Abstract
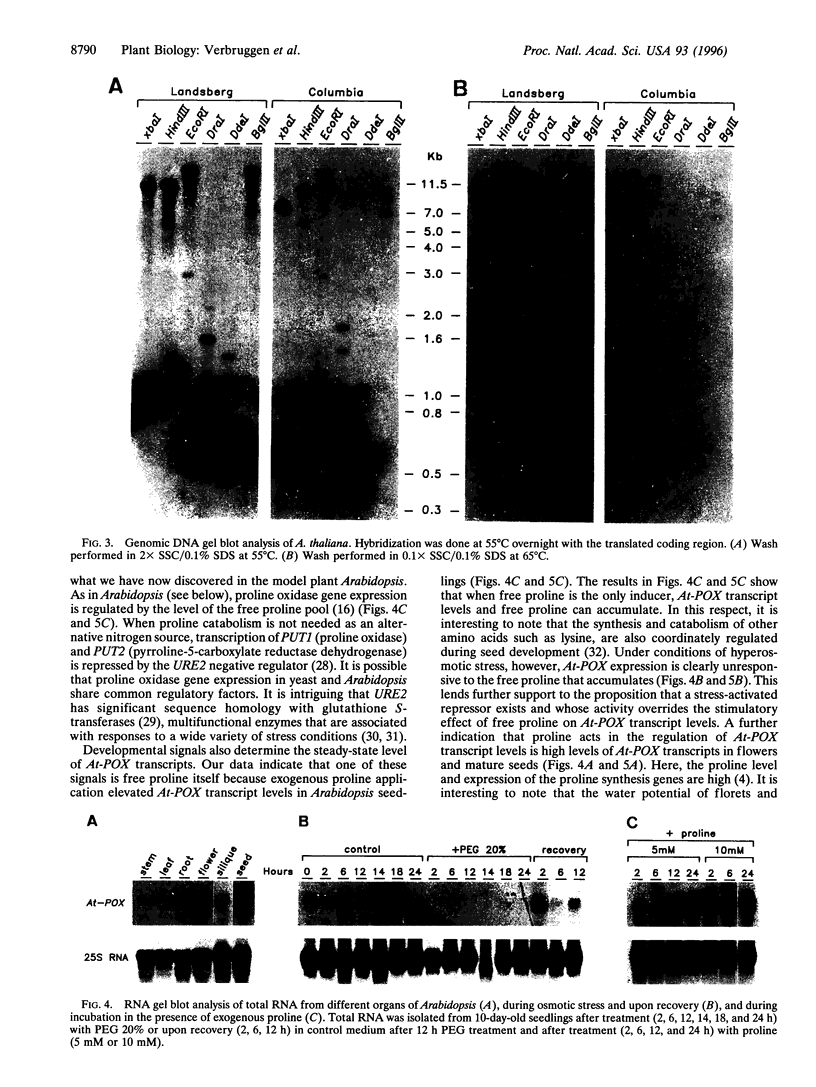
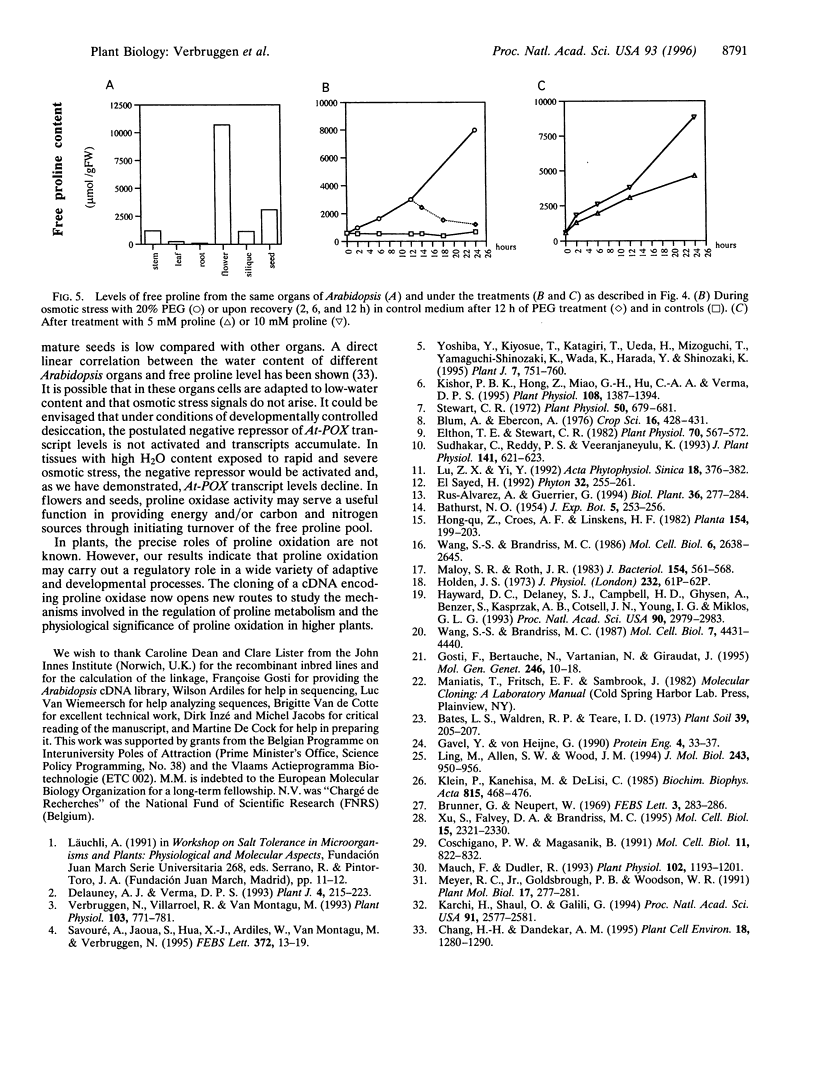
In many plants, osmotic stress induces a rapid accumulation of proline through de novo synthesis from glutamate. This response is thought to play a pivotal role in osmotic stress tolerance [Kishor, P. B. K., Hong, Z., Miao, G.-H., Hu, C.-A. A. and Verma, D. P. S. (1995) Plant Physiol. 108, 1387-1394]. During recovery from osmotic stress, accumulated proline is rapidly oxidized to glutamate and the first step of this process is catalyzed by proline oxidase. We have isolated a full-length cDNA from Arabidopsis thaliana, At-POX, which maps to a single locus on chromosome 3 and that encodes a predicted polypeptide of 499 amino acids showing significant similarity with proline oxidase sequences from Drosophila and Saccharomyces cerevisiae (55.5% and 45.1%, respectively). The predicted location of the encoded polypeptide is the inner mitochondrial membrane. RNA gel blot analysis revealed that At-POX mRNA levels declined rapidly upon osmotic stress and this decline preceded proline accumulation. On the other hand, At-POX mRNA levels rapidly increased during recovery. Free proline, exogenously added to plants, was found to be an effective inducer of At-POX expression; indeed, At-POX was highly expressed in flowers and mature seeds where the proline level is higher relative to other organs of Arabidopsis. Our results indicate that stress- and developmentally derived signals interact to determine proline homeostasis in Arabidopsis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunner G., Neupert W. Localisation of proline oxidase and Delta-pyrroline-5-carboxylic acid dehydrogenase in rat liver. FEBS Lett. 1969 Jun;3(4):283–286. doi: 10.1016/0014-5793(69)80159-6. [DOI] [PubMed] [Google Scholar]
- Coschigano P. W., Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione s-transferases. Mol Cell Biol. 1991 Feb;11(2):822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon T. E., Stewart C. R. Proline Oxidation in Corn Mitochondria : Involvement of NAD, Relationship to Ornithine Metabolism, and Sidedness on the Inner Membrane. Plant Physiol. 1982 Aug;70(2):567–572. doi: 10.1104/pp.70.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavel Y., von Heijne G. Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng. 1990 Oct;4(1):33–37. doi: 10.1093/protein/4.1.33. [DOI] [PubMed] [Google Scholar]
- Gosti F., Bertauche N., Vartanian N., Giraudat J. Abscisic acid-dependent and -independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol Gen Genet. 1995 Jan 6;246(1):10–18. doi: 10.1007/BF00290128. [DOI] [PubMed] [Google Scholar]
- Hayward D. C., Delaney S. J., Campbell H. D., Ghysen A., Benzer S., Kasprzak A. B., Cotsell J. N., Young I. G., Miklos G. L. The sluggish-A gene of Drosophila melanogaster is expressed in the nervous system and encodes proline oxidase, a mitochondrial enzyme involved in glutamate biosynthesis. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):2979–2983. doi: 10.1073/pnas.90.7.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden J. S. Free amino acid levels in the cockroach, Periplaneta americana. J Physiol. 1973 Jul;232(2):61P–62P. [PubMed] [Google Scholar]
- Karchi H., Shaul O., Galili G. Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proc Natl Acad Sci U S A. 1994 Mar 29;91(7):2577–2581. doi: 10.1073/pnas.91.7.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishor PBK., Hong Z., Miao G. H., Hu CAA., Verma DPS. Overexpression of [delta]-Pyrroline-5-Carboxylate Synthetase Increases Proline Production and Confers Osmotolerance in Transgenic Plants. Plant Physiol. 1995 Aug;108(4):1387–1394. doi: 10.1104/pp.108.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Ling M., Allen S. W., Wood J. M. Sequence analysis identifies the proline dehydrogenase and delta 1-pyrroline-5-carboxylate dehydrogenase domains of the multifunctional Escherichia coli PutA protein. J Mol Biol. 1994 Nov 11;243(5):950–956. doi: 10.1006/jmbi.1994.1696. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Roth J. R. Regulation of proline utilization in Salmonella typhimurium: characterization of put::Mu d(Ap, lac) operon fusions. J Bacteriol. 1983 May;154(2):561–568. doi: 10.1128/jb.154.2.561-568.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch F., Dudler R. Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 1993 Aug;102(4):1193–1201. doi: 10.1104/pp.102.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R. C., Jr, Goldsbrough P. B., Woodson W. R. An ethylene-responsive flower senescence-related gene from carnation encodes a protein homologous to glutathione S-transferases. Plant Mol Biol. 1991 Aug;17(2):277–281. doi: 10.1007/BF00039505. [DOI] [PubMed] [Google Scholar]
- Savouré A., Jaoua S., Hua X. J., Ardiles W., Van Montagu M., Verbruggen N. Isolation, characterization, and chromosomal location of a gene encoding the delta 1-pyrroline-5-carboxylate synthetase in Arabidopsis thaliana. FEBS Lett. 1995 Sep 18;372(1):13–19. doi: 10.1016/0014-5793(95)00935-3. [DOI] [PubMed] [Google Scholar]
- Stewart C. R. Proline Content and Metabolism during Rehydration of Wilted Excised Leaves in the Dark. Plant Physiol. 1972 Dec;50(6):679–681. doi: 10.1104/pp.50.6.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbruggen N., Villarroel R., Van Montagu M. Osmoregulation of a pyrroline-5-carboxylate reductase gene in Arabidopsis thaliana. Plant Physiol. 1993 Nov;103(3):771–781. doi: 10.1104/pp.103.3.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: analysis of the cloned PUT1 gene. Mol Cell Biol. 1986 Jul;6(7):2638–2645. doi: 10.1128/mcb.6.7.2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. S., Brandriss M. C. Proline utilization in Saccharomyces cerevisiae: sequence, regulation, and mitochondrial localization of the PUT1 gene product. Mol Cell Biol. 1987 Dec;7(12):4431–4440. doi: 10.1128/mcb.7.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Falvey D. A., Brandriss M. C. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1995 Apr;15(4):2321–2330. doi: 10.1128/mcb.15.4.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba Y., Kiyosue T., Katagiri T., Ueda H., Mizoguchi T., Yamaguchi-Shinozaki K., Wada K., Harada Y., Shinozaki K. Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J. 1995 May;7(5):751–760. doi: 10.1046/j.1365-313x.1995.07050751.x. [DOI] [PubMed] [Google Scholar]