Abstract
Regulated intercellular signaling is critical for the normal development and maintenance of multicellular organisms. Glypicans have been shown to regulate signaling by TGFβs, hedgehogs and Wnts, in several cellular contexts. Glypicans comprise a conserved family of heparan sulfated, glycosylphosphatidylinositol (GPI)-linked extracellular proteins. The structural complexity of glypicans may underlie their functional complexity. In a recent study31, we built on previous findings that one of the two C. elegans glypicans, LON-2, specifically inhibits signaling by the TGFβ superfamily member DBL-1. We tested the functional requirements of LON-2 protein core components and post-translational modifications for LON-2 activity. We provide the first evidence that two parts of a glypican can independently regulate TGFβ superfamily signaling in vivo: the N-terminal furin protease product and a C-terminal region containing heparan sulfate attachment sites. Furthermore, we show a protein-protein interaction motif is crucial for LON-2 activity in the N-terminal protein core, suggesting that LON-2 acts by serving as a scaffold for DBL-1 and an RGD-binding protein. In addition, we demonstrate specificity of glypican function by showing C. elegans GPN-1 does not functionally substitute for LON-2. This work reveals a molecular foundation for understanding the complexity and specificity of glypican function.
Keywords: LON-2, RGD-binding protein, TGFβ signaling, body size, glycosylphosphatidylinositol anchor, glypican, heparan sulfate proteoglycan
Introduction
Glypicans are complex proteins that play complex roles in the regulation of several intercellular signaling pathways, including transforming growth factor-β (TGF-β), fibroblast growth factor (FGF), Hedgehog (Hh) and Wnt family members.1-13 Glypicans are composed of a cylindrical, α-helical protein core that is modified with heparan sulfate glycosaminoglycan (HS-GAG) chains and glycosylphosphatidylinositol (GPI), which anchors the glypican to the external surface of the cell membrane.16-18 Glypicans can be shed into the intercellular space through cleavage of the protein core at a furin cleavage site or by removal of the GPI lipid anchor.19-21 The interactions of glypican with cell-cell signaling factors is sometimes apparently contradictory (Table 1), and to correlate this structural complexity of glypicans with their function is a significant problem relevant to both developmental and disease biology.4,14,15 Both core protein and post-translational modifications differentially mediate glypican activity in animal developmental models and in cell lines (Table 1). Glypican-3 (GPC3), both membrane-linked and shed, is associated with a variety of cancer types and with cell growth.1,22 Clinical trials are evaluating the use of a monoclonal antibody against GPC3 to treat hepatocellular carcinoma (ClinicalTrials.gov).23 Parsing the roles of glypicans in specific cell signaling pathways is important to better understand the basis of developmental events and disease states affected by this proteoglycan family.
Table 1. Comparison of glypican family member structure-function analyses.
| Pathway | Glypican | Model System | Protein core active |
Heparan sulfated C-terminus sufficient? |
RGD domain required? | Furin protease site required? | Heparan sulfate required? | GPI required? | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| to the furin protease site? | to the GPI cleavage site? | |||||||||
| TGF-β |
LON-2 |
C. elegans |
Yes |
Yes |
Yes |
Yes |
No |
No |
No |
Taneja-Bageshwar and Gumienny, 2012 |
| |
Dally |
Drosophila (in vivo, in vitro) |
- |
- |
- |
- |
- |
No |
- |
Kirkpatrick et al., 2006 |
| |
Dally |
Drosophila cell line |
- |
- |
- |
- |
- |
Yes |
- |
Dejima et al., 2011 |
| FGF |
OCI-5/ GPC3 |
Cell lines1 |
- |
- |
- |
- |
- |
Yes |
- |
Song et al., 1997 |
| Wnt |
Dally |
Drosophila |
- |
- |
- |
- |
- |
Yes |
- |
Tsuda et al., 1999 |
| |
Dally |
Drosophila |
- |
- |
- |
- |
- |
No |
- |
Kirkpatrick et al., 2006 |
| |
XGly4/ GPC4 |
Xenopus |
- |
- |
No |
- |
- |
- |
- |
Ohkawara et al., 2003 |
| |
GPC3 |
Cell lines1 |
- |
- |
- |
- |
- |
No |
Yes |
Capurro et al., 2005 |
| |
OCI-5/ GPC3 |
Cell lines1 |
- |
- |
- |
- |
- |
No |
- |
Song et al., 2005 |
| |
Dally-like |
Drosophila |
- |
- |
- |
- |
- |
- |
Yes |
Gallet et al., 2008 |
| |
Dally-like |
Drosophila |
- |
- |
- |
- |
- |
No |
No |
Yan et al., 2009 |
| |
GPC1 |
Chicken embryos |
- |
No |
- |
Not tested |
- |
Yes |
- |
Shiau et al., 2010 |
| |
GPC3 |
HCC2 |
- |
Yes |
- |
- |
- |
- |
Yes3 |
Zittermann et al., 2010 |
| Hh |
Dally |
Drosophila |
- |
- |
- |
- |
- |
- |
Yes |
Takeo et al., 2005 |
| |
OCI-5/ GPC3 |
Cell lines1 |
- |
No |
- |
- |
- |
No |
Yes |
Capurro et al., 2008 |
| |
Dally-like |
Drosophila |
- |
- |
- |
- |
- |
- |
Yes |
Gallet et al., 2008 |
| |
Dally-like |
Drosophila |
- |
- |
- |
- |
- |
No |
No |
Yan et al., 2009; Yan et al., 2010 |
| |
Dally-like |
Drosophila |
- |
- |
- |
- |
No |
No |
No |
Williams et al., 2010 |
| |
GPC5 |
Cell lines1 |
- |
- |
- |
- |
- |
Yes |
- |
Li et al., 2011 |
| Unknown |
OCI-5/ GPC3 |
Cell lines1 |
- |
- |
- |
- |
- |
No |
Yes |
Gonzalez et al., 1998 |
| GPC3 | Cell lines1 | Yes | - | - | - | - | - | - | De Cat et al., 2003 | |
1 Cell lines are derived from mammalian sources. 2HCC, hepatocellular carcinoma. 3GPC3 promotes Wnt signaling and cell growth, but GPC3 lacking GPI (sGPC3) inhibits Wnt signaling and cell growth of some HCC cell lines.
Genetic interactions have been shown between glypicans and TGF-β members in evolutionarily diverse systems from nematodes to mice, suggesting conservation of glypican function in TGF-β signaling.24-26 LON-2, one of two C. elegans glypicans, restricts the activity of TGF-β superfamily member DBL-1 and physically interacts with a mammalian DBL-1 homolog.24 The Drosophila glypican Dally also binds a TGF-β superfamily member and affects Dpp/TGF-β activity in a complex gradient-dependent manner.4 Members of a TGF-β subfamily that includes Dpp, C. elegans DBL-1 and vertebrate bone morphogenetic proteins (BMPs) contain a heparin-binding motif at the N-terminus of the mature protein.27-29 In the case of Drosophila Dpp, this motif binds glypican Dally.14 The heparan sulfate chain of Dally is not essential for Dally-Dpp interaction in vitro; however, heparan sulfation of Dally differentially contributes to Dally’s varied cellular roles through regulation of TGF-β and other pathways (Table 1).4,30 No other study to date has dissected the functional requirements for the glypican protein core or post-translational modifications in TGF-β superfamily signaling (Table 1).
Recently, we systematically analyzed these structural components of glypican in regulating TGF-β signaling.31 We used the C. elegans glypican LON-2 because it plays a clear role in specifically controlling TGF-β signaling at the level of the TGF-β ligand DBL-1 (Fig. 1A). This pathway regulates body length in a dose-dependent manner, so we used body length as a bioassay for activity of LON-2 variant transgene products. Loss of endogenous LON-2 function releases the “brakes” on DBL-1 body length-promoting activity, resulting in animals that are visibly longer than normal.24 We introduced transgenes into lon-2 mutant animals (caused by a deletion that removes most of the lon-2 coding sequence) and asked if transgene product could restore a more normal body length. We investigated the role of LON-2’s heparan sulfate side chain and GPI attachment sites. We also narrowed the region of the LON-2 core protein that regulates TGF-β signaling and investigated the role of a furin protease cleavage site and a protein-protein interaction motif, RGD. We discovered that membrane localization of LON-2 is not required for its activity. We have been able to uncover unexpected functional requirements and redundancies within the LON-2 structure related to its function as a TGF-β signaling regulator.
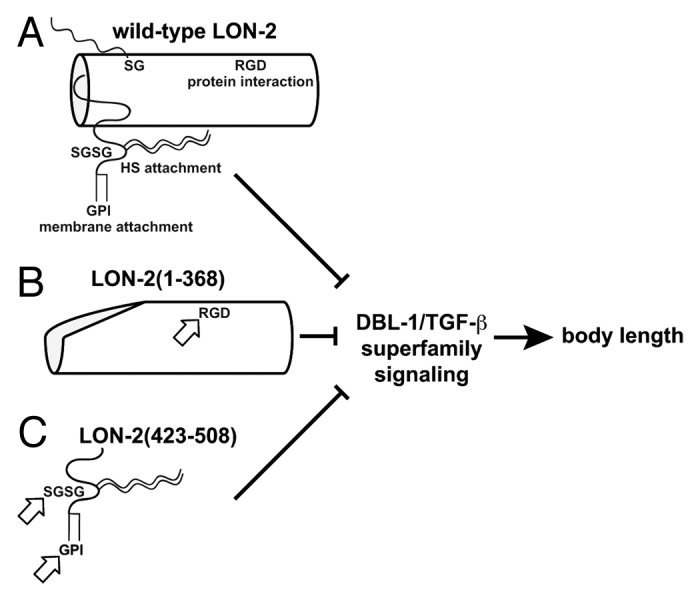
Figure 1. Model of LON-2 parts that regulate DBL-1/TGF-β superfamily signaling. (A) The full-length LON-2 inhibits DBL-1 signaling and prevents a long body length phenotype. Full-length LON-2 (shown in A) remains functional if it lacks any one of the sequences containing RGD, all heparan sulfate attachment sites or the GPI anchor site. (B) The cylindrical N-terminal core protein, LON-2(1–368), requires the RGD motif for function. (C) The unstructured C-terminal 86 amino acid region, LON-2(423–508), requires both heparan sulfate attachment sites and a GPI anchor sequence to inhibit DBL-1 signaling. Arrows indicate parts required for function of the LON-2 variants shown in B and C. This diagram is based on tertiary structure predictions from the crystal structures of Dally-like and human GPC1.17,18
LON-2 Does Not Require GAG Attachment Sites or GPI Anchored Membrane Attachment to Inhibit DBL-1 Signaling
HS-GAG attachment sites are a characteristic feature of all glypicans;16 however, the functional requirement for glycosylation varies from system to system (Table 1).1,2,4,7,9,11-13,21,30,32-34 GAG attachment sites are not essential for the function of the protein core of Drosophila Dally in wing development or to bind TGF-β superfamily members, but heparan sulfation of Dally is required during eye development and in in vitro systems.4,30 For regulating Hedgehog signaling, human GPC5 requires its heparan sulfate side chains, but Dally-like, the other Drosophila glypican, does not.2,11,13,21,33 Similarly, different glypicans have different heparan sulfation requirements to regulate Wnt signaling.1,4,6,7,9,11,34-36
C. elegans glypican LON-2 has three predicted sites for attachment of HS-GAG chains (Fig. 1A). We found that lon-2 constructs with all three GAG attachment sites altered could rescue the long body size phenotype of animals lacking endogenous LON-2, indicating that GAG sites are not required for LON-2 to inhibit DBL-1 signaling. Wild-type GFP-tagged LON-2 visibly localizes to cell surfaces.24 LON-2 lacking all three GAG attachment sites localizes to the cell surface similar to the wild-type protein. This result shows that the function and proper subcellular localization of LON-2 is not dependent on the sites for heparan sulfate side chain attachment.
Another defining feature of glypicans is their GPI glycolipid anchor, which links the protein to the extracellular membrane.37 Glypicans are shed from their GPI membrane anchor during development and disease states.36,38-42 In Drosophila, substituting a transmembrane domain for the GPI linkage site still allows Dally-like protein to inhibit Wg signaling.11 However, Drosophila Dally-like and mouse GPC3 require membrane tethering by GPI to function in Hedgehog signaling.2,21,35,43 Furthermore, a secreted form of Drosophila Dally has a weak dominant negative effect on signaling by TGF-β superfamily member Dpp.43 We found in C. elegans, the GPI-deleted form of LON-2 is as functional as full-length LON-2. This GFP-tagged construct fails to visibly localize to cell membranes at cell-cell junctions, an expected phenotype given its lack of membrane anchor. Though extracellular LON-2 lacking a GPI link was not visible, this LON-2 variant appeared as a bright haze within cells, suggesting that this variant affects protein secretion or stability. Our work indicates that membrane localization by GPI is not absolutely required for LON-2 to inhibit DBL-1 activity.
The N-Terminal LON-2 Protease Product Can Inhibit DBL-1 Activity
At least one furin proprotein convertase cleavage site is present or predicted in all glypicans.16,19,24 Inactivating the Drosophila Dally-like furin cleavage site does not alter the effect of Dally-like in Hedgehog signaling.21 Protease processing of GPC3 is required to repress both canonical and noncanonical Wnt signaling in cell lines and during zebrafish development, but is not required to stimulate canonical Wnt signaling in hepatocellular carcinoma.19,44 GPC3 protease fragments have been identified in the sera of hepatocellular carcinoma patients, but their activity is unknown.1,45 However, a role for proteolyzed glypican in TGF-β signaling has not been analyzed. To test if cleavage is important to glypican function, we altered the LON-2 furin cleavage site sequence RLGR to ALGA to prevent furin cleavage at this site. We found this LON-2 furin site variant rescued the long body phenotype. It could also properly localize to cell membranes at cell-cell junctions. Thus, processing at this furin cleavage site is not necessary for LON-2 to function as a negative regulator of body size.
We explored the importance of the N-terminal furin protease product of LON-2 in DBL-1 signaling. The furin protease cleavage product of LON-2, LON-2(1–368), is smaller than the non-GPI tethered form of LON-2 and lacks GAG attachment sites (Fig. 1B). This protein core fragment was functional in our bioassay, though it did not visibly associate with cell membranes.
We further found that by attaching the GPI anchor sequence to this rescuing N-terminal sequence, excluding the furin protease recognition site, the LON-2 minimal protein core does not need to be cleaved to be active. This GPI-anchored N-terminal fragment was weakly visible at the cell junctions but appeared to localize mainly in the cytoplasm. This result indicates that the N-terminal part of LON-2 is active in our bioassay, but it requires its full C-terminal protein core for normal levels of extracellular localization.
The LON-2 C-terminus Can Independently Inhibit DBL-1 Signaling
The predicted C-terminal portion of LON-2 produced by the cleavage at the consensus furin protease site is much smaller than the N-terminus and contains the heparan sulfate attachment sites and the GPI linkage site. The glycanated C-terminal protease product of any glypican has not been shown to contain glypican activity by itself.6
We made a construct, LON-2(423–508), that encodes the unstructured part of the predicted C-terminal furin protease product and tested its ability to inhibit DBL-1 activity (Fig. 1C). This 86 amino acid fragment, which contains two GAG attachment sites and the GPI attachment site, effectively inhibited DBL-1 activity in animals lacking functional LON-2. When GAG attachment sequences were altered in this construct, activity was lost. These results strongly support the model that the DBL-1-inhibitory function of LON-2(423–508) resides in the heparan sulfate chains attached to the LON-2 protein. Next, we asked if the GPI attachment site was required for activity of this glycanated C-terminal LON-2 fragment. Though the activity of the N-terminal LON-2 core protein does not depend on a membrane tether, activity of the C-terminal glycanated LON-2 fragment does depend on its GPI link to the outer cell membrane because C-terminal LON-2 constructs lacking the GPI anchor sequence fail to rescue the lon-2 mutant phenotype.
Glypicans Show Functional Specificity
We have previously shown that the Drosophila glypican Dally, which shares low sequence homology to LON-2, can functionally substitute for LON-2 in our C. elegans body length assay.24 Because TGF-β superfamily members bind heparin, and because the small heparan-sulfated C-terminal LON-2 fragment is sufficient to restore DBL-1 pathway inhibition, we asked if glypican inhibition of this heparin-binding cell-cell communication factor is specific or promiscuous. GPN-1 is the only other glypican in C. elegans. It is normally expressed at different times and locations than LON-2 and promotes some cell migrations, but does not affect body length.24,46 Structurally, GPN-1 shares the features common to all glypicans, but contains four heparan sulfate attachment sites, one more than LON-2 and has low sequence similarity to LON-2. We drove expression of GPN-1 from the lon-2 promoter in animals lacking lon-2 and discovered that GPN-1 did not inhibit the DBL-1-mediated body length defect. This result shows that LON-2 is not functionally replaceable by GPN-1 and supports a model whereby glypicans show specificity, possibly directed by the protein core, for binding heparan sulfate-binding ligand(s).
Soluble LON-2 Function Requires its RGD Tripeptide Motif or GPI Linkage
A tripeptide arginine-glycine-aspartic acid (RGD) motif can be found in many proteins of the extracellular matrix and regulates interactions between proteins and extracellular matrix constituents, particularly integrins.47-50 This sequence is shared within its family only by mammalian glypican-1 proteins, but it is common in many other extracellular TGF-β superfamily regulatory molecules. We previously showed that the RGD sequence is dispensable in the context of full-length, glycanated LON-2, but our recent work shows that heparan sulfation sites can confer activity independent of the N-terminal protein core, which includes the RGD site.24 Therefore, we tested the requirement of the RGD sequence for the function of the N-terminal protein core. We showed that N-terminal LON-2 lacking the RGD site was expressed but was not functional in our bioassay. We then asked if RGD was required in the context of the non-glycanated full length LON-2 variant. This variant lacking both RGD and GAG attachment sites is active, suppressing the DBL-1-mediated long body size defect of lon-2 mutant animals. This activity depends on the GPI anchor sequence, because modification of this construct to remove the GPI anchor sequence renders this variant inactive. These results show that when the full-length core protein is GPI linked to the membrane, the RGD motif is dispensable. Thus, RGD is not the sequence in the LON-2 core protein that directly regulates DBL-1 activity. Instead, this result indicates that the RGD binding to an unidentified protein facilitates DBL-1 inhibition by another part of the LON-2 N-terminus.
Conclusion and Future Directions
Glypicans play a complex role in regulating cell-to-cell signaling, but the mechanism of this regulation is not fully understood, especially for TGF-β superfamily signaling.15,16,51 By exploiting the long body phenotype of deregulated signaling by TGF-β superfamily member DBL-1 in C. elegans, we showed that LON-2 activity resides in two separable regions (Fig. 1B and C). These regions have specific functional requirements. First, we identified the smallest functional glypican protein core region to date (LON-2(1–386)) and showed that it requires either its RGD protein-protein interaction motif or membrane association through GPI (Fig. 1B). Second, we discovered that the disordered C-terminal region is also bioactive and showed that this activity requires both heparan sulfate attachment sites and a GPI linkage sequence (Fig. 1C). Our work shows that soluble glypican products are active in our system. Based on our studies and the work of others, we propose that glypicans in higher systems differentially regulate TGF-β and other signaling pathways in various tissues using both N- and C-terminal parts, membrane attached or shed.
Furthermore, these studies are the first to suggest the involvement of an additional player that interacts with the glypican protein core to control growth factor signaling. This result supports a model that LON-2 acts as a scaffold in a DBL-1-regulatory complex by binding both DBL-1 and an RGD-binding protein. LON-2 may act like the secreted glycoprotein fibrillin-1, an RGD-containing protein that regulates TGF-β signaling by scaffolding TGF-β and latent TGF-β binding protein (LTBP), integrins and other extracellular matrix proteins.52 It will be exciting to identify the LON-2-interacting protein. Determining if—and possibly how—other glypicans recruit accessory proteins to regulate signaling by TGF-βs and other pathways will be critical to understand glypican function and specificity during development and cancer progression.
Acknowledgments
We would like to thank the members of our lab for comments on this manuscript. This work was supported by a grant from the American Heart Association (#11BGIA5700014) and by MCMD departmental start-up funds. Microscopy equipment was purchased with funds from MCMD and the TAMHSC College of Medicine Office of the Dean.
Glossary
Abbreviations:
- BMP
bone morphogenetic protein
- DBL-1
Dpp and BMP-like
- Dpp
decapentaplegic
- FGF
fibroblast growth factor
- GPC1
glypican 1
- GPC3
glypican 3
- GPC4
glypican 4
- GPC5
glypican 5
- GPI
glycosylphosphatidylinositol
- GPN-1
glypican
- HCC
hepatocellular carcinoma
- Hh
hedgehog
- HS-GAG
heparan sulfate glycosaminoglycan
- LON-2
long
- RGD
arginine glycine aspartic acid
- TGF-β
transforming growth factor-β
- Wnt
wingless and Int
Submitted
01/10/2013
Accepted
01/31/2013
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/23843
References
- 1.Capurro MI, Xiang YY, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res. 2005;65:6245–54. doi: 10.1158/0008-5472.CAN-04-4244. [DOI] [PubMed] [Google Scholar]
- 2.Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits Hedgehog signaling during development by competing with patched for Hedgehog binding. Dev Cell. 2008;14:700–11. doi: 10.1016/j.devcel.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 3.Hagihara K, Watanabe K, Chun J, Yamaguchi Y. Glypican-4 is an FGF2-binding heparan sulfate proteoglycan expressed in neural precursor cells. Dev Dyn. 2000;219:353–67. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1059>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 4.Kirkpatrick CA, Knox SM, Staatz WD, Fox B, Lercher DM, Selleck SB. The function of a Drosophila glypican does not depend entirely on heparan sulfate modification. Dev Biol. 2006;300:570–82. doi: 10.1016/j.ydbio.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 5.Lin X, Perrimon N. Dally cooperates with Drosophila Frizzled 2 to transduce Wingless signalling. Nature. 1999;400:281–4. doi: 10.1038/22343. [DOI] [PubMed] [Google Scholar]
- 6.Ohkawara B, Yamamoto TS, Tada M, Ueno N. Role of glypican 4 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 2003;130:2129–38. doi: 10.1242/dev.00435. [DOI] [PubMed] [Google Scholar]
- 7.Song HH, Shi W, Xiang YY, Filmus J. The loss of glypican-3 induces alterations in Wnt signaling. J Biol Chem. 2005;280:2116–25. doi: 10.1074/jbc.M410090200. [DOI] [PubMed] [Google Scholar]
- 8.Topczewski J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, et al. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev Cell. 2001;1:251–64. doi: 10.1016/S1534-5807(01)00005-3. [DOI] [PubMed] [Google Scholar]
- 9.Tsuda M, Kamimura K, Nakato H, Archer M, Staatz W, Fox B, et al. The cell-surface proteoglycan Dally regulates Wingless signalling in Drosophila. Nature. 1999;400:276–80. doi: 10.1038/22336. [DOI] [PubMed] [Google Scholar]
- 10.Yan D, Lin X. Drosophila glypican Dally-like acts in FGF-receiving cells to modulate FGF signaling during tracheal morphogenesis. Dev Biol. 2007;312:203–16. doi: 10.1016/j.ydbio.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan D, Wu Y, Feng Y, Lin SC, Lin X. The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev Cell. 2009;17:470–81. doi: 10.1016/j.devcel.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song HH, Shi W, Filmus J. OCI-5/rat glypican-3 binds to fibroblast growth factor-2 but not to insulin-like growth factor-2. J Biol Chem. 1997;272:7574–7. doi: 10.1074/jbc.272.12.7574. [DOI] [PubMed] [Google Scholar]
- 13.Yan D, Wu Y, Yang Y, Belenkaya TY, Tang X, Lin X. The cell-surface proteins Dally-like and Ihog differentially regulate Hedgehog signaling strength and range during development. Development. 2010;137:2033–44. doi: 10.1242/dev.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akiyama T, Kamimura K, Firkus C, Takeo S, Shimmi O, Nakato H. Dally regulates Dpp morphogen gradient formation by stabilizing Dpp on the cell surface. Dev Biol. 2008;313:408–19. doi: 10.1016/j.ydbio.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umulis D, O’Connor MB, Blair SS. The extracellular regulation of bone morphogenetic protein signaling. Development. 2009;136:3715–28. doi: 10.1242/dev.031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9:224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim MS, Saunders AM, Hamaoka BY, Beachy PA, Leahy DJ. Structure of the protein core of the glypican Dally-like and localization of a region important for hedgehog signaling. Proc Natl Acad Sci U S A. 2011;108:13112–7. doi: 10.1073/pnas.1109877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson G, Awad W, Håkansson M, Mani K, Logan DT. Crystal structure of N-glycosylated human glypican-1 core protein: structure of two loops evolutionarily conserved in vertebrate glypican-1. J Biol Chem. 2012;287:14040–51. doi: 10.1074/jbc.M111.322487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Cat B, Muyldermans SY, Coomans C, Degeest G, Vanderschueren B, Creemers J, et al. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol. 2003;163:625–35. doi: 10.1083/jcb.200302152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fico A, Maina F, Dono R. Fine-tuning of cell signaling by glypicans. Cell Mol Life Sci. 2011;68:923–9. doi: 10.1007/s00018-007-7471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams EH, Pappano WN, Saunders AM, Kim MS, Leahy DJ, Beachy PA. Dally-like core protein and its mammalian homologues mediate stimulatory and inhibitory effects on Hedgehog signal response. Proc Natl Acad Sci U S A. 2010;107:5869–74. doi: 10.1073/pnas.1001777107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho M, Kim H. Glypican-3: a new target for cancer immunotherapy. Eur J Cancer. 2011;47:333–8. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao W, Ho M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011;1:14–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Gumienny TL, MacNeil LT, Wang H, de Bono M, Wrana JL, Padgett RW. Glypican LON-2 is a conserved negative regulator of BMP-like signaling in Caenorhabditis elegans. Curr Biol. 2007;17:159–64. doi: 10.1016/j.cub.2006.11.065. [DOI] [PubMed] [Google Scholar]
- 25.Jackson SM, Nakato H, Sugiura M, Jannuzi A, Oakes R, Kaluza V, et al. dally, a Drosophila glypican, controls cellular responses to the TGF-β-related morphogen, Dpp. Development. 1997;124:4113–20. doi: 10.1242/dev.124.20.4113. [DOI] [PubMed] [Google Scholar]
- 26.Paine-Saunders S, Viviano BL, Zupicich J, Skarnes WC, Saunders S. glypican-3 controls cellular responses to Bmp4 in limb patterning and skeletal development. Dev Biol. 2000;225:179–87. doi: 10.1006/dbio.2000.9831. [DOI] [PubMed] [Google Scholar]
- 27.Morita K, Chow KL, Ueno N. Regulation of body length and male tail ray pattern formation of Caenorhabditis elegans by a member of TGF-β family. Development. 1999;126:1337–47. doi: 10.1242/dev.126.6.1337. [DOI] [PubMed] [Google Scholar]
- 28.Ohkawara B, Iemura S, ten Dijke P, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr Biol. 2002;12:205–9. doi: 10.1016/S0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, et al. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–50. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- 30.Dejima K, Kanai MI, Akiyama T, Levings DC, Nakato H. Novel contact-dependent bone morphogenetic protein (BMP) signaling mediated by heparan sulfate proteoglycans. J Biol Chem. 2011;286:17103–11. doi: 10.1074/jbc.M110.208082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taneja-Bageshwar S, Gumienny TL. Two functional domains in C. elegans glypican LON-2 can independently inhibit BMP-like signaling. Dev Biol. 2012;371:66–76. doi: 10.1016/j.ydbio.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez AD, Kaya M, Shi W, Song H, Testa JR, Penn LZ, et al. OCI-5/GPC3, a glypican encoded by a gene that is mutated in the Simpson-Golabi-Behmel overgrowth syndrome, induces apoptosis in a cell line-specific manner. J Cell Biol. 1998;141:1407–14. doi: 10.1083/jcb.141.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li F, Shi W, Capurro M, Filmus J. Glypican-5 stimulates rhabdomyosarcoma cell proliferation by activating Hedgehog signaling. J Cell Biol. 2011;192:691–704. doi: 10.1083/jcb.201008087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiau CE, Hu N, Bronner-Fraser M. Altering Glypican-1 levels modulates canonical Wnt signaling during trigeminal placode development. Dev Biol. 2010;348:107–18. doi: 10.1016/j.ydbio.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallet A, Staccini-Lavenant L, Thérond PP. Cellular trafficking of the glypican Dally-like is required for full-strength Hedgehog signaling and wingless transcytosis. Dev Cell. 2008;14:712–25. doi: 10.1016/j.devcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Zittermann SI, Capurro MI, Shi W, Filmus J. Soluble glypican 3 inhibits the growth of hepatocellular carcinoma in vitro and in vivo. Int J Cancer. 2010;126:1291–301. doi: 10.1002/ijc.24941. [DOI] [PubMed] [Google Scholar]
- 37.Filmus J, Selleck SB. Glypicans: proteoglycans with a surprise. J Clin Invest. 2001;108:497–501. doi: 10.1172/JCI13712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–77. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 39.Brandan E, Carey DJ, Larraín J, Melo F, Campos A. Synthesis and processing of glypican during differentiation of skeletal muscle cells. Eur J Cell Biol. 1996;71:170–6. [PubMed] [Google Scholar]
- 40.Carey DJ, Stahl RC, Asundi VK, Tucker B. Processing and subcellular distribution of the Schwann cell lipid-anchored heparan sulfate proteoglycan and identification as glypican. Exp Cell Res. 1993;208:10–8. doi: 10.1006/excr.1993.1217. [DOI] [PubMed] [Google Scholar]
- 41.Feng M, Kim H, Phung Y, Ho M. Recombinant soluble glypican 3 protein inhibits the growth of hepatocellular carcinoma in vitro. Int J Cancer. 2011;128:2246–7. doi: 10.1002/ijc.25549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishihara M, Fedarko NS, Conrad HE. Involvement of phosphatidylinositol and insulin in the coordinate regulation of proteoheparan sulfate metabolism and hepatocyte growth. J Biol Chem. 1987;262:4708–16. [PubMed] [Google Scholar]
- 43.Takeo S, Akiyama T, Firkus C, Aigaki T, Nakato H. Expression of a secreted form of Dally, a Drosophila glypican, induces overgrowth phenotype by affecting action range of Hedgehog. Dev Biol. 2005;284:204–18. doi: 10.1016/j.ydbio.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Capurro MI, Shi W, Sandal S, Filmus J. Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma growth. J Biol Chem. 2005;280:41201–6. doi: 10.1074/jbc.M507004200. [DOI] [PubMed] [Google Scholar]
- 45.Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, et al. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res. 2004;64:2418–23. doi: 10.1158/0008-5472.CAN-03-2191. [DOI] [PubMed] [Google Scholar]
- 46.Hudson ML, Kinnunen T, Cinar HN, Chisholm AD. C. elegans Kallmann syndrome protein KAL-1 interacts with syndecan and glypican to regulate neuronal cell migrations. Dev Biol. 2006;294:352–65. doi: 10.1016/j.ydbio.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 47.Margadant C, Sonnenberg A. Integrin-TGF-β crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010;11:97–105. doi: 10.1038/embor.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munger JS, Sheppard D. Cross talk among TGF-β signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3:a005017. doi: 10.1101/cshperspect.a005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saharinen J, Taipale J, Monni O, Keski-Oja J. Identification and characterization of a new latent transforming growth factor-β-binding protein, LTBP-4. J Biol Chem. 1998;273:18459–69. doi: 10.1074/jbc.273.29.18459. [DOI] [PubMed] [Google Scholar]
- 50.Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J Cell Biol. 2007;179:1311–23. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robertson I, Jensen S, Handford P. TB domain proteins: evolutionary insights into the multifaceted roles of fibrillins and LTBPs. Biochem J. 2011;433:263–76. doi: 10.1042/BJ20101320. [DOI] [PubMed] [Google Scholar]


