Abstract
Electron detachment dissociation (EDD) has previously provided stereo-specific product ions that allow for the assignment of the acidic C-5stereochemistry in heparan sulfate glycosaminoglycans (GAGs), but application of the same methodology to an epimer pair in the chondroitin sulfate glycoform class does not provide the same result. A series of experiments have been conducted in which glycosaminoglycan precursor ions are independently activated by electron detachment dissociation (EDD), electron induced dissociation (EID), and negative electron transfer dissociation (NETD) to assign the stereochemistry in chondroitin sulfate (CS) epimers and investigate the mechanisms for product ion formation during EDD in CS glycoforms. This approach allows for the assignment of electronic excitation products formed by EID and detachment products to radical pathways in NETD, both of which occur simultaneously during EDD. The uronic acid stereochemistry in electron detachment spectra produces intensity differences when assigned glycosidic and cross-ring cleavages are compared. The variations in the intensities of the doubly deprotonated 0,2X3 and Y3 ions have been shown to be indicative of CS-A/DS composition during the CID of binary mixtures. These ions can provide insight into the uronic acid composition of binary mixtures in EDD, but the relative abundances, although reproducible, are low compared with those in a CID spectrum acquired on an ion trap. The application of principal component analysis (PCA) presents a multivariate approach to determining the uronic acid stereochemistry spectra of these GAGs by taking advantage of the reproducible peak distributions produced by electron detachment.
Keywords: Glycosaminoglycans, Carbohydrates, FTMS, Electron detachment dissociation, Chondroitin sulfate, Multivariate analysis
Introduction
G lycosaminoglycan (GAG) carbohydrates are acidic linear polysaccharides that participate in a variety of biological activities [1–7]. GAGs are biologically expressed as a heterogeneous mixture, varying in modifications to the hexuronic acid/hexosamine disaccharide unit and in degree of polymerization. These modifications include glycosidic bond linkage position and stereochemistry, position and extent of O- and N-sulfation and N-acetylation, and hexuronic acid stereochemistry. The analytical characterization of these modifications has been a standing challenge due to the natural polydispersity of these biomolecules arising from a nontemplate-based synthesis. Of the applied techniques, mass spectrometry [8–15] and tandem mass spectrometry [16–22] have proven to be the most useful due to their highly sensitive method of structural characterization.
The tandem mass spectrometry methods can be classified based upon the type of ion activation: threshold, which includes collisional induced dissociation (CID) [23] and infrared multiphoton dissociation (IRMPD) [24], and electron-based, which includes electron capture dissociation (ECD) [25], electron detachment dissociation (EDD) [26], electron transfer dissociation (ETD) [27], and negative electron transfer dissociation (NETD) [28]. Both classes of ion activation have been demonstrated for a variety of GAG glycoforms ranging in extent of sulfation and polymerization [16, 19–21, 29–35].
Chondroitin sulfate (CS) GAGs have largely been characterized by threshold activation [16–18, 29, 37–40] as electron-based methods have only recently been applied to CS GAG analysis [33, 34, 41]. Threshold activation is capable of locating the sulfate modification (4-O or 6-O dependent upon CS type) in pure samples as well as mixtures [16–18, 42]. Additionally, the ability to determine hexuronic acid stereochemistry has been demonstrated based upon fragment ion intensities and rationalized based upon the differences in oligomeric flexibility inferred by the incorporation of iduronic acid (IdoA) residues [18]. More recently, MSn has been shown to provide more structural detail than can be obtained by a single stage of tandem mass spectrometry, using collisional activation [43].
EDD analysis of CS glycoforms has presented experimental results that differ from prior results in heparan sulfate (HS). This outcome is not entirely unexpected as the two glycoforms are composed of differing hexosamine residues (glucosamine in HS with and galactosamine in CS), with a glycosidic bond linkage unique to each (alternating α1-4 and β1-4 in HS versus alternating β1-3 and β1-4 in CS) and have unique biologic function as chondroitin sulfate GAGs are found in connective tissue and serve a role in organization of the extracellular matrix [44], while heparan sulfate GAGs are found in the extracellular matrix where they regulate activities such as blood coagulation and cell adhesion [45]. Prior EDD results for HS have provided information relevant to the location of sulfate groups [32] as well as the assignment of hexuronic acid stereochemistry [46] based on the presence or absence of diagnostic fragment ions such as 0,2A3, B3′, and B3′-CO2. These ions are not sufficient for the assignment of hexuronic acid residue in CS glycoforms as some occur in both epimers and others are not present in either.
In the present work, EDD, as well as electron induced dissociation (EID) [41] and NETD [35] have been employed to activate the tetrasaccharides of the CS-A and DS epimer pair to fully characterize the electron-based ion activation. The tandem mass spectra generated from each allow the assignment of products due to electronic excitation (EID) or radical-based mechanisms (NETD). Similar to threshold activation, the stereochemistry of the hexuronic acid residue can be inferred by the intensities of cleavages at the non-reducing terminus. Although their occurrence can provide insight into the composition, the intensities are low, albeit reproducible, compared with CID products generated in an ion trap. Multivariate statistical methods, namely principal component analysis (PCA), have previously been applied to HS tetrasaccharide epimer differentiation [47] and are employed here to take advantage of the highly reproducible nature of EDD spectra and quantitatively assign the stereochemistry of hexuronic acid residues in 4-O sulfated CS epimers.
Experimental
Preparation of GAG Oligosaccharides
Chondroitin sulfate A (CS-A) and dermatan sulfate (DS) oligosaccharides were independently prepared by partial enzymatic depolymerization of bovine trachea chondroitin sulfate A (Celsus Laboratories, Cincinnati, OH, USA) and porcine intestinal mucosa dermatan sulfate (Celsus Laboratories). A 20 mg/mL solution of each, in 50 mM Tris-HCl/60 mM sodium acetate buffer, pH 8 was incubated at 37 °C with chondroitin ABC lyase from Proteus vulgaris, EC 4.2.2.4. (Associates of Cape Cod Inc., East Falmouth, MA, USA). After the absorbance at 232 nm indicated the digestion was 50 % completed, the digestion mixture was heated at 100 °C for 3 min. The resulting oligosaccharide mixture was filtered by a 0.22 μm unit (Millipore, Billerica, MA, USA) and fractionated by low pressure GPC on a Bio-Gel P10 (Bio-Rad, Richmond, CA, USA) column. Fractions containing oligosaccharides of interest were desalted by GPC on a Bio-Gel P2 column and freeze-dried [48]. Further purification of compounds was carried out using strong anion exchange high-pressure liquid chromatography (SAX-HPLC) on a semi-preparative SAX S5 Spherisorb column (Waters Corp, Milford, MA, USA). The SAX-HPLC fractions containing >90 % of oligosaccharides were collected, desalted by GPC, and freeze-dried. The solid was reconstituted in water and purified a second time by SAX-HPLC. Only the top 30 % of the chromatographic peak was collected, desalted, and freeze-dried. Concentration of the oligosaccharide solutions was determined by measuring the absorbance at 232 nm (e=3800M−1 cm−1). The resulting fractions containing individual oligosaccharides were characterized by PAGE, ESI-MS, and high-field nuclear magnetic resonance (NMR) spectroscopy [49].
Oligosaccharide Desulfation
The pyridinium salts of CS-A and DS dp4 were independently dissolved in 10 % aqueous MeOH and heated for 6 h at 60 °C [50] to produce the desulfated tetrasaccharides ΔUA-GalNAc-GlcA-GalNAc and ΔUA-GalNAc-IdoA-GalNAc, respectively. The mass-selective quadrupole of the FT-ICR MS instrument was employed to isolate the desulfated compounds in lieu of additional laboratory based purification. The desulfation reaction does not affect the stereochemistry of the hexuronic acid residue, and so the chirality of the product is the same as that of the reactant as established by NMR analysis.
Mass Spectrometry Analysis
Experiments were performed with a 9.4 T Bruker Apex Ultra QeFTMS (Billerica, MA, USA) fitted with an Apollo II dual source, 25 W CO2 laser (Synrad model J48-2; Mukilteo, WA, USA) for IRMPD, and an indirectly heated hollow cathode (HeatWave, Watsonville, CA, USA) to generate electrons for EDD. The sample solutions were infused at a rate of 120 μL/h and ionized by electrospray using a metal capillary (#G2427A; Agilent Technologies, Santa Clara, CA, USA). Solutions of each oligosaccharide were introduced at a concentration of 0.1 mg/mL in 50:50:0.1 methanol:H2O:formic acid (Sigma, St. Louis, MO, USA) to generate doubly deprotonated ions and 0.2 mg/mL in 50:50 methanol:H2O with 1 % 100 μM NaOH to generate triply deprotonated ions and sodium adduct ions. All oligosaccharides were examined in negative ion mode.
For the EDD experiments, precursor ions were isolated in the external quadrupole and accumulated for 1–3s before injection into the FT-ICR MS cell. One quadrupole isolation/cell fill was utilized per scan. The selection of the precursor ion was further refined by using in-cell isolation with a coherent harmonic excitation frequency (CHEF) event [51]. For electron irradiation the cathode bias was set to −19 V and the extraction lens was varied from −18.4 V to −18.6 V to optimize the electron current entering the analyzer cell. The cathode heater was set to 1.5 A. The precursor ions were then irradiated with electrons for 1s. EID experiments are performed with the same parameters as EDD, except a singly charged species is irradiated. 24 acquisitions were signal averaged per mass spectrum. For each mass spectrum, 512 K points were acquired, padded with one zero fill, and apodized using a sine bell window. Background spectra were acquired by leaving all parameters the same but setting the cathode bias to 0 V to ensure that no electrons reached the analyzer cell. External calibration of mass spectra produced mass accuracy of 5 ppm. Internal calibration was also performed using confidently assigned glycosidic bond cleavage products as internal calibrants, providing mass accuracy of <1 ppm. Due to the larger number of low intensity products formed by EDD, only peaks with S/N>10 are reported. Product ions were assigned using accurate mass measurement and Glycoworkbench [52]. All products are reported using the annotation proposed by Wolff and Amster [34], derived from the Domon and Costello nomenclature [53], in which a prime or double prime next to the product type indicates the loss of one or two atoms of hydrogen from the conventional glycosidic fragment. Fragmentation is illustrated in the molecular structure drawings with dashed lines. A slanted line at the top or bottom of the dashed line is used to indicate reducing end or nonreducing end products, respectively. Single and double hash marks across the slanted line indicate an additional loss of 1 or 2 hydrogen atoms from the conventional glycosidic fragments. To denote sulfate decomposition accompanying glycosidic or cross-ring cleavage, an open circle indicates the loss of one SO3 from a sulfate half ester, while a filled circle is used to show a loss of 2 or more equivalents of SO3.
NETD experiments were performed on a 12.0 T Bruker solariX FTMS instrument (Bruker Daltonics, Billerica, MA, USA) fitted with an ESI/MALDI dual source. For NETD experiments, precursor ions were isolated in the external quadrupole and accumulated for 1–3s in the hexapole collision cell before reaction with the reagent radical cation. The fluoranthene radical cation was generated in the chemical ionization (CI) source and reacted for 500–1000 ms in the hexapole collision cell. The precursor ion and NETD products were then injected into the mass analyzer through an RF-only ion guide. One analyzer cell fill was utilized per scan; 24–36 acquisitions were signal averaged per mass spectrum. For each mass spectrum, 1 M points were acquired, padded with one zero fill, and apodized using a sine bell window. Background spectra were acquired by leaving all parameters the same without the generation of the radical cation. Data reduction was conducted in the same manner described above.
Multivariate Statistical Analysis
Principal component analysis (PCA) was performed using PLS Toolbox (Eigenvector Research, Inc., Wenatchee, WA, USA). The abundances of 34 assigned fragment ions were normalized with respect to total ion abundance in each EDD spectrum. An input data matrix was constructed with each row containing the mass spectrum of a single tetrasaccharide epimer (samples) and each column, the normalized abundance of an assigned fragment ion (variables). For each tetrasaccharide, five EDD spectra were obtained in the same day. Prior to PCA, each data set was mean-centered and cross-validated. For comparison, the abundances of 19 assigned peaks from IRMPD spectra were also treated in the same manner as the EDD data.
Results and Discussion
EDD of Chondroitin Sulfate A (CS-A) Oligosaccharides
Electron detachment dissociation (EDD) spectra of dermatan sulfate (DS) oligosaccharides ranging in degree of polymerization (dp) 4 to 10 have been previously reported [33]. The examined chondroitin sulfate A (CS-A) oligosaccharides only differ from those samples in the stereochemistry of the hexuronic acid residues. Both DS and CS-A have a uniform disaccharide repeat in reference to 4-O-sulfation on the hexosamine (GalNAc4S-HexA), but differ in the hexuronic acid constituent as the DS oligosaccharides contain IdoA and the CS-A contain glucuronic acid (GlcA). Included in the Supplementary Material are the annotated structures for the EDD activation of CS-A dp4-10.
Across the degrees of polymerization examined for CS-A, results consistent with the EDD of DS are observed. In summary, cleavage of all glycosidic bonds is observed except for C2 and Z2 in the dp6 oligomer. Due to the uniform repeat unit, the Cn/2 and Zn/2 cleavages, where n is the degree of polymerization, are isobaric with the precursor ion and are not uniquely assigned. The loss of CO2 is observed from the Z2 cleavage and occurs in dp4-10. The loss of SO3 is observed from both glycosidic, with losses of H or 2 H, and cross-ring cleavages, typically assigned as 1,5Xn or 0,2Xn of GlcA residues. Consistent with observations in DS [33], cross-ring cleavages are largely observed on hexuronic acid residues, indicating a propensity for proton mobility which transfers the ionized site from the more acidic sulfate group and allows for radical site formation at the carboxylate, leading to increased cross-ring fragment generation through electron detachment based mechanisms.
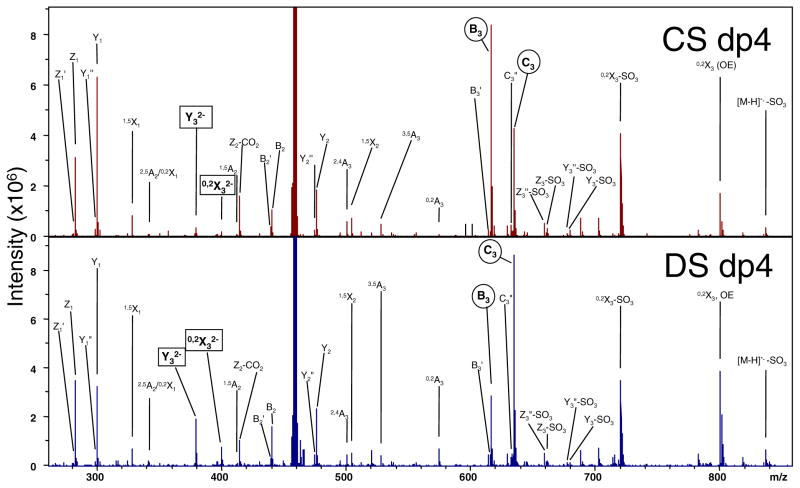
As the only difference between CS-A and DS is the stereochemistry of the hexuronic acid, it is not entirely unexpected that the EDD mass spectra look remarkably similar and no product ions are unique to one epimer when the activation of precursor ions of the same charge state and degree of polymerization are compared. Shown in Figure 1, the EDD mass spectra of the [M – 2H]2− precursor ions for the CS-A and DS dp4 oligomers are compared. (Note: the EDD mass spectrum of DS has been previously published [33] and is shown here for the benefit of direct comparison.) Activation of each precursor ion under nearly the same experimental conditions produces mass spectra that contain the same suite of product ions but differing in intensities in some cases. These differences, specifically that of the charge conserved 0,2Xn and Yn product ions on the nonreducing terminus, were exploited by Zaia and co-workers to identify the hexuronic acid stereochemistry in CID spectra generated in a quadrupole ion trap [18] and in a quadrupole-time-of-flight hybrid mass spectrometer [39]. In the doubly deprotonated dp4 oligomers shown, these ions occur as 0,2X32− and Y32− and reproducible intensity differences occur between the two epimers (denoted by squares in Figure 1).
Figure 1.
Comparison of the EDD activation for the [M – 2H]2− precursor ion of the tetrasaccharide epimer pair, CS-A and DS
Hexuronic Acid Determination by EDD
Direct tandem mass spectrometric evidence of hexuronic acid stereochemistry has previously been achieved in heparan sulfate (HS) tetrasaccharides by EDD [46]. The product ions diagnostic of GlcA containing tetrasaccharides were 0,2A3, B3′, and B3′-CO2 and secondary confirmation of IdoA was indicated by a more intense C3″ than C3. The generation of these product ions is thought to result from electron detachment from a carboxylate anionic site. As the number of sulfates is increased on an oligosaccharide, the charge state must exceed the number of sulfate groups by at least one in order for the weaker acid (carboxyl) to exist in ionized form [54]. Based on the hypothesis of mobile protons in CS glycoforms, the presence of an ionized carboxyl group may be inferred for the [M – 2H]2−- precursor, but is not certain unless the [M – 3H]3–ion is generated.
Comparison of the EDD generated fragments depicted in Figure 1 for the [M – 2H]2− precursor reveals that the diagnostic 0,2A3 ion (m/z 574.072), previously found to be indicative of GlcA in HS tetramers [46], occurs in both epimers and is in fact more intense in the IdoA-containing DS. This result was observed in the initial EDD study of DS oligosaccharides, as well as by IRMPD. Additionally, B3′ and C3″ are also assigned in both, but B3′-CO2 is not assigned in either. Examination of the relative intensities in each spectrum, though, can provide initial information for differentiation. When the intensities of the B3 and C3 glycosidic cleavages (denoted by circles in Figure 1) are compared within each epimer, B3 > C3 in CS-A and B3 < C3 inDS. B3′ and C3″ remain minor components in each epimer. Comparison between the two epimers reveals the 0,2A3 is more intense in DS than CS-A. A more quantitative method for epimer differentiation for epimer differentiation will be presented later in the text.
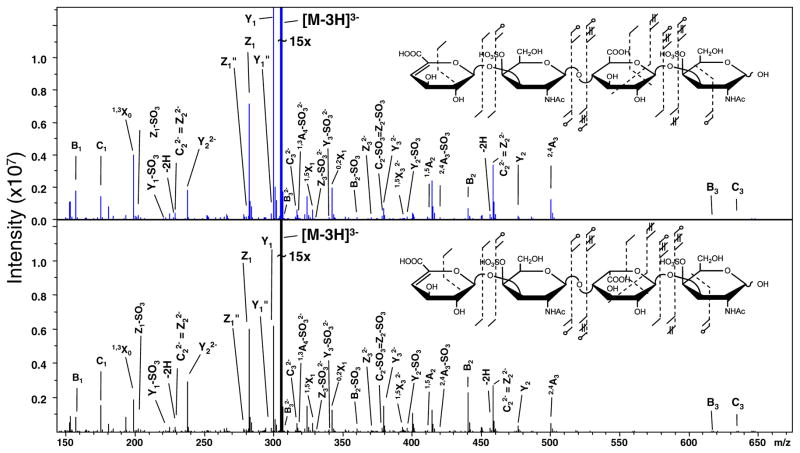
The EDD activation of the [M – 3H]3− precursor was conducted to insure the presence of an ionized carboxyl group and generated the tandem mass spectra shown in Figure 2. A unique stereo-specific ion does not occur in either spectrum. Subtle intensity differences are observed when 0,2X32− and Y32− are compared between epimers yet the 0,2A3, B3′ and B3′-CO2 cleavages do not occur.
Figure 2.
Comparison of the EDD activation for the [M–3H]3− precursor ion of the tetrasaccharide epimer pair, CS-A and DS
Mechanistic Aspects
The source of product ions observed during the electron-based ion activation of multiply de-protonated GAG ions can be rationalized by the schematic included as Supplementary Material. The activation path during EDD is initiated by irradiation with moderate energy electrons (typically 19 eV), which produces an excited state intermediate. From this state, two independent processes, electron detachment and electron induced dissociation (EID) lead to the products observed in an EDD tandem mass spectrum. Electron detachment results in product ions, which are reduced in absolute charge (formally oxidized) and can be assigned as either odd-electron species or even electron species. EID products result from the direct electronic excitation of the precursor ion and may either be charge conserved or reduced, existing largely as even electron species but in some cases odd-electron species have been observed. To determine the activation channel responsible for a specific product ion, it is possible to conduct negative electron transfer dissociation (NETD) for the assignment of products due to detachment as the excited state intermediate is avoided or the irradiation of a mass-selected singly de-protonated GAG anion by moderate energy electrons during which observed products will be solely due to EID processes. Additionally, the extent of ionized sites in the anion can be manipulated by the ESI spray solvent conditions as well as Na/H exchange [34].
Negative Electron Transfer Dissociation (NETD) of Doubly Deprotonated CS GAG Tetrasaccharides
NETD has previously been applied to GAG structural characterization in ion traps [35], but only recently coupled to the FT-ICR mass analyzer for GAG analysis [36]. Briefly, ion–ion reactions between multiply deprotonated GAG analytes and radical cations of fluoranthene occur in the high-pressure region of the instrument, and the products are transported by an rf-only ion guide to the ICR mass analyzer. A detailed account of the NETD FT-ICR MS of GAGs the subject of a recent publication [36].
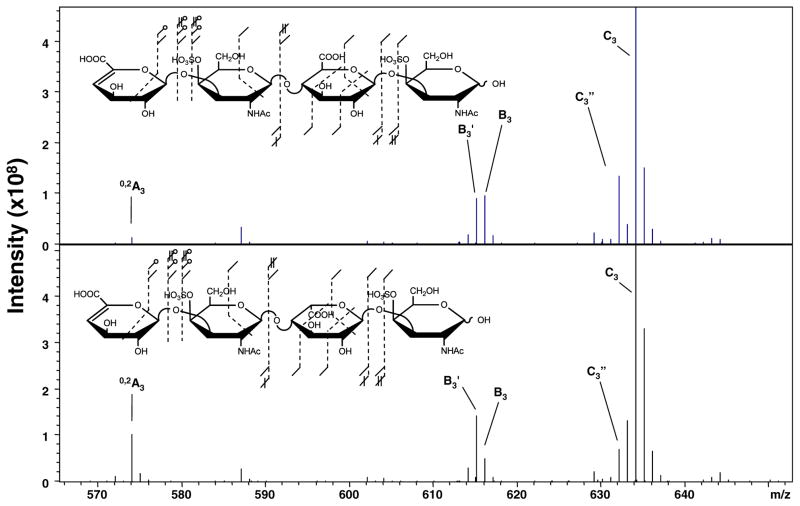
The NETD of the [M – 2H]2−- CS-A/DS dp4 epimer pair is shown in Figure 3. Comparison of the product ions in NETD with those from EDD experimentally confirms that they are a subset of EDD. Inspection of the previously stereo-specific region of the spectrum reveals that the 0,2A3, B3′, and C3″ ions are present in both epimers; therefore, radical detachment mechanisms are involved in their generation but exist in both diastereomers. Interestingly, the B3′ ion is more intense when compared to B3 in DS, which contains the IdoA residue, and 0,2A3 is more intense in DS than CS-A. This result is opposite that found in HS and may indicate a more facile hydrogen rearrangement in the IdoA case for CS glycoforms.
Figure 3.
NETD of the [M – 2H]2− precursor ion of the tetrasaccharide epimer pair, CS-A and DS, from m/z 470 to 560. Insets denote the assigned product ions. (Note: additional O18 labeling experiments since the publication of references 33 and 35 have resulted in the assignment of 0,2X1 and not 2,5A2)
Electron Induced Dissociation (EID) of Singly Deprotonated CS GAG Tetrasaccharides
The mass selection of the singly deprotonated precursor ion, [M – H]1−, for each tetrasaccharide and subsequent electron irradiation under the same conditions as the EDD experiment will produce product ions due solely to direct electronic excitation of the precursor as detachment results in a neutral species. Comparison of the EID activation (included in the Supplementary Material) reveals that the B3′ and C3″ are also generated by electronic excitation and occur in both epimers with similar intensities. Interestingly, the 0,2A3 ion is not present in either, nor in the IRMPD of the [M – H]1− precursor (data not shown). The occurrence of this ion in both the EDD and IRMPD of the [M – 2H]2− precursor and not the [M – H]1− indicates both a non-radical required mechanism and necessity for two ionized sites.
Manipulation of Na/H Heterogeneity
The ability to control the fragmentation of GAG anions by selection of an appropriate precursor ion charge state and the addition of metal counter-ions, specifically Na+, is well known within the threshold activation community and has been utilized to minimize and/or eliminate the loss of SO3 from sulfated GAGs [55]. These techniques have also been utilized to minimize the loss of SO3 by increasing the number of ionized sites above the total number of sulfates during the EDD of DS and synthetic HS GAGs, providing stereo-specific products in increasingly sulfated HS by previously proposed mechanisms. Application of this technique for CS epimer characterization introduces the possibility of an ionized carboxyl, yet no stereo-specific product ions are generated in either the [M – 3H + 1Na]2− nor [M – 4H + 2Na]2− precursor ion (Supplemental Material).
Oligosaccharide Desulfation
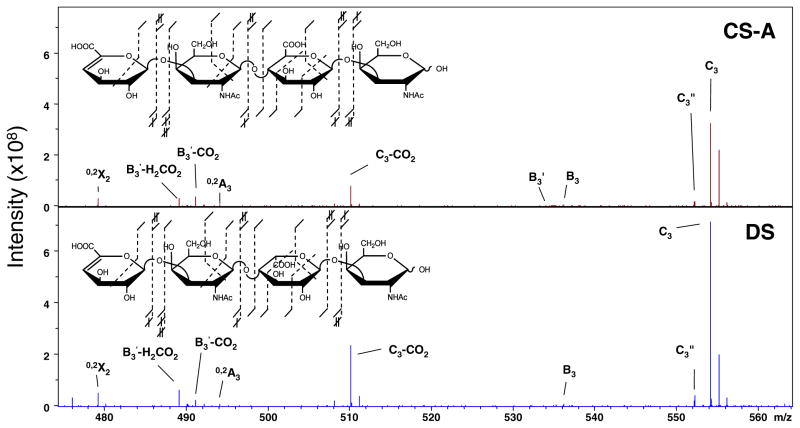
The di-sulfated tetrasaccharide epimer pair of CS-A and DS were solvolytically desulfated by the method described in the Experimental section to remove the participation of sulfate groups and potentially identify stereo-specific product ions due solely to electron detachment from a carboxylate. The doubly deprotonated precursor ion, [M – 2H]2−, of each produces unambiguous ionization of both carboxyl groups. The spectral region, which had previously been found to contain useful product for HS epimers, was closely examined for CS-A and DS, and is shown in Figure 4, with insets displaying all of the observed fragment ions. When compared, the two regions are nearly identical in terms of product ions observed and relative intensities. Also, the overall fragmentation depicted in the inset shows the occurrence of the same product ion suite for the epimers. This result provides direct evidence that the difference in the glycosidic linkage between HS and CS/DS glycoforms negates the ability to identify the hexuronic acid stereochemistry as GlcA based on the occurrence of 0,2A3 and B3′-CO2 in CS/DS glycoforms and other product ions must be assigned for this differentiation.
Figure 4.
EDD of the desulfated chondroitin epimer pair from m/z 470 to 570. Insets denote the assigned product ions
Multivariate Statistical Analysis
Based upon the systematic investigation of electron-based CS/DS GAG ion activation, the presence or absence of diagnostic products is not a reliable manner to identify the hexuronic acid stereochemistry by EDD for this GAG class. Multivariate (MVA) statistical methods are employed to quantitatively make this assignment. MVA methods have previously demonstrated the ability to separate synthetic HS GAG epimers at the tetrasaccharide oligomer length without the need for the identification of stereo-specific products [47]. The method selected for this study is principal component analysis (PCA). The most readily obtained charge state for CS/DS GAGs corresponds to the number of repeat units, for a mono-sulfated disaccharide repeat. In the examined tetrasaccharides, the [M – 2H]2− precursor satisfies this condition. Based on earlier qualitative descriptions of product ion intensity differences between epimers, the selection of this charge state for additional statistical examination is further validated.
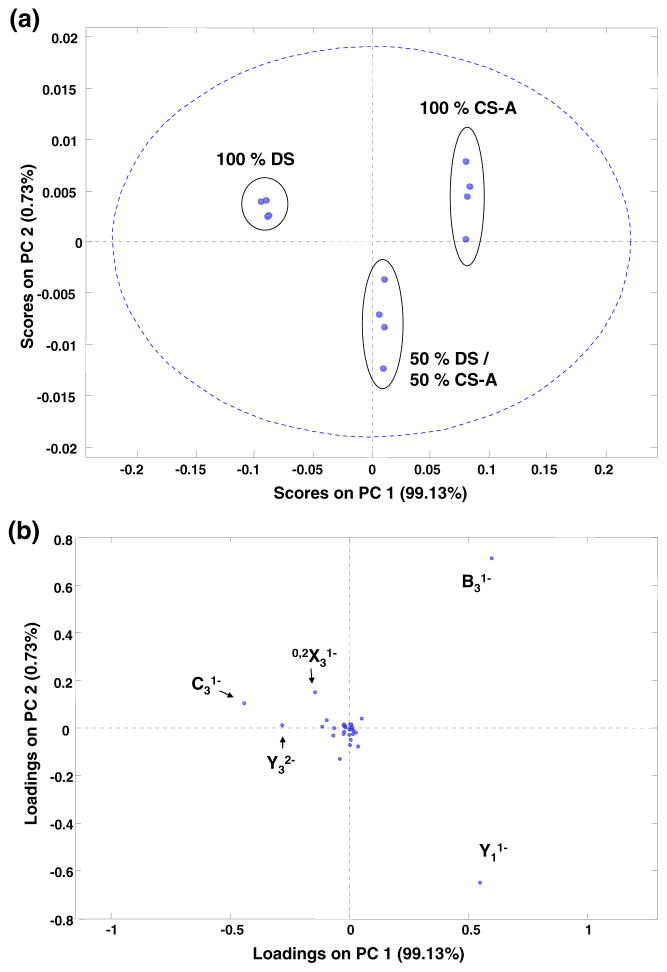
EDD activation of the [M – 2H]2− precursor for each epimer results in 34 assigned product ions. The application of PCA to replicate data sets for three binary mixtures is demonstrated in Figure 5. Separation of the pure components is obtained by the first principal component plotted as the abscissa. Replicates for each mixture are well grouped, and the projection of the 50/50 mixture lies at approximately the mid-point of the plot. Similar to results in synthetic HS tetrasaccharides, this result is due to the explanation of the majority of the data set variance by PC1 (99.13 %). Examination of the loading values allows for the assignment of component contributions to the separation. Similar to CID analysis, the Y32− ion contributes highly to the separation. Interestingly, the singly deprotonated 0,2X3 1− ion contributes more to the epimer distinction in EDD than the doubly deprotonated as in CID. Other significant loadings are the B3, C3, and Y1 ions generated by glycosidic cleavage adjacent to the stereocenter. Although the losses of H from B3 and C3 resulting in the B3′ and C3″ ions are observed in CS, these do not contribute to the statistical differentiation.
Figure 5.
PCA results for binary mixture analysis of the EDD product ions from the activation of the [M – 2H]2− precursor ion for the tetrasaccharide epimers, CS-A and DS. The plot of PC2 versus PC1 is shown in (a) and the respective loadings values are shown in (b)
For sake of comparison to threshold methods, the IRMPD activation of the [M – 2H]2− precursor was also performed. These spectra contained fewer assigned fragment ions (19), but PCA based differentiation was still possible. Shown in the Supplemental Material, the first two principal components are plotted. Similar to results in EDD, PC1 results in an approximate binary mixture scale. Examination of the PC2 axis reveals a spread within each replicate group. This result is likely due to variation in the overlap between IR photons and the ion cloud as the ions undergo magnetron motion within the FT-ICR analyzer cell. Examination of the loading values for the PCA analysis of IRMPD spectra generates a similar list of product ions with high contributions, namely, Y32−, B3, and Y1, with additional contributions due to 0,2X3 2−, C3, and Y1. The negative loadings of the doubly deprotonated 0,2X3 and Y3 contribute to the separation of DS whereas Y1 and B3 denote CS-A. These results are consistent with the prior CID ion trap results based on peak intensity analysis and serve to validate the application of MVA methods to epimer characterization.
Conclusions
Although CS and HS GAGs are structurally similar, the introduction of small stereochemical/linkage changes has a profound effect on the outcome of a tandem mass spectrometry experiment. Electron detachment dissociation (EDD) has previously shown dual utility in application to HS tetrasaccharides by providing increased cross-ring cleavage, which allows the location of the labile sulfate modification, and stereo-specific ions, which allows the identification of the hexuronic acid stereochemistry. When applied to CS/DS tetrasaccharides, the extent of cross-ring cleavage is diminished in comparison and GlcA is not differentiated from IdoA by a detachment-initiated radical-mechanism. Instead, hexuronic stereochemistry can be inferred by the relative intensities of charge-conserved products arising from electron excitation. The similarity of the diagnostic ions to those observed in collision activation experiments suggests that EDD may impart vibrational excitation to some fraction of the precursor ions. Due to the highly reproducible nature of EDD spectra, the application of multivariate statistical methods can provide assignment of uronic acid stereochemistry and a quantitative measure in mixture analysis.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge financial support from the National Institutes of Health grant GM038060-22.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s13361-012-0428-5) contains supplementary material, which is available to authorized users.
References
- 1.Fannon M, Forsten KE, Nugent MA. Potentiation and inhibition of bFGF binding by heparin: a model for regulation of cellular response. Biochemistry. 2000;39:1434–1445. doi: 10.1021/bi991895z. [DOI] [PubMed] [Google Scholar]
- 2.Wu ZL, Zhang L, Yabe T, Kuberan B, Beeler DL, Love A, Rosenberg RD. The involvement of heparan sulfate (HS) in FGF1/HS/FGFR1 signaling complex. J Biol Chem. 2003;278:17121–17129. doi: 10.1074/jbc.M212590200. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl U, Backstrom G, Hook M, Thunberg L, Fransson LA, Linker A. Structure of the antithrombin-binding site in heparin. Proc Nat Acad Sci USA. 1979;76:3198–3202. doi: 10.1073/pnas.76.7.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Maguire T, Hileman RE, Fromm JR, Esko JD, Linhardt RJ, Marks RM. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nature Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 5.Hoogewerf AJ, Kuschert GSV, Proudfoot AEI, Borlat F, Clark-Lewis I, Power CA, Wells TNC. Glycosaminoglycans mediate cell surface oligomerization of chemokines. Biochemistry. 1997;36:13570–13578. doi: 10.1021/bi971125s. [DOI] [PubMed] [Google Scholar]
- 6.Liu DF, Shriver Z, Gi YW, Venkataraman G, Sasisekharan R. Dynamic regulation of tumor growth and metastasis by heparan sulfate glycosaminoglycans. Semin Thromb Hemostasis. 2002;28:67–78. doi: 10.1055/s-2002-20565. [DOI] [PubMed] [Google Scholar]
- 7.Williams RK, Straus SE. Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J Virol. 1997;71:1375–1380. doi: 10.1128/jvi.71.2.1375-1380.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai Y, Whittal RM, Bridges CA, Isogai Y, Hindsgaul O, Li L. Matrix-assisted laser desorption ionization mass spectrometry for the analysis ofmonosulfated oligosaccharides. Carbohydr Res. 1997;304:1–9. doi: 10.1016/s0008-6215(97)00195-x. [DOI] [PubMed] [Google Scholar]
- 9.Juhasz P, Biemann K. Utility of noncovalent complexes in the matrix-assisted laser desorption ionization mass spectrometry of heparin-derived oligosaccharides. Carbohydr Res. 1995;270:131–147. doi: 10.1016/0008-6215(94)00012-5. [DOI] [PubMed] [Google Scholar]
- 10.Schiller J, Arnhold J, Benard S, Reichl S, Arnold K. Cartilage degradation by hyaluronate lyase and chondroitin ABC lyase: a MALDI-TOF mass spectrometric study. Carbohydr Res. 1999;318:116–122. doi: 10.1016/s0008-6215(99)00063-4. [DOI] [PubMed] [Google Scholar]
- 11.Takagaki K, Kojima K, Majima M, Nakamura T, Kato I, Endo M. Ion-spray mass-spectrometric analysis of glycosaminoglycan oligosaccharides. Glycoconj J. 1992;9:174–179. doi: 10.1007/BF00731162. [DOI] [PubMed] [Google Scholar]
- 12.Reinhold VN, Carr SA, Green BN, Petitou M, Choay J, Sinay P. Structural characterization of sulfated glycosaminoglycans by fast atom bombardment mass spectrometry: application to heparin fragments prepared by chemical synthesis. Carbohydr Res. 1987;161:305–313. doi: 10.1016/s0008-6215(00)90088-0. [DOI] [PubMed] [Google Scholar]
- 13.Lamb DJ, Wang HM, Mallis LM, Linhardt RJ. Negative ion fast-atom bombardment tandem mass spectrometry to determine sulfate and linkage position in glycosaminoglycan-derived disaccharides. J Am Soc Mass Spectrom. 1992;3:797–803. doi: 10.1016/1044-0305(92)80002-3. [DOI] [PubMed] [Google Scholar]
- 14.Laremore TN, Murugesan S, Park TJ, Avci FY, Zagorevski DV, Linhardt RJ. Matrix-assisted laser desorption/ionization mass spectrometric analysis of uncomplexed highly sulfated oligosaccharides using ionic liquid matrices. Anal Chem. 2006;78:1774–1779. doi: 10.1021/ac051121q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaia J, Costello CE. Compositional analysis of glycosaminoglycans by electrospray mass spectrometry. Anal Chem. 2001;73:233–239. doi: 10.1021/ac000777a. [DOI] [PubMed] [Google Scholar]
- 16.Zaia J, McClellan JE, Costello CE. Tandem mass spectrometric determination of the 4S/6S sulfation sequence in chondroitin sulfate oligosaccharides. Anal Chem. 2001;73:6030–6039. doi: 10.1021/ac015577t. [DOI] [PubMed] [Google Scholar]
- 17.McClellan JE, Costello CE, O’Connor PB, Zaia J. Influence of charge state on product ion mass spectra and the determination of 4S/6S sulfation sequence of chondroitin sulfate oligosaccharides. Anal Chem. 2002;74:3760–3771. doi: 10.1021/ac025506+. [DOI] [PubMed] [Google Scholar]
- 18.Zaia J, Li XQ, Chan SY, Costello CE. Tandem mass spectrometric strategies for determination of sulfation positions and uronic acid epimerization in chondroitin sulfate oligosaccharides. J Am Soc Mass Spectrom. 2003;14:1270–1281. doi: 10.1016/S1044-0305(03)00541-5. [DOI] [PubMed] [Google Scholar]
- 19.Zaia J, Costello CE. Tandem mass spectrometry of sulfated heparin-like glycosaminoglycan oligosaccharides. Anal Chem. 2003;75:2445–2455. doi: 10.1021/ac0263418. [DOI] [PubMed] [Google Scholar]
- 20.Saad OM, Leary JA. Delineating mechanisms of dissociation for isomeric heparin disaccharides using isotope labeling and ion trap tandem mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1274–1286. doi: 10.1016/j.jasms.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Saad OM, Leary JA. Heparin sequencing using enzymatic digestion and ESI-MS with HOST: a heparin/HS oligosaccharide sequencing tool. Anal Chem. 2005;77:5902–5911. doi: 10.1021/ac050793d. [DOI] [PubMed] [Google Scholar]
- 22.Viseux N, de Hoffmann E, Domon B. Structural assignment of permethylated oligosaccharide subunits using sequential tandem mass spectrometry. Anal Chem. 1998;70:4951–4959. doi: 10.1021/ac980443+. [DOI] [PubMed] [Google Scholar]
- 23.Cody RB, Freiser BS. Collision-induced dissociation in a Fourier-transform mass-spectrometer. Int J Mass Spectrom Ion Processes. 1982;41:199–204. [Google Scholar]
- 24.Little DP, Speir JP, Senko MW, O’Connor PB, McLafferty FW. Infrared Multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal Chem. 1994;66:2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 25.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 26.Budnik BA, Haselmann KF, Zubarev RA. Electron detachment dissociation of peptide di-anions: an electron-hole recombination phenomenon. Chem Phys Lett. 2001;342:299–302. [Google Scholar]
- 27.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Nat Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herron WJ, Goeringer DE, McLuckey SA. Gas-phase electron transfer reactions from multiply-charged anions to rare gas cations. J Am Chem Soc. 1995;117:11555–11562. [Google Scholar]
- 29.Desaire H, Sirich TL, Leary JA. Evidence of block and randomly sequenced chondroitin polysaccharides: sequential enzymatic digestion and quantification using ion trap tandem mass spectrometry. Anal Chem. 2001;73:3513–3520. doi: 10.1021/ac010385j. [DOI] [PubMed] [Google Scholar]
- 30.Saad OM, Leary JA. Compositional analysis and quantification of heparin and heparan sulfate by electrospray ionization ion trap mass spectrometry. Anal Chem. 2003;75:2985–2995. doi: 10.1021/ac0340455. [DOI] [PubMed] [Google Scholar]
- 31.Sweeney MD, Yu Y, Leary JA. Effects of sulfate position on heparin octasaccharide binding to CCL2 examined by tandem mass spectrometry. J Am Soc Mass Spectrom. 2006;17:1114–1119. doi: 10.1016/j.jasms.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 32.Wolff JJ, Amster IJ, Chi L, Linhardt RJ. Electron detachment dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2007;18:234–244. doi: 10.1016/j.jasms.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Electron detachment dissociation of dermatan sulfate oligosaccharides. J Am Soc Mass Spectrom. 2008;19:294–304. doi: 10.1016/j.jasms.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolff JJ, Laremore TN, Busch AM, Linhardt RJ, Amster IJ. Influence of charge state and sodium cationization on the electron detachment dissociation and infrared multiphoton dissociation of glycosaminoglycan oligosaccharides. J Am Soc Mass Spectrom. 2008;19:790–798. doi: 10.1016/j.jasms.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolff JJ, Leach FE, Laremore TN, Kaplan DA, Easterling ML, Linhardt RJ, Amster IJ. Negative electron transfer dissociation of glycosaminoglycans. Anal Chem. 2010;82:3460–3466. doi: 10.1021/ac100554a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leach FE, III, Wolff JJ, Xiao Z, Ly M, Laremore TN, Arungundram S, Al-Mafraji K, Venot A, Boons GJ, Linhardt RJ, Amster IJ. Negative electron transfer dissociation fourier transform mass spectrometry of glycosaminoglycan carbohydrates. Eur J Mass Spectrom. 2011;17:167–176. doi: 10.1255/ejms.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desaire H, Leary JA. Detection and quantification of the sulfated disaccharides in chondroitin sulfate by electrospray tandem mass spectrometry. J Am Soc Mass Spectrom. 2000;11:916–920. doi: 10.1016/S1044-0305(00)00168-9. [DOI] [PubMed] [Google Scholar]
- 38.Hitchcock AM, Costello CE, Zaia J. Glycoform quantification of chondroitin/dermatan sulfate using a liquid chromatography-tandem mass spectrometry platform. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang R, Pomin VH, Sharp JS. LC-MSn sequencing of isomeric chondroitin sulfate oligosaccharides using a chemical derivatization strategy. J Am Soc Mass Spectrom. 2011;22:1577–1587. doi: 10.1007/s13361-011-0174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller MJC, Costello CE, Malmstrom A, Zaia J. A tandem mass spectrometric approach to determination of chondroitin/dermatan sulfate oligosaccharide glycoforms. Glycobiology. 2006;16:502–513. doi: 10.1093/glycob/cwj093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19:1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ly M, Leach FE, Laremore TN, Toida T, Amster IJ, Linhardt RJ. The proteoglycan bikunin has a defined sequence. Nat Chem Biol. 2011;7:827–833. doi: 10.1038/nchembio.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bielik AM, Zaia J. Multistage tandem mass spectrometry of chondroitin sulfate and dermatan sulfate. Int J Mass Spectrom. 2011;305:131–137. doi: 10.1016/j.ijms.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hardingham TE, Fosang AJ. Proteoglycans: many forms and many functions. FASEB J. 1992;6:861–870. [PubMed] [Google Scholar]
- 45.Bernfield M, Gotte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, Zako M. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 46.Wolff JJ, Chi LL, Linhardt RJ, Amster IJ. Distinguishing glucuronic from iduronic acid in glycosaminoglycan tetrasaccharides by using electron detachment dissociation. Anal Chem. 2007;79:2015–2022. doi: 10.1021/ac061636x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oh HB, Leach FE, III, Arungundram S, Al-Mafraji K, Venot A, Boons GJ, Amster IJ. Multivariate analysis of electron detachment dissociation mass spectra for synthetic heparan sulfate tetrasaccharides differing only in hexuronic acid stereochemistry. J Am Soc Mass Spectrom. 2011;22(3):582–590. doi: 10.1007/s13361-010-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pervin A, Gallo C, Jandik KA, Han XJ, Linhardt RJ. Preparation and structural characterization of large heparin-derived oligosaccharides. Glycobiology. 1995;5:83–95. doi: 10.1093/glycob/5.1.83. [DOI] [PubMed] [Google Scholar]
- 49.Munoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic synthesis of heparin related polysaccharides on sensor chips: rapid screening of heparin–protein interactions. Biochem Biophys Res Commun. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagasawa K, Inoue Y, Kamata T. Solvolytic desulfation of glycosaminoglycuronan sulfates with dimethyl sulfoxide containing water or methanol. Carbohydr Res. 1977;58:47–55. doi: 10.1016/s0008-6215(00)83402-3. [DOI] [PubMed] [Google Scholar]
- 51.Heck AJR, de Koning LJ, Pinkse FA, Nibbering NMM. Mass-specific selection of ions in Fourier-transform ion cyclotron resonance mass spectrometry. Unintentional off-resonance cyclotron excitation of selected ions. Rapid Commun Mass Spectrom. 1991;5:406–414. [Google Scholar]
- 52.Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7:1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- 53.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- 54.Bartmess JE. In: Negative ion energetics data. Linstrom PJ, Mallard WG, editors. National Institute of Standards and Technology; Gaithersburg, MD: 2009. [Google Scholar]
- 55.Zaia J. Principles of mass spectrometry of glycosaminoglycans. J Biomol Mass Spectrom. 2005;1:3–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.