of direction during long-range communication on DNA. Biochem. Soc. Trans. 2010;38:404–409. doi: 10.1042/BST0380404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tuteja N, Tuteja R. Prokaryotic and eukaryotic DNA helicases. Essential molecular motor proteins for cellular machinery. Eur. J. Biochem. 2004;271:1835–1848. doi: 10.1111/j.1432-1033.2004.04093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Umate P, Tuteja N, Tuteja R. Genome-wide comprehensive analysis of human helicases. Commun. Integr. Biol. 2011;4:118–137. doi: 10.4161/cib.4.1.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ramanathan A, Agarwal PK. Evolutionarily conserved linkage between enzyme fold, flexibility, and catalysis. PLoS. Biol. 2011;9:e1001193. doi: 10.1371/journal.pbio.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mahdi AA, Briggs GS, Sharples GJ, Wen Q, Lloyd RG. A model for dsDNA translocation revealed by a structural motif common to RecG and Mfd proteins. EMBO J. 2003;22:724–734. doi: 10.1093/emboj/cdg043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudolph CJ, Upton AL, Briggs GS, Lloyd RG. Is RecG a general guardian of the bacterial genome? DNA Repair (Amst) 2010;9:210–223. doi: 10.1016/j.dnarep.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 89.Sisakova E, Stanley LK, Weiserova M, Szczelkun MD. A RecB-family nuclease motif in the Type I restriction endonuclease EcoR124I. Nucleic Acids Res. 2008;36:3939–3949. doi: 10.1093/nar/gkn333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sisakova E, Weiserova M, Dekker C, Seidel R, Szczelkun MD. The interrelationship of helicase and nuclease domains during DNA translocation by the molecular motor EcoR124I. J. Mol. Biol. 2008;384:1273–1286. doi: 10.1016/j.jmb.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bullas LR, Colson C, Van PA. DNA restriction and modification systems in Salmonella. SQ, a new system derived by recombination between the SB system of Salmonella typhimurium and the SP system of Salmonella potsdam. J. Gen. Microbiol. 1976;95:166–172. doi: 10.1099/00221287-95-1-166. [DOI] [PubMed] [Google Scholar]
- 92.Fuller-Pace FV, Bullas LR, Delius H, Murray NE. Genetic recombination can generate altered restriction specificity. Proc. Natl Acad. Sci. USA. 1984;81:6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nagaraja V, Shepherd JC, Bickle TA. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985;316:371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- 94.Fuller-Pace FV, Murray NE. Two DNA recognition domains of the specificity polypeptides of a family of type I restriction enzymes. Proc. Natl Acad. Sci. USA. 1986;83:9368–9372. doi: 10.1073/pnas.83.24.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schouler C, Gautier M, Ehrlich SD, Chopin MC. Combinational variation of restriction modification specificities in Lactococcus lactis. Mol. Microbiol. 1998;28:169–178. doi: 10.1046/j.1365-2958.1998.00787.x. [DOI] [PubMed] [Google Scholar]
- 96.Dybvig K, Sitaraman R, French CT. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl Acad. Sci. USA. 1998;95:13923–13928. doi: 10.1073/pnas.95.23.13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 98.Szybalski W, Kim SC, Hasan N, Podhajska AJ. Class-IIS restriction enzymes—a review. Gene. 1991;100:13–26. doi: 10.1016/0378-1119(91)90345-c. [DOI] [PubMed] [Google Scholar]
- 99.Bujnicki JM. Molecular Phylogenetics of Restriction Enzymes. In: Pingoud A, editor. Restriction Enzymes. Vol. 14. Berlin; New York: Springer; 2004. pp. 63–93. [Google Scholar]
- 100.Kong H, Smith CL. Substrate DNA and cofactor regulate the activities of a multi-functional restriction-modification enzyme, BcgI. Nucleic Acids Res. 1997;25:3687–3692. doi: 10.1093/nar/25.18.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith RM, Marshall JJ, Jacklin AJ, Retter SE, Halford SE, Sobott F. Organization of the BcgI restriction-modification protein for the cleavage of eight phosphodiester bonds in DNA. Nucleic Acids Res. 2013;41:391–404. doi: 10.1093/nar/gks1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Smith RM, Jacklin AJ, Marshall JJ, Sobott F, Halford SE. Organization of the BcgI restriction-modification protein for the transfer of one methyl group to DNA. Nucleic Acids Res. 2013;41:405–417. doi: 10.1093/nar/gks1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lacks S, Greenberg B. A deoxyribonuclease of Diplococcus pneumoniae specific for methylated DNA. J. Biol. Chem. 1975;250:4060–4066. [PubMed] [Google Scholar]
- 104.Friedhoff P, Lurz R, Luder G, Pingoud A. Sau3AI, a monomeric type II restriction endonuclease that dimerizes on the DNA and thereby induces DNA loops. J. Biol. Chem. 2001;276:23581–23588. doi: 10.1074/jbc.M101694200. [DOI] [PubMed] [Google Scholar]
- 105.Greene PJ, Gupta M, Boyer HW, Brown WE, Rosenberg JM. Sequence analysis of the DNA encoding the Eco RI endonuclease and methylase. J. Biol. Chem. 1981;256:2143–2153. [PubMed] [Google Scholar]
- 106.Newman AK, Rubin RA, Kim SH, Modrich P. DNA sequences of structural genes for Eco RI DNA restriction and modification enzymes. J. Biol. Chem. 1981;256:2131–2139. [PubMed] [Google Scholar]
- 107.Bougueleret L, Schwarzstein M, Tsugita A, Zabeau M. Characterization of the genes coding for the Eco RV restriction and modification system of Escherichia coli. Nucleic Acids Res. 1984;12:3659–3676. doi: 10.1093/nar/12.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tao T, Blumenthal RM. Sequence and characterization of pvuIIR, the PvuII endonuclease gene, and of pvuIIC, its regulatory gene. J. Bacteriol. 1992;174:3395–3398. doi: 10.1128/jb.174.10.3395-3398.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brooks JE, Nathan PD, Landry D, Sznyter LA, Waite-Rees P, Ives CL, Moran LS, Slatko BE, Benner JS. Characterization of the cloned BamHI restriction modification system: its nucleotide sequence, properties of the methylase, and expression in heterologous hosts. Nucleic Acids Res. 1991;19:841–850. doi: 10.1093/nar/19.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.McClarin JA, Frederick CA, Wang BC, Greene P, Boyer HW, Grable J, Rosenberg JM. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986;234:1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- 111.Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 112.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cheng X, Balendiran K, Schildkraut I, Anderson JE. Structure of PvuII endonuclease with cognate DNA. EMBO J. 1994;13:3927–3935. doi: 10.1002/j.1460-2075.1994.tb06708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 115.Anderson JE. Restriction endonucleases and modification methylases. Curr. Opin. Struct. Biol. 1993;3:24–30. [Google Scholar]
- 116.Aggarwal AK. Structure and function of restriction endonucleases. Curr. Opin. Struct. Biol. 1995;5:11–19. doi: 10.1016/0959-440x(95)80004-k. [DOI] [PubMed] [Google Scholar]
- 117.Kovall RA, Matthews BW. Structural, functional, and evolutionary relationships between lambda-exonuclease and the type II restriction endonucleases. Proc. Natl Acad. Sci. USA. 1998;95:7893–7897. doi: 10.1073/pnas.95.14.7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hickman AB, Li Y, Mathew SV, May EW, Craig NL, Dyda F. Unexpected structural diversity in DNA recombination: the restriction endonuclease connection. Mol. Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]
- 119.Roberts RJ. Restriction endonucleases. CRC Crit. Rev. Biochem. 1976;4:123–164. doi: 10.3109/10409237609105456. [DOI] [PubMed] [Google Scholar]
- 120.Roberts RJ. Restriction and modification enzymes and their recognition sequences. Nucleic Acids Res. 1985;13(Suppl):r165–r200. doi: 10.1093/nar/13.suppl.r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roberts RJ, Macelis D. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 1993;21:3125–3137. doi: 10.1093/nar/21.13.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Orlowski J, Bujnicki JM. Structural and evolutionary classification of Type II restriction enzymes based on theoretical and experimental analyses. Nucleic Acids Res. 2008;36:3552–3569. doi: 10.1093/nar/gkn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bujnicki JM, Rychlewski L. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001;3:69–72. [PubMed] [Google Scholar]
- 125.Sokolowska M, Czapinska H, Bochtler M. Hpy188I-DNA pre- and post-cleavage complexes—snapshots of the GIY-YIG nuclease mediated catalysis. Nucleic Acids Res. 2011;39:1554–1564. doi: 10.1093/nar/gkq821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Cymerman IA, Obarska A, Skowronek KJ, Lubys A, Bujnicki JM. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006;65:867–876. doi: 10.1002/prot.21156. [DOI] [PubMed] [Google Scholar]
- 127.Chan SH, Opitz L, Higgins L, O'loane D, Xu SY. Cofactor requirement of HpyAV restriction endonuclease. PLoS One. 2010;5:e9071. doi: 10.1371/journal.pone.0009071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mak AN, Lambert AR, Stoddard BL. Folding, DNA recognition, and function of GIY-YIG endonucleases: crystal structures of R.Eco29kI. Structure. 2010;18:1321–1331. doi: 10.1016/j.str.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sasnauskas G, Connolly BA, Halford SE, Siksnys V. Site-specific DNA transesterification catalyzed by a restriction enzyme. Proc. Natl Acad. Sci. USA. 2007;104:2115–2120. doi: 10.1073/pnas.0608689104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Murray IA, Stickel SK, Roberts RJ. Sequence-specific cleavage of RNA by Type II restriction enzymes. Nucleic Acids Res. 2010;38:8257–8268. doi: 10.1093/nar/gkq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–1323. doi: 10.1534/genetics.105.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hadi SM, Bachi B, Iida S, Bickle TA. DNA restriction—modification enzymes of phage P1 and plasmid p15B. Subunit functions and structural homologies. J. Mol. Biol. 1983;165:19–34. doi: 10.1016/s0022-2836(83)80240-x. [DOI] [PubMed] [Google Scholar]
- 133.Redaschi N, Bickle TA. Posttranscriptional regulation of EcoP1I and EcoP15I restriction activity. J. Mol. Biol. 1996;257:790–803. doi: 10.1006/jmbi.1996.0202. [DOI] [PubMed] [Google Scholar]
- 134.Kruger DH, Kupper D, Meisel A, Reuter M, Schroeder C. The significance of distance and orientation of restriction endonuclease recognition sites in viral DNA genomes. FEMS Microbiol. Rev. 1995;17:177–184. doi: 10.1111/j.1574-6976.1995.tb00200.x. [DOI] [PubMed] [Google Scholar]
- 135.Meisel A, Bickle TA, Kruger DH, Schroeder C. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature. 1992;355:467–469. doi: 10.1038/355467a0. [DOI] [PubMed] [Google Scholar]
- 136.Humbelin M, Suri B, Rao DN, Hornby DP, Eberle H, Pripfl T, Kenel S, Bickle TA. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequence of the EcoP1 operon, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 1988;200:23–29. doi: 10.1016/0022-2836(88)90330-0. [DOI] [PubMed] [Google Scholar]
- 137.Bachi B, Reiser J, Pirrotta V. Methylation and cleavage sequences of the EcoP1 restriction-modification enzyme. J. Mol. Biol. 1979;128:143–163. doi: 10.1016/0022-2836(79)90123-2. [DOI] [PubMed] [Google Scholar]
- 138.Piekarowicz A, Brzezinski R. Cleavage and methylation of DNA by the restriction endonuclease HinfIII isolated from Haemophilus influenzae Rf. J. Mol. Biol. 1980;144:415–429. doi: 10.1016/0022-2836(80)90329-0. [DOI] [PubMed] [Google Scholar]
- 139.Kruger DH, Schroeder C, Reuter M, Bogdarina IG, Buryanov YI, Bickle TA. DNA methylation of bacterial viruses T3 and T7 by different DNA methylases in Escherichia coli K12 cells. Eur. J. Biochem. 1985;150:323–330. doi: 10.1111/j.1432-1033.1985.tb09024.x. [DOI] [PubMed] [Google Scholar]
- 140.Meisel A, Mackeldanz P, Bickle TA, Kruger DH, Schroeder C. Type III restriction endonucleases translocate DNA in a reaction driven by recognition site-specific ATP hydrolysis. EMBO J. 1995;14:2958–2966. doi: 10.1002/j.1460-2075.1995.tb07296.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dryden DT, Edwardson JM, Henderson RM. DNA translocation by type III restriction enzymes: a comparison of current models of their operation derived from ensemble and single-molecule measurements. Nucleic Acids Res. 2011;39:4525–4531. doi: 10.1093/nar/gkq1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Crampton N, Roes S, Dryden DT, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Crampton N, Yokokawa M, Dryden DT, Edwardson JM, Rao DN, Takeyasu K, Yoshimura SH, Henderson RM. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl Acad. Sci. USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ramanathan SP, van AK, Sears A, Peakman LJ, Diffin FM, Szczelkun MD, Seidel R. Type III restriction enzymes communicate in 1D without looping between their target sites. Proc. Natl Acad. Sci. USA. 2009;106:1748–1753. doi: 10.1073/pnas.0807193106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schwarz FW, van AK, Toth J, Seidel R, Szczelkun MD. DNA cleavage site selection by Type III restriction enzymes provides evidence for head-on protein collisions following 1D bidirectional motion. Nucleic Acids Res. 2011;39:8024–8051. doi: 10.1093/nar/gkr502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Szczelkun MD. Translocation, switching and gating: potential roles for ATP in long-range communication on DNA by Type III restriction endonucleases. Biochem. Soc. Trans. 2011;39:589–594. doi: 10.1042/BST0390589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gupta YK, Yang L, Chan SH, Samuelson JC, Xu SY, Aggarwal AK. Structural insights into the assembly and shape of Type III restriction-modification (R-M) EcoP15I complex by small-angle X-ray scattering. J. Mol. Biol. 2012;420:261–268. doi: 10.1016/j.jmb.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wyszomirski KH, Curth U, Alves J, Mackeldanz P, Moncke-Buchner E, Schutkowski M, Kruger DH, Reuter M. Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res. 2012;40:3610–3622. doi: 10.1093/nar/gkr1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Revel HR, uria SE. DNA-glucosylation in T-even phage: genetic determination and role in phagehost interaction. Annu. Rev. Genet. 1970;4:177–192. doi: 10.1146/annurev.ge.04.120170.001141. [DOI] [PubMed] [Google Scholar]
- 150.Revel HR. DNA modification: glucosylation. In: Mathews CK, Kutter EM, Mosig G, Berget P, editors. Bacteriophage T4. Washington DC: American Society of Microbiology; 1983. pp. 156–165. [Google Scholar]
- 151.Hattman S, Fukasawa T. Host-induced modification of T-even phages due to defective glucosylation of their DNA. Proc. Natl Acad. Sci. USA. 1963;50:297–300. doi: 10.1073/pnas.50.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Shedlovsky A, Brenner S. A chemical basis for the host-induced modification of T-even bacteriophages. Proc. Natl Acad. Sci. USA. 1963;50:300–305. doi: 10.1073/pnas.50.2.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Revel HR, Hattman S, Luria SE. Mutants of bacteriophages T2 and T6 defective in alpha-glucosyl transferase. Biochem. Biophys. Res. Commun. 1965;18:545–550. doi: 10.1016/0006-291x(65)90788-6. [DOI] [PubMed] [Google Scholar]
- 154.Georgopoulos CP. Isolation and preliminary characterization of T4 mutants with nonglucosylated DNA. Biochem. Biophys. Res. Commun. 1967;28:179–184. doi: 10.1016/0006-291x(67)90426-3. [DOI] [PubMed] [Google Scholar]
- 155.Georgopoulos CP, Revel HR. Studies with glucosyl transferase mutants of the T-even bacteriophages. Virology. 1971;44:271–285. doi: 10.1016/0042-6822(71)90259-5. [DOI] [PubMed] [Google Scholar]
- 156.Revel HR. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967;31:688–701. doi: 10.1016/0042-6822(67)90197-3. [DOI] [PubMed] [Google Scholar]
- 157.Noyer-Weidner M, Diaz R, Reiners L. Cytosine-specific DNA modification interferes with plasmid establishment in Escherichia coli K12: involvement of rglB. Mol. Gen. Genet. 1986;205:469–475. doi: 10.1007/BF00338084. [DOI] [PubMed] [Google Scholar]
- 158.Raleigh EA, Wilson G. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl Acad. Sci. USA. 1986;83:9070–9074. doi: 10.1073/pnas.83.23.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Blumenthal RM. The PvuII restriction-modification system: cloning, characterization, and use in revealing an E. coli barrier to certain methylases or methylated DNA. In: Chirikjian JG, editor. Gene Amplification and Analysis. Vol. 5. Elsevier, New York; 1987. pp. 227–245. [PubMed] [Google Scholar]
- 160.Ravi RS, Sozhamannan S, Dharmalingam K. Transposon mutagenesis and genetic mapping of the rglA and rglB loci of Escherichia coli. Mol. Gen. Genet. 1985;198:390–392. doi: 10.1007/BF00332928. [DOI] [PubMed] [Google Scholar]
- 161.Raleigh EA, Trimarchi R, Revel H. Genetic and physical mapping of the mcrA (rglA) and mcrB (rglB) loci of Escherichia coli K-12. Genetics. 1989;122:279–296. doi: 10.1093/genetics/122.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Heitman J, Model P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 1987;169:3243–3250. doi: 10.1128/jb.169.7.3243-3250.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Janosi L, Yonemitsu H, Hong H, Kaji A. Molecular cloning and expression of a novel hydroxymethylcytosine-specific restriction enzyme (PvuRts1I) modulated by glucosylation of DNA. J. Mol. Biol. 1994;242:45–61. doi: 10.1006/jmbi.1994.1556. [DOI] [PubMed] [Google Scholar]
- 164.Bair CL, Black LW. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 2007;366:768–778. doi: 10.1016/j.jmb.2006.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhou X, He X, Liang J, Li A, Xu T, Kieser T, Helmann JD, Deng Z. A novel DNA modification by sulphur. Mol. Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 166.Liu G, Ou HY, Wang T, Li L, Tan H, Zhou X, Rajakumar K, Deng Z, He X. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLoS Genet. 2010;6:e1001253. doi: 10.1371/journal.pgen.1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Raleigh EA, Murray NE, Revel H, Blumenthal RM, Westaway D, Reith AD, Rigby PW, Elhai J, Hanahan D. McrA and McrB restriction phenotypes of some E. coli strains and implications for gene cloning. Nucleic Acids Res. 1988;16:1563–1575. doi: 10.1093/nar/16.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Woodcock DM, Crowther PJ, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith SS, Michael MZ, Graham MW. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Graham MW, Doherty JP, Woodcock DM. Efficient construction of plant genomic libraries requires the use of mcr- host strains and packaging mixes. Plant Mol. Biol. Rep. 1990;8:33–42. [Google Scholar]
- 170.Blumenthal RM, Gregory SA, Cooperider JS. Cloning of a restriction-modification system from Proteus vulgaris and its use in analyzing a methylase-sensitive phenotype in Escherichia coli. J. Bacteriol. 1985;164:501–509. doi: 10.1128/jb.164.2.501-509.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Palmer LE, Rabinowicz PD, O'Shaughnessy AL, Balija VS, Nascimento LU, Dike S, de la Bastide M, Martienssen RA, McCombie WR. Maize genome sequencing by methylation filtration. Science. 2003;302:2115–2117. doi: 10.1126/science.1091265. [DOI] [PubMed] [Google Scholar]
- 172.Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem. J. 1972;126:781–790. doi: 10.1042/bj1260781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Penn NW. Modification of brain deoxyribonucleic acid base content with maturation in normal and malnourished rats. Biochem. J. 1976;155:709–712. doi: 10.1042/bj1550709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ordway JM, Bedell JA, Citek RW, Nunberg A, Garrido A, Kendall R, Stevens JR, Cao D, Doerge RW, Korshunova Y, et al. Comprehensive DNA methylation profiling in a human cancer genome identifies novel epigenetic targets. Carcinogenesis. 2006;27:2409–2423. doi: 10.1093/carcin/bgl161. [DOI] [PubMed] [Google Scholar]
- 177.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Loenen WA, Daniel AS, Braymer HD, Murray NE. Organization and sequence of the hsd genes of Escherichia coli K-12. J. Mol. Biol. 1987;198:159–170. doi: 10.1016/0022-2836(87)90303-2. [DOI] [PubMed] [Google Scholar]
- 179.Szomolanyi E, Kiss A, Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980;10:219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- 180.Liu Y, Ichige A, Kobayashi I. Regulation of the EcoRI restriction-modification system: Identification of ecoRIM gene promoters and their upstream negative regulators in the ecoRIR gene. Gene. 2007;400:140–149. doi: 10.1016/j.gene.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 181.Liu Y, Kobayashi I. Negative regulation of the EcoRI restriction enzyme gene is associated with intragenic reverse promoters. J. Bacteriol. 2007;189:6928–6935. doi: 10.1128/JB.00127-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mruk I, Liu Y, Ge L, Kobayashi I. Antisense RNA associated with biological regulation of a restriction-modification system. Nucleic Acids Res. 2011;39:5622–5632. doi: 10.1093/nar/gkr166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ward DF, Murray NE. Convergent transcription in bacteriophage lambda: interference with gene expression. J. Mol. Biol. 1979;133:249–266. doi: 10.1016/0022-2836(79)90533-3. [DOI] [PubMed] [Google Scholar]
- 184.Heitman J, Zinder ND, Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc. Natl Acad. Sci. USA. 1989;86:2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ives CL, Nathan PD, Brooks JE. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J. Bacteriol. 1992;174:7194–7201. doi: 10.1128/jb.174.22.7194-7201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sohail A, Ives CL, Brooks JE. Purification and characterization of C.BamHI, a regulator of the BamHI restriction-modification system. Gene. 1995;157:227–228. doi: 10.1016/0378-1119(94)00698-r. [DOI] [PubMed] [Google Scholar]
- 188.Bart A, Dankert J, van der Ende A. Operator sequences for the regulatory proteins of restriction modification systems. Mol. Microbiol. 1999;31:1277–1278. doi: 10.1046/j.1365-2958.1999.01253.x. [DOI] [PubMed] [Google Scholar]
- 189.Anton BP, Heiter DF, Benner JS, Hess EJ, Greenough L, Moran LS, Slatko BE, Brooks JE. Cloning and characterization of the Bg/II restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. doi: 10.1016/s0378-1119(96)00638-5. [DOI] [PubMed] [Google Scholar]
- 190.Rimseliene R, Vaisvila R, Janulaitis A. The eco72IC gene specifies a trans-acting factor which influences expression of both DNA methyltransferase and endonuclease from the Eco72I restriction-modification system. Gene. 1995;157:217–219. doi: 10.1016/0378-1119(94)00794-s. [DOI] [PubMed] [Google Scholar]
- 191.Zheleznaya LA, Kainov DE, Yunusova AK, Matvienko NI. Regulatory C protein of the EcoRV modification-restriction system. Biochemistry (Mosc.) 2003;68:105–110. doi: 10.1023/a:1022105804578. [DOI] [PubMed] [Google Scholar]
- 192.Cesnaviciene E, Mitkaite G, Stankevicius K, Janulaitis A, Lubys A. Esp1396I restriction-modification system: structural organization and mode of regulation. Nucleic Acids Res. 2003;31:743–749. doi: 10.1093/nar/gkg135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Heidmann S, Seifert W, Kessler C, Domdey H. Cloning, characterization and heterologous expression of the SmaI restriction-modification system. Nucleic Acids Res. 1989;17:9783–9796. doi: 10.1093/nar/17.23.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Semenova E, Minakhin L, Bogdanova E, Nagornykh M, Vasilov A, Heyduk T, Solonin A, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 196.Sawaya MR, Zhu Z, Mersha F, Chan SH, Dabur R, Xu SY, Balendiran GK. Crystal structure of the restriction-modification system control element C.Bcll and mapping of its binding site. Structure. 2005;13:1837–1847. doi: 10.1016/j.str.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 197.Bogdanova E, Djordjevic M, Papapanagiotou I, Heyduk T, Kneale G, Severinov K. Transcription regulation of the type II restriction-modification system AhdI. Nucleic Acids Res. 2008;36:1429–1442. doi: 10.1093/nar/gkm1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Callow P, Sukhodub A, Taylor JE, Kneale GG. Shape and subunit organisation of the DNA methyltransferase M.AhdI by small-angle neutron scattering. J. Mol. Biol. 2007;369:177–185. doi: 10.1016/j.jmb.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marks P, McGeehan J, Wilson G, Errington N, Kneale G. Purification and characterisation of a novel DNA methyltransferase, M.AhdI. Nucleic Acids Res. 2003;31:2803–2810. doi: 10.1093/nar/gkg399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Marks P, McGeehan J, Kneale G. A novel strategy for the expression and purification of the DNA methyltransferase, M.AhdI. Protein Expr. Purif. 2004;37:236–242. doi: 10.1016/j.pep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 201.McGeehan JE, Papapanagiotou I, Streeter SD, Kneale GG. Cooperative binding of the C.AhdI controller protein to the C/R promoter and its role in endonuclease gene expression. J. Mol. Biol. 2006;358:523–531. doi: 10.1016/j.jmb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 202.McGeehan JE, Streeter S, Cooper JB, Mohammed F, Fox GC, Kneale GG. Crystallization and preliminary X-ray analysis of the controller protein C.AhdI from Aeromonas hydrophilia. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:323–325. doi: 10.1107/S0907444903026143. [DOI] [PubMed] [Google Scholar]
- 203.Papapanagiotou I, Streeter SD, Cary PD, Kneale GG. DNA structural deformations in the interaction of the controller protein C.AhdI with its operator sequence. Nucleic Acids Res. 2007;35:2643–2650. doi: 10.1093/nar/gkm129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Streeter SD, Papapanagiotou I, McGeehan JE, Kneale GG. DNA footprinting and biophysical characterization of the controller protein C.AhdI suggests the basis of a genetic switch. Nucleic Acids Res. 2004;32:6445–6453. doi: 10.1093/nar/gkh975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ball N, Streeter SD, Kneale GG, McGeehan JE. Structure of the restriction-modification controller protein C.Esp1396I. Acta Crystallogr. D. Biol. Crystallogr. 2009;65:900–905. doi: 10.1107/S0907444909020514. [DOI] [PubMed] [Google Scholar]
- 206.McGeehan JE, Streeter SD, Thresh SJ, Ball N, Ravelli RB, Kneale GG. Structural analysis of the genetic switch that regulates the expression of restriction-modification genes. Nucleic Acids Res. 2008;36:4778–4787. doi: 10.1093/nar/gkn448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Ball NJ, McGeehan JE, Streeter SD, Thresh SJ, Kneale GG. The structural basis of differential DNA sequence recognition by restriction-modification controller proteins. Nucleic Acids Res. 2012;40:10532–10542. doi: 10.1093/nar/gks718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.McGeehan JE, Ball NJ, Streeter SD, Thresh SJ, Kneale GG. Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex. Nucleic Acids Res. 2012;40:4158–4167. doi: 10.1093/nar/gkr1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Brooks JE, Benner JS, Heiter DF, Silber KR, Sznyter LA, Jager-Quinton T, Moran LS, Slatko BE, Wilson GG, Nwankwo DO. Cloning the BamHI restriction modification system. Nucleic Acids Res. 1989;17:979–997. doi: 10.1093/nar/17.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Moran CP, Jr, Lang N, LeGrice SF, Lee G, Stephens M, Sonenshein AL, Pero J, Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 1982;186:339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- 211.Nakayama Y, Kobayashi I. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl Acad. Sci. USA. 1998;95:6442–6447. doi: 10.1073/pnas.95.11.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annu. Rev. Genomics Hum. Genet. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 213.Rambach A, Tiollais P. Bacteriophage lambda having EcoRI endonuclease sites only in the nonessential region of the genome. Proc. Natl Acad. Sci. USA. 1974;71:3927–3930. doi: 10.1073/pnas.71.10.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Thomas M, Cameron JR, Davis RW. Viable molecular hybrids of bacteriophage lambda and eukaryotic DNA. Proc. Natl Acad. Sci. USA. 1974;71:4579–4583. doi: 10.1073/pnas.71.11.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kelley WS, Chalmers K, Murray NE. Isolation and characterization of a lambdapolA transducing phage. Proc. Natl Acad. Sci. USA. 1977;74:5632–5636. doi: 10.1073/pnas.74.12.5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Murray NE, Kelley WS. Characterization of lambdapolA transducing phages; effective expression of the E. coli polA gene. Mol. Gen. Genet. 1979;175:77–87. doi: 10.1007/BF00267858. [DOI] [PubMed] [Google Scholar]
- 217.Wilson GG, Murray NE. Molecular cloning of the DNA ligase gene from bacteriophage T4. I. Characterisation of the recombinants. J. Mol. Biol. 1979;132:471–491. doi: 10.1016/0022-2836(79)90270-5. [DOI] [PubMed] [Google Scholar]
- 218.Murray NE, Bruce SA, Murray K. Molecular cloning of the DNA ligase gene from bacteriophage T4. II. Amplification and preparation of the gene product. J. Mol. Biol. 1979;132:493–505. doi: 10.1016/0022-2836(79)90271-7. [DOI] [PubMed] [Google Scholar]
- 219.Midgley CA, Murray NE. T4 polynucleotide kinase; cloning of the gene (pseT) and amplification of its product. EMBO J. 1985;4:2695–2703. doi: 10.1002/j.1460-2075.1985.tb03989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Sutcliffe JG. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978;5:2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sutcliffe JG. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb. Symp. Quant. Biol. 1979;43(Pt.1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- 222.Klein B, Murray K. Phage lambda receptor chromosomes for DNA fragments made with restriction endonuclease I of Bacillus amyloliquefaciens H. J. Mol. Biol. 1979;133:289–294. doi: 10.1016/0022-2836(79)90537-0. [DOI] [PubMed] [Google Scholar]
- 223.Loenen WA, Brammar WJ. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980;10:249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- 224.Karn J, Brenner S, Barnett L, Cesareni G. Novel bacteriophage lambda cloning vector. Proc. Natl Acad. Sci. USA. 1980;77:5172–5176. doi: 10.1073/pnas.77.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Maniatis T, Hardison RC, Lacy E, Lauer J, O'Connell C, Quon D, Sim GK, Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 226.Maniatis T, Fritsch A, Sambrook J. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 227.Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. [Google Scholar]
- 228.Stahl FW, Crasemann JM, Stahl MM. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating rec-mediated recombination. J. Mol. Biol. 1975;94:203–212. doi: 10.1016/0022-2836(75)90078-9. [DOI] [PubMed] [Google Scholar]
- 229.Moir A, Brammar WJ. The use of specialised transducing phages in the amplification of enzyme production. Mol. Gen. Genet. 1976;149:87–99. doi: 10.1007/BF00275963. [DOI] [PubMed] [Google Scholar]
- 230.Hohn B, Murray K. Packaging recombinant DNA molecules into bacteriophage particles in vitro. Proc. Natl Acad. Sci. USA. 1977;74:3259–3263. doi: 10.1073/pnas.74.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sternberg N, Tiemeier D, Enquist L. In vitro packaging of a lambda Dam vector containing EcoRI DNA fragments of Escherichia coli and phage P1. Gene. 1977;1:255–280. doi: 10.1016/0378-1119(77)90049-x. [DOI] [PubMed] [Google Scholar]
- 232.Collins J, Hohn B. Cosmids: a type of plasmid gene-cloning vector that is packageable in vitro in bacteriophage lambda heads. Proc. Natl Acad. Sci. USA. 1978;75:4242–4246. doi: 10.1073/pnas.75.9.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Muller-Hill B, Kania J. Lac repressor can be fused to beta-galactosidase. Nature. 1974;249:561–563. doi: 10.1038/249561a0. [DOI] [PubMed] [Google Scholar]
- 234.Kalnins A, Otto K, Ruther U, Muller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2:593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Messing J, Crea R, Seeburg PH. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981;9:309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Messing J, Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982;19:269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- 237.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 238.Roberts TM, Kacich R, Ptashne M. A general method for maximizing the expression of a cloned gene. Proc. Natl Acad. Sci. USA. 1979;76:760–764. doi: 10.1073/pnas.76.2.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Guarente L, Roberts TM, Ptashne M. A technique for expressing eukaryotic genes in bacteria. Science. 1980;209:1428–1430. doi: 10.1126/science.6158095. [DOI] [PubMed] [Google Scholar]
- 240.Guarente L, Lauer G, Roberts TM, Ptashne M. Improved methods for maximizing expression of a cloned gene: a bacterium that synthesizes rabbit beta-globin. Cell. 1980;20:543–553. doi: 10.1016/0092-8674(80)90640-6. [DOI] [PubMed] [Google Scholar]
- 241.O'Farrell P, Polisky B, Gelfand DH. Regulated expression by readthrough translation from a plasmid-encoded beta-galactosidase. J. Bacteriol. 1978;134:645–654. doi: 10.1128/jb.134.2.645-654.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Casadaban MJ, Chou J, Cohen SN. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J. Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Fried L, Lassak J, Jung K. A comprehensive toolbox for the rapid construction of lacZ fusion reporters. J. Microbiol. Methods. 2012;91:537–543. doi: 10.1016/j.mimet.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 244.Linn T, Ralling G. A versatile multiple- and single-copy vector system for the in vitro construction of transcriptional fusions to lacZ. Plasmid. 1985;14:134–142. doi: 10.1016/0147-619x(85)90073-3. [DOI] [PubMed] [Google Scholar]
- 245.Linn T, St PR. Improved vector system for constructing transcriptional fusions that ensures independent translation of lacZ. J. Bacteriol. 1990;172:1077–1084. doi: 10.1128/jb.172.2.1077-1084.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Pourcel C, Marchal C, Louise A, Fritsch A, Tiollais P. Bacteriophage lambda-E. coli K12 vector-host system for gene cloning and expression under lactose promoter control: I. DNA fragment insertion at the lacZ EcoRI restriction site. Mol. Gen. Genet. 1979;170:161–169. doi: 10.1007/BF00337792. [DOI] [PubMed] [Google Scholar]
- 247.Uhlich GA, Chen CY. A cloning vector for creation of Escherichia coli lacZ translational fusions and generation of linear template for chromosomal integration. Plasmid. 2012;67:259–263. doi: 10.1016/j.plasmid.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 248.Windle BE. Phage lambda and plasmid expression vectors with multiple cloning sites and lacZ alpha-complementation. Gene. 1986;45:95–99. doi: 10.1016/0378-1119(86)90136-8. [DOI] [PubMed] [Google Scholar]
- 249.Yu D, Court DL. A new system to place single copies of genes, sites and lacZ fusions on the Escherichia coli chromosome. Gene. 1998;223:77–81. doi: 10.1016/s0378-1119(98)00163-2. [DOI] [PubMed] [Google Scholar]
- 250.Huynh TV, Young RA, Davis RW. Constructing and screening cDNA libraries in lambda gt10 and lambda gt11. In: Glover DM, editor. DNA Cloning. Vol. 1. Oxford, U.K.: IRL Press; 1985. pp. 49–78. [Google Scholar]
- 251.Ruther U, Koenen M, Sippel AE, Muller-Hill B. Exon cloning: immunoenzymatic identification of exons of the chicken lysozyme gene. Proc. Natl Acad. Sci. USA. 1982;79:6852–6855. doi: 10.1073/pnas.79.22.6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Cupples CG, Miller JH. Effects of amino acid substitutions at the active site in Escherichia coli beta-galactosidase. Genetics. 1988;120:637–644. doi: 10.1093/genetics/120.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Cupples CG, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl Acad. Sci. USA. 1989;86:5345–5349. doi: 10.1073/pnas.86.14.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Cupples CG, Cabrera M, Cruz C, Miller JH. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gossen JA, de Leeuw WJ, Vijg J. LacZ transgenic mouse models: their application in genetic toxicology. Mutat. Res. 1994;307:451–459. doi: 10.1016/0027-5107(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 256.Shwed PS, Crosthwait J, Douglas GR, Seligy VL. Characterisation of MutaMouse lambdagt10-lacZ transgene: evidence for in vivo rearrangements. Mutagenesis. 2010;25:609–616. doi: 10.1093/mutage/geq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Grodzicker T, Williams J, Sharp P, Sambrook J. Physical mapping of temperature-sensitive mutations of adenoviruses. Cold Spring Harb. Symp. Quant. Biol. 1975;39(Pt 1):439–446. doi: 10.1101/sqb.1974.039.01.056. [DOI] [PubMed] [Google Scholar]
- 258.Hutchison CA, III, Newbold JE, Potter SS, Edgell MH. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974;251:536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- 259.Potter SS, Newbold JE, Hutchison CA, III, Edgell MH. Specific cleavage analysis of mammalian mitochondrial DNA. Proc. Natl Acad. Sci. USA. 1975;72:4496–4500. doi: 10.1073/pnas.72.11.4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Heller R, Arnheim N. Structure and organization of the highly repeated and interspersed 1.3 kb EcoRI-Bg1II sequence family in mice. Nucleic Acids Res. 1980;8:5031–5042. doi: 10.1093/nar/8.21.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Jeffreys AJ. DNA sequence variants in the G gamma-, A gamma-, delta- and beta-globin genes of man. Cell. 1979;18:1–10. doi: 10.1016/0092-8674(79)90348-9. [DOI] [PubMed] [Google Scholar]
- 262.Jeffreys AJ, Craig IW, Francke U. Localisation of the G gamma-, A gamma-, delta- and beta-globin genes on the short arm of human chromosome 11. Nature. 1979;281:606–608. doi: 10.1038/281606a0. [DOI] [PubMed] [Google Scholar]
- 263.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc. Natl Acad. Sci. USA. 1978;75:5631–5635. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kan YW, Dozy AM. Antenatal diagnosis of sickle-cell anaemia by D.N.A. analysis of amniotic-fluid cells. Lancet. 1978;2:910–912. doi: 10.1016/s0140-6736(78)91629-x. [DOI] [PubMed] [Google Scholar]
- 265.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 266.Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, Watkins PC, Ottina K, Wallace MR, Sakaguchi AY, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- 267.Wexler NS. Huntington's disease: advocacy driving science. Annu. Rev. Med. 2012;63:1–22. doi: 10.1146/annurev-med-050710-134457. [DOI] [PubMed] [Google Scholar]
- 268.Gill P, Jeffreys AJ, Werrett DJ. Forensic application of DNA ‘fingerprints'. Nature. 1985;318:577–579. doi: 10.1038/318577a0. [DOI] [PubMed] [Google Scholar]
- 269.Lindsay JA. Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 2010;300:98–103. doi: 10.1016/j.ijmm.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 270.Otto M. MRSA virulence and spread. Cell Microbiol. 2012;14:1513–1521. doi: 10.1111/j.1462-5822.2012.01832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Arber W. What is the function of restriction enzymes? Trends Biochem. Sci. 1977;2:N176–N178. [Google Scholar]
- 272.Ishikawa K, Handa N, Kobayashi I. Cleavage of a model DNA replication fork by a Type I restriction endonuclease. Nucleic Acids Res. 2009;37:3531–3544. doi: 10.1093/nar/gkp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ishikawa K, Handa N, Sears L, Raleigh EA, Kobayashi I. Cleavage of a model DNA replication fork by a methyl-specific endonuclease. Nucleic Acids Res. 2011;39:5489–5498. doi: 10.1093/nar/gkr153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.McNeil EM, Melton DW. DNA repair endonuclease ERCC1-XPF as a novel therapeutic target to overcome chemoresistance in cancer therapy. Nucleic Acids Res. 2012;40:9990–10004. doi: 10.1093/nar/gks818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.