Abstract
Lymphoid Enhancer Binding Factor (Lef) 1 is a transcriptional effector of the Wnt/Lrp5/β-catenin signaling cascade, which regulates osteoblast differentiation, bone density and skeletal strength. In this study, we describe the expression and function of an alternative Lef1 isoform in osseous cells. Lef1ΔN is a naturally occurring isoform driven by a promoter (p2) within the intron between exons 3 and 4 of Lef1. Lef1ΔN is induced during late osteoblast differentiation. This is converse to the expression pattern of the full-length Lef1 protein, which as we previously showed, decreases during differentiation. Agonists of osteoblast maturation differentially affected Lef1ΔN expression. BMP2 stimulated Lef1ΔN expression, whereas Wnt3a blocked basal and BMP2-induced expression of Lef1ΔN transcripts during osteoblast differentiation. We determined that the Lef1ΔN p2 promoter is active in osteoblasts and Runx2 regulates its activity. Stable overexpression of Lef1ΔN in differentiating osteoblasts induced the expression of osteoblast differentiation genes, osteocalcin and type 1 collagen. Taken together, our results suggest Lef1ΔN is a crucial regulator of terminal differentiation in osseous cells.
Keywords: Lef1ΔN, BMP2, Wnt3a, osteocalcin, Cbfa1
Introduction
The canonical Wnt pathway dictates osteoblast specification from osteo/chondroprogenitors, stimulates osteoblast proliferation, regulates synthesis of matrix proteins, enhances osteoblast and osteocyte survival, and transmits mechanical loading signals to bone lining cells (reviewed in (Bonewald and Johnson, 2008; Khosla et al., 2008)). Canonical Wnt signaling is initiated when Wnts bind to receptor complexes consisting of Lrp5/6 and Frizzled proteins. This stabilizes β–catenin and induces its translocation to the nucleus where it displaces corepressors from Tcf7 and Lef1 transcription factors, leading to the activation of genes involved in cell proliferation, including cyclin D and c-myc (Westendorf et al., 2004). If unchecked or rampant, the Wnt pathway is often dangerous and oncogenic because of this pro-proliferative effect (Polakis, 2007).
Lef1 is one of four transcription factors (the others are Tcf7 (a.k.a. Tcf1), Tcf7L1 (a.k.a. Tcf3) and Tcf7L2 (a.k.a. Tcf4)) that bind β–catenin and regulate gene expression (Arce et al., 2006; Hoppler and Kavanagh, 2007). First discovered in lymphocytes, Lef1 binds one of the most common motifs in human promoters, CTTTGT (Xie et al., 2005), and is a context-dependent regulator of gene expression. When Lef1 interacts with DNA, it induces a sharp 130° bend in the double helix and alters the DNA binding of other transcription factors to regulate gene transcription (Carlsson et al., 1993; Giese et al., 1991; Giese and Grosschedl, 1993; Giese et al., 1995; Giese et al., 1997; Love et al., 1995; van de Wetering and Clevers, 1992). In the absence of nuclear β–catenin, Lef1 interacts weakly with chromatin (Tutter et al., 2001) and associates with transcriptional co-repressors (e.g. CtBP, TLE and histone deacetylases) (Billin et al., 2000; Brannon et al., 1999; Brantjes et al., 2001; Levanon et al., 1998). Nuclear β–catenin displaces these co-repressors and recruits co-activators (e.g. p300, Bcl9, and Pygopus) to facilitate chromatin binding and gene expression (Behrens et al., 1996; Billin et al., 2000; Daniels and Weis, 2005; Hecht et al., 2000; Hsu et al., 1998; Huber et al., 1996; Kramps et al., 2002; Miyagishi et al., 2000; Sun et al., 2000; Tutter et al., 2001). Interestingly, in some contexts Wnt signaling and β–catenin facilitate Lef1/Tcf7-mediated repression through poorly defined mechanisms (Baker et al., 1999; Cadigan et al., 1998; Cadigan et al., 2002; Jamora et al., 2003; Kahler and Westendorf, 2003; Payre et al., 1999; Piepenburg et al., 2000; Willert et al., 2002; Yang et al., 2000).
Lef1 and Tcf7 proteins share many molecular and biochemical characteristics; however, functional diversity exists between Tcf7/Lef factors during development (Arce et al., 2006; Brugmann et al., 2007; Hoppler and Kavanagh, 2007; Schroeder et al., 2004). In skeletal structures of mouse embryos (E14.5), Lef1 is detectable in tail prevertebrae, osteogenic cells of the hipbone, and the mesenchymal cells around the cochlea. By comparison, Tcf7 (previously referred to as Tcf1) is found in the pre-cartilagenous cells of the palate, maxilla, mandible, nasal and basioccipital bones, the thoracic pre-vertebrae and ribs at E14.5 (Oosterwegel et al., 1993). Knockout mice also reveal specific functions for Lef1 and Tcf7. Lef1-deficient mice are smaller than normal littermates, display multiple defects in tissues formed by epithelial and mesenchymal interactions (e.g. they lack teeth, body hair and whiskers), and die within two weeks of birth (Oosterwegel et al., 1993). A recent report examining the role of the Lef1 in the regulation of bone mass found that Lef1+/− female mice had decreased trabecular bone mass due to reduced osteoblast activity (Noh et al., 2009). Tcf7-deficient animals are viable, but they lack early thymocyte progenitors (Verbeek et al., 1995) and have low bone mass caused by decreased OPG expression and increased osteoclastogenesis (Glass et al., 2005). This bone loss is enhanced when Tcf7-null mice are crossed with β–catenin-deficient mice (Glass et al., 2005). Animals lacking both Lef1 and Tcf7 most resemble Wnt3a−/− mice (Galceran et al., 1999). Thus, Lef1 and Tcf7 act in concert during development to mediate canonical Wnt3a signaling.
In adults, the expression of the Lef1/Tcf7 transcription factors is generally restricted to mitotically active cells in renewable tissues. Examples of such tissues include lymphoid follicles, skin, hair follicles, colon, intestine, testis and human tumors (Mayer et al., 1997; Porfiri et al., 1997). Bone is also a renewable tissue, and accordingly, Lef1/Tcf7 activity is increased in areas of bone remodeling and regeneration, and in proliferating osteogenic cells (de Jong et al., 2002; Hadjiargyrou et al., 2002; Kahler and Westendorf, 2003; Kato et al., 2002; Qi et al., 2003). Lef1/Tcf7 expression is usually downregulated as cells stop proliferating and is lost in terminally differentiated cells (Hadjiargyrou et al., 2002; Kato et al., 2002; Kratochwil et al., 1996; Mariadason et al., 2001; Oosterwegel et al., 1993; Shibamoto et al., 2004; Travis et al., 1991; Zhou et al., 1995) including osteoblasts (Kahler et al., 2006).
Each of the four Lef1/Tcf7 genes generate functional diversity by encoding multiple isoforms through differential promoter usage and alternative splicing (Hovanes et al., 2001; Hovanes et al., 2000; Van de Wetering et al., 1996). Isoforms lacking the N-terminal high affinity β–catenin binding domain sometimes act as competitive inhibitors of full-length Lef1/Tcf7 proteins (Arce et al., 2006; Hoppler and Kavanagh, 2007). Lef1ΔN lacks the first 113 amino acids found in Lef1. Its 2.3 kb transcript is driven by a promoter (p2) within the intron between exons 3 and 4 (Arce et al., 2006; Hovanes et al., 2001). Differential expression of the full-length and shorter ΔN transcripts is now extensively documented in lymphocytes, myeloid cells, tumors, skin and the intestine (Merrill et al., 2001; Niemann et al., 2002; Skokowa et al., 2006; Takeda et al., 2006; Wang et al., 2005). In this report, we demonstrate that Lef1ΔN is induced in osteoblasts during the terminal differentiation process and accelerates the expression of genes involved in osteoblast differentiation when introduced into pre-osteoblasts.
Materials and Methods
Cell lines, primary calvarial osteoblast isolation, and osteoblast differentiation
C2C12 cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS), 200 mM L-glutamine, 50 U/ml penicillin, and 50 mg/ml streptomycin. C3H10T1/2, MC3T3-E1, ROS 17/2.8, and primary osteoblast cells were cultured in Minimal Essential Medium (MEM) containing the supplements listed above. Primary calvarial osteoblasts were isolated from C57Bl/6 mice as previously described (Schroeder et al., 2005). To induce osteogenic differentiation, cells were cultured in α-MEM supplemented with 10% FBS, 50 μg/ml ascorbic acid and 10 mM β-glycerolphosphate after reaching confluency. Where indicated, media were supplemented with 300 ng/ml BMP2 (Sofamor Danek) and 67 ng/ml Wnt3a (R&D Biosystems). Media were replaced every three days. Transient transfections were performed in 12 well plates with Lipofectamine (Invitrogen) according to the manufacturer's instructions.
RNA Isolation, Northern Blotting and Reverse Transcriptase-PCR
RNA was harvested from cells using Trizol Regent (Invitrogen). RNA (northern) blotting was performed as described by Kahler et al (Kahler et al., 2006). Quantitative RT-PCR was performed using the QuantiTech SYBR Green RT-PCR kit (Qiagen) and reactions were run on an iCycler (BioRad). RNA (10 ng) was used in a 20 μl reaction with QuantiTech SYBR Green RT mastermix, QuantiTech RT mix, and 0.5 pmol/μl of each of the primers for mouse Lef1 (Primer Pair #1), forward: GAT CCC CTT CAA GGA CGA AG, reverse: GGC TTG TCT GAC CAC CTC AT; mouse Lef1 + Lef1ΔN (Primer Pair #2), forward: TCA CTG TCA GGC GAC ACT TC, reverse: TGA GGC TTC ACG TGC ATT AG; mouse alkaline phosphatase, forward: TGT TGA CAA GGC AGA CAA GC, reverse: CAG GAC CGT TGC CGT ATA GT; mouse osteocalcin, forward: AAG CAG GAG GGC AAT AAG GT, reverse: TTT GTA GGC GGT CTT CAA GC; mouse actin, forward: AAG GAA GGC TGG AAA AGA GC, reverse: GCT ACA GCT TCA CCA CCA CA. Data were normalized to mouse actin levels. The reverse transcriptase reaction to convert the RNA to cDNA was performed at 50°C for 30 minutes. Initial denaturation was made at 95°C for 15 minutes followed by 40 cycles of three-step PCR: 95°C denaturation for 1 minute, 57°C (59.1°C for Lef1 primers) annealing for 1 minute, and 72°C elongation for 1 minute. The Lef1 primer pairs were used for differential quantitative RT-PCR. The efficiencies of Lef1 primer pairs 1 and 2 were determined by amplifying serial dilutions of Lef1 plasmid DNA. The efficiency (E) of each primer pair was taken into account when calculating the relative amounts of cDNA amplified through the equation, E−ΔΔCt (Pfaffl, 2001).
Electrophoretic Mobility Shift Assays (EMSA) and Chromatin Immunoprecipitation (ChIP)
EMSA were performed with a double stranded probe with a 24 bp sequence from p2 that surrounds and includes the Runx2 binding element as previously described (Kahler and Westendorf, 2003). ChIP analyses for Runx2 binding were performed on lysates from ROS17/2.8 cells as previously described (Schroeder et al., 2004) using antibodies against Runx2 (Santa Cruz, M-70) or normal rabbit IgG (Santa Cruz). PCR was performed using the following primers: 5' GGC CTT GCA TGT TAA CAC CT and 5' GGC TCA ACA GCA AAC CAG AT.
For histone and RNA polymerase II ChIP analyses, lysates were collected from differentiated MC3T3 cells as previously described (Lambert and Nordeen, 2003), except the cultures were treated with collagenase type 2 (Worthington; 1 mg/ml)/BSA (Sigma Aldrich; 1 mg/ml) at 37°C for 1 hour (to remove extracellular collagen that accumulates in osteoblast cultures) before adding 1% formaldehyde to crosslink intracellular proteins and DNA. Fractions of the lysate (20%) from a 10 cm plate were incubated with antibodies (2 μg) recognizing histone H3 (Upstate, 05-928), phosphorylated RNA polymerase II (GeneTex, GTX44758) or normal rabbit IgG (Santa Cruz). After reversing the crosslinks, DNA was purified and analyzed for enrichment of the Lef1 p2 promoter with real-time PCR (see above). Primers used to amplify the Lef1 P2 promoter were the “proximal” primer set: 5' TGCTGTTAAAGCCATTGAGG 3' and 5' AGACAGCCCAGACTTTACGC 3', and with the “distal” primer set: 5' GTCCTCTCAGGAGCCCTACC 3' and 5' GTATGTGGGGAAGTCTTGCAT 3'. Nonspecific background was eliminated in data by subtracting the relative amount of product detected in the control IgG immunoprecipitates from that detected in the H3 and RNA polymerase II immunoprecipitates.
Promoter/Reporter Plasmids
The Lef1 p2 promoter (−447–+35) was amplified from Balb/c mouse genomic DNA using gene specific oligonucleotides (primer sequences available upon request) and PCR. PCR products were cloned into the TOPO vector (Invitrogen) and then subcloned into the pGL2 plasmid (Promega) using HindIII restriction sites. The Lef1 promoter 1 (p1) was obtained from Dr. Marian Waterman. The murine osteocalcin gene (mOG) 2-luciferase plasmid (Montecino et al., 1996) was obtained from Dr. Jane Lian. The proximal Runx2 site within mOG2 was replaced with a GAL4 site, UAS (CGGAGTACTGT CCTCCG), with overlapping PCR.
Transcription Assays
Cells were transfected with Lipofectamine (Invitrogen) in 12-well plates with the indicated amounts of luciferase reporter plasmids and 50 ng pRL-null. pCMV-Runx2 expression plasmids (300 ng unless otherwise specified) were added as indicated. pcDNA3.1 was added to transfections to maintain a uniform amount of total DNA per transfection. Luciferase activity was measured 24 or 48 hours after transfection using the dual-luciferase assay system (Promega). Each transfection was performed in triplicate, and normalized to Renilla-luciferase activity.
Retroviral Subcloning and Transduction
To produce retroviruses, 293T cells were co-transfected with 5 μg of pMSCV-puro-Lef1-Flag or pMSCV-puro-Lef1ΔN-Flag and 5 μg of pCL2 using calcium phosphate precipitation. Virus-containing supernatants were collected after 48 and 72 hours, filtered, and added to C2C12 cells in the presence of 8 μg/ml polybrene. The transduction was repeated 8 hours and 24 hours later. Transduced cells were selected with 5 μg/ml puromycin for a minimum of 3 days.
Immunoblotting
Cells were treated with proteasome inhibitor MG132 (Sigma; 10 μM) for 4 hours prior to lysis. Cells were washed twice with PBS. Whole cell lysates were collected by incubating cells with modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, 0.25% NaDOC, protease inhibitors). Nuclear lysates were obtained from indicated cells by scraping the cells off the plate in 1 ml of PBS, centrifuging at 2000 rpm for 5 minutes, suspending the pellet in 100 μl Iso-Hi Buffer (10 mM Tris-HCl, pH 7.8, 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, protease inhibitors), incubating on ice for 5 minutes, centrifuging at 5000 rpm for 5 minutes at 4°C, and suspending the pellet in modified RIPA buffer. Lysates were sonicated and insoluble material was removed by centrifugation. Equal amounts of protein lysates were resolved by SDS-12% PAGE. Proteins were transferred to PVDF membrane (Immobilon-P, Millipore). Membranes were blotted with the indicated antibody diluted 1:1000 in 3% non-fat dried milk in TBST (TBS + 0.04% Tween-20). The antibodies used are: anti-Lef1 (Cell Signaling, C18A7) and anti-Lamin B (Santa Cruz).
Results
Osteoblasts Express Multiple Lef1 mRNA Transcripts
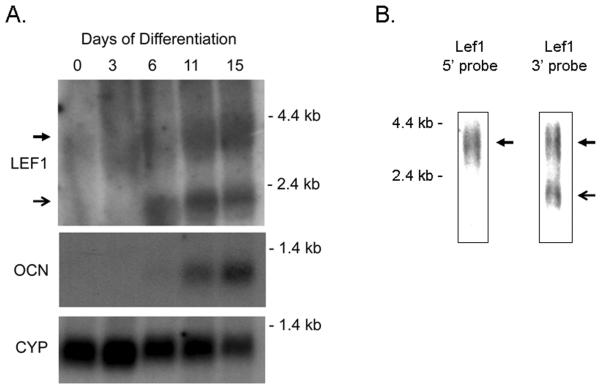
Lef1 mRNA expression levels were analyzed during MC3T3 osteoblast maturation by northern blotting. A 3.6 kb transcript was induced during the differentiation time course (Figure 1A). This transcript encodes a full-length isoform of Lef1 containing the high-affinity β–catenin binding domain at the N-terminus and the DNA binding domain at the C-terminus (Hovanes et al., 2000). A 2.3 kb transcript was also detected MC3T3 cells after six days of differentiation. This transcript appeared before the osteocalcin transcript (Figure 1A) and was only detected with a probe directed to the 3' region of the Lef1 gene (Figure 1B), whereas a probe recognizing the 5' region of Lef1 detected only the full-length 3.6 kb transcript (Figure 1B). These results indicate that maturing osteoblasts produce an alternative Lef1 transcript that resembles one previously described in T lymphocytes (Hovanes et al., 2001).
Figure 1. Northern Blot analysis for Lef1 transcripts in differentiating MC3T3 osteoblasts.
A. Northern blot analysis of MC3T3 cells differentiated in ascorbic acid and β-glycerolphosphate for 0 to 15 days. Blots were incubated with probes recognizing the 3' region of the Lef1 gene, osteocalcin (Ocn) or cyclophilin (Cyp). The 3.6 and 2.3 kb bands are indicated by closed and open arrows, respectively. B. Northern blot analysis of MC3T3 cells that had been differentiated for 9 days. Blots were incubated with probes recognizing the 5' or 3' regions of the Lef1 gene as indicated.
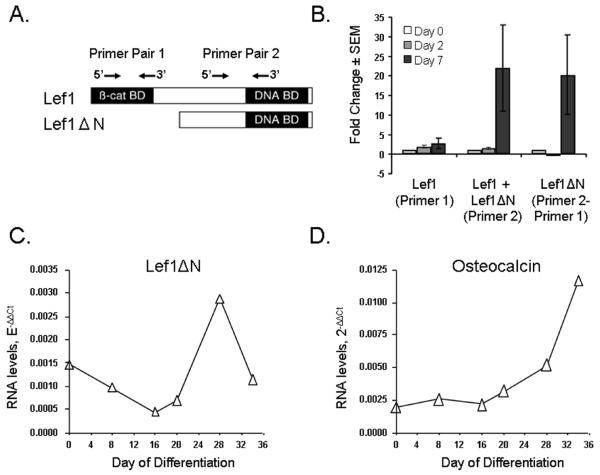
Based on the results of Hovanes and colleagues (Hovanes et al., 2001), we hypothesized that the 2.3 kb transcript encodes a truncated protein that lacks the N-terminal β–catenin binding domain, but retains the DNA binding domain (Figure 2A). To test this and verify the northern blotting analysis, we developed a differential PCR approach, similar to ones described by others (Wang et al., 2005; Willinger et al., 2006), to examine the relative amount of the shorter transcript in cells. Two primers pairs were synthesized (Figure 2A) and their relative efficiencies in amplifying Lef1 cDNA were calculated. Primer pair 1 amplifies sequences only in the 3.6 kb transcript, whereas primer pair 2 amplifies both the 3.6 and 2.3 kb mRNAs. The difference between the two products is the amount of the shorter transcript in the cells. Using this differential PCR approach, results similar to those obtained by northern blotting were obtained (Figure 2B). Thus, the longer transcript, Lef1 (recognized by primer pair 1), did not change significantly during the first 7 days of differentiation. However, the amount of product recognized by primer pair 2 increased more than 20 fold by day 7. No changes were observed at days 2 (Figure 2B) or 5 (data not shown). We derived the amount of the transcript only recognized by primer pair 2 by subtracting primer pair 1 products from primer pair 2 amplicons and called this Lef1ΔN transcript.
Figure 2. Detection of short Lef1ΔN transcripts by PCR in osteoblasts.
A. Schematic of the relative location of primer pairs for differential PCR. B. Results of differential quantitative RT-PCR assays. Primer pair 1 exclusively detects full-length Lef1, whereas primer pair 2 amplifies both full-length Lef1 and Lef1ΔN. To calculate the relative amount of Lef1ΔN present, yield of primer pair 1 product is subtracted from yield of primer pair 2 product. C–D. RNA was isolated from mouse primary calvarial cells after the indicated number of days in culture. Lef1ΔN was measured using differential quantitative real-time PCR (C), while osteocalcin was quantitated using standard gene-specific quantitative RT-PCR (D). Data were normalized to actin products. As in MC3T3 cells, Lef1ΔN transcripts appear immediately prior to osteocalcin transcripts.
We verified the induction of Lef1ΔN in mouse primary calvarial cells. As in MC3T3 cells, Lef1ΔN transcripts (Figure 2C) were detected prior to the time when osteocalcin transcripts (Figure 2D) were detectable in these cultures (days 28 and 34, respectively). Due to the longer differentiation period required for primary osteoblasts, Lef1ΔN levels peaked at 28 days, while osteocalcin levels continued to rise during days 28 to 34.
BMP2 and Wnt3a Differentially Affect Lef1ΔN Expression
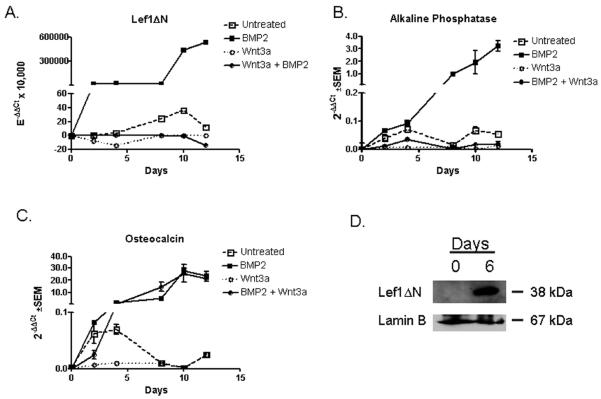
BMP2 and Wnt3a are agonists of osteoblast proliferation. BMP2 binds receptor complexes and activates numerous signaling pathways to regulate osteoblast gene expression, proliferation and differentiation (Katagiri et al., 1990; Katagiri et al., 1994; Wang et al., 1990; Yamaguchi et al., 1991). Wnt3a initiates canonical Wnt signaling by binding to receptor complexes consisting of Lrp5/6 and Frizzled on the cell surface, which results in nuclear translocation of β-catenin and activation of Lef1/Tcf transcription factors (Bhanot et al., 1996; Billin et al., 2000; Cadigan et al., 1998; Hecht et al., 2000; Shtutman et al., 1999; Sun et al., 2000; Wehrli et al., 2000; Yang-Snyder et al., 1996). Using differential quantitative PCR to evaluate Lef1ΔN transcript levels, we found that BMP2 is a very potent inducer of Lef1ΔN expression during osteoblast differentiation (Figure 3A). Thus, while Lef1ΔN was induced 35-fold by osteogenic medium lacking exogenous BMP2 (open boxes), it was stimulated 5×105 fold by BMP2 (closed boxes). Conversely, Wnt3a repressed basal and BMP2-induced Lef1ΔN expression (circles, Figure 3A). Alkaline phosphatase (AP) and osteocalcin transcripts were measured to monitor the differentiation process. AP, a relatively early marker of osteoblast differentiation, was induced by BMP2, while Wnt3a repressed basal and BMP2-induced alkaline phosphatase expression (Figure 3B). Basal expression of osteocalcin, a late osteoblast differentiation marker, was also upregulated by BMP2 and repressed by Wnt3a (Figure 3C). However, Wnt3a was unable to repress BMP2-induced expression of osteocalcin (Figure 3C). These results are consistent with other reports showing that Wnt3a blocks early osteogenesis of mesenchymal progenitor cells (Boland et al., 2004). To confirm the induction of Lef1ΔN during osteoblast differentiation, immunoblotting was preformed on lysates collected during osteoblast differentiation in the presence of BMP2. Consistent with the increase of Lef1ΔN transcript levels, Lef1ΔN protein levels increased during osteoblast differentiation in the presence of BMP2 (Figure 3D).
Figure 3. BMP2 and Wnt3a Affect Lef1ΔN Transcript Expression.
A–C. MC3T3 cells were differentiated in osteogenic media for 12 days in the presence of BMP2, Wnt3a, Wnt3a and BMP2, or endogenous cytokines. RNA was collected at regular intervals for quantitative PCR. All data were normalized to murine actin. Lef1ΔN transcript levels were determined by differential quantitative real-time PCR as described in Figure 2. D. MC3T3 cells were differentiated in osteogenic media containing BMP2 for 6 days. Nuclear protein lysates were collected at the indicated days, separated by SDS-PAGE electrophoresis, transferred to Immobilon-P membrane, probed with anti-Lef1, and reprobed with anti-Lamin B.
The Lef1ΔN Promoter, p2, is Active in Osteoblasts
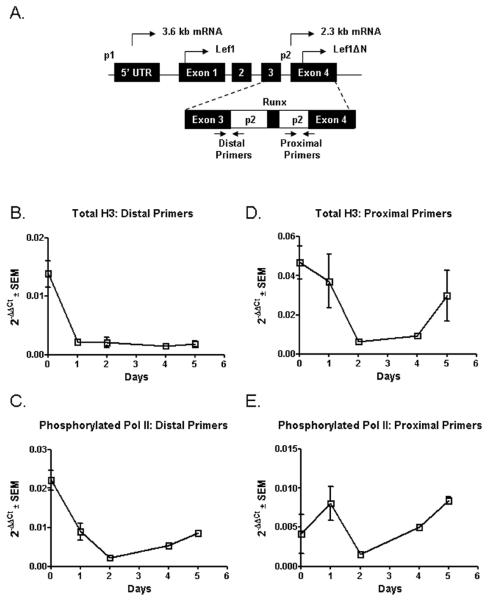
The temporal and inducible expression of Lef1ΔN indicates that it is important for late osteoblast differentiation. To understand how expression of Lef1ΔN is regulated, we examined the murine Lef1ΔN promoter, referred to as p2. In human cells, the transcriptional start site is ten base pairs 5' of exon 4, and translation begins with a methionine in the middle of exon 4 (Hovanes et al., 2001). To determine whether p2 is active in murine osteoblasts, we used chromatin immunoprecipitation (ChIP) to evaluate levels of histone 3 (H3) and phosphorylated RNA polymerase II at distal and proximal regions of the p2 promoter (Figure 4A). We first examined the levels of post-translationally modified H3 at the promoter. Surprisingly, levels of both acetylated (an activation mark) and methylated (a repressive mark) H3 were reduced (data not shown). Thus, we hypothesized that nucleosomes in this region were being disassembled. We tested this possibility by investigating total H3 levels. During the first 1 to 2 days of osteoblast differentiation, the amount of total H3 present at the distal and proximal regions of the p2 promoter decreased and remained low throughout the first 5 days of osteoblast differentiation (Figures 4B & 4D). This loss of total H3 suggests nucleosome disassembly and is expected to correspond with increased accessibility for transcriptional machinery. To determine if the promoter was being actively transcribed, we monitored the recruitment of phosphorylated RNA polymerase II to the p2 promoter in differentiating osteoblasts. From day 0 to day 2 of osteoblast differentiation, phosphorylated RNA polymerase II levels at the promoter decreased. However, between days 2–5 of differentiation, levels of phosphorylated RNA polymerase II steadily increased suggesting that the Lef1ΔN p2 promoter was becoming active at both the distal and proximal regions (Figure 4C & 4E). Our observations that total H3 levels were low and that phosphorylated RNA polymerase II levels were increasing at distal and proximal regions of p2 during the first 5 days of osteoblast differentiation correspond with our findings that Lef1ΔN message becomes detectable at days 6 to 7 of osteoblast differentiation (Figure 1A & 2B) and indicate that the Lef1 p2 promoter is activated in differentiating osteoblasts.
Figure 4. Identification of Active Elements in the Murine p2 Promoter.
A. Schematic of the relative location of distal and proximal primers used for chromatin immunoprecipitation. B–E. Lysates were collected from differentiated MC3T3 cells, intracellular proteins and DNA were crosslinked and incubated with antibodies recognizing Histone H3, phosphorylated RNA polymerase II, or IgG. After reversing the crosslinks, DNA was purified and used for qPCR with the indicated primer pairs. Nonspecific background was eliminated in data by subtracting the relative amount of product detected in the control IgG immunoprecipitates from that detected in the H3 and RNA polymerase II immunoprecipitates.
Runx2 Regulates p2 Activity
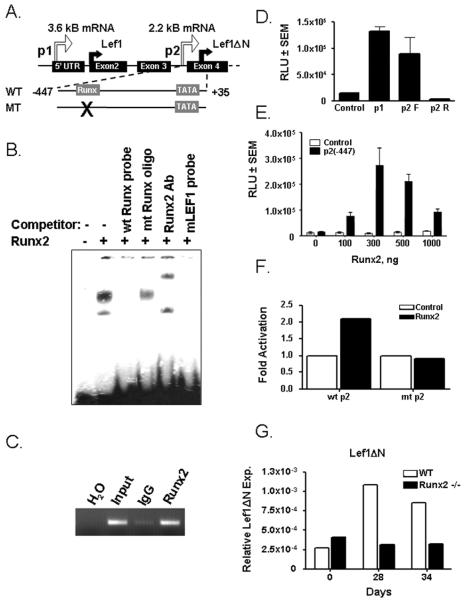
To understand how Lef1 p2 is regulated in osteoblasts, we scanned the sequence for transcription factor binding sites common in osteoblast promoters. Runx2 is an essential transcription factor for osteoblast development and regulates the expression of numerous osteoblast genes. We identified a potential Runx2 binding element in the p2 promoter at position −215 (Figure 5A). Runx2 bound a double-stranded probe containing this sequence of the promoter in electrophoretic mobility shift assays (Figure 5B, lane 2). Formation of the labeled DNA-Runx2 complex was specifically competed with an unlabeled double-stranded oligonucleotide containing a wildtype Runx2 binding element, but not by an unlabeled oligonucleotide containing a mutant Runx2 binding site (Figure 5B, lanes 3 and 4). Moreover, the complex was supershifted by a Runx2 antibody (Figure 5B, lane 5). Chromatin immunoprecipitation assays in ROS17/2.8 osseous cells demonstrated that Runx2 bound the Lef1 p2 promoter in vivo (Figure 5C). To determine the effects of Runx2 on the promoter, we amplified the p2 promoter (−447–+35) and placed it in a vector containing a luciferase reporter gene. This p2 reporter was as active as the Lef1 p1 promoter in C3H10T1/2 cells when subcloned in the forward (F) but not reverse (R) orientation (Figure 5D). Runx2 activated the reporter in a concentration-dependent fashion (Figure 5E); however, Runx2 did not activate a mutant p2 construct lacking a functional Runx2 binding site (Figure 5F). To determine whether Runx2 is required for Lef1 p2 activity, we differentiated primary calvarial cells from wildtype and Runx2−/− mice in osteoblast differentiation medium and evaluated Lef1ΔN transcript levels by differential qPCR. As previously shown (Figure 2C), Lef1ΔN rose in wildtype primary osteoblasts during late osteoblast differentiation (days 28 and 34). In contrast, Lef1ΔN levels did not change in the Runx2-deficient cells (Figure 5G). These data demonstrate that Runx2 binds the Lef1 p2 promoter and positively regulates its expression.
Figure 5. Runx2 Regulates the Lef1 p2 Promoter.
A. Schematic of the 5' portion of murine Lef1. Wildtype (wt) and a mutant (mt) p2 promoter wherein the Runx2 site is eliminated were cloned into the pGL2 luciferase plasmid and used in D–E. Transcriptional and translational start sites are indicated by the thin and thick arrows, respectively. B–C. Runx2 binds a consensus-binding element in the Lef1 p2 promoter as determined by EMSA (B) and chromatin immunoprecipitation (C). D. The p2 promoter (−447-+35) is active in C3H10T1/2 cells when placed in the forward (F) but not reverse (R) orientation. E. Runx2 activates the p2 promoter in a concentration-dependent manner in C3H10T1/2 cells. F. Mutating the Runx2 binding element prevents Runx2-dependent activation of the p2 promoter. G. Primary osteoblasts derived from wildtype (WT) and Runx2−/− mice were differentiated in osteogenic media for 34 days. RNA was collected for quantitative PCR. All data were normalized to murine actin. Lef1ΔN transcript levels were determined by differential quantitative real-time PCR as described in Figure 2.
Overexpression of Lef1ΔN Induces Expression of Late Osteoblast Differentiation Genes
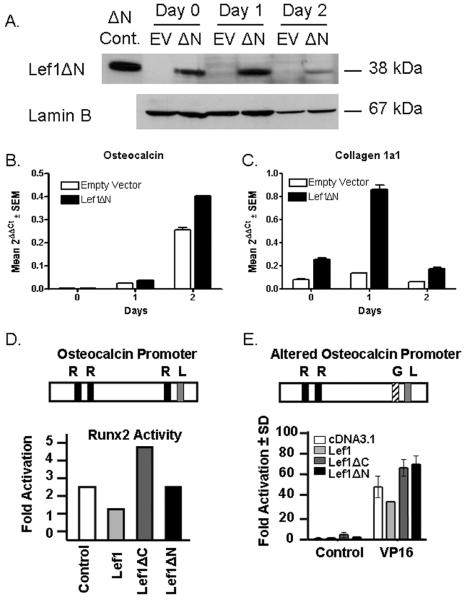
Given that the Lef1 p2 promoter is active in maturing osteoblasts and regulated by Runx2, we sought to understand the role of Lef1ΔN in osteoblast differentiation. To this end, we stably overexpressed Lef1ΔN in C2C12 myo-osteoblast progenitor cells using retroviral transduction and induced the cells to undergo osteoblast differentiation by culturing them in osteogenic medium containing BMP2. We verified exogenous Lef1ΔN protein and transcript expression by immunoblotting (Figure 6A) and differential qPCR (data not shown), respectively. To elucidate the role of Lef1ΔN in osteoblast differentiation, we evaluated the effects of Lef1ΔN overexpression on markers of osteoblast differentiation. Exogenous Lef1ΔN augmented the expression of osteocalcin and type 1 collagen in cells cultured in osteogenic medium (Figure 6B and 6C). Thus, overexpression of Lef1ΔN in myo-osteoblast progenitor cells accelerated the expression of genes involved in osteoblast differentiation.
Figure 6. Overexpression of Lef1ΔN in C2C12 Cells.
A−C. C2C12 cells were transduced with retrovirus containing supernatants derived from 293T cells transfected with MSCV-Lef1ΔN or empty MSCV vector. Lef1ΔN and empty vector (EV) cells were differentiated in osteogenic medium in the presence of BMP2. A. Protein lysates were collected after the indicated days in culture, separated by SDS-PAGE electrophoresis, transferred to Immobilon-P membrane, probed with anti-Lef1, and reprobed with anti-Lamin B. B−C. RNA was collected at regular intervals for quantitative PCR. All data were normalized to murine actin. D−E. Schematics of the osteocalcin promoters show the location of Runx2 (R), Lef1 (L), and GAL4 (G) binding elements. C2C12 cells were transfected with luciferase reporter plasmids driven by osteocalcin promoters, Runx2 (D) or VP16 (E), and pcDNA3.1 (control), Lef1, Lef1ΔC, or Lef1ΔN as indicated. The proximal Runx2 binding element in the osteocalcin promoter was mutated to a GAL4 binding site (E). Luciferase activity was measured 48 hours after transfection.
Lef1, but not Lef1ΔN, inhibits Runx2 activation of the osteocalcin promoter
We previously demonstrated that the full length isoform of Lef1 blocks Runx2-dependent activation of the osteocalcin promoter, which contains a Lef1 binding site just 4 base pairs away from the proximal Runx2 element (Kahler and Westendorf, 2003). To further understand the role of Lef1ΔN in osteoblast differentiation, we evaluated whether Lef1ΔN was also capable of blocking Runx2-dependent activation of the osteocalcin promoter. We found that Lef1ΔN, which lacks the β-catenin binding domain, does not repress Runx2-mediated osteocalcin expression, even though it retains a functional DNA binding domain (Figure 6D). Lef1ΔC, which lacks the DNA binding domain, also did not repress Runx2 and in fact activated the osteocalcin promoter (Figure 6D). To determine whether Lef1 could sterically block the access of Runx2 to the neighboring sequence in the osteocalcin promoter, we changed the proximal Runx2 site to a GAL4 binding site. VP16 strongly activated this mutant promoter (Figure 6E). Full-length Lef1 repressed VP16-driven activation of the osteocalcin promoter, but neither Lef1ΔN nor Lef1ΔC repressed VP16 activation (Figure 6E). These data suggest that full-length Lef1 can sterically block assess of transcription factors, specifically Runx2, to adjacent regions on the osteocalcin promoter and that the N-terminus of Lef1 is necessary for this repression.
Discussion
The Human Genome Project revealed that phenotypic variability is the result of genome complexity and versatility that creates many alternative gene products, rather than the presence of more genes (Tress et al., 2007). In this report, we show that an alternative Lef1 transcript appears during osteoblast maturation just prior to osteocalcin gene expression. This transcript is induced by BMP2 and Runx2, but is suppressed by Wnt3a. To the best of our knowledge, this is the first time that alternate Lef1 transcripts have been described in osseous cells. Lef1 was identified as a differentially regulated gene in several osteoblast microarray studies (James et al., 2006; Vaes et al., 2002), but it is not known which transcript(s) was affected because probes on microarray chips are heavily weighted to 3' regions of genes, which would detect both transcripts.
The four Lef1/Tcf7 genes encode multiple isoforms through differential promoter usage and alternative splicing (Hovanes et al., 2001; Hovanes et al., 2000; Van de Wetering et al., 1996). Lef1ΔN lacks the first 113 amino acids found in Lef1. Its 2.3 kb transcript is driven by a promoter (p2) within the intron between exons 3 and 4 (Hovanes et al., 2001; Li et al., 2006). Differential expression of the full-length Lef1 and short Lef1ΔN transcripts is well documented in lymphocytes, myeloid cells, tumors, skin and the intestine (Merrill et al., 2001; Niemann et al., 2002; Skokowa et al., 2006; Takeda et al., 2006; Wang et al., 2005). In T cells, both Lef1 isoforms are high in resting cells, but the short transcript is downregulated upon activation and proliferation (Willinger et al., 2006). In murine epidermal stem cells, Lef1ΔN suppresses hair cell differentiation, but stimulates sebocyte differentiation, epidermal cysts and skin tumors (Merrill et al., 2001; Niemann et al., 2002); moreover, point mutations in the N-terminus of Lef1 that prevent beta-catenin binding are found in human sebaceous tumors (Takeda et al., 2006). Interestingly, Lef1ΔN is sufficient to rescue the differentiation block induced by Lef1-deficiency in granulocytes (Skokowa et al., 2006). Thus, Lef1ΔN acts as an inhibitor of proliferative signals emanating from the canonical Wnt pathway in some cells, but it promotes the differentiation of other cells and is sufficient to rescue Lef1 functions in certain tissues. In this report, we demonstrate that Lef1ΔN is induced in osteoblasts during the terminal differentiation process and can increase the expression of osteoblast genes (Figure 6).
We previously reported that the full length isoform of Lef1 blocks Runx2-dependent activation of the osteocalcin promoter (Kahler and Westendorf, 2003). Here, we demonstrate that Lef1ΔN, unlike the full-length isoform of Lef1, does not to repress Runx2-mediated activation of the osteocalcin promoter. Correspondingly, Lef1ΔN exogenously expressed in differentiating osteoblasts induces the expression of osteocalcin. We also previously showed that Lef1 protein levels decrease during osteoblast differentiation (Kahler et al., 2006), but the current results indicate mRNA levels encoding the full length protein do not change and might increase during osteoblast maturation. These data and those within Kahler et al. (Kahler et al., 2006) support the notion that Lef1 levels are regulated posttranscriptionally. The discordance between Lef1 mRNA and protein levels was previously observed in differentiating cells at the hair follicle bases (DasGupta and Fuchs, 1999; Jimenez et al., 2005). The mechanisms regulating Lef1 protein levels in osteoblasts are unclear. However, Nrarp (Notch-regulated ankyrin repeat protein) has been shown to block Lef1 ubiquitination and stabilize Lef1 protein (Ishitani et al., 2005). We have found that Nrarp is expressed in osteoblasts (unpublished data). Taken together, these data suggest that Lef1 isoforms have distinguishable activities during osteoblast maturation, particularly with regards to regulation of the osteocalcin gene, and their regulation is complex.
BMP2 induced expression of Lef1ΔN, as well as alkaline phosphatase and osteocalcin, two markers of osteoblast differentiation. Interestingly, Wnt3a suppressed basal and BMP2-induced Lef1ΔN and alkaline phosphatase expression. These data are consistent with a previous study demonstrating that Wnt3a blocks early osteogenesis of mesenchymal progenitor cells (Boland et al., 2004). The inability of Wnt3a to suppress BMP2-induced osteocalcin expression indicates that Wnt3a might not suppress late osteoblast differentiation genes as potently as those expressed earlier.
Regulation of the Lef1ΔN promoter (p2) closely paralleled the observed timeframe of Lef1ΔN mRNA expression. Thus, the Lef1ΔN transcript (Figures 1 & 2) and protein (Figure 3D) become detectable at days 6 to 7 of osteoblast differentiation, while total histone H3 levels decreased and phosphorylated RNA polymerase II levels increased at distal and proximal regions of the Lef1 p2 promoter within the first 5 days of osteoblast differentiation. The short delay between detection of p2 activity and detection of Lef1ΔN transcript suggests that cofactors required for Lef1ΔN expression must be recruited. We identified a Runx2 binding element in p2 and demonstrated that Runx2 positively regulates the expression of Lef1ΔN. Others have demonstrated that Lef/Tcf binding sites exist within p2; however, overexpressed β-catenin and Tcf only weakly activated p2 due to a repressor element within the promoter (Li et al., 2006). A database search for putative transcription factor binding sites within the p2 promoter revealed that cofactors such as AP-1, c-myc, CREB, and Sp1 may be recruited to the p2 promoter to regulate transcription of Lef1ΔN.
In summary, we describe the expression pattern and function of an alternative Lef1 isoform in osteoblasts. Lef1ΔN transcription and translation increases during osteoblast maturation and is induced by Runx2 and BMP2. Lef1ΔN overexpression increases osteocalcin expression, a measure of osteoblast maturation. This contrasts the effect of full-length Lef1, which represses Runx2-dependent osteocalcin expression. We conclude that Lef1ΔN is an induced regulator of osteoblast maturation.
Acknowledgments
Contract grant sponsor and number: NIH AR050074
Literature Cited
- Arce L, Yokoyama NN, Waterman ML. Diversity of LEF/TCF action in development and disease. Oncogene. 2006;25(57):7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- Baker JC, Beddington RS, Harland RM. Wnt signaling in Xenopus embryos inhibits bmp4 expression and activates neural development. Genes Dev. 1999;13(23):3149–3159. doi: 10.1101/gad.13.23.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature. 1996;382(6592):638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382(6588):225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Billin AN, Thirlwell H, Ayer DE. Beta-catenin-histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol. 2000;20(18):6882–6890. doi: 10.1128/mcb.20.18.6882-6890.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93(6):1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- Bonewald LF, Johnson ML. Osteocytes, mechanosensing and Wnt signaling. Bone. 2008;42(4):606–615. doi: 10.1016/j.bone.2007.12.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M, Brown JD, Bates R, Kimelman D, Moon RT. XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development. 1999;126(14):3159–3170. doi: 10.1242/dev.126.14.3159. [DOI] [PubMed] [Google Scholar]
- Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucleic Acids Res. 2001;29(7):1410–1419. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Goodnough LH, Gregorieff A, Leucht P, ten Berge D, Fuerer C, Clevers H, Nusse R, Helms JA. Wnt signaling mediates regional specification in the vertebrate face. Development. 2007;134(18):3283–3295. doi: 10.1242/dev.005132. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Fish MP, Rulifson EJ, Nusse R. Wingless repression of Drosophila frizzled 2 expression shapes the Wingless morphogen gradient in the wing. Cell. 1998;93(5):767–777. doi: 10.1016/s0092-8674(00)81438-5. [DOI] [PubMed] [Google Scholar]
- Cadigan KM, Jou AD, Nusse R. Wingless blocks bristle formation and morphogenetic furrow progression in the eye through repression of Daughterless. Development. 2002;129(14):3393–3402. doi: 10.1242/dev.129.14.3393. [DOI] [PubMed] [Google Scholar]
- Carlsson P, Waterman ML, Jones KA. The hLEF/TCF-1 alpha HMG protein contains a context-dependent transcriptional activation domain that induces the TCR alpha enhancer in T cells. Genes Dev. 1993;7(12A):2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12(4):364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126(20):4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- de Jong DS, van Zoelen EJ, Bauerschmidt S, Olijve W, Steegenga WT. Microarray analysis of bone morphogenetic protein, transforming growth factor beta, and activin early response genes during osteoblastic cell differentiation. J Bone Miner Res. 2002;17(12):2119–2129. doi: 10.1359/jbmr.2002.17.12.2119. [DOI] [PubMed] [Google Scholar]
- Galceran J, Farinas I, Depew MJ, Clevers H, Grosschedl R. Wnt3a−/−-like phenotype and limb deficiency in Lef1(−/−)Tcf1(−/−) mice. Genes Dev. 1999;13(6):709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K, Amsterdam A, Grosschedl R. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 1991;5(12B):2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- Giese K, Grosschedl R. LEF-1 contains an activation domain that stimulates transcription only in a specific context of factor-binding sites. Embo J. 1993;12(12):4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCR alpha enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9(8):995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- Giese K, Pagel J, Grosschedl R. Functional analysis of DNA bending and unwinding by the high mobility group domain of LEF-1. Proc Natl Acad Sci U S A. 1997;94(24):12845–12850. doi: 10.1073/pnas.94.24.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, Taketo MM, Long F, McMahon AP, Lang RA, Karsenty G. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8(5):751–764. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- Hadjiargyrou M, Lombardo F, Zhao S, Ahrens W, Joo J, Ahn H, Jurman M, White DW, Rubin CT. Transcriptional profiling of bone regeneration. Insight into the molecular complexity of wound repair. J Biol Chem. 2002;277(33):30177–30182. doi: 10.1074/jbc.M203171200. [DOI] [PubMed] [Google Scholar]
- Hecht A, Vleminckx K, Stemmler MP, van Roy F, Kemler R. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. Embo J. 2000;19(8):1839–1850. doi: 10.1093/emboj/19.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppler S, Kavanagh CL. Wnt signalling: variety at the core. J Cell Sci. 2007;120(Pt 3):385–393. doi: 10.1242/jcs.03363. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Munguia JE, Truong T, Milovanovic T, Lawrence Marsh J, Holcombe RF, Waterman ML. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat Genet. 2001;28(1):53–57. doi: 10.1038/ng0501-53. [DOI] [PubMed] [Google Scholar]
- Hovanes K, Li TW, Waterman ML. The human LEF-1 gene contains a promoter preferentially active in lymphocytes and encodes multiple isoforms derived from alternative splicing. Nucleic Acids Res. 2000;28(9):1994–2003. doi: 10.1093/nar/28.9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Galceran J, Grosschedl R. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol Cell Biol. 1998;18(8):4807–4818. doi: 10.1128/mcb.18.8.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber O, Korn R, McLaughlin J, Ohsugi M, Herrmann BG, Kemler R. Nuclear localization of beta-catenin by interaction with transcription factor LEF-1. Mech Dev. 1996;59(1):3–10. doi: 10.1016/0925-4773(96)00597-7. [DOI] [PubMed] [Google Scholar]
- Ishitani T, Matsumoto K, Chitnis AB, Itoh M. Nrarp functions to modulate neural-crest-cell differentiation by regulating LEF1 protein stability. Nat Cell Biol. 2005;7(11):1106–1112. doi: 10.1038/ncb1311. [DOI] [PubMed] [Google Scholar]
- James MJ, Jarvinen E, Wang XP, Thesleff I. Different roles of Runx2 during early neural crest-derived bone and tooth development. J Bone Miner Res. 2006;21(7):1034–1044. doi: 10.1359/jbmr.060413. [DOI] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422(6929):317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez J, Jang GM, Semler BL, Waterman ML. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. Rna. 2005;11(9):1385–1399. doi: 10.1261/rna.7226105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahler RA, Galindo M, Lian J, Stein GS, van Wijnen AJ, Westendorf JJ. Lymphocyte enhancer-binding factor 1 (Lef1) inhibits terminal differentiation of osteoblasts. J Cell Biochem. 2006;97(5):969–983. doi: 10.1002/jcb.20702. [DOI] [PubMed] [Google Scholar]
- Kahler RA, Westendorf JJ. Lymphoid enhancer factor-1 and beta-catenin inhibit Runx2-dependent transcriptional activation of the osteocalcin promoter. J Biol Chem. 2003;278(14):11937–11944. doi: 10.1074/jbc.M211443200. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Ikeda T, Yoshiki S, Wozney JM, Rosen V, Wang EA, Tanaka H, Omura S, Suda T. The non-osteogenic mouse pluripotent cell line, C3H10T1/2, is induced to differentiate into osteoblastic cells by recombinant human bone morphogenetic protein-2. Biochem Biophys Res Commun. 1990;172(1):295–299. doi: 10.1016/s0006-291x(05)80208-6. [DOI] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney JM, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127(6 Pt 1):1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, 2nd, Hartmann C, Li L, Hwang TH, Brayton CF, Lang RA, Karsenty G, Chan L. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157(2):303–314. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosla S, Westendorf JJ, Oursler MJ. Building bone to reverse osteoporosis and repair fractures. J Clin Invest. 2008;118(2):421–428. doi: 10.1172/JCI33612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Zullig S, Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109(1):47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- Kratochwil K, Dull M, Farinas I, Galceran J, Grosschedl R. Lef1 expression is activated by BMP-4 and regulates inductive tissue interactions in tooth and hair development. Genes Dev. 1996;10(11):1382–1394. doi: 10.1101/gad.10.11.1382. [DOI] [PubMed] [Google Scholar]
- Lambert JR, Nordeen SK. CBP recruitment and histone acetylation in differential gene induction by glucocorticoids and progestins. Mol Endocrinol. 2003;17(6):1085–1094. doi: 10.1210/me.2001-0183. [DOI] [PubMed] [Google Scholar]
- Levanon D, Goldstein RE, Bernstein Y, Tang H, Goldenberg D, Stifani S, Paroush Z, Groner Y. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc Natl Acad Sci U S A. 1998;95(20):11590–11595. doi: 10.1073/pnas.95.20.11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TW, Ting JH, Yokoyama NN, Bernstein A, van de Wetering M, Waterman ML. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol Cell Biol. 2006;26(14):5284–5299. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376(6543):791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- Mariadason JM, Bordonaro M, Aslam F, Shi L, Kuraguchi M, Velcich A, Augenlicht LH. Down-regulation of beta-catenin TCF signaling is linked to colonic epithelial cell differentiation. Cancer Res. 2001;61(8):3465–3471. [PubMed] [Google Scholar]
- Mayer K, Hieronymus T, Castrop J, Clevers H, Ballhausen WG. Ectopic activation of lymphoid high mobility group-box transcription factor TCF-1 and overexpression in colorectal cancer cells. Int J Cancer. 1997;72(4):625–630. doi: 10.1002/(sici)1097-0215(19970807)72:4<625::aid-ijc13>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Merrill BJ, Gat U, DasGupta R, Fuchs E. Tcf3 and Lef1 regulate lineage differentiation of multipotent stem cells in skin. Genes Dev. 2001;15(13):1688–1705. doi: 10.1101/gad.891401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishi M, Fujii R, Hatta M, Yoshida E, Araya N, Nagafuchi A, Ishihara S, Nakajima T, Fukamizu A. Regulation of Lef-mediated transcription and p53-dependent pathway by associating beta-catenin with CBP/p300. J Biol Chem. 2000;275(45):35170–35175. doi: 10.1074/jbc.C000258200. [DOI] [PubMed] [Google Scholar]
- Montecino M, Frenkel B, Lian J, Stein J, Stein G. Requirement of distal and proximal promoter sequences for chromatin organization of the osteocalcin gene in bone-derived cells. J Cell Biochem. 1996;63(2):221–228. doi: 10.1002/(SICI)1097-4644(19961101)63:2%3C221::AID-JCB9%3E3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Niemann C, Owens DM, Hulsken J, Birchmeier W, Watt FM. Expression of DeltaNLef1 in mouse epidermis results in differentiation of hair follicles into squamous epidermal cysts and formation of skin tumours. Development. 2002;129(1):95–109. doi: 10.1242/dev.129.1.95. [DOI] [PubMed] [Google Scholar]
- Noh T, Gabet Y, Cogan J, Shi Y, Tank A, Sasaki T, Criswell B, Dixon A, Lee C, Tam J, Kohler T, Segev E, Kockeritz L, Woodgett J, Muller R, Chai Y, Smith E, Bab I, Frenkel B. Lef1 haploinsufficient mice display a low turnover and low bone mass phenotype in a gender- and age-specific manner. PLoS ONE. 2009;4(5):e5438. doi: 10.1371/journal.pone.0005438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Differential expression of the HMG box factors TCF-1 and LEF-1 during murine embryogenesis. Development. 1993;118(2):439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- Payre F, Vincent A, Carreno S. ovo/svb integrates Wingless and DER pathways to control epidermis differentiation. Nature. 1999;400(6741):271–275. doi: 10.1038/22330. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O, Vorbruggen G, Jackle H. Drosophila segment borders result from unilateral repression of hedgehog activity by wingless signaling. Mol Cell. 2000;6(1):203–209. [PubMed] [Google Scholar]
- Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17(1):45–51. doi: 10.1016/j.gde.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Porfiri E, Rubinfeld B, Albert I, Hovanes K, Waterman M, Polakis P. Induction of a beta-catenin-LEF-1 complex by wnt-1 and transforming mutants of beta-catenin. Oncogene. 1997;15(23):2833–2839. doi: 10.1038/sj.onc.1201462. [DOI] [PubMed] [Google Scholar]
- Qi H, Aguiar DJ, Williams SM, La Pean A, Pan W, Verfaillie CM. Identification of genes responsible for osteoblast differentiation from human mesodermal progenitor cells. Proc Natl Acad Sci U S A. 2003;100(6):3305–3310. doi: 10.1073/pnas.0532693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75(3):213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Kahler RA, Li X, Westendorf JJ. Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J Biol Chem. 2004;279(40):41998–42007. doi: 10.1074/jbc.M403702200. [DOI] [PubMed] [Google Scholar]
- Shibamoto S, Winer J, Williams M, Polakis P. A blockade in Wnt signaling is activated following the differentiation of F9 teratocarcinoma cells. Exp Cell Res. 2004;292(1):11–20. doi: 10.1016/j.yexcr.2003.08.009. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A. 1999;96(10):5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Cario G, Uenalan M, Schambach A, Germeshausen M, Battmer K, Zeidler C, Lehmann U, Eder M, Baum C, Grosschedl R, Stanulla M, Scherr M, Welte K. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12(10):1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- Sun Y, Kolligs FT, Hottiger MO, Mosavin R, Fearon ER, Nabel GJ. Regulation of beta - catenin transformation by the p300 transcriptional coactivator. Proc Natl Acad Sci U S A. 2000;97(23):12613–12618. doi: 10.1073/pnas.220158597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda H, Lyle S, Lazar AJ, Zouboulis CC, Smyth I, Watt FM. Human sebaceous tumors harbor inactivating mutations in LEF1. Nat Med. 2006;12(4):395–397. doi: 10.1038/nm1386. [DOI] [PubMed] [Google Scholar]
- Travis A, Amsterdam A, Belanger C, Grosschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor alpha enhancer function [corrected] Genes Dev. 1991;5(5):880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, Reeves GA, Wesselink JJ, Yeats C, Olason PI, Albrecht M, Hegyi H, Giorgetti A, Raimondo D, Lagarde J, Laskowski RA, Lopez G, Sadowski MI, Watson JD, Fariselli P, Rossi I, Nagy A, Kai W, Storling Z, Orsini M, Assenov Y, Blankenburg H, Huthmacher C, Ramirez F, Schlicker A, Denoeud F, Jones P, Kerrien S, Orchard S, Antonarakis SE, Reymond A, Birney E, Brunak S, Casadio R, Guigo R, Harrow J, Hermjakob H, Jones DT, Lengauer T, Orengo CA, Patthy L, Thornton JM, Tramontano A, Valencia A. The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci U S A. 2007;104(13):5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutter AV, Fryer CJ, Jones KA. Chromatin-specific regulation of LEF-1-beta-catenin transcription activation and inhibition in vitro. Genes Dev. 2001;15(24):3342–3354. doi: 10.1101/gad.946501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaes BL, Dechering KJ, Feijen A, Hendriks JM, Lefevre C, Mummery CL, Olijve W, van Zoelen EJ, Steegenga WT. Comprehensive microarray analysis of bone morphogenetic protein 2-induced osteoblast differentiation resulting in the identification of novel markers for bone development. J Bone Miner Res. 2002;17(12):2106–2118. doi: 10.1359/jbmr.2002.17.12.2106. [DOI] [PubMed] [Google Scholar]
- Van de Wetering M, Castrop J, Korinek V, Clevers H. Extensive alternative splicing and dual promoter usage generate Tcf-1 protein isoforms with differential transcription control properties. Mol Cell Biol. 1996;16(3):745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Clevers H. Sequence-specific interaction of the HMG box proteins TCF-1 and SRY occurs within the minor groove of a Watson-Crick double helix. Embo J. 1992;11(8):3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374(6517):70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- Wang EA, Rosen V, D'Alessandro JS, Bauduy M, Cordes P, Harada T, Israel DI, Hewick RM, Kerns KM, LaPan P, et al. Recombinant human bone morphogenetic protein induces bone formation. Proc Natl Acad Sci U S A. 1990;87(6):2220–2224. doi: 10.1073/pnas.87.6.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ji P, Steffen B, Metzger R, Schneider PM, Halfter H, Schrader M, Berdel WE, Serve H, Muller-Tidow C. Alterations of lymphoid enhancer factor-1 isoform expression in solid tumors and acute leukemias. Acta Biochim Biophys Sin (Shanghai) 2005;37(3):173–180. [PubMed] [Google Scholar]
- Wehrli M, Dougan ST, Caldwell K, O'Keefe L, Schwartz S, Vaizel-Ohayon D, Schejter E, Tomlinson A, DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407(6803):527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. doi: 10.1016/j.gene.2004.06.044. [DOI] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willinger T, Freeman T, Herbert M, Hasegawa H, McMichael AJ, Callan MF. Human naive CD8 T cells down-regulate expression of the WNT pathway transcription factors lymphoid enhancer binding factor 1 and transcription factor 7 (T cell factor-1) following antigen encounter in vitro and in vivo. J Immunol. 2006;176(3):1439–1446. doi: 10.4049/jimmunol.176.3.1439. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3' UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A, Katagiri T, Ikeda T, Wozney JM, Rosen V, Wang EA, Kahn AJ, Suda T, Yoshiki S. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J Cell Biol. 1991;113(3):681–687. doi: 10.1083/jcb.113.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, van Beest M, Clevers H, Jones T, Hursh DA, Mortin MA. decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development. 2000;127(17):3695–3702. doi: 10.1242/dev.127.17.3695. [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Curr Biol. 1996;6(10):1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Zhou P, Byrne C, Jacobs J, Fuchs E. Lymphoid enhancer factor 1 directs hair follicle patterning and epithelial cell fate. Genes Dev. 1995;9(6):700–713. doi: 10.1101/gad.9.6.700. [DOI] [PubMed] [Google Scholar]