Abstract
Like many bacteria, yeast species can form biofilms on several surfaces. Candida albicans colonizes the surfaces of catheters, prostheses, and epithelia, forming biofilms that are extremely resistant to antifungal drugs. We have used transcript profiling to investigate the specific properties of C. albicans biofilms. Biofilm and planktonic cultures produced under different conditions of nutrient flow, aerobiosis, or glucose concentration were compared by overall gene expression correlation. Correlation was much higher between biofilms than planktonic populations irrespective of the growth conditions, indicating that biofilm populations formed in different environments display very similar and specific transcript profiles. A first cluster of 325 differentially expressed genes was identified. In agreement with the overrepresentation of amino acid biosynthesis genes in this cluster, Gcn4p, a regulator of amino acid metabolism, was shown to be required for normal biofilm growth. To identify biofilm-related genes that are independent of mycelial development, we studied the transcriptome of biofilms produced by a wild-type, hypha-producing strain and a cph1/cph1 efg1/efg1 strain defective for hypha production. This analysis identified a cluster of 317 genes expressed independently of hypha formation, whereas 86 genes were dependent on mycelial development. Both sets revealed the activation of the sulfur-amino acid biosynthesis pathway as a feature of C. albicans biofilms.
The ability to grow in close association with surfaces is widespread among microorganisms. Despite the prevalence of so-called biofilm-like growth in most aqueous environments, it is only recently that the properties of surface-attached communities have been compared to those of free-living (planktonic) populations. Within biofilms, microorganisms lose their motility and produce an intercellular matrix of secreted materials and a degree of specialization is seen among individual cells (6, 32).
While bacteria have been the focus of many biofilm studies, little attention has been paid to fungal biofilms. Because many fungi are able to colonize nonaqueous environments, surface attachment is essential and free-living planktonic forms are usually restricted to dissemination processes. However, both forms are common in budding yeasts. The model yeast Saccharomyces cerevisiae adheres to plastic surfaces under low glucose concentrations and initiates biofilm formation (27). Pathogenic Candida species also establish well-developed biofilms. For example, Candida albicans colonizes several tissues such as vaginal and oral epithelia, developing a biofilm that, in immunocompromised patients, can disseminate into the bloodstream and cause fatal systemic infections (22). This species also forms biofilms on inert surfaces such as urinary or central venous catheters, dental prostheses, and other indwelling devices. These communities are often resistant to drug therapy and act as a source of reinfections (7).
C. albicans biofilm formation is initiated when planktonic cells adhere to a surface, presumably through cell wall-located adhesion molecules, and begin to aggregate into a microcolony. Candidate adhesion molecules are the proteins encoded by the ALS gene family, homologous to the mating-associated agglutinin of S. cerevisiae and involved in adhesion of C. albicans to host tissues (5, 15). Once a basal layer of cells is formed, an extracellular matrix of proteins and polysaccharides is produced to consolidate the early biofilm (5). C. albicans biofilms contain small amounts of extracellular material under static conditions, but copious quantities are secreted in dynamic models (12). The contribution of yeast or hyphal forms to the different layers varies with substrate, medium, or strain, but generally the inner layers are composed of yeast-like cells while the external ones include mostly hyphal forms (7).
Some properties of C. albicans biofilms differ markedly from those of planktonic populations. Their resistance to therapeutic drugs is remarkably high and remains high even when the biofilm is disrupted (16). Concomitantly increased expression of some drug efflux pumps (MDR1, CDR1, and CDR2) has been reported (24).
Even though some differential properties of biofilms have been observed and expression changes in a few genes have been reported, it is still unclear whether biofilm-like growth itself imposes a significant change in C. albicans physiology and whether this change is reflected in a particular pattern of gene expression. Genome-wide approaches are now available that allow this question to be fully investigated (2). Here we present for the first time evidence of a gene expression pattern specific to C. albicans biofilms, which is only marginally influenced by environmental parameters. We studied biofilm populations developed under different conditions of flow, oxygenation, and glucose concentration and compared their global transcript profiles to those of planktonic populations. Biofilm transcriptomes were remarkably similar under different environmental conditions and differed dramatically from those of planktonic populations. Hence, a detailed study of the expression patterns that accompany biofilm growth in C. albicans is presented.
MATERIALS AND METHODS
Strains and media.
The following strains of C. albicans were used: SC5314 (wild-type isolate), CAF2-1 (URA3/ura3::λimm434), and CAI4 (ura3::λimm434/ura3::λimm434) (9), GTC41 (CAI4 GCN4/gcn4::hisG-URA3-hisG), GTC43 (CAI4 gcn4::hisG-URA3-hisG/gcn4::hisG), and GTC65 (CAI4 gcn4::hisG/gcn4::hisG URA3 GCN4) (30); and CDB1 (CAI4 cph1::hisG/cph1::hisG efg1::hisG/efg1::hisG) (3).
The C. albicans strains were grown in 0.67% yeast nitrogen base (YNB; Difco) with 0.4 or 2% glucose. Strains CAF2-1, GTC41, GTC43, and GTC65 were cultured in YPD1/3 (0.33% yeast extract, 0.66% Bacto Peptone, 0.4% glucose) supplemented with histidine and arginine (20 mg/liter). Dilution of standard YPD was necessary to grow these strains in microfermenters. All media were supplemented with uridine (40 mg/liter) and adenine (20 mg/liter).
Biofilm and planktonic cultures.
An inoculum was prepared from an early-stationary-phase culture grown in flasks at 30°C in an orbital shaker and diluted to an optical density at 600 nm (OD600) of 1. Biofilms b1, b2, b6, Bwt, and Bm were produced in microfermentors (11). These consist of a glass vessel with a 40-ml incubation chamber where two glass tubes are inserted to drive the entry of medium and air. Used medium is evacuated through a third tube. Medium flow is controlled by a recirculation pump (Ismatec) and pushed by the pressured air. Plastic slides (Thermanox; Nunc) glued to a glass spatula were immersed in the inoculum for 1 h at room temperature. After this adhesion period, the spatula was transferred to the chamber and incubated at 37°C (except for b6, which was incubated at 30°C), with the medium flow set to 0.6 ml/min and air supplied at 105 Pa; under these conditions, the growth of the planktonic phase is minimized and most of the cells remain on the spatula. Biofilm b3 was produced in wells of 96-well microtiter plates (Techno Plastic Products) as previously described (25). A 100-μl volume of the inoculum were incubated for 1 h in wells at room temperature; 200 μl of fresh medium was added, and the plates were incubated at 37°C for 48 h with shaking at 40 rpm. The planktonic phase was discarded, and the wells were washed with YNB before the biofilms were collected. Biofilm b4 was obtained using disks of Exacanal (MERCK-Eurolab) preadsorbed in calf serum for 24 h (5), rinsed, and incubated with the inoculum for 72 h at 37°C with shaking at 40 rpm. Disks were collected by filtration through gauze and washed with YNB, and cells were recovered. Planktonic cultures were produced in flasks as described for the biofilm preinoculum, except for p5, where cultures were incubated in microtiter plates. Cells collected from several microfermentors, microtiter plates, or flasks were pooled to obtain enough RNA for replicated transcript profiling experiments.
Biofilm formation in microtiter plates was quantified by the method of Ramage et al. (25). The XTT [2,3-bis(2-methoxy-4-nitro-5-sulfo-phenyl)-2H-tetrazolium-5-carboxanilide sodium salt] stock solution was prepared at 0.5 g/liter in phosphate-buffered saline PBS (pH 7.4), filter sterilized, and stored at −20°C. A colorimetric change resulting from XTT reduction was measured in a microtiter plate reader (Labsystem Multiscan RC) at 492 nm.
RNA preparation, array hybridization, data capture, and analysis.
Total-RNA preparation, [α-33P]dCTP labeling, Northern blot hybridizations, and hybridization of macroarrays were carried out as previously described (19). Each RNA sample was independently labeled and hybridized a minimum of three times against swapped filters. Preliminary normalization, logarithmic transformation, and determination of correlation coefficients were performed with Excel 2000 (Microsoft). Data were loaded into GeneSpring version 5.0 (Silicon Genetics) and normalized in a per-chip (to the 50th percentile) and per-gene (to the median) normalization. Microarrays spotted in duplicate with probes for 5,907 C. albicans genes (information available at www.pasteur.fr/Galar_Fungail) were obtained from Eurogentec (Seraing, Belgium). For each of the four pair comparisons described in the text, RNA preparations were independently labeled with the two dyes and dyes were swapped in two replicate arrays. A total of 16 paired hybridizations were performed. Cy3/Cy5 labeling and hybridization were carried out as recommended by Eurogentec. Sixty pmoles of each dye were hybridized onto the array overnight at 42°C in a Corning chamber. Data were captured by GenePix Pro version 4.0.1.12. Each microarray was inspected to identify low-quality spots that were not included in the analysis. Low-quality hybridizations were excluded on the basis of the GenePix vital-statistic table. Median F635, F560, B635, and B560 of each spot were loaded into GeneSpring for calculation of signal and background intensities. Signals were log transformed and per-chip normalized by an intensity-dependent method (Lowess) applied to the print-tip region (35). Significance analysis was performed using GeneSpring version 5.0 as indicated above. Gene annotations were from http://genolist.pasteur.fr/CandidaDB. Expression data are available at http://www.pasteur.fr/recherche/unites/Galar_Fungail/Biofilm/biofilm.html.
Microscopy and image analysis.
Biofilm development in microfermentors was recorded with a Nikon Coolpix digital camera. Biofilms developed on Thermanox slides (wild type) or glass coverslips (cph1/cph1 efg1/efg1 strain) were examined by scanning electron microscopy (SEM) as described previously (23). Briefly, biofilm samples were fixed for 1 h in 0.07 M sodium cacodylate buffer (pH 7.3) containing 1.2% glutaraldehyde and 0.05% ruthenium red. The samples were then washed in the same buffer containing 0.05% ruthenium red and postfixed in 1% osmium tetroxide in cacodylate buffer, treated by the critical-point drying method, and observed on a Gemini DSM 982 scanning electron microscope. Transmission and scanning electron microscopy were performed by Brigitte Arbeille and Claude Lebos at the Laboratoire de Biologie Cellulaire et Microscopie Electronique, UFR Médecine, Tours, France.
RESULTS
Transcript profiles of biofilm populations produced under different conditions are strongly correlated.
A first objective of this study was to establish the extent to which C. albicans biofilm and planktonic populations differ at the transcriptome level. For this purpose, we performed multiple comparisons of biofilm and planktonic populations of C. albicans wild-type strain SC5314 obtained under different conditions of culture (Table 1).
TABLE 1.
Biofilm and planktonic cultures
| Culture | C. albicans strain | Vessel | Medium flow | Glucose (%) | Aeration | Time (h) | Temp (°C) |
|---|---|---|---|---|---|---|---|
| b1/b2 | SC5314 | Microfermentor | Continuous | 0.4 | +++ | 48 | 37 |
| b3 | SC5314 | Microtiter plate | Limited | 0.4 | ± | 48 | 37 |
| b4 | SC5314 | Catheter disks | Limited | 0.4 | ± | 72 | 37 |
| b6 | SC5314 | Microfermentor | Continuous | 2.0 | +++ | 72 | 30 |
| b7 | SC5314 | Microfermentor | Continuous | 0.4 | +++ | 72 | 37 |
| p2 | SC5314 | Flask | Limited | 0.4 | ++ | 20 | 37 |
| p3 | SC5314 | Flask | Limited | 0.4 | ++ | 48 | 37 |
| p4 | SC5314 | Flask | Limited | 2.0 | ++ | 48 | 30 |
| p5 | SC5314 | Microtiter plate | Limited | 0.4 | + | 48 | 37 |
| Bwt | CAI4 | Microfermentor | Continuous | 0.4 | +++ | 48 | 37 |
| Bm | CDB1 | Microfermentor | Continuous | 0.4 | +++ | 48 | 37 |
| Pwt | CAI4 | Flask | Limited | 0.4 | ++ | 48 | 37 |
| Pm | CDB1 | Flask | Limited | 0.4 | ++ | 48 | 37 |
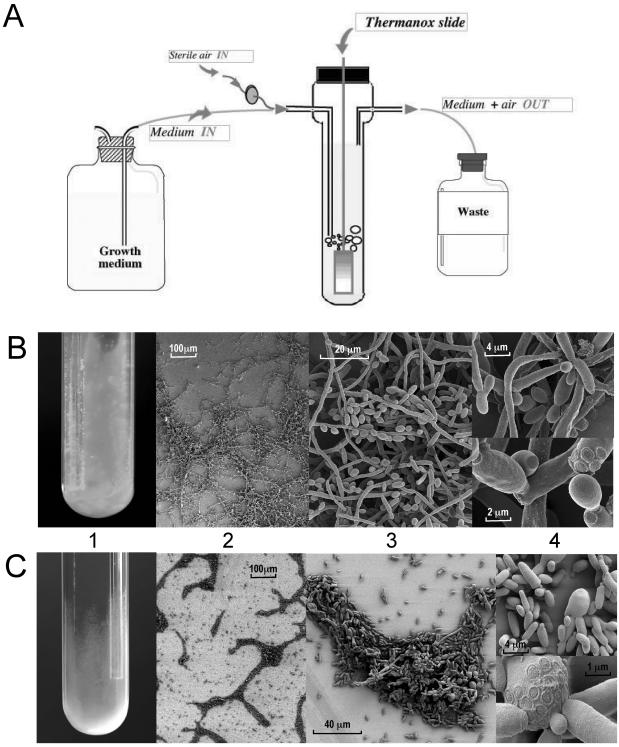
Biofilm populations b1 and b2 were produced in a microfermentor (11). This model permits the recovery of pure biofilm cultures in milligram quantities, in a single and rapid way that does not interfere with RNA purification. Briefly, following adhesion of planktonic cells to plastic slides, the slides were incubated vertically at 37°C in a continuous flow of YNB-0.4% glucose (Fig. 1A). After 48 h, the slides were covered by a ca. 3-mm-thick biofilm of fungal cells, developing a “cottony” surface with depressions and some filamentous projections (Fig. 1B). Biofilm development continued for about 72 h. Therefore, the biofilms collected in samples b1 and b2 were representative of a growing population. Another biofilm (b6; Table 1) was produced in microfermentors under different conditions of nutrient supply (YNB-2.0% glucose), time (72 h), and temperature (30°C).
FIG. 1.
Structure of the biofilm formed by C. albicans wild-type and cph1/cph1 efg1/efg1 strains. (A) Schematic representation of the microfermentor model used to produce C. albicans biofilms. C. albicans yeast cells are contacted with a Thermanox slide glued to a glass spatula, which is subsequently introduced into the microfermentor. A continuous flow of air and fresh growth medium is applied at a rate such that planktonic growth is not observed in the microfermentor. (B) Wild-type (CAI4) biofilm on a plastic slide (C) cph1/cph1 efg1/efg1 (CDB1) biofilm on a glass surface. (Panels 1) Wild-type biofilm was formed on the surface of the plastic slide, in the microfermentor; mutant biofilm was formed on the glass wall. (Panels 2) SEM showing the reticulated and compact structures. (Panels 3) Different proportions of each cell morphology. (Panels 4) Budding pattern during biofilm-like growth: bouquet-like structures (top) and coaxial location of the budding scar (bottom panel). Pictures were produced by Brigitte Arbeille (Biologie Cellulaire et Microscopie Electronique, Tours, France) and are reprinted with permission.
In microfermentors, biofilms develop under a flow that supplies unlimited nutrients; air bubbling provides agitation and strongly aerobic conditions. Since flow parameters influence the secretion of matrix material (12), we also studied biofilms produced in static and more anaerobic environments. These were the surface of microtiter plates for sample b3 (Table 1) (25) and serum-treated catheters for sample b4 (Table 1) (5). The proportions of yeast and hyphal forms were variable in each model, with longer incubation times, higher temperature, and the inclusion of serum leading to increased hyphal content.
To rule out genes whose differential gene expression was due to differences in growth rate, we collected exponential (p2) and stationary-phase (p3) populations of planktonic cultures (0.4% glucose). Two further planktonic populations were produced in microtiter plates (p5) or in YNB-2% glucose (p4).
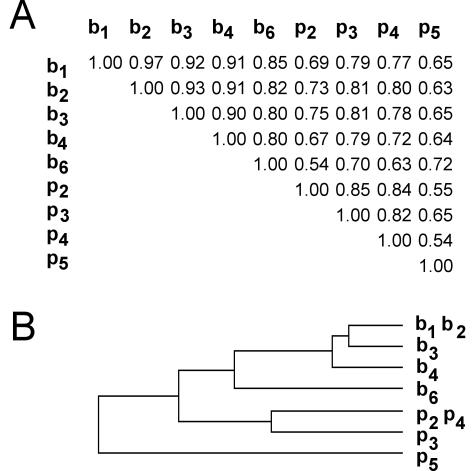
Transcript profiles were determined using macroarrays with probes for 2,002 C. albicans open reading frames that are a random representation of the whole genome (19). These arrays were the only tools available at the start of this study, but they also offer advantages for multiple transcriptome comparisons in that hybridizations are independently performed for each sample, thus limiting the bias due to the choice of pairs, as is the case in two-color experiments. To evaluate variations inherent in cell culture and RNA isolation, we first calculated the correlation between two replicate biofilms, b1 and b2. The Pearson coefficient for this comparison was 0.97, which is in agreement with previously reported correlations for two independent hybridizations of the same RNA preparation (19). This value also represents the limit for detecting similarity between two related populations. Correlation coefficients for the biofilm-versus-biofilm comparisons were greatest (0.97 to 0.80), followed by planktonic-versus-planktonic (0.85 to 0.54) and planktonic-versus-biofilm (0.81 to 0.54) comparisons (Fig. 2A). Planktonic and biofilm cultures under the same glucose concentration (b6 versus p4) or produced in the same microtiter plate model (b3 versus p5) showed low correlation (0.63 and 0.65, respectively); however, two biofilms produced under such different conditions as in b2 and b4 were highly correlated (0.91). For most of the comparisons, differences in the growth conditions did not significantly affect the correlation between biofilms (6 of 10 comparisons produced correlation coefficients over 0.90), whereas they produced a noticeable dispersion between planktonic populations (none of them over 0.90). The relationships are illustrated in a dendrogram for clarity (Fig. 2B). A cluster containing the biofilm populations was clearly separated from planktonic branches.
FIG. 2.
Correlation between transcript profiles of biofilm and planktonic populations. (A) Correlation coefficients between transcript profiles. Normalized expression values were log transformed and plotted; correlation was measured by Pearson's correlation coefficient. (B) Dendrogram representing the linkage between populations, as measured by Pearson's correlation coefficient. Biofilms: b1, b2, continuous-flow microfermentor, 0.4% glucose, 37°C, 48 h; b3, microtiter plate, 0.4% glucose, 37°C, 48 h; b4, serum-preadsorbed catheter disks, 0.4% glucose, 37°C, 72 h; b6, microfermentor, 2% glucose, 30°C, 72 h. Planktonic populations: p2, exponential growth 0.4% glucose, 37°C, 20 h; p3, stationary phase, 0.4% glucose, 37°C, 48 h; p4, 2% glucose, 30°C, 48 h; p5, microtiter plate, 0.4% glucose, 37°C, 48 h.
A set of 325 genes is differentially expressed.
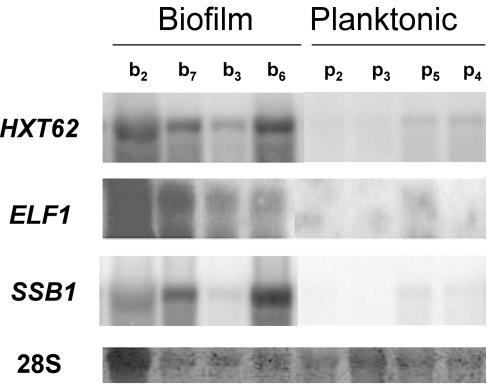
We applied significance analysis to detect genes with differential expression between biofilm and planktonic conditions. Normalized gene expression values for all the biofilm samples were averaged and compared to the average value for all the planktonic samples. A combination of nonparametric tests (P ≤ 0.02) was used for ranking the genes, and Benjamini's correction for the false discovery rate (FDR) was applied (31). Of 1,850 genes, 325 were found to be differentially expressed between biofilm and planktonic conditions (Table 2). The 325-gene cluster was divided by K-means into two subsets; the first contained 214 genes that were overexpressed in biofilms, and the second contained 111 genes that were underexpressed (see Supplementary Table A1 available at http://www.pasteur.fr/recherche/unites/Galar_Fungail/Biofilm/TableA1.xls). Differential expression of HXT62, ELF1, and SSB1 was confirmed by Northern hybridization (Fig. 3). To clarify the way in which regulated expression of these 325 genes contributes to the biofilm phenotype, C. albicans genes represented on the array were first assigned to 25 functional categories based on homology to S. cerevisiae genes (http://mips.gsf.de/proj/yeast/catalogues/funcat/index.html). The expected frequency of genes belonging to each category in a random group of 325 genes was compared to the observed frequency in the set of differentially expressed genes. The distribution of expected and observed frequencies differed significantly, as shown by a χ2 test. Rather than a nonspecific effect on all cell processes, the χ2 test suggests that some cellular functions are differentially expressed in biofilms (Table 2). In particular, the cluster is significantly enriched in genes involved in the following activities (categories are from Table 2).
TABLE 2.
Frequency distribution of differentially expressed genes among 25 functional categories
| Functional category
|
325-gene set
|
317-gene set
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Description | No. of genes on macroarraya | fob | fec | Significanced | No. of genes on microarraya | fob | fec | Significanced |
| 1 | Amino acid metabolism | 57 | 16 | 9 | * | 156 | 18 | 9 | * |
| 2 | Nitrogen and sulfur metabolism | 19 | 2 | 3 | 51 | 5 | 3 | ||
| 3 | Nucleotide metabolism | 42 | 11 | 7 | * | 104 | 18 | 6 | * |
| 4 | Phosphate metabolism | 11 | 1 | 2 | 26 | 5 | 2 | * | |
| 5 | C-compound and carbohydrate metabolism | 101 | 12 | 17 | 248 | 29 | 15 | * | |
| 6 | Lipid, fatty acid, and isoprenoid metabolism | 71 | 16 | 12 | * | 159 | 10 | 10 | |
| 7 | Metabolism of vitamins | 30 | 2 | 5 | 71 | 2 | 4 | ||
| 8 | Secondary metabolism | 1 | 1 | 0 | 5 | 0 | 0 | ||
| 9 | Energy | 57 | 6 | 9 | 163 | 20 | 10 | * | |
| 10 | Cell cycle and DNA processing | 151 | 19 | 25 | 452 | 9 | 27 | ||
| 11 | Transcription | 188 | 44 | 31 | * | 562 | 25 | 34 | |
| 12 | Protein synthesis | 79 | 34 | 13 | * | 254 | 71 | 15 | * |
| 13 | Protein fate | 162 | 14 | 26 | 479 | 16 | 29 | ||
| 14 | Intracellular transport | 109 | 18 | 18 | 362 | 13 | 22 | ||
| 15 | Cellular communication and signal transduction | 16 | 3 | 3 | 43 | 2 | 3 | ||
| 16 | Cell rescue | 64 | 8 | 10 | 143 | 7 | 9 | ||
| 17 | Regulation of and interaction with the environment | 48 | 3 | 8 | 136 | 5 | 8 | ||
| 18 | Cell fate | 112 | 4 | 18 | * | 290 | 9 | 17 | |
| 19 | Transposons and others | 1 | 0 | 0 | 4 | 0 | 0 | ||
| 20 | Control of cellular organization | 56 | 13 | 9 | * | 153 | 4 | 9 | |
| 21 | Subcellular localization | 606 | 106 | 99 | * | 1,675 | 145 | 101 | * |
| 22 | Protein activity regulation | 2 | 0 | 0 | 8 | 0 | 0 | ||
| 23 | Transport facilitation | 60 | 10 | 10 | 184 | 10 | 11 | ||
| 24 | Unclassified | 276 | 60 | 45 | * | 990 | 43 | 60 | |
| 25 | No homology to S. cerevisiae | 741 | 97 | 121 | 2,366 | 80 | 142 | ||
Number of genes spotted on the macro- or microarray that belong to each category. Many genes are included in several categories according to the MIPS classification of S. cerevisiae.
Observed frequency of genes falling into a given functional category.
Expected frequency of genes falling into a given functional category in a random set of 325 or 317 genes, respectively.
Categories that are significantly over or underrepresented (calculated on the basis of the F-Snedecor approximation with P = 0.975) are indicated by an asterisk. For both sets, distribution of observed and expected frequencies differs significantly, as tested by a χ[24;0.01]2 test.
FIG. 3.
Confirmation of differential expression by Northern bloting. A 10-μg portion of total RNA was blotted onto nylon membranes and hybridized to 33P-labeled probes for the HXT62, ELF1, and SSB1 genes. A control line showing the methylene blue staining of the 28S rRNA is included. b, biofilm samples, as described in Table 1; p, planktonic samples, as described in Table 1. b7 was not used for the transcript-profiling experiments.
(i) Protein synthesis (category 12).
The largest difference between observed and expected frequencies was found in the protein synthesis category. The 34 genes included here were overexpressed. They encode 14 ribosomal proteins, several translation factors, aminoacyl-tRNA synthases, and protein turnover factors. This suggests that increased ribosome production and protein translation occur in biofilms.
(ii) Amino acid and nucleotide metabolism (categories 1 and 3).
Categories 1 and 3 are the metabolic categories most strongly represented in the overexpressed subset. Many of those genes are involved in the synthesis of aromatic (TRP4, TRP5, ARO3, and ARO4) or sulfur (MET3, ECM17/MET5 CYS4, CYS3, and SER33) amino acids. S. cerevisiae homologues of many genes (MET3, ILV3, SER33, HIS1, ARO3, ARO4, TRP4, TRP5, ADE8, and ADE12) in this category are subject to regulation by the transcriptional activator Gcn4p, a general regulator of amino acid metabolism (21). All these genes were overexpressed.
(iii) Lipid metabolism (category 6).
The overexpressed genes are involved in the biosynthesis of membrane-associated lipids such as ergosterol (ERG25 and ERG16), sphingolipids (SUR2 and FAT1), or mitochondrial phospholipids (PEL1). SEC14 encodes a phospholipid transfer protein that regulates several aspects of membrane traffic and is involved in the production of secretory vesicules from Golgi apparatus.
(iv) Transcription (category 11).
Many overexpressed genes encode general components of the transcriptional machinery, in particular RNA polymerase subunits. Other genes are involved in mRNA or rRNA processing and stability. Some specific transcriptional regulators were also overexpressed, e.g., MIG1. This gene plays a decisive role in glucose-mediated repression in S. cerevisiae, but its function in C. albicans is not fully equivalent (36).
(v) Control of cellular organization (category 20).
Of the 13 differentially expressed genes classified here, 6 (ODC1, HCA4, ECM17, CIS2, PLC1, and SPT14) directly control cell wall biogenesis. ODC1 is a key regulator of polyamine biosynthesis and plays a pivotal role in cell wall organization, since polyamines compensate for anionic charges in this fungal structure. C. albicans ODC1 deletion leads to a defect in hyphal formation (13). Interestingly, expression of PTK2, encoding a protein kinase required for efficient polyamine uptake, was also significantly elevated in biofilms. IPF3518 (VPS16 homologue) and VPS35 are both included in the underexpressed cluster and encode vacuolar sorting proteins. In contrast, IPF13042, which encodes the C. albicans homologue of S. cerevisiae Sys3p involved at the vesicle-docking stage between endosome and distal Golgi, is overexpressed.
We analyzed the expression profiles of CDR, MDR, and ALS genes whose differential expression was previously demonstrated in biofilms (5, 24). CDR1, CDR2, CDR3, CDR4, and MDR1 (BMR1) were not differentially expressed in our study. Transcription of ALS genes has been found in a polymethylmethacrylate model using a probe that recognizes several members of the family (5). Our analysis identified ALS1 as the major overexpressed gene. ALS1 was dramatically overexpressed in all the biofilm populations (average B/P expression ratio = 19.04; see Supplementary Table A1 for detailed values), in agreement with the observation by Chandra et al. (5) and with the role of similar adhesins (FLO11) in S. cerevisiae biofilms (27). In contrast, ALS7 was underexpressed and expression of ALS10 and ALS5 was not significantly altered. Thus, each ALS family member makes independent contributions to the biofilm transcriptome.
GCN4 is necessary for efficient biofilm formation by C. albicans.
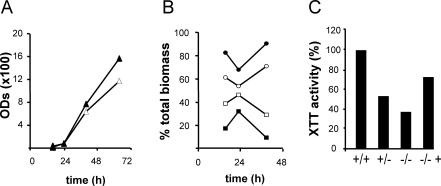
Based on the fact that a high proportion of the 214-gene set are amino acid biosynthesis genes and homologues of genes that are Gcn4p targets in S. cerevisiae (14, 21), a role for general control of amino acid biosynthesis in biofilm development was envisaged. Like S. cerevisiae Gcn4p, C. albicans Gcn4p controls many amino acid starvation responses, some of which lead to hyphal development (30). We tested the ability of a C. albicans gcn4/gcn4 strain to form biofilms in a microfermentor. Since we observed decreased planktonic growth of the gcn4/gcn4 strain in minimal medium (data not shown), presumably because this medium imposes amino acid starvation, we assayed biofilm formation in the microfermentor under conditions in which the growth rate and morphology of the two strains are equivalent. In rich medium (YPD1/3), the total biomass produced by the two strains was similar (Fig. 4A) and so were the proportions between yeast and hyphal forms (data not shown). However, the proportion of biofilm-growing cells was significantly lower in the mutant (Fig. 4B). Quantification of biofilm growth in microtiter plates (Fig. 4C) also confirmed the requirement for a functional Gcn4p for normal biofilm formation.
FIG. 4.
Biofilm formation in a C. albicans gcn4/gcn4 strain. (A) Total biomass was collected from microfermentors for 64 h and measured as OD600. Symbols: Δ, CAF2-1 GCN4/GCN4; ▴, GTC43 gcn4/gcn4. (B) Biofilm and planktonic growth were monitored in the microfermentor; cells on the plastic slide (biofilm) and those in the liquid medium (planktonic) were collected and subjected to several rounds of sonication and the OD600 was measured. OD units are expressed as a percentage of total biomass (biofilm plus planktonic). □, CAF2-1 biofilm; ○, CAF2-1 planktonic; ▪, GTC43 biofilm; •, GTC43 planktonic. (C) Quantification in a microtiter plate by the XTT reduction assay (25). Data are expressed as a percentage of wild-type activity. +/+, CAF2-1; +/−, GTC41 gcn4/GCN4 heterozygote; −/−, GTC43; −/− +, GTC65 GCN4-complemented GTC43.
A C. albicans cph1/cph1 efg1/efg1 strain is able to form biofilms on glass surfaces.
Our next objective was to study overlaps in the cellular functions required for hyphal and biofilm development. Dimorphism is not an absolute prerequisite for biofilm formation but can influence the structure of C. albicans biofilms (1). Since Efg1p and Cph1p are two major regulators of the yeast-to-hypha transition and their simultaneous inactivation prevents switching under most hypha-inducing conditions (8), we decided to use the cph1/cph1 efg1/efg1 mutant for a biofilm formation assay.
The C. albicans CPH1/CPH1 EFG1/EFG1 CAI4 and cph1/cph1 efg1/efg1 CDB1 strains were precontacted against plastic slides and incubated in microfermentors. Surprisingly, after 48 h the mutant had developed a thin, haze-like biofilm on the microfermentor glass wall and the glass spatula rather than the plastic slide itself. In contrast, the wild-type strain produced a thick, cottony biofilm which colonized the plastic surface (Fig. 1B and C, panels 1). When incubated directly in contact with the spatula, the wild-type strain was unable to colonize the glass surface.
SEM of the biofilms formed by both strains showed that wild-type biofilms are composed of microcolonies connected by “bridges” of hyphae (Fig. 1B, panel 2) and contain blastospores, hyphae, and pseudohyphae in variable proportions (Fig. 1B, panel 3). In contrast, cph1/cph1 efg1/efg1 biofilms are less reticulated, as a reflection of the lack of hyphal bridges, and contain mostly blastospores (Fig. 1C, panels 2 and 3). Hyphae emerged occasionally as short and nonbranched tips, but most cells remained as slightly elongated yeasts, as observed for the mutant under hypha-inducing conditions (17). The structure of the first layer of cells appears highly organized: cells orientate very close to each other, in a parallel manner (Fig. 1C, panel 3). All of these traits result in a quite “compact” but thin biofilm, whose resistance to detachment is similar to that of the wild type.
For both strains, budding of the new blastospores (Fig. 1B and C, panels 4) was frequently observed adjacent to the first bud (coaxial pattern), similar to that in planktonic cultures. Remarkably, however, in biofilms some hyphae also emerge coaxially while in planktonic cultures hyphal budding is bipolar or lateral. This often results in the production of bouquet-like structures (Fig. 1B, panel 4) that might facilitate the three-dimensional development of the biofilm.
Similar to Ramage et al. (26), we have observed that the cph1/cph1 efg1/efg1 strain is unable to form a biofilm on a plastic surface. However, our results suggest that the CPH1 and EFG1 genes are not essential for biofilm formation since this can progress on a glass surface.
Identification of biofilm-related genes independent of mycelial development.
We decided to exploit the altered cph1/cph1 efg1/efg1 biofilm phenotype to extend our transcriptome analysis of biofilm-intrinsic expression patterns that are unrelated to hyphal development. Indeed, a minimum set of genes required for biofilm development should be expressed in the mutant biofilm despite the filamentation defect. RNAs were prepared from duplicate cultures of wild-type and mutant biofilms as in b1 (Bwt and Bm, respectively [Table 1]) and from duplicate planktonic cultures as in p3 (Pwt and Pm [Table 1]). Four independent comparisons, Bwt/Pwt, Bm/Pm, Bwt/Bm, and Pwt/Pm, were performed in which two samples for each comparison were simultaneously hybridized on a microarray with duplicated probes for 5,907 C. albicans genes.
Significance analysis was performed using GeneSpring version 5.0 only on genes with a log ratio above 1.5 in at least one hybridization. This cutoff was introduced to control the stringency of the FDR correction, which is directly influenced by the number of genes analyzed (31). The number of differentially expressed genes is shown in Table 3 (see also Supplementary Table A2, available at http://www.pasteur.fr/recherche/unites/Galar_Fungail/Biofilm/TableA2.xls). As expected, a considerable number of genes (748 and 856 genes) were affected in biofilm-versus-planktonic comparisons. Since the cph1/cph1 efg1/efg1 mutation has a remarkable impact on biofilm structure, expression of many genes (386 genes) was also altered in the biofilm-versus-biofilm comparison. Thirty-three of these genes were further affected by the mutation under planktonic conditions, indicating a global effect of the mutation rather than a biofilm-dependent effect.
TABLE 3.
Significance analysis of paired microarray comparisonsa
| Paired-slide comparisonb | No. of genes with log r > 1.5 | No. of differentially expressed genes |
|---|---|---|
| Bwt vs Pwt | 966 | 748 |
| Bm vs Pm | 1,007 | 856 |
| Bwt vs Bm | 579 | 386 |
| Pwt vs Pm | 319 | 125 |
Wilcoxon-Mann-Whitney tests (P ≤ 0.02) were applied to the genes whose log ratios were > 1.5. The FDR correction was then applied to identify the most significant differentially expressed genes
B, biofilm; P, planktonic; wt, wild type; m, cph1/cph1 efg1/efg1.
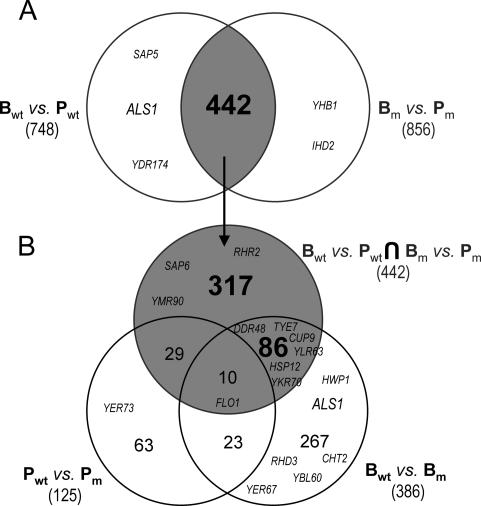
We used Venn diagrams to delimit each group of differentially expressed genes. The intersection between the Bwt/Pwt and the Bm/Pm sets comprised 442 genes (Fig. 5A) whose planktonic-versus-biofilm expression changed in both strains. However, some of these genes may also alter their expression in response to other conditions. We therefore excluded from the 442-gene set a group of 39 genes whose expression was also dependent on the cph1/cph1 efg1/efg1 mutation during planktonic growth (Fig. 5B). The intersection of the remaining 403 genes with the set of genes changing between biofilms (Bwt/Bm, 386 genes) identified 86 shared genes and 317 unshared genes. The two subsets are proposed as potential biofilm-specific candidates since they are (i) differentially expressed in biofilms independently of whether these contain hyphae and (ii) not affected by the mutation under planktonic conditions. Besides, expression of the 317-gene set was not significantly altered when mycelial development in the biofilm was impaired. In contrast, the 86-gene set identified genes whose expression changed between biofilms that contained hyphae and those that did not. The pertinence of this approach was confirmed by the exclusion from the 317-gene subset of some of the biofilm-related genes mentioned above that have also been identified as hypha specific or Efg1p or Cph1p dependent. For instance, ALS1, a downstream effector of the Efg1p filamentation pathway (10), was not included in either the 317- or 86-gene set, since its expression is not induced in mutant biofilms; some other hypha-specific genes (20) are discussed below.
FIG. 5.
Overlaps between the significant sets of differentially expressed genes. (A) Intersection between the two sets arising from biofilm-versus-planktonic comparisons. This intersection identifies a 442-gene set. (B) The 442-gene set is crossed against the planktonic-versus-planktonic and biofilm-versus-biofilm sets. B, biofilm; P, plankton; wt, wild type; m, cph1/cph1 efg1/efg1 mutant. Numbers indicate the size of each Venn set. The positions of several effectors of filamentation pathways are shown. From the 46 hypha-related genes identified by Nantel et al. (20), all were found in Candida. DB and 21 were included in the significant sets. Candida DB name is given to genes with different nomenclatures. FLO1 = IPF5185; CUP9 = IPF3912; YDR174 = HMO1, YLR63 = IFU3; YER67 = IPF20056; YBL060W = IPF6654; YER73 = ALD5; YMR90 = IPF351; YKR70 = IPF5915; IHD2 = IPF10761; RHD3 = IPF8527.
The intersection between the two B/P sets also recognized many biofilm-related genes identified by our first approach: 72 genes from the 325-gene set were included in the intersection, 49 of them were included in the 317-gene set, and 17 were included in the 86-gene set (Supplementary Table A2).
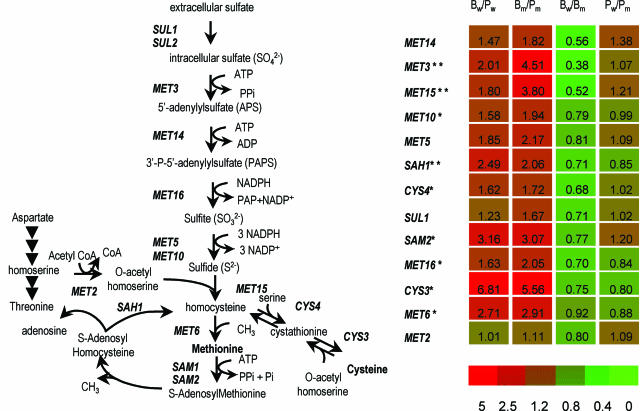
A χ2 test revealed significant alterations in the distribution of functional categories in the 317-gene set (Table 2). As observed for the 325-gene set, the most overrepresented categories are protein synthesis (category 12) and amino acid and nucleotide metabolism (categories 1 and 3). The group includes six genes (MET16, MET10, CYS4, CYS3, MET6, and SAM2) involved in sulfur amino acid biosynthesis and salvage pathways (Fig. 6). Carbohydrate metabolism and energy were overrepresented with respect to the 325-gene set, mostly due to the presence of several genes for the glycolytic pathway (PGI1, FBA1, TPI1, PGK1, GPM1, ENO1, and PDC1), 2 genes responsible for the conversion of the glycolytic intermediate dihydroxyacetone-phosphate to glycerol (GPD1 and RHR2), and the catabolic repressor MIG1. All these genes were overexpressed. Consistent with this, the underexpressed genes include known targets of glucose repression (SNF3, MLS1, and TPS3). The study of the representation of functional categories also underlined several genes related to phosphate transport (category 4; PHO84, PHO87, and PHO88).
FIG. 6.
Differential expression of the genes involved in sulfur amino acid biosynthesis and salvage. Expression ratios of the four paired comparisons (log scale) are represented on the right. B, biofilm; P, planktonic; w, wild type; m, cph1/cph1 efg1/efg1 strain. Single or double asterisks indicate the genes included in the 317- and 86-gene sets, respectively. SUL2 and SAM1 do not have homologues in Candida DB. MET5 = ECM17. The pathway is adapted from reference 29.
Inspection of the 86-gene set revealed another 3 genes in the sulfur amino acid pathways (MET3, MET15, and SAH1), completing the biosynthesis and salvage route (Fig. 6). These genes, like those in the 317-gene set, were overexpressed in both B/P comparisons and their expression was higher in mutant than in wild-type biofilms. In contrast, the agglutinin genes ALS12 and ALS4 and the TUP1-repressed genes RBT2 and RBT5 were overexpressed in both B/P comparisons but lower in the cph1/cph1 efg1/efg1 biofilm than in the hypha-producing wild-type biofilm. Some of these genes respond to several types of hypha-switching stimuli under planktonic conditions (4). Of the 46 genes identified by Nantel et al. (20) as modulated during the yeast-to-hypha transition induced by serum and temperature, 6 [DDR48, IPF3912(CUP9), TYE7, IFU3(YLR63), IPF5915(YKR70), and HSP12] are included in the 86-gene set (Fig. 5). Since we did not expose B and P samples to serum, elevated temperature, or other environmental conditions known to induce hyphal switching, the inclusion of known hypha-modulated genes (20) in the 86-gene cluster and, more broadly, in the set of genes that show differential expression between wild-type and mutant biofilms (Fig. 5; Bwt-versus-Bm set) is likely to reflect a response to a switching stimulus that is specific for biofilms.
DISCUSSION
In most natural environments, microorganisms follow a sessile way of life in which they remain surface attached in close contact with each other. As a result, such populations exhibit a number of features that are very different from those of planktonic communities. Many forms of candidiasis develop on organ or implant surfaces. Consequently, C. albicans populations can follow a surface-attached, biofilm-like way of life that confers, among other characteristics, an increased resistance to antifungal drugs (7).
In this study we used transcript profiling to perform a global analysis of the biofilm-specific features of C. albicans. We produced biofilms in three laboratory models that involved very different environmental conditions. Transcript profile comparisons of these biofilms with planktonic populations led us to our initial observation that biofilm transcriptomes are highly correlated (Fig. 2A) and that nutrient flow, aerobiosis, and other factors that strongly influence the transcriptome of planktonic populations do not have a noticeable effect on biofilm-growing communities. Hence, by comparison to planktonic populations, transcriptome invariance is proposed as a specific feature of biofilm populations. This observation is unlikely to reflect a slower growth of all biofilm populations than of planktonic populations, since both actively growing biofilms and stationary planktonic populations were included in the study.
The second observation is the identification of genes encoding biofilm-associated traits. As expected from the low correlation between biofilm and planktonic populations, the number of differentially expressed genes is considerable (Table 3). This is also true when both populations arise from a cph1/cph1 efg1/efg1 strain unable to develop hyphae (Table 3), indicating that the differential expression is not due only to differences in the hyphal content of planktonic and biofilm populations. We identified three significant sets of genes: the 325-gene set contains biofilm-related genes whose expression is independent of extrinsic conditions, the 317-gene set contains biofilm-related genes that are expressed independently of the intrinsic ability of the cell to form mycelia, and the 86-gene set identifies genes whose expression varies between hypha-containing and hypha-noncontaining biofilms. The classification of these genes into functional categories and the study of their representation in each set revealed that expression changes specifically affect some cellular functions (Fig. 3; Table 2).
Genes involved in protein synthesis are the most highly represented in the three sets (Fig. 2; Table 2), and they are exclusively overexpressed. One reason for this activation of the protein synthesis machinery could be that biofilms grow faster than planktonic populations. However, genes related to cell-cycle and DNA processing are underrepresented. Thus, the increase of protein synthesis is independent of cell division and unlikely to be caused by differences in growth rate. On the other hand, this excess protein might be used for the production of extracellular matrix, which is not present in planktonic cells. This is in keeping with the inclusion in the 325-gene set of several components of the secretion machinery, such as SEC14 or SYS3, and the dramatic increase in the expression of the extracellular agglutinin gene ALS1. Enhanced production and secretion of certain proteins with agglutination properties may facilitate the cohesion of cells within the biofilm. In addition to ALS1, several new genes that encode proteins with secretion signals and adhesin-like domains, such as IPF20161, IFP5185, or IPF20008, were found to be significantly overexpressed.
Among metabolic activities, amino acid biosynthetic pathways are strongly represented. We have demonstrated that GCN4 is required for efficient biofilm-like growth. In C. albicans, this transcription factor induces two different processes in response to amino acid starvation: the activation of amino acid biosynthesis and the triggering of morphogenesis (30). The latter is unlikely to be related to the biofilm-forming defect exhibited by the gcn4/gcn4 strain, since the proportion of yeast and hyphal cells was roughly the same in wild-type and gcn4/gcn4 biofilms (data not shown). Therefore, the effect of GCN4 on biofilm formation resembles a GCN-like response, although it was not observed under conditions that are known to impose amino acid starvation on planktonic populations. The situation might be different in biofilms that seem to have an increased need for protein synthesis, and amino acids might therefore become limiting faster. Under these conditions, the regulatory role of GCN4 could be decisive for biofilm progression.
A more specific role for the sulfur amino acid biosynthesis/salvage pathway is revealed by the 317- and 86-gene subsets, which identify overexpression of 9 of the 11 key genes in this route (Fig. 6). The route leads to the production of S-adenosylmethionine (SAM), with a separate branch for the production of cysteine (29). Since SAM is the precursor of polyamines and since ODC1 and PTK2 are also included in the 325-gene set, all of these changes could be associated with cell wall rearrangements that allow closer interaction of cells within the biofilm (13). Alternatively, biofilms may have a special requirement for methionine- and cysteine-rich proteins, an idea supported by the overexpression of genes in the cysteine biosynthesis branch. Finally, activation of the genes for SAM biosynthesis might be related to the production of a quorum-sensing molecule associated with biofilm formation. Indeed, the bacterial autoinducer AI-2 is produced from SAM, which has been proposed to serve as a “universal signal” for interspecies communication (34).
The activation of glycolytic flux and catabolic repression is underscored by the 317-gene set. In the wild-type-versus-cph1/cph1 efg1/efg1 experiment, biofilms were produced under a continuous flow of nutrients, in contrast to planktonic cultures (Table 1). Thus, activation of glycolysis could be merely a reflection of unlimited sugar availability in the microfermentor. However, activation of MIG1 in biofilms is revealed by the 325-gene set as well and is therefore independent of whether biofilms were produced under continuous flow. Overexpression of MIG1 in S. cerevisiae is associated with flocculation (28). Whether overexpression of MIG1 in C. albicans biofilms could promote flocculation and hence increase the aggregation of the cells in biofilms remains to be investigated.
Biofilm resistance to antifungals has been associated with increased expression of the MDR and CDR genes (24). Expression of these genes was studied previously in a single microtiter plate model, whereas we report a multiple-model comparison. Here, changes in the expression of these genes are not considered significant. However, CDR1 is included in the 86-gene set arising from the biofilm produced in the microfermenter model. Thus, expression of this gene could be significant under certain conditions of biofilm production, and it is also predicted to be influenced by the triggering of hyphal development concomitant with biofilm formation. Instead, we observed differential expression of ERG25 and ERG16 (in the 325-gene set) and ERG6 (in the 317-gene set). ERG16 mRNA levels correlate with azole resistance in clinical isolates C. albicans (33). An enrichment or redistribution of sterols in biofilm membranes could explain their resistance to azole-derived antifungal agents. This is consistent with the recent data of Mukherjee et al. (18) showing that expression of MDR and CDR genes in biofilms is phase specific and contributes to azole resistance only during the early phase of biofilm development whereas changes in sterol composition are involved in the resistance of the mature phase. Hence, our laboratory models for biofilm production and transcript profiling analysis converge with previous approaches to the identification of differentially expressed genes related to pathogenesis and underline the importance of the study of biofilm-like growth for a full understanding of virulence in C. albicans.
Acknowledgments
We thank J.-Y. Coppée, O. Sismeiro, M.-A. Dillies, A. Maitournam, and other members of the Institut Pasteur DNA microarray platform for their excellent assistance; F. Dromer and members of our respective laboratories for constant support; the Brown and Ernst laboratories for C. albicans strains; C. Lebos and B. Arbeille for SEM; and K. A. Charlton, C. Munro, and T. Pugsley for proofreading the manuscript. Sequence data from C. albicans were obtained from the Stanford Genome Technology Center (http://www.sequence.stanford.edu/group/candida).
Sequencing of C. albicans was accomplished with the support of the NIDR and the Burroughs Wellcome Fund. This work was supported by grants from Institut Pasteur (PTR 50), the Ministère de la Recherche et de la Technologie (P.R.F.M.M.I.P. ‘Réseau Infections Fongiques'), and the European Commission (QLK2-2000-00795; ‘Galar Fungail consortium’).
REFERENCES
- 1.Baillie, G. S., and L. J. Douglas. 1999. Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48:671-679. [DOI] [PubMed] [Google Scholar]
- 2.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 3.Bockmuhl, D. P., S. Krishnamurthy, M. Gerads, A. Sonneborn, and J. F. Ernst. 2001. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Mol. Microbiol. 42:1243-1257. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., and A. D. Johnson. 2000. TUP1, CPH1 and EFG1 make independent contributions to filamentation in Candida albicans. Genetics 155:57-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandra, J., D. M. Kuhn, P. K. Mukherjee, L. L. Hoyer, T. McCormick, and M. A. Ghannoum. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183:5385-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas, L. J. 2003. Candida biofilms and their role in infection. Trends Microbiol. 11:30-36. [DOI] [PubMed] [Google Scholar]
- 8.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 9.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y. C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards, Jr. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61-72. [DOI] [PubMed] [Google Scholar]
- 11.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 12.Hawser, S. P., G. S. Baillie, and L. J. Douglas. 1998. Production of extracellular matrix by Candida albicans biofilms. J. Med. Microbiol. 47:253-256. [DOI] [PubMed] [Google Scholar]
- 13.Herrero, A. B., M. C. Lopez, S. Garcia, A. Schmidt, F. Spaltmann, J. Ruiz-Herrera, and A. Dominguez. 1999. Control of filament formation in Candida albicans by polyamine levels. Infect. Immun. 67:4870-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinnebusch, A. G. 1992. General and pathway-specific regulatory mechanisms controlling the synthesis of amino acid biosynthetic enzymes in Saccharomyces cerevisiae, p. 319-414. In J. R. Broach, E. W. Jones, and J. R. Pringle (ed.), The molecular and cellular biology of the yeast Saccharomyces cerevisiae. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 16.Kumamoto, C. A. 2002. Candida biofilms. Curr. Opin. Microbiol. 5:608-611. [DOI] [PubMed] [Google Scholar]
- 17.Lo, H. J., J. R. Köhler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 18.Mukherjee, P. K., J. Chandra, D. M. Kuhn, and M. A. Ghannoum. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect. Immun. 71:4333-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murad, A. M., C. d'Enfert, C. Gaillardin, H. Tournu, F. Tekaia, D. Talibi, D. Marechal, V. Marchais, J. Cottin, and A. J. Brown. 2001. Transcript profiling in Candida albicans reveals new cellular functions for the transcriptional repressors CaTup1, CaMig1 and CaNrg1. Mol. Microbiol. 42:981-993. [DOI] [PubMed] [Google Scholar]
- 20.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natarajan, K., M. R. Meyer, B. M. Jackson, D. Slade, C. Roberts, A. G. Hinnebusch, and M. J. Marton. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 21:4347-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odds, F. C. 1998. Candida and candidosis, 2nd edn. ed. Baillière Tindall, London, United Kingdom.
- 23.Prigent-Combaret, C., G. Prensier, T. T. Le Thi, O. Vidal, P. Lejeune, and C. Dorel. 2000. Developmental pathway for biofilm formation in curli-producing Escherichia coli strains: role of flagella, curli and colanic acid. Environ. Microbiol. 2:450-64. [DOI] [PubMed] [Google Scholar]
- 24.Ramage, G., S. Bachmann, T. F. Patterson, B. L. Wickes, and J. L. Lopez-Ribot. 2002. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J. Antimicrob. Chemother. 49:973-980. [DOI] [PubMed] [Google Scholar]
- 25.Ramage, G., K. Vande Walle, B. L. Wickes, and J. L. Lopez-Ribot. 2001. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45:2475-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramage, G., K. VandeWalle, J. L. Lopez-Ribot, and B. L. Wickes. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 214:95-100. [DOI] [PubMed] [Google Scholar]
- 27.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 28.Shankar, C. S., M. S. Ramakrishnan, and S. Umesh-Kumar. 1996. MIG1 overexpression causes flocculation in Saccharomyces cerevisiae. Microbiology 142:2663-2667. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, D., and Y. Surdin-Kerjan. 1997. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61:503-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tripathi, G., C. Wiltshire, S. Macaskill, H. Tournu, S. Budge, and A. J. Brown. 2002. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 21:5448-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watnick, P., and R. Kolter. 2000. Biofilm, city of microbes. J. Bacteriol. 182:2675-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, T. C. 1997. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 41:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xavier, K. B., and B. L. Bassler. 2003. LuxS quorum sensing: more than just a numbers game. Curr. Opin. Microbiol. 6:191-197. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Y. H., S. Dudoit, P. Luu, and T. P. Speed. 2001. Normalization for cDNA microarray data, p. 141-152. In M. L. Bittner, Y. Chen, A. N. Dorsel, and E. R. Dougherty (ed.), Microarrays: optical technologies and informatics. SPIE, San Jose, Calif.
- 36.Zaragoza, O., C. Rodriguez, and C. Gancedo. 2000. Isolation of the MIG1 gene from Candida albicans and effects of its disruption on catabolite repression. J Bacteriol 182:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]