Abstract
Tumor protein D52 (TPD52) is a coiled-coil motif bearing hydrophilic polypeptide known to be overexpressed in cancers of diverse cellular origins. Increased TPD52 expression is associated with increased proliferation and invasive capacity in different cell types. Recent studies have reported a correlation between TPD52 transcript levels and G2 chromosomal radiosensitivity in lymphocytes of women at risk of hereditary breast cancer, and that TPD52 knockdown significantly reduced the radiation sensitivity of multiple cancer cell lines. In this study, we investigated possible roles for TPD52 in DNA damage response, and found that increased TPD52 expression in breast cancer and TPD52-expressing BALB/c 3T3 cells compromised ATM-mediated cellular responses to DNA double-strand breaks induced by γ-ray irradiation, which was associated with downregulation of steady-state ATM protein, but not transcript levels, regardless of irradiation status. TPD52-expressing 3T3 cells also showed significantly increased radiation sensitivity compared with vector cells evaluated by clonogenic assays. Furthermore, direct interactions between exogenous and endogenous ATM and TPD52 were detected by GST pull-down and co-immunoprecipitation assays. We also identified the interaction domains involved in this binding as TPD52 residues 111–131, and ATM residues 1–245 and 772–1102. Taken together, our results suggest that TPD52 may represent a novel negative regulator of ATM protein levels.
Keywords: TPD52, ATM, DNA damage response, cancer, protein-protein interaction
Introduction
It is well known that genomic stability and integrity are maintained through sophisticated cellular DNA damage response (DDR) networks that alter chromatin organization, switch on cell cycle checkpoints, activate DNA repair, and modulate numerous cellular processes. Defects in DDR lead to genetic instability, which, in turn, may enhance the rate of cancer development.1 The DNA repair ability of tumor cells also prevents the accumulation of lethal DNA damage from cytotoxic agents and ionizing radiation (IR), leading to tumor resistance.2 Therefore, DDR plays a central role in tumorigenesis and cancer therapy. Among the many types of damage, DNA double-strand breaks (DSBs) induced by IR and radiomimetic chemicals are most deleterious to cell survival. In response to DSBs, activation of ATM (ataxia telangiectasia mutated) protein, a member of the phosphatidylinositol 3-kinase-related protein kinase (PIKK) family, plays a central role in the recognition and signaling of DNA damage through rapid phosphorylation of numerous substrates.3,4 The hallmark of ATM’s response to DSBs is a rapid increase in kinase activity initiated by autophosphorylation on S1981 immediately following DSB formation.5 The rapid phosphorylation of S139 on H2AX by ATM is essential for focus formation at DNA DSB sites and for further recruitment of repair factors, such as the MRE11-RAD50-NBN (MRN) complex, RAD51, and BRCA1.6 p53, a main component in the G1–S cell cycle checkpoint, is rapidly phosphorylated on S15 by ATM following DNA damage, contributing primarily to p53 activation.7,8 ATM also phosphorylates and activates CHK2 on T68, which regulates several cell cycle checkpoints.9 Loss of functional ATM is associated with both increased genomic instability and cancer risk.1
Tumor protein D52 (TPD52) is the founding member of the TPD52-like protein family, which are coiled-coil motif-bearing small hydrophilic polypeptides conserved from lower organisms to human.10 The coiled-coil motif is required for homo- and heteromeric interactions between family members.11 TPD52-like proteins also include N- and C-terminally located PEST domains, a proteolytic signal, as well as the D52-motif, which is shared among family members and duplicated in TPD52L2.12,13 TPD52 was initially cloned through differential screening of a breast carcinoma cDNA library, and subsequent studies have identified overexpression of TPD52 in many different types of malignancies.14 TPD52 also represents a candidate target of 8q21 amplification/gain in different types of cancer, including breast, prostate, ovarian, and other cancers.14 Interestingly, while a number of studies have associated increased TPD52 expression with poor survival in breast15-17 and prostate cancers,18 increased TPD52 expression has been reported as a favorable independent prognostic marker in ovarian carcinoma, which is usually treated with DNA damaging agents.19 TPD52 promotes proliferation, migration/invasion, and metastasis in different cell models,17,20,21 whereas knockdown of TPD52 induced apoptosis in breast and prostate cancer cell lines.17,22 The underlying mechanisms are not yet fully understood, apart from the regulation of Akt/protein kinase B and Stat3/Bcl-2 signaling pathways shown in prostate cancer cell lines.21,22
Studies have recently indicated a possible role of TPD52 in regulating DDR, where TPD52 levels positively correlated with G2 chromosomal radiosensitivity in lymphocytes from women with or at risk of hereditary breast cancer.23 A genome-wide association study combined with radiation cytotoxicity assays using human lymphoblastoid cell lines again identified TPD52 levels to be positively associated with radiation sensitivity, and TPD52 knockdown in multiple tumor cell lines significantly de-sensitized these cell lines to radiation treatment.24 However, the molecular mechanisms explaining the association of TPD52 expression with DDR are unknown.
In this study, we investigated possible roles for TPD52 in DDR within cell lines and found that expression of TPD52 compromised ATM-mediated cellular response to DSBs induced by IR, and increased cell radiation sensitivity, which may be explained by the negative regulation of steady-state ATM protein but not transcript levels by TPD52. Decreased ATM levels were not restored by proteasome inhibitor MG132 treatment in TPD52-expressing BALB/c 3T3 cells. Direct interactions between exogenous and endogenous ATM and TPD52 were detected. Taken together, our results indicate that TPD52 might represent a novel negative regulator of ATM protein levels.
Results
Overexpression of TPD52 compromised DNA repair capacity in SK-BR-3 cells
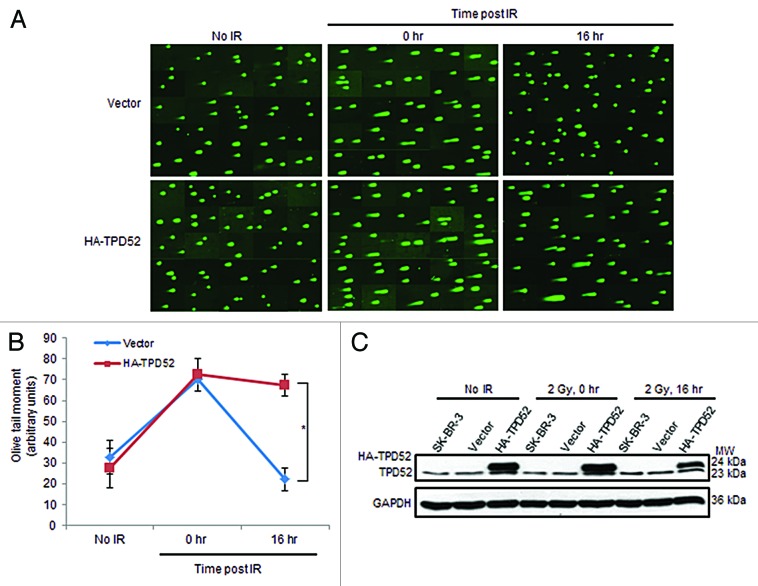
We first investigated the effects of TPD52 expression on cellular responses to DNA damage induced by γ-ray irradiation. The breast cancer cell line SK-BR-3, which has TPD52 amplification and high expression levels, as well as a TP53 mutation,25,26 was transfected with pHM6 vector or HA-TPD52, encoding human tumor protein D52 (TPD52) transcript variant 3 (GenBank NM_005079.2). After 72 h, untreated or 2 Gy irradiated cells were harvested immediately (0 h) or 16 h post-IR and subjected to neutral comet assays. Although exposure to IR increased comet tails in both vector and HA-TPD52 transfected cell populations at time point 0 h compared with no IR controls, the comet tails in vector-transfected populations returned to baseline levels at 16 h, whereas cells within the HA-TPD52-transfected populations retained long comet tails at the same time point (Fig. 1A). Measuring olive tail moments confirmed that there was a significant increase in the mean olive tail moment in HA-TPD52-transfected vs. vector-transfected populations at 16 h post-IR, compared with no significant differences in the absence of treatment, or immediately after irradiation (Fig. 1B). Levels of endogenous TPD52 and HA-TPD52 at the indicated time points in each cell line are shown in Figure 1C. These results suggest that TPD52 reduces SK-BR-3 cell capacity to repair DNA DSBs.
Figure 1. Transient HA-tagged TPD52 transfection of SK-BR-3 cells disrupted DNA damage repair post-IR. (A) SYBR Green-stained DNA in un-irradiated cells (no IR), cells harvested immediately (0 h) or 16 h post 2 Gy IR (16 h). Composite images shown are from one of 3 independent experiments. (B) DNA damage was quantified by comparing mean ± SE olive tail moments (y-axis, arbitrary units) at different time points. Experiments were performed in triplicate and images from 3 independent experiments (47–54 comet specimens per condition per experiment) were quantified. *P = 0.016, Student t test. (C) Representative western blots indicate endogenous TPD52 and HA-TPD52 levels in SK-BR-3, vector-, or HA-TPD52-transfected cells. GAPDH served as a loading control. MW, molecular weight in kDa (right).
Ectopic expression of TPD52 downregulated ATM-mediated DNA repair signaling
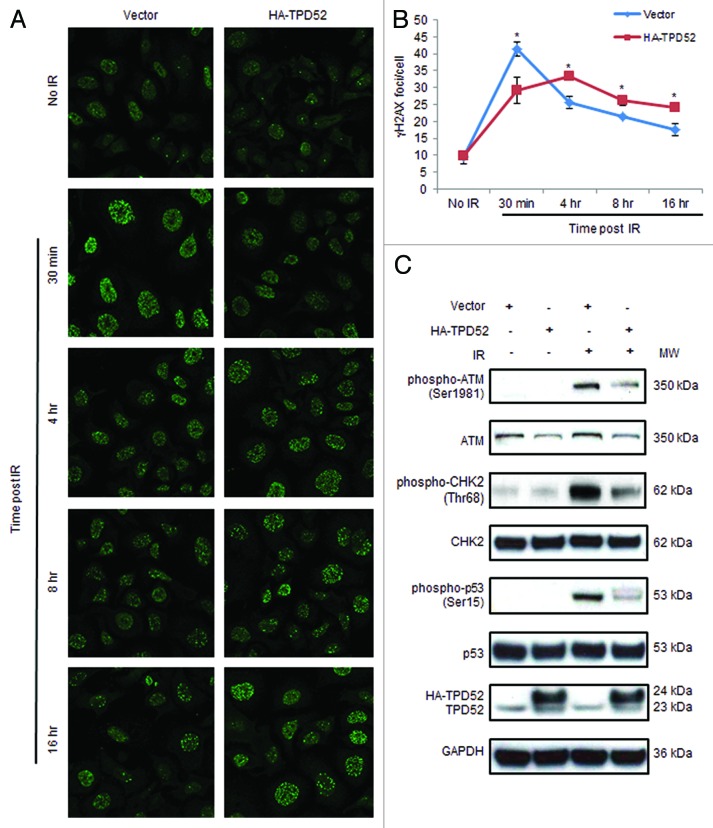
Since γH2AX foci are early markers of DSB sites undergoing DNA damage repair,4,6 and the rate of γH2AX clearance represents an important factor associated with radiosensitivity,27 we examined the effects of HA-TPD52 transfection on γH2AX focus formation. In vector-transfected SK-BR-3 cells, the intensity (Fig. 2A) and number (Fig. 2B) of γH2AX foci were maximal at 30 min and gradually returned toward baseline levels at 16 h post 4 Gy IR. In contrast, in HA-TPD52-transfected cells, γH2AX focus formation peaked at 4 h post-IR, at a lower level than that noted at 30 min in vector-transfected populations. The mean number of γH2AX foci/cell at 30 min post-IR in HA-TPD52-transfected cells was significantly less than in control cells (Fig. 2B). At 8 h and 16 h post-IR, HA-TPD52-transfected cells maintained significantly increased numbers of γH2AX foci/cell, compared with control cells (Fig. 2B). Western blot analyses showed that ectopic expression of HA-TPD52 in SK-BR-3 cells reduced IR-induced phospho-ATM, phospho-CHK2, and phospho-p53 levels 15 min post-IR (Fig. 2C). Similar results were obtained in MDA-MB-231 cells, which show low endogenous TPD52 levels (data not shown). These results indicate that TPD52 expression downregulates ATM-mediated DNA repair signaling in response to DSBs. Interestingly, we noticed an approximate 50% reduction in total ATM protein levels (normalized to GAPDH) in HA-TPD52-transfected cells compared with control cells, regardless of irradiation status, whereas total CHK2 and p53 levels were unaffected (Fig. 2C).
Figure 2. HA-tagged TPD52 expression in SK-BR-3 cells compromised ATM-mediated DDR post-IR. (A) Cells were untreated or treated with 4 Gy IR 72 h post-transfection with pHM6 vector or HA-TPD52, and γH2AX foci were detected at the indicated time points using immunofluorescence. Images shown are representative of those obtained from one of 3 independent experiments. (B) Quantitation of γH2AX foci/cell from 3 independent experiments (70–100 cells per condition per experiment) was performed using Metamorph. Mean ± SE of γH2AX foci/cell (y-axis) at different time points are shown. *P = 0.05 (30 min); P = 0.02 (4 h); P = 0.03 (8 h); P = 0.02 (16 h); Student t test. (C) Total protein was extracted from SK-BR-3 cells treated as described in (A) at 15 min post-IR and subjected to western blot analyses with antisera to the proteins shown (left). GAPDH served as a loading control. At least 3 independent experiments were performed.
Transient knockdown of endogenous TPD52 in SK-BR-3 cells increased ATM-mediated DNA repair signaling
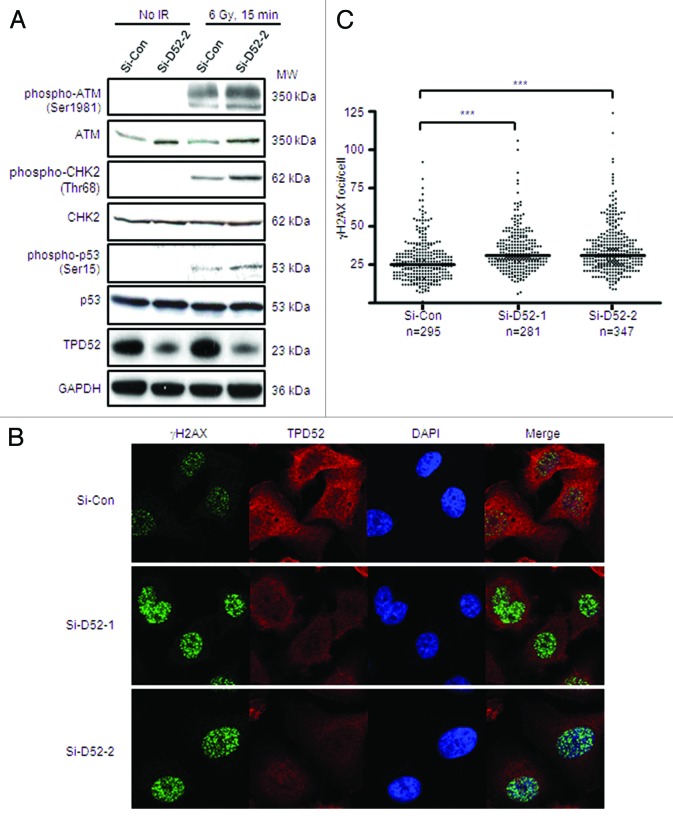
Next we examined whether knockdown of endogenous TPD52 could improve cellular responses to DSBs. A significant reduction (>80%) in TPD52 levels was achieved in TPD52 siRNA-transfected cells (Si-D52-2) compared with non-targeting control siRNA-treated cells (Si-Con) (Fig. 3A). Similar results were obtained with TPD52 siRNA Si-D52-1 (data not shown). Total ATM levels were increased approximately 2-fold in TPD52 knockdown cells compared with control cells, with no change in CHK2 or p53 levels (Fig. 3A). Western blot analyses showed increased detection of phospho-ATM, phospho-CHK2, as well as phospho-p53 levels 15 min post 6 Gy IR in TPD52-siRNA vs. non-targeting siRNA-treated cells (Fig. 3A). Immunofluorescence analyses also detected a statistically significant increase in γH2AX foci/cell in TPD52 siRNA-transfected cells compared with control cells (Figs. 3B and C).
Figure 3. Knockdown of endogenous TPD52 in SK-BR-3 cells improved ATM-mediated DDR post-IR. Cells were untreated or treated with 6 Gy IR 72 h post-transfection with non-targeting (Si-Con) or TPD52 (Si-D52-1, Si-D52-2) siRNAs. (A) Total protein were harvested and subjected to western blot analyses using antisera indicated. (B) γH2AX foci were visualized using immunofluorescence with co-staining of TPD52. Nuclei were counterstained using DAPI. Representative images from one of 3 independent experiments are shown. (C) γH2AX foci were quantitated using Metamorph. Scatter plots of the numbers of γH2AX foci/cell from 3 independent experiments obtained using GraphPad Prism 4.03. Horizontal bars indicate the median γH2AX foci/cell for each siRNA. ***P < 0.0001 (Si-Con v. Si-D52-1, n = 576; Si-Con v. Si-D52-2, n = 642); Mann–Whitney U test.
Stable TPD52 expression decreased Atm protein but not transcript levels in mouse BALB/c 3T3 cells
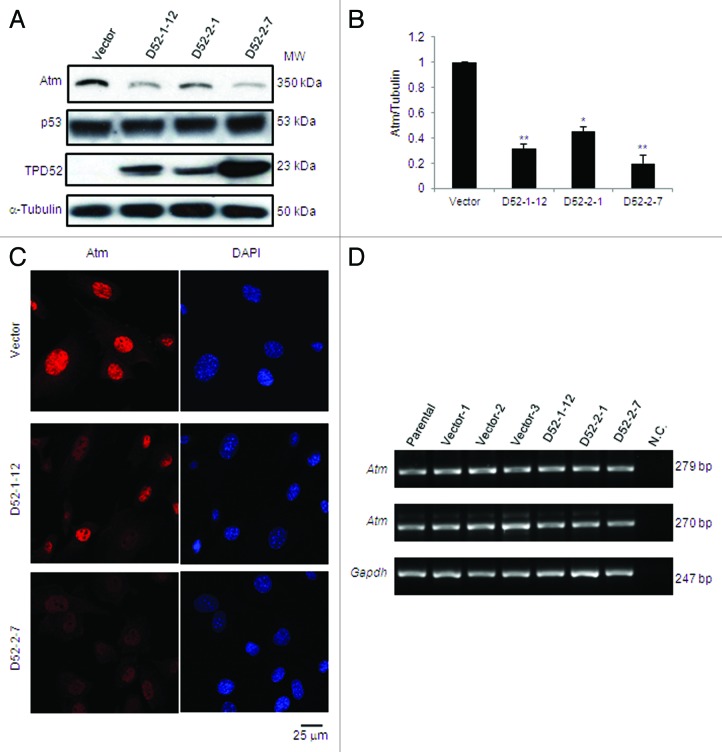
To further address the effects of TPD52 expression on ATM protein levels, we evaluated endogenous Atm levels in vector- or TPD52-transfected stable 3T3 cell lines. Western blot analyses detected a striking reduction in Atm levels in all 3 TPD52-expressing cell lines relative to vector control, whereas p53 levels in these cells were unchanged (Fig. 4A). Quantification of Atm levels revealed an 80% reduction in D52-2-7 cell line, which shows the highest TPD52 levels, while 70% and 50% reductions were measured in the D52-1-12 and D52-2-1 cell lines, respectively (Fig. 4B).
Figure 4. Stable TPD52 expression decreased Atm protein but not transcript levels in mouse 3T3 cells. (A) Western blot analyses of vector- and TPD52-transfected 3T3 cell lines with antisera to the proteins shown (left). α-Tubulin served as a loading control. Results are representative of 3 independent experiments. (B) Densitometry analyses of western blots obtained in (A). Mean ± SD of Atm levels relative to α-tubulin are plotted on Y-axis with vector control set as 1. **P = 0.0060 (D52-1-12); P = 0.0033 (D52-2-7); *P = 0.0159 (D52-2-1); Student t test. (C) Immunofluorescence analyses of endogenous Atm (left). Nuclei were counterstained using DAPI (right). Scale bar = 25 μm. (D) Total RNA from 3T3 cells (parental), 3 vector control (Vector-1, Vector-2, Vector-3) and 3 TPD52-expressing cell lines (D52-1-12, D52-2-1, D52-2-7) was subjected to cDNA synthesis and RT-PCR to amplify Atm products 279 bp or 270 bp. Gapdh was amplified as a control gene. N.C., negative control in which cDNA was replaced by water.
We also examined immunofluorescent staining of endogenous Atm in these cell lines. In contrast to the strong nuclear detection of Atm in vector control, the intensity of Atm staining was reduced by approximately 50% in D52-1-12 cells, while a lack of distinctive Atm nuclear staining was observed in most D52-2-7 cells (Fig. 4C).
We further queried whether reduced Atm levels resulted from reduced Atm transcript levels in TPD52-expressing 3T3 cell lines. We performed semi-quantitative reverse-transcriptase (RT)-PCR using cDNA from 3T3 parental cells, 3 vector-transfected, and 3 TPD52-expressing cell lines to amplify 2 Atm products of 279 bp or 270 bp. None showed significantly different RT-PCR product levels compared with the Gapdh control (Fig. 4D).
Stable TPD52 expression disrupted γH2AX focus formation and sensitized cells to DNA damage
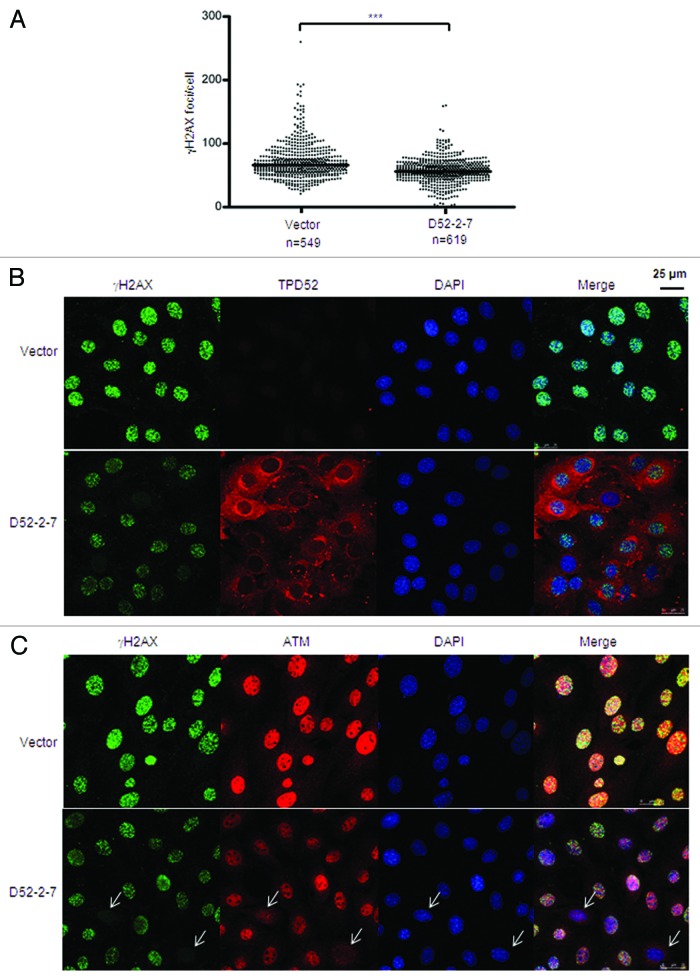
To determine whether the reduction of Atm levels alters cellular responses to DSBs, we assessed γH2AX focus formation in vector control and D52-2-7 cells 30 min post 2 Gy IR, which revealed a significant reduction in γH2AX foci/cell in D52-2-7 cells (Figs. 5A and B). We also compared Atm staining in these cells, and found that most D52-2-7 cells showed reductions in both Atm intensity and γH2AX focus formation. When there was no nuclear Atm staining, cells also showed a lack of γH2AX foci formed post-IR (Fig. 5C).
Figure 5. Stable TPD52 expression disrupted γH2AX focus formation post-IR. Vector and D52-2-7 cells were treated with 2 Gy IR, fixed after 30 min and subjected to immunofluorescence analyses. (A) γH2AX foci were visualized using immunofluorescence and quantitated using Metamorph. Scatter plots of the numbers of γH2AX foci/cell from 3 independent experiments obtained as described in Figure 3. ***P < 0.0001 (n = 1168; Mann–Whitney U test). (B) Co-staining of γH2AX and TPD52, or (C) γH2AX and Atm, with nuclei counterstained using DAPI. Arrows highlight cells lacking nuclear Atm staining and γH2AX focus formation. Scale bar = 25 μm.
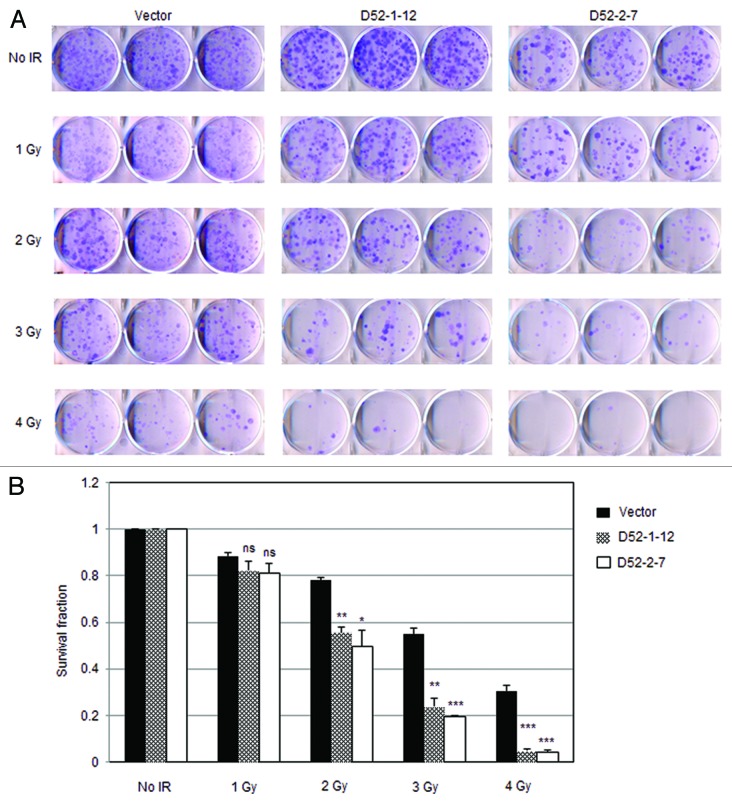
We further investigated whether reduced Atm levels in TPD52-expressing cells impairs cell survival post-IR. Indeed, both D52-1-12 and D52-2-7 cells displayed significantly reduced survival fractions in response to γ-ray irradiation compared with vector control cells (Figs. 6A and B). These results suggest that TPD52 expression downregulates Atm protein levels, which contributes to the defective cellular DDR.
Figure 6. Stable TPD52 expression sensitized cells to γ-ray irradiation. (A) 3T3 vector cell line and 2 TPD52-expressing cell lines (D52-1-12, D52-2-7) were untreated or exposed to the indicated doses of IR, and clonogenic survival assays were then performed. Images shown are representative of those obtained from 3 independent experiments. (B) Colonies with > 50 cells/ colony were counted and mean ± SE of survival fraction for each dose per cell line plotted on the Y-axis, with no treatment control set as 1. Vector v. D52-1-12: ns (not significant, 1 Gy); P = 0.0012 (2 Gy); P = 0.0015 (3 Gy); P = 0.0006 (4 Gy). Vector v. D52-2-7: ns (1 Gy); P = 0.0183 (2 Gy); P = 0.0002 (3 Gy); P = 0.0007 (4 Gy), Student t test.
TPD52 directly interacted with ATM
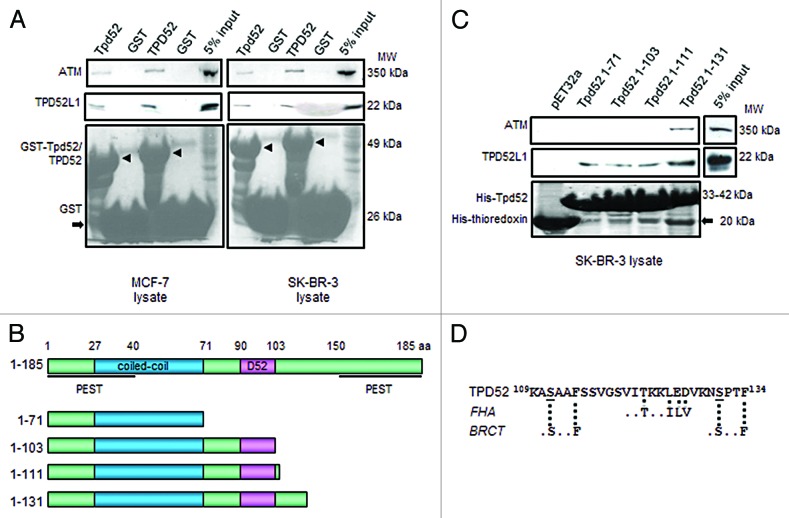
To explore the underlying mechanisms by which TPD52 regulates ATM levels, we examined whether TPD52 interacts with ATM. Using GST pull-down assays, ATM protein was recovered by GST-tagged mouse (Tpd52) or human TPD52, but not by the GST tag alone, from un-irradiated MCF-7 or SK-BR-3 cell lysates (Fig. 7A) and MDA-MB-231 cell lysates (data not shown), indicating a direct interaction between TPD52 and ATM. As a positive control, the binding of TPD52/Tpd52 with TPD52L1 was confirmed (Fig. 7A).28
Figure 7. Interactions between TPD52 and ATM. (A) GST pull-down assays using GST-tagged full-length mouse Tpd52 or human TPD52 or GST tag, and MCF-7 (left) or SK-BR-3 (right) cell lysates. Upper and middle panels show the results of western blot analyses using ATM antibody and TPD52L1 antisera, respectively. The lower panel shows Ponceau S staining to reveal GST-Tpd52/TPD52 (arrowheads) or GST tag (arrow). At least 3 independent experiments were performed. (B) Schematic representations of deleted Tpd52 recombinant proteins. Amino acid coordinates for each motif/domain are indicated. (C) Pull-down assays using thioredoxin-6His-tagged Tpd52-deleted proteins and SK-BR-3 cell lysates. Upper and middle panels show results of western blot analyses using ATM antibody and TPD52L1 antisera, respectively. The lower panel shows Ponceau S staining to reveal His-Tpd52 proteins or His-thioredoxin tag (arrow). Results shown are representative of those obtained from 3 independent experiments. (D) Sequence of TPD52 aa 109–134 with broken lines indicating FHA and BRCT motifs as predicted through ELM. S111 and S131 are underlined.
To identify the ATM interaction domain within TPD52, pull-down assays were performed using a series of thioredoxin-6His-tagged truncated Tpd52 proteins (Fig. 7B). ATM expressed in SK-BR-3 lysates bound to an N-terminal Tpd52 fragment representing amino acids (aa) 1–131, but not aa 1–71, aa 1–103 or aa 1–111, indicating that aa 111–131 are required to bind ATM (Fig. 7C). Re-probing the same membrane with TPD52L1 antisera revealed interactions between TPD52L1 and all Tpd52-truncated proteins, as predicted. The same results were also obtained using MCF-7 cell lysates (data not shown). Interestingly, functional site prediction through ELM (http://elm.eu.org/)29 identified a FHA (forkhead associated domain) motif ..(T)..[ILV] located at V120-E126 and 2 BRCT-BRCA1 domains (.[S]..F) located at A110-F114 and N130-F134, flanking the FHA motif (Fig. 7D).
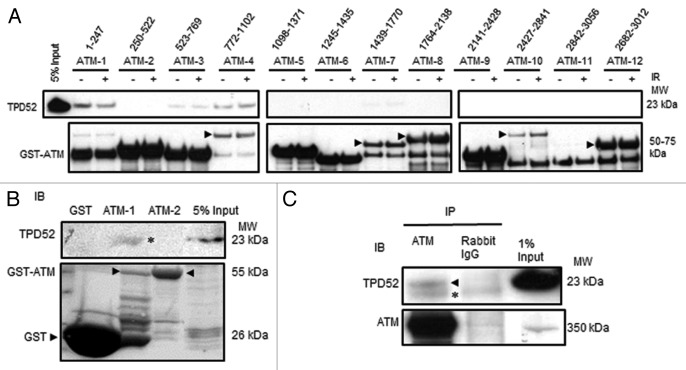
To determine which ATM domain(s) are required for interactions with TPD52, we employed a series of GST-tagged ATM proteins spanning the full-length of ATM and SK-BR-3 lysates prepared from cells with or without prior exposure to 6 Gy IR (Fig. 8A). GST pull-down assays identified fragments ATM-1 (aa 1–247) and ATM-4 (aa 772–1102) as binding to TPD52, with weaker binding detected with fragment ATM-3 (aa 523–769). After normalizing to the input of respective GST-ATM protein, the TPD52 binding capacity of ATM aa 772–1102 is about 1.5-fold higher compared with aa 1–247. Irradiation did not obviously affect the binding capacity in both cases (Fig. 8A).
Figure 8. ATM directly interacted with TPD52. (A) GST pull-down assays using GST-tagged deleted ATM recombinant proteins (1–12, amino acid coordinates for each protein are indicated) and SK-BR-3 cell lysates with or without prior exposure to 6 Gy IR. Arrowheads highlight GST-ATM proteins where multiple bands were detected. (B) GST pull-down assays using either GST, GST-ATM-1, or GST-ATM-2 and in vitro translated TPD52. The upper panel shows the results of western blot analyses using TPD52 antisera with TPD52 band indicated (*), and the lower panel shows Ponceau S staining of the same membrane, highlighting GST-ATM proteins, or the GST tag protein (arrowheads). Five % of synthesized TPD52 is shown. (C) Co-immunoprecipitations performed with SK-BR-3 cell lysates, using ATM antibody or rabbit IgG as a negative control. The arrowhead indicates TPD52. *IgG light chain. All results are representative of 3 independent experiments.
To confirm direct interactions between TPD52 and ATM, we synthesized TPD52 using the TnT® Quick Coupled Transcription/Translation System. The resulting TPD52 protein was subjected to GST pull-down assays using GST-tagged ATM-1 (aa 1–247), ATM-2 (aa 250–522), or GST tag alone (Fig. 8B). Only GST-tagged ATM-1, but not ATM-2 or GST tag, recovered synthesized TPD52 protein, further suggesting a direct interaction between ATM aa 1–247 and TPD52. We next examined whether endogenous ATM and TPD52 form a complex. As shown in Figure 8C, ATM-immunoprecipitation recovered TPD52 from SK-BR-3 cell lysates, whereas nonspecific immunoglobulin did not. Together, the results suggest that ATM and TPD52 directly interact in vitro and form a complex in cultured cells.
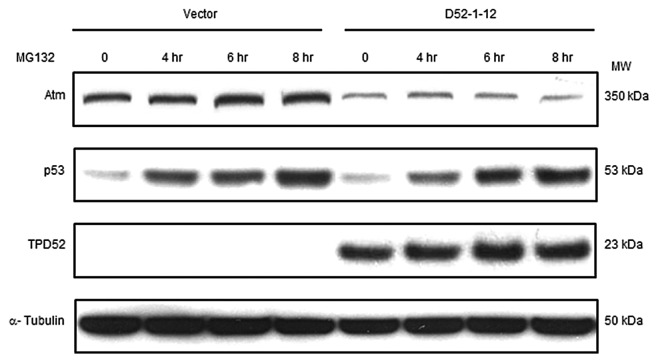
Proteasome inhibitor MG132 did not increase Atm protein levels in BALB/c 3T3 cell lines
As TPD52 decreased Atm protein but not transcript levels in 3T3 cell lines, and TPD52 directly interacted with ATM, we investigated whether TPD52 was involved in ATM degradation. We identified a potential PEST motif located at ATM residues R832-K892 using the PEST-FIND algorithm, which is considered to be a proteolytic signal.30 Therefore, vector- and TPD52-expressing cell lines (D52-1-12 and D52-2-1) were treated with 50 µM MG132 for up to 8 h, the Atm half-life determined in mouse embryonic fibroblasts (MEFs).31 Although western blot analyses revealed that MG132 treatment stabilized p53 protein levels in a time-dependent manner, Atm levels were unchanged post-MG132 treatment (Fig. 9 and data not shown). We also did not detect ubiquitinated forms of Atm following Atm immunoprecipitation (data not shown), which indicates that the ubiquitin-proteasome pathway, a major proteolytic pathway in the degradation of abnormal and short-lived regulatory proteins,32 might not contribute to this regulation.
Figure 9. MG132 treatment did not increase Atm levels in 3T3 cell lines. Vector and TPD52-expressing (D52-1-12) cell lines were treated with 50 μM MG132 (4 h, 6 h, 8 h) or DMSO vehicle (0 h) and harvested at the indicated time points. Total proteins were subjected to western blot analyses with the indicated antisera (left). Results shown are representative of 3 independent experiments.
Discussion
A swift and effective DDR is essential for the maintenance of genome stability and integrity, which is fundamental to cell survival. Defects in the DDR network result in a variety of diseases, ranging from severe genomic instability syndromes to chronic diseases, cancer predisposition, and accelerated aging.33 On the other hand, DNA repair ability of tumor cells prevents the accumulation of lethal DNA damage from cytotoxic agents and ionizing radiation, which leads to tumor resistance.34,35 Therefore, DDR plays a central role in tumorigenesis and cancer therapy.
The ATM protein kinase acts as an important safeguard to DNA damage, particularly in response to DNA double-strand breaks. While ATM represents a low-penetrance risk factor in hereditary breast and ovarian cancers,36,37 inactivation of ATM is a frequent event in the development of certain common sporadic cancers. ATM is frequently altered in hematological cancers, where T-cell prolymphocytic leukemia, B-cell chronic lymphocytic leukemia, and mantle cell lymphoma are all characterized by high rates of inactivating mutations, and/or deletion of ATM.38 Reduced ATM protein levels have also been reported in breast cancer,39 glioblastoma,40 gastric cancer,41 and colorectal cancer,42 yet the cellular mechanisms that lead to deregulation of ATM expression are poorly understood. In sporadic breast cancer, although loss of heterozygosity (LOH) in the region of ATM on chromosome 11q22.3 has been reported in up to 40% cases, somatic mutations of ATM are infrequent.38,43 Hypermethylation of the ATM promoter44 and gene-body ATM methylation have also been shown in breast cancer samples and associated with lower steady-state ATM mRNA levels.45 However, these mechanisms could only occur in a small proportion of breast cancers, whereas downregulation of ATM is observed in up to 75% cases.39
Interestingly, increased TPD52 expression has been identified in many different types of cancer, including breast cancer,46-48 colorectal cancer,49 leukemia, and lymphoma,50-52 which overlap with the spectrum of ATM-deficient malignancies. It is of particular interest that increased TPD52 expression has been associated with radiosensitivity in independent studies using lymphocytes or lymphoblastoid cell lines,23,24 indicating that variations in TPD52 levels could alter radiosensitivity in normal cells. It has also been reported that TPD52 expression was significantly upregulated in HeLa cells stably transfected with a specific BRCA1 mutant (S1841N),53 which produced microsatellite instability and impairment in the binding between BRCA1 and the mismatch repair protein MLH1 in the same cell line.54
To better understand the biological functions of TPD52 in DDR, we investigated the effects of TPD52 expression on the cellular response to DNA damage. Neutral comet assays demonstrated that DNA repair capacity was profoundly compromised in ectopic TPD52-expressing SK-BR-3 cells, indicated by a significant increase in the mean olive tail moments compared with vector-expressing cells at 16 h post-IR (Fig. 1). Consistent with these results, γH2AX focus formation following IR was significantly impaired in ectopic TPD52-expressing SK-BR-3 cells (Fig. 2). These results confirmed the association of TPD52 expression with cellular responses to DSBs.23,24
As ATM plays a central role in sensing and signaling DSBs induced by IR, we assessed ATM-mediated signaling pathways according to TPD52 expression status. Ectopic TPD52 expression significantly decreased phospho-ATM (S1981) levels, as well as that of its substrates phospho-p53 (S15) and phospho-CHK2 (T68) post-IR, while TPD52 knockdown showed opposite effects (Figs. 2C and 3A). Taken together with the effects on γH2AX foci formation detected by immunofluorescence analyses, our results suggest that TPD52 expression regulates cellular response to DSBs through an ATM-mediated pathway.
Notably, the steady-state levels of ATM but not CHK2 or p53 levels showed a negative association with TPD52 levels. Quantification indicated that an approximate 50% reduction in total ATM levels was detected in HA-TPD52 transfected SK-BR-3 cells, whereas TPD52 knockdown increased ATM levels by 1.8–2 fold. This association was further confirmed in 3 TPD52-expressing BALB/c 3T3 cells, with the cell line with the highest TPD52 expression (D52-2-7) showing the greatest reduction in Atm levels. Immunofluorescence analyses also highlighted a lack of distinctive Atm nuclear staining, and significantly reduced γH2AX focus formation post-IR in D52-2-7 cells. Accordingly, TPD52-expressing cells showed significantly increased radiation sensitivity in response to DSBs induced by IR, as evaluated by colony-formation assays. Therefore, reduced steady-state ATM protein levels in TPD52-expressing cells might be responsible for their compromised cellular responses to DNA damage.
The fact that no significant change in Atm levels was noted in TPD52-expressing BALB/c 3T3 cells, and interactions between TPD52 and ATM were identified by GST pull-down assays and co-immunoprecipitations suggested that TPD52 might be involved in ATM degradation. A potential PEST motif was also identified to be located at ATM residues R832-K892 using the PEST-FIND algorithm. However, the proteasome inhibitor MG132 did not restore Atm levels in TPD52-expressing BALB/c 3T3 cells. Interestingly, Takai et al. reported that TEL2 (telomere maintenance 2) regulates the stability of all 6 PIKK family members, including ATM, and also found that proteasome inhibitors (MG132 and lactacystin) did not stabilize PIKKs in Tel2-deficient MEFs.31
TPD52 detectably bound ATM on fragments aa 1–247 and aa 772–1102, which have been identified as an N-terminal substrate-binding site interacting with substrates such as NBN, p53, and BRCA1,4 and a binding site (aa 811–950) with β-adaptin, a component of the AP-2 adaptor complex involved in clathrin-mediated endocytosis of receptors,55 respectively. Interaction domain mapping also indicated that the fragment aa 111–131 in TPD52 but not the coiled-coil motif, was essential for TPD52 and ATM binding. This fragment is predicted to contain a FHA motif and 2 BRCT motifs flanking the FHA motif. FHA domains are exclusively phosphothreonine-binding domains found in proteins involved in transcription, DNA damage signaling, and cell cycle regulation.56 BRCT (BRCA1 C-terminal) domains are often found as tandem repeats within a single protein, and essentially all BRCT domain-containing proteins have roles in DNA damage signaling and DNA repair.56 Many DNA damage repair proteins, such as MDC1 and NBN, both ATM-interacting proteins, contain both domains, which allow these proteins to function as a core scaffolds and mediate events downstream.56 Moreover, since TPD52 could form homo- or heterodimers through its coiled-coil motif,11 TPD52 could serve as an adaptor to mediate the interactions between ATM and its substrates, or for ATM in vesicle-associated processes, irrespective of irradiation.
It has been shown that ATMIN (ATM interacting protein) interacts with ATM and stabilizes ATM reciprocally; however, the ATM/ATMIN complex is disrupted by IR.57 Recent studies have also reported that TEL2 forms a 2-MDa complex with TTI1, TTI2, and Hsp90 chaperones, which is required for PIKK stability.58,59 These studies indicate that TEL2 interacts with newly synthesized PIKKs and does not associate efficiently with the activated, mature form of ATM. Another group also reported that the (AAA+) family proteins RuvB-like 1 (RUVBL1) and RUVBL2 control PIKK abundance at least at the mRNA level, and also form a complex with Hsp90.60,61 In addition to these proteins which enhance ATM abundance, our results add TPD52 protein as a negative regulator of ATM protein levels irrespective of IR status.
TPD52 transcript and protein levels show wide variations in breast cancer samples and cell lines,17,28,47 and as a gene amplification target, TPD52 has been predicted to acquire oncogenic functions through overexpression. Our data here further support this prediction, with TPD52 reducing ATM levels in cell lines with varying TPD52 levels. While SK-BR-3 cells are TPD52-amplified,25 TPD52 has been previously detected immunohistochemically at higher levels in breast cancer cells in situ, compared with SK-BR-3 cells present on the same tissue microarrays,17 which supports the use of SK-BR-3 cells as a model for interrogating TPD52 function. Similarly, exogenous TPD52 levels achieved in the 3T3 cell lines employed in this study were comparable with those detected in MCF-7 breast cancer cells,17 which show moderate TPD52 levels.28 That variations in TPD52 levels in normal cells may also be significant is suggested by associations between increased TPD52 expression and radiosensitivity in independent studies using lymphocytes23 or lymphoblastoid cell lines,24 indicating that variations in TPD52 levels could also alter radiosensitivity in normal cells.
In summary, this study identified TPD52 as a novel negative regulator of ATM protein levels, which, in turn, impacts ATM-mediated cellular responses to DNA damage. This highlights the possibility that increased TPD52 expression might represent an alternative, possible widely used mechanism of inactivating ATM in different types of cancer, and might contribute to tumor development by facilitating genomic instability. Since cells with reduced ATM displayed increased sensitivity to radiation therapy62 and DNA damaging agents such as poly-ADP ribose polymerase (PARP) inhibitors, which have been shown effective in the treatment of human malignancies characterized by deficiencies in the DNA damage repair proteins BRCA1 and BRCA2,63,64 our results also indicate that elevated TPD52 levels in tumor may represent a marker of sensitivity to DNA damaging agents such as PARP inhibitor and/or radiation therapy.
Materials and Methods
Cell lines and cell culture
Human breast carcinoma cell lines (SK-BR-3, MCF-7, MDA-MB-231) were cultured in GIBCO® RPMI 1640 (Life technologies) medium supplemented with 10% FBS (Life technologies), 6 mM L-glutamine (Life technologies), and 10 μg/ml insulin (Sigma-Aldrich) (MCF-7) in a humidified atmosphere containing 5% CO2 at 37 °C. Cell line identities were confirmed through short tandem repeat profiling by CellBank Australia. The vector- or TPD52-transfected BALB/c 3T3 cell lines have been previously reported.17
Plasmid constructs
The pHM6 HA-tagged TPD52 and control vectors have been previously described.65 The pBluescript SK- vector encoding full-length TPD52 cDNA (GenBank NM_005079.2) was digested with SmaI and EcoRV to produce a 3.3 kb fragment which was subcloned into pcDNA3.1 (Life Technologies) using the EcoRV site. The pGEX-6P-1 TPD52 plasmid was constructed by ligating full-length TPD52 cDNA digested with BamHI and XhoI from pcDNA3.1-TPD52 into the same sites of pGEX-6P-1 (GE Healthcare). Glutathione S-transferase (GST)-tagged full-length mouse Tpd52 (pGEX-3X-6His) and a series of thioredoxin-6His-tagged truncated Tpd52 constructs (pET32a) were described elsewhere.11,66 The 12 GST-ATM deletion constructs that span the full-length ATM sequence have been previously described.67
Transient plasmid transfection
Cells were seeded into 6-well plates or 24-well plates 24 h before transfection and transfected at ~50–60% confluency by adding pHM6 vector or HA-TPD52 plasmid DNA with TransIT-LT1 transfection reagent (Mirus) at a ratio of 1:3, according to the manufacturer’s instructions. Cells were maintained at 37 °C in a 5% CO2 humidified atmosphere for 72 h before further analyses.
Single-cell DNA electrophoresis (neutral comet assay)
pHM6 vector- or HA-TPD52-transfected SK-BR-3 cells were untreated or treated with 2 Gy of γ-ray irradiation using a Gammacell 3000 Elan irradiator (MDS Nordion) at a dose rate of 1.3 Gy/min, harvested at the indicated time points, and subjected to Neutral Comet Assays using the CometAssay® kit (Trevigen) according to the manufacturer’s protocol. Slides were stained with SYBR green and visualized using an Olympus BX50 microscope with a 40× objective lens. Images were quantified using CometScore freeware (TriTek Corp). DNA damage was expressed as the olive tail moment (percentage of tail DNA x distance between the center of the head and the center of the tail).
Indirect immunofluorescence analyses and quantitation of γH2AX foci
Cells were untreated or treated with the indicated doses of IR, fixed in 3% paraformaldehyde in PBS at the indicated time points, then permeabilized in 0.2% Triton X-100 in PBS. Cells were stained using phospho-Histone H2A.X (S139) mouse monoclonal (JBW301) antibody (1:2000, Millipore), ATM rabbit polyclonal antisera (1:200, Millipore), and affinity-purified rabbit polyclonal TPD52 antisera (1:100).46 Alexa Fluor®488 anti-mouse IgG (Life Technologies) and Cy3-conjugated anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc) were utilized as secondary antibodies. DNA was counterstained using 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI, Sigma-Aldrich). Samples were viewed with a Leica SP5 II confocal microscope using a 63× objective lens. To allow for direct comparisons, all treatments for each experiment were processed simultaneously, and all images were captured using the same parameters. γH2AX foci numbers were quantitated using MetaMorph software 7.6.5 (Molecular Devices).
Transient small interfering RNA transfection
Two sets of TPD52 siRNA duplexes were synthesized by Dharmacon RNAi Technologies (Thermo Fisher Scientific).17 The targeted sequences were:
5′-GCGGAAACUUGGAAUCAAU-3′ (Si-D52-1), and 5′-GGAGAAGUCUUGAAUUCGG-3′ (Si-D52-2). Non-targeting siRNA (Allstar Negative Control siRNA) was purchased from QIAGEN.
SK-BR-3 cells were seeded at 8 × 104 cells/well in duplicate into 24-well plates, or at 5 × 105 cells/well onto glass coverslips in 6-well plates. After 24 h incubation, cells were transfected with 100 nM siRNA duplexes using TransIT-TKO transfection reagent (Mirus) in complete media according to the manufacturer’s instructions. After 72 h incubation, cells were treated with γ-irradiation, or not, and subjected to further analyses.
Western blot analyses and antibodies
Cells were lysed in 3% SDS lysis buffer as described previously28 or in NETN lysis buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris⋅HCl pH 7.5, 0.5% [v/v] Nonidet P-40) containing phosphatase and protease inhibitors (50 mM sodium fluoride, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and EDTA-free Protease Inhibitor Cocktail Tablets from Roche Applied Science).68 Between 30–60 μg total protein were resolved by SDS-PAGE on 12.5% mini polyacrylamide gels, 6% large polyacrylamide gels, or gradient NuPAGE® Novex 4–12% Bis-Tris or NuPAGE® Novex 3–8% Tris-Acetate gels (Life Technologies). Densitometry analyses employed ImageJ 1.45s freeware to quantify fold changes in protein levels relative to loading controls.
ATM (D2E2) rabbit monoclonal antibody, phospho-p53 (S15) (16G8) mouse monoclonal antibody, CHK2 (1C12) mouse monoclonal antibody, phospho-CHK2 (T68) rabbit polyclonal antibody, and p53 (1C12) mouse monoclonal antibody were purchased from Cell Signaling Technology and used at a dilution of 1:1000. Phospho-ATM (S1981) (10H11.E12) mouse monoclonal antibody (1:1000) was purchased from Millipore. Pantropic p53 (DO-1, 1:500) and GST (1:10 000) antibodies were purchased from Calbiochem and Novagen, respectively. Affinity-purified rabbit polyclonal TPD52 (1:100) and TPD52L1 (1:100) antisera were raised in-house.28,46 Mouse monoclonal α-tubulin antibody (DM1A) (1:5000; Sigma-Aldrich) and mouse monoclonal glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibody (6C5) (1:5000; Life Technologies) were used as loading controls.
Total RNA extraction, cDNA synthesis, and reverse-transcriptase (RT)-PCR
Total RNA was extracted from 3T3 cell lines using TRIzol LS reagent (Life Technologies) and 1 μg RNA was subjected to cDNA synthesis using the SuperScript III First-Strand Synthesis System for RT-PCR kit (Life Technologies) according to the manufacturer’s instructions. The resulting cDNA was amplified using Taq DNA polymerase (QIAGEN) and 2 sets of Atm primers: Atm-2, sense 5′-TGCTAGATCTTCTGAGAGCG-3′ and antisense 5′-TATTGTTGAGGGCAGTCAGC-3′; Atm-3, sense 5′-CTGTATCTACAGCAGAGACC-3′ and antisense 5′-TCACACCCAAGCTTTCCATC-3′, which amplified mouse Atm (GenBank NM_007499) mRNA sequence fragments at nt 5040–5318 and nt 9071–9340, respectively. Gapdh (GenBank NM_008084) was amplified using sense primer 5′-TGCACCACCAACTGCTTAGC-3′ and antisense primer 5′-GGAAGGCCATGCCAGTGA-3′. RT-PCR reactions were performed using a Veriti® 60-well Thermal Cycler (Applied Biosystems). Thermal cycling conditions comprised an initial denaturation step at 94 °C for 3 min, 30 cycles of (94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s), followed by 7 min at 72 °C.
Clonogenic survival assays
Cell survival was evaluated by colony formation assays. 3T3 cell lines (Vector, D52-1-12, and D52-2-7) were seeded at 500 cells/well in triplicate into 6-well plates. After 24 h incubation at 37 °C in a 5% CO2 humidified atmosphere, cells were untreated or treated with indicated doses of γ-ray irradiation. Cells were grown for 14 d and resulting colonies were fixed, stained with 0.4% crystal violet in 20% ethanol, and colonies with >50 cells/colony were counted. Data from 3 independent experiments were subjected to statistical analyses.
Pull-down assays
Mouse or human TPD52 fusion proteins were produced in the BL21 E. coli strain following the induction of log-phase cultures with 100 μM IPTG (isopropyl-β-D-thiogalactopyranoside, Sigma-Aldrich) for 3 h at 29 °C. Proteins were isolated by lysing bacterial samples in lysis buffer (20 mM Tris⋅HCl pH 7.4, 250 mM NaCl, 1 mg/ml lysozyme, 1 mM PMSF, 20 μg/ml leupeptin, 1 mg/ml DNase I, 1% [v/v] Triton X-100) and bound to GSH-agarose (Glutathione Sepharose 4B, GE Healthcare) or Ni-NTA agarose (Qiagen).11,66 GST-ATM fusion proteins were isolated as previously described.69 Purified proteins were assessed by SDS-PAGE. SK-BR-3 or MCF-7 cells were lysed in high-salt lysis buffer (250 mM NaCl, 5 mM EDTA, 50 mM Tris⋅HCl pH 7.5, 0.1% [v/v] Nonidet P-40) or NETN buffer containing phosphatase and protease inhibitors as described above, with the addition of 25 u/ml Benzonase nuclease (Sigma-Aldrich). Recombinant Tpd52/TPD52 proteins (5–10 μg/100 μl GSH-agarose or Ni-NTA agarose) were incubated with cell lysates as indicated. Purified ATM proteins were incubated with SK-BR-3 cell lysates prepared from non-irradiated cells or from cells irradiated with 6 Gy IR, and harvested 15 min after irradiation. Matrices were washed extensively, and bound proteins eluted into SDS sample buffer.
In vitro transcription/translation
Coupled in vitro transcription/translation of TPD52 protein was performed using the TnT® Quick Coupled Transcription/Translation System (Promega) according to the manufacturer’s instructions. Briefly, 1 μg pcDNA3.1-TPD52 plasmid was mixed with TNT T7 Quick Master Mix and incubated at 30°C for 90 min. Synthesized TPD52 protein was then subjected to GST pull-down assays with GSH-agarose conjugated ATM proteins (ATM-1, ATM-2), or GST only.
Immunoprecipitation
SK-BR-3 cells were lysed in NETN lysis buffer and 5 mg total protein extract were immunoprecipitated using 2 μg ATM (Ab-3) polyclonal rabbit antibody (Calbiochem) or 2 μg normal rabbit IgG (Sigma-Aldrich). Immunoprecipitates were washed, then resuspended in SDS sample buffer, and analyzed by SDS-PAGE.
Proteasome inhibition
3T3 cells (Vector, D52-1-12, and D52-2-1) were cultured to 80–90% confluence and split into 6-well plates at 1:4 or 1:5 ratios. After 2 h incubation at 37°C, 50 μM MG132 (Sigma-Aldrich) in DMSO or equal volumes of DMSO (0.5% v/v) were added. Protein samples were extracted at the indicated time points using NETN lysis buffer and subjected to western blot analyses.
Statistical analysis
GraphPad Prism 4.03 (GraphPad Software) was used for graph generation and statistical analyses. Comparisons of olive tail moment and the kinetics of γH2AX foci formation between vector- and HA-TPD52 transfected cells, cell survival fractions, as well as fold changes in protein levels were made using 2-tailed, unpaired Student t test. The Mann–Whitney U test was used to compare the numbers of γH2AX foci/cell between groups.
Acknowledgments
We thank Dr Mona Shehata and Dr Laurence Cantrill for expert advice and discussions, Dr Erdahl Teber and A/Prof Jonathan Arthur (Children’s Medical Research Institute, CMRI) for bioinformatics assistance, and CMRI (Westmead, NSW, AU) for providing access to the irradiator.
This work was supported by funding from the University of Sydney Cancer Research Fund (to JB), an National Health and Medical Research Council (NHMRC) postdoctoral fellowship, and a Priority-driven Collaborative Cancer Research Scheme (PdCCRS) co-funded by Cancer Australia and Cure Cancer Australia Foundation (to YC), and by donations to the Children’s Cancer Research Unit of the Children’s Hospital at Westmead.
The Leica SP5 in the CLEM Suite at the Kids Research Institute was supported by the following grants: Cancer Institute New South Wales Research Equipment [10/REG/1-23], NHMRC [2009-02759], the Ian Potter Foundation [20100508], the Perpetual Foundation [730], Ramaciotti Foundation [3037/2010], and the Sydney Medical School Research Infrastructure Major Equipment Scheme.
Glossary
Abbreviations:
- ATM
ataxia telangiectasia mutated
- BRCT
BRCA1 C-terminal domain
- DDR
DNA damage response
- DSBs
double-strand breaks
- FHA
forkhead associated domain
- IR
ionizing radiation
- PIKK
phosphatidylinositol 3-kinase-related protein kinase family
- TPD52
Tumor protein D52
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/26146
References
- 1.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–54. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 2.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12:801–17. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 3.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–68. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 4.Bhatti S, Kozlov S, Farooqi AA, Naqi A, Lavin M, Khanna KK. ATM protein kinase: the linchpin of cellular defenses to stress. Cell Mol Life Sci. 2011;68:2977–3006. doi: 10.1007/s00018-011-0683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 6.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12:162–9. doi: 10.1016/S0959-437X(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 7.Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–7. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 8.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 9.Matsuoka S, Rotman G, Ogawa A, Shiloh Y, Tamai K, Elledge SJ. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc Natl Acad Sci U S A. 2000;97:10389–94. doi: 10.1073/pnas.190030497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutros R, Fanayan S, Shehata M, Byrne JA. The tumor protein D52 family: many pieces, many puzzles. Biochem Biophys Res Commun. 2004;325:1115–21. doi: 10.1016/j.bbrc.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 11.Sathasivam P, Bailey AM, Crossley M, Byrne JA. The role of the coiled-coil motif in interactions mediated by TPD52. Biochem Biophys Res Commun. 2001;288:56–61. doi: 10.1006/bbrc.2001.5721. [DOI] [PubMed] [Google Scholar]
- 12.Byrne JA, Mattei MG, Basset P. Definition of the tumor protein D52 (TPD52) gene family through cloning of D52 homologues in human (hD53) and mouse (mD52) Genomics. 1996;35:523–32. doi: 10.1006/geno.1996.0393. [DOI] [PubMed] [Google Scholar]
- 13.Nourse CR, Mattei MG, Gunning P, Byrne JA. Cloning of a third member of the D52 gene family indicates alternative coding sequence usage in D52-like transcripts. Biochim Biophys Acta. 1998;1443:155–68. doi: 10.1016/S0167-4781(98)00211-5. [DOI] [PubMed] [Google Scholar]
- 14.Shehata M, Weidenhofer J, Thamotharampillai K, Hardy JR, Byrne JA. Tumor protein D52 overexpression and gene amplification in cancers from a mosaic of microarrays. Crit Rev Oncog. 2008;14:33–55. doi: 10.1615/CritRevOncog.v14.i1.30. [DOI] [PubMed] [Google Scholar]
- 15.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet. 2006;38:421–30. doi: 10.1038/ng1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–26. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 17.Shehata M, Bièche I, Boutros R, Weidenhofer J, Fanayan S, Spalding L, Zeps N, Byth K, Bright RK, Lidereau R, et al. Nonredundant functions for tumor protein D52-like proteins support specific targeting of TPD52. Clin Cancer Res. 2008;14:5050–60. doi: 10.1158/1078-0432.CCR-07-4994. [DOI] [PubMed] [Google Scholar]
- 18.Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, Kutok J, Aster JC, Tang J, Kuefer R, et al. Defining aggressive prostate cancer using a 12-gene model. Neoplasia. 2006;8:59–68. doi: 10.1593/neo.05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Byrne JA, Maleki S, Hardy JR, Gloss BS, Murali R, Scurry JP, Fanayan S, Emmanuel C, Hacker NF, Sutherland RL, et al. MAL2 and tumor protein D52 (TPD52) are frequently overexpressed in ovarian carcinoma, but differentially associated with histological subtype and patient outcome. BMC Cancer. 2010;10:497. doi: 10.1186/1471-2407-10-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis JD, Payton LA, Whitford JG, Byrne JA, Smith DI, Yang L, Bright RK. Induction of tumorigenesis and metastasis by the murine orthologue of tumor protein D52. Mol Cancer Res. 2007;5:133–44. doi: 10.1158/1541-7786.MCR-06-0245. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Wang J, Pang B, Liang RX, Li S, Huang PT, Wang R, Chung LW, Zhau HE, Huang C, et al. PC-1/PrLZ contributes to malignant progression in prostate cancer. Cancer Res. 2007;67:8906–13. doi: 10.1158/0008-5472.CAN-06-4214. [DOI] [PubMed] [Google Scholar]
- 22.Zhang D, He D, Xue Y, Wang R, Wu K, Xie H, Zeng J, Wang X, Zhau HE, Chung LW, et al. PrLZ protects prostate cancer cells from apoptosis induced by androgen deprivation via the activation of Stat3/Bcl-2 pathway. Cancer Res. 2011;71:2193–202. doi: 10.1158/0008-5472.CAN-10-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sims AH, Finnon P, Miller CJ, Bouffler SD, Howell A, Scott D, Clarke RB. TPD52 and NFKB1 gene expression levels correlate with G2 chromosomal radiosensitivity in lymphocytes of women with and at risk of hereditary breast cancer. Int J Radiat Biol. 2007;83:409–20. doi: 10.1080/09553000701317366. [DOI] [PubMed] [Google Scholar]
- 24.Niu N, Qin Y, Fridley BL, Hou J, Kalari KR, Zhu M, Wu TY, Jenkins GD, Batzler A, Wang L. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–92. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–4. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasielewski M, Elstrodt F, Klijn JG, Berns EM, Schutte M. Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast Cancer Res Treat. 2006;99:97–101. doi: 10.1007/s10549-006-9186-z. [DOI] [PubMed] [Google Scholar]
- 27.MacPhail SH, Banáth JP, Yu TY, Chu EH, Lambur H, Olive PL. Expression of phosphorylated histone H2AX in cultured cell lines following exposure to X-rays. Int J Radiat Biol. 2003;79:351–8. doi: 10.1080/0955300032000093128. [DOI] [PubMed] [Google Scholar]
- 28.Boutros R, Bailey AM, Wilson SH, Byrne JA. Alternative splicing as a mechanism for regulating 14-3-3 binding: interactions between hD53 (TPD52L1) and 14-3-3 proteins. J Mol Biol. 2003;332:675–87. doi: 10.1016/S0022-2836(03)00944-6. [DOI] [PubMed] [Google Scholar]
- 29.Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van Roey K, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, et al. ELM--the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40(Database issue):D242–51. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–8. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 31.Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–59. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- 32.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 33.Shiloh Y, Lehmann AR. Maintaining integrity. Nat Cell Biol. 2004;6:923–8. doi: 10.1038/ncb1004-923. [DOI] [PubMed] [Google Scholar]
- 34.Barakat K, Gajewski M, Tuszynski JA. DNA repair inhibitors: the next major step to improve cancer therapy. Curr Top Med Chem. 2012;12:1376–90. doi: 10.2174/156802612801319070. [DOI] [PubMed] [Google Scholar]
- 35.Srinivasan A, Gold B. Small-molecule inhibitors of DNA damage-repair pathways: an approach to overcome tumor resistance to alkylating anticancer drugs. Future Med Chem. 2012;4:1093–111. doi: 10.4155/fmc.12.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed M, Rahman N. ATM and breast cancer susceptibility. Oncogene. 2006;25:5906–11. doi: 10.1038/sj.onc.1209873. [DOI] [PubMed] [Google Scholar]
- 37.Thorstenson YR, Roxas A, Kroiss R, Jenkins MA, Yu KM, Bachrich T, Muhr D, Wayne TL, Chu G, Davis RW, et al. Contributions of ATM mutations to familial breast and ovarian cancer. Cancer Res. 2003;63:3325–33. [PubMed] [Google Scholar]
- 38.Hall J. The Ataxia-telangiectasia mutated gene and breast cancer: gene expression profiles and sequence variants. Cancer Lett. 2005;227:105–14. doi: 10.1016/j.canlet.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 39.Angèle S, Treilleux I, Brémond A, Tanière P, Hall J. Altered expression of DNA double-strand break detection and repair proteins in breast carcinomas. Histopathology. 2003;43:347–53. doi: 10.1046/j.1365-2559.2003.01713.x. [DOI] [PubMed] [Google Scholar]
- 40.Tribius S, Pidel A, Casper D. ATM protein expression correlates with radioresistance in primary glioblastoma cells in culture. Int J Radiat Oncol Biol Phys. 2001;50:511–23. doi: 10.1016/S0360-3016(01)01489-4. [DOI] [PubMed] [Google Scholar]
- 41.Kang B, Guo RF, Tan XH, Zhao M, Tang ZB, Lu YY. Expression status of ataxia-telangiectasia-mutated gene correlated with prognosis in advanced gastric cancer. Mutat Res. 2008;638:17–25. doi: 10.1016/j.mrfmmm.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Grabsch H, Dattani M, Barker L, Maughan N, Maude K, Hansen O, Gabbert HE, Quirke P, Mueller W. Expression of DNA double-strand break repair proteins ATM and BRCA1 predicts survival in colorectal cancer. Clin Cancer Res. 2006;12:1494–500. doi: 10.1158/1078-0432.CCR-05-2105. [DOI] [PubMed] [Google Scholar]
- 43.Prokopcova J, Kleibl Z, Banwell CM, Pohlreich P. The role of ATM in breast cancer development. Breast Cancer Res Treat. 2007;104:121–8. doi: 10.1007/s10549-006-9406-6. [DOI] [PubMed] [Google Scholar]
- 44.Vo QN, Kim WJ, Cvitanovic L, Boudreau DA, Ginzinger DG, Brown KD. The ATM gene is a target for epigenetic silencing in locally advanced breast cancer. Oncogene. 2004;23:9432–7. doi: 10.1038/sj.onc.1208092. [DOI] [PubMed] [Google Scholar]
- 45.Flanagan JM, Munoz-Alegre M, Henderson S, Tang T, Sun P, Johnson N, Fletcher O, Dos Santos Silva I, Peto J, Boshoff C, et al. Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum Mol Genet. 2009;18:1332–42. doi: 10.1093/hmg/ddp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balleine RL, Fejzo MS, Sathasivam P, Basset P, Clarke CL, Byrne JA. The hD52 (TPD52) gene is a candidate target gene for events resulting in increased 8q21 copy number in human breast carcinoma. Genes Chromosomes Cancer. 2000;29:48–57. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1005>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 47.Scanlan MJ, Gout I, Gordon CM, Williamson B, Stockert E, Gure AO, Jäger D, Chen YT, Mackay A, O’Hare MJ, et al. Humoral immunity to human breast cancer: antigen definition and quantitative analysis of mRNA expression. Cancer Immun. 2001;1:4. [PubMed] [Google Scholar]
- 48.Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–75. [PubMed] [Google Scholar]
- 49.Petrova DT, Asif AR, Armstrong VW, Dimova I, Toshev S, Yaramov N, Oellerich M, Toncheva D. Expression of chloride intracellular channel protein 1 (CLIC1) and tumor protein D52 (TPD52) as potential biomarkers for colorectal cancer. Clin Biochem. 2008;41:1224–36. doi: 10.1016/j.clinbiochem.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 50.Tiacci E, Orvietani PL, Bigerna B, Pucciarini A, Corthals GL, Pettirossi V, Martelli MP, Liso A, Benedetti R, Pacini R, et al. Tumor protein D52 (TPD52): a novel B-cell/plasma-cell molecule with unique expression pattern and Ca(2+)-dependent association with annexin VI. Blood. 2005;105:2812–20. doi: 10.1182/blood-2004-07-2630. [DOI] [PubMed] [Google Scholar]
- 51.Barbaric D, Byth K, Dalla-Pozza L, Byrne JA. Expression of tumor protein D52-like genes in childhood leukemia at diagnosis: clinical and sample considerations. Leuk Res. 2006;30:1355–63. doi: 10.1016/j.leukres.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Kang H, Wilson CS, Harvey RC, Chen IM, Murphy MH, Atlas SR, Bedrick EJ, Devidas M, Carroll AJ, Robinson BW, et al. Gene expression profiles predictive of outcome and age in infant acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2012;119:1872–81. doi: 10.1182/blood-2011-10-382861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crugliano T, Quaresima B, Gaspari M, Faniello MC, Romeo F, Baudi F, Cuda G, Costanzo F, Venuta S. Specific changes in the proteomic pattern produced by the BRCA1-Ser1841Asn missense mutation. Int J Biochem Cell Biol. 2007;39:220–6. doi: 10.1016/j.biocel.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Quaresima B, Faniello MC, Baudi F, Crugliano T, Cuda G, Costanzo F, Venuta S. In vitro analysis of genomic instability triggered by BRCA1 missense mutations. Hum Mutat. 2006;27:715. doi: 10.1002/humu.9427. [DOI] [PubMed] [Google Scholar]
- 55.Lim DS, Kirsch DG, Canman CE, Ahn JH, Ziv Y, Newman LS, Darnell RB, Shiloh Y, Kastan MB. ATM binds to beta-adaptin in cytoplasmic vesicles. Proc Natl Acad Sci U S A. 1998;95:10146–51. doi: 10.1073/pnas.95.17.10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–17. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanu N, Behrens A. ATMIN defines an NBS1-independent pathway of ATM signalling. EMBO J. 2007;26:2933–41. doi: 10.1038/sj.emboj.7601733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takai H, Xie Y, de Lange T, Pavletich NP. Tel2 structure and function in the Hsp90-dependent maturation of mTOR and ATR complexes. Genes Dev. 2010;24:2019–30. doi: 10.1101/gad.1956410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hurov KE, Cotta-Ramusino C, Elledge SJ. A genetic screen identifies the Triple t complex required for DNA damage signaling and ATM and ATR stability. Genes Dev. 2010;24:1939–50. doi: 10.1101/gad.1934210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Izumi N, Yamashita A, Hirano H, Ohno S. Heat shock protein 90 regulates phosphatidylinositol 3-kinase-related protein kinase family proteins together with the RUVBL1/2 and Tel2-containing co-factor complex. Cancer Sci. 2012;103:50–7. doi: 10.1111/j.1349-7006.2011.02112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Izumi N, Yamashita A, Iwamatsu A, Kurata R, Nakamura H, Saari B, Hirano H, Anderson P, Ohno S. AAA+ proteins RUVBL1 and RUVBL2 coordinate PIKK activity and function in nonsense-mediated mRNA decay. Sci Signal. 2010;3:ra27. doi: 10.1126/scisignal.2000468. [DOI] [PubMed] [Google Scholar]
- 62.Fang Z, Kozlov S, McKay MJ, Woods R, Birrell G, Sprung CN, Murrell DF, Wangoo K, Teng L, Kearsley JH, et al. Low levels of ATM in breast cancer patients with clinical radiosensitivity. Genome Integr. 2010;1:9. doi: 10.1186/2041-9414-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson CT, Muzik H, Turhan AG, Zamò A, O’Connor MJ, Bebb DG, Lees-Miller SP. ATM deficiency sensitizes mantle cell lymphoma cells to poly(ADP-ribose) polymerase-1 inhibitors. Mol Cancer Ther. 2010;9:347–57. doi: 10.1158/1535-7163.MCT-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Weston VJ, Oldreive CE, Skowronska A, Oscier DG, Pratt G, Dyer MJ, Smith G, Powell JE, Rudzki Z, Kearns P, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116:4578–87. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 65.Thomas DD, Martin CL, Weng N, Byrne JA, Groblewski GE. Tumor protein D52 expression and Ca2+-dependent phosphorylation modulates lysosomal membrane protein trafficking to the plasma membrane. Am J Physiol Cell Physiol. 2010;298:C725–39. doi: 10.1152/ajpcell.00455.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Byrne JA, Nourse CR, Basset P, Gunning P. Identification of homo- and heteromeric interactions between members of the breast carcinoma-associated D52 protein family using the yeast two-hybrid system. Oncogene. 1998;16:873–81. doi: 10.1038/sj.onc.1201604. [DOI] [PubMed] [Google Scholar]
- 67.Khanna KK, Keating KE, Kozlov S, Scott S, Gatei M, Hobson K, Taya Y, Gabrielli B, Chan D, Lees-Miller SP, et al. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat Genet. 1998;20:398–400. doi: 10.1038/3882. [DOI] [PubMed] [Google Scholar]
- 68.Ye R, Goodarzi AA, Kurz EU, Saito S, Higashimoto Y, Lavin MF, Appella E, Anderson CW, Lees-Miller SP. The isoflavonoids genistein and quercetin activate different stress signaling pathways as shown by analysis of site-specific phosphorylation of ATM, p53 and histone H2AX. DNA Repair (Amst) 2004;3:235–44. doi: 10.1016/j.dnarep.2003.10.014. [DOI] [PubMed] [Google Scholar]
- 69.Frangioni JV, Neel BG. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal Biochem. 1993;210:179–87. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]