Abstract
To study growth and development of ocular tissues, gene expression patterns in normal human fetal versus adult eyes were compared. Human retina/retinal pigment epithelium, choroid, sclera, optic nerve* and cornea* tissues were dissected from fetal (24 week gestational age) (N = 9; *N = 6), and adult (N = 6) normal donor eyes. The Illumina® whole genome expression microarray platform was used to assess differential expression. Statistical significance for all comparisons was determined using the Benjamin and Hochberg False Discovery Rate (FDR, 5%). Significant gene expression fold changes > 1.5 were found in adult versus fetal retina/RPE (N = 1185), choroid (N = 6446), sclera (N = 1349), and cornea (N = 3872), but not optic nerve. Genes showing differential expression were assessed using Ingenuity Pathway Analysis (IPA) for enriched functions and canonical pathways. In all tissues, development, cell death/growth, cancer functions, and signaling canonical pathways were enriched. There was also a general trend of down-regulation of collagen genes in adult tissues.
Keywords: gene expression, microarray, retina, retinal pigment epithelium, choroid, sclera, optic nerve, cornea
1. Introduction
The human visual system is complex and requires numerous tissue and cell types to communicate with each other and the brain throughout the process of development (Kolb et al., 2011). In humans, rapid axial growth of the eye globe is seen in fetal development with slowing toward the end of gestation (Fledelius and Christensen, 1996). However, despite this rapid fetal growth, the human eye is not full sized at birth nor is the visual system fully developed; visual signals and nutritional factors contribute to postnatal ocular development in the early years of life (Bremond-Gignac et al., 2011). During childhood development, an active regulatory process of ocular growth, emmetropization, aims to match the optical power of the cornea and lens to the axial length of the eye (Gordon and Donzis, 1985). Failure of emmetropization in postnatal ocular development commonly results in either myopic or hyperopic refractive error, or blurred vision (Gordon and Donzis, 1985).
Given the impracticality of human sample collection, researchers have utilized animal models to study developmental and late-onset ocular diseases. These animal models have demonstrated that visual cues interpreted by the retina and signaled through the choroid and sclera locally control the shape and size of the eye through unknown mechanisms (Faulkner et al., 2007; Tkatchenko et al., 2009; Wallman and Winawer, 2004; Wildsoet and Wallman, 1995). Human eyes with high degrees of refractive error have distinct clinical phenotypes including changes to their size, shape and tissue structure (Xu et al., 2007). Highly myopic eyes tend to be larger than their emmetropic counterparts, with the elongation primarily localized axially (Atchison et al., 2004), suggesting that changes in any or all of those tissues (central retinal, retinal pigment epithelium [RPE] and sclera) may be responsible for failed emmetropization and myopic development.
A better understanding of the tissue-specific expression differences during normal growth and development is key to identify ocular growth and development mechanisms in human tissues. As collection of postnatal eyes undergoing emmetropization is impractical, fetal ocular tissue types of central retina/RPE, choroid, sclera, optic nerve and cornea were compared to their adult counterparts from normal donor eyes to study patterns of ocular growth during development. Although it is unclear to what extent the mechanisms controlling postnatal emmetropization are active in prenatal development, one gene identified in a study of extreme hyperopic refractive error, MFRP (MIM 606227), has been demonstrated to be necessary for both prenatal ocular growth and postnatal emmetropization (Sundin et al., 2008). To our knowledge this is the first whole genome expression analysis comparing human adult versus fetal ocular tissues. This data can provide an understanding of normal changes these tissues undergo during prenatal growth and development in humans and it also may contain clues to understand how diseases such as myopia may result from disruptions to normal growth processes.
2. Materials and methods
2.1. Ocular sample selection
The tissues selected for this study were central retina/RPE, choroid, sclera, optic nerve and cornea. Selection of tissues was based on relevance to disorders studied in our lab such as micro-phthalmia and myopic exaggerated eye growth, and feasibility of collection. The posterior wall tissues were selected and prioritized for larger sample sizes based on their relation to a focal disease studied in our lab, high myopia. Additionally, the cornea and optic nerve tissues were selected due to their relations with other diseases under study in our lab including glaucoma and corneal abnormalities.
To compare growth and development gene expression in ocular tissue types, normal samples from two age groups were used: fetal eyes and adult eyes. The fetal donor eyes were obtained from Advanced Biosciences Resources (Alameda, CA, USA), while the adult eyes were obtained from the North Carolina Eye Bank (Winston–Salem, North Carolina, USA). Fetal gestational age was determined by most recent menstruation in addition to fetal foot measurements. The group of fetal eyes consisted of late prenatal fetal eyes of approximately 24-weeks gestational age from elective abortions with no known defects or abnormalities. 24-weeks gestational eyes are the oldest prenatal eyes readily available and are undergoing rapid growth and axial elongation (Fledelius and Christensen, 1996). Nine fetal donor eyes (four male and five female samples), and six fully grown adult donor eyes (three of each gender) were used for microarray analyses. Space and cost limitations required that optic nerve and cornea sample sizes were reduced to six adult and six fetal samples each. All adult donors were Caucasian, and donors with known ocular disorders were excluded (Table 1). Ethnic and health information was not available for fetal donors. The study was approved by Duke University's Institutional Review Board and adhered to the tenets of the Declaration of Helsinki guidelines.
Table 1. Donor Information for Adult Ocular Whole Globes Used in Tissue Expression Analyses.
Preservation time interval (PTI) is listed in hours. Age is listed in years.
| Individual | Race | Sex | Age | PTI | Cause of death |
|---|---|---|---|---|---|
| 2809 | Caucasian | Female | 76 | 4:00 | Leukemia/acute myeloid leukemia |
| 2835 | Caucasian | Female | 55 | 4:55 | Dementia |
| 2828 | Caucasian | Male | 80 | 5:20 | Pneumonia |
| 2834 | Caucasian | Male | 56 | 5:22 | Abdominal aortic aneurysm rupture/heart disease |
| 2217 | Caucasian | Female | 77 | 6:24 | Chronic obstructive pulmonary disease/emphysema |
| 2836 | Caucasian | Male | 67 | 5:10 | Heart disease |
2.2. Ocular dissection
All adult whole globes were immersed in RNAlater® (Qiagen, Hilden, Germany) within 6.5 h (Table 1) of collection, and shipped overnight on ice. Several studies have shown that the postmortem delay in preservation of samples within this time frame have limited effects on RNA integrity from brain tissue (Durrenberger et al., 2010; Ervin et al., 2007). Fetal whole globes were collected and preserved in RNAlater® within minutes of collection and shipped overnight on ice. Prior to immersion in RNAlater®, a 2 mm incision was cut equatorially into all whole globes to allow permeation of the solution to the inner tissues while minimizing unwanted physical changes to the tissue, such as retinal tearing. All whole globes were dissected on the same day as arrival. The retina, RPE, choroid and scleral tissues were isolated at the posterior pole using a circular, double embedded technique using round 7 mm and 5 mm biopsy punches. To reduce contamination of retina to the other tissues samples, the second biopsy punch of 5 mm was used in the center of the 7 mm punch after retinal removal. The adult RPE was collected in RNAlater® by gentle brushing from the choroid, pipetting the solution, and centrifuging at 4°C to remove the RNAlater®. Fetal eye samples proved difficult to separate the RPE from the retina. Consequently, the retina and RPE were collected in total. Additionally, optic nerve and corneal samples were isolated from each eye in each age group. Central corneal samples were isolated using a clean 5 mm biopsy punch. The whole optic nerve was collected using clean dissection scissors. The fibrous sclera, optic nerve, and cornea samples were cut into smaller portions (about 1 mm2) using a scalpel to aid in subsequent homogenization. After dissection all tissues were immediately frozen in liquid nitrogen for storage at −80°C until RNA extraction.
2.3. RNA extraction and whole genome expression processing
RNA was extracted from each tissue sample using the mirVanaTM total RNA extraction kit (Ambion, Austin, Texas, USA) following the manufacturer's protocol. The tissue samples were homogenized at 4°C in Ambion lysis buffer using a Bead Ruptor Tissue Homogenizer (Omni, Kennesaw, Georgia, USA) with 2.38 mm metal bead tubes using the following machine settings: 4 cycles × 30 s at speed 4.0 with 30 s break between cycles. Quality control for each sample included measuring RNA concentration and 260/280 nm ratios using a Nanodrop® (Invitrogen, Carlsbad, California, USA). The ocular tissue RNA samples were labeled and amplified using the Illumina® Total Prep kit (Ambion, Austin, Texas, USA). RNA samples were hybridized to Illumina® HumanHT-12 v4 Expression BeadChips (San Diego, California, USA), which target over 25,000 annotated genes (over 48,000 probes). All protocols were performed following the manufacturer's recommendations. Twelve samples per chip were processed and chips were run in two batches, each containing samples of at least two tissue types. The first batch (N = 51) included all adult and fetal retina/RPE, choroid and sclera samples. The second batch included all adult and fetal cornea and optic nerve samples (N = 24).
2.4. Whole genome expression analyses
The microarray data preprocessing procedures were conducted separately for each of the two batches (See Section 2.3). After the data was generated for each batch, background noise was subtracted from the intensity values using the Illumina® GenomeStudio program. The data was exported from GenomeStudio and log2 transformed. Sample outliers were determined by principle component analyses using the Hoteling's T2 test (Hotelling, 1931) (at 95% confidence interval) and removed from further analyses. The data intensity was normalized by Quantile normalization followed by Multichip Averaging (Irizarry et al., 2003) to reduce chip effects. The exact Wilcoxon rank sum test (Wilcoxen, 1945) was used to identify differentially expressed genes because of the relatively small sample size. Fetal ocular tissues were compared to their adult counterparts, and assessed for relative fold changes per probe. Adult retina and RPE samples were averaged for comparison to fetal retina/RPE. The Benjamin and Hochberg False Discovery Rate (Benjamini and Hochberg, 1995) (FDR) procedure was applied, and FDR was controlled at 0.05 to determine statistical significance for all comparisons.
2.5. Quantitative real-time PCR (qPCR)
A total of 1.5 μg of RNA for each sample was converted into double –stranded cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystem, Austin, Texas) according to the manufacturer's instructions. qPCR reactions were carried out with TaqMan primers and probe sets from Applied Biosystem, (Austin, Texas). GAPDH was used as the endogenous control house-keeping gene. PCR reactions were carried out with three technical duplicates for each sample. The comparative threshold cycle (CT) method (McCurdy et al., 2008), was used to calculate the relative gene expression difference between adult and 24-week groups, and the results were expressed as fold changes in gene expression.
2.6. Pathway analyses
In total, five adult versus fetal expression comparisons were made, representing each layer of tissue. Functional and pathway enrichment assessment was conducted using Ingenuity® Pathway Analysis ([IPA], Ingenuity® Systems, www.ingenuity.com) for each tissue comparison using lists of genes with differentially expressed probes meeting the following criteria. In the retina/RPE, choroid, sclera and cornea comparisons, only probes meeting FDR significance (See Section 2.4) were included. The adult versus fetal optic nerve comparisons did not produce any probes meeting FDR significance; consequently, probes with raw p-values <0.05 were included. To focus on changes that we were confident were biologically important, only those probes with fold changes ≥1.5 in each tissue were included. For each tissue comparison, Table 2 contains the number of probes, representative unique genes, as well as the range and average fold changes.
Table 2. Filtered Number of Probes and Unique Genes.
Retina/RPE, Choroid, Sclera and Cornea were filtered by False Discovery Rate (FDR) <0.05. *Optic Nerve were filtered by raw p-value <0.05. All tissues were filtered by a fold change (FC) >1.5.
| Tissue | FC range | Average FC | # probes | # unique genes |
|---|---|---|---|---|
| Retina/RPE | −8.50 ↔ 21.09 | 1.86 | 1236 | 1185 |
| Choroid | −617.57 ↔ 550.49 | 5.56 | 7124 | 6446 |
| Sclera | −125.11 ↔ 6.40 | 2.28 | 1423 | 1349 |
| Optic nerve* | −205.22 ↔ 1695.34 | 5.90 | 2399 | 2179 |
| Cornea | −1252.45 ↔ 205.77 | 7.34 | 4354 | 3872 |
For each tissue comparison, the lists of differentially expressed genes, their significance and fold change were imported into IPA. Each gene was mapped to its corresponding object In the Ingenuity® Knowledge Database (www.ingenuity.com). Functional analysis of these genes identified biological functions and/or diseases that were most significant to the molecules (genes) in the group. A right-tailed Fischer's exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned was due to chance alone. Additionally, IPA calculated a z-score, or standard score, for each functional classification. A z-score of <−2.00 indicates that function has a significantly decreased predicted activation in the data set. Conversely, a z-score of >2.0 indicates a predicted increased activation of that functional group in the data set.
Canonical pathway analysis identified pathways from the IPA library of canonical pathways that were most significant to the group. The significance of the association between the data set and the canonical pathway was measured in two ways: 1) A ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the pathway; 2) Fischer's exact test was used to calculate a p-value determining the probability that the association between the genes in the data set and the canonical pathway was due to chance (Ingenuity® Systems, www.ingenuity.com).
3. Results
3.1. Microarray analyses
The number and fold change of genes differentially expressed varied by tissue type. The retina/RPE had the lowest average fold change and the fewest number of differentially expressed genes, while the cornea had the largest fold changes and the highest number of differentially expressed genes (Table 2). The optic nerve comparisons failed to yield any FDR significantly differentially expressed genes.
The microarray chips were redundant, thus, many genes had multiple tagging probes within them. Given the stringent quality control and fold change criteria used in our testing, all probes for a given gene did not always meet that threshold. Consequently, some genes have differing numbers of probes in different tissue types. Additionally, probes with near/below background intensity readings in one or both age groups tended to have very high fold changes (Table 3). In particular, the most differentially expressed genes in the retina/RPE and sclera included a number of genes whose expression detection levels were low (bottom 5% intensity of differentially expressed genes) for one or both age groups (Table 3). These low readings may have resulted either from failed probes or absent expression. The degree of expression fold changes in these genes (noted in Table 3) with low readings in both groups may appear higher due to the relative calculation and the normalization process and should be considered when interpreting the data.
Table 3. Highest Fold Changes.
Genes (represented by probes) from each tissue with the highest fold changes (FC). Negative FC indicates increased expression in fetal relative to adult tissues.
| Retina/RPE | Choroid | Sclera | Optic nerve | Cornea | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||||
| Gene | FC | P-value | Gene | FC | P-value | Gene | FC | P-value | Gene | FC | P-value | Gene | FC | P-value |
| CRHR1b | −8.5 | 0.0003 | HBG1 | −617.57 | 0.0007 | ACOX1b | −125.11 | 0.0065 | COL1A1 | −205.22 | 0.0043 | LOC100134134b | −1252.45 | 0.0043 |
| FLJ40244a | −8.11 | 0.0012 | COL9A1b | −165.29 | 0.0007 | EYA2a | −96.93 | 0.0016 | COL3A1 | −171.4 | 0.0043 | OGNb | −612.53 | 0.0043 |
| LOC100133528a | −5.25 | 0.0025 | HBG2 | −159.85 | 0.0007 | KRTAP5-6a | −91.07 | 0.0003 | COL1A2 | −100.87 | 0.0043 | MEG3b | −451.89 | 0.0043 |
| CYP24A1a | −4.65 | 0.0026 | HCG2P7 | −117.26 | 0.0013 | GRIA1 | −32.41 | 0.0012 | C5ORF13 | −97.75 | 0.0043 | COL5A1 | −321.1 | 0.0043 |
| ETFBb | −4.31 | 0.000027 | HBA1 | −98.16 | 0.0007 | CDONa | −9.68 | 0.0011 | CPNE4b | −73.78 | 0.0075 | FAM180Ab | −277.35 | 0.0043 |
| C13ORF23b | −4.1 | 0.0059 | GRIK1 | −92.66 | 0.0007 | CDC45Lb | −9.61 | 0.000037 | MMP15b | −63.99 | 0.0067 | OGN | −274.14 | 0.0043 |
| NUP155a | −3.49 | 0.0036 | APLNR | −75.93 | 0.0007 | MRI1 | −9.43 | 0.0009 | PRNDb | −63.09 | 0.0043 | THBS2 | −265.3 | 0.0043 |
| ZNF92b | −3.4 | 0.0002 | HBM | −74.74 | 0.0007 | SNIPa | −8.94 | 0.0001 | SEMA5Bb | −60.69 | 0.008 | DPTb | −256.95 | 0.0043 |
| MAGED4b | −3.38 | 0.0001 | DUXAP3 | −66.52 | 0.0027 | MAL2 | −8.59 | 0.000037 | COL1A2 | −60.34 | 0.0043 | SOCS1 | −250.99 | 0.0043 |
| C20ORF118b | −3.36 | 0.0055 | DMC1 | −65 | 0.0027 | FOXL2a | −8.04 | 0.0007 | CDH7b | −60.03 | 0.0067 | THRA | −228.18 | 0.0043 |
| KRT6Ab | −3.28 | 0.0011 | NLRP8 | −62.8 | 0.0027 | KCNIP2 | −7.92 | 0.0016 | C20ORF103b | −59.18 | 0.008 | PXDN | −217.78 | 0.0043 |
| SNIPa | −3.2 | 0.0068 | GRIK1b | −62.17 | 0.0007 | COL11A1a | −7.81 | 0.0044 | PCDH12b | −46.03 | 0.0075 | IGDCC4b | −211.63 | 0.0043 |
| ASB6b | −3.17 | 0.0064 | LOC100128084 | −55.69 | 0.0013 | UBASH3Aa | −7.43 | 0.0001 | COL5A1 | −45.92 | 0.0043 | COL1A1 | −209.79 | 0.0043 |
| KIF4Ab | −3.16 | 0.0008 | TOP2A | −55.64 | 0.0007 | S100A12a | −7.15 | 0.000037 | TBC1D10Cb | −44.02 | 0.0067 | COL3A1 | −161.78 | 0.0043 |
| FAM19A1b | −3.09 | 0.0042 | ARMCX6 | −54.79 | 0.0024 | NEU3 | −7.05 | 0.000037 | KAL1 | −43.22 | 0.0043 | RASL11Bb | −157.22 | 0.0043 |
| MMD2b | −3.08 | 0.0001 | LOC645452 | −50.32 | 0.0027 | KIAA1549 | −6.56 | 0.000037 | APOA1b | −36.38 | 0.0075 | MMP23B | −154.42 | 0.0043 |
| LRRIQ3b | −3.05 | 0.0008 | IGF1b | −45.62 | 0.0007 | DEFA4a | −6.3 | 0.0027 | HS.204481b | −35.77 | 0.0055 | KCNIP4b | −148.3 | 0.0043 |
| SGOL2a | −3.01 | 0.0018 | FCAR | −45.56 | 0.0027 | TCF2a | −6.26 | 0.0046 | TNC | −34.88 | 0.0043 | PCDH17b | −147.06 | 0.0043 |
| KLK12b | −2.97 | 0.0068 | KCNK4b | −45.45 | 0.0007 | PTGER3a | −6.14 | 0.0011 | FNDC1 | −33.21 | 0.0043 | MFAP2 | −143.13 | 0.0043 |
| TDGF1b | −2.96 | 0.0001 | CRTAP | −43.96 | 0.0007 | DDX25 | −6.01 | 0.0021 | RSPO4b | −31.26 | 0.0055 | NTMb | −131.38 | 0.0043 |
| NNAT | −2.94 | 0.000027 | CXCL12 | −43.05 | 0.0007 | RASSF5a | −5.98 | 0.0046 | FOXL2b | −29.21 | 0.0054 | COCHb | −131.04 | 0.0043 |
| LOC651746b | −2.92 | 0.0002 | UBE2C | −42.86 | 0.0007 | BUB1b | −5.94 | 0.0005 | COL5A2 | −26.92 | 0.0043 | UNQ1940b | −126.31 | 0.0043 |
| OTOSb | −2.92 | 0.0008 | CDKN2AIPNL | −41.93 | 0.0047 | RPEa | −5.92 | 0.0046 | TNFRSF25b | −24.92 | 0.0043 | SRPX | −126.27 | 0.0043 |
| COL6A6b | −2.89 | 0.0046 | COL4A5b | −39.77 | 0.0007 | MATN4a | −5.91 | 0.0027 | CDC20Bb | −24.3 | 0.0043 | CAPN6 | −103.79 | 0.0043 |
| ZWILCHb | −2.88 | 0.0008 | KIAA1751 | −36.97 | 0.0027 | KLK3b | −5.75 | 0.0004 | SLIT1b | −23.74 | 0.008 | HBG1b | −99.54 | 0.0043 |
| SSX2b | −2.86 | 0.0068 | SFRP4 | −36.3 | 0.0007 | CACNA1G | −5.73 | 0.0021 | THSD1b | −23.04 | 0.008 | FBN2 | −93.03 | 0.0043 |
| NSBP1b | −2.84 | 0.000027 | TRIM24b | −36.18 | 0.0007 | METTL1a | −5.71 | 0.0004 | SLC16A14b | −22.9 | 0.008 | HBA2b | −89.18 | 0.0043 |
| RIPPLY2b | −2.81 | 0.000027 | CCT7 | −35.79 | 0.0024 | HBMa | −5.71 | 0.0027 | OCA2b | −22.81 | 0.0075 | THY1b | −86.78 | 0.0043 |
| COL2A1b | −2.74 | 0.0002 | NCAPG | −35.44 | 0.0007 | ARPP-21 | −5.67 | 0.0054 | ARSEa | −21.69 | 0.0066 | SOX11 | −81.66 | 0.0043 |
| TTTY17Ba | −2.74 | 0.005 | MCART1 | −34.88 | 0.0027 | AGR2b | −5.61 | 0.0012 | SPON1 | −20.66 | 0.0043 | SMOb | −80.53 | 0.0043 |
| TTTY9Aa | −2.73 | 0.0036 | COL1A2 | −34.43 | 0.0024 | ALDH4A1 | −5.44 | 0.0035 | MYH7b | −20.65 | 0.008 | C1QTNF1 | −79.43 | 0.0043 |
| BPY2Ca | −2.68 | 0.0072 | LRRC37B2 | −34.37 | 0.0027 | TOP2Ab | −5.43 | 0.000037 | UNC13C | −20.63 | 0.0043 | MMP23A | −78.59 | 0.0043 |
| KIAA0514b | −2.65 | 0.000027 | LOC100128505 | −34.36 | 0.0027 | HRH3 | −5.37 | 0.0072 | FBN2 | −20.57 | 0.0043 | COL11A1b | −75.57 | 0.0043 |
| LOC653889b | −2.64 | 0.0002 | ARHGAP28 | − 33.11 | 0.0007 | TBC1D29a | −5.35 | 0.0016 | NKD2 | −20.31 | 0.0043 | PMEPA1b | −74.45 | 0.0043 |
| MATKb | −2.62 | 0.0006 | AP1S1b | −32 | 0.0024 | OR1E2a | −5.31 | 0.0033 | MMP25b | −20 | 0.0043 | CCND2 | −73.66 | 0.0043 |
| TREML2Pa | −2.6 | 0.0025 | GPC3 | −31.98 | 0.0007 | MIR1258b | −5.3 | 0.000074 | MXRA5 | −19.37 | 0.0043 | HBG2b | −70.49 | 0.0043 |
| BTBD17 | −2.59 | 0.000027 | LOC399900 | −31.51 | 0.0027 | GPR44a | −5.22 | 0.0072 | C20ORF46b | −18.85 | 0.0075 | SCXAb | −69.87 | 0.0043 |
| SLC18A2b | −2.54 | 0.0008 | HBZ | −30.9 | 0.0007 | COL6A6b | −5.18 | 0.000037 | VCAN | −18.8 | 0.0043 | SNCAIPb | −63.58 | 0.0043 |
| PAMR1a | −2.54 | 0.0036 | LAMA1 | −30.8 | 0.0007 | TRIM29a | −5.17 | 0.0062 | DACT1 | −18.02 | 0.0043 | PYCR1 | −62.75 | 0.0043 |
| ABHD11b | −2.53 | 0.0019 | IP6K2b | −30.51 | 0.0007 | RHBDF2a | −5.12 | 0.0007 | NMUa | −18.02 | 0.0067 | C1QTNF5 | −61.52 | 0.0043 |
| INMTb | 21.09 | 0.000027 | SERPINA3 | 550.49 | 0.0007 | MYOCb | 6.4 | 0.000037 | BCAS1b | 1695.34 | 0.0067 | MAMDC2 | 205.77 | 0.0043 |
| C5ORF48a | 9.79 | 0.0004 | SAA1 | 344.94 | 0.0007 | ABCF2a | 5.47 | 0.0004 | BCAS1b | 1105.48 | 0.0075 | AKR1C2 | 192.43 | 0.0043 |
| GCGRb | 5.33 | 0.000053 | MT1Mb | 221.47 | 0.0007 | FMO3a | 4.1 | 0.0021 | MAGb | 111.21 | 0.0043 | KRT24 | 181.18 | 0.0043 |
| ASCL4a | 5.17 | 0.0046 | PLA2G2A | 220.99 | 0.0007 | SLPIa | 3.92 | 0.000037 | S1PR5b | 79.37 | 0.0055 | ADH7 | 174.83 | 0.0043 |
| HAND1b | 4.37 | 0.0018 | SNTNb | 179.32 | 0.0027 | PLA2G2A | 3.64 | 0.000037 | ZNF488b | 50.7 | 0.0075 | ZBTB16b | 156.59 | 0.0039 |
| LOC399706b | 4.08 | 0.0003 | C19ORF12 | 174.68 | 0.0007 | GLDN | 3.26 | 0.000037 | GPX3 | 47.64 | 0.0043 | ANGPTL7 | 142.31 | 0.0043 |
| HNMTa | 3.9 | 0.0044 | LOC100128693 | 139.44 | 0.0013 | AKR1B10b | 3.14 | 0.000037 | RAB11FIP4b | 43.7 | 0.008 | CA3b | 114 | 0.0039 |
| KIR2DL3b | 3.8 | 0.000053 | LOC100132620 | 137.85 | 0.0007 | SLC26A4a | 3.14 | 0.000037 | FA2Hb | 38.01 | 0.0075 | IL20RAb | 111.17 | 0.0043 |
| NRP2b | 3.72 | 0.0001 | TTC21B | 123.34 | 0.0013 | SCNN1Aa | 3.08 | 0.000037 | MAL | 25.43 | 0.0043 | AKR1C4 | 109.18 | 0.0043 |
| DLX6b | 3.52 | 0.0005 | ZNF273 | 112.7 | 0.0027 | CCL15b | 2.94 | 0.0012 | CENTA1 | 24.84 | 0.0043 | SLURP1b | 99.65 | 0.0039 |
| FOXR1b | 3.42 | 0.0015 | LOC100132518 | 108.2 | 0.0007 | KCNE1b | 2.9 | 0.0001 | SEPT4 | 22.55 | 0.0043 | SLC47A1b | 89.77 | 0.0039 |
| OR52A1a | 3.41 | 0.0001 | IL1RL1b | 96.02 | 0.0007 | COMP | 2.78 | 0.000037 | COBLb | 19.78 | 0.0043 | IFI27 | 82.06 | 0.0043 |
| LOC100127983b | 3.36 | 0.0068 | GTF2IRD2B | 86.41 | 0.0007 | CD69 | 2.72 | 0.0021 | MAP6D1 | 18.84 | 0.0043 | INMTb | 81.8 | 0.0039 |
| HERV-FRDb | 3.35 | 0.0005 | LOC100132540 | 85.5 | 0.0024 | C10ORF81b | 2.59 | 7.37E-05 | SEPT4 | 18.7 | 0.0043 | GJB4 | 81.42 | 0.0043 |
| 40610b | 3.33 | 0.0025 | NNMTb | 84.35 | 0.0007 | TCEAL6 | 2.58 | 0.000037 | COL4A3 | 17.72 | 0.0067 | MME | 74.43 | 0.0043 |
| IL1RNb | 3.3 | 0.0036 | PPIL3 | 80.97 | 0.0007 | SCGB3A2 | 2.57 | 0.000037 | APOD | 16.26 | 0.0043 | MYOC | 74.2 | 0.0043 |
| DEFB119b | 3.14 | 0.000027 | RUNDC2B | 80.95 | 0.0007 | CHI3L2b | 2.56 | 0.000037 | QDPR | 14.35 | 0.0043 | EFCBP1b | 73.56 | 0.0039 |
| EDNRBb | 3.12 | 0.0002 | LOC729652 | 80.46 | 0.0007 | COL4A3 | 2.56 | 0.0011 | PRODH | 12.89 | 0.0043 | TMEM45Bb | 68.26 | 0.0039 |
| MAGEA11b | 3.1 | 0.0005 | STAG3L1 | 74.06 | 0.0007 | LIN28b | 2.54 | 0.0007 | MIR1974 | 12.05 | 0.0173 | STEAP4b | 66.43 | 0.0039 |
| MT1M | 3.02 | 0.000027 | PRIM2 | 69.86 | 0.0013 | SAA1 | 2.54 | 0.0007 | APLP1 | 12.02 | 0.0043 | PLK5Pb | 66.16 | 0.0039 |
| LOC389834b | 3 | 0.000027 | SNHG10 | 68.88 | 0.0007 | MMP3 | 2.53 | 0.000037 | CAPN3 | 11.78 | 0.0087 | HSD11B1b | 63.84 | 0.0039 |
| FAM55A | 2.99 | 0.0003 | BCAS4 | 67.49 | 0.0007 | ABCC2 | 2.51 | 0.000037 | PPP1R14A | 11.55 | 0.0043 | ECM1 | 63.1 | 0.0043 |
| DCST2b | 2.97 | 0.000053 | LOC100134172 | 66.89 | 0.0007 | KRTAP9-3b | 2.49 | 0.0003 | PCTK3 | 11.43 | 0.0043 | NQO1 | 58.54 | 0.0043 |
| LOC646085b | 2.94 | 0.0003 | LOC100129445 | 65.3 | 0.0007 | MMP12a | 2.43 | 0.0011 | MIR1978 | 11.06 | 0.0043 | CRTAC1 | 54.78 | 0.0043 |
| RHO | 2.93 | 0.000027 | PTERb | 65.12 | 0.0027 | MT1Mb | 2.42 | 0.000037 | SLC7A2 | 9.89 | 0.0043 | KRT27 | 47.54 | 0.0043 |
| CAMPb | 2.93 | 0.0037 | GNPTAB | 64.42 | 0.0047 | CYP4F12b | 2.39 | 0.000037 | TP53INP2 | 9.58 | 0.0043 | CALML3 | 46.96 | 0.0043 |
| BEST3b | 2.91 | 0.0003 | TRIM16L | 60.82 | 0.0047 | OR7D2 | 2.39 | 0.0003 | C7ORF41 | 9.54 | 0.0043 | PTPN22b | 46.1 | 0.0039 |
| MICBb | 2.86 | 0.000027 | CHI3L2 | 59.38 | 0.0007 | SERPINA3 | 2.39 | 0.0005 | MT1X | 8.93 | 0.0043 | COL8A1b | 43.63 | 0.0039 |
| LGSNb | 2.86 | 0.005 | LOC202134 | 59.22 | 0.0007 | ASB11b | 2.35 | 0.0027 | BCYRN1 | 8.92 | 0.0043 | PSCA | 40.4 | 0.0043 |
| HUS1b | 2.82 | 0.005 | LOC100128562 | 58.77 | 0.0007 | GPR88 | 2.34 | 0.000037 | COL4A4a | 8.89 | 0.0067 | ZBTB16b | 40.19 | 0.0043 |
| LRRC25b | 2.8 | 0.0068 | TAF8 | 58.48 | 0.0007 | PDPN | 2.34 | 0.0024 | DBNDD2 | 8.63 | 0.0043 | ECM1 | 38.83 | 0.0043 |
| FAM55Db | 2.77 | 0.0025 | PPID | 58.47 | 0.0007 | LOC100130988b | 2.29 | 0.0046 | FAM107A | 8.21 | 0.0043 | SOD3 | 37.61 | 0.0043 |
| EPYCb | 2.76 | 0.0012 | LOC651192 | 53.93 | 0.0007 | ANGPTL7 | 2.27 | 0.000037 | CDKN1A | 8.09 | 0.0173 | MICALCLb | 37.6 | 0.0043 |
| KCNJ14b | 2.74 | 0.0008 | LOC728054 | 52.7 | 0.0013 | NNMTa | 2.27 | 0.000037 | SYNM | 7.74 | 0.0087 | AGXT2L1b | 36.92 | 0.0039 |
| VCAM1 | 2.73 | 0.005 | LOC647592 | 52.36 | 0.0007 | GBP2 | 2.25 | 0.000037 | ENPP2 | 7.67 | 0.0173 | CYP26A1 | 35.79 | 0.0043 |
| GJA8b | 2.69 | 0.0008 | STAP2 | 49.61 | 0.0027 | INMTb | 2.25 | 0.000037 | C10ORF116 | 7.64 | 0.0043 | FAM164Cb | 35.05 | 0.0039 |
| KRTAP19-4b | 2.65 | 0.0018 | ZNF100 | 49.02 | 0.0007 | MYOC | 2.22 | 0.000037 | CHN2 | 7.47 | 0.0043 | ALDH3A1 | 34.93 | 0.0043 |
| OR13F1b | 2.64 | 0.0068 | TEX11 | 48.84 | 0.0013 | KRT28b | 2.22 | 0.0062 | S100A8b | 7.38 | 0.0353 | SYT8 | 34.61 | 0.0043 |
| LOC653075b | 2.63 | 0.0005 | SLA2 | 47.56 | 0.0007 | ZBTB16 | 2.21 | 0.000037 | ENPP2 | 7.22 | 0.0173 | RGS7BPb | 34.48 | 0.0039 |
| CYP2A7b | 2.63 | 0.0068 | ST20 | 46.86 | 0.0007 | KGFLP1a | 2.21 | 0.0007 | RGS1b | 7.05 | 0.0043 | CLCA4 | 34 | 0.0043 |
|
| ||||||||||||||
| Gene | FC | P-value | Gene | FC | P-value | |||||||||
|
| ||||||||||||||
| Adult versus fetal retina/RPE | ||||||||||||||
| CRHR1b | −8.5 | 0.0003 | INMTb | 21.09 | 2.65E-05 | |||||||||
| FLJ40244a | −8.11 | 0.0012 | C5ORF48a | 9.79 | 0.0004 | |||||||||
| LOC100133528a | −5.25 | 0.0025 | GCGRb | 5.33 | 5.29E-05 | |||||||||
| CYP24A1a | −4.65 | 0.0026 | ASCL4a | 5.17 | 0.0046 | |||||||||
| ETFBb | −4.31 | 2.65E-05 | HAND1b | 4.37 | 0.0018 | |||||||||
| C13ORF23b | −4.1 | 0.0059 | LOC399706b | 4.08 | 0.0003 | |||||||||
| NUP155a | −3.49 | 0.0036 | HNMTa | 3.9 | 0.0044 | |||||||||
| ZNF92b | −3.4 | 0.0002 | KIR2DL3b | 3.8 | 5.29E-05 | |||||||||
| MAGED4b | −3.38 | 0.0001 | NRP2b | 3.72 | 0.0001 | |||||||||
| C20ORF118b | −3.36 | 0.0055 | DLX6b | 3.52 | 0.0005 | |||||||||
| KRT6Ab | −3.28 | 0.0011 | FOXR1b | 3.42 | 0.0015 | |||||||||
| SNIPa | −3.2 | 0.0068 | OR52A1a | 3.41 | 0.0001 | |||||||||
| ASB6b | −3.17 | 0.0064 | LOC100127983b | 3.36 | 0.0068 | |||||||||
| KIF4Ab | −3.16 | 0.0008 | HERV-FRDb | 3.35 | 0.0005 | |||||||||
| FAM19A1b | −3.09 | 0.0042 | 40610b | 3.33 | 0.0025 | |||||||||
| MMD2b | −3.08 | 0.0001 | IL1RNb | 3.3 | 0.0036 | |||||||||
| LRRIQ3b | −3.05 | 0.0008 | DEFB119b | 3.14 | 2.65E-05 | |||||||||
| SGOL2a | −3.01 | 0.0018 | EDNRBb | 3.12 | 0.0002 | |||||||||
| KLK12b | −2.97 | 0.0068 | MAGEA11b | 3.1 | 0.0005 | |||||||||
| TDGF1b | −2.96 | 0.0001 | MT1M | 3.02 | 2.65E-05 | |||||||||
| NNAT | −2.94 | 2.65E-05 | LOC389834b | 3 | 2.65E-05 | |||||||||
| LOC651746b | −2.92 | 0.0002 | FAM55A | 2.99 | 0.0003 | |||||||||
| OTOSb | −2.92 | 0.0008 | DCST2b | 2.97 | 5.29E-05 | |||||||||
| COL6A6b | −2.89 | 0.0046 | LOC646085b | 2.94 | 0.0003 | |||||||||
| ZWILCHb | −2.88 | 0.0008 | RHO | 2.93 | 2.65E-05 | |||||||||
| SSX2b | −2.86 | 0.0068 | CAMPb | 2.93 | 0.0037 | |||||||||
| NSBP1b | −2.84 | 2.65E-05 | BEST3b | 2.91 | 0.0003 | |||||||||
| RIPPLY2b | −2.81 | 2.65E-05 | MICBb | 2.86 | 2.65E-05 | |||||||||
| COL2A1b | −2.74 | 0.0002 | LGSNb | 2.86 | 0.005 | |||||||||
| TTTY17Ba | −2.74 | 0.005 | HUS1b | 2.82 | 0.005 | |||||||||
| TTTY9Aa | −2.73 | 0.0036 | LRRC25b | 2.8 | 0.0068 | |||||||||
| BPY2Ca | −2.68 | 0.0072 | FAM55Db | 2.77 | 0.0025 | |||||||||
| KIAA0514b | −2.65 | 2.65E-05 | EPYCb | 2.76 | 0.0012 | |||||||||
| LOC653889b | −2.64 | 0.0002 | KCNJ14b | 2.74 | 0.0008 | |||||||||
| MATKb | −2.62 | 0.0006 | VCAM1 | 2.73 | 0.005 | |||||||||
| TREML2Pa | −2.6 | 0.0025 | GJA8b | 2.69 | 0.0008 | |||||||||
| BTBD17 | −2.59 | 2.65E-05 | KRTAP19-4b | 2.65 | 0.0018 | |||||||||
| SLC18A2b | −2.54 | 0.0008 | OR13F1b | 2.64 | 0.0068 | |||||||||
| PAMR1a | −2.54 | 0.0036 | LOC653075b | 2.63 | 0.0005 | |||||||||
| ABHD11b | −2.53 | 0.0019 | CYP2A7b | 2.63 | 0.0068 | |||||||||
| Adult versus fetal choroid | ||||||||||||||
| HBG1 | −617.57 | 0.0007 | SERPINA3 | 550.49 | 0.0007 | |||||||||
| COL9A1b | −165.29 | 0.0007 | SAA1 | 344.94 | 0.0007 | |||||||||
| HBG2 | −159.85 | 0.0007 | MT1Mb | 221.47 | 0.0007 | |||||||||
| HCG2P7 | −117.26 | 0.0013 | PLA2G2A | 220.99 | 0.0007 | |||||||||
| HBA1 | −98.16 | 0.0007 | SNTNb | 179.32 | 0.0027 | |||||||||
| GRIK1 | −92.66 | 0.0007 | C19ORF12 | 174.68 | 0.0007 | |||||||||
| APLNR | −75.93 | 0.0007 | LOC100128693 | 139.44 | 0.0013 | |||||||||
| HBM | −74.74 | 0.0007 | LOC100132620 | 137.85 | 0.0007 | |||||||||
| DUXAP3 | −66.52 | 0.0027 | TTC21B | 123.34 | 0.0013 | |||||||||
| DMC1 | −65 | 0.0027 | ZNF273 | 112.7 | 0.0027 | |||||||||
| NLRP8 | −62.8 | 0.0027 | LOC100132518 | 108.2 | 0.0007 | |||||||||
| GRIK1b | −62.17 | 0.0007 | IL1RL1b | 96.02 | 0.0007 | |||||||||
| LOC100128084 | −55.69 | 0.0013 | GTF2IRD2B | 86.41 | 0.0007 | |||||||||
| TOP2A | −55.64 | 0.0007 | LOC100132540 | 85.5 | 0.0024 | |||||||||
| ARMCX6 | −54.79 | 0.0024 | NNMTb | 84.35 | 0.0007 | |||||||||
| LOC645452 | −50.32 | 0.0027 | PPIL3 | 80.97 | 0.0007 | |||||||||
| IGF1b | −45.62 | 0.0007 | RUNDC2B | 80.95 | 0.0007 | |||||||||
| FCAR | −45.56 | 0.0027 | LOC729652 | 80.46 | 0.0007 | |||||||||
| KCNK4b | −45.45 | 0.0007 | STAG3L1 | 74.06 | 0.0007 | |||||||||
| CRTAP | −43.96 | 0.0007 | PRIM2 | 69.86 | 0.0013 | |||||||||
| CXCL12 | −43.05 | 0.0007 | SNHG10 | 68.88 | 0.0007 | |||||||||
| UBE2C | −42.86 | 0.0007 | BCAS4 | 67.49 | 0.0007 | |||||||||
| CDKN2AIPNL | −41.93 | 0.0047 | LOC100134172 | 66.89 | 0.0007 | |||||||||
| COL4A5b | −39.77 | 0.0007 | LOC100129445 | 65.3 | 0.0007 | |||||||||
| KIAA1751 | −36.97 | 0.0027 | PTERb | 65.12 | 0.0027 | |||||||||
| SFRP4 | −36.3 | 0.0007 | GNPTAB | 64.42 | 0.0047 | |||||||||
| TRIM24b | −36.18 | 0.0007 | TRIM16L | 60.82 | 0.0047 | |||||||||
| CCT7 | −35.79 | 0.0024 | CHI3L2 | 59.38 | 0.0007 | |||||||||
| NCAPG | −35.44 | 0.0007 | LOC202134 | 59.22 | 0.0007 | |||||||||
| MCART1 | −34.88 | 0.0027 | LOC100128562 | 58.77 | 0.0007 | |||||||||
| COL1A2 | −34.43 | 0.0024 | TAF8 | 58.48 | 0.0007 | |||||||||
| LRRC37B2 | −34.37 | 0.0027 | PPID | 58.47 | 0.0007 | |||||||||
| LOC100128505 | −34.36 | 0.0027 | LOC651192 | 53.93 | 0.0007 | |||||||||
| ARHGAP28 | −33.11 | 0.0007 | LOC728054 | 52.7 | 0.0013 | |||||||||
| AP1S1b | −32 | 0.0024 | LOC647592 | 52.36 | 0.0007 | |||||||||
| GPC3 | −31.98 | 0.0007 | STAP2 | 49.61 | 0.0027 | |||||||||
| LOC399900 | −31.51 | 0.0027 | ZNF100 | 49.02 | 0.0007 | |||||||||
| HBZ | −30.9 | 0.0007 | TEX11 | 48.84 | 0.0013 | |||||||||
| LAMA1 | −30.8 | 0.0007 | SLA2 | 47.56 | 0.0007 | |||||||||
| IP6K2b | −30.51 | 0.0007 | ST20 | 46.86 | 0.0007 | |||||||||
| Adult versus fetal sclera | ||||||||||||||
| ACOX1b | −125.11 | 0.0065 | MYOCb | 6.4 | 3.69E-05 | |||||||||
| EYA2a | −96.93 | 0.0016 | ABCF2a | 5.47 | 0.0004 | |||||||||
| KRTAP5-6a | −91.07 | 0.0003 | FMO3a | 4.1 | 0.0021 | |||||||||
| GRIA1 | −32.41 | 0.0012 | SLPIa | 3.92 | 3.69E-05 | |||||||||
| CDONa | −9.68 | 0.0011 | PLA2G2A | 3.64 | 3.69E-05 | |||||||||
| CDC45Lb | −9.61 | 3.69E-05 | GLDN | 3.26 | 3.69E-05 | |||||||||
| MRI1 | −9.43 | 0.0009 | AKR1B10b | 3.14 | 3.69E-05 | |||||||||
| SNIPa | −8.94 | 0.0001 | SLC26A4a | 3.14 | 3.69E-05 | |||||||||
| MAL2 | −8.59 | 3.69E-05 | SCNN1Aa | 3.08 | 3.69E-05 | |||||||||
| FOXL2a | −8.04 | 0.0007 | CCL15b | 2.94 | 0.0012 | |||||||||
| KCNIP2 | −7.92 | 0.0016 | KCNE1b | 2.9 | 0.0001 | |||||||||
| COL11A1a | −7.81 | 0.0044 | COMP | 2.78 | 3.69E-05 | |||||||||
| UBASH3Aa | −7.43 | 0.0001 | CD69 | 2.72 | 0.0021 | |||||||||
| S100A12a | −7.15 | 3.69E-05 | C10ORF81b | 2.59 | 7.37E-05 | |||||||||
| NEU3 | −7.05 | 3.69E-05 | TCEAL6 | 2.58 | 3.69E-05 | |||||||||
| KIAA1549 | −6.56 | 3.69E-05 | SCGB3A2 | 2.57 | 3.69E-05 | |||||||||
| DEFA4a | −6.3 | 0.0027 | CHI3L2b | 2.56 | 3.69E-05 | |||||||||
| TCF2a | −6.26 | 0.0046 | COL4A3 | 2.56 | 0.0011 | |||||||||
| PTGER3a | −6.14 | 0.0011 | LIN28b | 2.54 | 0.0007 | |||||||||
| DDX25 | −6.01 | 0.0021 | SAA1 | 2.54 | 0.0007 | |||||||||
| RASSF5a | −5.98 | 0.0046 | MMP3 | 2.53 | 3.69E-05 | |||||||||
| BUB1b | −5.94 | 0.0005 | ABCC2 | 2.51 | 3.69E-05 | |||||||||
| RPEa | −5.92 | 0.0046 | KRTAP9-3b | 2.49 | 0.0003 | |||||||||
| MATN4a | −5.91 | 0.0027 | MMP12a | 2.43 | 0.0011 | |||||||||
| KLK3b | −5.75 | 0.0004 | MT1Mb | 2.42 | 3.69E-05 | |||||||||
| CACNA1G | −5.73 | 0.0021 | CYP4F12b | 2.39 | 3.69E-05 | |||||||||
| METTL1a | −5.71 | 0.0004 | OR7D2 | 2.39 | 0.0003 | |||||||||
| HBMa | −5.71 | 0.0027 | SERPINA3 | 2.39 | 0.0005 | |||||||||
| ARPP-21 | −5.67 | 0.0054 | ASB11b | 2.35 | 0.0027 | |||||||||
| AGR2b | −5.61 | 0.0012 | GPR88 | 2.34 | 3.69E-05 | |||||||||
| ALDH4A1 | −5.44 | 0.0035 | PDPN | 2.34 | 0.0024 | |||||||||
| TOP2Ab | −5.43 | 3.69E-05 | LOC100130988b | 2.29 | 0.0046 | |||||||||
| HRH3 | −5.37 | 0.0072 | ANGPTL7 | 2.27 | 3.69E-05 | |||||||||
| TBC1D29a | −5.35 | 0.0016 | NNMTa | 2.27 | 3.69E-05 | |||||||||
| OR1E2a | −5.31 | 0.0033 | GBP2 | 2.25 | 3.69E-05 | |||||||||
| MIR1258b | −5.3 | 7.37E-05 | INMTb | 2.25 | 3.69E-05 | |||||||||
| GPR44a | −5.22 | 0.0072 | MYOC | 2.22 | 3.69E-05 | |||||||||
| COL6A6b | −5.18 | 3.69E-05 | KRT28b | 2.22 | 0.0062 | |||||||||
| TRIM29a | −5.17 | 0.0062 | ZBTB16 | 2.21 | 3.69E-05 | |||||||||
| RHBDF2a | −5.12 | 0.0007 | KGFLP1a | 2.21 | 0.0007 | |||||||||
| Adult versus fetal optic nerve | ||||||||||||||
| COL1A1 | −205.22 | 0.0043 | BCAS1b | 1695.34 | 0.0067 | |||||||||
| COL3A1 | −171.4 | 0.0043 | BCAS1b | 1105.48 | 0.0075 | |||||||||
| COL1A2 | −100.87 | 0.0043 | MAGb | 111.21 | 0.0043 | |||||||||
| C5ORF13 | −97.75 | 0.0043 | S1PR5b | 79.37 | 0.0055 | |||||||||
| CPNE4b | −73.78 | 0.0075 | ZNF488b | 50.7 | 0.0075 | |||||||||
| MMP15b | −63.99 | 0.0067 | GPX3 | 47.64 | 0.0043 | |||||||||
| PRNDb | −63.09 | 0.0043 | RAB11FIP4b | 43.7 | 0.008 | |||||||||
| SEMA5Bb | −60.69 | 0.008 | FA2Hb | 38.01 | 0.0075 | |||||||||
| COL1A2 | −60.34 | 0.0043 | MAL | 25.43 | 0.0043 | |||||||||
| CDH7b | −60.03 | 0.0067 | CENTA1 | 24.84 | 0.0043 | |||||||||
| C20ORF103b | −59.18 | 0.008 | SEPT4 | 22.55 | 0.0043 | |||||||||
| PCDH12b | −46.03 | 0.0075 | COBLb | 19.78 | 0.0043 | |||||||||
| COL5A1 | −45.92 | 0.0043 | MAP6D1 | 18.84 | 0.0043 | |||||||||
| TBC1D10Cb | −44.02 | 0.0067 | SEPT4 | 18.7 | 0.0043 | |||||||||
| KAL1 | −43.22 | 0.0043 | COL4A3 | 17.72 | 0.0067 | |||||||||
| APOA1b | −36.38 | 0.0075 | APOD | 16.26 | 0.0043 | |||||||||
| HS.204481b | −35.77 | 0.0055 | QDPR | 14.35 | 0.0043 | |||||||||
| TNC | −34.88 | 0.0043 | PRODH | 12.89 | 0.0043 | |||||||||
| FNDC1 | −33.21 | 0.0043 | MIR1974 | 12.05 | 0.0173 | |||||||||
| RSPO4b | −31.26 | 0.0055 | APLP1 | 12.02 | 0.0043 | |||||||||
| FOXL2b | −29.21 | 0.0054 | CAPN3 | 11.78 | 0.0087 | |||||||||
| COL5A2 | −26.92 | 0.0043 | PPP1R14A | 11.55 | 0.0043 | |||||||||
| TNFRSF25b | −24.92 | 0.0043 | PCTK3 | 11.43 | 0.0043 | |||||||||
| CDC20Bb | −24.3 | 0.0043 | MIR1978 | 11.06 | 0.0043 | |||||||||
| SLIT1b | −23.74 | 0.008 | SLC7A2 | 9.89 | 0.0043 | |||||||||
| THSD1b | −23.04 | 0.008 | TP53INP2 | 9.58 | 0.0043 | |||||||||
| SLC16A14b | −22.9 | 0.008 | C7ORF41 | 9.54 | 0.0043 | |||||||||
| OCA2b | −22.81 | 0.0075 | MT1X | 8.93 | 0.0043 | |||||||||
| ARSEa | −21.69 | 0.0066 | BCYRN1 | 8.92 | 0.0043 | |||||||||
| SPON1 | −20.66 | 0.0043 | COL4A4a | 8.89 | 0.0067 | |||||||||
| MYH7b | −20.65 | 0.008 | DBNDD2 | 8.63 | 0.0043 | |||||||||
| UNC13C | −20.63 | 0.0043 | FAM107A | 8.21 | 0.0043 | |||||||||
| FBN2 | −20.57 | 0.0043 | CDKN1A | 8.09 | 0.0173 | |||||||||
| NKD2 | −20.31 | 0.0043 | SYNM | 7.74 | 0.0087 | |||||||||
| MMP25b | −20 | 0.0043 | ENPP2 | 7.67 | 0.0173 | |||||||||
| MXRA5 | −19.37 | 0.0043 | C10ORF116 | 7.64 | 0.0043 | |||||||||
| C20ORF46b | −18.85 | 0.0075 | CHN2 | 7.47 | 0.0043 | |||||||||
| VCAN | −18.8 | 0.0043 | S100A8b | 7.38 | 0.0353 | |||||||||
| DACT1 | −18.02 | 0.0043 | ENPP2 | 7.22 | 0.0173 | |||||||||
| NMUa | −18.02 | 0.0067 | RGS1b | 7.05 | 0.0043 | |||||||||
| Adult versus fetal cornea | ||||||||||||||
| LOC100134134b | −1252.45 | 0.0043 | MAMDC2 | 205.77 | 0.0043 | |||||||||
| OGNb | −612.53 | 0.0043 | AKR1C2 | 192.43 | 0.0043 | |||||||||
| MEG3b | −451.89 | 0.0043 | KRT24 | 181.18 | 0.0043 | |||||||||
| COL5A1 | −321.1 | 0.0043 | ADH7 | 174.83 | 0.0043 | |||||||||
| FAM180Ab | −277.35 | 0.0043 | ZBTB16b | 156.59 | 0.0039 | |||||||||
| OGN | −274.14 | 0.0043 | ANGPTL7 | 142.31 | 0.0043 | |||||||||
| THBS2 | −265.3 | 0.0043 | CA3b | 114 | 0.0039 | |||||||||
| DPTb | −256.95 | 0.0043 | IL20RAb | 111.17 | 0.0043 | |||||||||
| SOCS1 | −250.99 | 0.0043 | AKR1C4 | 109.18 | 0.0043 | |||||||||
| THRA | −228.18 | 0.0043 | SLURP1b | 99.65 | 0.0039 | |||||||||
| PXDN | −217.78 | 0.0043 | SLC47A1b | 89.77 | 0.0039 | |||||||||
| IGDCC4b | −211.63 | 0.0043 | IFI27 | 82.06 | 0.0043 | |||||||||
| COL1A1 | −209.79 | 0.0043 | INMTb | 81.8 | 0.0039 | |||||||||
| COL3A1 | −161.78 | 0.0043 | GJB4 | 81.42 | 0.0043 | |||||||||
| RASL11Bb | −157.22 | 0.0043 | MME | 74.43 | 0.0043 | |||||||||
| MMP23B | −154.42 | 0.0043 | MYOC | 74.2 | 0.0043 | |||||||||
| KCNIP4b | −148.3 | 0.0043 | EFCBP1b | 73.56 | 0.0039 | |||||||||
| PCDH17b | −147.06 | 0.0043 | TMEM45Bb | 68.26 | 0.0039 | |||||||||
| MFAP2 | −143.13 | 0.0043 | STEAP4b | 66.43 | 0.0039 | |||||||||
| NTMb | −131.38 | 0.0043 | PLK5Pb | 66.16 | 0.0039 | |||||||||
| COCHb | −131.04 | 0.0043 | HSD11B1b | 63.84 | 0.0039 | |||||||||
| UNQ1940b | −126.31 | 0.0043 | ECM1 | 63.1 | 0.0043 | |||||||||
| SRPX | −126.27 | 0.0043 | NQO1 | 58.54 | 0.0043 | |||||||||
| CAPN6 | −103.79 | 0.0043 | CRTAC1 | 54.78 | 0.0043 | |||||||||
| HBG1b | −99.54 | 0.0043 | KRT27 | 47.54 | 0.0043 | |||||||||
| FBN2 | −93.03 | 0.0043 | CALML3 | 46.96 | 0.0043 | |||||||||
| HBA2b | −89.18 | 0.0043 | PTPN22b | 46.1 | 0.0039 | |||||||||
| THY1b | −86.78 | 0.0043 | COL8A1b | 43.63 | 0.0039 | |||||||||
| SOX11 | −81.66 | 0.0043 | PSCA | 40.4 | 0.0043 | |||||||||
| SMOb | −80.53 | 0.0043 | ZBTB16b | 40.19 | 0.0043 | |||||||||
| C1QTNF1 | −79.43 | 0.0043 | ECM1 | 38.83 | 0.0043 | |||||||||
| MMP23A | −78.59 | 0.0043 | SOD3 | 37.61 | 0.0043 | |||||||||
| COL11A1b | −75.57 | 0.0043 | MICALCLb | 37.6 | 0.0043 | |||||||||
| PMEPA1b | −74.45 | 0.0043 | AGXT2L1b | 36.92 | 0.0039 | |||||||||
| CCND2 | −73.66 | 0.0043 | CYP26A1 | 35.79 | 0.0043 | |||||||||
| HBG2b | −70.49 | 0.0043 | FAM164Cb | 35.05 | 0.0039 | |||||||||
| SCXAb | −69.87 | 0.0043 | ALDH3A1 | 34.93 | 0.0043 | |||||||||
| SNCAIPb | −63.58 | 0.0043 | SYT8 | 34.61 | 0.0043 | |||||||||
| PYCR1 | −62.75 | 0.0043 | RGS7BPb | 34.48 | 0.0039 | |||||||||
| C1QTNF5 | −61.52 | 0.0043 | CLCA4 | 34 | 0.0043 | |||||||||
Indicates fold probes/genes whose intensity values for both adult and fetal samples were in the lowest 5% among the differentially expressed genes for that tissue.
Indicates the adult (negative FC) or fetal (positive FC) intensity value was in the lowest 5% among the differentially expressed genes for that tissue.
In each tissue, some of the most differentially expressed genes have near background readings in one of the two age groups, which may indicate expression being turned on or off (Table 3). Also the most differentially expressed genes per tissue type included many genes that have either previously been implicated or suggested for involvement in ocular diseases particular to these tissues. As a group, extracellular matrix collagens were among the most differentially expressed genes during growth and development in all tissue types tested (Table 3). Fig. 1 is a heat map of 31 extracellular matrix, ocular developmental, and glaucoma-associated genes comparatively expressed in fetal relative to adult ocular tissues.
Fig. 1.
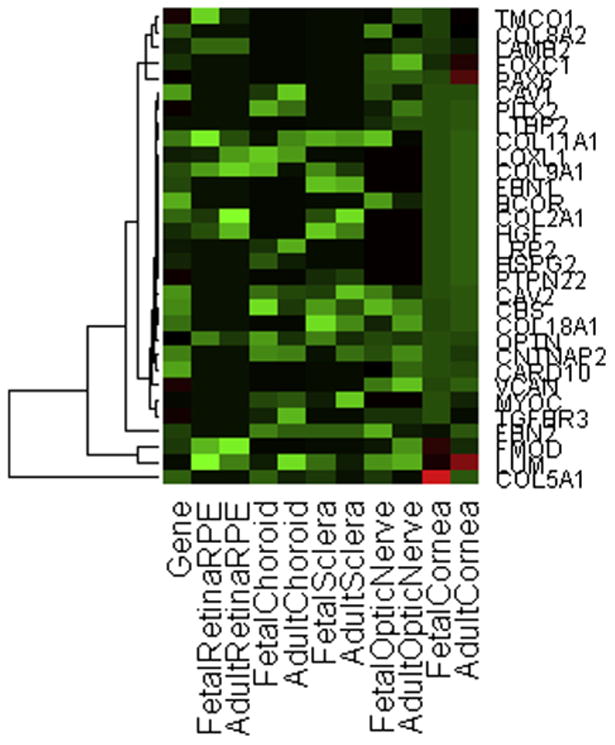
Heat map of 31 extracellular matrix, ocular development, and glaucoma-associated genes comparatively expressed in fetal relative to adult ocular tissues.
3.2. Real-time quantitative PCR
Real time quantitative PCR was performed on 6 genes for cornea, 7 genes for optic nerve, and three genes for retina/retinal pigment epithelium, choroid, and sclera. All demonstrated verification of the microarray results with respect to up- or down-regulation (Fig. 2).
Fig. 2.
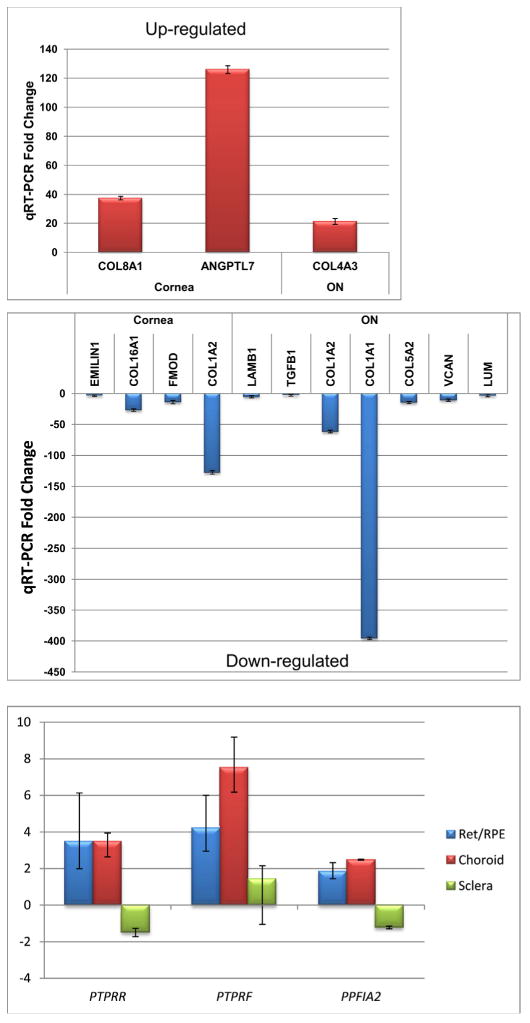
Quantitative real-time polymerase chain reaction validation results of select genes.
3.3. Pathway analyses
IPA functional and canonical pathway analysis was performed separately for each tissue comparison. In all tissue comparisons there was overlap in the most significant functional categories identified (Table 4). Conversely, the most significant canonical pathways identified were more variable between tissue types (Table 5).
Table 4. Ten Most Significant Functional Assignments.
Ten most significant functional groups for adult versus fetal differentially expressed genes.
| Tissue | Functions annotation | P-value | # genes |
|---|---|---|---|
| Retina/RPE | Differentiation | 7.15E-16 | 156 |
| Tumorigenesis | 2.48E-12 | 260 | |
| Proliferation of cells | 8.05E-12 | 182 | |
| Cell movement | 8.36E-11 | 133 | |
| Tissue development | 9.28E-11 | 182 | |
| Function of blood cells | 4.33E-10 | 57 | |
| Immune response | 1.14E-09 | 116 | |
| Cell death of blood cells | 3.18E-09 | 60 | |
| Development of lymphocytes | 4.35E-09 | 60 | |
| Quantity of cells | 1.06E-08 | 118 | |
| Choroid | Tumorigenesis | 3.19E-28 | 1317 |
| Cell death | 1.37E-23 | 1058 | |
| Cell cycle progression | 3.81E-20 | 376 | |
| Proliferation of cells | 1.07E-16 | 835 | |
| Growth of cells | 1.22E-14 | 588 | |
| Expression of RNA | 2.95E-14 | 658 | |
| Transcription | 1.63E-13 | 610 | |
| Differentiation | 2.10E-13 | 617 | |
| Transcription of RNA | 2.37E-13 | 599 | |
| Organization of cytoplasm | 6.20E-13 | 329 | |
| Sclera | Tumorigenesis | 6.17E-18 | 365 |
| Cell division of chromosomes | 3.73E-12 | 43 | |
| Cell cycle progression | 1.14E-10 | 110 | |
| Tissue development | 1.35E-10 | 237 | |
| Proliferation of cells | 2.13E-10 | 231 | |
| Differentiation | 4.42E-10 | 179 | |
| Cell death | 2.39E-09 | 272 | |
| Cell movement | 6.59E-09 | 164 | |
| Interphase | 1.30E-08 | 73 | |
| Morphology of organ | 8.54E-08 | 132 | |
| Optic Nerve | Tumorigenesis | 5.22E-10 | 548 |
| Tissue development | 1.31E-08 | 371 | |
| Expression of RNA | 8.54E-08 | 288 | |
| Proliferation of cells | 9.32E-08 | 357 | |
| Microtubule dynamics | 8.38E-07 | 100 | |
| Cell cycle progression | 9.49E-07 | 152 | |
| Transcription | 9.68E-07 | 263 | |
| Morphology of basement membrane | 1.24E-06 | 9 | |
| Immortalization | 4.28E-06 | 17 | |
| Morphology of blood vessel | 1.18E-05 | 44 | |
| Cornea | Tumorigenesis | 9.15E-28 | 1060 |
| Tissue development | 6.71E-19 | 702 | |
| Cell death | 7.63E-19 | 831 | |
| Proliferation of cells | 1.51E-18 | 684 | |
| Growth of cells | 1.05E-17 | 492 | |
| Encephalopathy | 3.18E-17 | 363 | |
| Cell movement | 1.96E-16 | 483 | |
| Invasion of cells | 2.00E-16 | 199 | |
| Movement disorder | 2.52E-14 | 289 | |
| Organization of cytoplasm | 3.77E-14 | 275 |
Table 5. Top Five Canonical Pathways.
Significance (p-value) of pathways is determined by Fischer's exact test of probability of genes in the dataset fit into the pathway by chance alone and by proportion (ratio) of genes in pathway that are included in the dataset (# genes).
| Tissue | Ingenuity canonical pathways | –log(p) | Ratio | # genes |
|---|---|---|---|---|
| Retina/RPE | Atherosclerosis signaling | 3.99 | 1.09E-01 | 14 |
| LXR/RXR activation | 3.25 | 9.56E-02 | 13 | |
| p38 MAPK signaling | 3.25 | 1.13E-01 | 12 | |
| MIF regulation of innate immunity | 3.15 | 1.40E-01 | 7 | |
| Hepatic fibrosis/hepatic stellate cell activation | 3.14 | 9.52E-02 | 14 | |
| Choroid | Axonal guidance signaling | 7.03 | 2.98E-01 | 128 |
| Molecular mechanisms of cancer | 5.55 | 2.88E-01 | 109 | |
| CXCR4 signaling | 5.33 | 3.39E-01 | 57 | |
| Ephrin receptor signaling | 5.09 | 3.12E-01 | 62 | |
| EIF2 signaling | 4.85 | 3.22E-01 | 66 | |
| Sclera | ATM signaling | 5.31 | 2.20E-01 | 13 |
| Role of CHK proteins in cell cycle Checkpoint control | 3.68 | 2.29E-01 | 8 | |
| Atherosclerosis signaling | 3.53 | 1.24E-01 | 16 | |
| Mitotic roles of polo-like kinase | 2.37 | 1.38E-01 | 9 | |
| Cell cycle: G2/M DNA damage Checkpoint regulation | 2.17 | 1.43E-01 | 7 | |
| Optic Nerve | Wnt/β-catenin signaling | 5.61 | 2.03E-01 | 35 |
| Hereditary breast cancer signaling | 4.99 | 2.05E-01 | 26 | |
| Human embryonic stem cell pluripotency | 4.58 | 1.80E-01 | 27 | |
| Cell cycle: G1/S checkpoint regulation | 3.64 | 2.31E-01 | 15 | |
| RAR activation | 3.47 | 1.60E-01 | 30 | |
| Cornea | Breast cancer regulation by Stathmin1 | 6.67 | 2.93E-01 | 61 |
| Mitochondrial dysfunction | 6.35 | 2.64E-01 | 46 | |
| Wnt/β-catenin signaling | 6.16 | 3.14E-01 | 54 | |
| Axonal guidance signaling | 5.89 | 2.37E-01 | 102 | |
| Molecular mechanisms of cancer | 5.79 | 2.41E-01 | 91 |
3.3.1. Functional annotation enrichment
To identify overrepresentation of genes within known functional pathways involved in growth and development, we performed IPA functional annotation analyses. Probes including their associated gene, p-value, and fold change were analyzed separately for each comparison. The significance of each functional group is given as a p-value. The ten most significant functional assignments for each tissue type by p-value are shown in Table 4. For presentation purposes, functional assignments containing redundant genes and functions were consolidated. Full data is available in Supplementary Table S1.
In all comparisons of ocular tissues, the most significant functional assignments included development, cell growth/death and cancer categories. Tumorigenesis was one of the two most significantly associated functional assignments in all tissues. Other common categories included cell cycle, movement, function, morphology and expression. These significant functional assignments showed that genes involved in basic cellular regulations were differentially activated in rapidly growing and developing tissues relative to their adult counterparts.
In the retina/RPE, the most significant functional assignments also included differentiation, cell proliferation, cell movement and tissue development. In the choroid, cell death, proliferation and growth were among the most significantly associated functional annotations. In the sclera, the most significant functional groups included cell division and progression, as well as tissue development. In the optic nerve, tissue development, RNA expression, cell proliferation and microtubule dynamics were some of the most significantly enriched functions. The lower significance for the optic nerve in the array data was reflected in its lower significance for functional classifications. Despite large variances in the number and significance of genes between tissue type comparisons, the most significant functional assignments across all adult versus fetal comparisons, including the optic nerve, were comparable (Table 4). Lastly in the cornea, the most significantly associated functional annotations included tissue development, cell death, proliferation and growth.
Supplementary Table S2 contains the significant functional assignments with predicted activation changes for each adult versus fetal tissue comparison (absolute value [z-score] > 2.0). Adult versus fetal compared tissues still shared major functional categories in common, but with some differences from those most significantly present. When taking direction of fold changes and their predicted effects on a functional assignment, all tissues had predicted activation changes for cancer and cell movement categories. Many tissue types also had development and cell growth/death categories in common. Even within the same tissue type these shared categories were not consistently predicted to have the same directional activation; however, those shared categories had differing functional assignments and contained distinct genes. Functionally assigned groups containing redundant genes which were removed from the data for publication did not contradict one another in the direction of predicted activation. In the adult retina/RPE, choroid and sclera, functions predicted to have increased activation included cell proliferation, while those predicted to have decreased activation included lymphocyte movement and/or proliferation. In the adult optic nerve, predicted increased functions included cell growth, formation, and organization. Lastly in the adult cornea, predicted functional increases included development, differentiation, and cell cycle progression, while decreased included cell invasion and survival.
3.3.2. Canonical pathway enrichment
To identify specific pathways involved in the rapid growth and development seen in prenatal development we performed canonical pathway analyses using IPA. Probes including their associated gene, p-value, and fold change were analyzed simultaneously for each comparison to identify overrepresentation of genes within known canonical pathways. The significance of each pathway was assigned a p-value. The five most significant canonical pathways by p-value are shown in Table 5. Additionally, IPA calculates a ratio of genes differentially expressed within each canonical pathway. Full data is available in Supplementary Table S3.
There was some overlap in pathways between tissue types and all tissues included signaling pathways among the most significantly associated (Table 5). The retina/RPE and sclera both included atherosclerosis signaling among pathways with the highest significance. The most significant pathways in the choroid and cornea both included axonal guidance signaling. In the sclera and optic nerve tissues both included significant pathways involving cell cycle regulations. Other significant pathways in retina/RPE involved expression, regulation and immune responses of the cell. In the choroid, significant pathways also included growth, expression and immune responses. In the sclera, many of the most significantly associated pathways involved cell cycle regulation. In the optic nerve, the most significant pathways were cell fate, growth, regulation and expression. For the cornea, the pathways with the highest significance were cell fate and growth.
3.4. Disease gene overlap
The most differentially expressed genes in each of the tissues included genes previously implicated in ocular diseases such as myopia, glaucoma and syndromes with an ocular component (Table 3). Examples from the 40 most differentially expressed probes in each direction for each tissue include: MYOC (MIM 601652), which has been implicated in the development of myopia (Tang et al., 2007; Vatavuk et al., 2009), had probes with 6.40 and 2.22 fold higher expression in the adult relative to fetal scleral tissue and 74.2 fold higher expression in the adult corneal tissue. LAMA1 (MIM 150320), which has also previously been implicated in myopic development (Zhao et al., 2011), had 30.80 lower fold expression in the adult (relative to fetal) choroid. NTM (MIM 607938), which has been implicated in central corneal thickness in primary open angle glaucoma (Ulmer et al., 2012), had −131.38 lower fold expression in adult corneal tissue relative to the fetal tissue. PTPN22 (MIM 600716), implicated in Behçet disease (Baranathan et al., 2007; Sahin et al., 2007), has 46.10 higher fold expression in the adult cornea. Also, all tissues tested included collagens in the most differentially expressed genes (Table 3), many of which have been implicated in ocular diseases.
Additionally, a number of genes previously identified as involved with ocular disease belonged to the most significantly enriched functional annotations for all tissues tested. In particular, there were a large number of genes implicated in either non-syndromic or syndromic high myopia in various degrees, differentially expressed in these tissues and present in the most significantly enriched functional annotations. Fourteen differentially expressed genes associated with non-syndromic myopia were present in the ten most significant functional classifications for the retina/RPE, choroid and/or sclera (Table S4). Many of these myopia-associated genes were found in the same functional annotations with concentrations in those groups related to cell growth and proliferation (including tumorigenesis).
4. Discussion
To our knowledge, this is the first whole genome expression study comparing human adult and fetal retina/RPE, choroid, sclera, optic nerve and cornea. This study investigated gene expression patterns during ocular growth and development in humans. We assessed differentially expressed genes between rapidly growing fetal tissues relative to their mature adult counterparts. Then, we examined the functions and canonical pathways of those differentially expressed genes to extrapolate information regarding the mechanisms of prenatal ocular growth and development. This study identified genes, biological functions and canonical pathways that were most active or inactive during normal growth and development in human ocular tissues. In addition to not having been investigated in this way before, some of the tissues investigated in this study also are involved in clinical features of highly myopic individuals. Investigation of normal growth and development mechanisms in these human tissues may also help to interpret biological mechanisms of ocular disease by providing a reference framework.
4.1. Microarray analyses
Whole genome expression analysis comparing human fetal and adult ocular tissues revealed over 1000 genes that were FDR significantly differentially expressed in each of the tissues tested, except the optic nerve. Tissues-specific gene expression patterns that are likely attributable to their distinct functions were identified for growth and development, such as rhodopsin up-regulation in the adult retina/RPE or hemoglobin down-regulation in the adult vascular choroid. Despite having different biological functions, the ocular tissues examined in this study had overlap in their most differentially expressed genes (Table 3). Some of the overlap shared by all or most tissue types, such as up-regulation of methyltransferases in adult tissues, may be attributed to the fundamental similarities in development between tissues. Other overlaps, such as extracellular matrix genes in the fibrous tissues, may reflect structural similarities between specific tissues. In all tissues examined – especially the fibrous sclera, optic nerve and cornea – there was a general trend of down-regulation of collagen genes in adult tissues, which is consistent with previous findings in mouse postnatal versus adult sclera (Zhou et al., 2006).
4.2. Pathway analyses
All tissues examined in this study had overlap in biological functions the differentially expressed genes were most significantly associated with. While tumorigenesis was one of the most significantly enriched biological functions in all tissues, this finding does not indicate abnormal growth in the tissues samples under investigation. Rather, we believe it suggests that there was an enrichment of genes that regulate normal cell proliferation and/or death mechanisms (also shared by all tissues) that are frequently targeted in the development of cancer. There was considerable variation between tissue types in the most significantly enriched canonical pathways in the sets of differentially expressed genes (Table 5). Yet despite these differences, there was broad consistency in the presence of signaling pathways among the tissues. As with the enrichment for tumorigenesis functional genes, these significantly enriched canonical pathways included disease pathways. The genes involved in these diseases (as reported in the literature) were likely enriched in the data sets not because these tissues are diseased, but because these genes may have normal functional roles in these tissues.
In the retina/RPE, the enrichment of biological functions relating to blood cells and the immune system may be due to differences in the proportion of foveal vs. non-foveal tissues and consequently differences in vasculature in the collected tissue, or indicative of vascular formation in the fetal tissue. A deeper analysis of the direction of expression of these genes and/or differential expression analyses of foveal versus non-foveal retina/RPE may clarify this finding. Similarly, the most significant canonical pathways in the retina/RPE broadly related to vasculature. Again, further study would be necessary to determine the cause of these findings. Interestingly, axonal guidance signaling was the most significantly enriched pathway in the choroid. It is possible that known signaling pathways such as this may also function as the inter-tissue signaling pathways that locally regulate ocular growth in postnatal emmetropization. In the sclera and optic nerve, the enrichment of canonical pathways involving the regulation of cell growth may be because these fibrous tissues undergo substantial growth and remodeling as they develop. Finally, the cornea had the most variation within a tissue in the most significant canonical pathways; though similarly to the other fibrous tissues examined, three of the top five involved the regulation of cell growth.
It is not surprising that all tissues shared functional categories involving cell growth, death and development given that they were in a state of rapid growth and development. These significantly enriched functional categories were identified from IPA's network derived from scientific findings, suggesting that many of the prenatal growth and development mechanisms in the eye are similar to other mechanisms found in other tissues and systems. Likewise, these same genes and pathways involved in normal growth may contribute to abnormal growth when dysfunctional. This abnormal growth could arise in the form of a tumor when restricted to a specific tissue or cell type, or could accompany corresponding changes in nearby tissues. Our findings suggest that the prenatal growth and development seen in these ocular tissues is largely influenced by standard mechanisms and this data provides a useful framework to study candidates. However, we are still unsure how these tissues are communicating with one another and how prenatal and postnatal ocular growth signals may differ. While, these inter-tissue signaling molecules may not be among the most significant functional categories if they are indeed specific to the eye and not well established in the literature; other molecules in the mechanistic pathway could still be identified as a starting point.
4.3. Disease gene overlap
In all tissues, a number of genes previously implicated in ocular disease were identified among the most differentially expressed genes and the most significantly associated biological functions. It is not unreasonable to think that the disruption of these genes in their normal functions within these tissues may contribute to or cause disease. For example, as a gene that regulates cellular growth may be mutated to allow unrestricted cancerous growth, a mutation in a gene regulating local control of eye growth may result in failure of the emmetropization process leading to refractive error. While these differentially expressed genes presented were not identified in diseased versus normal tissue, our findings suggest that many disease genes also have roles in normal prenatal growth and development of the eye. This information on the normal roles of these genes in the tissues with phenotypic changes in diseases eyes may help to elucidate their mechanistic roles in disease development.
4.4. Consideration of limitations
Since the fetal and adult eyes were obtained from different sources, expression differences could in part be attributed to different processing methods and time frames for procurement.
None of the genes in the optic nerve met FDR statistical significance-However, other qualitative metrics such as probe concordance and pathway assessment were comparable to the other tissues examined in this study, lending some credibility to their validity. It is unlikely that there are no truly differentially expressed genes during growth and development in the optic nerve given the physical and developmental changes the tissue undergoes, the fold changes seen in expression, and the overlap with other tissues in genes and enriched functional annotations of the set of genes. The optic nerve may have failed to reach statistical significance for differential expression of any of its genes for several reasons, such as its small sample size (due to sample drop out) or biological variation (due to variation in the proportion of optic nerve stem to head in biological replicates). It is more likely that the high variability between samples was the major contributor, as we did note large expression changes (Table 2).
There were also potential confounding factors in the assessment of differentially expressed genes during growth and development in the retina/RPE in this study. Combining the retina and RPE may have reduced the power to detect expression differences, which may account for the relatively low number of differentially expressed genes and their corresponding fold changes. Another potential caveat is the possible differences in contribution of foveal tissues to the posterior retina/RPE in the fetal versus adult tissues. Given that the same size biopsy punch was used in both age groups, it may be assumed that the fetal tissue included a higher proportion of non-foveal retina/RPE than the adult tissue.
4.5. Conclusions
We present the first whole genome expression analysis comparing human adult and fetal retina/RPE, choroid, sclera, optic nerve and cornea. This data provides a wealth of information regarding the genes, biological functions and canonical pathways that are up- or down-regulated during growth and development. A deeper understanding of the expression patterns in the developing human eye, as gained by this information, was a missed early step in understanding how genetic defects or damage may manifest as diseases. This information is not only beneficial for congenital diseases arising from developmental failures, but may also be used to study other diseases resulting from failures to the normal visual system such as myopia. This data set provides a network with which to study the normal expression, function and pathway interactions of genes in ocular tissues that are relevant to disease development. This information could help generate testable hypotheses for the mechanistic roles of these genes in disease development. Although not emphasized in this manuscript, there were limited microRNAs (approximately 200) that were incidentally expressed in various ocular tissues. A future study of a purposeful microRNA microarray or RNASeq study of ocular tissues would be useful for subsequent microRNA target identification and functional validation of predicted targets or to study the regulatory effects of miRNAs on target genes. We propose that the information in this data set may be used to prioritize candidate genes by providing insights into how they might function in disease development as well as confirming their presence in the relevant tissues. However, further genetic and/or functional validation of candidate genes would be necessary to make specific claims regarding their involvement in the development of myopia.
This expression information may be particularly useful when other genetic means, such as association mapping or sequencing, fail to reduce the number of candidate genes to a cost and resource effective number. The expression data presented in this study for the retina/RPE, choroid and sclera was also used to prioritize candidate genes from a genetic association mapping study for the high myopia locus MYP3 (MIM 603221) (Hawthorne et al., 2013). While expression data alone is not sufficient to determine candidates for ocular disease, it may be used interpret and prioritize candidates when validation of a large number of genes in not feasible or practical, as was the case with the MYP3 locus. The candidates identified in our MYP3 association mapping study were found to be nominally significantly associated in an independent cohort and one of the candidates, PTPRR (MIM 602853), was also recently identified in a large scale meta-analysis of myopia GWA study (Verhoeven et al., 2013). This replication of a candidate gene for myopic development supports its validity and also provides preliminary support of the utilization of this expression data for disease candidate prioritization.
4.6. Future directions
While these hypotheses require further testing, we have provided a novel framework (normal ocular growth and development) in which to interpret and contextualize candidate genes for ocular disease. Early analyses of the functional classifications of these genes may help elucidate their potential roles in disease progression, particularly their tissue-specific roles. Next, a deeper understanding of the mechanisms connecting these tissues may be better studied using the enriched signaling pathways identified. Genes whose mutations may confer an increased risk for ocular disease via known or undetermined pathways may be components of these established pathways regardless of their differential expression in the tissues. This data could be used to prioritize candidate genes within previously identified disease loci containing large numbers of genes. Supplemental Table S5 contains a list of genes within MIM recognized myopia loci (a focus disease in our lab) which are differentially expressed in the retina/RPE, choroid, and/or sclera. We have only scratched the surface of the possibilities with this vast resource by interpreting and prioritizing candidate genes for one disease study of myopia. The use of this information for other loci and common ocular diseases associated with these tissues should be explored further.
Supplementary Material
Acknowledgments
We acknowledge and thank the donors who made this study possible. We thank Janeen Morgan for assistance in data acquisition. We thank Catherine Bowes Rickman for technical advice. We also acknowledge the following sources of funding: NIH Grant R01-EY014685 and Research To Prevent Blindness Inc (both to TLY).
Footnotes
Accession numbers: The accession number(s) for the microarray data reported in the paper will be deposited in the Gene Expression Omnibus database.
Web resource: The URL for data presented herein is as follows:
Online Mendelian Inheritance in Man (OMIM), http://ncbi.nlm.nih.gov/Omim.
Appendix A. Supplementary data: Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.exer.2013.08.009.
References
- Atchison DA, Jones CE, Schmid KL, Pritchard N, Pope JM, Strugnell WE, Riley RA. Eye shape in emmetropia and myopia. Invest Ophthalmol Vis Sci. 2004;45:3380–3386. doi: 10.1167/iovs.04-0292. [DOI] [PubMed] [Google Scholar]
- Baranathan V, Stanford MR, Vaughan RW, Kondeatis E, Graham E, Fortune F, Madanat W, Kanawati C, Ghabra M, Murray PI, Wallace GR. The association of the PTPN22 620W polymorphism with Behcet's disease. Ann Rheum Dis. 2007;66:1531–1533. doi: 10.1136/ard.2007.073866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc Ser B (Methodol) 1995;57:289–300. [Google Scholar]
- Bremond-Gignac D, Copin H, Lapillonne A, Milazzo S. Visual development in infants: physiological and pathological mechanisms. Curr Opin Ophthalmol. 2011;22(Suppl):S1–S8. doi: 10.1097/01.icu.0000397180.37316.5d. [DOI] [PubMed] [Google Scholar]
- Durrenberger PF, Fernando S, Kashefi SN, Ferrer I, Hauw JJ, Seilhean D, Smith C, Walker R, Al-Sarraj S, Troakes C, Palkovits M, Kasztner M, Huitinga I, Arzberger T, Dexter DT, Kretzschmar H, Reynolds R. Effects of antemortem and postmortem variables on human brain mRNA quality: a BrainNet Europe study. J Neuropathol Exp Neurol. 2010;69:70–81. doi: 10.1097/NEN.0b013e3181c7e32f. [DOI] [PubMed] [Google Scholar]
- Ervin JF, Heinzen EL, Cronin KD, Goldstein D, Szymanski MH, Burke JR, Welsh-Bohmer KA, Hulette CM. Postmortem delay has minimal effect on brain RNA integrity. J Neuropathol Exp Neurol. 2007;66:1093–1099. doi: 10.1097/nen.0b013e31815c196a. [DOI] [PubMed] [Google Scholar]
- Faulkner AE, Kim MK, Iuvone PM, Pardue MT. Head-mounted goggles for murine form deprivation myopia. J Neurosci Methods. 2007;161:96–100. doi: 10.1016/j.jneumeth.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Fledelius HC, Christensen AC. Reappraisal of the human ocular growth curve in fetal life, infancy, and early childhood. Br J Ophthalmol. 1996;80:918–921. doi: 10.1136/bjo.80.10.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon RA, Donzis PB. Refractive development of the human eye. Arch Ophthalmol. 1985;103:785–789. doi: 10.1001/archopht.1985.01050060045020. [DOI] [PubMed] [Google Scholar]
- Hawthorne F, Feng S, Metlapally R, Li YJ, Tran-Viet KN, Guggenheim JA, Malecaze F, Calvas P, Rosenberg T, Mackey DA, Venturini C, Hysi PG, Hammond CJ, Young TL. Association mapping of the high-grade myopia MYP3 locus reveals novel candidates UHRF1BP1L, PTPRR, and PPFIA2. Invest Ophthalmol Vis Sci. 2013 Mar 21;54(3):2076–2086. doi: 10.1167/iovs.12-11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotelling H. The generalization of student's ratio. Ann Math Statist. 1931;2:360–378. [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Kolb H, Nelson R, Fernandez E, Jones B. Kolb H, Nelson R, Fernandez E, Jones B, editors. Webvision – the Organization of the Retina and the Visual System. 2011 http://webvision.med.utah.edu/ [PubMed]
- McCurdy RD, McGrath JJ, Mackay-Sim A. Gene Ther Mol Biol. 2008;12:15–24. [Google Scholar]
- Sahin N, Bicakcigil M, Atagunduz P, Direskeneli H, Saruhan-Direskeneli G. PTPN22 gene polymorphism in Behcet's disease. Tissue Antigens. 2007;70:432–434. doi: 10.1111/j.1399-0039.2007.00928.x. [DOI] [PubMed] [Google Scholar]
- Sundin OH, Dharmaraj S, Bhutto IA, Hasegawa T, McLeod DS, Merges CA, Silval ED, Maumenee IH, Lutty GA. Developmental basis of nano-phthalmos: MFRP is required for both prenatal ocular growth and postnatal emmetropization. Ophthalmic Genet. 2008;29:1–9. doi: 10.1080/13816810701651241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WC, Yip SP, Lo KK, Ng PW, Choi PS, Lee SY, Yap MK. Linkage and association of myocilin (MYOC) polymorphisms with high myopia in a Chinese population. Mol Vis. 2007;13:534–544. [PMC free article] [PubMed] [Google Scholar]
- Tkatchenko TV, Shen Y, Tkatchenko AV. Mouse experimental myopia has features of primate myopia. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.09-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer M, Li J, Yaspan BL, Ozel AB, Richards JE, Moroi SE, Hawthorne F, Budenz DL, Friedman DS, Gaasterland D, Haines J, Kang JH, Lee R, Lichter P, Liu Y, Pasquale LR, Pericak-Vance M, Realini A, Schuman JS, Singh K, Vollrath D, Weinreb R, Wollstein G, Zack DJ, Zhang K, Young T, Allingham RR, Wiggs JL, Ashley-Koch A, Hauser MA. Genome-wide analysis of central corneal thickness in primary open-angle glaucoma cases in the NEIGHBOR and GLAUGEN Consortia. Invest Ophthalmol Vis Sci. 2012;53:4468–4474. doi: 10.1167/iovs.12-9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatavuk Z, Skunca Herman J, Bencic G, Andrijevic Derk B, Lacmanovic Loncar V, Petric Vickovic I, Bucan K, Mandic K, Mandic A, Skegro I, Pavicic Astalos J, Merc I, Martinovic M, Kralj P, Knezevic T, Barac-Juretic K, Zgaga L. Common variant in myocilin gene is associated with high myopia in isolated population of Korcula Island, Croatia. Croat Med J. 2009;50:17–22. doi: 10.3325/cmj.2009.50.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, Höhn R, Macgregor S, Hewitt AW, Nag A, Cheng CY, Yonova-Doing E, Zhou X, Ikram MK, Buitendijk GH, McMahon G, Kemp JP, Pourcain BS, Simpson CL, Mäkelä KM, Lehtimäki T, Kähönen M, Paterson AD, Hosseini SM, Wong HS, Xu L, Jonas JB, Pärssinen O, Wedenoja J, Yip SP, Ho DW, Pang CP, Chen LJ, Burdon KP, Craig JE, Klein BE, Klein R, Haller T, Metspalu A, Khor CC, Tai ES, Aung T, Vithana E, Tay WT, Barathi VA, Consortium for refractive error and myopia (CREAM) Chen P, Li R, Liao j, Zheng Y, Ong RT, Döring a, Diabetes control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Research group. Evans DM, Timpson NJ, Verkerk AJ, Meitinger T, Raitakari O, Hawthorne F, Spector TD, Karssen LC, Pirastu M, Murgia F, Ang W, Wellcome Trust case control Consortium 2 (WTCCC2) Mishra a, Montgomery GW, Pennell CE, Cumberland PM, Cotlarciuc I, Mitchell P, Wang JJ, Schache M, Janmahasatian S, RP, Jr, Lass JH, Chew E, Iyengar SK, Fuchs' genetics Multi-center study group. Gorgels TG, Rudan I, Hayward C, Wright AF, Polasek O, Vatavuk z, Wilson JF, Fleck B, Zeller T, Mirshahi a, Müller C, Uitterlinden AG, Rivadeneira F, Vingerling JR, Hofman a, Oostra BA, Amin N, Bergen AA, Teo YY, Rahi JS, Vitart V, Williams C, Baird PN, Wong TY, Oexle K, Pfeiffer N, Mackey DA, Young TL, van Duijn CM, Saw SM, Bailey-Wilson jE, Stambolian D, Klaver CC, Hammond CJ. Genome-wide meta-analyses of multi-ancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013 Jun;45(6):712. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43:447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Wilcoxen F. Individual comparisons by ranking methods. Biometr Bull. 1945;1:80–83. [PubMed] [Google Scholar]
- Wildsoet C, Wallman J. Choroidal and scleral mechanisms of compensation for spectacle lenses in chicks. Vis Res. 1995;35:1175–1194. doi: 10.1016/0042-6989(94)00233-c. [DOI] [PubMed] [Google Scholar]
- Xu L, Li Y, Wang S, Wang Y, Jonas JB. Characteristics of highly myopic eyes: the Beijing Eye Study. Ophthalmology. 2007;114:121–126. doi: 10.1016/j.ophtha.2006.05.071. [DOI] [PubMed] [Google Scholar]
- Zhao YY, Zhang FJ, Zhu SQ, Duan H, Li Y, Zhou ZJ, Ma WX, Li Wang N. The association of a single nucleotide polymorphism in the promoter region of the LAMA1 gene with susceptibility to Chinese high myopia. Mol Vis. 2011;17:1003–1010. [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Rappaport EF, Tobias JW, Young TL. Differential gene expression in mouse sclera during ocular development. Invest Ophthalmol Vis Sci. 2006;47:1794–1802. doi: 10.1167/iovs.05-0759. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


