ABSTRACT
Regulation of organelle transport along microtubules is important for proper distribution of membrane organelles and protein complexes in the cytoplasm. RNAi-mediated knockdown in cultured Drosophila S2 cells demonstrates that two microtubule-binding proteins, a unique isoform of Darkener of apricot (DOA) protein kinase, and its substrate, translational elongation factor EF1γ, negatively regulate transport of several classes of membrane organelles along microtubules. Inhibition of transport by EF1γ requires its phosphorylation by DOA on serine 294. Together, our results indicate a new role for two proteins that have not previously been implicated in regulation of the cytoskeleton. These results further suggest that the biological role of some of the proteins binding to the microtubule track is to regulate cargo transport along these tracks.
KEY WORDS: Darkener of apricot; DOA Translation elongation factor EF1γ, EF1γ; Organelle transport; Drosophila melanogaster S2 cells
INTRODUCTION
Organization of the cytoplasm of eukaryotic cells is defined by molecular motors that move organelles and other cellular cargoes along microtubules and microfilaments to their correct destinations. A large number of motor proteins of the kinesin, dynein and myosin superfamilies have been identified using genetic and cell biological techniques. Many of these motors have been well characterized by biochemical and biophysical methods (Vale, 2003) and found to be tightly regulated in the cell so that cargoes are transported to specific locations.
Microtubules are copolymers of the main structural protein tubulin and multiple associated proteins that are bound to the microtubule lattice. It is thought likely that some microtubule-associated proteins (MAPs) regulate transport along microtubules. Indeed, a number of reports suggests that cargo transport or the activity of microtubule motors can be modulated by major MAPs such as tau, MAP2 or ensconsin (MAP7) (Barlan et al., 2013; Dixit et al., 2008; Reed et al., 2006; Vershinin et al., 2007; Vershinin et al., 2008). In addition, post-translational modifications of tubulin itself, such as acetylation, can either directly or indirectly promote interaction of kinesin with microtubules both in vivo and in vitro (Friedman et al., 2010; Reed et al., 2006), supporting the idea that the state of microtubule tracks can regulate transport.
In our search for pathways that regulate organelle movement along microtubules, we performed a systematic analysis of factors associated with microtubules that can modify intracellular transport. For this analysis we used Drosophila S2 cells because they are highly sensitive to RNAi-mediated protein knockdown. Analysis of microtubule-dependent transport in S2 cells is facilitated by their ability to form long microtubule-filled processes after treatment with actin-depolymerizing drugs (Ling et al., 2004). Microtubules in these processes have uniform polarity (plus-ends out) and transport along microtubules can be studied in this simplified model system without the influence of other cytoskeletal elements. The role of MAPs in transport in Drosophila cells has been strengthened by proteomic analysis that identified over 250 Drosophila proteins binding to microtubules in vitro (Hughes et al., 2008). Many of these proteins are probably bona fide MAPs that are associated with microtubules not only in vitro but also in cells.
To identify potential regulators of cargo transport along microtubules, we conducted a systematic RNAi analysis of the effect of MAP knockdown on movement of organelles along microtubules. Here we report the first results of this analysis. Our data demonstrate that two microtubule-binding proteins, a unique isoform of Darkener of apricot (DOA) protein kinase and its substrate, translational elongation factor-1γ (EF1γ), negatively regulate motor-mediated transport of membrane organelles along microtubules. Furthermore, the effect of DOA on transport is mediated by phosphorylation of EF1γ.
RESULTS
DOA negatively regulates organelle transport along microtubules
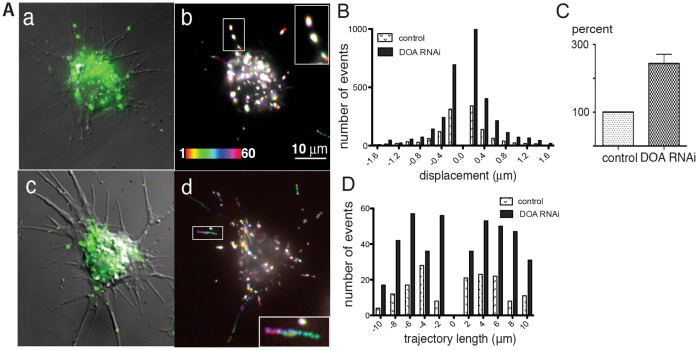
In thus study we used Drosophila S2 cells treated with cytochalasin D (CytoD) as a model system. When plated on a coverslip coated with concanavalin A (ConA), S2 cells form thin processes containing uniformly polarized (plus-ends out) microtubules (Kim et al., 2007a). Furthermore, because CytoD destroys the network of actin filaments, this treatment enables the assessment of microtubule-based transport without interference from other cytoskeletal systems. S2 cells expressing GFP targeted to peroxisomes (GFP-SKL) (Kim et al., 2007a) were incubated for 5 days with RNAi directed against the catalytic domain of DOA, then treated with CytoD and plated on coverslips coated with ConA. Both control cells and cells treated with dsRNA attached to the substrate and grew long unbranched processes filled with microtubules. GFP-tagged peroxisomes moved bidirectionally in the processes (Fig. 1Aa,b; supplementary material Movie 1, left panel). At any given moment, ∼10% of peroxisomes in control cells underwent bidirectional movement. Visual inspection of the RNAi-treated cells indicated that knocking down DOA increased peroxisome motility (Fig. 1Ac,d; supplementary material Movie 1, right panel). Quantitative analysis of peroxisome motility performed with Diatrack software confirmed our visual impression and demonstrated that treatment with RNAi against DOA dramatically affected the movement of peroxisomes by both increasing the number of motile organelles and the distance they travelled without stopping or changing direction. Characteristically, both movement toward the process tips (toward the microtubule plus-ends) and to the cell body were equally stimulated (Fig. 1B–D).
Fig. 1.
DOA knockdown stimulates peroxisome transport in S2 cells. (A) Peroxisomes were visualized by expression of peroxisome-targeted GFP (Kim et al., 2007a) and imaged for 60 seconds at a rate of 1 frame per second. (a,b) Control cell; (c,d), DOA RNAi-treated cell. Panels a and c show superposition of the DIC image and the first frame of the fluorescent stack; panels b and d show a merge of 60 frames from the GFP fluorescence sequence, color-coded according to the bar on the lower left in b). Owing to the superposition of multiple colors, stationary particles appear white, whereas moving particles appear as rainbow-colored paths. Boxed regions are enlarged in the insets. Scale bar: 10 µm. (B–D) Quantification of peroxisome transport. (B) Histogram of vector lengths in control and DOA RNAi-treated cells. (C) Relative number of vectors larger than 0.2 µm (as a percentage of the control). The number of events in each group was normalized by the number of tracked organelles (see Materials and Methods). (D) Length distribution of uninterrupted trajectories (processivity of movement).
Drosophila DOA kinase is expressed as multiple isoforms generated through alternative promoter usage (Fig. 2A). These isoforms have the same C-terminal kinase catalytic domain but different N-termini that have at least three non-redundant functions (Kpebe and Rabinow, 2008). In order to determine which isoform is responsible for modification of microtubule-based transport, we designed isoform-specific dsRNAs (shown as red bars in Fig. 2A) and tested the effect of individual isoform depletion on peroxisome transport. Our results (Fig. 2B) clearly show that organelle transport was stimulated only by RNAi directed against the 227 kDa DOA isoform (DOA227), whereas RNAi against other isoforms had no effect. We were unable to test the effect of the 91 kDa isoform knockdown because the mRNA sequence of this isoform is entirely contained within that of the DOA227.
Fig. 2.
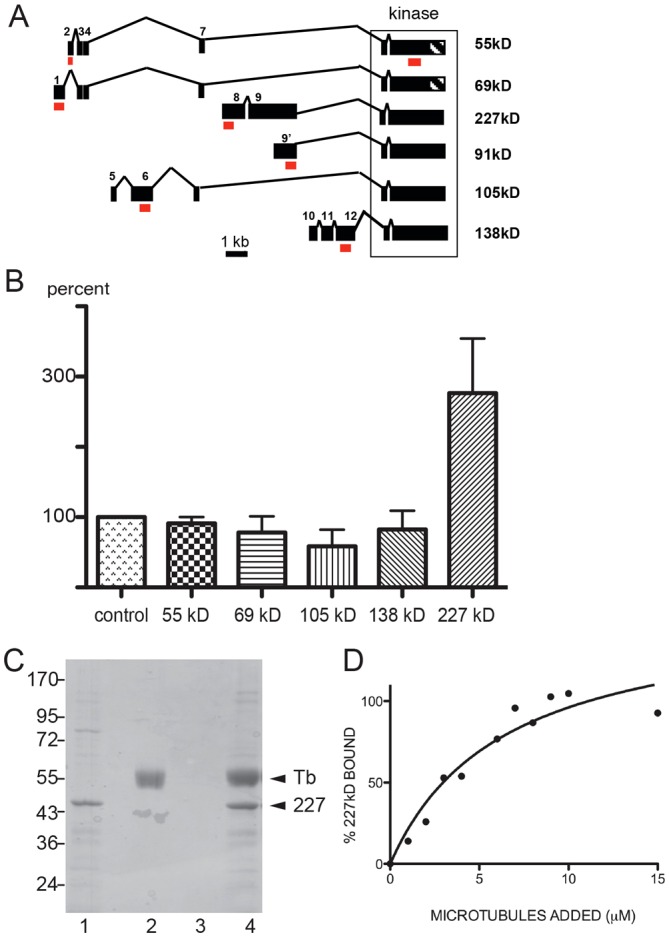
Effect of DOA isoforms on peroxisome transport. (A) Schematic representation of six verified DOA isoforms. As defined by its exons (solid black boxes), the locus spans 34,699 bp. Each protein isoform is named according to the molecular mass of the transcript. Red boxes show target sites for isoform-specific dsRNA. (B) Relative motility of peroxisomes in S2 cells treated with dsRNA targeting specific DOA isoforms (as a percentage of the untreated control). (C) The unique N-terminal fragment of DOA227 encoded by exon 8 (purified as a GST fusion) binds to microtubules. Lane 1, GST–DOA227 fragment; lane 2, tubulin; lane 3, control pellet (GST–DOA227 without microtubules); lane 4, DOA227+microtubule pellet. (D) The binding curve. The percentage of the DOA227 fragment bound to microtubules was determined by densitometry of Coomassie-Blue-stained gels. The curve is a best fit for the data, which corresponds to a single type of binding site with a Kd≈0.6×10−6 M.
Analysis of DOA peptides that were identified in microtubule-binding experiments (Hughes et al., 2008) showed that these peptides are all derived from the DOA227 isoform. To confirm that this isoform indeed can bind to microtubules, we expressed a GST fusion of a portion of the unique N-terminal fragment of DOA227 (residues 915–1197) in Escherichia coli and purified it. Recombinant protein was tested for its ability to interact with microtubules polymerized from bovine brain tubulin. Fig. 2C shows that the bacterially expressed fragment of DOA227 could be pelleted with microtubules through a glycerol cushion. Analysis of binding over a range of microtubule concentrations demonstrated that the peptide bound to microtubules with the affinity of ≈0.6 µM (Fig. 2D). Therefore, the unique N-terminal extension present in DOA227 mediates its binding to microtubules. Interestingly, this peptide fragment contains an amino-acid motif (THVPGGG) identical to one found in mammalian microtubule-binding proteins tau and MAP2.
In order to test whether the effect of DOA227 on transport is restricted to peroxisomes, we analyzed the effects of knockdown on transport of two other organelles, mitochondria and lysosomes. Lysosomes and mitochondria in S2 cells were labeled with the fluorescent dyes, LysoTracker Red and MitoTracker Red, respectively. Both types of organelles move along microtubules in S2 cells in a bidirectional fashion (supplementary material Movies 2 and 3, left panels). Mitochondria, like peroxisomes, are transported along microtubules by conventional kinesin (kinesin-1) and cytoplasmic dynein (Ally et al., 2009), whereas motility of lysosomes is completely abolished by dynein knockdown but is not inhibited by treatment of cells by dsRNA against kinesin-1 heavy chain (Fig. 3C). Depletion of DOA227 stimulates transport of both organelles. Similarly to transport of peroxisomes, this effect was isoform specific; motility of lysosomes and mitochondria was stimulated by depletion of DOA227, but not by any other isoform (Fig. 3A,B). The results of this experiment show that DOA227 regulates bidirectional organelle transport along microtubules by several plus-end motors and by cytoplasmic dynein. Thus, this indicates that DOA227 probably affects microtubule tracks rather than individual motors.
Fig. 3.
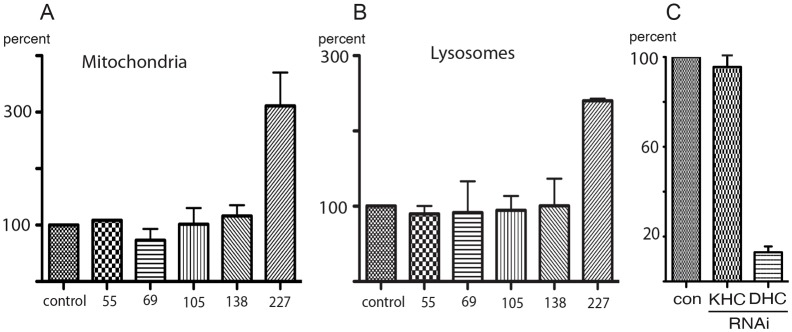
Effect of DOA isoform knockdown on transport of mitochondria and lysosomes. Knockdown of DOA227 but not other DOA isoforms stimulates transport of mitochondria (A) and lysosomes (B) in S2 cells. Bars show the relative numbers of vectors larger than 0.2 µm (as a percentage of the control). (C) Knockdown of kinesin-1 does not affect lysosome transport. Cells were treated with RNAi against kinesin or dynein heavy chains; lysosomes were visualized by staining with LysoTracker Red and imaged for 60 seconds at a rate of 1 frame per second. The relative number of vectors larger than 0.2 µm (as a percentage of the control) is shown.
If DOA227 is a negative regulator of transport, its overexpression should inhibit movement of organelles. We tested this hypothesis using lysosomes, as these organelles move in S2 cells more robustly than peroxisomes and mitochondria, two other membrane organelles used in this study. Full-length cDNA encoding DOA227 was tagged at the N-terminus with GFP and the fusion protein was transiently expressed in S2 cells. Fig. 4A demonstrates that indeed, GFP-DOA227 expression significantly suppressed lysosome transport. To further demonstrate the specificity of the effects of DOA227 on organelle transport, we depleted all endogenous DOA using dsRNA targeting 3′-UTR sequence (that is shared by all isoforms). This treatment, as expected, stimulated lysosome transport, consistent with the knockdown results using the coding sequence (Fig. 1C). However, overexpression of GFP-DOA227 lacking 3′-UTR sequence, and, therefore, resistant to this dsRNA, suppressed motility in the cells treated with dsRNA (Fig. 4B) similarly to cells that had not been treated with dsRNA (Fig. 4A). These overexpression results, together with isoform-specific knockdown data, demonstrated that DOA227 regulates transport of organelles along microtubules, and stimulation of transport by dsRNA treatments are not explained by off-target effects.
Fig. 4.
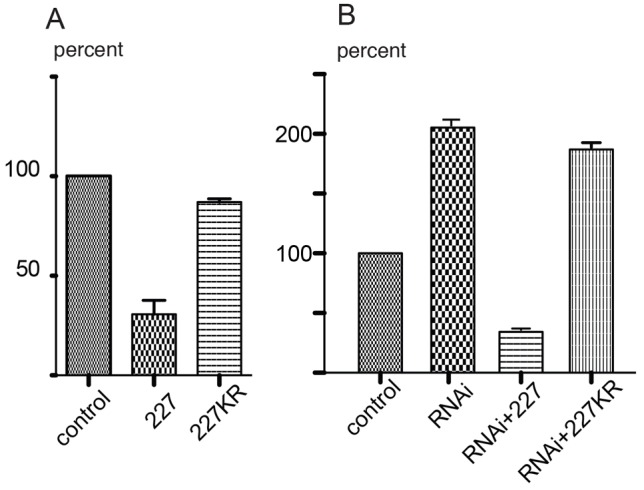
Overexpression of DOA227 but not its kinase-dead mutant inhibits organelle transport. (A) DOA227 (227) or its kinase-dead version (Lys1728Arg; 227KR) were tagged with GFP and overexpressed in S2 cells. Only the catalytically active form inhibits lysosome transport. (B) Effect of DOA knockdown on lysosome motility can be reversed by the wild-type but not kinase-dead DOA227. DOA knockdown was produced using dsRNA against the 3′-UTR region common to all DOA isoforms; overexpression was induced by transfection with cDNA encoding GFP-tagged DOA227 or its catalytically inactive version. Bars in A and B show the relative numbers of vectors larger than 0.2 µm (as a percentage of the control).
DOA227 regulates transport through phosphorylation of microtubule-binding protein EF1γ
One potential mechanism of transport inhibition by DOA227 is through its binding to microtubules and blocking cargo movement by motor proteins. Alternatively, DOA227 could phosphorylate downstream effector(s), which are important for transport, rather than physically blocking organelles moving on microtubules. To distinguish between these two possibilities we compared the effect of overexpression of the wild-type DOA227 and the kinase-dead Lys1728Arg mutant [see Lee et al. (Lee et al., 1996) for characterization of the kinase activity of the recombinant catalytic domain and its Lys to Arg mutant]. Fig. 4A,B shows that the wild-type form of DOA227, but not its catalytically inactive version inhibits movement of lysosomes along microtubules. In addition, overexpression of the kinase-dead mutant of DOA227 does not rescue the effect of DOA RNAi (Fig. 4B), further demonstrating the necessity of kinase activity for transport regulation.
Which downstream transport regulators are phosphorylated by DOA227? DOA kinase interacts with and phosphorylates the γ-subunit of translation elongation factor 1 gamma (EF1γ; also known as eEF1γ) (Fan et al., 2010). Interestingly, similar to DOA227, EF1γ binds to microtubules in vitro (Hughes et al., 2008; Janssen and Möller, 1988). We therefore tested whether EF1γ would also have an effect on microtubule-based organelle transport. Similar to DOA, treatment of S2 cells with dsRNA against EF1γ resulted in stimulation of transport along microtubules of all three classes of membrane organelles, peroxisomes, lysosomes and mitochondria (Fig. 5A, for efficiency of knockdown see Fig. 5B)
Fig. 5.
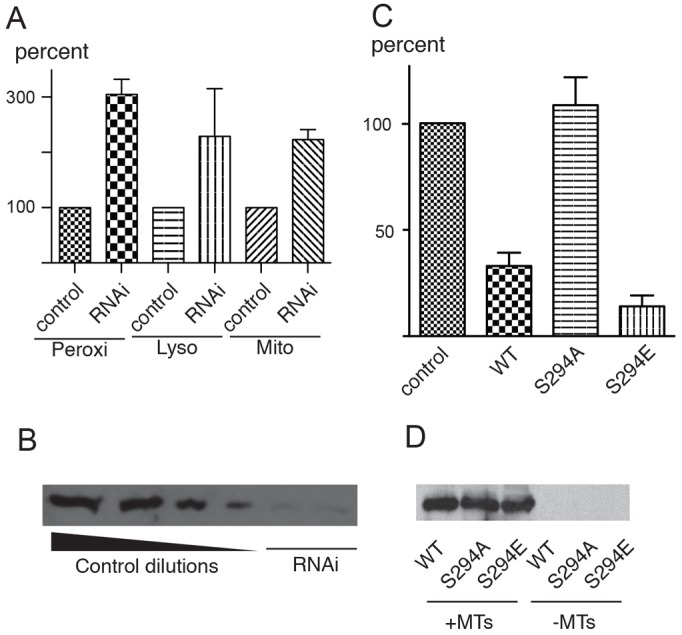
Effects of EF1γ on organelle transport. (A) Depletion of EF1γ by RNAi stimulates transport of peroxisomes, lysosomes and mitochondria. (B) Efficiency of RNAi against EF1-γ. Blot with the anti-EF1γ antibody. Left four lanes: serial twofold dilutions of the control extract; two right lanes: extracts of cells treated with dsRNA directed at the coding sequence (left) and 3′-UTR (right). (C) Overexpression of mCherry–EF1γ or its phosphomimetic mutant S294E inhibits transport whereas the non-phosphorylatable mutant S294A does not affect peroxisome transport. (D) Mutations of S294 do not affect the ability of mCherry-EF1γ to bind microtubules. Extracts were prepared from S2 cell expressing mCherry–EF1γ or its phosphomimetic mutants and centrifuged. The pellets were analyzed by western blotting with anti-mCherry antibody. Bars in A and C show the relative numbers of vectors larger than 0.2 µm (as a percentage of the control).
To further analyze the effects of EF1γ, we tagged it at the C-terminus with mCherry and transiently transfected this construct into S2 cells expressing GFP-labeled peroxisomes. Although depletion of EF1γ stimulated transport, its overexpression dramatically inhibited movement of peroxisomes along microtubules (Fig. 5C).
To confirm that EF1γ binds to microtubules, we performed microtubule-pelleting experiments using S2 cell extracts. Pre-polymerized bovine brain microtubules were added to extracts of cells expressing mCherry-EF1γ and after 30 minutes of incubation, microtubules were pelleted through a 30% glycerol cushion to separate them from soluble proteins. The microtubule-bound fraction was analyzed using western blotting with an antibody against mCherry. The results of this experiment confirmed the published data (Hughes et al., 2008; Janssen and Möller, 1988) and show that EF1γ is indeed a microtubule-binding protein (Fig. 5D).
The Ser294 LAMMER kinase phosphorylation site of Drosophila EF1γ is conserved among EF1γ homologs in all eukaryotes. We therefore investigated whether the phosphorylation state of EF1γ had an effect on either microtubule binding or on its ability to modify microtubule-mediated organelle transport. Constructs expressing an mCherry-tagged non-phosphorylatable form of the protein (S294A) and a phosphomimetic form (S294E), were generated and used to express recombinant protein in S2 cells for microtubule-pelleting experiments. Similar to the wild-type protein, both S294A and S294E were found in microtubule-containing pellets (only in the presence of microtubules), demonstrating that the phosphorylation state of Ser294 did not affect its affinity for microtubules (Fig. 5D). In contrast, the effect of EF1γ on microtubule-based transport was dramatically affected by these amino-acid substitutions. When expressed in S2 cells, the S294A mutant of EF1γ failed to inhibit transport of peroxisomes, whereas the S294E mutant had a strong inhibitory effect (Fig. 5C). Therefore, activity of EF1γ as a regulator of organelle transport requires its phosphorylation on Ser294, consistent with the inefficient rescue of EF1γ Drosophila mutant lethality by expression of EF1γ-S294A (Fan et al., 2010).
We next sought to establish whether DOA227 regulates organelle transport upstream of EF1γ by phosphorylating it on Ser294. We therefore combined overexpression of EF1γ, or its phosphorylation-site mutant EF1γ-Ser294E, with depletion of DOA227. We argued that if phosphorylation of EF1γ by DOA227 is required for its inhibitory activity, overexpression of wild-type EF1γ in the absence of DOA227 would have no effect on transport. On the other hand, overexpression of the Ser294E mutant mimicking the phosphorylated state of EF1γ would affect transport independently of DOA227. The results (Fig. 6) demonstrated that indeed inhibition of transport by the phosphomimetic mutant of EF1γ does not require DOA227, whereas wild-type EF1γ inhibits transport only when DOA227 is present in the cells. These results strongly argue for the necessity of a kinase–substrate relationship between DOA and EF1γ in the regulation of microtubule-based organelle transport.
Fig. 6.
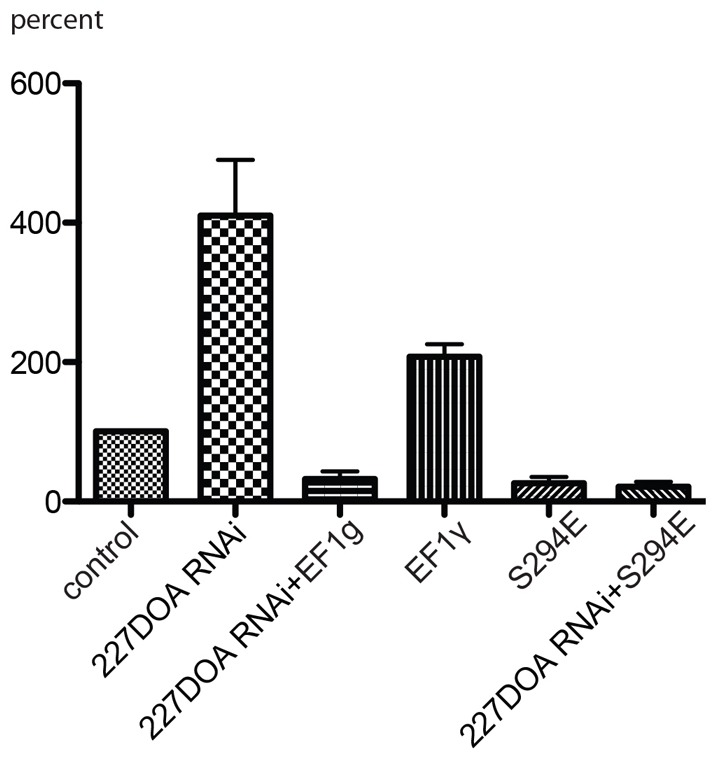
EF1γ inhibits organelle transport downstream of DOA227. Depletion of DOA227 abolishes the inhibitory effect of mCherry-tagged EF1γ on peroxisome transport but does not affect inhibition by the phosphomimetic mutant S294E. Bars show the relative numbers of vectors larger than 0.2 µm (as a percentage of the control).
DISCUSSION
The data presented in this study show that two microtubule-associated proteins, DOA227 protein kinase and EF1γ negatively regulate transport of organelles along microtubules. The effects of DOA227 requires its catalytic activity and therefore cannot be explained simply by its binding to microtubules. Analysis of a phosphomimetic mutant of EF1γ demonstrates that inhibition of organelle transport requires its phosphorylation on conserved Ser294 and that DOA227 acts upstream of EF1γ by phosphorylating this amino acid residue.
Regulation of organelle transport is a new function for both DOA and EF1γ proteins. Both proteins have numerous other roles in processes unrelated to cytoskeleton function and intracellular transport. LAMMER/CLK kinases, such as DOA, are found ubiquitously among eukaryotes and classed as eukaryotic ‘founder’ proteins (Hartman and Fedorov, 2002). Although best known for their role in the phosphorylation of SR splicing factors and the ensuing effects on alternative splicing of pre-mRNAs (Colwill et al., 1996; Du et al., 1998; Nikolakaki et al., 2002), the presence of a LAMMER kinase ortholog in S. cerevisiae (Padmanabha et al., 1991) demonstrates that the ancestral function of these kinases is not linked to the regulation of alternative splicing, since there is none described in this species. Characterization of the Drosophila LAMMER kinase, DOA, showed that multiple isoforms function in at least three different processes (Kpebe and Rabinow, 2008), including alternative splicing. Other data demonstrate a role for DOA kinase in the activation of Hedgehog signaling, through the direct phosphorylation of the GLI protein CI (Hurtado, R. R., Du, C., Sledd, M., Holmgren, R. A. and Rabinow, L., personal communication). Here we have described the regulation of organelle transport along microtubules as a novel function specifically attributable to the microtubule-binding isoform DOA227. Interestingly, RNAi knockdown of general DOA activity reduces protein secretion in S2 cells (Bard et al., 2006), an effect that may be related to the role of DOA in regulation of transport along microtubules.
In addition to finding that a specific DOA isoform regulates microtubule-based organelle transport, we discovered that EF1γ, a previously described DOA-interacting protein and microtubule-binding protein (Fan et al., 2010) possesses the same properties as DOA in the regulation of transport. Moreover, we showed that EF1γ activity requires its phosphorylation by DOA on a LAMMER kinase phosphorylation site that is conserved among EF1γ orthologs in all eukaryotes; this strongly suggests that this function of EF1γ is conserved throughout Eukaryota.
Although it was initially thought to stimulate the nucleotide exchange activity on EF1 (Fan et al., 2010), direct evidence of EF1γ function in translation is largely lacking, because it is dispensable for translation in vitro. The identification of the protein as an ‘elongation factor’ is largely due to the fact that it co-isolates with EF1γ. Several alternative roles for the protein have been previously suggested (Ejiri, 2002). For example, Artemia salina EF1γ binds to tubulin (Janssen and Möller, 1988). Drosophila EF1γ (CG11901) was identified in a screen for microtubule-associated proteins (Hughes et al., 2008), and it is also colocalized with microtubules in Drosophila embryos (Fan et al., 2010). EF1γ also interacts with keratin (Kim et al., 2007b). Interactions with nucleic acids are also included in the repertoire of EF1γ functions: it specifically binds to the 3′-UTR of vimentin mRNA (Al-Maghrebi et al., 2002), and also binds its promoter region, contacting RNA Pol II (Corbi et al., 2010). Interestingly, the latter report also describes mislocalization of mitochondria and vimentin protein upon EF1γ depletion, consistent with our findings of a role for the protein in the regulation of microtubule-based transport. EF1γ was also identified as participating in the pre-mRNA 3′-end cleavage complex (Shi et al., 2009).
At present we do not know the molecular link between the EF1γ–DOA227 pathway and the organelle-transporting machinery. One possibility is that this link is a motor-binding protein Unc-76 (and its mammalian ortholog fasciculation and elongation protein zeta-1, FEZ1). Yeast two-hybrid data demonstrate that human FEZ1 interacts with EF1γ (Ishii et al., 2001), and FEZ1 can modulate organelle transport by directly interacting with motors (Blasius et al., 2007; Gindhart et al., 2003). If EF1γ interaction with Unc-76 depends on EF1γ phosphorylation it would explain how DOA227 can affect organelle trafficking.
In conclusion, our results demonstrate a role for EF1γ and its kinase DOA as regulators of organelle transport along microtubules. The fact that EF1γ (and its Ser294 phosphorylation site) are evolutionarily conserved suggests that this mechanism is likely to operate not only in Drosophila but also in higher eukaryotes.
MATERIALS AND METHODS
Cell culture
Drosophila S2 cells were maintained in serum-free Insect-Xpress medium (Lonza, Walkersville, MD) in a humidified incubator at 28°C. Cell transfection was performed using Effectene Transfection Reagent (Qiagen, Germantown, MD) according to the manufacturer's protocol. For RNAi, cells were plated in a 12-well plate (106 cells/well). To deplete a protein of interest, cells were treated with 15–20 µg dsRNA per well twice during a 5-day period. Efficiency of RNAi was checked by western blotting. For imaging, cells from a 12-well plate were plated onto ConA-coated coverslips in the presence of CytoD.
dsRNA synthesis
dsRNA was generated by in vitro transcription reactions. In a typical 100 µl reaction 5 µg of T7-appended PCR product was mixed with transcription buffer [80 mM HEPES, pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT)], 5 mM rNTP mix, 0.75 units of pyrophosphatase, 150 units of RNasin (Promega, Madison, WI) and 25 µl purified T7 polymerase (100 µg/ml). The transcription mixture was incubated at 37°C for 2 hours. dsRNA was precipitated with lithium chloride and iso-propanol. The pellet was washed once with 75% ethanol and resuspended in diethylpyrocarbonate (DEPC)-treated water.
For amplification of the in vitro transcription templates the PCR primers were appended with the T7 promoter sequences. Sequences of primers are listed below. DOA catalytic domain: 5′-AGGGAACTTTTGGACGTGTG-3′ and 5′-TTCTCCGGCTTGAGATCTGT-3′; 55 kDa isoform of DOA: 5′-CACAACACTGTCTCCCAGAC-3′ and 5′-CTTGCAATTTGCTGCTGATA-3′; 69 kDa isoform of DOA: 5′-CATCACTAACCAAGTGCAAT-3′ and 5′-CTCTTTCTTCGCGCGGCGCT-3′; 91+227 kDa isoform of DOA: 5′-TGTTGCACCACCCACCTGCC-3′ and 5′-TGCTGTTCCTGATGTTGCTG-3′; 105 kDa isoform of DOA: 5′-TCGACTTCCTCCATCCATCA-3′ and 5′-GCAGCGGTGGCTGCGGTGC-3′; 138 kDa isoform of DOA: 5′-CGGATAGAGTGGCTCTAATG-3′ and 5′-ATCTCTTCGCTGAGGAATC-3′; 227 kDa isoform of DOA: 5′-ATGTCCAACGAACTTAGCGA-3′ and 5′-ATGTCCAACGAACTTAGCGA-3′; 3′-UTR for all isoforms of DOA: 5′-ACTCATGCAAACATACACTC-3′ and 5′-TGACAGGTGTAGACATGGGG-3′; EF1γ: 5′-ACAACGAGATTGTGCCTGC-3′ and 5′-AACTCGGCGTACTTCTTGG-3′.
Cloning
EF1γ was digested with EcoRI–NotI and mCherry with NotI–XbaI and they were then cloned into pMT/V5 His-A (Invitrogen, Grand Island, NY) to produce pMT-EF1γ-mCherry. Ser294A and Ser294E mutations were introduced into the EF1γ construct using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA)
The p227 kDa kinase expression plasmid was reconstructed from the full-length cDNA LD31161, which encodes the partially overlapping 91 kDa DOA isoform, which was fully sequenced prior to further modifications. Genomic DNA fragments encompassing the entire 227 kDa-isoform coding region were added by PCR and cloning, to reconstitute the entire 227 kDa DOA isoform region. An in-frame GFP cassette was added at the 5′-end of the construct, which was then entirely sequenced and inserted into the pMT vector for expression in S2 cells.
Organelle labeling
To study peroxisome movement we used S2 cells, stably transfected with pAC-GFP-SKL plasmid, encoding peroxisome-targeted GFP (Kural et al., 2005). For analysis of mitochondria and lysosome movement, S2 cells were stained with 0.1 µM MitoTracker Red CMXRos or LysoTracker Red DND-99 (Molecular Probes/Life Technologies, Eugene, OR), respectively, for 10 minutes and imaged immediately after staining.
Cell imaging
Images were acquired using an inverted TE-2000 microscope (Nikon Instruments, Melville, NY) equipped with a Perfect Focus attachment using a 100× 1.49 NA objective. Either a 100 W mercury bulb or a 100 W halogen bulb was used for fluorescence excitation. The camera was a Cascade II EM CCD camera (Roper Scientific) controlled by Metamorph software (Molecular Devices, Downingtown, PA)
Analysis of organelle movement
Movement of organelles in cell processes was tracked and analyzed using DIATRACK software (version 3.01; Semasopht, North Epping, Australia). A single vector was defined as the distance moved by an organelle in 1 second. Vectors were separated into two categories: plus (toward the process tip) and minus (toward the cell body) and all vectors smaller than 0.2 µm were discarded. For all conditions, the number of vectors was normalized to the total number of analyzed particles to account for variability in the number of particles in cell processes and number of processes between different cells. For analysis of trajectories presented in Fig. 1D an uninterrupted movement in one direction was considered as a single trajectory. All particles that could not be tracked for at least ten frames were discarded. Only particles in cell processes were tracked, as direction of movement along microtubules in the cell body cannot be established. Each experiment was repeated at least three times and the data were expressed as a percentage of the control and pooled.
Recombinant DOA fragment expression and purification
DNA encoding the N-terminal fragment of DOA227 (residues 915–1197) was subcloned into the pRSET expression vector. His6-tagged protein was expressed in BL21(DE3)pLysS E. coli. Expression of the protein was induced by 1 mM isopropyl β-D-1-thiogalactopyranoside at 37°C for 4 hours.
Bacterial pellets were resuspended in phosphate buffer (50 mM NaHPO4, 300 mM NaCl, pH 8.0), supplemented with 1 mM PMSF and 10 µM/ml of protease inhibitors (pepstatin, leupeptin and chymostatin), sonicated and centrifuged at 200,000 g for 30 minutes at 4°C. The supernatant was incubated with TALON Resin (Clontech Labs, Inc., Mountain View, CA) for 1 hour at 4°C; the resin was washed three times with phosphate buffer. Protein was eluted using 300 mM imidazole in phosphate buffer at pH 8.0 and dialyzed against BRB 80 buffer (80 mM PIPES pH 6.9, 0.2 mM EGTA, 0.2 mM MgCl2).
Pelleting of the DOA227 fragment and mCherry–EF1-γ with microtubules
Purified bovine brain tubulin was polymerized in the presence of 1 mM GTP, 1 mM DTT and 20 µM Taxol in BRB 80 buffer for 20 minutes at 37°C. S2 cells were lysed in the same buffer, containing 1 mM DTT, 1 mM PMSF, 10 µg/ml of proteinase inhibitors, 1% NP-40. The extract was clarified by centrifugation at 200,000 g for 30 minutes at 4°C and the supernatant was used for microtubule binding. S2 cell extracts or purified recombinant proteins were incubated with microtubules for 30 minutes at room temperature. The mixture was loaded on the top of 30% glycerol cushions in BRB 80 buffer with 2 µM Taxol and pelleted at 150,000 g at 4°C for 40 minutes in a SW-55Ti rotor. Control samples were prepared identically but without addition of microtubules. The pellets were dissolved in SDS sample buffer for electrophoresis and western blotting. The amount of the DOA227 fragment bound to microtubules was measured by densitometry of Coomassie-Blue-stained gels.
Supplementary Material
Acknowledgments
We thank Laetitia Philippe for cloning of the DOA227 N-terminal fragment.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
A.S.S. and K.T. performed experiments. Experiments were conceived and designed by V.I.G. and L.R. Data were analysed and interpreted and the paper was written by A.S.S., L. R. and V.I.G.
Funding
This work was supported by the National Institute of General Medical Science of the National Institutes of Health [grant number R01GM052111 to V.I.G.]; and support from Universite Paris Sud and Centre national de recherche scientifique to L.R. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.123885/-/DC1
References
- Al-Maghrebi M., Brulé H., Padkina M., Allen C., Holmes W. M., Zehner Z. E. (2002). The 3′ untranslated region of human vimentin mRNA interacts with protein complexes containing eEF-1gamma and HAX-1. Nucleic Acids Res. 30, 5017–5028 10.1093/nar/gkf656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ally S., Larson A. G., Barlan K., Rice S. E., Gelfand V. I. (2009). Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol. 187, 1071–1082 10.1083/jcb.200908075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R. et al. (2006). Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439, 604–607 10.1038/nature04377 [DOI] [PubMed] [Google Scholar]
- Barlan K., Lu W., Gelfand V. I. (2013). The microtubule-binding protein ensconsin is an essential cofactor of kinesin-1. Curr. Biol. 23, 317–322 10.1016/j.cub.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius T. L., Cai D., Jih G. T., Toret C. P., Verhey K. J. (2007). Two binding partners cooperate to activate the molecular motor Kinesin-1. J. Cell Biol. 176, 11–17 10.1083/jcb.200605099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill K., Feng L. L., Yeakley J. M., Gish G. D., Cáceres J. F., Pawson T., Fu X. D. (1996). SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J. Biol. Chem. 271, 24569–24575 10.1074/jbc.271.40.24569 [DOI] [PubMed] [Google Scholar]
- Corbi N., Batassa E. M., Pisani C., Onori A., Di Certo M. G., Strimpakos G., Fanciulli M., Mattei E., Passananti C. (2010). The eEF1γ subunit contacts RNA polymerase II and binds vimentin promoter region. PLoS ONE 5, e14481 10.1371/journal.pone.0014481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit R., Levy J. R., Tokito M., Ligon L. A., Holzbaur E. L. (2008). Regulation of dynactin through the differential expression of p150Glued isoforms. J. Biol. Chem. 283, 33611–33619 10.1074/jbc.M804840200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C., McGuffin M. E., Dauwalder B., Rabinow L., Mattox W. (1998). Protein phosphorylation plays an essential role in the regulation of alternative splicing and sex determination in Drosophila. Mol. Cell 2, 741–750 10.1016/S1097-2765(00)80289-0 [DOI] [PubMed] [Google Scholar]
- Ejiri S. (2002). Moonlighting functions of polypeptide elongation factor 1: from actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 66, 1–21 10.1271/bbb.66.1 [DOI] [PubMed] [Google Scholar]
- Fan Y., Schlierf M., Gaspar A. C., Dreux C., Kpebe A., Chaney L., Mathieu A., Hitte C., Grémy O., Sarot E. et al. (2010). Drosophila translational elongation factor-1gamma is modified in response to DOA kinase activity and is essential for cellular viability. Genetics 184, 141–154 10.1534/genetics.109.109553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J. R., Webster B. M., Mastronarde D. N., Verhey K. J., Voeltz G. K. (2010). ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 190, 363–375 10.1083/jcb.200911024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gindhart J. G., Chen J., Faulkner M., Gandhi R., Doerner K., Wisniewski T., Nandlestadt A. (2003). The kinesin-associated protein UNC-76 is required for axonal transport in the Drosophila nervous system. Mol. Biol. Cell 14, 3356–3365 10.1091/mbc.E02-12-0800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H., Fedorov A. (2002). The origin of the eukaryotic cell: a genomic investigation. Proc. Natl. Acad. Sci. USA 99, 1420–1425 10.1073/pnas.032658599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J. R., Meireles A. M., Fisher K. H., Garcia A., Antrobus P. R., Wainman A., Zitzmann N., Deane C., Ohkura H., Wakefield J. G. (2008). A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 6, e98 10.1371/journal.pbio.0060098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii H., Vecchione A., Murakumo Y., Baldassarre G., Numata S., Trapasso F., Alder H., Baffa R., Croce C. M. (2001). FEZ1/LZTS1 gene at 8p22 suppresses cancer cell growth and regulates mitosis. Proc. Natl. Acad. Sci. USA 98, 10374–10379 10.1073/pnas.181222898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. M., Möller W. (1988). Elongation factor 1 beta gamma from Artemia. Purification and properties of its subunits. Eur. J. Biochem. 171, 119–128 10.1111/j.1432-1033.1988.tb13766.x [DOI] [PubMed] [Google Scholar]
- Kim H., Ling S. C., Rogers G. C., Kural C., Selvin P. R., Rogers S. L., Gelfand V. I. (2007a). Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176, 641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Kellner J., Lee C. H., Coulombe P. A. (2007b). Interaction between the keratin cytoskeleton and eEF1Bgamma affects protein synthesis in epithelial cells. Nat. Struct. Mol. Biol. 14, 982–983 10.1038/nsmb1301 [DOI] [PubMed] [Google Scholar]
- Kpebe A., Rabinow L. (2008). Alternative promoter usage generates multiple evolutionarily conserved isoforms of Drosophila DOA kinase. Genesis 46, 132–143 10.1002/dvg.20374 [DOI] [PubMed] [Google Scholar]
- Kural C., Kim H., Syed S., Goshima G., Gelfand V. I., Selvin P. R. (2005). Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science 308, 1469–1472 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- Lee K., Du C., Horn M., Rabinow L. (1996). Activity and autophosphorylation of LAMMER protein kinases. J. Biol. Chem. 271, 27299–27303 10.1074/jbc.271.44.27299 [DOI] [PubMed] [Google Scholar]
- Ling S. C., Fahrner P. S., Greenough W. T., Gelfand V. I. (2004). Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl. Acad. Sci. USA 101, 17428–17433 10.1073/pnas.0408114101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolakaki E., Du C., Lai J., Giannakouros T., Cantley L., Rabinow L. (2002). Phosphorylation by LAMMER protein kinases: determination of a consensus site, identification of in vitro substrates, and implications for substrate preferences. Biochemistry 41, 2055–2066 10.1021/bi011521h [DOI] [PubMed] [Google Scholar]
- Padmanabha R., Gehrung S., Snyder M. (1991). The KNS1 gene of Saccharomyces cerevisiae encodes a nonessential protein kinase homologue that is distantly related to members of the CDC28/cdc2 gene family. Mol. Gen. Genet. 229, 1–9 10.1007/BF00264206 [DOI] [PubMed] [Google Scholar]
- Reed N. A., Cai D., Blasius T. L., Jih G. T., Meyhofer E., Gaertig J., Verhey K. J. (2006). Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16, 2166–2172 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Shi Y., Di Giammartino D. C., Taylor D., Sarkeshik A., Rice W. J., Yates J. R., III, Frank J., Manley J. L. (2009). Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33, 365–376 10.1016/j.molcel.2008.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D. (2003). The molecular motor toolbox for intracellular transport. Cell 112, 467–480 10.1016/S0092-8674(03)00111-9 [DOI] [PubMed] [Google Scholar]
- Vershinin M., Carter B. C., Razafsky D. S., King S. J., Gross S. P. (2007). Multiple-motor based transport and its regulation by Tau. Proc. Natl. Acad. Sci. USA 104, 87–92 10.1073/pnas.0607919104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vershinin M., Xu J., Razafsky D. S., King S. J., Gross S. P. (2008). Tuning microtubule-based transport through filamentous MAPs: the problem of dynein. Traffic 9, 882–892 10.1111/j.1600-0854.2008.00741.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.