Abstract
In a recent study, we demonstrated that structurally compact fluorophores incorporated into the side chains of amino acids could be introduced into dihydrofolate reductase from E. coli (ecDHFR) with minimal disruption of protein structure or function, even where the site of incorporation was within a folded region of the protein. The modified proteins could be employed for FRET measurements, providing sensitive monitors of changes in protein conformation. The very favorable results achieved in that study encouraged us to prepare additional fluorescent amino acids of potential utility for studying protein dynamics. Presently, we describe the synthesis and photophysical characterization of four positional isomers of biphenyl-phenylalanine, all of which were found to exhibit potentially useful fluorescent properties. All four phenylalanine derivatives were used to activate suppressor tRNA transcripts, and incorporated into multiple positions of ecDHFR. All phenylalanine derivatives were incorporated with good efficiency into position 16 of ecDHFR, and afforded modified proteins which consumed NADPH at rates up to about twice the rate measured for wild type. This phenomenon has been noted on a number of occasions previously and shown to be due to an increase in the off-rate of tetrahydrofolate from the enzyme, altering a step that is normally rate limiting. When introduced into sterically accessible position 49, the four phenylalanine derivatives afforded DHFRs having catalytic function comparable to wild type. The four phenylalanine derivatives were also introduced into position 115 of ecDHFR, which is known to be a folded region of the protein less tolerant of structural alteration. As anticipated, significant differences were noted in the catalytic efficiencies of the derived proteins. The ability of two of the sizeable biphenyl-phenylalanine derivatives to be accommodated at position 115 with minimal perturbation of DHFR function is attributed to rotational flexibility about the biphenyl bonds.
Förster resonance energy transfer (FRET) has greatly facilitated the study of protein structure and function, as well as the interaction between proteins.1,2 Excitation of the fluorescence donor results in energy transfer to an acceptor, the latter of which can be another fluorescent molecule or a quencher.2–6 In the case of a fluorescent acceptor, the overlap in the emission spectrum of the donor and excitation spectrum of the acceptor results in acceptor emission at a longer wavelength. A widely used FRET system for the visualization of protein is based on two different color variants of the green fluorescent protein (GFP), in which the blue or cyan mutant of GFP is used as the donor and the green or yellow mutant GFP is employed as the fluorescence acceptor (emitter).1,7–9 Two selected fluorescent proteins can be fused to the C- and N- termini, respectively, of a single protein to study changes in its folding;7–9 or fused to two interacting partners to study protein complex formation.10 While the FRET approach based on fluorescent proteins is a powerful strategy for studying protein interactions, the large size of the fluorophores imposes significant constraints on the observations of relatively small conformational changes in single proteins.
Chemical methods have also been used to introduce FRET donor/acceptor pairs into a single protein at two desired positions, e.g. through the coupling of cysteine residues with maleimide derivatives.11,12 However, since chemically accessible cysteines on a protein surface tend to be of comparable reactivity, the fluorophore and quencher are typically not introduced uniquely at the reactive sites. This need not affect ensemble studies due to signal averaging; however, the mixed population of labeled species can complicate the interpretation of the two signals observed in single molecule studies.12 Additionally, because the fluorophores/quenchers are often large polycyclic aromatic molecules attached to the proteins via flexible linkers, they are capable of a range of motion even in the absence of any protein conformational change. Again, this renders them poorly suited to measure small conformational changes in proteins, such as those involved in the catalytic cycle of many enzymes.
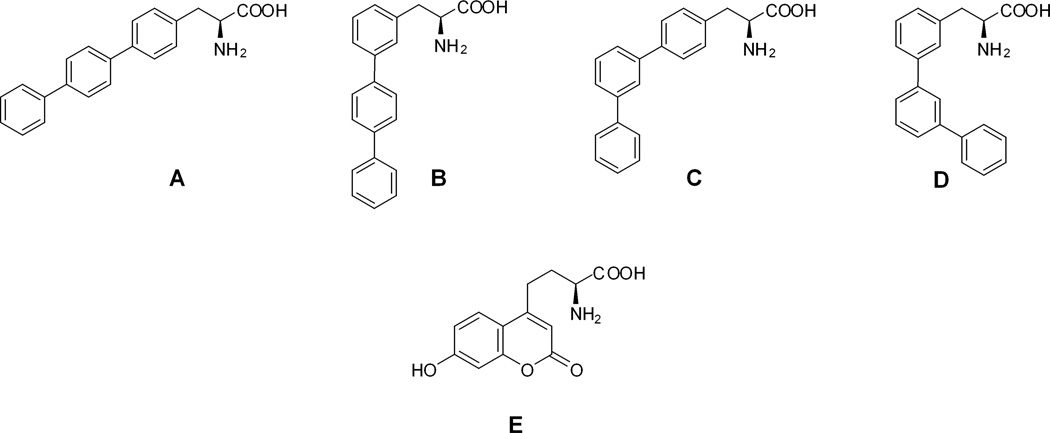
A biosynthetic method involving the use of misacylated suppressor transfer RNAs has also been employed to incorporate fluorescent amino acids into proteins in a site-specific fashion.13–18 Studies of this type involving the incorporation of donor and acceptor amino acids has enabled the measurement of protein backbone cleavage19 and relatively large changes in protein structure.20,21 Recently, we described the preparation of modified dihydrofolate reductases (DHFRs) containing two fluorescent amino acids with relatively compact side chains, namely 4-biphenyl-L-phenylalanine (A) and L-(7-hydroxycoumarin-4-yl)ethylglycine (E)22 (Figure 1), as fluorescence donor and acceptor, respectively. We demonstrated that both amino acids were well tolerated at sterically accessible position 17, but that the DHFR containing 4-biphenyl-L-phenylalanine at position 115 (which is within a folded region of the protein) functioned better catalytically, gave more efficient energy transfer, and provided more information concerning small conformational changes in the protein.23 This finding suggested the importance of utilizing fluorescent amino acids capable of minimal perturbation of protein structure to enable sensitive monitoring of small changes in protein structure. The conformational flexibility about the biphenyl linkages in A seemed of great interest in this regard.
Figure 1.
Biphenyl-phenylalanine (A – D) and L-(7-hydroxycoumarin-4-yl)ethylglycine (E) amino acids prepared and incorporated into dihydrofolate reductase.
Presently, we describe the synthesis and characterization of three novel isomeric biphenyl-L-phenylalanine derivatives (B – D) which differ from A only in the orientation of the biphenyl rings. These new fluorescent amino acids have somewhat different photophysical properties in comparison with A, and were anticipated to be able to accommodate different steric requirements in replacing proteinogenic amino acids in folded regions of proteins. It was found that all four afforded DHFRs having comparable catalytic properties when incorporated into sterically accessible positions 16 and 49, but that catalytic function was affected differently when A – D were introduced into sterically encumbered position 115. Also assessed was the ability of these four fluorophores to transfer energy to L-(7-hydroxycoumarin-4-yl)ethylglycine (E) at DHFR position 17.
EXPERIMENTAL PROCEDURES
Synthesis of Biphenyl-L-phenylalanyl-pdCpA Derivatives
The synthesis of the aminoacylated pdCpA derivative of amino acid A has been reported previously.12 The syntheses of the pdCpA derivatives of amino acids B, C and D are outlined in Schemes 1, S1 and S2, respectively, and the experimental procedures and compound characterizations are provided in the Supporting Information.
Scheme 1.
Synthetic Route Employed for the Preparation of Biphenyl-phenylalanyl-pdCpA 6
Biochemical Experiments
Ligation of Suppressor tRNACUA-COH with Biphenyl-phenylalanine Analogues and Deprotection of N-Pentenoyl Group
The yeast suppressor tRNAPheCUA was prepared as previously reported.24 Activation of suppressor tRNACUA was carried out in 100 µL (total volume) of 100 mM Hepes buffer, pH 7.5, containing 2.0 mM ATP, 15 mM MgCl2, 100 µg of suppressor tRNA-COH, 2.0 A260 units of N-pentenoyl protected aminoacyl-pdCpA (5–10 fold molar excess), 15% DMSO and 200 units of T4 RNA ligase. After incubation at 37 °C for 1 h, the reaction was quenched by the addition of 10 µL of 3 M NaOAc, pH 5.2, followed by 300 µL of ethanol. The reaction mixture was incubated at −20 °C for 30 min, then centrifuged at 15,000 × g at 4 °C for 30 min. The supernatant was carefully decanted and the tRNA pellet was washed with 100 µL of 70% ethanol, and dissolved in 80 µL of RNase free H2O. The efficiency of ligation was estimated by 8% denaturing PAGE (pH 5.2).25 The pentenoyl protecting group was removed by treatment with 5 mM I2 at room temperature for 10 min.26,27 The reaction mixture was treated with 10 µL of 0.3 M NaOAc, pH 5.2. followed by 300 µL of ethanol. After centrifugation at 15,000 × g at 4 °C for 30 min, the supernatant was carefully decanted and the tRNA pellet was washed with 100 µL of 70% ethanol, and dissolved in 35 µL of RNase free H2O.
In vitro Translation of DHFR Containing Biphenyl-phenylalanine Analogues at Positions 16, 49 and 115
The modified DHFR plasmid was obtained by site-directed mutagenesis as described as previously using the wild-type DHFR plasmid as the template.28 The DNA primer for mutagenesis at position 16 was 5′-GCGGTAGATCGCGTTATCGGCTAGGAAAACGCCATGCCGTGGAAC-3′; the primer for mutagenesis at position 49 was 5′-GGCCGCCATACCTGGGAATAGATCGGTCGTCCGTTGCCAG-3′; the primer for mutagenesis at position 115 was 5′-CGCAAAAACTGTATCTGACGCATTAGGACGCAGAAGTGGAAGGCGACAC-3′.
The in vitro expression mixture (300 µL total volume) contained 30 µg of modified DHFR (TAG at position 16, 49 or 115) plasmid DNA, 120 µL of premix (35 mM Tris-acetate, pH 7.0, 190 mM potassium glutamate, 30 mM ammonium acetate, 2.0 mM dithiothreitol, 11 mM maganesium acetate, 20 mM phospho(enol)pyruvate, 0.8 mg/mL of E. coli tRNA, 0.8 mM IPTG, 20 mM ATP and GTP, 5 mM CTP and UTP and 4 mM cAMP), 100 µM of each of the 20 amino acids, 30 µCi of [35S]-L-methionine, 10 µg/µL rifampicin, 90 µg of deprotected misacylated tRNACUA and 90 µL of S-30 extract from E. coli strain BL21(DE3). The reaction mixture was incubated at 37 °C for 45 min. Plasmid DNA containing the gene for wild-type DHFR was used as the positive control, and an abbreviated tRNA (tRNA-COH) lacking any amino acid was used as the negative control. An aliquot containing 2 µL of reaction mixture was removed, treated with 2 µL of loading buffer and heated at 90 °C for 2 min. The sample was analyzed by 15% SDS-PAGE at 100 V for 2 h.
Purification of DHFR Analogues
The analogues of DHFR containing an N-terminal hexahistidine fusion peptide were purified by Ni-NTA chromatography. The in vitro translation reaction mixture (300 µL) was diluted with 900 µL of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl and 10 mM imidazole, and mixed gently with 100 µL of a 50% slurry of Ni-NTA resin at 4 °C for 2 h. Then the mixture was applied to a column and washed with 600 µL of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl and 20 mM imidazole. Finally, the DHFR analogue was washed three times with 200 µL of 50 mM Tris-HCl, pH 8.0, containing 30 mM NaCl and 150 mM imidazole. The final three Ni-NTA column eluates were combined and applied to a 100-µL DEAE-Sepharose CL-6B column. The column was washed with 300 µL of 50 mM Tris-HCl, pH 8.0, containing 100 mM NaCl, 300 µL of 50 mM Tris-HCl, pH 8.0, containing 200 mM NaCl, and then three 100-µL portions of 50 mM Tris-HCl, pH 8.0, containing 300 mM NaCl. Aliquots of each fraction were analyzed by 15% SDS-PAGE.
Enzymatic Activities of DHFR Analogues
The enzymatic activities of wild-type and modified DHFRs were measured in 1 mL of MTEN buffer (containing 50 mM MES, 25 mM Tris, 25 mM ethanolamine, 100 mM NaCl, 0.1 mM EDTA and 10 mM β-mercaptoethanol, pH 7.0). MTEN buffer (0.97 mL) was mixed with 10 µL of 10 mM NADPH and 100 ng of protein.29 The mixture was incubated at 37 °C for 3 min. Then 20 µL of 5 mM dihydrofolate in MTEN buffer, pH 7.0, was added. The A340 value was monitored over a period of 10 min.
Inhibition of Wild-type and Modified DHFRs by Methotrexate and Trimethoprim
The enzymatic activities of wild-type DHFR and modified DHFRs (biphenyl-phenylalanines at position 115) were measured in 1 mL of MTEN buffer (containing 50 mM MES, 25 mM trizma base, 25 mM ethanolamine, 100 mM NaCl, 0.1 mM EDTA and 10 mM 2-mercaptoethanol, pH 7.0). MTEN buffer (0.97 mL) was mixed with 10 µL of 10 mM NADPH, 100 ng of DHFR and 0.5 – 50 nM MTX or TMP. The reaction mixture was incubated at 37 °C for 3 min. Then 20 µL of 5 mM dihydrofolate in MTEN buffer, pH 7.0, was added. The OD value at 340 nm was monitored over a period of 10 min.
Fluorescence Spectra of DHFR Analogues
The fluorescence spectra of DHFR analogues were measured using a Varian Cary Eclipse Fluorescence Spetrophotometer with the excitation slit as 10 nm and emission slit as 10 nm. The protein samples (0.2 – 1.0 µM) were excited with ultraviolet light close to the excitation maxima, and the emission spectra were recorded.
Quantum Yields of Fluorescent Compounds
Fluorescence quantum yields of fluorescent compounds were determined using the gradient method.30 The N-pentenoyl derivatives of biphenyl-phenylalanines A – D were dissolved in acetonitrile. Solutions of each compound were made such that the UV absorption at the maximum wavelength were 0.02, 0.04, 0.06, 0.08 and 0.1. Two standards, 2-aminopyridine (ΦF 0.60, λex 295 nm) and anthracene (ΦF 0.27, λex 340 nm), were used to calculate the fluorescent quantum yield of the N-pentenoyl biphenyl-phenylalanine analogues according to the formula Φx= Φs× (Gradx × nx2)/(Grads × ns2), where Grad is gradient of the plot of integrated intensity versus absorbance, n is the refractive index of the solvent, s is the standard of known ΦF, and x is the tested sample.30
RESULTS
Synthesis of Fluorescent Aminoacyl pdCpAs Esters
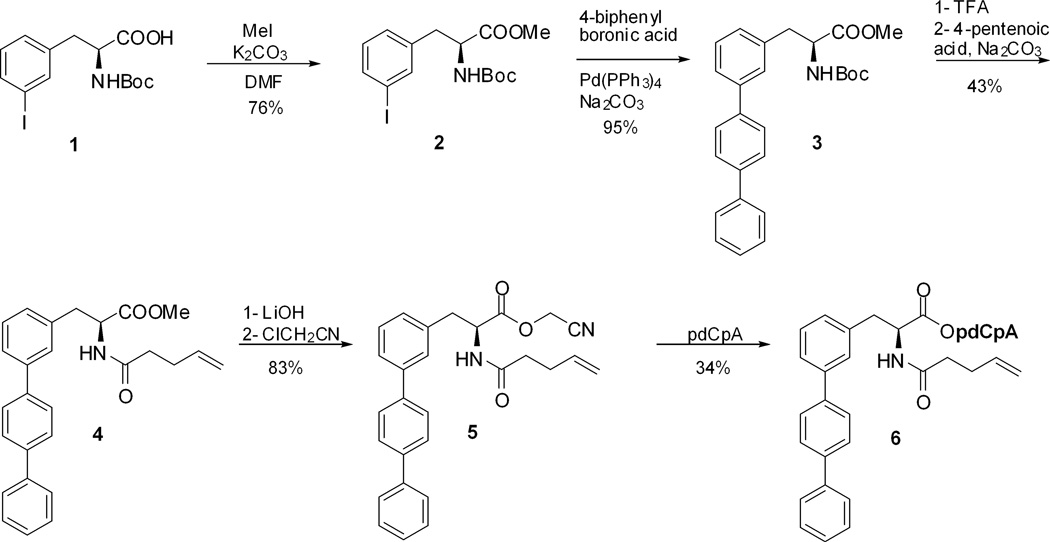
The preparation of the fluorescent amino acids (A – D, Figure 1) was based on a Suzuki coupling of two different biphenyl boronic acids with protected 3- and 4-iodophenylalanine.31 The coupling product was obtained in nearly quantitative yield in each case. These terphenylalanine analogues were activated as cyanomethyl esters and coupled with the dinucleotide pdCpA to afford the corresponding aminoacyl pdCpAs. The synthesis of the aminoacylated pdCpA derivative of amino acid A has been reported previously.23 The synthesis of biphenyl-phenylalanyl-pdCpA 6, containing amino acid B, was accomplished starting from commercially available N-Boc-3-iodophenylalanine (1), which was esterified using iodomethane in DMF in the presence of K2CO3.32,33 N-Boc-3-iodophenylalanine methyl ester (2) was obtained in 76% yield (Scheme 1). Treatment of compound 2 with 4-biphenylboronic acid in the presence of Pd(PPh3)4 gave N-(tert-butoxycarbonyl)-[4-(1,1’,3’,1”)-terphenyl]-L-alanine methyl ester (3) in 95% yield. Removal of the Boc protecting group from compound 3 using TFA, followed by treatment with 4-pentenoic acid succinimide ester26,27 in the presence of Na2CO3, afforded the N-pentenoyl protected amino acid 4 in 43% overall yield. The conversion of 4 to the corresponding cyanomethyl ester 5 was achieved following hydrolysis of the methyl ester using LiOH by treatment with chloroacetonitrile in the presence of triethylamine (83% yield). Treatment of cyanomethyl ester 5 with a solution of tris-(tetrabutylammonium) salt of pdCpA34 in dry DMF afforded the corresponding pdCpA ester 6 in 34% yield.
The syntheses of the remaining two aminoacylated pdCpA derivatives, incorporating amino acids C and D were prepared starting from N-Boc-4-iodophenylalanine and N-Boc-3-iodophenylalanine, respectively (Schemes S1 and S2). For amino acid C, the fully protected 4-iodophenylalanine was treated with 3-biphenylboronic acid in the presence of Pd(PPh3)4 to afford the biphenyl-phenylalanine derivative in 95% yield (Scheme S1). Replacement of the Boc-protecting group by an N-pentenoyl group was achieved by treatment with TFA followed by treatment of the resulting amine intermediate with 4-pentenoic acid succinimidyl ester (71% overall yield). Hydrolysis of the methyl ester followed by treament with chloroacetonitrile in the presence of triethylamine gave the cyanomethyl ester in 76% yield. Treatment of the cyanomethyl ester with a solution of tris-(tetrabutylammonium) salt of pdCpA in dry DMF gave the corresponding aminoacylated pdCpA containing amino acid C in 38% yield.
The synthesis of the pdCpA derivative activated with amino acid D was carried out analogously by treating fully protected 3-iodophenylalanine with 3-phenylboronic acid in the presence of Pd(PPh3)4 to afford the biphenyl-phenylalanine derivative in 93% yield (Scheme S2). The remaining functional group manipulations were the same as for amino acid C; each of the intermediates and the final pdCpA derivative incorporating amino acid D were also obtained in yields roughly comparable to those shown in Scheme S1.
Activation of Suppressor tRNACUA and Synthesis of Modified DHFRs
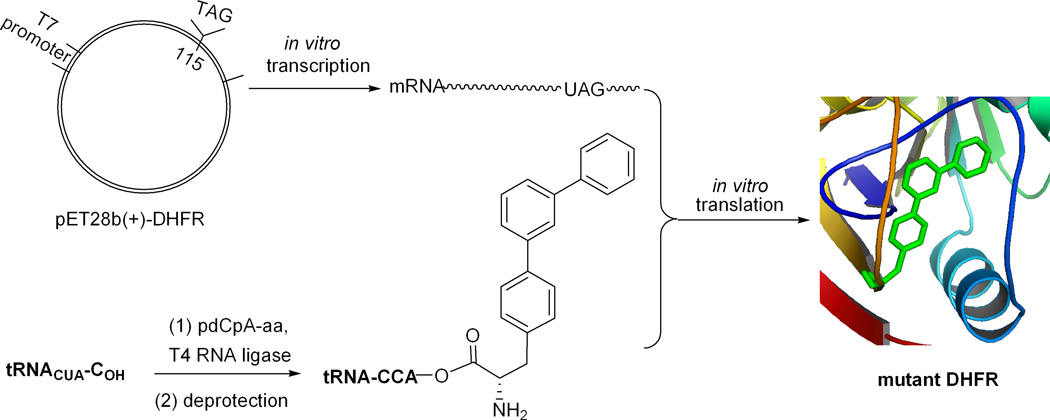
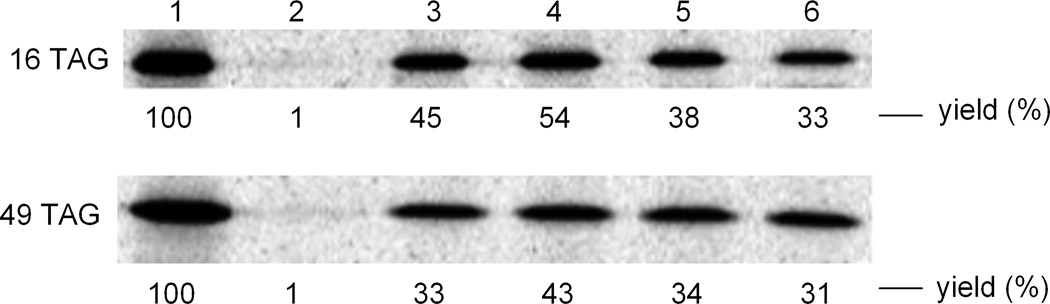
The individual N-pentenoyl protected aminoacylated pdCpA derivatives were ligated via the agency of T4 RNA ligase to an in vitro RNA transcript having the sequence of a suppressor tRNACUA lacking its 3′-terminal cytidine and adenosine residues (tRNACUA-COH24; illustrated in Scheme 1 for the suppressor tRNA transcript activated with biphenyl-phenylalanine B). As shown in Figure S1, this afforded full length tRNA transcripts activated with amino acids A, B, C and D. The N-pentenoyl protecting groups were then removed by brief treatment with aqueous iodine,26,27 after which the aminoacyl-tRNAs were employed in a cell free prokaryotic protein synthesizing system programmed with DNA plasmids encoding DHFR and having TAG codons at the positions corresponding to amino acids 16, 49 or 115 in DHFR. This is illustrated in Scheme 2 for the incorporation of biphenyl-phenylalanine derivative C into position 115 of DHFR. Protein translation was carried out in a coupled transcription-translation system in the presence of a single activated suppressor tRNA. As shown in Figure 2, each of the four biphenyl-phenylalanyl-tRNAs afforded good suppression of the UAG codons at positions 16 and 49 of DHFR mRNAs, with suppression yields ranging from 31 to 54%. Each of the modified DHFRs contained an N-terminal hexahistidine fusion peptide,24 enabling initial purification on a Ni-NTA column,35 illustrated in Figure S2 for the DHFR containing amino acid A at position 16. Final purification was then accomplished on a DEAE-Sepharose CL-6B column.
Scheme 2.
Strategy Employed for Incorporation of Biphenyl-phenylalanine C into DHFR at Position 115.
Figure 2.
Autoradiogram of a 15% SDS-polyacrylamide gel (100 V, 2 h) illustrating the incorporation of biphenyl-phenylalanine derivatives into positions 16 (upper panel) and 49 (lower panel) of DHFR. Lane 1, wild-type DHFR expression; lane 2, modified DHFR DNA in the presence of abbreviated suppressor tRNACUA-COH; lane 3, incorporation of amino acid A; lane 4, incorporation of amino acid B; lane 5, incorporation of amino acid C; lane 6, incorporation of amino acid D. Phosphorimager analysis was performed using an Amersham Biosciences Storm 820 equipped with ImageQuant version 5.2 software from Molecular Dynamics.
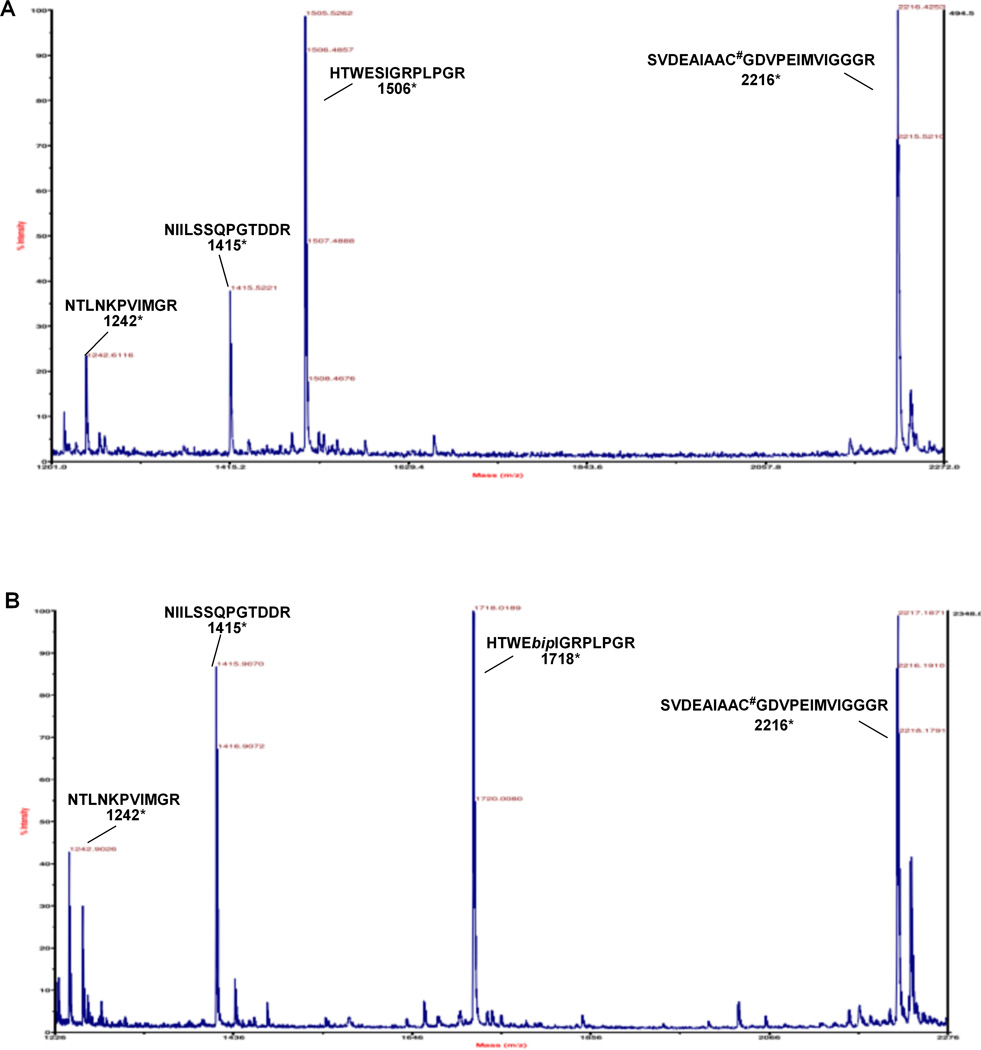
In order to verify the incorporation of the biphenyl-phenylalanine derivatives into DHFR at the intended sites, the DHFR analogue putatively containing biphenyl-phenylalanine derivative C at position 49 was prepared on a larger scale. Following purification on a polyacrylamide gel, the modified DHFR was subjected to in-gel digestion as described previously,36 and then analyzed by MALDI mass spectrometry. As shown in Figure 3A, the peptide fragment containing amino acids 45–57 was present in the digest prepared from wild type at m/z 1506, consistent with the anticipated presence of serine at position 49. In comparison, the mass spectrum of the modified DHFR putatively containing biphenyl-phenylalanine at position 49 lacked this fragment ion, but contained the expected fragment ion at m/z 1718 (Figure 3B). All of the other observable fragment ions in the two mass spectra were the same (Table 1).
Figure 3.
MALDI-MS of tryptic fragments of wild-type (panel A) and modified DHFR containing biphenyl-phenylalanine C at position 49 (panel B) (*: calculated value in Da; #: cysteine was alkylated with 2-iodoacetamide; bip: biphenyl-phenylalanine derivative C). Figure 4.
Table 1.
MALDI-MS Analysis of Tryptic Digests of Wild-type and Modified DHFRs (Biphenyl-phenylalanine C in Position 49). Position 49 is Denoted in ![]()
| position | peptide sequence | MALDI-MS analysis, molecular mass, Da |
|||
|---|---|---|---|---|---|
| wild-type | modified | ||||
| Est | MS | Est | MS | ||
| 13– 32 | VIGMENAMPWNLPADLAWFK | 2304 | 2304.1 | 2304 | 2304.3 |
| 34–44 | NTLNKPVIMGR | 1242 | 1242.8 | 1242 | 1242.9 |
| 45–57 | 1506 | 1505.5 | 1718 | 1718.0 | |
| 59–71 | NIILSSQPGTDDR | 1415 | 1415.5 | 1415 | 1415.9 |
| 72–76 | VTWVK | 632 | 632.5 | 632 | 634.3 |
| 77–98 | SVDEAIAAC#GDVPEIMVIGGGR | 2216 | 2216.4 | 2216 | 2217.1 |
| 99–106 | VYEQFLPK | 1023 | 1023.3 | 1023 | 1023.7 |
cysteine was alkylated with 2-iodoacetamide.
The enzymatic activities of the individual DHFRs were measured (Table 2). Replacement of Met16, which is in the Met20 loop subdomain of DHFR,37 with biphenyl-phenylalanines A – D afforded DHFRs which produced 1.70 to 1.97 times more tetrahydrofolate than wild-type DHFR under the assay conditions. Substitution of amino acids A – D at position 49 in lieu of serine resulted in DHFRs which functioned 67–77% as well as wild type. While positions 16 and 49 are relatively exposed in DHFR,38 position 115 is in a folded region of the protein that is significantly more crowded. Two of the four biphenyl-phenylalanines (A and C) were well tolerated at position 115, affording DHFR analogues having 91 and 82%, respectively, of the activity of wild type. In contrast, the DHFR containing amino acid B at position 115 functioned only 25% as well as wild type, and introduction of D into this position reduced activity by a factor of 8–9. Thus para-substitution of the aromatic ring of phenylalanine, which might be expected to introduce the least steric hindrance in proximity to the protein backbone, was the best tolerated in a crowded region of DHFR, while meta-substitution was the less well tolerated, as might have been expected. This was especially true for amino acid D, which has two meta-substituted phenyl moieties.
Table 2.
Enzymatic Activities of Modified DHFRs
| DHFR | position 16 (Met) (%) |
position 49 (Ser) (%) |
position 115 (Ile) (%) |
|---|---|---|---|
| wild type DHFR | 100 | 100 | 100 |
| A | 170 ± 12a | 67 ± 5 | 91 ± 3 |
| B | 197 ± 12 | 68 ± 5 | 25 ± 2 |
| C | 171 ± 9 | 77 ± 6 | 82 ± 3 |
| D | 192 ± 11 | 76 ± 6 | 12 ± 2 |
Standard deviation based on data from three experiments.
The findings of differences in activities for modified DHFRs substituted with individual biphenyl-phenylalanines prompted a study of the effects of DHFR inhibitors on these modified enzymes. As shown in Table 3, methotrexate (MTX) inhibited all four modified enzymes to about the same extent as wild type. In contrast, inhibition of the modified DHFRs by trimethoprim (TMP) (IC50 values 8.5 – 35.2 nM) differed significantly from that of wild type (IC50 2.1 nM), and the diminution of inhibition relative to wild type appeared unrelated to the relative activities of the modified DHFRs. Thus the modified DHFRs were clearly able to distinguish the binds of MTX from that of TMP, and the orientation of the biphenyl moiety at position 115 significantly affected enzyme binding by TMP.
Table 3.
Inhibition of DHFRs Modified at Position 115 by TMP and MTX
| DHFR | relative activity (%) |
trimethoprim (IC50) (nM) |
methotrexate (IC50) (nM) |
|---|---|---|---|
| wild type DHFR | 100 | 2.1 ± 0.2a | 1.0 ± 0.1 |
| A | 91 ± 3 | 26 ± 2.2 | 1.1 ± 0.1 |
| B | 25 ± 2 | 8.5 ± 0.7 | 1.4 ± 0.2 |
| C | 82 ± 3 | 10.6 ± 0.9 | 1.3 ± 0.2 |
| D | 12 ± 2 | 35.2 ± 2.9 | 1.2 ± 0.1 |
Standard deviation based on data from three experiments.
Also determined were the UV and fluorescence spectral properties of the four biphenyl-phenylalanine derivatives, measured in acetonitrile using the N-pentenoyl protected amino acids. As shown in Table 4, λmax for the amino acids ranged from 249 to 283 nm, although as might have been expected, the values for A and B were quite close, as were those for C and D. When excited at their respective λmax values, the λem values for all four amino acids were within 5 nm, although the quantum yields for A and B (0.73 and 0.67, respectively) were quite close, as were those for C and D (0.12 and 0.10, respectively).
Table 4.
Ultraviolet and Fluorescent Spectral Properties of Four Amino Acids in theirNPentenoyl Protected Formsa
| Amino Acid | λmax (nm) |
ε | λex (nm) |
λem (nm) |
ΦF |
|---|---|---|---|---|---|
| A | 283 | 38,250 | 283 | 344 | 0.73 |
| B | 280 | 28,800 | 280 | 342 | 0.67 |
| C | 251 | 45,800 | 251 | 340 | 0.12 |
| D | 249 | 27,200 | 249 | 339 | 0.10 |
Measured in acetonitrile.
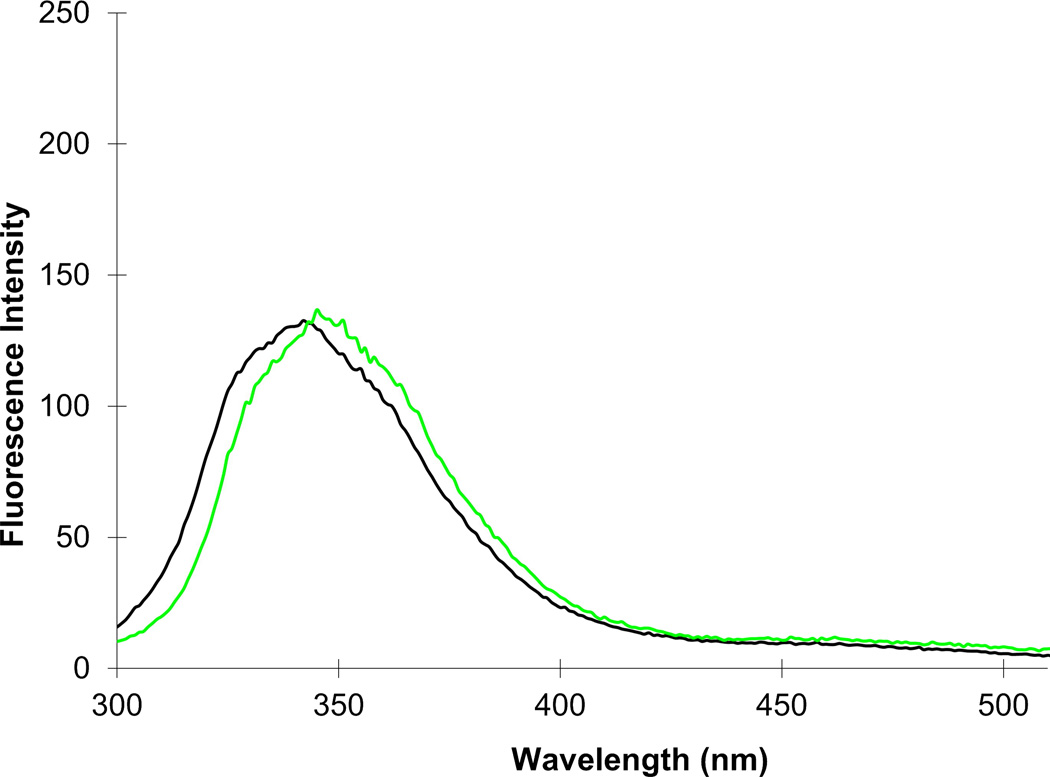
The fluorescence spectra of the DHFRs containing the biphenyl-phenylalanines at positions 16 and 49 are shown in Figures 4 and S3. Roughly comparable results were obtained for the emission spectra of the four biphenyl-phenylalanine derivatives at positions 16 and 49, although small differences were apparent both among individual biphenyl-phenylalanine derivatives, and between positions 16 and 49.
Figure 4.
Fluorescence emission spectra of DHFRs containing amino acid B at positions 16 (black trace) and 49 (green trace).
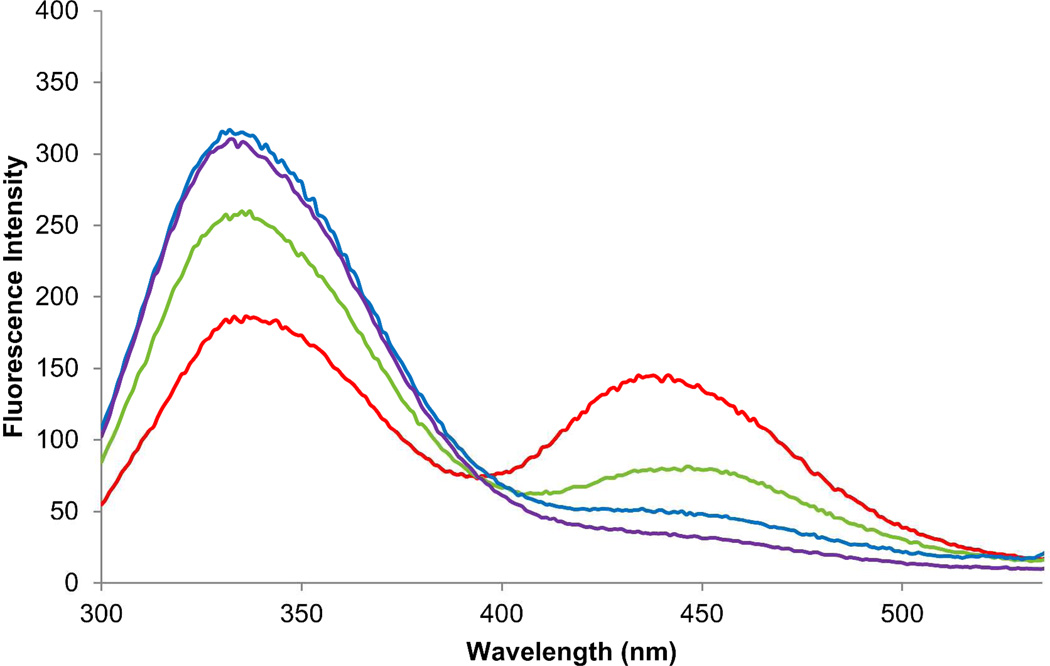
Also prepared were modified DHFRs containing the fluorescent amino acid L-(7-hydroxycoumarin-4-yl)ethylglycine (E) at position 17 and biphenyl-phenylalanine A, B, C or D at position 115. These were prepared as described previously for the construct containing amino acids A and E.23 As shown in Figure 5, the modified DHFR containing amino acids A and E exhibited the most efficient FRET. The modified DHFR containing amino acids B and E exhibited less efficient energy transfer, in parallel with the lesser enzymatic activity of this construct (Table 2). The remaining two modified DHFRs exhibited minimal energy transfer, possibly reflecting the lesser quantum yield of their common fluorophore (Table 4).
Figure 5.
Fluorescence of modified DHFRs (0.3 µM) containing L-(7-hydroxycoumarin-4-yl)ethylglycine (E) at position 17 and biphenyl-phenylalanine A (red curve), B (green curve), C (blue curve) or D (purple curve) at position 115. The spectra were recorded at pH 8.0 with λex = 280 nm.
DISCUSSION
Conformational changes in macromolecules, including nucleic acids and proteins, can be monitored conveniently by Förster resonance energy transfer (FRET).39–42 A fluorophore donor, covalently attached to a known position in the macromolecule, is irradiated to produce an electronically excited state, from which it may transfer its absorbed energy to an acceptor chromophore in a non-radiative process via dipole–dipole coupling. Typical fluorophores are molecules with multiple aromatic rings, which are connected to the macromolecule of interest by means of flexible tethers. This permits free rotation of the fluorophores during the lifetime of the excited state, thus approximating isotropic orientation and obviating issues related to dipole orientation. The distances measured in this fashion by FRET typically involve tens of Angströms.30 Measurements of smaller conformational changes in macromolecules using FRET are problematic for a number of reasons, notably the ability of the large chromophores attached to flexible tethers to traverse significant distances even in the absence of any conformational change in the macromolecule. Additionally, interactions between donor and acceptor chromophores, which often have large aromatic and hydrophobic surfaces, has the potential to cause macromolecule refolding.43,44
Recently, we have described the use of a biphenyl-phenylalanine FRET donor to measure smaller conformational changes in DHFR.23 This fluorophore had been used successfully in an earlier study as one of a number of fluorescent nucleobases to facilitate measurements of DNA conformation.45 We perceived that the rotational freedom about the biphenyl bonds in this fluorophore might well permit an amino acid having this fluorophore as a side chain to be incorporated into a folded protein structure with minimal perturbation of protein structure. In fact, biphenyl-phenylalanine A could be incorporated into position 115 of DHFR, known to be a folded region of the protein, without significant perturbation of DHFR function.23 In comparison the smaller fluorophore L-(7-hydroxycoumarin-4-yl)ethylglycine (E), containing two fused rings, significantly diminished DHFR function when incorporated into position 115. Biphenyl-phenylalanine A also functioned well when incorporated into a DHFR position (17) believed to be sterically unencumbered.23 The success of the introduction of biphenyl-phenylalanine A into DHFR prompted us to prepare three additional isomeric biphenyl-phenylalanines (B – D) for study. These were prepared and used to acylate the dinucleotide pdCpA using synthetic routes (Schemes 1, S1 and S2) analogous to the one employed for the elaboration of A. The similar fluorescence emission characteristics of A – D (Table 3) supported the idea that the isomeric species might find utility in modifying proteins at individual positions in proteins having specific steric requirements. The aminoacyl-pdCpAs were then ligated to a tRNACUA-COH transcript with T4 RNA ligase (Scheme 1), and the derived activated tRNAs were employed to incorporate amino acids A – D into DHFR at each of three positions (16, 49 and 115) (illustrated for the incorporation of biphenyl-phenylalanine C into position 115 in Scheme 2).
DHFR catalyzes the reductive conversion of 7,8-dihydrofolate to 5,6,7,8-tetrahydrofolate by stereospecific transfer of the pro-R hydrogen of NADPH to the C6 atom of the pterin nucleus, accompanied by concomitant protonation at N5.46 E. coli DHFR is an ∼18 kDa protein with an α/β structure having a central 8-stranded β-sheet (β-strands A – H) and four flanking α-helices (αB, αC, αE and αF).37 The active site cleft divides the protein into two structural subdomains, i.e. an adenosine binding subdomain and a loop subdomain. The smaller adenosine binding subdomain in E. coli DHFR encompasses residues 38–88; it contains the binding site for the NADPH adenosine moiety. The larger loop subdomain consists of ∼100 amino acids and has a set of three loops (Met 20 [residues 9–24]; F–G [116–132]; G–H [142–150]) which are near the active site. Changes in the structure and dynamics of the active site loops of DHFR have been associated with enzyme complexes containing substrate, product and cofactor present during the catalytic cycle.47–49 There is considerable evidence suggesting that the Met20 loop of E. coli DHFR exhibits conformational changes on the same time scale as the processes of substrate and cofactor binding and product release.50,51 Loop fluctuations may well play an important role in the catalytic cycle of E. coli DHFR, and may contribute to catalysis through compression of the active site and transition state stabilization.52
Based on the X-ray crystallographic structure of DHFR (1RX5),47 the biphenyl-phenylalanine moiety was close to the site of substrate binding when it was incorporated into position 16 of the Met20 loop. Substitutions close to this loop and elsewhere in DHFR have been shown previously53,54 to accelerate the release of tetrahydrofolate following dihydrofolate reduction by the enzyme, thereby altering what is normally the rate-limiting step in the catalytic cycle. This is entirely consistent with the observation that the introduction of biphenyl-phenylalanines A – D all resulted in nearly two-fold increases in product formation by the enzyme (Table 2). In comparison, only a modest reduction in activity accompanied introduction of the same four species into position 49. This portion of the adenosine binding subdomain is associated with the binding of a structural water molecule rather than substrate or cofactor.47
We have reported previously that replacement of Ile115 with biphenyl-phenylalanine A had no significant effect on the rate at which ecDHFR consumed NADPH in the presence of dihydrofolate.23 In comparison, introduction of L-(7-hydroxycoumarin-4-yl)ethylglycine (E), containing two fused rings, into the same position resulted in a rate of NADPH consumption about 25% that of wild type. We suggest that the ability of the biphenyl-phenylalanine side chain to rotate about the biphenyl bonds, and the para-connectivity of the phenyl moieties in the side chain, permitted this amino acid to be incorporated at position 115 with minimal disruption of the DHFR structure. In the present study, we carried out the same substitution using biphenyl-phenylalanines B – D. As shown in Table 2, the introduction of biphenyl-phenylalanine C at position 115 modestly reduced enzyme activity to 82% of the wild type. In common with A, this amino acid has the first biphenyl ring attached at the para position of phenylalanine, but with the phenyl ring farthest from the protein attached through the meta position of the penultimate phenyl ring. The small diminution in enzyme activity must be due to the orientation of the third ring, suggesting its interaction with the folded protein structure. The introduction of biphenyl-phenylalanine B at position 115 reduced activity to 25% of wild type, due undoubtedly to attachment of the first biphenyl ring to phenylalanine at the meta position, with the potential for greater steric interactions closer to the protein backbone. Logically enough, the modified DHFR containing biphenyl-phenylalanine D (with two phenyl moieties connected at their meta positions), has activity 8–9-fold lower than wild type.
The DHFRs having biphenyl-phenylalanines at position 115 all bound the inhibitor methotrexate with IC50 values little altered from that determined for wild type, indicating that those structural domains in DHFR recognized by MTX were essentially unaltered. In contrast, the IC50 values for TMP were all quite different, and uncorrelated with the relative enzymatic activities of the individual enzymes, suggesting strongly that the orientations of the biphenyl residues at position 115 in these modified DHFRs must differ significantly from one modified enzyme to another, and interact differently with TMP.
An analogous result was obtained for modified DHFRs containing L-(7-hydroxycoumarin-4-yl)ethylglycine (E) at position 17 and either biphenyl-phenylalanine A or B at position 115. As reported previously,23 the construct containing amino acid A exhibited efficient FRET when irradiated at 280 nm (Figure 5). In comparison, the construct containing amino acid B, which has the same fluorophore as A, exhibited much less efficient energy transfer. The construct containing amino acid B also exhibited lower enzymatic activity (Table 2), consistent with the interpretation that amino acid B causes a change in the conformation of DHFR when introduced into position 115. It seems unlikely that the significant reduction in FRET efficiency E noted for these two modified DHFRs was primarily a consequence of a change in the distance between the fluorophores. A more likely source of diminished energy transfer is a change in the dipole orientation factor (κ2), reflecting differences in the way in which amino acids A and B are accommodated at position 115. In fact, in an earlier study, it was shown by emission anisotropy that amino acid A had restricted rotational freedom when introduced into position 115.
The foregoing results indicate that biphenyl-phenylalanines can be incorporated into sterically unencumbered positions in DHFR with minimal impact on the structure or function of the enzyme. Even for a position (115) known to be within a folded region of the enzyme, two of the four modified DHFRs retained essentially full activity. These findings are consistent with the potential utility of the biphenyl-phenylalanines as fluorescent probes for proteins that do not substantially alter protein structure. It may be noted, however, that to the extent that the individual regioisomers must rotate their phenyl rings away from phenyl-ring coplanarity to achieve favorable interactions within the folded protein structure, the quantum yield of the fluorophore may be reduced.
Supplementary Material
Acknowledgments
This study was supported by research grant GM 092946 from the National Institutes of Health.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Additional experimental data for the synthesis, purification and characterization of DHFRs containing biphenyl-phenylalanine derivatives, and experimental procedures for the synthesis and characterization of the biphenyl-phenylalanine amino acid derivatives themselves. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Tsien RY. The green fluorescent protein. Annu. Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 2.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 3.Phillips SR, Wilson LJ, Borkman RF. Acrylamide and iodide fluorescence quenching as a structural probe of tryptophan microenvironment in bovine lens crystallins. Curr. Eye Res. 1986;5:611–619. doi: 10.3109/02713688609015126. [DOI] [PubMed] [Google Scholar]
- 4.Zhang P, Beck T, Tan W. Design of a molecular beacon DNA probe with two fluorophores. Angew. Chem. Int. Ed. 2001;40:402–405. doi: 10.1002/1521-3773(20010119)40:2<402::AID-ANIE402>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 5.Marti AA, Jockusch S, Li Z, Ju J, Turro NJ. Molecular beacons with intrinsically fluorescent nucleotides. Nucleic Acids Res. 2006;34:e50–e57. doi: 10.1093/nar/gkl134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jockusch S, Marti AA, Turro NJ, Li Z, Li X, Ju J, Stevens N, Akins DL. Spectroscopic investigation of a FRET molecular beacon containing two fluorophores for probing DNA/RNA sequences. Photochem. Photobiol. Sci. 2006;5:493–498. doi: 10.1039/b600213g. [DOI] [PubMed] [Google Scholar]
- 7.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki Y, Yasunaga T, Ohkura R, Wakabayashi T, Sutoh K. Swing of the lever arm of a myosin motor at the isomerization and phosphate-release steps. Nature. 1998;396:380–383. doi: 10.1038/24640. [DOI] [PubMed] [Google Scholar]
- 9.Tsien RY, Miyawaki A. Biochemical imaging. Seeing the machinery of live cells. Science. 1998;280:1954–1955. doi: 10.1126/science.280.5371.1954. [DOI] [PubMed] [Google Scholar]
- 10.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by β-galactosidase complementation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z, Rajagopalan PT, Selzer T, Benkovic SJ, Hammes GG. Single-molecule and transient kinetics investigation of the interaction of dihydrofolate reductase with NADPH and dihydrofolate. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2764–2769. doi: 10.1073/pnas.0400091101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antikainen NM, Smiley RD, Benkovic SJ, Hammes GG. Conformation coupled enzyme catalysis: Single-molecule and transient kinetics investigation of dihydrofolate reductase. Biochemistry. 2005;44:16835–16843. doi: 10.1021/bi051378i. [DOI] [PubMed] [Google Scholar]
- 13.Cornish VW, Benson DR, Altenbach CA, Hideg K, Hubbell WL, Schultz PG. Site-specific incorporation of biophysical probes into proteins. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2910–2914. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendel D, Cornish VW, Schultz PG. Site-directed mutagenesis with an expanded genetic code. Annu. Rev. Biophys. Biomol. Struct. 1995;24:435–462. doi: 10.1146/annurev.bb.24.060195.002251. [DOI] [PubMed] [Google Scholar]
- 15.Steward LE, Collins CS, Gilmore MA, Gilmore MA, Carlson JE, Ross JB, Chamberlin AR. In vitro site-specific incorporation of fluorescent probes into β-galactosidase. J. Am. Chem. Soc. 1997;119:6–11. [Google Scholar]
- 16.Hohsaka T, Kajihara D, Ashizuka Y, Murakami H, Sisido M. Efficient incorporation of nonnatural amino acids with large aromatic groups into streptavidin in in vitro protein synthesizing systems. J. Am. Chem. Soc. 1999;121:34–40. [Google Scholar]
- 17.Hohsaka T, Muranaka N, Komiyama C, Matsui K, Takaura S, Abe R, Murakami H, Sisido M. Position-specific incorporation of dansylated non-natural amino acids into streptavidin by using a four-base codon. FEBS Lett. 2004;560:173–177. doi: 10.1016/S0014-5793(04)00099-7. [DOI] [PubMed] [Google Scholar]
- 18.Hamada H, Kameshima N, Szymańska A, Wegner K, Łankiewicz L, Shinohara H, Taki M, Sisido M. Position-specific incorporation of a highly photodurable and blue-laser excitable fluorescent amino acid into proteins for fluorescence sensing. Bioorg. Med. Chem. 2005;13:3379–3384. doi: 10.1016/j.bmc.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 19.Anderson RD, III, Zhou J, Hecht SM. Fluorescence resonance energy transfer between unnatural amino acids in a structurally modified dihydrofolate reductase. J. Am. Chem. Soc. 2002;124:9674–9675. doi: 10.1021/ja0205939. [DOI] [PubMed] [Google Scholar]
- 20.Murakami H, Hohsaka T, Ashizuka Y, Hashimoto K, Sisido M. Site-directed incorporation of fluorescent nonnatural amino acids into streptavidin for highly sensitive detection of biotin. Biomacromolecules. 2000;1:118–125. doi: 10.1021/bm990012g. [DOI] [PubMed] [Google Scholar]
- 21.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat. Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Xie J, Schultz PG. A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 2006;128:8738–8739. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 23.Chen S, Fahmi NE, Wang L, Bhattacharya C, Benkovic SJ, Hecht SM. Detection of dihydrofolate reductase conformational change by FRET using two fluorescent amino acids. J. Am. Chem. Soc. 2013;135:12924–12927. doi: 10.1021/ja403007r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karginov VA, Mamaev SV, An H, Van Cleve MD, Hecht SM, Komatsoulis GA, Abelson JN. Probing the role of an active site aspartic acid in dihydrofolate reductase. J. Am. Chem. Soc. 1997;119:8166–8176. [Google Scholar]
- 25.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 26.Lodder M, Golovine S, Hecht SM. A chemical deprotection strategy for the elaboration of misacylated transfer RNA's. J. Org. Chem. 1997;62:778–779. [Google Scholar]
- 27.Lodder M, Golovine S, Laikhter AL, Karginov VA, Hecht SM. Misacylated transfer RNAs having a chemically removable protecting group. J. Org. Chem. 1998;63:794–803. doi: 10.1021/jo971692l. [DOI] [PubMed] [Google Scholar]
- 28.Sawano A, Miyawaki A. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 2000;28:E78. doi: 10.1093/nar/28.16.e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maglia G, Javed MH, Allemann RK. Hydride transfer during catalysis by dihydrofolate reductase from Thermotoga maritima. Biochem. J. 2003;374:529–535. doi: 10.1042/BJ20030412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. New York, NY: Springer; 2006. [Google Scholar]
- 31.Kotha S, Lahiri K. A new approach for modification of phenylalanine peptides by Suzuki-Miyaura coupling reaction. Bioorg. Med. Chem. Lett. 2001;11:2887–2890. doi: 10.1016/s0960-894x(01)00582-0. [DOI] [PubMed] [Google Scholar]
- 32.Gu W, Liu S, Silverman RB. Solid-phase, Pd-catalyzed silicon-aryl carbon bond formation. Synthesis of sansalvamide A peptide. Org. Lett. 2002;4:4171–4174. doi: 10.1021/ol0269392. [DOI] [PubMed] [Google Scholar]
- 33.Wilbur DS, Hamlin DK, Srivastava RR, Burns HD. Synthesis and radioiodination of N-Boc-p-(tri-n-butylstannyl)-L-phenylalanine tetrafluorophenyl ester: Preparation of a radiolabeled phenylalanine derivative for peptide synthesis. Bioconj. Chem. 1993;4:574–580. doi: 10.1021/bc00024a024. [DOI] [PubMed] [Google Scholar]
- 34.Robertson SA, Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. The use of 5’-phospho-2 deoxyribocytidylylriboadenosine as a facile route to chemical aminoacylation of tRNA. Nucleic Acids Res. 1989;17:9649–9660. doi: 10.1093/nar/17.23.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janknecht R, de Martynoff G, Lou J, Hipskind RA, Nordheim A, Stunnenberg HG. Rapid and efficient purification of native histidine-tagged protein expressed by recombinant vaccinia virus. Proc. Natl. Acad. U.S.A. 1991;88:8972–8976. doi: 10.1073/pnas.88.20.8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maini R, Nguyen D, Chen S, Dedkova LM, Roy Chowdhury S, Alcana-Torano R, Hecht SM. Incorporation of β-amino acids into dihydrofolate reductase by ribosomes having modifications in the peptidyltransferase center. Bioorg. Med. Chem. 2013;21:1088–1096. doi: 10.1016/j.bmc.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Bolin JT, Filman DJ, Matthews DA, Hamlin RC, Kraut J. Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 Å resolution. I. General features and binding of methotrexate. J. Biol. Chem. 1982;257:13650–13652. [PubMed] [Google Scholar]
- 38.Chen S, Wang L, Fahmi NE, Benkovic SJ, Hecht SM. Two pyrenylalanines in dihydrofolate reductase form an excimer enabling the study of protein dynamics. J. Am. Chem. Soc. 2012;134:18883–18885. doi: 10.1021/ja307179q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich A, Buschmann V, Müller C, Sauer M. Fluorescence resonance energy transfer (FRET) and competing processes in donor-acceptor substituted DNA strands: a comparative study of ensemble and single-molecule data. Rev. Mol. Biotech. 2002;82:211–231. doi: 10.1016/s1389-0352(01)00039-3. [DOI] [PubMed] [Google Scholar]
- 40.Jares-Erijman EA, Jovin TM. FRET imaging. Nat. Biotech. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 41.Dickenson NE, Picking WD. Forster resonance energy transfer (FRET) as a tool for dissecting the molecular mechanisms for maturation of the Shigella type III secretion needle tip complex. Int. J. Mol. Sci. 2012;13:15137–15161. doi: 10.3390/ijms131115137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okumoto S, Jones A, Frommer WB. Quantitative imaging with fluorescent biosensors. Annu. Rev. Plant Biol. 2012;63:663–706. doi: 10.1146/annurev-arplant-042110-103745. [DOI] [PubMed] [Google Scholar]
- 43.Murakami H, Hohsaka T, Ashizuka Y, Hashimoto K, Sisido M. Site-directed incorporation of fluorescent nonnatural amino acids into streptavidin for highly sensitive Detection of biotin. Biomacromolecules. 2000;1:118–125. doi: 10.1021/bm990012g. [DOI] [PubMed] [Google Scholar]
- 44.Kajihara D, Abe R, Iijima I, Komiyama C, Sisido M, Hohsaka T. FRET analysis of protein conformational change through position-specific incorporation of fluorescent amino acids. Nat. Methods. 2006;3:923–929. doi: 10.1038/nmeth945. [DOI] [PubMed] [Google Scholar]
- 45.Gao J, Watanabe S, Kool ET. Modified DNA analogues that sense light exposure with color changes. J. Am. Chem. Soc. 2004;126:12748–12749. doi: 10.1021/ja046910o. [DOI] [PubMed] [Google Scholar]
- 46.Charlton PA, Young DW, Birdsall B, Feeney J, Roberts GC. Stereochemistry of reduction of folic acid using dihydrofolate reductase. J. Chem. Soc. Chem. Commun. 1979;20:922–924. [Google Scholar]
- 47.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 48.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: Role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 49.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Defining the role of active-site loop fluctuations in dihydrofolate reductase catalysis. Proc. Natl. Acad. Sci. U S.A. 2005;102:5032–5037. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnell JR, Dyson HJ, Wright PE. Effect of cofactor binding and loop conformation on side chain methyl dynamics in dihydrofolate reductase. Biochemistry. 2004;43:374–383. doi: 10.1021/bi035464z. [DOI] [PubMed] [Google Scholar]
- 51.Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu. Rev. Biophys. Biomol. Struct. 2004;33:119–140. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 52.Cannon WR, Garrison BJ, Benkovic SJ. Electrostatic characterization of enzyme complexes: evaluation of the mechanism of catalysis of dihydrofolate reductase. J. Am. Chem. Soc. 1997;119:2386–2395. [Google Scholar]
- 53.Wagner CR, Huang Z, Singleton SF, Benkovic SJ. Molecular basis for nonadditive mutational effects in Escherichia coli dihydrofolate reductase. Biochemistry. 1995;34:15671–15680. doi: 10.1021/bi00048a011. [DOI] [PubMed] [Google Scholar]
- 54.Posner BA, Li L, Bethell R, Tsuji T, Benkovic SJ. Engineering specificity for folate into dihydrofolate reductase from Escherichia coli. Biochemistry. 1996;35:1653–1663. doi: 10.1021/bi9518095. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.