Abstract
In the yeast Kluyveromyces lactis, the telomeres are composed of perfect 25-bp repeats copied from a 30-nucleotide RNA template defined by 5-nucleotide terminal repeats. A genetic dissection of the K. lactis telomere was performed by using mutant telomerase RNA (TER1) alleles to incorporate mutated telomeric repeats. This analysis has shown that each telomeric repeat contains several functional regions, some of which may physically overlap. Mutations in the terminal repeats of the template RNA typically lead to telomere shortening, as do mutations in the right side of the Rap1p binding site. Mutations in the left half of the Rap1p binding site, however, lead to the immediate formation of long telomeres. When mutated, the region immediately 3′ of the Rap1p binding site on the TG-rich strand of the telomere leads to telomeres that are initially short but eventually undergo extreme telomere elongation. Mutations between this region and the 3′ terminal repeat cause elevated recombination despite the presence of telomeres of nearly wild-type length. Mutants with highly elongated telomeres were further characterized and exhibit signs of telomere capping defects, including elevated levels of subtelomeric recombination and the formation of extrachromosomal and single-stranded telomeric DNA. Lengthening caused by some Rap1 binding site mutations can be suppressed by high-copy-number RAP1. Mutated telomeric repeats from a delayed elongation mutant are shown to be defective at regulating telomere length in cells with wild-type telomerase, indicating that the telomeric repeats are defective at telomere length regulation.
Telomeres are complexes composed of proteins and repetitive, typically GT-rich, DNA sequences that cap the ends of linear chromosomes, protecting them from fusion and nucleolytic attack. Defects in telomere maintenance and function have been implicated in human disease and aging. The telomeric tract contains both double-stranded and single-stranded components, the lengths of which are regulated by a number of proteins. In yeast, the tracts of duplex telomeric repeats are typically a few hundred base pairs in length, with a region of single-stranded DNA at the telomeric tip that is seen in a cell-cycle-specific manner (65). It is at the single-stranded telomeric tip that the addition or loss of new telomeric repeats occurs. Telomeric DNA is normally maintained by telomerase, a ribonucleoprotein containing the template for new telomeric repeat synthesis in its RNA component (reviewed in references 3 and 24).
The length of the duplex telomeric tract is regulated via the effects of several proteins (reviewed in references 14 and 51), the most important of which appears to be Rap1p, an essential gene product that is also involved in transcriptional regulation (reviewed in references 42 and 47). Rap1p binds directly to double-stranded telomeric repeats in both the budding yeasts Saccharomyces cerevisiae and Kluyveromyces lactis (26, 31), where it recruits other proteins such as Rif1p and Rif2p (23, 66). The number of Rap1p molecules bound to the telomere also appear to be “counted,” such that new telomeric repeats are not added when the appropriate number of Rap1p molecules are bound at the telomere (34). The tethering of additional Rap1p C termini to the telomere leads to the shortening of the telomeric repeat tract, indicating that the number of Rap1p molecules is counted as part of the mechanism that regulates telomere length. Rap1p is also known to be able to bend DNA, which may be important to its telomere function (18, 43, 63). Cdc13p, on the other hand, is a single-strand telomeric repeat binding protein that binds telomeres with high affinity and interacts with other proteins that act on telomeric ends (6, 13, 30, 45, 46). Cdc13p positively regulates telomere length through interactions with Est1p and the telomerase complex. It also serves a negative regulatory role through interactions with Stn1p. Est1p may function as a cell cycle regulator of telomerase activity (56). Cdc13p, Stn1p, and Ten1p are essential in S. cerevisiae, where they play crucial roles in forming a protein complex that provides a protective capping function at telomeres (19, 20).
Although telomeric repeats in most organisms are 5 to 8 nucleotides long, some yeast species have telomeric repeats that are much longer (10, 37). In K. lactis, the telomeric DNA is composed of perfect copies of a 25-nucleotide repeat. The long, homogeneous repeats make this species ideal for genetic dissection of the functions occurring at the telomere. In order to synthesize the 25-nucleotide K. lactis repeat, a 30-nucleotide template region of the Ter1 RNA is copied. The first 5 nucleotides (terminal repeat positions 1 to 5) are identical to the last 5 nucleotides (terminal repeat positions 26 to 30), which provides telomerase with the ability to base pair with the telomeric DNA for new telomeric repeat synthesis (16). A mutation made in the telomerase template will typically be copied into the telomeric repeat during DNA synthesis (38, 48, 52, 68). It is thus possible to examine the effects of mutating specific nucleotides of the telomeric repeat. This type of analysis is complicated by the fact that both the telomerase template RNA and the telomeric DNA (and thus potentially the complex of telomere-associated proteins) are altered; determining which is the cause of any defects seen can be difficult. However, experiments with S. cerevisiae (49), as well as with K. lactis (38, 40), have shown that altering the telomeric sequence can have dramatic consequences for the cell in terms of telomere length regulation.
In K. lactis, point mutations in the TER1 template that produce telomeric repeats with the corresponding change have been shown to cause several different telomere length phenotypes. However, the extent of the domains producing these phenotypes has not yet been determined. Mutations in part of the Rap1p binding site cause telomere elongation and in some instances telomere fusions. Other mutations lead to telomere shortening and elevated levels of subtelomeric recombination (39). Two classes of mutations, to our knowledge, have been reported only for K. lactis: delayed telomere elongation (38), in which telomeres in the mutant cells initially shorten but eventually become very long, and phenotypically silent mutations (41, 50). Both the immediate and delayed telomere elongation phenotypes were recessive, indicating that the elongation is due not to an overactive telomerase but rather to telomeric defects. In this study, point mutations made at each position of the K. lactis TER1 template were used to determine the extents of various telomeric domains. Telomerase template mutations leading to long, uncapped telomeres, were examined in more detail. Some of these mutants are known to have telomeres that are defective at binding to Rap1p, while others are outside the Rap1p binding site. The stability of these long telomeres and their response to high copy number of RAP1 were investigated.
MATERIALS AND METHODS
Mutagenesis and strain construction.
The plasmid pTER-BX:UA (38), containing TER1 and URA3, was used to construct all ter1 mutations described in these here. Mutations were made via one of two methods. Some mutations were constructed by using a single-stranded template and a mutagenic oligonucleotide (28). The majority of the mutations were made by using the Quik-change mutagenesis kit as described by the manufacturer (Stratagene, La Jolla, Calif.).
The strains used in this study are derivatives of haploid K. lactis 7B520 (ura3 his3 trp1) (67). Previously reported ter1 template mutants (38-41) were constructed directly in the 7B520 strain. New mutants were constructed primarily in dhBcl+His1, a His+ revertant of 7B520 containing the phenotypically silent TER1-Bcl(7C) allele, although ter1-13G, ter1-21C, and ter1-28A were constructed in the 7B520 strain, using the plasmid loop-in, loop-out method as described previously (38). The dhBcl+His1 strain is used as the control and is designated as such in Fig. 1 and as TER1-Bcl in subsequent figures. Transformation was carried out by using a modified lithium acetate electroporation procedure identical to that used for S. cerevisiae (62). When two telomerase molecules were present in the same cell, they existed as “heteroalleles” and were the product of a pTER-BX:UA plasmid loop-in.
FIG. 1.
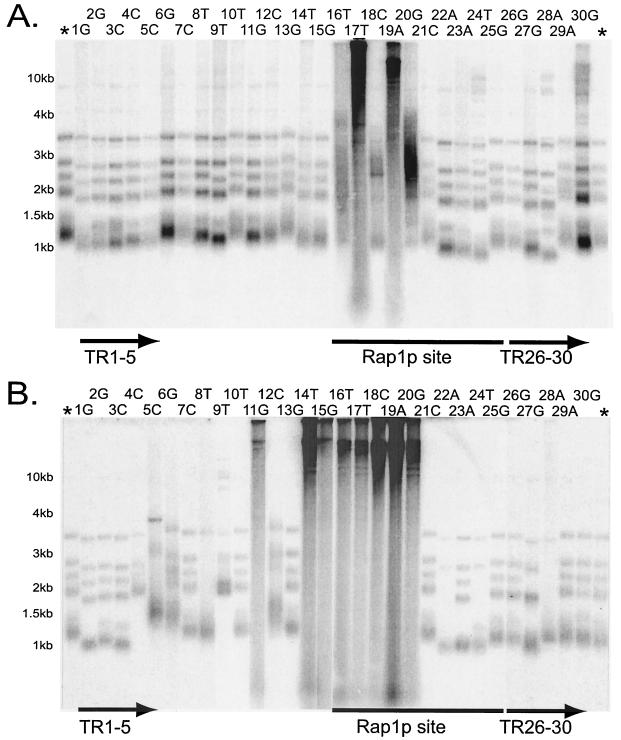
Most point mutations in the TER1 template cause changes in telomere length that worsen after passaging. (A) Each of the 30 positions in the K. lactis telomerase template was mutated independently to create a collection of point mutants. A Southern blot of EcoRI-digested genomic DNA from newly created mutants, probed with a telomeric oligonucleotide, is shown. *, parental TER1 control. Labeled lanes indicate the template position that is mutated in the strain shown in that lane. Strains are shown in order, with the leftmost lane being position 1 and the rightmost lane being position 30. The Rap1p binding site is underlined; the terminal repeats are underlined with arrows. Size markers are shown. (B) Southern blot of the telomere length at the 50th streak, except for the strain with a mutation at position 3, for which the 29th streak is shown.
In order to be confident that any phenotypes observed were due to the introduction of the intended mutation, the experimental design included redundancy at two steps. For the majority of the mutations discussed in this work, two independent plasmids containing the desired mutation were utilized. Two independently derived strains containing the mutation were then isolated from each plasmid. Thus, four strains containing each mutation were isolated. Typically, two of the four clones were then used for long-term passaging, which was carried out by serial restreaking to single colonies on rich medium (yeast extract-peptone-dextrose [YPD]plates) every 3 to 4 days. Each streak is estimated to represent 20 to 25 cell divisions.
ter1 rad52Δ double mutants were constructed by mating the ter1-14T, ter1-16T, and ter1-17T mutants with TAQ-STU-19 (ura3 his3 rad52Δ) containing one telomere with a subtelomeric URA3 gene (39). Diploids were selected on medium lacking histidine and uracil and passaged on YPD medium for 14 serial restreaks to allow the telomeres to shorten. The diploids were then streaked on sporulation plates, and the resulting tetrads were dissected. DNA was prepared from haploid ter1 rad52 cells restreaked onto YPD after tetrad dissection. As the haploid segregants are not fully isogenic, TER1 RAD52 strains from the same mating were used as controls.
Construction of Bcl- and Kpn-STU telomeres.
The Bcl- and Kpn-STU (subtelomeric URA3) telomeres were made by mutating a derivative of the pAK25 plasmid (39) as described previously (61). Briefly, by using a variation on the mutagenesis procedure described by Kunkel et al. (28) one to three oligonucleotides were used to mutate each of the 11 telomeric repeats in a cloned telomere. An EcoRI-SacII fragment that contains a URA3 gene inserted into the subtelomeric region and 11.5 mutated or wild-type telomeric repeats was transformed into yeast strains as described above. Genomic DNA was prepared and digested with XhoI to release the STU telomere as a fragment separable from all other telomeres in the cells. Fragments were visualized on 2 or 4% agarose gels.
Subtelomeric recombination assay.
An EcoRI-SacII fragment from the Bcl-STU plasmid that contains a URA3 gene inserted into the subtelomeric region and 11.5 Bcl telomeric repeats was transformed into K. lactis cells. Construction of the Bcl-STU telomere was previously described (61). As described in Results, in some cases this fragment was transformed directly into the ter1 strains to be assayed. In other cases, a rare URA3-tagged telomere was first recovered in the UHA strain (ura3 his3 ade2 rad52Δ). This STU transformant was then mated with the ter1 strain to be assayed; the diploids were then sporulated, and the tetrads were dissected. The resulting strains were screened for the presence of RAD52, the URA3-marked telomere, and the presence of the mutated telomerase. In the Bcl-STU strain used for mating, there appears to be a small subtelomeric duplication at the Bcl-STU telomere. As this might affect levels of subtelomeric recombination and because the strains produced through mating were not fully isogenic, TER1 strains resulting from the same diploids were used as controls. Rates of URA3 loss by gene conversion were determined by using plates containing 5-fluoro-orotic acid as previously described (39). Rates of gene conversion on a per-cell basis were estimated by the method of the median (54). Standard errors were calculated as the standard deviation divided by the square root of n, the number of repetitions.
Hybridizations and quantification.
Restriction enzyme-digested yeast genomic DNA was run on 0.8 or 1% agarose gels, stained with ethidium bromide, and Southern blotted onto Hybond N+ membranes (Amersham Biosciences, Piscataway, N.J.). Hybridizations were carried out in Na2HPO4 and sodium dodecyl sulfate (SDS) (9). Telomeric oligonucleotides used were the G-strand oligonucleotide Klac1-25 (ACGGATTTGATTAGGTATGTGGTGT) and its reverse complement C-strand oligonucleotide (ACACCACATACCTAATCAAATCCGT). Telomeric hybridizations and washes were performed at 45 or 55°C. Washes were done with 200 mM Na2HPO4 and 2% SDS. The subtelomeric probe was an ∼600-bp EcoRI-SacI fragment from the plasmid pAK25. Hybridizations and washes were performed at 60 or 65°C; washes were done with 100 mM Na2HPO4 and 2% SDS.
The quantification of broken DNA was determined by PhosphorImager analysis. Calculations were based on 12 telomeres per cell, each with a maximum of 20 telomeric repeats in the TER1-7C(Bcl) strain. Each telomere would therefore be less than 500 bp long. The total telomeric signal in the uncut TER1-7C(Bcl) sample was then compared with the telomeric signal seen in the broken DNA running at 450 to 550 bp in the mutant strains. Comparable amounts of DNA were used in the samples; the relative intensities of telomeric signal between the uncut TER1-7C(Bcl) DNA and the broken pieces of mutant DNA running at approximately 450 to 550 bp indicated the relative numbers of molecules in each strain. The numbers calculated were likely to be an underestimate of the number of broken telomeric molecules in the mutant cells. Because the probe was to a wild-type sequence, there might have been somewhat weaker hybridization intensity in the mutant telomeric repeats.
In-gel hybridization.
In-gel hybridizations were performed by using a variation of a previously described method (12). Thin (5 to 7 mm thick) low-percent-agarose gels (0.7%) were run at 25 or 30 V for 16 h. The gel was stained with ethidium bromide for photographing and then soaked in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for at least 30 min. The gel was then dried for 4 to 5 min per side until it was thicker than plastic wrap but thinner than Whatman 3MM paper. The gels were then placed in 10× SSC and hybridized with the desired probe at 23°C overnight. The gels were washed two or three times for at least 1.5 h per wash in 0.25× SSC.
Transformation and growth of high-copy-number RAP1 strains.
Mutant ter1 strains were transformed with either plasmid pCXJ3 (8) or pCXJ3+RAP, which contains the K. lactis RAP1 gene (a gift from A. Krauskopf). The transformants were selected for on medium lacking uracil. The strains were subsequently grown on plates containing 125 μg of G418 per ml in order to select for higher plasmid copy number. Previous results indicated that at this concentration, there are more than 100 copies of the plasmid per cell (8). In order to maintain selection for the plasmid, these strains were not grown in overnight culture before DNA extraction. Instead, scoops of cells were taken from the plates and prepared by the normal protocol.
Cloning of long telomeres.
Total yeast genomic DNA was treated with T4 DNA polymerase and deoxynucleoside triphosphates to fill in any overhangs and make the ends blunt. A SacI linker was ligated onto the blunt ends, and the construct was cut with SacI. The T4 DNA polymerase-treated genomic DNA was then ligated into pBluescript that had been linearized by SacI digestion. Plasmids extracted from the Escherichia coli transformants were screened by hybridizing Southern blots of digested plasmid DNA with telomeric oligonucleotides. Those hybridizing with the telomeric probe were sequenced.
RESULTS
Mutational analysis of the K. lactis TER1 template.
The long telomeric repeat in K. lactis provides an excellent system for genetic dissection of a telomere. By making point mutations in the template of the K. lactis telomerase RNA (TER1), it was possible to generate yeast cells having specific sequence changes in the newly added telomeric repeats of every chromosome end in the cell. At least one of the three possible point mutations was made at each of the 30 positions in the telomerase template. The mutants were named according to the position of the base change within the template, such that the change that would cause position 1 of the newly synthesized telomeric strand to be changed from a T to a G is called ter1-1G. Previously published mutations, renamed to be more consistent with the nomenclature used in this paper, will be described by both names, such as ter1-18C(Bsi).
Two clones containing each mutation, typically independently constructed (see Materials and Methods), were isolated, and their telomere lengths were examined every 10 serial restreaks (200 to 250 generations) for 60 restreaks. Figure 1 shows a Southern blot of telomeric restriction fragments from a representative mutation at each template position. Results from two time points (the 1st and 50th restreaks) are shown in Fig. 1A and B, respectively; the results from all mutants constructed are summarized in Table 1. Telomere lengths in the mutants varied widely, and in many mutants the telomere length changed significantly over time. In addition, some mutants exhibited elevated levels of subtelomeric recombination as judged from loss of one or more telomeric EcoRI restriction fragments, a phenomenon that was previously shown to occur in several ter1 mutants that had short telomeres (39) and was found to involve subtelomeric gene conversion. The subtelomeric sequences of at least 11 of the 12 telomeres in K. lactis 7B520 share substantial homology; thus, subtelomeric recombination commonly led to the loss of subtelomeric polymorphisms. Increased levels of subtelomeric recombination are therefore a good indicator of compromised telomeric function. Abnormal colony phenotypes, which arise from aberrant cellular morphologies and cell division defects (38, 53), were observed only in strains having very long telomeres. In these strains, the colonies had a rough appearance and irregular edges compared to the smooth colonies observed in wild-type strains.
TABLE 1.
Specific TER1 template mutations lead to telomeric length and capping defects
| Position | Basea
|
Telomeric length phenotypeb
|
Recombinationc
|
|||
|---|---|---|---|---|---|---|
| Wild type | Mutation | Early | Late | Band loss | STU assay | |
| 1 | T | G | Short | Short | + | |
| 2 | T | G | Short | Short | ND | |
| 3 | T | C | Short | Short | ND | |
| 4 | G | C | Slightly short | Wild type | ++ | Elevated |
| 5 | A | C | Slightly short | Slightly long | ++ | |
| 6 | T | G | Wild type | Long | ND | |
| 7 | T | C | Wild type | Wild type | ND | Wild-type |
| 8 | A | T | Wild type | Slightly long | ++ | |
| 9 | G | T | Short | Wild type | ++ | |
| 10 | G | T | Slightly long | Wild type | ND | |
| 11 | T | G | Wild type | Very long | ++ | |
| 12 | A | C | Wild type | Slightly long | + | |
| 13 | T | G | Wild type | Wild type | ND | |
| 14 | G | T | Short | Very long | ? | Elevated |
| 15 | T | G | Short | Very long | ++ | |
| 16 | G | T | Long | Very long | ? | |
| A | Long | Very long | ? | |||
| 17 | G | T | Very long | Very long | ? | |
| A | Wild type | Wild type | ND | |||
| 18 | T | C | Long | Very long | ? | Elevated |
| 19 | G | A | Very long | Very long | ? | Elevated |
| C | Very long | Very long | ? | |||
| 20 | T | G | Very long | Very long | ? | |
| C | Wild type | Slightly short | ND | |||
| 21 | A | C | Slightly long | Slightly long | ND | |
| 22 | C | A | Short | Short | + | |
| 23 | G | A | Short | Slightly short | + | |
| 24 | G | T | Short | Short | ++ | |
| C | Short | Short | ++ | |||
| 25 | A | C | Slightly long | Slightly long | ND | |
| 26 | T | G | Wild type | Wild type | ND | |
| 27 | T | G | Short | Short | ++ | |
| A | Short | Slightly long | + | |||
| 28 | T | A | Short | Short | ++ | |
| G | Wild type | Wild type | ND | |||
| C | Short | Short | ++ | Elevated | ||
| 29 | G | A | Slightly long | Slightly long | ND | |
| 30 | A | G | Wild type | Slightly long | ND | |
The specific base changes made at each position are shown. The changes are shown in relation to the wild-type sequence of the telomeric G-strand.
The telomere length phenotypes caused by each group of mutations at two time points are shown. Early indicates phenotypes seen in the first 30 restreaks; late indicates phenotypes seen in the restreaks 40 to 60. Length data are derived from two to four clones of each mutant.
Recombination defects are shown when known. Band loss indicates loss of telomeric restriction fragments. +, loss of one or two restriction fragments; ++, indicates loss of three or more restriction fragments; ND, band loss was not detected; ?, due to extreme telomere length, it was not possible to determine whether telomeric bands were lost. When subtelomeric recombination assays have been done, results are shown.
Interestingly, only 5 of the 38 mutations examined (7C, 13G, 17A, 26G, and 28A) showed no evidence of telomere dysfunction; at all time points examined, these five mutants had telomeres of wild-type length and exhibited no evidence of subtelomeric recombination as judged by loss of telomeric restriction fragments (Table 1). The strains were examined at many time points, so some subtle phenotypes may not be visible at the times shown. Mutants with mutations at two of those positions (17 and 28), however, had wild-type-length telomeres when one point mutation was made but long telomeres when a different mutation was made at the same site. This indicates that at most three positions in the TER1 template have no base-specific function. It remains possible that mutations at those positions (positions 7, 13, and 26) might cause telomeric defects if different base substitutions were examined. One of the phenotypically silent strains, the TER1-7C(Bcl) mutant, has been used extensively to examine nontemplate TER1 mutations and telomeric turnover (41, 50, 60).
Mutations in the 5-nucleotide terminal repeats of the TER1 template often led to telomere shortening and subtelomeric recombination. At early time points, 9 of the 13 strains with mutations in these regions had short telomeres; 7 of these had also lost EcoRI telomeric fragments by the 50th streak. Many mutations in this region have been shown to cause aberrant telomerase translocation events, leading to the synthesis of telomeric repeats that were sizes other than the normal 25 bp in length (62). This may, at least in part, underlie the telomere length defects seen in these mutants. As discussed below, however, it is possible that the 5-bp telomeric sequence encoded by the terminal repeats of the TER1 template has additional telomeric functions.
Mutations in template positions 4 to 9 led to telomeres that were initially wild type in length or slightly short (Fig. 1A and Table 1). However, after 50 streaks, five of the six mutants had lost many of their subtelomeric restriction fragments. In the ter1-4C and ter1-9T mutants shown, the major group of telomeric EcoRI fragments, which are normally 1 kb in length and comprised of 7 of the 12 telomeres in the cell, was lost. This suggested that these mutants experienced high levels of subtelomeric recombination. Consistent with this, the ter1-4C mutant was found to have a subtelomeric gene conversion rate 100 times higher than that of control cells (Table 2). The telomeres in the ter1-5C and ter1-6G mutants had elongated significantly and, along with the ter1-8T mutant, had a smeary appearance. Although it is unclear whether this region encodes a discrete telomeric function, these results clearly show that it does have an important role.
TABLE 2.
Subtelomeric recombination is elevated in strains with long telomeres
| Strain background | Gene conversion ratea
|
Method of STU introductionb | nc | ||
|---|---|---|---|---|---|
| Median | SE | Relative | |||
| TER1-7C(Bcl) | 2.7 × 10−6 | 3.5 × 10−6 | 1 | Transformation | 24 |
| ter1-18C(Bsi) | 4.2 × 10−5 | 2.4 × 10−4 | 15.4 | Transformation | 21 |
| ter1-14T | 7.8 × 10−5 | 3.0 × 10−5 | 28.6 | Mating | 15 |
| ter1-19A(Acc) | 1.2 × 10−4 | 3.1 × 10−3 | 45.3 | Transformation or mating | 19 |
| ter1-4C | 2.8 × 10−4 | 4.6 × 10−5 | 103.5 | Transformation | 12 |
| TER1 [ter1-19A(Acc) background] | 1.3 × 10−5 | 2.5 × 10−6 | 4.8 | Mating | 14 |
Recombination rates were measured in three strains with ter1 template mutations causing telomere elongation and one strain (ter1-4C mutant) that showed evidence of elevated subtelomeric recombination as seen by Southern blotting (Fig. 1). The median and standard error were calculated for each. The median relative to the TER1-7C(Bcl) control rate is shown.
Method used to introduce the marked telomere (STU).
Number of assays performed with each mutant.
It was previously shown that some mutations within the Rap1p binding site found in the K. lactis telomeric repeat led to immediately elongated telomeres and a significant reduction in Rap1p binding (26, 38, 40). However, two other mutations in the Rap1p binding site led to telomere shortening. The more extensive mutational analysis presented here confirmed that mutations in the left and right halves of the Rap1p binding site have opposite effects on telomere length. At least one of the mutations tested at each position in the left side of the consensus Rap1p binding site (positions 16 to 20) led to immediate or gradual telomere elongation (Table 1 and Fig. 1), consistent with Rap1p's role as a negative regulator of telomere length (reviewed in reference 35). Although mutations in the right side of the putative Rap1p binding site (positions 21 to 25) were also expected to cause defective Rap1p binding, four of the six strains with point mutations in this region had telomeres that remained short throughout the entire time course examined. Unlike yeast with mutations in the left side of the Rap1p binding site, they displayed no signs of telomere elongation (40) (Fig. 1 and Table 1). These data clearly show that template positions 21 to 25 encode a positive telomeric function.
Mutants containing ter1-17T, ter1-19C, and ter1-19A(Acc) showed immediate severe telomere elongation, in which many of the telomeres ran at limit mobility in cells from the first streak, the earliest time point that could be examined (38, 40) (Fig. 1A). The telomeric hybridization patterns of these mutants also had a smeary appearance that extended the length of the lane; this point is discussed in more detail below. These mutants will henceforth be described as immediate elongation mutants. In contrast, the ter1-16T, ter1-16A, ter1-18C(Bsi), and ter1-20G mutants initially had a more modest lengthening phenotype (41) (Fig, 1A); by the 5th or 10th streak, however, their telomere lengths approached or equaled those exhibited by the immediate elongation mutants (Fig. 1B and data not shown). These mutants will be referred to as gradual elongation mutants. It is likely that the immediate and gradual elongation mutants represent the same defect, such as a decrease in Rap1p binding, and vary only by the severity of the resulting phenotype (26). Telomeric fragments from the ter1-16T and ter1-17T mutants were cloned and shown to contain only the expected point mutation (data not shown), suggesting that aberrant telomerase translocation does not typically occur in these mutants.
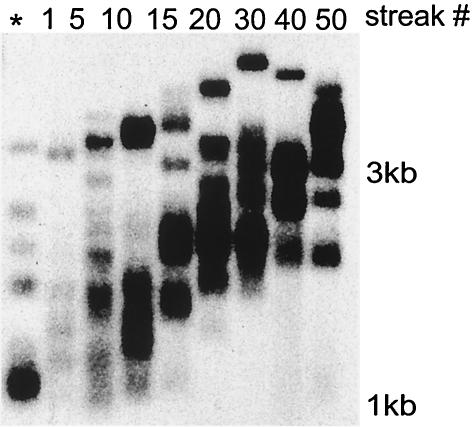
In cases where heteroallelic K. lactis cells (plasmid loop-in; see Materials and Methods) containing both a wild-type TER1 allele and a mutated ter1 allele were examined, the mutated allele was typically recessive (reference 38 and data not shown). This would generally be expected, since repeats added by the wild-type telomerase could presumably provide the necessary telomeric functions. It was found, however, that mutations causing immediate telomere elongation can lead to gradual telomere elongation even in the presence of wild-type TER1. In heteroallelic strains containing both ter1-19A(Acc) and TER1, the telomeres gradually lengthened (Fig. 2). After the cells were passaged for 50 streaks, the telomeres were typically several times longer than the 250- to 500-bp telomeres seen in wild-type cells. Elongation was very slow, however, compared to the much faster elongation typically observed in strains containing only the mutant allele (Fig. 1A). The ter1-19A(Acc)/TER1 heteroallelic strain lacked the long smear of telomeric DNA and the abnormal colony phenotype characteristic of the ter1-19A(Acc) mutant (Fig. 2). The semidominant telomere elongation is likely due to the incoropration of ter1-19A(Acc)-templated repeats that are not counted as normal telomeric repeats, while the wild-type repeats that are present provide partial capping function.
FIG. 2.
The telomere elongation phenotype of the ter1-19A(Acc) mutant is not completely recessive. A heteroallelic strain containing both TER1 and ter1-19A(Acc) was grown for 50 restreaks, and DNA samples were periodically isolated. Shown is a telomeric probe of a Southern blot of EcoRI-digested genomic DNA from several time points. DNA from the TER1-7C(Bcl) strain was used as a control (*). Size markers are shown.
Several of the mutants with changes in the positions adjacent to the left side of the Rap1p binding site [ter1-11G, ter1-14T, ter1-15G, ter1-10A/12C(Bgl), and ter1-13C/14C(Kpn)] led to delayed telomere elongation (reference 38 and this work). This unusual phenotype was characterized by telomeres that initially shortened but eventually became extremely long (Fig. 1). Some of the mutations led to elongation within 150 generations (5 streaks) while others took more than 1,000 generations (50 streaks) to elongate. Although the telomeres became long only after repeated passaging, the elongation itself was often abrupt, frequently occurring within only a few streaks. In the ter1-14T mutant, elongation occurred between the 5th and 10th streaks, similar to the timing of elongation previously observed in the ter1-13C/14C(Kpn) mutant (38). The ter1-11G and ter1-15T mutants, on the other hand, exhibited a smeary hybridization pattern after 30 streaks but did not become extremely long until after 50 streaks. Similarly, the previously reported ter1-10A/12C(Bgl) mutant also elongated only after extensive passaging (38). Once the telomeres did elongate, however, they remained very long through repeated passaging. The smeary hybridization pattern and slightly elongated telomeres in the ter1-12C mutant, which initially had short telomeres, indicate that it may have also undergone extreme delayed telomere elongation had it been passaged further. These results confirm that in addition to the Rap1p binding site, there is another discrete region of the K. lactis telomeric repeat that serves a crucial role in proper telomere length regulation.
Rap1p overexpression suppresses some ter1 lengthening alleles.
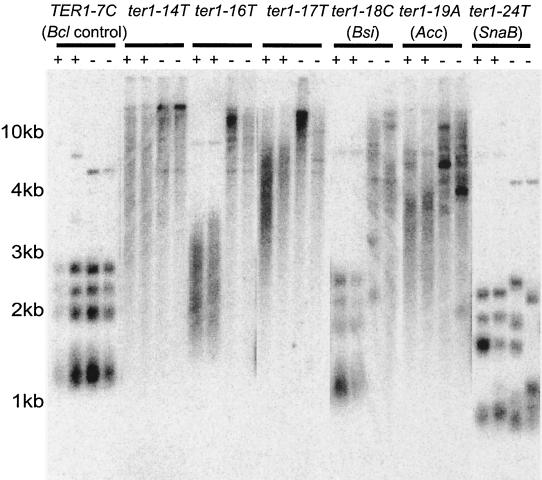
It was previously shown that telomeric repeats containing the ter1-19A(Acc) or ter1-18C(Bsi) mutations had weakened Rap1p binding in vitro (26). The repeat templated by ter1-19A(Acc) had the most severe defect of the repeats examined, while ter1-18C(Bsi) led to a less severe binding defect. In stark contrast, the telomeric repeats made by two delayed elongation mutants, ter1-10A/12C(Bgl) and ter1-13C/14C(Kpn), which have mutations outside the Rap1p binding site appeared to bind Rap1p at least as well as wild-type repeats (26). It was therefore predicted that telomere elongation caused by weakened Rap1p binding could be suppressed by the presence of more Rap1p in the cells. K. lactis RAP1 was introduced on a high-copy-number plasmid into cells of the three types of telomere elongation mutants. In addition, cells containing either TER1-7C(Bcl) or ter1-24C(SnaB) were also transformed. TER1-7C(Bcl) has no telomere length defect and thus served as a control, while the ter1-24C(SnaB)-templated repeats contain a mutation on the right side of the Rap1p binding site and lead to telomere shortening. Transformants were streaked on plates containing 125 μg of G418 per ml to select for high plasmid copy number; this drug concentration was shown to select for K. lactis cells with more than 100 copies of the parental plasmid per cell (8).
Telomere length was examined by Southern blotting at the 1st, 3rd, and 10th streaks; results for the 10th streak are shown in Fig. 3). In strains containing ter1-16T and ter1-18C(Bsi), which had a gradual lengthening phenotype, the telomere elongation was strongly suppressed by the introduction of the RAP1-containing plasmid. In the ter1-18C(Bsi) mutant, the telomeres became essentially wild type in length within 10 streaks, while in the ter1-16T mutant, which had initially elongated more rapidly, the telomeres shortened significantly although they remained somewhat longer than wild-type telomeres. Conversely, the ter1-17T and ter1-19A(Acc) immediate elongation mutants showed little suppression of telomere lengthening when in the presence of the high-copy-number RAP1 plasmid. Although the amount of telomeric hybridization signal seen at limit mobility in the Southern blots was reduced, the majority of the telomeres remained very long, as seen with both telomeric and subtelomeric probes (data not shown and Fig. 3). The RAP1 plasmid had little or no effect on the telomere length in the ter1-24C(SnaB) mutant or the TER1-7C(Bcl) strain. There was also little effect seen in the ter1-14Tdelayed elongation mutant, consistent with the idea that the primary defect in delayed elongation mutants was not the inability of the telomeric repeats to bind to Rap1p. These results support the idea that the long telomeres produced by mutations in the left side of the Rap1p binding site are caused by a defect in Rap1p binding. Therefore, the disruption of a function other than Rap1p binding is responsible for the short telomeres caused by mutations on the right side of the Rap1p binding site.
FIG. 3.
High-copy-number RAP1 suppresses telomere elongation in some ter1 template mutants. Strains with mutated TER1 alleles were transformed with plasmids with or without the gene encoding Rap1p. DNA was prepared from the strains after 10 serial streaks on 125 μg of G418 per ml, and the blot, probed with a subtelomeric probe, is shown. For each mutant line, two independent transformants are shown for the high-copy-number RAP1 (+) and vector-only control (−).
Delayed elongation mutant repeats fail to regulate telomere length in TER1 cells.
The unusual delayed elongation phenotype seen in strains with certain ter1 alleles was thought to be due the inability of the mutant telomeric repeats to perform the function(s) required of repeats at the base of the telomere (38). If this was the case, introducing the mutant repeats at the base of the telomere would be expected to cause elongation of that telomere even in the presence of wild-type TER1. It is possible to mutate every repeat of a telomere in vitro and then replace one telomere in a yeast strain with the totally mutated telomere (61), and this experiment was performed with two types of total telomere mutants. Telomeres composed entirely of either Bcl repeats or Kpn repeats were created. Their names refer to the one- or two-base substitutions in the repeats; Bcl repeats created a BclI restriction site and were the type of repeat templated by the phenotypically silent allele TER1-7C(Bcl), while Kpn repeats created a KpnI restriction site and were of the type predicted to be synthesized by the delayed elongation mutant ter1-13C/14C(Kpn). These constructs will henceforth be referred to as Bcl-STU or Kpn-STU (subtelomeric URA3) telomeres.
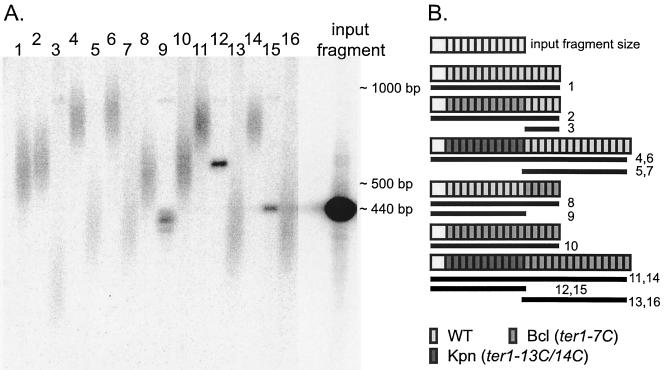
Digestion with XhoI cleaves a site next to the URA3 gene in the marked telomere to yield a telomeric restriction fragment that can be separated from all other XhoI telomeric restriction fragments. This allowed the lengths of the marked telomeres to be readily examined. While a typical wild-type K. lactis telomere was composed of 10 to 20 telomeric repeats, the STU construct contained only 11.5 telomeric repeats (Fig. 4A, input fragment). Therefore, when a wild-type STU or Bcl-STU was introduced into strains with either wild-type or TER1-7C(Bcl) telomerase alleles, it was not unexpected that a small number of telomeric repeats were added by the endogenous telomerase. This yielded a telomeric fragment of the size expected for a wild-type telomere, which was somewhat larger than the input fragment (Fig. 4A, compare lanes 1, 2, 8, and 10 to the input fragment length). Introduction of a wild-type STU telomere into a TER1-7C(Bcl) strain followed by cleavage with XhoI and BclI released the wild-type STU telomere that was introduced. As seen in Fig. 4A, lane 9, the fragment released was somewhat shorter than the input fragment, likely due to replacement of a small number of wild-type repeats with Bcl repeats via the normal process of telomeric turnover. Conversely, when a Bcl-STU was introduced into a wild-type TER1 strain, digestion with BclI cleaved the STU telomere and released a small telomeric fragment composed of the wild-type telomeric repeats added to the tip of the Bcl-STU (Fig. 4A, lane 3). In contrast to the results observed when the Bcl-STU and wild-type STU were used, when a telomere composed entirely of Kpn repeats was introduced into either a wild-type strain or the TER1-7C(Bcl) strain, a full array of 10 to 20 wild-type repeats was added onto the Kpn-STU fragment. The telomeric fragment became significantly longer than the input fragment (Fig. 4A, lanes 4, 6, 11, and 14 [compare with input fragment]) and was elongated to a greater extent than either the wild-type or Bcl-STU telomere construct (Fig. 4A, compare lanes 4, 6, 11, and 14 with lanes 1, 2, 8, and 10). When the lengths of wild-type TER1- or TER1-7C(Bcl)-templated telomeric tracts added to the Kpn-STU telomere were examined by KpnI digestion, they were found to be approximately as long as the input fragment (Fig. 4A, lanes 5, 7, 13, and 16). The input fragment, which resulted from cleavage at a subtelomeric restriction site, contains ∼120 bp of subtelomeric sequence, making it longer than just 11.5 telomeric repeats. This indicates that the telomeric tracts added to the Kpn-STU telomere contained at least 12 telomeric repeats. The size of the Kpn repeat-containing region of the Kpn-STU telomere was examined by XhoI-BclI digestion. In one clone it had become longer than the input fragment (Fig. 4A, lane 12), while the fragment from the other clone was the same size as the original input fragment (Fig. 4A, lane 15). Possible reasons for the difference are addressed in Discussion. These results indicate that the mutant repeats do not perform the basal functions required for normal telomere length regulation, despite their ability to bind Rap1p very well.
FIG. 4.
Basal telomeric repeats play a role in telomere length regulation. (A) Wild-type-, Kpn-, and Bcl-STU constructs the length of the input fragment, each containing 11.5 telomeric repeats, were transformed into TER1 and TER1-7C(Bcl) strains. The URA3-marked STU telomere was separated from the other cellular telomeres by digestion with XhoI, which specifically cleaves the STU telomere. The other 11 telomeres run at sizes well above 1 kb and are not shown. A telomeric probe was used. Lanes 1, 2, 8, and 10, full-length wild-type- or Bcl-STUs in wild-type or TER1-7c(Bcl) backgrounds (shown diagrammatically in (panel B). Lane 3, digestion with XhoI plus BclI to release the newly added wild-type repeats. Lanes 4, 6, 11, and 14, total length of the telomere in the presence of the Kpn-STU. Lanes 5, 7, 13, and 16, digestion with XhoI plus KpnI to release the newly added wild-type or Bcl repeats. Lane 9, digestion with XhoI plus BclI to release the wild-type-STU input fragment. Lanes 12 and 15, digestion with XhoI plus BclI to release the Kpn-STU. (B) Schematic diagram of the fragments examined in panel A. The input fragment is shown in white. In all other telomeres, the white box indicates the ∼120-bp subtelomeric region (not shown to scale). WT, wild type.
Subtelomeric recombination is elevated in mutants with highly elongated telomeres.
Because increased subtelomeric recombination has been shown to associate with the telomere dysfunction caused by short telomeres (39), it was of interest to see if it also occurred in long telomere mutants. The large size and smeary appearance of telomeric restriction fragments in long telomere mutants, however, made it infeasible to detect subtelomeric gene conversion as telomeric band loss in Southern blots. We instead assayed subtelomeric gene conversion by using a telomere tagged with a subtelomeric URA3 insert (a STU telomere). Rates of URA3 loss via subtelomeric gene conversion could be determined by plating the cells on medium containing 5-fluoro-orotic acid (39). Representative delayed, gradual, and immediate elongation mutants [ter1-14T, ter1-18C(Bsi), and ter1-19A(Acc), respectively] were assayed by this method. A strain containing the phenotypically silent TER1-7C(Bcl) was used as a control. A Bcl-STU telomere was first transformed into the TER1-7C(Bcl), ter1-18C(Bsi), and ter1-19A(Acc) strains; transformants in which a Bcl-STU telomere had replaced a single native telomere were chosen for further study. In other experiments, the Bcl-STU telomere was transformed into a strain of the opposite mating type and mated into the ter1-14T and ter1-19A(Acc) strains (see Materials and Methods), and the spores resulting from the diploid were screened to find the progeny that contained the URA3 gene and the mutated telomerase allele. ter1-14T cells were grown for several streaks to permit elongation of telomeres before assays were performed. TER1 strains arising from the same diploids were also examined to confirm that there were no effects due to the outcrossing required to make the strain.
The mutant strains with long telomeres were observed to have rates of subtelomeric recombination that were 15- to 45-fold higher than the rate of 2.7 × 10−6 observed in the TER1-Bcl control (Table 2). TER1-7C(Bcl) cells were previously shown to have the same subtelomeric recombination rate as TER1 cells, although the rate that we obtained here was approximately twofold lower than that observed previously (39). A somewhat elevated rate [∼5-fold above TER1-7C(Bcl) level and ∼2-fold above previously published TER1 level] was observed in the TER1 strains that resulted from the mating that produced the ter1-14T and ter1-19A(Acc) stains used in this experiment. This may have been due to the presence of long telomeres containing mutated repeats that were acquired from the TER1/ter1-14T or TER1/ter1-19A(Acc) diploids. However, this difference is quite small when compared with the 28- and 45-fold increases in recombination, respectively, observed in the mutants produced from the same diploids. Recombination rates also appear to correlate with the severity of the telomeric defect in mutants with very long telomeres; the recombination rate for the more mild ter1-18C(Bsi) mutant was lower than that for the ter1-19A(Acc) mutant. The ter1-14T mutant exhibited an intermediate rate. Experiments done with ter1-14T mutants at an earlier time point, likely prior to significant telomere elongation, showed an elevated recombination rate that was slightly lower than that seen at the later time point (data not shown). A very high recombination rate was also seen in the ter1-4C mutant, which does not have a severe telomere length defect. Therefore, it is possible that a capping defect caused by mutant telomeric repeats, rather than telomere length per se, led to elevated levels of subtelomeric recombination. However, it is clear that severe telomere elongation mutants of each type examined have defects in telomere protection that led to elevated levels of subtelomeric recombination.
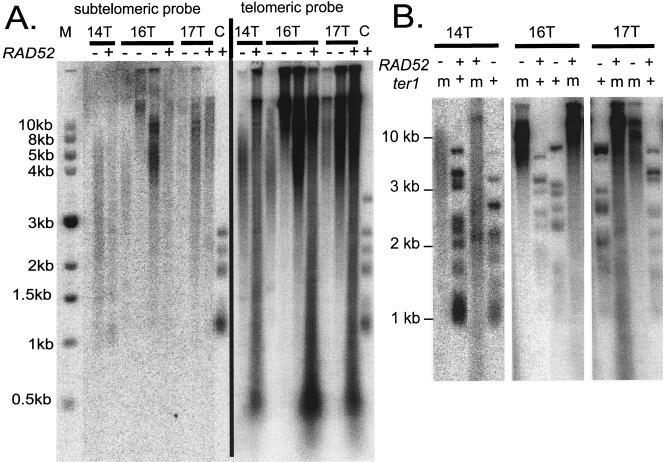
RAD52 is not required for the formation of very long telomeres.
In the absence of telomerase, yeast cells can maintain telomeres by recombinational mechanisms (7, 32, 36, 58, 59). Therefore, it was of interest to know whether recombination played a role in the formation of the very long telomeres seen in strains containing certain TER1 template mutations. Strains containing ter1-14T, ter1-16T, and ter1-17T, representing the delayed, gradual, and immediate lengthening classes, respectively, were mated with a RAD52 deletion strain in order to make ter1 rad52Δ double mutants. Diploids were made which contained both normal-length and elongated telomeres from the two parental strains. The cells were passaged for 14 streaks to allow the telomeres to shorten. Once the long telomeres had appreciably shortened and all of the telomeres were of comparable lengths, the cells were sporulated. The telomeres in the diploid strains did not shorten quickly, although the smear of DNA seen in the mutant strains was significantly reduced (data not shown). This was consistent with the idea that the telomeres in the diploid cells were capped and generally shortened gradually over time by normal telomeric turnover, as previously described (27, 53). The progeny haploid strains were examined by Southern blotting to determine whether they had wild-type RAD52 or the rad52Δ allele and to examine the lengths of the telomeres (Fig. 5A). For each of the three mutations examined, strains that contained long telomeres and lacked RAD52 were isolated; both RAD52 and the telomere elongation phenotype segregated meiotically 2:2, as expected for single genes. Because the telomeres in the diploid were not extremely long and lacked the smeary appearance of the mutants and because TER1 haploid strains created from the same sporulation event had telomeres that were neither severely elongated nor smeary (Fig. 5B), the long telomeres seen in the mutants were clearly formed after sporulation. One minor difference was seen in the ter1-14T mutant. Although the telomeres had lengthened considerably, those in the rad52Δ ter1-14T strain were not quite as long as those in the RAD52 ter1-14T strain, even after 10 restreaks (Fig. 5A). Because only one rad52Δ ter1-14T clone was examined, it is possible that the difference is due to strain variability. These results indicated that while RAD52-dependent recombination could be partially responsible the formation of long telomeres in the delayed elongation mutant, it was not required for the telomere elongation seen in any of the mutants examined.
FIG. 5.
RAD52 is not required for extreme telomere elongation. (A) EcoRI-digested genomic DNAs from ter1-14T, ter1-16T, and ter1-17T mutants and a wild-type control (lane C) was hybridized with a subtelomeric probe (left panel). After the membrane was stripped to remove the subtelomeric probe, the filter was rehybridized to the telomeric probe (right panel). −, rad52 strains; +, RAD52 strains. Telomere lengths are most clearly visualized with the subtelomeric probe. Size markers are shown. DNA was extracted after five serial restreaks, except for the ter1-14T mutant, where the yeast was restreaked 10 times. Lanes 3, 6, and 7 are underloaded relative to other lanes from the same background. (B) To confirm that the telomeres had elongated, telomere lengths from ter1 rad52, ter1 RAD52, TER1 rad52, and TER1 RAD52 spores, each derived from the same diploid, are shown for each mutant. Maintenance of the elongated telomeres was dependent on the presence of the mutated telomerase allele (m).
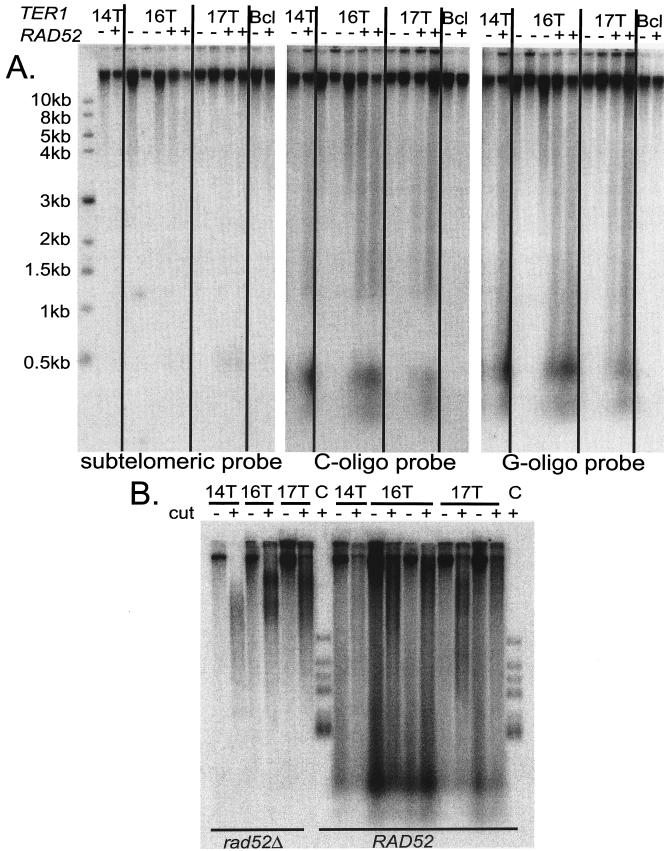
Extrachromosomal DNA is produced in strains with extreme telomere elongation in a RAD52-dependent manner.
One unusual phenotype of K. lactis ter1 mutants with very long telomeres was that all of them contained a smear of telomeric DNA extending from the top of the blot (greater than 20 kb) to the bottom (less than 200 bp) (Fig. 1). Although all telomeric signal below 0.7 kb would be expected to result from extrachromosomal telomeric DNA, as the size of the subtelomeric sequence and a wild-type telomere is 0.7 kb, the contents of the entire smear were unclear. To further characterize this DNA, Southern blots of uncut yeast genomic DNA were serially probed with a subtelomeric probe and probes specific to each telomeric strand (Fig. 6A). Because the DNA was uncut, chromosomal DNA was expected to run at limit mobility. In samples from both wild-type and mutant cells, essentially all of the visible subtelomeric signal was seen at limit mobility. This indicates that the subtelomeric DNA was not generally degraded or broken during preparation. When the gel was subsequently reprobed with the telomeric probes, a different pattern was seen. With either strand-specific telomeric probe, the majority of the signal was also seen at limit mobility. However, in the mutants with long telomeres, 10 to 25% of the signal seen with telomeric probes to either strand was present as molecules smaller than 3 kb. Included in this is the large peak of hybridization signal migrating at positions smaller than approximately 0.5 kb. This peak was estimated to represent ∼2 to 10 molecules per mutant cell (see Materials and Methods). In contrast, in the phenotypically silent TER1-Bcl control only 3% of the telomeric DNA was below 3 kb in length. These results indicate that in the mutants examined, a significant percentage of the telomeric DNA is extrachromosomal. However, it was not possible to determine how much of the telomeric signal above 3 kb in the mutants was also extrachromosomal. The large peak of hybridization signal migrating at positions smaller than ∼0.5 kb ran slightly lower when visualized with the C-strand telomeric probe. The gap in telomeric signal seen in the blot probed with the C-oligonucleotide is not reproducible.
FIG. 6.
A significant amount of the telomeric DNA is present as extrachromosomal pieces in strains with very long telomeres. (A) Uncut genomic DNAs from ter1-14T, ter1-16T, and ter1-17T mutants and the phenotypically wild-type TER1-7C(Bcl) control were blotted and serially probed with a subtelomeric probe, a C-strand telomeric oligonucleotide probe, and a G-strand telomeric oligonucleotide probe. − and +, absence and presence of RAD52 in the strain, respectively. Size markers are shown. (B) Genomic DNA that is either cut with EcoRI (+) or uncut (−) is shown for each mutant in the absence or presence of RAD52. A C-strand telomeric oligonucleotide was used. Lanes C, DNA from the wild-type parental 7B520 strain.
Although cells with mutated TER1 alleles could elongate in the absence of RAD52, the character of the telomeric DNA present in the cell was different in ter1 rad52Δ double mutants. The long broken smear of telomeric DNA seen in the long telomere mutants was significantly reduced in the rad52Δ strains (Fig. 6B), and the material below 3 kb in length was essentially eliminated (Fig. 6 and unpublished data). These data indicate that recombination was involved in the formation of most of the broken DNA.
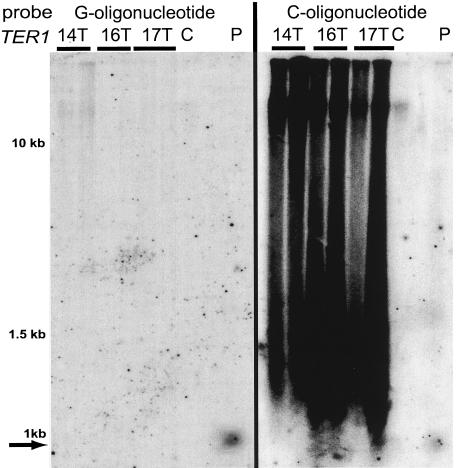
Long telomere mutants have increased amounts of single-stranded telomeric DNA.
In S. cerevisiae, extensive 3′ single-stranded overhangs can be generated at double-strand breaks (DSBs) and uncapped telomeres (17, 55). In order to see if the broken DNA in ter1 mutants with long telomeres contained single stranded telomeric DNA, nondenaturing in-gel hybridizations were done (12) (Fig. 7). When probing was with the G-strand oligonucleotide, which hybridizes with the C-rich telomeric DNA, little signal was seen. In contrast, when the C-strand telomeric oligonucleotide was used, the lane showed a smear of strong signal. In the wild-type TER1 control, only a trace amount of hybridization signal was visible at the top of the gel. Because the DNA examined was uncut, only the very large pieces running at limit mobility were likely to be G-strand overhangs attached to the chromosome. Therefore, a majority of the single-stranded telomeric DNA in the long telomere mutants was extrachromosomal. The lack of telomeric hybridization signal seen with the G-strand telomeric oligonucleotide in the in-gel hybridization indicates that the signal seen with this probe under denaturing conditions (as shown in Fig. 6) was double stranded. The telomeric signal in the in-gel hybridization migrated between limit mobility and the double-stranded 1-kb size marker. The diminished signal below ∼1.2 kb seen with the C-strand probe (Fig. 7) does not necessarily indicate an absence of DNA fragments with single-stranded telomeric DNA in this part of the gel. DNA smaller than about that size is often simply lost during the in-gel drying and hybridization steps. Other experiments indeed have shown that a substantial amount of small single-stranded G-strand telomeric DNA is present in ter1-16T cells (S. Natarajan, C. Groff-Vindman, and M. McEachern, unpublished data). When the in-gel hybridization was done with a subtelomeric probe derived from sequence within 700 bp of the base of the telomeres, no signal was seen (data not shown), indicating that the single-stranded character did not extend appreciably into the subtelomeric region of the chromosome. The smear of single-stranded DNA was considerably diminished in mutants with RAD52 deleted (Fig. 6B). Because RAD52 would not be expected to be required for the formation of 3′ tails, the reduced level of single-stranded telomeric DNA could be explained by the decrease in the amount of extrachromosomal molecules in the cells. The single-stranded telomeric DNA appeared to only be partially sensitive to digestion with ExoI (data not shown). This suggests that it may not exist entirely as 3′ overhangs.
FIG. 7.
Mutants with long telomeres show large amounts of single-stranded telomeric G-strand DNA. Uncut genomic DNAs from ter1-14T, ter1-16T, and ter1-17T mutants and an equivalent amount of the phenotypically wild-type TER1-7C(Bcl) control (lane C) were run on a 0.7% gel. Two samples from each mutant were examined. An in-gel hybridization was used to visualize the single-stranded DNA. Lane P, ∼500-bp fragment from a plasmid that has telomeric sequence (arrow). The denatured plasmid control does not run at the expected position for a 500-bp sequence. The positions of double-stranded DNA size markers are shown.
DISCUSSION
The K. lactis telomeric repeat contains several functional domains.
Mutagenesis of the entire K. lactis telomerase template was used to incorporate mutations into the telomeric repeats. This analysis has revealed that the telomeric repeat contains multiple functional regions. One is a 5-bp region of the repeat that is templated by two regions in the TER1 template, the terminal repeats, which are 5 nucleotides in length and mark the ends of the template. These regions are critical for proper translocation, the process by which telomerase dissociates and realigns with the template to make new telomeric repeats. In work presented elsewhere, many mutations in this region were shown to cause aberrant translocation events, leading to the synthesis of telomeric repeats that are longer or shorter than the 25-bp repeat synthesized by wild-type telomerase (62). The elimination of specific base-pairing interactions between the telomere and the telomerase template, which disrupts normal translocation, is likely to be largely responsible for the telomere shortening often seen when mutations are made in the TER1 terminal repeats. However, some data suggest that telomeric nucleotides copied from the terminal repeats of the TER1 template may serve another function. The short-telomere phenotype seen in both the ter1-3C and ter1-28C point mutants could have been entirely due to the inability of telomerase to translocate properly, as both alleles lead to the synthesis of abnormally sized telomeric repeats. However, a double point mutant that was altered at positions 3 and 28, the ter1-3C/28C mutant, contained identical mutations in both TER1 template terminal repeats and synthesized 25-bp repeats with the expected single-base substitution. The telomere length, however, remained quite short (data not shown), indicating that the sequence copied into the telomere from the terminal repeats may play an important role in telomere length regulation. Additional experiments will be needed to separate the effects of altered telomerase function from the effects of disrupted telomere function in these mutants.
The Rap1p binding site, the most highly conserved feature of the telomeric repeat among many related yeasts (10, 37), was a predicted functional domain within the telomeric repeat, but the analysis of this region was not as simple as might be expected. Rap1 has two distinct DNA binding domains, and the two halves of the Rap1 binding site can contribute unequally to overall affinity (25, 57). However, from alignment of the K. lactis Rap1p binding site with the S. cerevisiae site, it can be inferred that ter1 mutations leading to telomere elongation also disrupt specific required contacts between the telomeric repeat and Rap1p. Some mutations would be predicted to disrupt specific molecular interactions, while other mutations may preserve the necessary contact sites. This may explain why, for example, the ter1-17T allele leads to telomere elongation, whereas ter1-17A does not. It is possible that changing position 17 to a thymine disrupts a specific contact between Rap1p and its site, while a change to adenine does not. Interestingly, the ter1-16A/17A double mutant had normal-length telomeres, even after passaging for 130 streaks, when introduced into cells by the plasmid loop-in, loop-out method, but the mutation caused telomere elongation when introduced into a ter1Δ strain with short telomeres (38). In the latter protocol, basal wild-type repeats that could influence telomere length are largely gone before the mutant telomerase is expressed. As all of the mutants described in this study were made by the plasmid loop-in, loop-out method, it is possible that some of the mutants with telomeres of wild-type length, such as the ter1-17A mutant, could also have a “cryptic” telomere elongation phenotype that would be detectable only if few or no wild-type repeats remained in the telomeres containing the mutant repeats.
In S. cerevisiae, Rap1p has been shown to be a negative regulator of telomere length (34). The results presented here provide further evidence that K. lactis Rap1p is also a negative regulator of telomere length. Mutations in the left side of the Rap1p binding site lead to telomere elongation. The presence of RAP1 on a high-copy-number plasmid was able to suppress telomere elongation caused by some of these mutations. This strongly argues that the immediate and gradual telomere elongation phenotypes were due to defects in Rap1p binding and also indicates that the presence of additional copies of Rap1p leads to an increase in cellular Rap1p levels. An increased level of Rap1p might suppress the long telomere phenotype by forcing the mutant repeats with reduced Rap1p binding affinity to bind Rap1p. The lack of effect on telomere length in wild-type cells may indicate that the K. lactis telomeres are normally fully saturated with Rap1p. The presence of high-copy-number RAP1 in S. cerevisiae can actually lead to modest telomere elongation through a mechanism that is unclear (11). The extent of suppression of telomere lengthening by high-copy-number RAP1 is consistent with the degree of disruption of Rap1p binding observed in vitro for the ter1-19A(Acc) and ter1-18C(Bsi) mutations (26). It is also interesting that the presence of high-copy-number RAP1 in wild-type cells did not lead to an obvious growth defect. This is in contrast to the results seen for S. cerevisiae, where high levels of Rap1p are toxic (5, 15). Whether this difference is due to differences in Rap1p overexpression levels in the two species or to some other reason is unknown.
Mutations in the right side of the Rap1p binding site were typically found to cause telomere shortening; none caused the extreme telomere elongation seen when the left side of the site was mutated. This surprising result could be explained by proposing that this region encodes two functional domains: a Rap1p binding site and also a site, which partially overlaps with the Rap1p site, that positively modulates telomere length. It has been shown that the telomeric tip has special sequence requirements independent of Rap1p binding (22), and one likely explanation is that this region is a binding site for either Cdc13p or Est1p, proteins that bind single-stranded telomeric DNA and activate telomerase or recruit it to the telomeric tip (reviewed in reference 14). It is also possible that the mutations affect telomerase translocation (62). Although some mutations in the right side of the Rap1p binding site would also be expected to disrupt Rap1p binding, the inability to recruit telomerase to the telomere would limit or prevent the extensive telomere lengthening typically seen with mutations in the Rap1p binding site. Therefore, the identification of K. lactis CDC13 and EST1, and the examination of their DNA binding specificity, will be of great interest.
Another functional domain of the K. lactis telomeric repeat is the region adjacent to the left side of the Rap1p binding site. Mutations in at least four different positions cause the telomeres to become short initially but eventually lead to severe telomere elongation. The initial shortening of the telomeres in these mutants strongly suggests that this region of the telomere serves a positive function in telomere length regulation, which is disrupted by the mutations. It is not clear if this effect is due to a defect separate from that leading to telomere elongation or if the two phenotypes are different manifestations of the same defect. The delay in formation of the long telomeres appears to be due to the requirement for telomeric turnover to replace some percentage of the telomere with mutant repeats before telomere length regulation is critically disrupted (38). For this reason, the role that the internal telomeric repeats play in length regulation was examined. Mutated telomeric repeats containing the mutation specified by ter1-13C/14C(Kpn) were introduced into the base of a single telomere in a TER1 background. This led to the addition of a full array of 10 to 20 wild-type repeats onto the 11.5 mutant repeats. This indicates that the Kpn telomeric repeats are not capable of fulfilling the length regulation functions of basal telomeric repeats. It remains unclear whether telomeric repeats at different positions within the basal part of the telomere have equal effects on length regulation. Interestingly, in three of the six Kpn-STU mutants in a TER1-7C(Bcl) background examined (Fig. 4 and data not shown), the Kpn-STU fragment recovered after BclI cleavage was longer than the input fragment. This elongation could not be telomerase mediated, as any repeats added by TER1-7C(Bcl) would contain the BclI restriction site and thus be cleaved. It is therefore likely that the three Kpn-STU telomeres acquired wild-type repeats by recombining with the wild-type base of another telomere in the cell. Although only a small number of clones were examined, these results suggest that in addition to a defect in telomere length regulation, the mutant repeats of the Kpn-STU may also have a capping defect that makes them prone to recombination.
The presence of the high-copy-number RAP1 plasmid had little or no effect on the delayed elongation mutant examined, indicating that the primary defect of this mutant is not in Rap1p binding. There are, however, several possible explanations for the telomere elongation phenotype seen in the delayed elongation mutants. First, it is possible that the mutations increase the strength of binding for a positive regulatory protein. A second possibility is that the mutations may directly alter interactions between Rap1p and the telomeric DNA. Rap1p is known to cause a bend in the telomeric DNA when bound. The nucleotides adjacent to the Rap1p binding site may provide additional contacts with Rap1p that affect DNA bending and thus the telomeric chromatin structure. Third, mutations in the delayed elongation domain of the telomere could cause a defect in binding a protein that interacts with the telomeric DNA, perhaps only in the presence of Rap1p, which also serves a negative regulatory function. Possible candidates include Rif1p and Rif2p, which interact with the telomere via Rap1p and may be part of the mechanism by which telomeric repeats are counted by the cell (34; D. Levy and E. H. Blackburn, personal communication).
A final region of the template was defined by mutations in template positions 4 to 9. Mutations in this region led to modest changes in telomere length and unexpectedly high levels of subtelomeric recombination, as determined by loss of subtelomeric restriction fragments in Southern blots. In several of the mutants, the telomeric hybridizations developed a smeary appearance over time. Mutations leading to delayed elongation often developed a smeary appearance before elongating, indicating that these mutants might also elongate if given sufficient time. This region might therefore be a less critical component of the delayed elongation domain of the telomeric repeat. Alternatively, it could delineate another domain that contributes to proper telomere function.
Mutants with severe telomere elongation exhibit telomere capping defects.
One of the main roles of telomeres is to prevent the chromosome ends from being recognized as DSBs. Telomeres composed of defective telomeric repeats could potentially be treated similarly to DSBs. TER1 template mutants with long telomeres exhibit defects in both colony and cellular morphology, reminiscent of yeast cells arrested with unrepaired DSBs (53, 64). They are also likely to be subject to the same repair by homologous recombination and/or nonhomologous end joining that occurs at DSBs. Short telomeres in K. lactis were previously shown to have elevated levels of subtelomeric recombination (39). In this work K. lactis delayed, gradual, and immediate telomere elongation mutants were examined by recombination assays, and all three were shown to have elevated levels of subtelomeric recombination. The frequency of recombination did not correlate directly with the length of the telomeres. Indicative of this, the ter1-14T delayed elongation mutant displayed elevated rates of subtelomeric recombination at both time points examined, despite the expectation that the telomeres were very different in length. This is consistent with the idea that high rates of subtelomeric recombination result from compromised telomere capping and can occur in the presence of long or short telomeres with capping defects. Although the STU recombination assays indicate that subtelomeric recombination is elevated in the mutants, they do not address whether recombination within the telomeric repeat tracts is also elevated.
TER1 template mutants with very long telomeres also exhibit a change in the composition of telomeric DNA in the cell. Not only are the telomeres very long, but a large amount of the telomeric DNA is extrachromosomal, existing as broken fragments that run from the top of the gel to sizes of less than 100 bp. When wild-type TER1 is introduced into a ter1 mutant with long telomeres, the telomeres remain much longer than wild type. However, the telomere lengths are more sharply defined, and the smear of telomeric fragments is not present (references 26, 33, and 53 and data not shown). A similar response is seen in the TER1/ter1-19A(Acc) heteroallelic strain (Fig. 2). These data indicate that the extrachromosomal telomeric DNA is an effect of telomere uncapping that can be reversed by the presence of wild-type repeats near the telomeric termini. Interestingly, although telomere elongation does not depend on RAD52, formation of the broken DNA in the ter1-14T, ter1-16T, ter1-17T, and ter1-19A(Acc) mutants is largely RAD52 dependent (Fig. 6 and data not shown). This result strongly suggests that recombination within telomeric repeat tracts is highly elevated in the long telomere mutants. Recombination between or within the long tracts of telomeric DNA could thus result in the formation of extrachromosomal telomeric DNA molecules. These extrachromosomal fragments of telomeric DNA may potentially be subject to further degradation and telomerase-mediated telomere addition.
Highly elevated rates of recombination within the telomeric repeat tracts have been shown to occur in ter1Δ mutants (Z. Topcu and M. J. McEachern, unpublished data). Telomeric recombination may also explain why, when the long telomeres in the mutants shortened upon introduction of wild-type TER1 or high-copy-number RAP1, shortening was more rapid than the 5 bp per end per cell division that would be expected from replicative sequence loss (38). The high rate of telomere shortening could be related to the telomere rapid deletion that has been observed in S. cerevisiae (4, 29). When significant telomere shortening occurred in some of the high-copy-number RAP1 experiments described in this work, the telomeres initially shortened by as much as 4.5 kb in only three restreaks (60 to 75 generations) (data not shown); this is more than 10 times the telomere shortening rate in ter1Δ cells (38). Turnover of telomeric repeats close to the base of the telomere has previously been shown to occur in TER1 template mutants with highly elongated telomeres (38, 41). This further supports the idea that very long telomeres in the TER1 template mutants examined are highly dynamic and prone to high rates of telomeric deletion, recombination, and unregulated telomeric repeat addition by telomerase.
Normal telomere function in most eukaryotes involves the formation of small single-stranded 3′ tails at telomeric termini. In S. cerevisiae, the short single-stranded G overhangs have been shown to form in a cell-cycle-dependent manner, independently of telomerase (12, 65). One consequence of telomere uncapping can be the formation of much larger 3′ G-strand tails at the telomeric tip through the resection of the telomeric C-rich strand. Single-stranded overhangs that can extend thousands of base pairs from the telomere are found throughout the cell cycle in some cdc13 mutants (17). In the TER1 template mutants with long telomeres examined in this work, there is a substantial amount of single-stranded telomeric DNA that is composed almost entirely of the telomeric G-rich strand. Our data do not address the lengths of the single-stranded overhangs in these mutants; however, they do not appear to routinely extend into the subtelomere (data not shown). The large amount of single-strand hybridization signal seen in mutants with long telomeres is likely due to the presence of 3′ tails on both the chromosomal and extrachromosomal telomeric ends. Despite the extreme length of the duplex telomeres in the mutants examined, the single-stranded component at the telomeric tip may be somewhat regulated. The single-stranded component of the telomere may form a structure, such as a T loop (21, 44) or G-quartet structure (1, 2), that limits its size. Alternatively, if the low rate of 5′ end resectioning seen at DSBs (55) also occurs at uncapped telomeres, relatively short telomeric 3′ tails may initiate recombination events before the tails become large. Whatever the length of the single-stranded overhangs, it is clear that the formation of single-stranded DNA occurs at a high level in each type of long telomere mutant.
Acknowledgments
We thank Teresa Monteith and Brianne Ray for their help in passaging the yeast mutants and Will McRae for technical assistance. We also are grateful to Raymund Wellinger for help with the in-gel hybridization protocol and to Anat Krauskopf for the pCXJ3 and pCXJ3+RAP plasmids.
This work has been supported by grants from the American Cancer Society (RPG-GMC-99746 and RSG-02-253-01-GMC) and from the National Institutes of Health (GM61645-01).
REFERENCES
- 1.Balagurumoorthy, P., and S. K. Brahmachari. 1995. Intra- and interloop interactions in the folded G quartet structure of Oxytricha telomeric sequence. Indian J. Biochem. Biophys. 32:385-390. [PubMed] [Google Scholar]
- 2.Balagurumoorthy, P., S. K. Brahmachari, D. Mohanty, M. Bansal, and V. Sasisekharan. 1992. Hairpin and parallel quartet structures for telomeric sequences. Nucleic Acids Res. 20:4061-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackburn, E. H. 2001. Switching and signaling at the telomere. Cell 106:661-673. [DOI] [PubMed] [Google Scholar]
- 4.Bucholc, M., Y. Park, and A. J. Lustig. 2001. Intrachromatid excision of telomeric DNA as a mechanism for telomere size control in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers, A. 1996. Highly conserved toxicity of Saccharomyces cerevisiae Rap1p. Mol. Microbiol. 22:449-458. [DOI] [PubMed] [Google Scholar]
- 6.Chandra, A., T. R. Hughes, C. I. Nugent, and V. Lundblad. 2001. Cdc13 both positively and negatively regulates telomere replication. Genes Dev. 15:404-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Q., A. Ijpma, and C. W. Greider. 2001. Two survivor pathways that allow growth in the absence of telomerase are generated by distinct telomere recombination events. Mol. Cell. Biol. 21:1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, X. J. 1996. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131-136. [DOI] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohn, M., M. J. McEachern, and E. H. Blackburn. 1998. Telomeric sequence diversity within the genus Saccharomyces. Curr. Genet. 33:83-91. [DOI] [PubMed] [Google Scholar]
- 11.Conrad, M. N., J. H. Wright, A. J. Wolf, and V. A. Zakian. 1990. RAP1 protein interacts with yeast telomeres in vivo: overproduction alters telomere structure and decreases chromosome stability. Cell 63:739-750. [DOI] [PubMed] [Google Scholar]
- 12.Dionne, I., and R. J. Wellinger. 1996. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc. Natl. Acad. Sci. USA 93:13902-13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, S. K., and V. Lundblad. 1999. Est1 and Cdc13 as comediators of telomerase access. Science 286:117-120. [DOI] [PubMed] [Google Scholar]
- 14.Evans, S. K., and V. Lundblad. 2000. Positive and negative regulation of telomerase access to the telomere. J. Cell Sci. 113:3357-3364. [DOI] [PubMed] [Google Scholar]
- 15.Freeman, K., M. Gwadz, and D. Shore. 1995. Molecular and genetic analysis of the toxic effect of RAP1 overexpression in yeast. Genetics 141:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulton, T. B., and E. H. Blackburn. 1998. Identification of Kluyveromyces lactis telomerase: discontinuous synthesis along the 30-nucleotide-long templating domain. Mol. Cell. Biol. 18:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garvik, B., M. Carson, and L. Hartwell. 1995. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 15:6128-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilson, E., M. Roberge, R. Giraldo, D. Rhodes, and S. M. Gasser. 1993. Distortion of the DNA double helix by RAP1 at silencers and multiple telomeric binding sites. J. Mol. Biol. 231:293-310. [DOI] [PubMed] [Google Scholar]
- 19.Grandin, N., C. Damon, and M. Charbonneau. 2001. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20:1173-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grandin, N., S. I. Reed, and M. Charbonneau. 1997. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11:512-527. [DOI] [PubMed] [Google Scholar]
- 21.Griffith, J. D., L. Comeau, S. Rosenfield, R. M. Stansel, A. Bianchi, H. Moss, and T. deLange. 1999. Mammalian telomeres end in a large duplex loop. Cell 97:503-514. [DOI] [PubMed] [Google Scholar]
- 22.Grossi, S., A. Bianchi, P. Damay, and D. Shore. 2001. Telomere formation by Rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol. Cell. Biol. 21:8117-8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hardy, C. F., L. Sussel, and D. Shore. 1992. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6:801-814. [DOI] [PubMed] [Google Scholar]
- 24.Harrington, L. 2003. Biochemical aspects of telomerase function. Cancer Lett. 194:139-154. [DOI] [PubMed] [Google Scholar]
- 25.Konig, P., R. Giraldo, L. Chapman, and D. Rhodes. 1996. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell 85:125-136. [DOI] [PubMed] [Google Scholar]
- 26.Krauskopf, A., and E. H. Blackburn. 1996. Control of telomere growth by interactions of RAP1 with the most distal telomeric repeats. Nature 383:354-357. [DOI] [PubMed] [Google Scholar]
- 27.Krauskopf, A., and E. H. Blackburn. 1998. Rap1 protein regulates telomere turnover in yeast. Proc. Natl. Acad. Sci. USA 95:12486-12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 29.Li, B., and A. J. Lustig. 1996. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 10:1310-1326. [DOI] [PubMed] [Google Scholar]
- 30.Lin, J. J., and V. A. Zakian. 1996. The Saccharomyces CDC13 protein is a single-strand TG1-3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc. Natl. Acad. Sci. USA 93:13760-13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Longtine, M. S., N. M. Wilson, M. E. Petracek, and J. Berman. 1989. A yeast telomere binding activity binds to two related telomere sequence motifs and is indistinguishable from RAP1. Curr. Genet. 16:225-239. [DOI] [PubMed] [Google Scholar]
- 32.Lundblad, V., and E. H. Blackburn. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73:347-360. [DOI] [PubMed] [Google Scholar]
- 33.Maddar, H., N. Ratzkovsky, and A. Krauskopf. 2001. Role for telomere cap structure in meiosis. Mol. Biol. Cell 12:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marcand, S., E. Gilson, and D. Shore. 1997. A protein-counting mechanism for telomere length regulation in yeast. Science 275:986-990. [DOI] [PubMed] [Google Scholar]
- 35.Marcand, S., D. Wotton, E. Gilson, and D. Shore. 1997. Rap1p and telomere length regulation in yeast. Ciba Found. Symp. 211:76-93. [DOI] [PubMed] [Google Scholar]
- 36.McEachern, M. J., and E. H. Blackburn. 1996. Cap-prevented recombination between terminal telomeric repeat arrays (telomere CPR) maintains telomeres in Kluyveromyces lactis lacking telomerase. Genes Dev. 10:1822-1834. [DOI] [PubMed] [Google Scholar]
- 37.McEachern, M. J., and E. H. Blackburn. 1994. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc. Natl. Acad. Sci. USA 91:3453-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McEachern, M. J., and E. H. Blackburn. 1995. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature 376:403-409. [DOI] [PubMed] [Google Scholar]
- 39.McEachern, M. J., and S. Iyer. 2001. Short telomeres in yeast are highly recombinogenic. Mol. Cell 7:695-704. [DOI] [PubMed] [Google Scholar]
- 40.McEachern, M. J., S. Iyer, T. B. Fulton, and E. H. Blackburn. 2000. Telomere fusions caused by mutating the terminal region of telomeric DNA. Proc. Natl. Acad. Sci. USA 97:11409-11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McEachern, M. J., D. H. Underwood, and E. H. Blackburn. 2002. Dynamics of telomeric DNA turnover in yeast. Genetics 160:63-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morse, R. H. 2000. RAP, RAP, open up! New wrinkles for RAP1 in yeast. Trends Genet. 16:51-53. [DOI] [PubMed] [Google Scholar]
- 43.Muller, T., E. Gilson, R. Schmidt, R. Giraldo, J. Sogo, H. Gross, and S. M. Gasser. 1994. Imaging the asymmetrical DNA bend induced by repressor activator protein 1 with scanning tunneling microscopy. J. Struct. Biol. 113:1-12. [DOI] [PubMed] [Google Scholar]
- 44.Munoz Jordan, J. L., G. A. Cross, T. de Lange, and J. D. Griffith. 2001. t-loops at trypanosome telomeres. EMBO J. 20:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nugent, C. I., T. R. Hughes, N. F. Lue, and V. Lundblad. 1996. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274:249-252. [DOI] [PubMed] [Google Scholar]
- 46.Pennock, E., K. Buckley, and V. Lundblad. 2001. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104:387-396. [DOI] [PubMed] [Google Scholar]
- 47.Pina, B., J. Fernandez-Larrea, N. Garcia-Reyero, and F. Z. Idrissi. 2003. The different (sur)faces of Rap1p. 268:791-798. [DOI] [PubMed]
- 48.Prescott, J., and E. H. Blackburn. 1997. Telomerase RNA mutations in Saccharomyces cerevisiae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11:528-540. [DOI] [PubMed] [Google Scholar]
- 49.Prescott, J. C., and E. H. Blackburn. 2000. Telomerase RNA template mutations reveal sequence-specific requirements for the activation and repression of telomerase action at telomeres. Mol. Cell. Biol. 20:2941-2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy, J., T. B. Fulton, and E. H. Blackburn. 1998. Specific telomerase RNA residues distant from the template are essential for telomerase function. Genes Dev. 12:3286-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saldanha, S. N., L. G. Andrews, and T. O. Tollefsbol. 2003. Assessment of telomere length and factors that contribute to its stability. Eur. J. Biochem. 270:389-403. [DOI] [PubMed] [Google Scholar]
- 52.Singer, M. S., and D. E. Gottschling. 1994. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science 266:404-409. [DOI] [PubMed] [Google Scholar]
- 53.Smith, C. D., and E. H. Blackburn. 1999. Uncapping and deregulation of telomeres lead to detrimental cellular consequences in yeast. J. Cell Biol. 145:203-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snedecor, G., and W. Cochran. 1980. Statistical methods, 7th ed. The Iowa State University Press, Ames.
- 55.Sugawara, N., and J. E. Haber. 1992. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol. 12:563-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taggart, A. K., S. C. Teng, and V. A. Zakian. 2002. Est1p as a cell cycle-regulated activator of telomere-bound telomerase. Science 297:1023-1026. [DOI] [PubMed] [Google Scholar]
- 57.Taylor, H. O., M. O'Reilly, A. G. Leslie, and D. Rhodes. 2000. How the multifunctional yeast Rap1p discriminates between DNA target sites: a crystallographic analysis. J. Mol. Biol. 303:693-707. [DOI] [PubMed] [Google Scholar]
- 58.Teng, S. C., J. Chang, B. McCowan, and V. A. Zakian. 2000. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent, Rif-inhibited recombinational process. Mol. Cell 6:947-952. [DOI] [PubMed] [Google Scholar]
- 59.Teng, S. C., and V. A. Zakian. 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:8083-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tzfati, Y., T. B. Fulton, J. Roy, and E. H. Blackburn. 2000. Template boundary in a yeast telomerase specified by RNA structure. Science 288:863-867. [DOI] [PubMed] [Google Scholar]
- 61.Underwood, D. H., and M. J. McEachern. 2001. Totally mutant telomeres: single-step mutagenesis of tandem repeat DNA sequences. BioTechniques 30:934-935, 938. [DOI] [PubMed] [Google Scholar]
- 62.Underwood, D. H., R. P. Zinzen, and M. J. McEachern. 2004. Template requirements for telomerase translocation in Kluyveromyces lactis. Mol. Cell. Biol. 24:912-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vignais, M. L., and A. Sentenac. 1989. Asymmetric DNA bending induced by the yeast multifunctional factor TUF. J. Biol. Chem. 264:8463-8466. [PubMed] [Google Scholar]
- 64.Weinert, T. A., and L. H. Hartwell. 1993. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134:63-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wellinger, R. J., A. J. Wolf, and V. A. Zakian. 1993. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell 72:51-60. [DOI] [PubMed] [Google Scholar]
- 66.Wotton, D., and D. Shore. 1997. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11:748-760. [DOI] [PubMed] [Google Scholar]
- 67.Wray, L. V., M. M. Witte, R. C. Dickson, and M. I. Riley. 1987. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:1111-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu, G. L., J. D. Bradley, L. D. Attardi, and E. H. Blackburn. 1990. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature 344:126-132. [DOI] [PubMed] [Google Scholar]