Abstract
Nuclear receptors are transcription factors that are activated by physiological stimuli to bind DNA in the context of chromatin and regulate complex biological pathways. Major advances in nuclear receptor biology have been aided by genome scale examinations of receptor interactions with chromatin. In this review, we summarize the roles of the chromatin landscape in regulating nuclear receptor function. Chromatin acts as a central integrator in the nuclear receptor-signaling axis, operating in distinct temporal modalities. Chromatin effects nuclear receptor action by specifying its genomic localization and interactions with regulatory elements. On receptor binding, changes in chromatin operate as an effector of receptor signaling to modulate transcriptional events. Chromatin is therefore an integral component of the pathways that guide nuclear receptor action in cell-type-specific and cell state-dependent manners.
The DNA of eukaryotic genomes is complexed with histones to form higher-order chromatin structures, the basic unit of which is the nucleosome, which consists of ∼146 bp of DNA wrapped around a histone octamer (1). Once considered a mechanism to compact DNA into a small nuclear space, evidence over the past 25 years has emphasized the importance of chromatin as a regulatory feature in diverse biological processes including DNA replication, DNA repair, cell division, and transcription (2–5). DNA-binding factors, such as insulators, activating or repressing transcription factors (TF), including nuclear receptors, interact with and modify chromatin to regulate target gene expression. Chromatin structure can be modified through a multitude of mechanisms, including chemical modifications to DNA or histones, substitution of canonical histones for histone variants in nucleosomes, and repositioning of nucleosomes along regulatory DNA (6).
Nuclear receptors are a highly evolutionarily conserved class of TF that act as molecular sensors of physiological and environmental stimuli. In response to diverse signals that include steroids, metabolic intermediates, and chemical toxins, nuclear receptors regulate reproductive, metabolic, and circadian pathways and contribute to pathological states in inflammation, obesity, and cancer. Nuclear receptors such as the androgen, estrogen, glucocorticoid, retinoic acid, and peroxisome proliferator-activated receptors regulate transcription of target genes by binding to specific DNA recognition sequences and mediate responses to external cues. Classically, nuclear receptors have been shown to bind to specific DNA sequences and recruit cofactors that modify the local chromatin structure, which in turn modulate the recruitment and activity of RNA polymerases to repress or enhance transcription (7, 8).
The mechanisms that govern the selective activity of nuclear receptors in moderating cell- and signal-specific physiological programs have long been unclear. However, the emergence of genome scale technologies have helped uncover new mechanisms that regulate nuclear receptor binding and function, shedding additional light on their roles in health and disease. Here, we review nuclear receptor biology from a global perspective, highlighting the role of chromatin in regulating selective nuclear receptor binding and the role of nuclear receptor cofactors in regulating DNA methylation, histone posttranslational modifications, and chromatin remodeling.
Chromatin: The Regulator
Chromosomes are organized as a continuum of condensation states (9–12). These varying states of chromatin compaction in turn modulate the accessibility of the underlying DNA to TF. Generally, condensed heterochromatin is known to regulate the silencing of retroviral elements, restrict gene transcription, suppress chromosomal recombination, and stabilize centromeres and telomeres (13–15). In contrast, euchromatin is relatively decondensed and rich in gene coding regions and actively transcribed elements (15, 16). Within these broad euchromatic and heterochromatic domains exist discrete regions of highly decondensed chromatin associated with active regulatory elements, such as promoters, enhancers, silencers, insulators, and locus control regions (17–20). Numerous activities, specifically those of chromatin remodeling and modifying proteins, regulate both broad and discrete patterns of chromatin accessibility (21–24).
Nucleosomal histones, H2A, H2B, H3, and H4, undergo posttranslational modifications of residues predominantly in the N-terminal tails (25). These diverse modifications, including acetylation, methylation, phosphorylation, and ubiquitination, program activating or silencing signals depending on the specific chemical modification and histone residue (15, 26, 27). Genome-scale analyses have greatly contributed to uncovering correlations between gene activity and histone modifications. Acetylation is generally associated with active regulatory elements and includes the modification of lysines primarily on the tails of H3 and H4 (27, 28). Although histone lysine acetylation generally confers chromatin decondensation features and promotes transcription by permitting DNA-binding factors and the transcriptional machinery to gain access to regulatory elements in chromatin, histone methylation is associated with both activation and repression, depending on the residue modified and the number of methyl groups incorporated. Mono-, di-, and trimethylation of lysine 4 on H3 (H3K4me1, H3K4me2, and H3K4me3, respectively) are constitutive marks associated with active enhancers and promoters (16, 26). In contrast, trimethylation of H3 lysine 27 (H3K27me3) and di- and trimethylation of H3 lysine 9 (H3K9me2 and H3K9me3, respectively) are associated with silencing and heterochromatin (26, 29). Methylation also regulates silencing through cytosine methylation on DNA predominantly at CpG dinucleotides in mammalian cells. DNA methylation epigenetically promotes silencing of gene expression as has been observed in genomic imprinting, in heterochromatin formation at retroviral elements, and at the inactive X chromosome (30–32).
Chromatin: The Regulated
Steroid nuclear receptors are selectively expressed in distinct tissue types and regulate genes in a highly cell-specific manner. These receptors have the potential to interact with hundreds of thousands of the target cognate motif in mammalian genomes. However, genome-wide chromatin immunoprecipitation (ChIP) experiments demonstrate that steroid receptors show a much more restricted binding profile (33–37). A large fraction of nuclear receptor DNA binding sequences, therefore, remains unoccupied. Furthermore, the binding pattern of receptors is cell-specific, with pronounced differences in the distribution of binding events across the genome of different cell types (33, 38). In this context, chromatin accessibility has emerged as a critical regulator of cell-selective nuclear receptor occupancy.
Hormone activation of glucocorticoid receptor (GR) has been long known to be able to induce remodeling of chromatin de novo by recruiting chromatin remodeling complexes such as Swi/Snf (39) (Figure 1C) in a hormone-dependent manner. Recent whole-genome analysis using DNase-Seq has shown that in contrast to this accepted view, de novo induction of chromatin accessibility is not the predominant feature of GR activation (33). Rather, most GR binding events occur at accessible regulatory elements that preexist in the basal state (33). GR binding in mouse mammary epithelial, pituitary cells, human lung epithelial cells, and mouse liver was found to be globally associated with baseline accessible chromatin (ie, accessible in the unstimulated state) (33, 40–42). These studies have, therefore, revealed two distinct classes of GR interactions with chromatin: the first and predominant mechanism involves binding at accessible chromatin regions in the unstimulated cell state, whereas the second class is mediated via the “classic” mechanism of receptor-dependent remodeling of chromatin. In mammary cells, >70% of GR binding is associated with preexisting open chromatin, whereas ∼20% of GR binding was observed at hormone-induced remodeled chromatin (33). Such TF-chromain interaction classes have also been validated with other nuclear receptors (43).
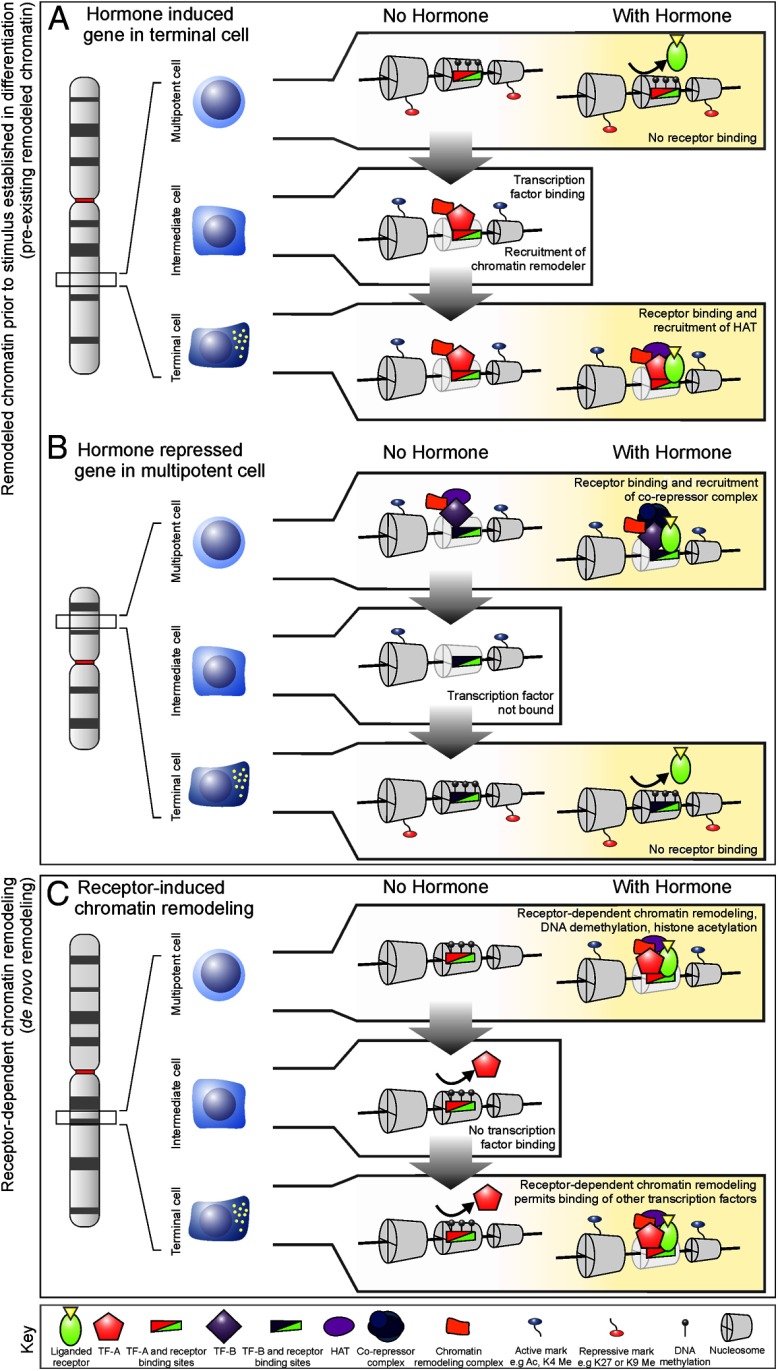
Figure 1.
Chromatin remodeling regulates nuclear receptor-mediated interactions with DNA across different cell types during differentiation. Cellular identity changes during differentiation concordant with changes in genes expression, chromatin structure, and expression of lineage-determining transcription. Chromatin structure is illustrated for three genomic loci (A, B, and C) across three stages of cellular differentiation. A and B represent regulatory elements within chromatin, which are dynamically remodeled during differentiation, mediated by cell-specific transcription factors (TF-A in A and TF-B in B). The occupancy of cell-specific factors creates a permissive chromatin environment for nuclear receptor binding by establishing a preexisting accessible chromatin domain for inducible factors such as nuclear receptors. These preexisting regulatory elements can drive differential outcomes in receptor-dependent gene transcription dependent on the cofactors involved. Genes regulated by a preexisting regulatory element at one locus can therefore be induced by hormone (A), while a separate locus is repressed by hormone (B). In C, nuclear receptors can remodel chromatin de novo without requiring preestablished open chromatin through the recruitment of cofactors including chromatin remodeling complexes. Nuclear receptors can therefore act on chromatin in two distinct ways: at preexisting accessible chromatin (A and B) and through receptor-dependent remodeling de novo (C). The majority (approximately 70%) of nuclear receptor interacts interact through preexisting open chromatin, while approximately 20% operate through de novo remodeling. A, Differentiation is marked by expression of TF-A, a lineage-determining factor, which changes chromatin structure at select regulatory elements. In the absence of TF-A expression, the regulatory element is silenced and precludes receptor binding. The expression and binding of TF-A to chromatin permits receptor interactions with DNA and subsequent recruitment of coactivators such as p300. B, The expression of TF-B maintains the multipotency state by activating select regulatory elements. The accessibility of chromatin at these elements in multipotent cells permits binding of receptors, which represses the regulatory element by recruitment of a corepressor complex. On differentiation, expression of TF-B is lost, silencing the regulatory elements and occluding receptor binding in the differentiated cell state. C, Receptor-dependent chromatin remodeling induces chromatin accessibility and DNA demethylation, activates gene expression, and facilitates the secondary binding of TF (TF-A, which is expressed during differentiation). In the absence of receptor binding, other transcription factors are excluded from interacting with the regulatory element.
Genome-scale profiling of androgen receptor (AR) binding in prostate cancer cells and estrogen receptor (ER) binding in breast cancer cells by ChIP with microarrays or sequencing has shown a substantial association of nuclear receptor binding with chromatin enriched for the active H3K4me2 mark at enhancers (36, 44). In macrophages and adipocytes, peroxisome proliferator-activated receptor γ (PPARγ) binding is strongly associated with H3K9ac (34, 38). The binding of ER is also coincident with open chromatin, demonstrating the same bimodal classes as GR, with ∼60%-70% of ER binding associated with preexisting open chromatin, whereas ∼10%-20% of ER binding sites at induced accessible chromatin (43). The mechanisms that promote discrete regions of open chromatin, however, are not fully clear and may extend beyond histone modifications to include other chromatin features such as histone variants (12, 45). Taken together, binding of PPARγ, AR, ER, and GR is dependent on preexisting regions of highly accessible and modified chromatin. The establishment of open chromatin in the unstimulated cell state is likely determined by the presence of multiprotein complexes that include TF, cofactors, and chromatin remodeling complexes that collectively modify chromatin accessibility (40, 46).
The open chromatin landscape in unstimulated cells demonstrates degrees of accessibility that vary across the genome, with certain regulatory elements displaying greater accessibility such as promoters, than that of enhancer elements when assayed by nuclease digestion (23, 33). This suggests that there is a continuum of accessibility that may be dictated by the presence of DNA-binding factors (46) but also by the presence of neighboring nucleosomes (23). Although nuclear receptors preferentially bind to regulatory elements that are preestablished in their chromatin accessibility, these regions show further enhancement in accessibility following receptor binding (33, 47). The mechanisms that contribute to the potentiation of the open chromatin signal on receptor binding to preexisting open chromatin are unclear. The progesterone receptor has been shown to bind at regulatory elements marked by preexisting open chromatin using nuclease digestion but paradoxically occupied by nucleosomes that undergo H1 and H2A/H2B dimer eviction following progesterone receptor binding (48).
In vitro, ER binding to histones marked by H3K4me3 can undergo selective acetylation of H3K9, which may contribute to enhanced chromatin accessibility on ER binding (49). As with GR, most ER binding in breast cancer cells occurs at accessible chromatin, with a subset showing additional accessibility on ER binding. However, for ER the evidence suggests that this increased accessibility is not associated with hormone-dependent nucleosome depletion (50). In contrast, the binding of AR to regulatory elements enhances global chromatin accessibility by modifying H3K4me2, potentially through the eviction of nucleosomes or destabilization of DNA-nucleosome contacts (44). Chromatin remodeling by nuclear receptors such as GR and AR can also be associated with depletion of nucleosomes containing H2A.Z, a histone variant thought to mark labile nucleosomes (40, 44, 45). Furthermore, the binding of nuclear receptors induces increased histone acetylation mediated by the enzymatic activity of receptor recruited histone acetyltransferases (HAT), such as p300 and CREB binding protein (CBP) (51, 52) (Figure 1A). DNA methylation, previously thought to be a stable epigenetic mark, can undergo hormone-dependent demethylation on ER and GR binding, demonstrating that these epigenetic marks are dynamic and amenable to regulator control (53–55).
Chromatin accessibility is, therefore, mediated by a multitude of mechanisms including demethylation of DNA, modifications of histones such as acetylation, and exchange of canonical histones for histone variants mediated by chromatin remodeling proteins (Figure 1, A and B). The regulation of these chromatin features is highly cell-specific and help delineate cell-type-restricted regulatory elements established during lineage determination (15, 29, 56, 57). Across the genome, chromatin acts as a regulator of selective nuclear receptor interactions with DNA to drive specific transcriptional programs. Understanding the role of TF and cofactors that regulate chromatin structure is crucial to identify novel pathways, biomarkers, and drug targets in nuclear receptor-dependent diseases such as inflammation, obesity, and cancer.
A Genomic View of Nuclear Receptor Cofactors
Our current understanding of nuclear receptor biology is being increasingly shaped by our ability to examine nuclear receptor action in a global manner using genome-wide approaches. Classically, nuclear receptors have been shown to interact with other TF, chromatin remodeling complexes, coactivators, and corepressors. Hundreds of nuclear receptor interacting proteins have been identified to date (58, 59). Indeed, the interaction of nuclear receptors with specific cofactors has been shown to control the outcome of nuclear receptor regulation, that is, regulating the induction or repression of the transcription of target genes (49, 52, 60). Genome-scale studies have also helped explain the selective binding of nuclear receptor cofactors in transcriptional regulation, differentiation, and specification of cell fate and identity (33, 61, 62). The mechanisms by which nuclear receptor cofactors regulate nuclear receptor function and integrate stimuli with biological outputs frequently converge on structural modifications of chromatin at promoters and enhancers.
Coactivator Recruitment and Histone Modifications
Histone acetylation and methylation
Nuclear receptor associated coactivators are important biological players and act as potent regulators of metabolic and hormonal stress responses (63–67). The recruitment of HAT, such as p300, CBP, and steroid receptor coactivator 1, by nuclear receptors through direct protein-protein interactions represents a general mechanism of inducing gene expression (52, 68, 69). These enzymatic activities have been shown to acetylate histones at regulatory elements and promoters of target genes in both hormone-dependent and -independent manners (51, 70, 71) (Figure 1A). Although a given nuclear receptor can have hundreds of target genes and bind to thousands of regulatory elements, the use of a single coactivator is not universal. Global transcription studies of the AR signaling pathway show that approximately 50% of androgen-regulated genes are dependent on the p300 HAT, whereas less than 1% of target genes use CBP (72). The regulation of gene transcription via cis-regulatory elements likely requires selective cofactor use to determine transcriptional outcomes of nuclear receptor activation.
In addition to HAT, nuclear receptors interact with and recruit histone methyltransferases. Steroid receptors recruit myeloid/lymphoid or mixed-lineage leukemia 1 (MLL1), which is associated with the active H3K4 methylation mark, and coactivator-associated arginine methyltransferase 1 (CARM1), which dimethylates H3 arginine residues, a mark also linked with active transcription (73, 74). Other methyltransferases have been shown to play dual roles in nuclear receptor action, for example, G9a, the H3K9 methyltransferase, is recruited by nuclear receptors to regulate both transcriptional activation and repression (75, 76). Genes silenced by the H3K9 or H3K27 methylation marks can be reactivated by nuclear receptors through the recruitment of demethylases, such as the H3K9 demethylase JHDM2A in androgen signaling (77) and the H3K27 demethylase JMJD3 in estrogen signaling (78). Interestingly, the roles of HAT and methyltransferases have been shown to converge. HAT activity has been shown to be required for CARM1 activity (73) and the methylation of CBP by CARM1 has been shown to mediate genome-wide CBP recruitment to chromatin by the ER (79). In addition, G9a has been found to act cooperatively with CARM1 and p300 in AR and ER function (75). Histone methyltransferases such as MLL1 are recruited by ER to modify chromatin and subsequently promote acetylation of H2A lysine 5 by the acetyltransferase TIP60 (80).
The mechanisms that govern the selective recruitment and action of coactivators to nuclear receptor-bound regulatory elements remain unclear. Phosphorylation of nuclear receptors is thought to modify interactions with cofactors and could underlie restricted recruitment of coactivators (73, 81, 82). The presence of distinct histone modifications could also provide signals to specific readers recruited with nuclear receptors. ER binding at actively marked H3K4me3 chromatin recruits distinct coregulators than ER binding sites associated with the repressive H3K9me3 (49). Selectivity could also be reflective of the underlying properties of individual regulatory elements, such as the presence of secondary TF, spatial positioning of regulatory elements, or stochastic interactions resulting from limited coactivator and corepressor molecules (83–85).
The DNA sequence of individual regulatory elements might also act to specify the recruitment of cofactors. Genome-scale analysis of nuclear receptor binding has shown that although there is a consensus binding motif for each nuclear receptor, the thousands of occurrences of the hormone response element harbor numerous DNA sequence variants. These variant hormone response elements have been shown to greatly influence the interactions of nuclear receptors with DNA by altering protein surfaces that coactivators interact with, thereby promoting variable cofactors interactions in a DNA sequence-dependent manner (86, 87). The DNA sequence of regulatory elements might therefore act to define the transcriptional outcome of nuclear receptor binding (88), suggesting that single-nucleotide polymorphisms could influence nuclear receptor binding to DNA and the cofactors that it interacts with (89–91).
Corepressor complexes
The recruitment of corepressors is a primary mechanism through which transcriptional repression by nuclear receptors is mediated. Multiple corepressor proteins are known to interact with nuclear receptors, including histone deacetylases (HDACs) and nuclear receptor corepressor/silencing mediator for retinoid or thyroid hormone receptors (NCoR/SMRT). HDACs and NCoR/SMRT are thought to form a corepressor complex that down-regulates transcription by promoting histone deacetylation and chromatin compaction (60, 92). GR has been shown to recruit corepressor complexes at DNA sequences associated with negative regulation (88) and at sites of transrepression, where GR acts in a DNA-independent manner, by tethering to other DNA binding proteins and recruiting histone deacetylases (93).
Nuclear receptors are known to play critical roles in the maintenance of circadian patterns. Some nuclear receptors in their unliganded basal state stabilize corepressor complexes on chromatin to silence target genes. For example, Rev-erbα, a regulator of circadian clocks, recruits NCoR/SMRT to repress Bmal1 transcription and modulate circadian rhythms (94). Genome analysis of HDAC3 occupancy inversely correlates with histone acetylation and displays a circadian pattern of occupancy similar to that of Rev-erbα (95), suggesting that nuclear receptor-associated corepressors play critical roles in the maintenance of the molecular circadian clock. Nuclear receptors also mediate transrepression through mechanisms distinct from the recruitment of the HDAC-NCoR/SMRT corepressor complex. Serine and threonine residues of proteins can be modified with O-linked N-acetylglucosamine (O-GlcNAc) monosaccharides by O-GlcNAc transferases (OGT). GR-mediated transrepression of nuclear factor κB (NF-κB) has been shown, in some instances, to recruit OGT in a hormone-dependent fashion, which in turn blocks Pol II elongation (96). OGT has also been shown to mediate repression of other TF through mSin3a (97). Paradoxically, OGT-dependent O-GlcNAc modification occurs on histone H2B at serine 112, which has been linked with active transcription (98). The role of OGT in transcription regulation and chromatin in regulating GR action remains intriguing, as GR is known to regulate glucose metabolism in liver while the active H2B S112 GlcNAcylation mark is thought to act as a glucose sensor. How these pathways converge on transactivation or transrepression could play an important role in our understanding of diabetes and associated metabolic syndromes.
DNA methylation is a well-studied epigenetic mechanism to silence genes. The global role of active DNA demethylation in nuclear receptor action is unclear. However, the binding of receptors to regulatory elements at open chromatin is coincident with hypomethylated DNA (43, 53). Recent evidence has shown that nuclear receptors can dynamically regulate DNA methylation status at enhancer and promoter elements (53–55). Although DNA methyltransferases (DNMTs) are known to be part of corepressor complexes (99), DNA demethylation on ER activation was found to be dependent on DNMTs themselves, suggesting a dual role for DNMTs in ER-dependent gene regulation (54). Nuclear receptors, therefore, use multiple mechanisms to mediate transcriptional repression and the regulation of major pathways in inflammation, metabolism, and circadian rhythms.
The Many Faces of Chromatin Remodelers
The large family of chromatin remodelers, which includes Swi/Snf, ISWI, INO80, and CHD subfamilies, modifies nucleosome-DNA contacts and regulates chromatin accessibility through mechanisms involving nucleosome sliding, nucleosome or histone eviction, and histone exchange accompanied by incorporation or eviction of histone variants (100). The activity of chromatin remodeling complexes is integral to numerous biological processes, including maintenance of pluripotency, cellular differentiation of lymphocytes and neurons, inflammation, DNA damage and repair, and tumor suppression. Frequent mutations occurring in the subunits of these remodeling complexes have been observed in lung, hepatocellular, ovarian, and bladder transitional cell carcinomas (101–109).
Chromatin remodelers are recruited to regulatory elements by nuclear receptors (39). Brg1, the ATPase subunit of the Swi/Snf chromatin remodeling complex, is required for the maintenance of chromatin accessibility at a subset of regulatory elements bound by GR (40, 47). The induction and repression of transcription by GR require the ATPase activity of Brg1 at a significant proportion of induced genes and a smaller fraction of repressed genes (40). The role of chromatin remodelers in silencing is not unique to nuclear receptors. SMARCAD1, a Swi/Snf-like remodeling complex, is required for reestablishing histone deacetylation and H3K9 methylation at silenced chromatin following DNA replication (110). In embryonic stem cells, Swi/Snf acts with PRC2 to silence the Hox locus to maintain stem cells identity (111). In differentiated cells, Swi/Snf is recruited to silencing elements by factors such as REST (repressor element 1-silencing transcription factor), which restricts the expression of neuronal genes in nonneuronal cells (112, 113).
The mechanisms through which chromatin remodelers interact with nucleosomes, however, are currently unclear. Proposed models suggest that chromatin remodelers directly interact with nucleosomes through modified histone residues (100, 114, 115). Alternatively, TF such as GR and ER could actively recruit remodelers to DNA in a conditional manner, or “priming” proteins (see Transcription Factors Define Chromatin Landscapes for Nuclear Receptor Binding) could stabilize the constitutive interaction of remodelers at regulatory elements (39, 40, 116). In nuclear receptor-mediated induction and repression, chromatin remodelers might, therefore, act to maintain accessible chromatin for inducible DNA-binding factors, whereby the activation or repression of transcription is mediated through the recruitment of activating or repressing cofactors that specify transcriptional outcomes. All these models implicate a critical role for chromatin remodeling complexes in the DNA binding of nuclear receptors. Loss of chromatin remodeling activity, by mutational inactivation in cancers, has been shown to reorganize the binding landscape of nuclear receptors, resulting in the silencing or activation of disease-related genes (108).
Nuclear Receptors Interact With Diverse Transcription Factors
As stimulus-inducible factors, nuclear receptors are known to interact directly or indirectly with constitutively bound TF and other inducible TF at regulatory elements (58, 84). These interactions regulate local chromatin structure and modulate transcriptional responses. Interactions between TF are a general principle of gene regulatory networks that extend beyond nuclear receptors and represent a general property of the chromatin environment in eukaryotic cells (117–122). For nuclear receptors, these interactions influence a broad range of biological and pathological functions, from circadian regulation, metabolism, and differentiation to inflammation and cancer.
Steroid hormone receptors, such as GR, AR, and ER, can homodimerize and bind to their cognate response elements and interact with TF bound near modulate gene transcription through additive, synergistic, or antagonistic interactions (123–125). Alternatively, these steroid receptors can bind as monomers and interact indirectly with TF bound to regulatory elements by tethering to DNA-bound TF (126). Nuclear receptors have been shown to interact with a single transcription factor through both modalities. For example, GR has been shown to interact with the proinflammatory factors AP1, NF-kB, and members of the signal transducer and activator of transcription (STAT) family (84, 126–128). Similarly, ER has also been shown to interact with proinflammatory factors such as AP1 in an analogous manner as GR (129). The current model supported by genome-wide data suggests that composite binding of nuclear receptors with proinflammatory factors can result in gene activation or repression, whereas the principal effect of nuclear receptor tethering is thought to be that of transcriptional down-regulation (transrepression) (84, 126–128, 130, 131). Other nuclear receptors such as peroxisome proliferator-activated receptor, liver X receptor, and retinoic acid receptor require the retinoid X receptor as a heterodimeric partner to form active transcription complexes (35, 132, 133). At regulatory elements, these nuclear receptor heterodimers interact with other DNA-bound TF to regulate gene expression. PPARγ, for example, serves as a potent regulator of inflammatory responses in lymphocytes through interactions with STAT6 in macrophages and dendritic cells (134).
Transcription Factors Define Chromatin Landscapes for Nuclear Receptor Binding
GR is capable of remodeling chromatin de novo on hormone activation and this in turn assists in the binding of other TF such as AP1 and ER (84, 135, 136). Surprisingly, most GR interactions are not de novo remodeling events but rather chromatin that is preprogrammed (ie, accessible in the basal cell state before GR activation). This suggests that other factors act to open and maintain a permissive chromatin state (33). Open chromatin, as measured by DNase I digestion of regulatory elements, is reflected by the cumulative and combinatorial binding of TF in basal, unstimulated cells (23). Transcription factors that act on chromatin in unstimulated cells could therefore contribute to determining the selective recruitment of nuclear receptors to regulatory elements. The occupancy of AP1 at regulatory elements has been shown to prime an accessible chromatin state and thereby dictates the genomic location of GR binding (84). These observations have been recapitulated in vivo using mouse liver tissue, where the CCAAT/enhancer-binding protein-β has been shown to prime chromatin for GR binding on hormone activation (42). Similarly, FOXA1, which has been shown to interact with ER and AR in breast and prostate cancer cells, respectively, acts to maintain chromatin in an active and permissive state for nuclear receptor binding (36, 137, 138). Indeed, the expression of FOXA1 in primary breast cancers reprograms ER binding. This genome-wide redistribution of ER binding has been shown to strongly correlate with poor clinical outcomes (139). Collectively, these results suggest that nonnuclear receptor TF act to prime discrete chromatin domains for stimulus-induced factors (Figure 1, A and B). It is, therefore, likely that the chromatin landscape is shaped by the action of TF that is established during differentiation (Figure 1A) or during various cellular and disease states to provide context-specific transcriptional responses to external signals.
The association of nuclear receptors with chromatin shows significant plasticity that is conditional to specific cellular states and external cues. The specific activating signal of nuclear receptors can alter its genomic binding landscape by changing its preference for interacting proteins. In breast cancer cells, estradiol-activated ER preferentially binds to regulatory elements harboring constitutively bound FOXA1; however, epidermal growth factor–activated ER shows a distinct genomic binding profile with preference for the AP1 family of factors (140). The activation of GR during signal-induced NF-kB binding has also been shown to remodel the distribution of GR at a number of regulatory sites (141). The expression and activation of TF, such as NF-kB and AP1, can therefore dynamically reorganize nuclear receptor binding to regulatory elements in a conditional and signal-specific manner. Similarly, in pituitary cells, GR and STAT3 have been shown to extensively interact with chromatin when both factors are activated by their stimuli, with the interactions occurring at regulatory elements preset as open chromatin in unstimulated cells (128). Interestingly, although GR and STAT3 cobound regulatory elements are marked by active histone modifications, the transcriptional outcome could be either that of gene repression or transcriptional synergism (128). Therefore, the open chromatin landscape in the basal cell state can lead to diverse transcriptional outcomes, suggesting that the binding of specific TF rather than the preexisting open chromatin dictates the transcriptional outcome (see Figure 1, A and B).
During differentiation, dynamic changes in chromatin structure occur through the expression and activation of lineage-determining factors (117, 122, 142–145). Nuclear receptors, such as the adipogenic regulator PPARγ, are known to play important roles in differentiation programs. During adipogenesis, PPARγ binding is associated with CCAAT/enhancer-binding protein-β and -δ, GR, and STAT5a through multiple stages of chromatin remodeling to establish adipocyte-specific active regulatory elements (34, 146, 147). In macrophages, however, PPARγ associates with the PU.1 factor and binds to distinct regulatory elements when compared with adipocytes. This suggests that the presence of PU.1 in macrophages (and not adipocytes) helps specify the activity of PPARγ (38). In macrophages and B lymphocytes, the PU.1 factor also regulates the binding of liver X receptor to chromatin (61). Lineage-determining factors are, therefore, critical in establishing the chromatin landscape that specifies nuclear receptor binding (Figure 1A). Through the priming of chromatin, cell-specific DNA-binding factors can orchestrate the recruitment of inducible factors, either through direct protein-protein interactions or indirectly through the maintenance of accessible chromatin.
Conclusions
Nuclear receptors act as dynamic sensors of external signals and mediate rapid regulatory responses in a chromatin context (148). However, upstream of receptor signaling, chromatin accessibility of baseline cell states act to specify the recruitment of receptors to regulatory elements. Chromatin accessibility at nuclear receptor binding sites is regulated by priming TF, coactivators, corepressors, and chromatin remodeling and by modifying enzymes. Although preexisting sites of open chromatin are frequently regulatory elements to which nuclear receptors are recruited, the induced binding of receptors confers further changes to chromatin accessibility through further remodeling of the underlying chromatin and associated nucleosomes, potentially through acetylation of specific histone residues or eviction of histones mediated by recruitment of cofactors (48, 49).
Changes in the chromatin landscape can inevitably alter the occupancy of receptors in acute and chronic disease states, such as inflammation, diabetes, and cancer. In disease states, the activation, silencing, or overexpression of nuclear receptor cofactors could promote altered or discordant accessibility patterns. This, in turn, could remodel the receptor binding landscape with consequential effects on transcription and disease outcomes (71, 139, 149). Illustrating this, mutation of the transcription factor FOXA1 has been shown to alter ER and AR signaling in breast and prostate cancers (150). Mutations of subunits of the Swi/Snf chromatin remodeling complex in liver, ovarian, and lung cancers and malignant rhabdoid tumors are implicated in aberrations of receptor interactions with chromatin (107, 108, 151, 152). Indeed, ligand-activated TF are known to directly interact with subunits of chromatin remodeling complexes (39, 153–157). In addition, mutations of coactivators and corepressors such as MLL and NCoR in breast cancers, p300 in B cell lymphomas, MLL, and DNMT3a in acute myeloid leukemia are thought to influence nuclear receptor binding and alter transcriptional responses to external stimuli (158, 159). Mutations of coregulators have been shown to be strongly associated with syndromic diseases. CBP and p300 mutations are linked with Rubenstein-Taybi syndrome, ARID1B of the Swi/Snf chromatin remodeler with Coffin-Siris syndrome, and SMARCA2 subunit of Swi/Snf with Nicolaides-Baraitser syndrome (160–164).
The emergence of genome-scale methods to study nuclear receptor function have provided general rules for receptor signaling mechanisms, validated previous models of receptor action, and expanded the repertoire of mechanisms that govern their actions on chromatin. In addition to sequencing technologies, other high-throughput technologies such as proteomics continue to expand the coregulator interactome, identifying hundreds of novel receptor-associated cofactors (165). Through whole genome studies, the contributions of cofactors have been shown to be of particular importance in nuclear receptor signaling by regulating the selective recruitment of receptors, influencing receptor binding to chromatin, and controlling transcriptional outputs. Although many cofactors are known to associate with nuclear receptors through direct or indirect interactions, the critical role of chromatin accessibility in the basal cell state as a determinant of selective receptor recruitment suggests that factors that regulate chromatin structure at regulatory elements act to facilitate receptor binding as a cofactor without a mandatory requirement of directly interacting with nuclear receptors (46). Understanding the role of cofactors and mechanisms regulating nuclear receptor actions on chromatin and transcription could provide novel druggable pathways, as epigenetic and chromatin regulators have emerged as promising targets in disease treatment (166–168).
Acknowledgments
The authors would like to thank Andre Nussenzweig, Stephanie A. Morris, Tina Miranda, Cathy Smith, and Michael R. Stallcup for helpful comments and critical reading of the manuscript. We also thank the reviewers for their helpful comments.
This research was supported, in part, by the Intramural Research Program of the National Institutes of Health.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AR
- androgen receptor
- CARM1
- coactivator-associated arginine methyltransferase 1
- CBP
- CREB binding protein
- ChIP
- chromatin immunoprecipitation
- DNMT
- DNA methyltransferases
- ER
- estrogen receptor
- GR
- glucocorticoid receptor
- HAT
- histone acetyltransferases
- HDACs
- histone deacetylases
- MLL1
- mixed-lineage leukemia 1
- NCoR/SMRT
- nuclear receptor corepressor/silencing mediator for retinoid or thyroid hormone receptors
- NF-κB
- nuclear factor κB
- OGT
- O-GlcNAc transferases
- O-GlcNAc
- O-linked N-acetylglucosamine
- PPARγ
- peroxisome proliferator-activated receptor γ
- STAT
- signal transducer and activator of transcription
- TF
- transcription factors.
References
- 1. Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389(6648):251–260 [DOI] [PubMed] [Google Scholar]
- 2. Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72(1):73–84 [DOI] [PubMed] [Google Scholar]
- 3. Miller KM, Tjeertes JV, Coates J, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu M, Long C, Chen X, Huang C, Chen S, Zhu B. Partitioning of histone H3–H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328(5974):94–98 [DOI] [PubMed] [Google Scholar]
- 5. Tachiwana H, Kagawa W, Shiga T, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476(7359):232–235 [DOI] [PubMed] [Google Scholar]
- 6. Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov. 2012;1(5):384–400 [DOI] [PubMed] [Google Scholar]
- 7. Scheidereit C, Geisse S, Westphal HM, Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983;304(5928):749–752 [DOI] [PubMed] [Google Scholar]
- 8. Zaret KS, Yamamoto KR. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38(1):29–38 [DOI] [PubMed] [Google Scholar]
- 9. Wu C, Bingham PM, Livak KJ, Holmgren R, Elgin SC. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979;16(4):797–806 [DOI] [PubMed] [Google Scholar]
- 10. Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306(5701):1571–1573 [DOI] [PubMed] [Google Scholar]
- 11. Teif VB, Vainshtein Y, Caudron-Herger M, et al. Genome-wide nucleosome positioning during embryonic stem cell development. Nat Struct Mol Biol. 2012;19(11):1185–1192 [DOI] [PubMed] [Google Scholar]
- 12. Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474(7352):516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301(5634):798–802 [DOI] [PubMed] [Google Scholar]
- 14. Peters AH, Kubicek S, Mechtler K, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589 [DOI] [PubMed] [Google Scholar]
- 15. Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39(3):311–318 [DOI] [PubMed] [Google Scholar]
- 17. Bell O, Schwaiger M, Oakeley EJ, et al. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol. 2010;17(7):894–900 [DOI] [PubMed] [Google Scholar]
- 18. Payvar F, DeFranco D, Firestone GL, et al. Sequence-specific binding of glucocorticoid receptor to MTV DNA at sites within and upstream of the transcribed region. Cell. 1983;35(2 Pt 1):381–392 [DOI] [PubMed] [Google Scholar]
- 19. Jongstra J, Reudelhuber TL, Oudet P, et al. Induction of altered chromatin structures by simian virus 40 enhancer and promoter elements. Nature. 1984;307(5953):708–714 [DOI] [PubMed] [Google Scholar]
- 20. Lyko F, Brenton JD, Surani MA, Paro R. An imprinting element from the mouse H19 locus functions as a silencer in Drosophila. Nat Genet. 1997;16(2):171–173 [DOI] [PubMed] [Google Scholar]
- 21. Boyle AP, Davis S, Shulha HP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sabo PJ, Kuehn MS, Thurman R, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3(7):511–518 [DOI] [PubMed] [Google Scholar]
- 23. Thurman RE, Rynes E, Humbert R, et al. The accessible chromatin landscape of the human genome. Nature. 2012;488(7414):75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grontved L, Bandle R, John S, et al. Rapid genome-scale mapping of chromatin accessibility in tissue. Epigenetics Chromatin. 2012;5(1):10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705 [DOI] [PubMed] [Google Scholar]
- 26. Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837 [DOI] [PubMed] [Google Scholar]
- 27. Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kan PY, Lu X, Hansen JC, Hayes JJ. The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol Cell Biol. 2007;27(6):2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y, van Essen D, Saccani S. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol Cell. 2012;46(4):408–423 [DOI] [PubMed] [Google Scholar]
- 30. Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–191 [DOI] [PubMed] [Google Scholar]
- 31. Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93(3):309–312 [DOI] [PubMed] [Google Scholar]
- 32. Sado T, Hoki Y, Sasaki H. Tsix silences Xist through modification of chromatin structure. Dev Cell. 2005;9(1):159–165 [DOI] [PubMed] [Google Scholar]
- 33. John S, Sabo PJ, Thurman RE, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43(3):264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lefterova MI, Zhang Y, Steger DJ, et al. PPARγ and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22(21):2941–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nielsen R, Pedersen TA, Hagenbeek D, et al. Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22(21):2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lupien M, Eeckhoute J, Meyer CA, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132(6):958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reddy TE, Pauli F, Sprouse RO, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19(12):2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lefterova MI, Steger DJ, Zhuo D, et al. Cell-specific determinants of peroxisome proliferator-activated receptor γ function in adipocytes and macrophages. Mol Cell Biol. 2010;30(9):2078–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393(6680):88–91 [DOI] [PubMed] [Google Scholar]
- 40. John S, Sabo PJ, Johnson TA, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29(5):611–624 [DOI] [PubMed] [Google Scholar]
- 41. Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32(18):3756–3767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Grøntved L, John S, Baek S, et al. C/EBP maintains chromatin accessibility in liver and facilitates glucocorticoid receptor recruitment to steroid response elements. EMBO J. 2013;32(11):1568–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gertz J, Savic D, Varley KE, et al. Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol Cell. 2013;52(1):25–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He HH, Meyer CA, Shin H, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42(4):343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jin C, Zang C, Wei G, et al. H3.3/H2A.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet. 2009;41(8):941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neph S, Vierstra J, Stergachis AB, et al. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;488(7414):83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burd CJ, Ward JM, Crusselle-Davis VJ, et al. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol Cell Biol. 2012;32(10):1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ballaré C, Castellano G, Gaveglia L, et al. Nucleosome-driven transcription factor binding and gene regulation. Mol Cell. 2013;49(1):67–79 [DOI] [PubMed] [Google Scholar]
- 49. Foulds CE, Feng Q, Ding C, et al. Proteomic analysis of coregulators bound to ERα on DNA and nucleosomes reveals coregulator dynamics. Mol Cell. 2013;51(2):185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome Res. 2012;22(6):1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Louie MC, Yang HQ, Ma AH, et al. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc Natl Acad Sci USA. 2003;100(5):2226–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chakravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383(6595):99–103 [DOI] [PubMed] [Google Scholar]
- 53. Wiench M, John S, Baek S, et al. DNA methylation status predicts cell type-specific enhancer activity. EMBO J. 2011;30(15):3028–3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Métivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45–50 [DOI] [PubMed] [Google Scholar]
- 55. Kangaspeska S, Stride B, Métivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452(7183):112–115 [DOI] [PubMed] [Google Scholar]
- 56. Shen Y, Yue F, McCleary DF, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Norris JD, Chang CY, Wittmann BM, et al. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol Cell. 2009;36(3):405–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. York B, O'Malley BW. Steroid receptor coactivator (SRC) family: masters of systems biology. J Biol Chem. 2010;285(50):38743–38750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nagy L, Kao HY, Chakravarti D, et al. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89(3):373–380 [DOI] [PubMed] [Google Scholar]
- 61. Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lupien M, Eeckhoute J, Meyer CA, et al. Coactivator function defines the active estrogen receptor α cistrome. Mol Cell Biol. 2009;29(12):3413–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Louet JF, Chopra AR, Sagen JV, et al. The coactivator SRC-1 is an essential coordinator of hepatic glucose production. Cell Metab. 2010;12(6):606–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Winnay JN, Xu J, O'Malley BW, Hammer GD. Steroid receptor coactivator-1-deficient mice exhibit altered hypothalamic-pituitary-adrenal axis function. Endocrinology. 2006;147(3):1322–1332 [DOI] [PubMed] [Google Scholar]
- 65. Han SJ, Hawkins SM, Begum K, et al. A new isoform of steroid receptor coactivator-1 is crucial for pathogenic progression of endometriosis. Nat Med. 2012;18(7):1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anzick SL, Kononen J, Walker RL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968 [DOI] [PubMed] [Google Scholar]
- 67. Liu Z, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Sequential recruitment of steroid receptor coactivator-1 (SRC-1) and p300 enhances progesterone receptor-dependent initiation and reinitiation of transcription from chromatin. Proc Natl Acad Sci USA. 2001;98(22):12426–12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736 [DOI] [PubMed] [Google Scholar]
- 69. Feng W, Ribeiro RC, Wagner RL, et al. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280(5370):1747–1749 [DOI] [PubMed] [Google Scholar]
- 70. Conway-Campbell BL, George CL, Pooley JR, et al. The HSP90 molecular chaperone cycle regulates cyclical transcriptional dynamics of the glucocorticoid receptor and its coregulatory molecules CBP/p300 during ultradian ligand treatment. Mol Endocrinol. 2011;25(6):944–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zwart W, Theodorou V, Kok M, Canisius S, Linn S, Carroll JS. Oestrogen receptor-co-factor-chromatin specificity in the transcriptional regulation of breast cancer. EMBO J. 2011;30(23):4764–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ianculescu I, Wu DY, Siegmund KD, Stallcup MR. Selective roles for cAMP response element-binding protein binding protein and p300 protein as coregulators for androgen-regulated gene expression in advanced prostate cancer cells. J Biol Chem. 2012;287(6):4000–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen D, Ma H, Hong H, et al. Regulation of transcription by a protein methyltransferase. Science. 1999;284(5423):2174–2177 [DOI] [PubMed] [Google Scholar]
- 74. Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. Methylation at arginine 17 of histone H3 is linked to gene activation. EMBO Rep. 2002;3(1):39–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J Biol Chem. 2006;281(13):8476–8485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bittencourt D, Wu DY, Jeong KW, et al. G9a functions as a molecular scaffold for assembly of transcriptional coactivators on a subset of glucocorticoid receptor target genes. Proc Natl Acad Sci USA. 2012;109(48):19673–19678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125(3):483–495 [DOI] [PubMed] [Google Scholar]
- 78. Svotelis A, Bianco S, Madore J, et al. H3K27 demethylation by JMJD3 at a poised enhancer of anti-apoptotic gene BCL2 determines ER-α ligand dependency. EMBO J. 2011;30(19):3947–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ceschin DG, Walia M, Wenk SS, et al. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 2011;25(11):1132–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jeong KW, Kim K, Situ AJ, Ulmer TS, An W, Stallcup MR. Recognition of enhancer element-specific histone methylation by TIP60 in transcriptional activation. Nat Struct Mol Biol. 2011;18(12):1358–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Duplessis TT, Williams CC, Hill SM, Rowan BG. Phosphorylation of estrogen receptor α at serine 118 directs recruitment of promoter complexes and gene-specific transcription. Endocrinology. 2011;152(6):2517–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Galliher-Beckley AJ, Williams JG, Cidlowski JA. Ligand-independent phosphorylation of the glucocorticoid receptor integrates cellular stress pathways with nuclear receptor signaling. Mol Cell Biol. 2011;31(23):4663–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Voss TC, John S, Hager GL. Single-cell analysis of glucocorticoid receptor action reveals that stochastic post-chromatin association mechanisms regulate ligand-specific transcription. Mol Endocrinol. 2006;20(11):2641–2655 [DOI] [PubMed] [Google Scholar]
- 84. Biddie SC, John S, Sabo PJ, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43(1):145–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Phillips-Cremins JE, Sauria ME, Sanyal A, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhang J, Chalmers MJ, Stayrook KR, et al. DNA binding alters coactivator interaction surfaces of the intact VDR-RXR complex. Nat Struct Mol Biol. 2011;18(5):556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324(5925):407–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Surjit M, Ganti KP, Mukherji A, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145(2):224–241 [DOI] [PubMed] [Google Scholar]
- 89. Kasowski M, Grubert F, Heffelfinger C, et al. Variation in transcription factor binding among humans. Science. 2010;328(5975):232–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. McDaniell R, Lee BK, Song L, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328(5975):235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet. 2010;11(2):109–123 [DOI] [PubMed] [Google Scholar]
- 93. Bilodeau S, Vallette-Kasic S, Gauthier Y, et al. Role of Brg1 and HDAC2 in GR trans-repression of the pituitary POMC gene and misexpression in Cushing disease. Genes Dev. 2006;20(20):2871–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Phelan CA, Gampe RT, Jr, Lambert MH, et al. Structure of Rev-erbα bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 2010;17(7):808–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Feng D, Liu T, Sun Z, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Li MD, Ruan HB, Singh JP, et al. O-GlcNAc transferase is involved in glucocorticoid receptor-mediated transrepression. J Biol Chem. 2012;287(16):12904–12912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yang X, Zhang F, Kudlow JE. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell. 2002;110(1):69–80 [DOI] [PubMed] [Google Scholar]
- 98. Fujiki R, Hashiba W, Sekine H, et al. GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature. 2011;480(7378):557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25(3):338–342 [DOI] [PubMed] [Google Scholar]
- 100. Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149(7):1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363(16):1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gui Y, Guo G, Huang Y, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43(9):875–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li M, Zhao H, Zhang X, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43(9):828–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes Dev. 2011;25(3):275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Takeuchi JK, Lou X, Alexander JM, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chia NY, Chan YS, Feng B, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468(7321):316–320 [DOI] [PubMed] [Google Scholar]
- 107. Jones S, Wang TL, Shih IM, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330(6001):228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Romero OA, Setien F, John S, et al. The tumour suppressor and chromatin remodelling factor BRG1 antagonizes Myc activity and promotes cell differentiation in human cancer. EMBO Mol Med. 2012;4(7):603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gaspar-Maia A, Alajem A, Polesso F, et al. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009;460(7257):863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rowbotham SP, Barki L, Neves-Costa A, et al. Maintenance of silent chromatin through replication requires SWI/SNF-like chromatin remodeler SMARCAD1. Mol Cell. 2011;42(3):285–296 [DOI] [PubMed] [Google Scholar]
- 111. Ho L, Miller EL, Ronan JL, Ho WQ, Jothi R, Crabtree GR. esBAF facilitates pluripotency by conditioning the genome for LIF/STAT3 signalling and by regulating polycomb function. Nat Cell Biol. 2011;13(8):903–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Watanabe H, Mizutani T, Haraguchi T, et al. SWI/SNF complex is essential for NRSF-mediated suppression of neuronal genes in human nonsmall cell lung carcinoma cell lines. Oncogene. 2006;25(3):470–479 [DOI] [PubMed] [Google Scholar]
- 113. Battaglioli E, Andrés ME, Rose DW, et al. REST repression of neuronal genes requires components of the hSWI.SNF complex. J Biol Chem. 2002;277(43):41038–41045 [DOI] [PubMed] [Google Scholar]
- 114. Liu CL, Kaplan T, Kim M, Buratowski S, et al. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3(10):e328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39(5):711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hassan AH, Prochasson P, Neely KE, et al. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111(3):369–379 [DOI] [PubMed] [Google Scholar]
- 117. Junion G, Spivakov M, Girardot C, et al. A transcription factor collective defines cardiac cell fate and reflects lineage history. Cell. 2012;148(3):473–486 [DOI] [PubMed] [Google Scholar]
- 118. Mullen AC, Orlando DA, Newman JJ, et al. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell. 2011;147(3):565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Ravasi T, Suzuki H, Cannistraci CV, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140(5):744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462(7269):65–70 [DOI] [PubMed] [Google Scholar]
- 121. Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Neph S, Stergachis AB, Reynolds A, Sandstrom R, Borenstein E, Stamatoyannopoulos JA. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150(6):1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Schüle R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242(4884):1418–1420 [DOI] [PubMed] [Google Scholar]
- 124. Shemshedini L, Knauthe R, Sassone-Corsi P, Pornon A, Gronemeyer H. Cell-specific inhibitory and stimulatory effects of Fos and Jun on transcription activation by nuclear receptors. EMBO J. 1991;10(12):3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Sakai DD, Helms S, Carlstedt-Duke J, Gustafsson JA, Rottman FM, Yamamoto KR. Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev. 1988;2(9):1144–1154 [DOI] [PubMed] [Google Scholar]
- 126. Schüle R, Rangarajan P, Kliewer S, et al. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62(6):1217–1226 [DOI] [PubMed] [Google Scholar]
- 127. Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249(4974):1266–1272 [DOI] [PubMed] [Google Scholar]
- 128. Langlais D, Couture C, Balsalobre A, Drouin J. The Stat3/GR interaction code: predictive value of direct/indirect DNA recruitment for transcription outcome. Mol Cell. 2012;47(1):38–49 [DOI] [PubMed] [Google Scholar]
- 129. Adler S, Waterman ML, He X, Rosenfeld MG. Steroid receptor-mediated inhibition of rat prolactin gene expression does not require the receptor DNA-binding domain. Cell. 1988;52(5):685–695 [DOI] [PubMed] [Google Scholar]
- 130. Reichardt HM, Kaestner KH, Tuckermann J, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93(4):531–541 [DOI] [PubMed] [Google Scholar]
- 131. Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14(18):2314–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-γ-RXR- nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Shen Q, Bai Y, Chang KC, et al. Liver X receptor-retinoid X receptor (LXR-RXR) heterodimer cistrome reveals coordination of LXR and AP1 signaling in keratinocytes. J Biol Chem. 2011;286(16):14554–14563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Szanto A, Balint BL, Nagy ZS, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33(5):699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Voss TC, Schiltz RL, Sung MH, et al. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146(4):544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Miranda TB, Voss TC, Sung MH, et al. Reprogramming the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res. 2013;73:5130–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43(1):27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wang D, Garcia-Bassets I, Benner C, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Ross-Innes CS, Stark R, Teschendorff AE, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481(7381):389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Lupien M, Meyer CA, Bailey ST, et al. Growth factor stimulation induces a distinct ERα cistrome underlying breast cancer endocrine resistance. Genes Dev. 2010;24(19):2219–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Rao NA, McCalman MT, Moulos P, et al. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21(9):1404–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Novershtern N, Subramanian A, Lawton LN, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144(2):296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23(4):796–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Samstein RM, Arvey A, Josefowicz SZ, et al. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell. 2012;151(1):153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Paige SL, Thomas S, Stoick-Cooper CL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151(1):221–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Siersbaek R, Nielsen R, John S, et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 2011;30(8):1459–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Steger DJ, Grant GR, Schupp M, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24(10):1035–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Stavreva DA, Wiench M, John S, et al. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat Cell Biol. 2009;11(9):1093–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Chng KR, Chang CW, Tan SK, et al. A transcriptional repressor co-regulatory network governing androgen response in prostate cancers. EMBO J. 2012;31(12):2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Barbieri CE, Baca SC, Lawrence MS, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44(6):685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Fujimoto A, Totoki Y, Abe T, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44(7):760–764 [DOI] [PubMed] [Google Scholar]
- 152. Jagani Z, Mora-Blanco EL, Sansam CG, et al. Loss of the tumor suppressor Snf5 leads to aberrant activation of the Hedgehog-Gli pathway. Nat Med. 2010;16:1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Nie Z, Xue Y, Yang D, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20(23):8879–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Yan Z. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19(14):1662–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928 [DOI] [PubMed] [Google Scholar]
- 156. Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279(16):16677–16686 [DOI] [PubMed] [Google Scholar]
- 157. Link KA, Burd CJ, Williams E, et al. BAF57 governs androgen receptor action and androgen-dependent proliferation through SWI/SNF. Mol Cell Biol. 2005;25(6):2200–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471(7337):189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Lonard DM, O'Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8(10):598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Petrij F, Giles RH, Dauwerse HG, et al. Rubinstein-Taybi syndrome caused by mutations in the transcriptional co-activator CBP. Nature. 1995;376:348–351 [DOI] [PubMed] [Google Scholar]
- 162. Santen GW, Aten E, Sun Y, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1B cause Coffin-Siris syndrome. Nat Genet. 2012;44(4):379–380 [DOI] [PubMed] [Google Scholar]
- 163. Tsurusaki Y, Okamoto N, Ohashi H, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44(4):376–378 [DOI] [PubMed] [Google Scholar]
- 164. Van Houdt JK, Nowakowska BA, Sousa SB, et al. Heterozygous missense mutations in SMARCA2 cause Nicolaides-Baraitser syndrome. Nat Genet. 2012;44(4):445–449 [DOI] [PubMed] [Google Scholar]
- 165. Malovannaya A, Lanz RB, Jung SY, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145(5):787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]