Abstract
Concanavalin A (ConA) kills the procyclic (insect) form of Trypanosoma brucei by binding to its major surface glycoprotein, procyclin. We previously isolated a mutant cell line, ConA 1-1, that is less agglutinated and more resistant to ConA killing than are wild-type (WT) cells. Subsequently we found that the ConA resistance phenotype in this mutant is due to the fact that the procyclin either has no N-glycan or has an N-glycan with an altered structure. Here we demonstrate that the alteration in procyclin N-glycosylation correlates with two defects in the N-linked oligosaccharide biosynthetic pathway. First, ConA 1-1 has a defect in activity of polyprenol reductase, an enzyme involved in synthesis of dolichol. Metabolic incorporation of [3H]mevalonate showed that ConA 1-1 synthesizes equal amounts of dolichol and polyprenol, whereas WT cells make predominantly dolichol. Second, we found that ConA 1-1 synthesizes and accumulates an oligosaccharide lipid (OSL) precursor that is smaller in size than that from WT cells. The glycan of OSL in WT cells is apparently Man9GlcNAc2, whereas that from ConA 1-1 is Man7GlcNAc2. The smaller OSL glycan in the ConA 1-1 explains how some procyclin polypeptides bear a Man4GlcNAc2 modified with a terminal N-acetyllactosamine group, which is poorly recognized by ConA.
Trypanosoma brucei is a protozoan parasite that causes sleeping sickness in Africa. Trypanosomes dwell in the bloodstream of their mammalian host and are transmitted from one mammal to another by a tsetse fly vector. The bloodstream stage of the parasite can be obtained in the laboratory either from infected rodents or from culture. The procyclic stage, which is the subject of this study, inhabits the tsetse midgut and is also easily cultured. Both stages have major glycosylphosphatidylinositol (GPI)-anchored glycoproteins that form a surface coat. Bloodstream parasites are coated with a variant surface glycoprotein that functions in immune evasion (6). The coat of procyclic trypanosomes is composed of procyclins, glycoproteins of unusual structure that are important for parasite development in the insect vector (3, 22, 32, 35).
There are two major forms of procyclin. EP-procyclins have an N-terminal domain of 27 to 35 residues and a C-terminal domain of 22 to 30 EP repeats followed by a single G. The N-terminal domain of GPEET-procyclin has 21 residues, and its C terminus has five or six GPEET repeats followed by EPEPEPG (2, 5, 21, 30, 31, 37). In addition to the GPI anchors, which are unique because of their exceptionally large and heterogeneous glycan side chains (8, 37), EP- but not GPEET-procyclin has a Man5GlcNAc2 glycan, which is unusual in that it has no microheterogeneity (11, 37).
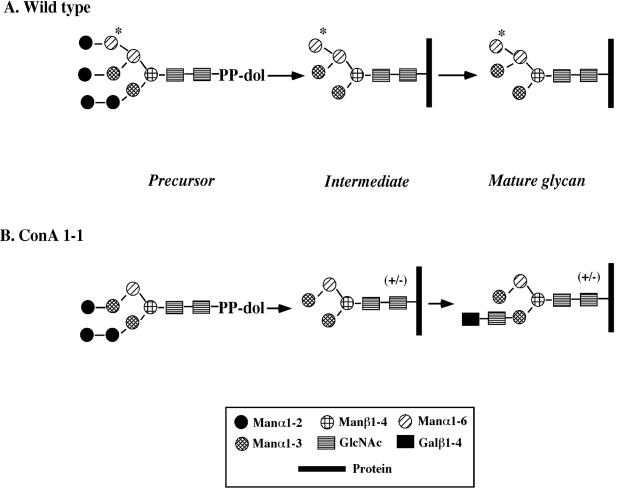
We recently isolated two mutants of procyclic T. brucei that are defective in N-glycosylation (11). We selected these mutants, ConA 1-1 and ConA 4-1, from chemically mutagenized cultures by incubation with concanavalin A (ConA). ConA not only agglutinates wild-type (WT) procyclic trypanosomes but also kills them by an undefined cell death mechanism (26, 38). Characterization of the N-glycans on ConA 1-1 (1, 11, 12) revealed that its procyclin (mainly the product of the EP1-3 gene) has a hybrid-type N-glycan containing a terminal N-acetyllactosamine (Fig. 1B shows its structure and a comparison with that of WT [Fig. 1A]). The ConA 1-1 N-glycan binds ConA with affinity lower than that of the WT. Furthermore, the occupancy of the ConA 1-1 procyclin glycosylation site is significantly less than that of procyclin in WT cells, which is virtually 100% (1). ConA 4-1 mutant cells, on the other hand, despite expressing altered N-glycans with the same structure as those found in ConA 1-1, are even more resistant to ConA. The major reason for their high resistance is their predominant expression of EP2-procyclin, the only member of the EP-procyclin family that lacks a site for N-glycosylation (1).
FIG. 1.
Structure of procyclin N-glycans from WT and mutant trypanosomes. (A) WT; (B) ConA 1-1 (1, 11, 12). “+/−” indicates heterogeneity in ConA 1-1 N-glycan.
In this paper we explore the biochemical defects of ConA 1-1 that account for the alteration in its N-glycosylation. We find that it has two defects: one in the reduction of polyprenol to dolichol and the other in synthesis of the N-glycan precursor. WT cells apparently synthesize Man9GlcNAc2-PP-dolichol, whereas ConA 1-1 makes a smaller precursor, Man7GlcNAc2-PP-dolichol. Both defects could result in altered glycosylation of procyclin.
MATERIALS AND METHODS
Cell culture.
WT procyclic T. brucei (strain 427-60) was a gift of Mary Lee (New York University). The ConA 1-1 mutant was obtained after mutagenesis and selection with ConA (11). Cells were maintained at 27°C in SDM-79 medium (4) supplemented with 10% fetal bovine serum (Life Technologies). Crithidia fasciculata was cultured at room temperature in brain heart infusion (Difco Laboratories, Inc.) containing 10 μg of hemin/ml.
Radiolabeling with mevalonolactone.
WT or mutant trypanosomes in SDM-79 medium were diluted to a concentration of about 106 cells/ml. Cells were centrifuged (4°C, 2,500 × g, 10 min) and resuspended in 40 ml of the same medium containing 1.5 μM [5-3H]mevalonolactone (NEN Life Science Products, Inc.; 20 to 40 Ci/mmol) at either 25 or 50 μCi/ml (cell concentration, 1.5 × 106/ml). Cells were then incubated at 27°C for at least 36 h. After centrifugation they were resuspended in 3 ml of methanol.
Analysis of total prenols.
After saponification of the cell suspension, the base-stable lipids were extracted into ether and neutralized, followed by enzymatic dephosphorylation (27). Lipids were then fractionated by size-exclusion chromatography on a Toyopearl HW-40S (Tosohaas, Philadelphia, Pa.) column as described previously (27) except that dolichol-11, polyprenol-11, coenzyme Q9, cholesteryl oleate, and cholesterol (Sigma) were used as unlabeled internal standards. Fractions from this column, which separates the larger prenols from sterols, were pooled based solely on the elution of the internal standards. Prenols were then analyzed by normal-phase silica high-pressure liquid chromatography (HPLC) (27).
Analysis of phosphorylated and neutral prenols.
[3H]mevalonate-labeled trypanosomes (5 × 108 cells) were extracted twice with 9 ml of chloroform-methanol (2:1, vol/vol) to obtain neutral prenols, prenyl phosphate, and monosaccharyl-phosphorylated prenols. The pellet was further extracted three times with 3 ml of chloroform-methanol-4 mM MgCl2 (10:10:3, vol/vol/vol) to obtain oligosaccharide lipid (OSL). The chloroform-methanol (2:1, vol/vol) extracts were applied to a DEAE-cellulose column equilibrated in chloroform-methanol (2:1, vol/vol). Neutral lipids were washed off the column in chloroform-methanol (2:1, vol/vol), and the phosphorylated lipids were then eluted by addition of chloroform-methanol (2:1, vol/vol) containing 250 mM ammonium acetate. Salt in the latter fractions was removed by partitioning with a 1/5 volume of saline. The neutral lipids, phosphorylated lipids, and OSLs were dried under nitrogen and resuspended in 3 ml of methanol. All lipids were then saponified by adding 1.5 ml of 60% KOH and heating them at 100°C for 1 h. Internal standards consisting of pig liver [14C]dolichol (a gift from W. J. Lennarz) and [14C]dolichyl phosphate (a gift from Adina Kaiden) were added to the neutral and phosphorylated lipid samples, respectively. The mixture was cooled, and the lipids were extracted with an equal volume of ether. The ether phases were neutralized by extraction with an equal volume of 5% acetic acid. The phosphorylated lipids were treated with acid phosphatase (27). The long-chain prenols were separated from other labeled lipids such as sterols and lower-molecular-weight prenols by size-exclusion chromatography on a Toyopearl HW-40S column as described previously (27) with cholesterol, cholesteryl oleate, pig liver dolichol, undecaprenol, dolichol (C55), and ubiquinone-9 as internal standards. The identity of the prenol, as polyprenol or dolichol, was determined by adsorptive HPLC (Dionex BioLC, Sunnyvale, Calif.) as described previously (33). The data from the HPLC were normalized per 108 cells and for the recovery of radiolabeled internal standard.
Preparation of cell lysates and radiolabeling of OSL.
Hypotonic lysates of WT and ConA 1-1 trypanosomes were prepared as described previously (18) except that the addition of tunicamycin was omitted during preparation of the membranes. For labeling OSL and other glycolipids, we used a protocol designed for synthesis of GPIs (18). Thawed cell lysates (0.4 to 0.8 ml) were washed twice with 10 ml of HKML buffer (50 mM HEPES [pH 7.4], 25 mM KCl, 5 mM MgCl2, 1 μg of leupeptin/ml) by centrifugation (5,800 × g for 10 min at 4°C). The membranes (8 × 107 cell equivalents) were suspended in HKML buffer (final volume, 120 μl) and further incubated for 1 to 2 min at 27°C with 5 mM MnCl2-1 mM dithiothreitol-1.2 mM ATP (tunicamycin at 1.6 μg/ml was added for some experiments). The Man-containing glycolipids were pulse-labeled by transferring the membranes (2 × 107 cell equivalents) into another tube (final volume, 20 μl) containing GDP-[3,4-3H]Man (Dupont; 15.5 Ci/mmol, 5 μCi/ml) and 2 mM UDP-GlcNAc for 5 min. As indicated, the reaction was chased with 1 mM nonradioactive GDP-Man for 20 min at 27°C. Reactions were terminated by adding CHCl3-CH3OH (1:1, vol/vol) to give a final CHCl3/CH3OH/H2O ratio of 10:10:3 (vol/vol/vol). Lipids were extracted for 10 min in a bath sonicator, and the insoluble debris was removed by centrifugation. The organic supernatant was dried under a stream of nitrogen, and the lipids were extracted by adding 100 μl of n-butanol and 100 μl of water. After a quick centrifugation, the organic upper phase was saved and the lower aqueous phase was reextracted twice with 100 μl of water-saturated n-butanol. The pooled organic phases were then dried in a Speed Vac concentrator and resuspended in 10 μl of CHCl3-CH3OH-H2O (10:10:3, vol/vol/vol) for loading on thin-layer chromatography (TLC) plates.
TLC analysis.
For the experiments shown in Fig. 3, [3H]Man-labeled lipids were loaded onto a predried silica gel 60 TLC plate (Merck) and resolved using the solvent CHCl3-CH3OH-H2O (10:10:3, vol/vol/vol). The plate was sprayed with En3Hance (Dupont) and exposed to preflashed X-Omat X-ray film (Kodak) at −80°C. For preparative TLC, labeled glycolipids were resolved in CHCl3-CH3OH-0.25% KCl (55:45:10), a solvent system yielding better resolution of mature OSL. Mature OSL was extracted from the silica with 200 μl of CHCl3-CH3OH-H2O (10:10:3, vol/vol/vol) followed by CH3OH-pyridine-H2O (2:1:2), and glycolipids were subjected to mild acid hydrolysis and P4 analysis as described below. For glycan analysis (Fig. 5), samples were fractionated on a predried TLC silica gel 60 (Merck) by being run three times with n-butanol-acetone-water (6:5:4). After development, plates were processed for autoradiography as indicated above.
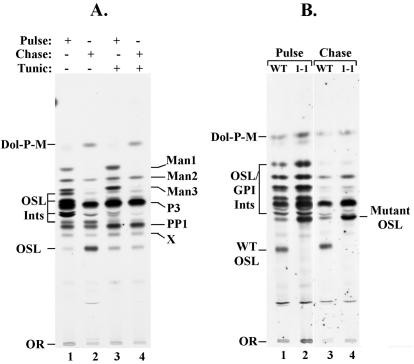
FIG. 3.
Cell-free synthesis of OSL. Glycolipids were synthesized in a cell-free system containing washed trypanosome membranes. The membranes were incubated with GDP-[3H]Man and UDP-GlcNAc and then chased with nonradioactive GDP-Man. In both cases, glycolipids were extracted with organic solvents, fractionated by TLC, and detected by autoradiography. (A) Synthesis of glycolipids in WT membranes in the absence or presence of 1.6 μg of tunicamycin/ml (Tunic). (B) Synthesis of glycolipids in WT and ConA 1-1 membranes under the same conditions as shown in panel A, except that tunicamycin was omitted. Dol-P-M, dolichol phosphoryl-Man; PP1 and P3, GPI precursors; Man1, Man2, and Man3, GPI biosynthetic intermediates (Ints) with the indicated number of Man residues; X, unknown species.
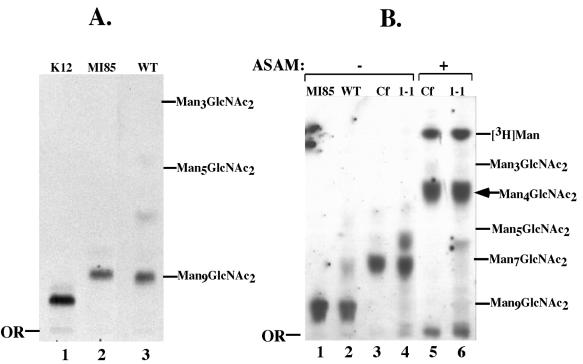
FIG. 5.
Structural characterization of OSL oligosaccharides. [3H]Man-labeled glycans were analyzed by TLC and autoradiography. (A) Lane 1, 3H-labeled Glc3Man9GlcNAc2 from WT CHO cells (K12); lane 2, 3H-labeled Man9GlcNAc2 from MI8-5 mutant CHO cells; lane 3, 3H-labeled OSL glycan from WT T. brucei. (B) 3H-labeled OSL glycans from MI8-5 CHO cells (lane 1), WT T. brucei (lane 2), C. fasciculata (lanes 3 and 5), and ConA 1-1 (lanes 4 and 6). Glycans in lanes 5 and 6 were treated with A. saitoi α-mannosidase (ASAM), and those in lanes 3 and 4 were mock treated. Positions of glycan standards (2 μg; Sigma) visualized by orcinol-H2SO4 staining are indicated on the right. Cf, C. fasciculata glycan.
Mild acid hydrolysis.
Dried [3H]Man-labeled lipids were resuspended in 100 μl of 0.1 M HCl and heated at 100°C. After 15 min the tubes were placed in ice-water, and the released glycans were separated from glycolipids by extraction. n-Butanol (100 μl) was added, and the sample was equilibrated twice with 100 μl of water-saturated n-butanol. Glycans, in the aqueous phase, were dried in the Speed Vac concentrator and dissolved in either 40% 1-propanol (for TLC analysis) or water (for P4 analysis).
In vivo labeling of C. fasciculata cells.
Log-phase cells (108) were harvested by centrifugation and washed once in phosphate-buffered saline and once in RPMI 1640 (Gibco) containing 25 mM HEPES (pH 7.4), 10 μg of hemin/ml, 2% bovine serum albumin, and 3 mg of glycerol/ml (medium A). Cells were then incubated for 10 min at 27°C in 1 ml of medium A and then labeled with 100 μCi of [3H]Man (Dupont; 22.2 Ci/mmol) for 20 min with gentle shaking. The parasites were then centrifuged for 30 s in a microcentrifuge, and the pellet was extracted three times with 250 μl of CHCl3-CH3OH-H2O (10:10:3, vol/vol/vol). The lipids were recovered by n-butanol-water partition as described above. Total labeled lipids were subjected to mild acid hydrolysis, and the released N-glycans were recovered as described above.
Gel filtration.
Glycans were analyzed by Bio-Gel P4 chromatography with an Oxford Glycosystem GlycoMap and detected by liquid scintillation counting of the fractions. The size of the glycans was determined as glucose units (GU), relative to the elution positions of coinjected unlabeled d-Glc dextran oligomers (Oxford Glycosystems), which were detected by monitoring the refractive index.
RESULTS
ConA 1-1 mutants have a defect in the conversion of polyprenol to dolichol.
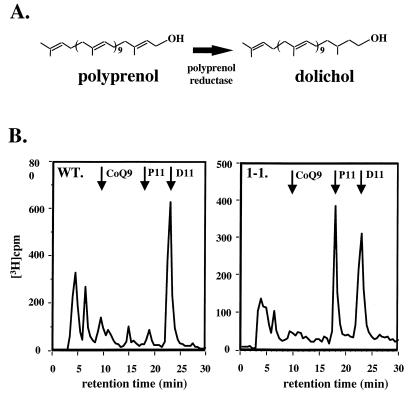
Since previous studies had shown that the glycosylation defect in a line of ConA-resistant CHO cells resulted from a lack of polyprenol reductase activity (13), we thought that a similar defect might be present in ConA 1-1. Polyprenol reductase converts polyprenol to dolichol by reducing the double bond in the isoprene unit proximal to the hydroxyl moiety (Fig. 2A). Lack of polyprenol reductase might contribute to an altered N-glycan structure, as it does in CHO cells (16). To test this possibility, we incubated cultures of WT and ConA 1-1 with [3H]mevalonolactone for four to five generations in order to achieve steady-state labeling of the prenyl lipids. We then isolated the total prenol fraction and analyzed the products by HPLC. We found that, whereas WT cells (Fig. 2B) synthesized predominantly (>95%) dolichol, ConA 1-1 had a striking defect in dolichol synthesis, making approximately equal amounts of dolichol and polyprenol (Fig. 2B). In both parental and mutant cells, radioactivity coeluted with the dolichol-11 internal standard, as expected from a previous study of trypanosome prenols (17). Based on the elution time of the radioactivity and internal standards, the prenol chain lengths in the WT and mutant were identical.
FIG. 2.
Identification of the prenol in WT T. brucei and ConA 1-1. (A) Synthesis of dolichol from polyprenol by polyprenol reductase. (B) Lipids, which were labeled during a 36-h incubation of growing cells with [3H]mevalonolactone, were extracted, saponified, and dephosphorylated. Prenols were then separated from sterols, and the prenols were analyzed on a silica column by normal-phase HPLC. The elution positions of coenzyme Q9 (CoQ9), polyprenol-11 (P11), and dolichol-11 (D11) were determined by absorbance (A210) of internal standards. The elution positions and the amounts of the radioactive products were determined by counting the entire fraction. The polar metabolites eluting at the breakthrough have not been characterized.
Since ConA 1-1 cells produced both prenol lipids, we next determined whether ConA 1-1 cells preferentially utilized dolichol or polyprenol for synthesis of oligosaccharide-PP lipid and Man-P lipid. We used CHCl3-CH3OH (2:1, vol/vol) to extract neutral and phosphorylated lipids (e.g., prenol, prenol ester, prenyl phosphate, Man-P lipid, and small oligosaccharide-PP lipids) and CHCl3-CH3OH-H2O (10:10:3) to extract larger oligosaccharide-PP lipids. Identification of the prenols in these fractions showed clearly that both WT and ConA 1-1 cells almost exclusively utilized dolichol as the prenol for phosphorylated lipids (Table 1).
TABLE 1.
Distribution of dolichol and polyprenol in phosphorylated and neutral prenols of parental and mutant cellsa
| Cell line | Prenol (%)
|
|||||
|---|---|---|---|---|---|---|
| Total
|
Phosphorylated
|
Neutral
|
||||
| Phosphorylated | Neutral | Dolichol | Polyprenol | Dolichol | Polyprenol | |
| WT | 81 ± 4 | 20 ± 4 | 98 ± 1 | 2.5 ± 1 | 77 ± 16 | 23 ± 16 |
| ConA 1-1 | 23 ± 19 | 77 ± 19 | 91 ± 7 | 9 ± 7 | 35 ± 9 | 65 ± 9 |
Cells were incubated at 27°C with [3H] mevalonolactone for at least 36 h. Labeled lipids were sequentially extracted into chloroform-methanol (2:1) and then into chloroform-methanol-water (10:10:3) (for analysis of total prenol). Lipids extracted into chloroform-methanol (2:1) were resolved into phosphorylated and neutral fractions by chromatography on a DEAE-cellulose column (for analysis of dolichol and polyprenol). All fractions were saponified, and the alkali-resistant lipids were extracted into ether. Phosphorylated lipids were treated with acid phosphatase. Labeled lipids were fractionated by gel filtration chromatography, and the labeled prenols were analyzed by normal-phase silica HPLC. The amount of radioactivity was normalized for cell number and in the majority of experiments for the recovery of labeled internal standards. There were four determinations (three experiments) with WT cells and three determinations (three experiments) with ConA 1-1 cells.
The data in Table 1 illustrate another important phenotype of the ConA 1-1 mutant. Whereas about 80% of the prenol (dolichol) in WT cells is phosphorylated (as either oligosaccharide-PP lipid, dolichyl phosphate, or Man-P-dolichol), only about 23% of the prenol is phosphorylated in ConA 1-1 cells. The majority of the neutral prenol in WT cells is dolichol, whereas most of the neutral prenol in ConA 1-1 cells is polyprenol (Table 1). Thus, even though ConA 1-1 synthesizes both dolichol and phosphorylated, glycosylated derivatives of dolichol, most of the polyprenol in these cells is not activated for use in glycosylation reactions.
ConA 1-1 mutants make N-glycan precursors with an oligosaccharide smaller than that of the WT.
The defect in polyprenol reductase could influence the glycan structure of the OSL precursor, and this, in turn, could account for the altered glycan structure on procyclin (16). We therefore studied the OSL precursor to determine whether it differed in structure between the WT and mutant cells. Our initial approach involved synthesis of glycolipids in a cell-free system consisting of washed trypanosome membranes. Incubation with UDP-GlcNAc and GDP-[3H]Man radiolabels both OSL and GPI precursors as well as their biosynthetic intermediates, and these species can be resolved by TLC (18). As shown in Fig. 3A, synthesis of the most polar [3H]Man-labeled species (marked OSL) was inhibited by tunicamycin. Since this species accumulated during a chase with nonradioactive GDP-Man and was sensitive to mild acid hydrolysis (data not shown), we concluded that it was a candidate N-glycan precursor. The major species synthesized in the presence of tunicamycin are PP1 and P3, the well-characterized GPI anchor precursors of procyclic trypanosomes (9, 10).
We next conducted a similar experiment to compare the glycolipid products from WT and ConA 1-1 cells (Fig. 3B). Although OSL was synthesized as expected in the WT cell-free system, it was not produced in the ConA 1-1 system. Instead, a less polar species, designated mutant OSL, accumulated. Consistent with mutant OSL being an N-glycan precursor, it was the most polar [3H]Man-labeled species, it accumulated during the chase, it was sensitive to mild acid hydrolysis, and its synthesis was inhibited by tunicamycin (data not shown). Based on all of these experiments, we tentatively concluded that mutant OSL was an oligosaccharide-PP-dolichol whose structure differed from that of the WT.
Structural characterization of OSL from WT and ConA 1-1 cells.
Since the TLC system in Fig. 3 could not distinguish whether the WT and mutant OSLs differed in their lipid or in their glycan component, we decided to study the glycans alone, i.e., free of the lipid. We labeled the glycans with [3H]Man in cell-free systems like those in Fig. 3 and purified the OSLs by TLC. The [3H]Man-labeled oligosaccharide from each OSL was cleaved using mild acid hydrolysis and then subjected to Bio-Gel P4 gel filtration (Fig. 4A) and TLC analysis (Fig. 5A).
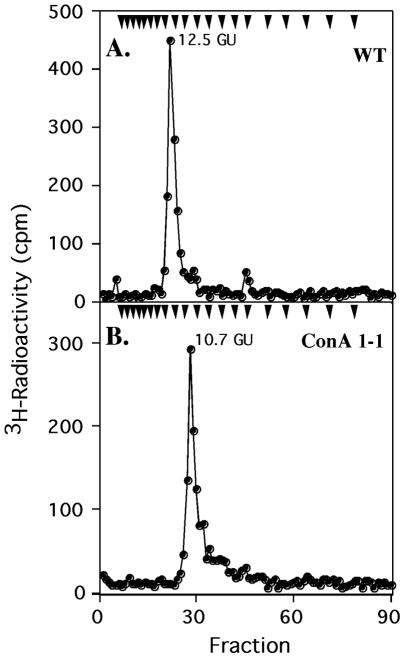
FIG. 4.
Bio-Gel P4 analyses of WT and ConA 1-1 OSL glycans. [3H]Man-labeled glycans (labeled in the cell-free system with GDP-[3H]Man and isolated from hydrolysates of TLC-purified OSL) from WT and ConA 1-1 cells were mixed with unlabeled dextran Glc oligomers and analyzed by P4 gel filtration. Arrowheads indicate elution positions of Glc oligomers, starting with Glc1 on the right.
In contrast to those from other trypanosomatid parasites (24), the OSL glycan structure of WT T. brucei had not been determined previously. Gel filtration on Bio-Gel P4 demonstrated that the WT glycan had a size of 12.5 GU, which is close to the reported value (12.3 GU) for a Man9GlcNAc2 glycan (39) (Fig. 4A). Similarly, TLC analysis (Fig. 5A, lane 3) revealed that it migrated considerably faster than Glc3Man9GlcNAc2, the well-characterized OSL species from WT K12 Chinese hamster ovary (CHO) cells (Fig. 5A, lane 1). Instead, it comigrated with an Rf similar to that of Man9GlcNAc2 (Fig. 5A, lane 2), the species found in MI8-5, a CHO mutant defective in glucosylation of the N-glycan precursor (28) and with an authentic nonradioactive Man9GlcNAc2 standard. The lack of OSL glucosylation is consistent with OSL structures in other trypanosomatids (24). Taken together, all these results indicate that the OSL in WT procyclic T. brucei is Man9GlcNAc2-PP-dolichol (model in Fig. 6).
FIG. 6.
Proposed structure and processing of WT and ConA 1-1 OSLs. After formation of WT OSL (containing Man9GlcNAc2) (A) and ConA 1-1 OSL (containing Man7GlcNAc2) (B), glycans are transferred to EP-procyclins. Whereas the WT glycan is efficiently transferred to procyclins, those from mutants are only partially transferred, forming underglycosylated proteins. Subsequently, all the α1-2Man residues are removed from glycans by specific mannosidases, forming procyclins bearing Man5GlcNAc2 (WT) or Man4GlcNAc2 (ConA 1-1). Whereas WT glycans are not further processed, most glycans in ConA 1-1 EP-procyclins are modified to hybrid types by the addition of an N-acetyllactosamine group to the terminal α1-3Man residue. *, Man residue in WT whose transfer is defective in ConA 1-1. “(+/−)” indicates that some procyclin glycosylation sites are unmodified in ConA 1-1 cells.
In contrast to the OSL glycan of WT cells, the ConA 1-1 mutant glycan eluted from the P4 column with an approximate size of 10.7 GU (Fig. 4B). This finding raised the possibility that the ConA 1-1 glycan is two hexoses smaller than that of the WT. Supporting that possibility is the observation that the glycan from ConA 1-1 OSL migrated on TLC with that from the C. fasciculata OSL, known to have a Man7GlcNAc2 structure (25) (Fig. 5B, compare lanes 3 and 4). To study the structures of these glycans in more detail, we then used Aspergillus saitoi α-mannosidase, an enzyme that selectively cleaves Manα1-2 linkages. This enzyme cleaved the OSL glycan from C. fasciculata (lane 5) or ConA 1-1 (lane 6) to produce the same product, Man4GlcNAc2.
DISCUSSION
Deficiency in polyprenol reductase.
Our first objective in this study was to evaluate whether a T. brucei ConA mutant is deficient in polyprenol reductase, as a defect in that enzyme resulted in ConA resistance in CHO cells (13). It is well established that the mutant CHO cells accumulate polyprenol, the immediate precursor of dolichol, rather than dolichol itself, and that they utilize polyprenol as the lipid carrier of oligosaccharides involved in N-glycosylation. As a result of having the incorrect lipid carrier, the major OSL intermediate in ConA-resistant CHO cells is Man5GlcNAc2-PP-polyprenol rather than Glc3Man9GlcNAc2-PP-dolichol. They also synthesize less OSL and transfer less glycan to protein. ConA-resistant CHO cells also accumulate larger amounts of neutral polyprenol relative to the level of neutral dolichol found in parental cells (13, 16). We find that ConA 1-1 cells are also deficient in polyprenol reductase activity, as they make equal amounts of dolichol and polyprenol (Fig. 2B). This finding contrasts with that for the CHO mutants, which make only polyprenol (16). In CHO cells, which are functionally haploid in many loci (36), the mutation is apparently in the one functional allele for polyprenol reductase. T. brucei, on the other hand, is functionally diploid at most loci. Therefore, if ConA 1-1 has a mutation in one allele, it would presumably have half the normal level of polyprenol reductase activity. Unfortunately, we could not measure activity directly, as an in vitro assay for this enzyme is not available. Previous cell fusion studies with mammalian cells suggested that polyprenol reductase activity is a rate-limiting step in dolichol synthesis (33). The same may be true in T. brucei, as ConA 1-1, presumably a heterozygote, makes equal amounts of polyprenol and dolichol. Interestingly, human cells with a similar phenotype, making both dolichol and polyprenol, have been isolated from patients with carbohydrate-deficient glycoprotein syndrome type I (23).
Although ConA 1-1 cells synthesize oligosaccharide-PP-dolichol and Dol-P-Man (Fig. 3B), they have a lower fraction of phosphorylated prenyl intermediates than do WT cells (Table 1). A reduced amount of the dolichol intermediates could be the cause of reduced protein glycosylation and the altered oligosaccharide-PP lipid in ConA 1-1 (Fig. 3B). Unfortunately, we could not provide proof that the phosphorylated dolichols are at a lower concentration in the ConA 1-1 mutant because of uncertainty of the specific radioactivities of the radiolabeled lipids. In mammalian cells, an inhibitor of high-mobility-group protein coenzyme A reductase (mevinolin or lovastatin) had been used during the labeling protocol in order to ensure that the specific activity of mevalonate was the same in different cell lines during the labeling reaction (34). Lovastatin, up to 50 μM, had no effect on the growth rate of parental T. brucei cells, and thus, it appeared that this drug was not taken up by these cells or did not inhibit the production of mevalonate or that prenol synthesis is not essential for growth. Thus, in our labeling experiments, we can compare the percentages of lipids in the various fractions from different cell types but we cannot compare the absolute amounts of lipids from one cell type to another.
The reason for accumulation of neutral prenol in ConA 1-1 cells is not known. However, it is possible that the accumulation is due to the substrate specificities of dolichol kinase, which prefers dolichol over polyprenol (15). Dolichyl phosphatase seems to hydrolyze both polyprenyl phosphate (14, 15) and dolichyl phosphate, and N-acetylglucosaminyl 1-phosphate transferase (19) and mannosylphosphoryldolichol synthase (20) both strongly prefer dolichyl phosphate over polyprenyl phosphate.
The N-glycan precursor in WT and ConA 1-1 cells.
The OSL in WT cells appeared to be Man9GlcNAc2, rather than the Glc3Man9GlcNAc2 found in higher eukaryotes. This assignment was based on TLC of the product of the cell-free system (Fig. 3) and the sensitivity of its synthesis to tunicamycin (Fig. 3). Furthermore, analysis of its oligosaccharide component (produced by mild acid hydrolysis) by TLC (Fig. 5) and P4 gel filtration provided evidence for the Man9GlcNAc2 structure shown in Fig. 6. A weakness of this conclusion is that a glucosyl transferase in our cell-free system could have been inactivated, even though we conducted some experiments in the presence of 1.5 mM UDP-glucose (data not shown). We tried to radiolabel the OSL precursor with [3H]Man in cultured trypanosomes, in hopes that we could compare the in vivo OSL with that produced in the cell-free system. However, we were unsuccessful in this labeling reaction despite multiple attempts under different conditions. Nevertheless, there are three reasons why it is highly unlikely that the OSL precursor in procyclic T. brucei is glucosylated. These are (i) we reproducibly produced Man9GlcNAc2 in the cell-free system, (ii) none of the trypanosomatids studied so far seem to glucosylate their OSL precursors (24), and (iii) Alg6, whose product (UDP-glucose OSL-glucosyltransferase) transfers the first αGlc to OSLs (29), is apparently absent in the nearly complete T. brucei genome database.
In contrast, experiments like those mentioned in the previous paragraph (Fig. 3, 4, and 5) provide strong evidence that the OSL precursor in ConA 1-1 cells has a truncated oligosaccharide Man7GlcNAc2 (see proposed structure in Fig. 6B). This structure suggests that the second α1-6-linked mannose (marked by an asterisk in the WT glycan, Fig. 6A) is not added to the growing oligosaccharide of the OSL. Without this mannose, and given the ordered pathway for assembly of the glycan, a terminal Manα1,2-Manα1,6 branch is not formed. This would yield the Man7GlcNAc2 structure shown. Subsequent processing of this mutant oligosaccharide after transfer to procyclin would involve removal of the three α1-2-linked mannose residues, resulting in the Man4GlcNAc2 intermediate shown in Fig. 6B. Final processing of this species would involve addition of the terminal N-acetyllactosamine, forming the mature hybrid-type glycan detectable on ConA 1-1 procyclin (Fig. 6B). Consistent with this processing model, mass spectrometry of glycans from ConA 1-1 procyclin has revealed small amounts of Man4GlcNAc2 and Man4GlcNAc3 species as well as the major species containing terminal N-acetyllactosamine (11). Furthermore, in addition to these procyclin species with the three different glycans, about 50% of ConA 1-1 EP-procyclin is not glycosylated at all (1). The fact that the vast majority of ConA 1-1 procyclins either are nonglycosylated or have hybrid glycans that are poorly bound by ConA explains the resistance of ConA 1-1 cells to ConA killing. This deficiency in glycosylation is not found in WT cells, where the procyclins are almost 100% occupied by a single Man5GlcNAc2 (1, 37). In fact, only high-mannose oligosaccharides ranging from Man9GlcNAc2 to Man5GlcNAc2 (the major species) are found linked to total glycoproteins from WT procyclic cells (12). Thus, in contrast to bloodstream forms (40), procyclic trypanosomes do not normally process the Man5GlcNAc2 intermediate to make either hybrid- or complex-type glycans. Man5GlcNAc2 glycan is the natural substrate for the UDP-GlcNAc-glycoprotein GlcNAc transferase type I in all systems studied so far, but that does not seem to be the case for the trypanosome enzyme. Thus, it is puzzling how procyclic cells are still capable of adding an N-acetyllactosamine to a Man4GlcNAc2 N-glycan as found in ConA 1-1 procyclins.
The fact that ConA 1-1 cells make an OSL with a Man7GlcNAc2 glycan instead of Man9GlcNAc2 suggests that the trypanosome homolog of ALG12, which encodes a dolichyl-P-Man:Man7GlcNAc2-PP-dolichyl α6-mannosyltranferase, might be also altered by the mutagenesis strategy and/or the activity of its product is affected by the dolichol deficiency.
Underglycosylation of procyclins could be explained if the T. brucei oligosaccharyl transferase complex is inefficient in recognizing and transferring precursors with glycans shorter than the Man9GlcNAc2 present in WT cells. This situation differs in other trypanosomatids, such as T. cruzi trypomastigotes, which can efficiently transfer into proteins N-glycans containing either Man7GlcNAc2 or Man9GlcNAc2 (7). Alternatively, as mentioned above, underglycosylation could be explained simply by low levels of dolichol-containing OSL.
Conclusion.
We found two biochemical defects in the ConA 1-1 mutant, one involving a deficiency in polyprenol reductase activity and the other concerning the structure of the glycan in the OSL. One possible interpretation of these results is that the primary defect resides in the gene for polyprenol reductase and that this deficiency causes an alteration in the structure of the OSL, much as in the case with CHO cells (16). The dolichol deficiency could affect OSL structure either directly or by reducing the concentration of dolichol-P-Man, a substrate required for maturation of the OSL. Alternatively, there could be two independent mutations in these heavily mutagenized cells, one in polyprenol reductase and the other in a glycosyl transferase involved in N-glycan precursor synthesis. Further studies, probably involving genetic techniques, will be required to distinguish between these possibilities.
Acknowledgments
This work was supported by National Institutes of Health grants AI21334 to P.T.E. and CA20421 to S.S.K. A.A.-S. was supported in part by a postdoctoral fellowship from CONICIT (Venezuela) and a Wellcome Trust Traveling Fellowship.
We thank Helen Lei for providing [3H]Man-labeled OSL samples used as standards, Terry Smith for generous assistance with the P4 analysis, and Mike Ferguson for use of the GlycoMap.
Footnotes
We dedicate this work to the loving memory of Viiu Klein.
REFERENCES
- 1.Acosta-Serrano, A., R. N. Cole, and P. T. Englund. 2000. Killing of Trypanosoma brucei by concanavalin A: structural basis of resistance in glycosylation mutants. J. Mol. Biol. 304:633-644. [DOI] [PubMed] [Google Scholar]
- 2.Acosta-Serrano, A., R. N. Cole, A. Mehlert, M. G. Lee, M. A. Ferguson, and P. T. Englund. 1999. The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem. 274:29763-29771. [DOI] [PubMed] [Google Scholar]
- 3.Acosta-Serrano, A., E. Vassella, M. Liniger, C. K. Renggli, R. Brun, I. Roditi, and P. T. Englund. 2001. The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc. Natl. Acad. Sci. USA 98:1513-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brun, R., and M. Shonenberger. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 5.Clayton, C. E., and M. R. Mowatt. 1989. The procyclic acidic repetitive proteins of Trypanosoma brucei. Purification and post-translational modification. J. Biol. Chem. 264:15088-15093. [PubMed] [Google Scholar]
- 6.Cross, G. A. 1996. Antigenic variation in trypanosomes: secrets surface slowly. Bioessays 18:283-291. [DOI] [PubMed] [Google Scholar]
- 7.Doyle, P., L. de la Canal, J. C. Engel, and A. J. Parodi. 1986. Characterization of the mechanism of protein glycosylation and the structure of glycoconjugates in tissue culture trypomastigotes and intracellular amastigotes of Trypanosoma cruzi. Mol. Biochem. Parasitol. 21:93-101. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, M. A. J., P. Murray, H. Rutherford, and M. J. McConville. 1993. A simple purification of procyclic acidic repetitive protein and demonstration of a sialylated glycosyl-phosphatidylinositol membrane anchor. Biochem. J. 291:51-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Field, M. C., A. K. Menon, and G. A. M. Cross. 1992. Developmental variation of glycosylphosphatidylinositol membrane anchors in Trypanosoma brucei. In vitro biosynthesis of intermediates in the construction of the GPI anchor of the major procyclic surface glycoprotein. J. Biol. Chem. 267:5324-5329. [PubMed] [Google Scholar]
- 10.Field, M. C., A. K. Menon, and G. A. M. Cross. 1991. A glycosylphosphatidylinositol protein anchor from procyclic stage Trypanosoma brucei: lipid structure and biosynthesis. EMBO J. 10:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwa, K. Y., A. Acosta-Serrano, K. H. Khoo, T. Pearson, and P. T. Englund. 1999. Protein glycosylation mutants of procyclic Trypanosoma brucei: defects in the asparagine-glycosylation pathway. Glycobiology 9:181-190. [DOI] [PubMed] [Google Scholar]
- 12.Hwa, K. Y., and K. H. Khoo. 2000. Structural analysis of the asparagine-linked glycans from the procyclic Trypanosoma brucei and its glycosylation mutants resistant to concanavalin A killing. Mol. Biochem. Parasitol. 111:173-184. [DOI] [PubMed] [Google Scholar]
- 13.Kaiden, A., A. G. Rosenwald, R. Cacan, A. Verbert, and S. S. Krag. 1998. Transfer of two oligosaccharides to protein in a Chinese hamster ovary cell B211 which utilizes polyprenol for its N-linked glycosylation intermediates. Arch. Biochem. Biophys. 358:303-312. [DOI] [PubMed] [Google Scholar]
- 14.Keller, R. K., W. L. Adair, Jr., N. Cafmeyer, F. A. Simion, B. Fleischer, and S. Fleischer. 1986. Characterization of polyisoprenyl phosphate phosphatase activity in rat liver. Arch. Biochem. Biophys. 249:207-214. [DOI] [PubMed] [Google Scholar]
- 15.Keller, R. K., G. D. Rottler, N. Cafmeyer, and W. L. Adair, Jr. 1982. Subcellular localization and substrate specificity of dolichol kinase from rat liver. Biochim. Biophys. Acta 719:118-125. [DOI] [PubMed] [Google Scholar]
- 16.Krag, S. S. 1998. The importance of being dolichol. Biochem. Biophys. Res. Commun. 243:1-5. [DOI] [PubMed] [Google Scholar]
- 17.Low, P., G. Dallner, S. Mayor, S. Cohen, B. T. Chait, and A. K. Menon. 1991. The mevalonate pathway in the bloodstream form of Trypanosoma brucei. Identification of dolichols containing 11 and 12 isoprene residues. J. Biol. Chem. 266:19250-19257. [PubMed] [Google Scholar]
- 18.Masterson, W. J., T. L. Doering, G. W. Hart, and P. T. Englund. 1989. A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell 56:793-800. [DOI] [PubMed] [Google Scholar]
- 19.McLachlan, K. R., and S. S. Krag. 1992. Substrate specificity of N-acetylglucosamine 1-phosphate transferase activity in Chinese hamster ovary cells. Glycobiology 2:313-319. [DOI] [PubMed] [Google Scholar]
- 20.McLachlan, K. R., and S. S. Krag. 1994. Three enzymes involved in oligosaccharide-lipid assembly in Chinese hamster ovary cells differ in lipid substrate preference. J. Lipid Res. 35:1861-1868. [PubMed] [Google Scholar]
- 21.Mehlert, A., A. Treumann, and M. A. Ferguson. 1999. Trypanosoma brucei GPEET-PARP is phosphorylated on six out of seven threonine residues. Mol. Biochem. Parasitol. 98:291-296. [DOI] [PubMed] [Google Scholar]
- 22.Nagamune, K., T. Nozaki, Y. Maeda, K. Ohishi, T. Fukuma, T. Hara, R. T. Schwarz, C. Sutterlin, R. Brun, H. Riezman, and T. Kinoshita. 2000. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 97:10336-10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohkura, T., K. Fukushima, A. Kurisaki, H. Sagami, K. Ogura, K. Ohno, S. Hara-Kuge, and K. Yamashita. 1997. A partial deficiency of dehydrodolichol reduction is a cause of carbohydrate-deficient glycoprotein syndrome type I. J. Biol. Chem. 272:6868-6875. [DOI] [PubMed] [Google Scholar]
- 24.Parodi, A. J. 1993. N-glycosylation in trypanosomatid protozoa. Glycobiology 3:193-199. [DOI] [PubMed] [Google Scholar]
- 25.Parodi, A. J., L. A. Quesada Allue, and J. J. Cazzulo. 1981. Pathway of protein glycosylation in the trypanosomatid Crithidia fasciculata. Proc Natl. Acad. Sci. USA 78:6201-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearson, T. W., R. P. Beecroft, S. C. Welburn, S. Ruepp, I. Roditi, K. Hwa, P. T. Englund, C. W. Wells, and N. B. Murphy. 2000. The major cell surface glycoprotein procyclin is a receptor for induction of a novel form of cell death in African trypanosomes in vitro. Mol. Biochem. Parasitol. 111:333-349. [DOI] [PubMed] [Google Scholar]
- 27.Quellhorst, G. J., Jr., C. W. Hall, A. R. Robbins, and S. S. Krag. 1997. Synthesis of dolichol in a polyprenol reductase mutant is restored by elevation of cis-prenyl transferase activity. Arch. Biochem. Biophys. 343:19-26. [DOI] [PubMed] [Google Scholar]
- 28.Quellhorst, G. J., Jr., J. L. O'Rear, R. Cacan, A. Verbert, and S. S. Krag. 1999. Nonglucosylated oligosaccharides are transferred to protein in MI8-5 Chinese hamster ovary cells. Glycobiology 9:65-72. [DOI] [PubMed] [Google Scholar]
- 29.Reiss, G., S. te Heesen, J. Zimmerman, P. W. Robbins, and M. Aebi. 1996. Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology 6:493-498. [DOI] [PubMed] [Google Scholar]
- 30.Richardson, J. P., R. P. Beecroft, D. L. Tolson, M. K. Liu, and T. W. Pearson. 1988. Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol. Biochem. Parasitol. 31:203-216. [DOI] [PubMed] [Google Scholar]
- 31.Roditi, I., M. Carrington, and M. Turner. 1987. Expression of a polypeptide containing a dipeptide repeat is confined to the insect stage of Trypanosoma brucei. Nature 325:272-274. [DOI] [PubMed] [Google Scholar]
- 32.Roditi, I., A. Furger, S. Ruepp, N. Schurch, and P. Butikofer. 1998. Unravelling the procyclin coat of Trypanosoma brucei. Mol. Biochem. Parasitol. 91:117-130. [DOI] [PubMed] [Google Scholar]
- 33.Rosenwald, A. G., P. Stanley, K. R. McLachlan, and S. S. Krag. 1993. Mutants in dolichol synthesis: conversion of polyprenol to dolichol appears to be a rate-limiting step in dolichol synthesis. Glycobiology 3:481-488. [DOI] [PubMed] [Google Scholar]
- 34.Rosenwald, A. G., J. Stoll, and S. S. Krag. 1990. Regulation of glycosylation. Three enzymes compete for a common pool of dolichyl phosphate in vivo. J. Biol. Chem. 265:14544-14553. [PubMed] [Google Scholar]
- 35.Ruepp, S., A. Furger, U. Kurath, C. K. Renggli, A. Hemphill, R. Brun, and I. Roditi. 1997. Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J. Cell Biol. 137:1369-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siminovitch, L. 1976. On the nature of hereditable variation in cultured somatic cells. Cell 7:1-11. [DOI] [PubMed] [Google Scholar]
- 37.Treumann, A., N. Zitzmann, A. Hulsmeier, A. R. Prescott, A. Almond, J. Sheehan, and M. A. Ferguson. 1997. Structural characterisation of two forms of procyclic acidic repetitive protein expressed by procyclic forms of Trypanosoma brucei. J. Mol. Biol. 269:529-547. [DOI] [PubMed] [Google Scholar]
- 38.Welburn, S. C., C. Dale, D. Ellis, R. Beecroft, and T. W. Pearson. 1996. Apoptosis in procyclic Trypanosoma brucei rhodesiense in vitro. Cell Death Differ. 3:229-236. [PubMed] [Google Scholar]
- 39.Yamashita, K., T. Mizuochi, and A. Kobata. 1982. Analysis of oligosaccharides by gel filtration. Methods Enzymol. 83:105-126. [DOI] [PubMed] [Google Scholar]
- 40.Zamze, S. E., D. A. Ashford, E. W. Wooten, T. W. Rademacher, and R. A. Dwek. 1991. Structural characterization of the asparagine-linked oligosaccharides from Trypanosoma brucei type II and type III variant surface glycoproteins. J. Biol. Chem. 266:20244-20261. [PubMed] [Google Scholar]