Abstract
Macrophages are able to polarize to proinflammatory M1 or alternative M2 states with distinct phenotypes and physiological functions. How metabolic status regulates macrophage polarization remains not well understood, and here we examine the role of mTOR (Mechanistic Target of Rapamycin), a central metabolic pathway that couples nutrient sensing to regulation of metabolic processes. Using a mouse model in which myeloid lineage specific deletion of Tsc1 (Tsc1Δ/Δ) leads to constitutive mTOR Complex 1 (mTORC1) activation, we find that Tsc1Δ/Δ macrophages are refractory to IL-4 induced M2 polarization, but produce increased inflammatory responses to proinflammatory stimuli. Moreover, mTORC1-mediated downregulation of Akt signaling critically contributes to defective polarization. These findings highlight a key role for the mTOR pathway in regulating macrophage polarization, and suggest how nutrient sensing and metabolic status could be “hard-wired” to control of macrophage function, with broad implications for regulation of Type 2 immunity, inflammation, and allergy.
Introduction
Macrophages play a dynamic role in host defense and maintenance of tissue homeostasis. This necessitates a delicate balance between their proinflammatory and immunomodulatory functions to ensure appropriate responses to environmental stimuli. Macrophages can be broadly classified into M1 (classical) and M2 (alternative) subtypes based on function. M1 macrophages are activated by LPS and/or IFN-γ to elaborate proinflammatory cytokine production and tissue inflammation 1. Conversely, M2 macrophages are stimulated by Th2 cytokines IL-4 and/or IL-13 to promote helminthic immunity, fibrosis, allergy, and immunomodulation 2. Stimulation of macrophages with IL-4 and IL-13 leads to activation of the transcription factor STAT6, which is indispensable for M2 polarization 3. Additionally, activation of the nuclear receptors PPARγ and PPARδ is necessary for full implementation of the M2 program 4, 5. A hallmark of M2 macrophages is an increase in Arginase-1 gene expression and activity 2, which converts L-arginine to L-ornithine to promote polyamine synthesis and tissue repair 6. The M2 program is also characterized by upregulation of C-type lectins, mannose receptor, chitinase family proteins, resistin-like molecules, and Interleukin-10, all of which contribute to immunomodulatory function 7. Importantly, distinct metabolic programs are required to support energy demands of M1 and M2 macrophages. M1 macrophages rely primarily on glycolytic metabolism, mediated by HIF-1α, while M2 macrophages utilize fatty acid oxidation mediated by PPARγ and the transcriptional coactivator, PGC-1β 3, 8, 9. This suggests that macrophage metabolism and inflammatory phenotype are integrally linked, and hint at additional regulatory control of macrophage polarization by metabolic pathways.
The Mechanistic Target of Rapamycin (mTOR) is a key nutrient/energy sensor that couples nutrient availability to regulation of downstream metabolic processes such as protein synthesis, glycolysis, and de novo lipogenesis 10, 11. mTOR, a serine/threonine kinase, exists in a rapamycin-sensitive complex called mTORC1 that is negatively regulated by the tuberous sclerosis complex comprised of TSC1 and TSC2 12. Genetic loss of either TSC1 or TSC2 leads to constitutive mTORC1 activation 13. Importantly, recent studies demonstrate that mTOR controls multiple aspects of T-cell biology including quiescence, activation, and differentiation 14. However, little is known regarding the role of mTOR in regulating macrophage activation.
In the current study, we elucidate a role for mTOR in macrophage polarization. We demonstrate that Tsc1Δ/Δ macrophages have a marked defect in M2 polarization in response to IL-4, while the inflammatory response to LPS is enhanced. Aberrant polarization is due, at least in part, to mTORC1-mediated attenuation of Akt activity, which renders Tsc1Δ/Δ macrophages resistant to the immunomodulatory effects of Akt downstream of IL-4 and LPS signaling. Lastly, we show that IL-4 and chitin administration in Tsc1Δ/Δ mice recapitulates the defective M2 polarization in vivo.
Results
Constitutive mTORC1 Activity Impairs M2 Polarization
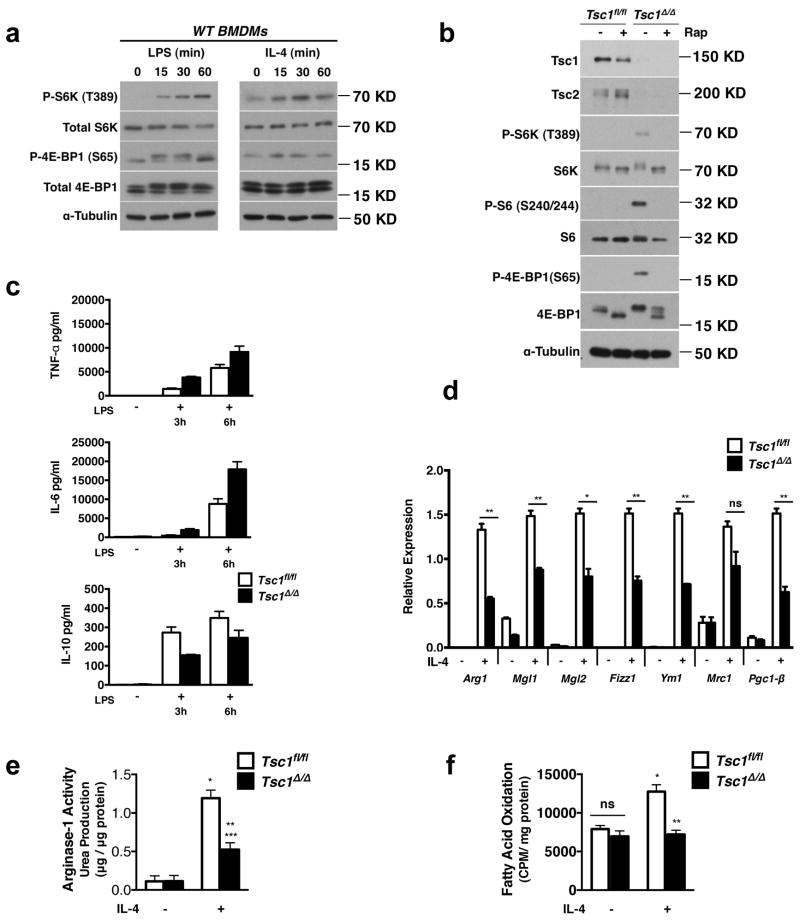
The mTOR pathway integrates a variety of inputs to regulate cell growth and to balance anabolic and catabolic processes 15. Interestingly, stimulation of bone marrow derived macrophages (BMDMs) with IL-4 or LPS resulted in mTORC1 activation as indicated by increased phosphorylation of the downstream targets S6K1 and 4E-BP1 (Fig. 1a), suggesting that the mTORC1 pathway may coordinate metabolic changes during macrophage activation. To examine this hypothesis, we utilized a model of myeloid-specific Tsc1-deficiency in which mTORC1 is constitutively active. Tsc1loxP/loxP LysMCre mice and Tsc1loxP/loxP controls are herein referred to asTsc1Δ/Δ and Tsc1fl/fl respectively. Immunoblotting confirmed that TSC1 is absent in the Tsc1Δ/Δ BMDMs (Fig. 1b). TSC2 protein level was also diminished (Fig. 1b), as TSC1 stabilizes TSC2 16. Tsc1Δ/Δ BMDMs displayed constitutive phosphorylation of the mTORC1 downstream targets S6K1 and 4E-BP1, as well as the S6K1 target ribosomal S6, all of which were sensitive to the mTORC1-specific inhibitor, rapamycin (Fig. 1b). Furthermore, Tsc1Δ/Δ BMDMs appeared to differentiate normally in vitro and expressed similar levels of the macrophage markers F4/80 and CD11b (Supplementary Fig. S1). As reported in other models of TSC-deficiency 17. Tsc1Δ/Δ BMDMs were larger (Supplementary Fig. S1) due likely to a role of mTORC1 in regulating cell size 18. These observations confirmed constitutive mTORC1 activation in Tsc1Δ/Δ BMDMs and established the validity of our genetic model.
Figure 1. Tsc1Δ/Δ BMDMs Have Defective M2 Polarization and Enhanced Responses to LPS stimulation.
a. Immunoblot analysis of WT BMDMs stimulated with LPS or IL-4 for 15–60 min as indicated. b. Immunoblot analysis of lysates from Tsc1fl/fl and Tsc1Δ/Δ BMDMs treated with or without rapamycin for 15h. c. Measurement of TNF-α, IL-6, and IL-10 secretion by ELISA after treatment with LPS for 3h and 6h, (n=2 representative experiments). d. Expression of M2 genes in Tsc1fl/fl and Tsc1Δ/Δ BMDMs after treatment with IL-4 for 24h (n=3). *p<0.05, **p<0.01, ***p<0.001.e. Urea production normalized to total protein in Tsc1fl/fl and Tsc1Δ/Δ BMDMs stimulated as in (c), (n=4), *p<0.001 for untreated vs IL-4 for Tsc1fl/fl, **p<0.01 for IL-4 treated Tsc1fl/fl vs Tsc1Δ/Δ, ***p<0.05 for untreated vs IL-4 for Tsc1Δ/Δ. f. Fatty acid oxidation of 3H-palmitic acid presented as counts per minute normalized to mg of total protein after 36h treatment with IL-4, (n=3). *p<0.01 for untreated vs IL-4 in Tsc1fl/fl, **p<0.01 for IL-4 treated Tsc1fl/fl vs Tsc1Δ/Δ. Graphs are shown as mean ± SEM. P-values were determined using Student’s t-tests.
To assess macrophage polarization in Tsc1Δ/Δ BMDMs, we used LPS treatment to promote an M1-like phenotype and IL-4 stimulation to induce an M2 phenotype. We found that LPS-treated Tsc1Δ/Δ BMDMs secreted more of the proinflammatory cytokines IL-6 and TNF-α, but less of the anti-inflammatory cytokine IL-10 (Fig. 1c). Given the enhanced responses of Tsc1Δ/Δ BMDMs to LPS, we postulated that M2 polarization could be defective. Indeed, Tsc1Δ/Δ BMDMs failed to fully upregulate the M2 program, with significant reductions in Arg-1, Mgl1, Mgl2, Ym1, Fizz1 and Pgc1-β expression (Fig. 1d). Arginase-1 activity assessed by urea production was reduced, correlating with lower levels of Arg-1 mRNA (Fig. 1e). IL-4-stimulated fatty acid oxidation, another hallmark feature of M2 macrophages, was also defective (Fig. 1f). This finding is consistent with the 3-fold reduction in Pgc1-β (Fig. 1d), a known mediator of fatty acid oxidation in M2 macrophages 9. Interestingly, M2c polarization triggered by IL-10 stimulation was also deficient in Tsc1Δ/Δ BMDMs (Supplementary Fig. S2a), which may indicate impaired orchestration of anti-inflammatory responses during tissue remodeling or wound healing 19. Macrophage activation to the M2b phenotype (by treatment with LPS/immune complexes), which has features of both M1 and M2 macrophages 20, 21, was not affected (Supplementary Fig. S2b). Taken together, our findings demonstrate that aberrant mTORC1 activation critically modulates macrophage polarization. Impaired induction of Pgc-1β and fatty acid oxidation also highlights a key role of mTOR in orchestrating macrophage cellular metabolism.
STAT6 and PPARγ Activity are Normal in Tsc1Δ/Δ BMDMs
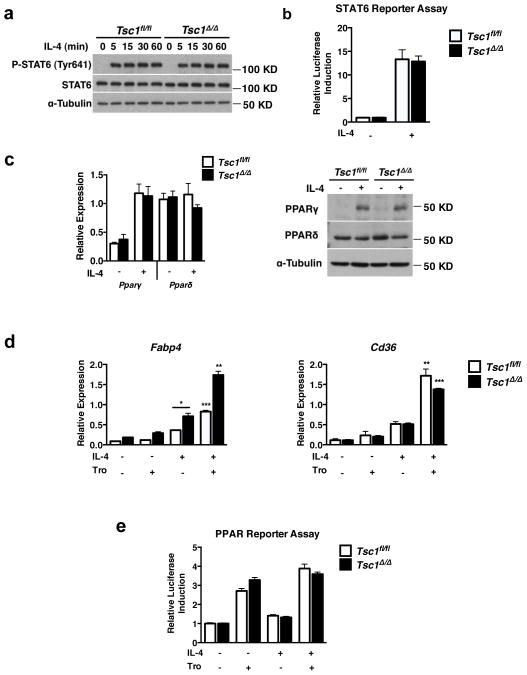
IL-4R signaling leads to activation of JAK1/JAK3 and tyrosine phosphorylation of the transcription factor STAT6, enabling nuclear translocation 2 and induction of target genes such as Pparγ and Arg-13. To interrogate the mechanism underpinning defective M2 polarization in Tsc1Δ/Δ BMDMs, we first examined signal transduction downstream of the IL-4R. Tsc1Δ/Δ BMDMs expressed comparable levels of Jak1, Jak3, and IL-4rα mRNAs as well as STAT6 protein at basal state (Fig. 2a, Supplementary Fig. S3a). Following IL-4 stimulation, STAT6 was tyrosine phosphorylated consistent with normal activation (Fig. 2a). Chromatin immunoprecipitation assays indicated comparable IL-4- induced recruitment of STAT6 to the promoter of the Arg1 gene (Supplementary Fig. S3b). Furthermore, IL-4-inducible STAT6 transcriptional activity as measured by a STAT6 reporter assay was unaffected (Fig. 2b). Finally, normal induction of some M2 genes inTsc1Δ/Δ BMDMs, including the STAT6-dependent gene Pparγ 22 (Fig. 2c), indicates no general defect in expression or activation of STAT6, the master regulator of M2 gene induction. IL-4 signaling can activate STAT1 in some contexts 23, 24, but not in macrophages (Supplementary Fig. S3c) consistent with a prior report, excluding a role for impaired STAT1 activation in the phenotype of Tsc1Δ/Δ BMDMs 25.
Figure 2. STAT6 and PPARγ Activity are Normal in Tsc1Δ/Δ BMDMs.
a. Immunoblot analysis of lysates from Tsc1fl/fl and Tsc1Δ/Δ BMDMs stimulated with IL-4 for 5–60min. b. STAT6 luciferase reporter assay in Tsc1fl/fl and Tsc1Δ/Δ BMDMs. Data shown as fold induction of firefly luciferase activity normalized to renilla luciferase for IL- 4 treatment relative to untreated sample (n=2 experiments performed in duplicate). c. Gene expression and immunoblots for PPARγ and PPARδ in Tsc1fl/fl and Tsc1Δ/Δ BMDMs after treatment with IL-4 for 24h. Gene expression data is shown as mean ± SEM (n=3). d. Expression of PPARγ-dependent genes in Tsc1fl/fl and Tsc1Δ/Δ BMDMs treated with IL-4 in the presence or absence of troglitazone for 24h. DMSO vehicle was used as control, ( n=3). *p<0.05 for IL-4 treated Tsc1fl/fl and Tsc1Δ/Δ, **p<0.01, ***p<0.001 for IL-4 versus IL-4+Tro. e. PPAR luciferase reporter assay in Tsc1fl/fl and Tsc1Δ/Δ BMDMs. Data shown as fold induction of firefly luciferase activity normalized to renilla luciferase for IL-4 or troglitazone treatment relative to untreated sample (representative of 3 experiments performed in triplicate). Graphs are shown as mean ± SEM. P-values determined using Student’s t-tests.
Because previous studies indicate a key role for the nuclear receptors PPARγ and PPARδ in M2 polarization, we next turned to an analysis of their expression and activity in Tsc1Δ/Δ BMDMs. First, we showed comparable expression of PPARγ and PPARδ in Tsc1fl/fl and Tsc1Δ/Δ BMDMs, at basal state and after IL-4 stimulation (Fig. 2c). Since expression does not necessarily reflect functional activity, we examined PPARγ activity in Tsc1Δ/Δ BMDMs. We found that IL-4 induced comparable expression of the canonical PPARγ-dependent genes Fabp4 22, 26 and Cd36 27 (Fig. 2d). Furthermore, the PPARγ agonist troglitazone synergized with IL-4 to a similar extent in Tsc1fl/fl and Tsc1Δ/Δ BMDMs (Fig. 2d). Analogous findings were obtained with the PPARδ-dependent gene Atgl 28 using the PPARδ agonist GW501516 (Supplementary Fig. S4). To corroborate gene expression data, we used a PPAR reporter assay to assess PPAR transcriptional activity, and found commensurate induction in Tsc1fl/fl and Tsc1Δ/Δ BMDMs (Fig. 2e). Taken together, these findings indicate a selective defect in M2 polarization in Tsc1Δ/Δ BMDMs that may not be due to defects in STAT6, PPARγ, or PPARδ expression or activity.
mTORC1 Activity Attenuates IL-4-Induced Akt Activation
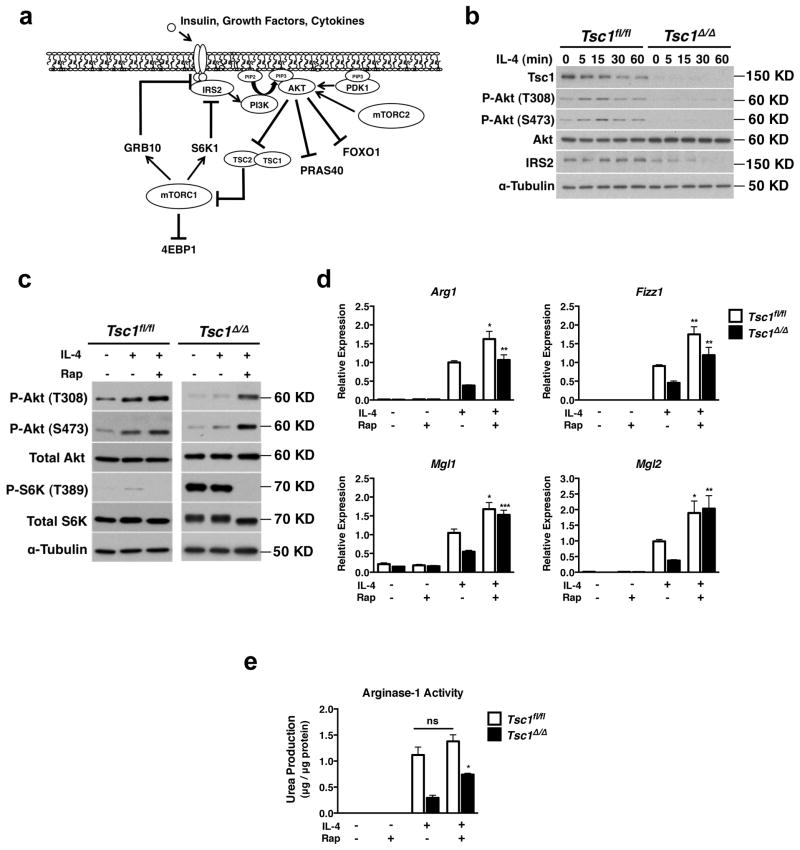
The data above suggests that Tsc1Δ/Δ BMDMs may not be able to activate some signals downstream of IL-4R. We turned our attention to IRS2/PI3K/Akt signaling, since this pathway is engaged by the IL-4R in parallel to the JAK/STAT6 pathway 29, 30. PI3K activation leads to increased activity of mTORC2, which phosphorylates Akt at S473 to activate the protein and promote membrane localization. In addition to S473, Akt is critically controlled by phosphorylation at T308, a step mediated by PDK1 (Fig. 3a). Thus we examined S473 and T308 phosphorylation as readouts of Akt activity, as well as phosphorylation of the downstream Akt targets FOXO1, PRAS40 and GSK-3 31 (Fig. 3a).
Figure 3. Constitutive mTORC1 Activity Attenuates IL-4-Induced Akt Activation.
a. Overview of mTORC1 signaling downstream of IL-4, insulin, and growth factors. Receptor activation engages the IRS/PI3K/Akt pathway. PI3K converts PIP2 to PIP3 thus recruiting PDK1 and Akt to the plasma membrane, enabling PDK1-mediated phosphorylation of Akt on T308. PI3K also activates mTORC2, which phosphorylates Akt on S473. Thus activated, Akt can phosphorylate downstream targets to regulate their activity. One consequence of Akt activation is increased mTORC1 activity, which feeds back to attenuate IRS2/PI3K/Akt signaling through multiple mechanisms, including reducing levels of IRS2 while increasing levels of GRB10. b. Immunoblot analysis of lysates from Tsc1fl/fl and Tsc1Δ/Δ BMDMs stimulated with IL-4 for 5–60 min. c. Immunoblot analysis of lysates from Tsc1fl/fl and Tsc1Δ/Δ BMDMs stimulated with IL-4 for 20 min in the presence or absence of rapamycin (20nM, 1h pretreatment). DMSO vehicle was used as control. d. Expression of M2 genes in Tsc1fl/fl and Tsc1Δ/Δ BMDMs after treatment with IL-4 for 15h in the presence or absence of rapamycin (20nM, 1h pretreatment). (n=5). *p<0.05, **p<0.01, ***p<0.001 for IL-4 versus IL-4+rap. e. Urea production normalized to total protein in Tsc1fl/fl and Tsc1Δ/Δ BMDMs stimulated with IL- 4 for 20h in the presence or absence of rapamycin (20nM, 1h pretreatment). (n=5),*p<0.001. Graphs are shown as mean ± SEM. P-values were determined using Student’s t-tests.
Interestingly, IL-4-stimulated Tsc1Δ/Δ BMDMs displayed a striking attenuation in Akt signaling as indicated by reduced phosphorylation of AktS473 and AktT308 (Fig. 3b). Consistently, phosphorylation of the Akt targets FOXO1 and PRAS40 was diminished (Supplementary Fig. S5a). GSK-3 phosphorylation was not reduced (Supplementary Fig. S5a), perhaps because of its regulation by multiple inputs 32, 33, 34, 35. Importantly, diminished Akt signaling has been noted in TSC-deficient cells during stimulation with insulin and other growth factors 36, 37, 38. Such reduction of Akt activity is due to mTORC1-mediated negative feedback that impinges on multiple targets, including but not limited to IRS1/2 degradation 39, 40 and phosphorylation and stabilization of GRB10 41, 42 (Fig. 3a). While such mTORC1-mediated negative feedback is well-defined for insulin signaling, little is known regarding its role in the regulation of cytokine signaling. Interestingly, we found that Tsc1Δ/Δ BMDMs display reduced IRS2 levels in response to IL-4 treatment (Fig. 3b). This is likely to contribute to mTORC1-mediated attenuation of Akt signaling, since IRS2 has been implicated in Akt activation during IL-4 stimulation 29. Increased levels of phosphorylated and total GRB10 (Supplementary Fig. S5b) may also play a role given that GRB10 inhibits signaling downstream of RTKs 43, 44, 45. Thus increased mTORC1 activity in our model is likely to attenuate Akt signaling at least in part through IRS2 and GRB10. PDK1, the Akt T308 kinase, has constitutive kinase activity and is critically regulated by PIP3-mediated recruitment to the plasma membrane 46, thus normal PDK1 activity in Tsc1Δ/Δ BMDMs (Supplementary Fig. S5c) also supports our model that attenuated Akt signaling in Tsc1Δ/Δ BMDMs occurs at a receptor proximal step upstream of PI3K mediated PIP3 production (Fig. 3a). Finally, LPS-mediated Akt activation was diminished in Tsc1Δ/Δ BMDMs, as evidenced by a defect in phosphorylation of Akt and the Akt target FOXO1 (Supplementary Fig. S5d). Collectively, these findings demonstrate that mTORC1- mediated negative feedback mechanisms converge to ultimately attenuate Akt signaling in Tsc1Δ/Δ BMDMs.
Having shown a defect in Akt activation in Tsc1Δ/Δ BMDMs, we addressed a potential role in impaired M2 polarization. Previous studies have used rapamycin treatment to alleviate mTORC1-mediated negative feedback of Akt signaling and to interrogate the role of attenuated Akt activation in TSC-deficient models 36, 37, 38. Accordingly, we found that rapamycin treatment of Tsc1Δ/Δ BMDMs rescued IL-4 inducible Akt signaling (Fig. 3c). Importantly, such treatment restored induction of M2 genes Arg1, Fizz1, and Mgl1/2 as well as Arginase activity (Fig. 3d,e). Together this suggests that deficient M2 polarization in Tsc1Δ/Δ BMDMs may be due to mTORC1-mediated negative feedback of Akt signaling. We note that rapamycin treatment of control Tsc1fl/fl BMDMs also modestly increased Akt signaling (Fig. 3c) as well as M2 responses (Fig. 3d,e), indicating that acute, signal-dependent activation of mTORC1 during IL-4 signaling can also feedback to inhibit Akt activation.
Attenuated Akt signaling underlies aberrant polarization
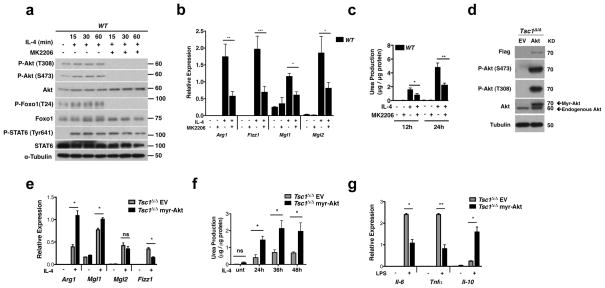
Our findings linking reduced Akt activation to impaired M2 polarization in Tsc1Δ/Δ BMDMs (Fig. 3b–e) are interesting given that little is known regarding the role of Akt in this process. To address this directly, we treated WT BMDMs with MK-2206, an allosteric inhibitor of Akt. This led to a decrease in IL-4-inducible phosphorylation of AktT308 and AktS473 as well as the Akt target FOXO1, but did not affect STAT6 phosphorylation (Fig. 4a). Importantly, pretreatment with MK-2206 reduced IL-4 mediated induction of Arg1, Fizz1, Mgl2, and Mgl1 (Fig. 4b), as well as Arginase-1 activity (Fig. 4c). Similar effects were observed with the structurally distinct Akt inhibitor Aktviii (Supplementary Fig. S6a–b), indicating the specificity of the inhibitors. These findings argue that Akt may play an important role in M2 polarization. Together with the data in Figure 3, they also support the idea that attenuated Akt signaling underpins defective M2 polarization in Tsc1Δ/Δ BMDMs.
Figure 4. Akt Signaling is Critical For Polarization in Tsc1Δ/Δ BMDMs.
a. Immunoblot analysis of WT BMDMs pretreated with MK-2206 for 1h and treated with IL-4 for the indicated time points. b and c. WT BMDMs were pretreated with MK-2206 or DMSO for 1h before stimulation with IL-4 for 24h and examination of (b) M2 gene expression (n=3) or (c) urea production (n=4). d. Immunoblot analysis of Tsc1Δ/Δ BMDMs transduced with myr-flag-Akt or empty vector (EV). e and f. Myr-Akt Tsc1Δ/Δ BMDMs and EV Tsc1Δ/Δ BMDMs were stimulated with IL-4 followed 24h later by analysis of (e) M2 gene expression (n=4 representative experiments) or (f) urea production (n=3). g. Cytokine gene expression in myr-Akt Tsc1Δ/Δ BMDMs and EV Tsc1Δ/Δ BMDMs stimulated with LPS for 6 hours (n=3). Graphs are shown as mean ± SEM,*p<0.05, **p<0.01, ***p<0.001. P-values were determined using Student’s t-tests.
Next, we took a genetic approach to rescue Akt signaling in Tsc1Δ/Δ BMDMs. As expected, retroviral transduction of Tsc1Δ/Δ BMDMs with myristylated-Akt1 (myr-Akt) led to constitutive Akt signaling as indicated by high basal state P-AktT308 and P-AktS473 (Fig. 4d). Importantly, this was associated with increased induction of Arg1 and Mgl1 (Fig. 4e), as well as Arginase-1 activity (Fig. 4f), following IL-4 stimulation. Although ectopic expression of myr-Akt was insufficient to rescue Fizz1 and Mgl2 expression, this could be achieved in the context of rapamycin co-treatment (Supplementary Fig. S6c). Thus, myr-Akt may not fully recapitulate IL-4 inducible Akt activation; or alternatively, rescue of Akt signaling is not sufficient for restoring Mgl2 and Fizz1 expression, and some other consequence of constitutive mTORC1 activation is critically relieved by rapamycin treatment. Finally, to extend these studies to the proinflammatory responses, we examined myr-Akt expressing Tsc1Δ/Δ BMDMs following LPS treatment. We observed significantly reduced expression of Il-6 and Tnfα but increased expression of Il-10 (Fig. 4g and Supplementary Fig. S6d).
Deficient M2 polarization in Tsc1Δ/Δ mice
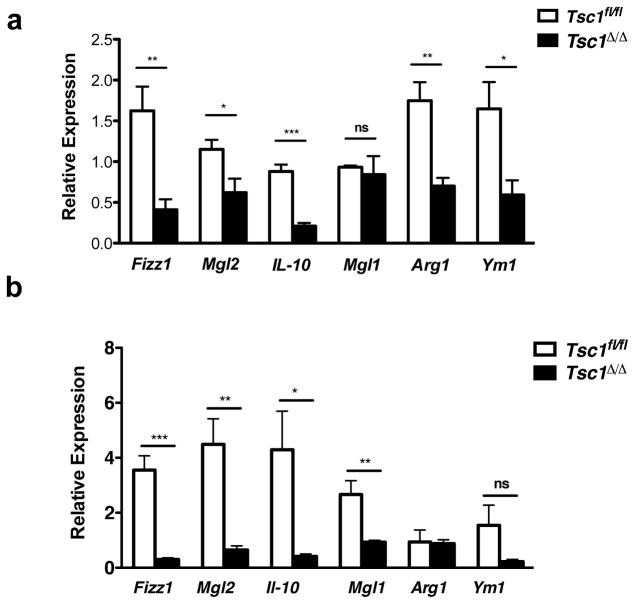
Finally, we asked if TSC1-deficiency would impair M2 polarization in vivo. We used intraperitoneal (IP) injection of an IL-4/IL-4 antibody complex to elicit IL-4-dependent recruitment and in situ proliferation of M2 macrophages 47 in Tsc1fl/fl and Tsc1Δ/Δ mice. Strikingly, induction of most M2 genes was decreased in peritoneal exudate cells (PECs) fromTsc1Δ/Δ mice (Fig. 5a). To corroborate these findings, we used a model of chitin administration that triggers IL-4-dependent recruitment and polarization of M2 macrophages 48, 49. We observed a near universal reduction of M2 gene expression in Tsc1Δ/Δ PECs (Fig. 5b), similar to the IL-4 injection. Collectively the findings support our model that constitutive mTORC1 activity can attenuate macrophage M2 polarization in vivo.
Figure 5. M2 Polarization of Tsc1Δ/Δ Mice is Impaired in vivo.
a. M2 gene expression in PECs from Tsc1Δ/Δ and Tsc1fl/fl mice 4 days post IP injection with IL-4 complex on days 0 and 2 (n= 4 mice per genotype). b. M2 gene expression in PECs from male Tsc1Δ/Δ and Tsc1fl/fl mice 48h post IP injection with chitin (n=5 mice per genotype). Data shown as mean ± SEM, *p<0.05, **p<0.01, ***p<0.001. P-values were determined using Student’s t-tests.
Discussion
In this study, we use a novel model of myeloid-specific Tsc1 deletion and constitutive mTORC1 activity to elucidate mTORC1 function in macrophages. We found that Tsc1Δ/Δ BMDMs have enhanced proinflammatory cytokine production while IL-10 secretion is reduced, in line with a recent analysis of Tsc1Δ/Δ BMDMs 17. Other studies reached conflicting conclusions, using shRNA knockdown in monocytes and dendritic cells 50, 51. Extending the analysis of mTORC1 function in macrophages, we showed that Tsc1Δ/Δ BMDMs are impaired in M2 polarization, expressing reduced levels of key M2 markers such as Arg-1, Fizz1, Mgl1, Mgl2, Ym1 and Pgc-1β (Fig. 1d). Interestingly, the defect in M2 activation seems to be selective, since induction of PPARγ and some PPARγ dependent genes (e.g. Cd36 and Fabp4) occur normally (Fig. 2c,d). Additionally we find that Tsc1Δ/Δ BMDMs have diminished levels of Arginase-1 activity and fatty acid oxidation, hallmark features of M2 macrophages (Fig. 1e,f). This suggests that mTORC1 may couple regulation of fatty acid oxidation to control of macrophage polarization, consistent with the emerging view that macrophage cellular metabolism is closely linked to activation status 3, 8, 52. Finally, we use models of IL-4 and chitin injection to show that constitutive mTORC1 activity in myeloid lineage cells results in broad defects in M2 polarization in vivo (Fig. 5).
Normal activation of known regulators of M2 polarization, including STAT6, PPARγ and PPARδ (Fig. 2), suggests that impaired M2 polarization in Tsc1Δ/Δ BMDMs may be due to block of a parallel signaling pathway. In addition to the JAK/STAT6 pathway, IL-4R signaling engages IRS2/PI3K signaling in parallel to mediate Akt activation 29, 30. While STAT6 is indispensable for M2 polarization 1, 53, the role of Akt signaling has not been well characterized. Our findings suggest a critical role for mTORC1-mediated feedback inhibition of Akt signaling in Tsc1Δ/Δ BMDMs. In support of this, Tsc1Δ/Δ BMDMs display decreased IL-4-inducible Akt activation, as indicated by diminished P-AktT308 and P-AktS473 (Fig. 3b) and phosphorylation of the Akt targets FOXO1 and PRAS40 (Supplementary Fig. S5a). Importantly, rapamycin treatment (Fig. 3c–e) and ectopic expression of myr-Akt (Fig. 4d–f, Supplementary Fig. S6c) restore Akt activation simultaneous with rescue of M2 gene expression and Arginase activity. We suggest that decreased IRS2 (Fig. 3b) but increased GRB10 (Supplementary Fig. S5b) levels may contribute to mTORC1-mediated attenuation of Akt signaling, since IRS2 has been implicated in IL-4R signaling 54, while GRB10 downregulates signaling downstream of RTKs 43. While other negative feedback mechanisms have been described in insulin signaling 40, 55 and may exist in our setting, they act synergistically and ultimately converge to attenuate Akt activation. Finally, we believe that feedback inhibition to Akt may also underlie the enhanced responsiveness of Tsc1Δ/Δ BMDMs to LPS stimulation. While we have not extensively characterized the underlying mechanism(s) in this context, Akt activation in Tsc1 Δ/Δ BMDMs is diminished following LPS signaling (Supplementary Fig. S5d), and its rescue with myr-Akt expression reduces the exaggerated responses (Fig. 4g).
Importantly, control of M2 polarization by Akt signaling is likely to extend beyond our genetic model to other settings, since pharmacological inhibition of Akt impairs M2 activation in wild-type BMDMs (Fig. 4b,c, Supplementary Fig. S6a,b). Moreover, rapamycin treatment of control BMDMs modestly increases Akt signaling and M2 responses (Fig. 3c–e). Thus our findings reveal a largely unappreciated role for Akt in synergizing with the STAT6 pathway to regulate full M2 polarization (Fig. 6a). Whether Akt promotes or inhibits inflammation downstream of LPS signaling is not entirely clear 56, but at least in Tsc1Δ/Δ BMDMs, simultaneous mTORC1 activation and Akt attenuation lead to enhanced proinflammatory responses that can be rescued by restoring Akt signaling (Fig. 4g). The relevant Akt targets that regulate macrophage polarization are not well defined, but could include FOXO1 and CEBPβ 57, 58. Interestingly, macrophages deficient in Rictor, a subunit of the mTORC2 complex that phosphorylates Akt on S473, were deficient in some but not all Akt-dependent activities (Supplementary Fig. S7a) consistent with previous models of mTORC2- deficiency 59, 60 but polarized normally to the M2 phenotype (Supplementary Fig. S7b–c), and could serve as a plausible model to identify the relevant Akt targets controlling M2 activation.
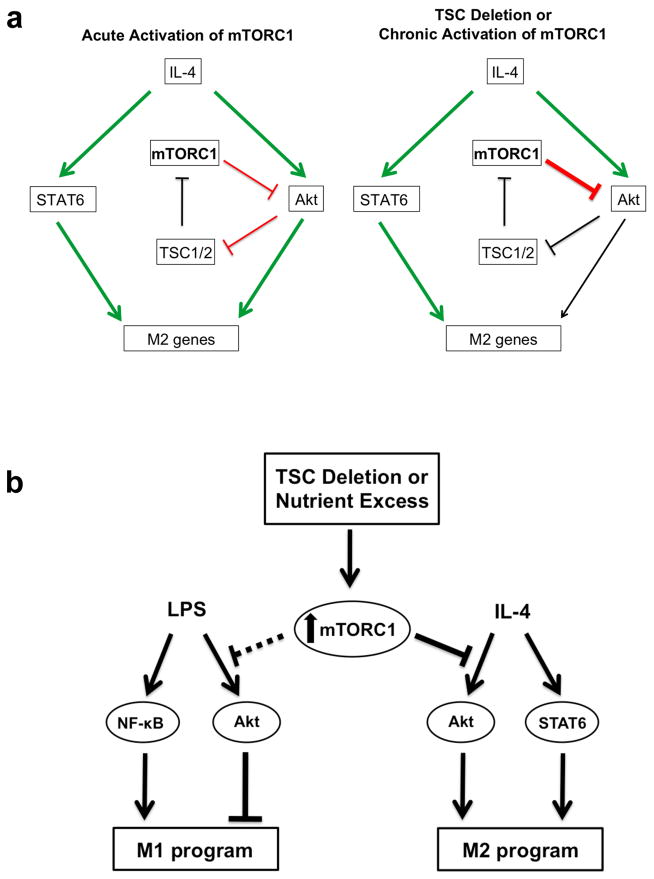
Figure 6. Model.
Proposed model for how mTORC1 activity controls macrophage polarization. a. Physiological induction of the Akt-mTORC1 signaling loop by IL-4 stimulation (left) allows for transient, inducible activation of the pathway, and enables Akt to synergize with the JAK/STAT pathway for M2 polarization. mTORC1 activity is also regulated by nutrient availability (not shown here), so such wiring of the signaling pathway may allow calibration of M2 activation to metabolic status (left). In contrast, constitutive or aberrant mTORC1 activation corrupts this signaling pathway and modulation of macrophage activation by metabolic inputs (right). (Green = activation; Red = inhibition; Black = attenuated) b. Constitutive or aberrant activation of mTORC1 impairs the ability of macrophages to respond appropriately to polarizing stimuli. A critical mediator of this process is Akt, whose activity is downregulated by increased mTORC1 activity.
Our studies indicate the existence of a mTORC1-Akt regulatory loop in the IL-4 signaling pathway that parallels that of the insulin pathway. In the latter, a feedback loop between mTORC1 and Akt—in which receptor engagement of the IRS/PI3K/Akt pathway leads to mTORC1 activation that feeds back to attenuate Akt signaling—is critical for transient, signal-dependent activation of these two signaling modules (Fig. 3a). Similarly, activation of the IRS2/PI3K/Akt pathway by IL-4 mediates mTORC1 activation (Fig. 1a, Supplementary Fig. S8) and as shown here, results in feedback inhibition of Akt signaling (Fig. 3a–c, 6a). Importantly, mTORC1 activity is critically modulated by nutrient/energy availability 61, thus we propose that integration of the mTORC1-Akt regulatory loop into the IL-4 signaling pathway may allow macrophages to calibrate their activation and function to metabolic status and nutrient availability (Fig. 6a). In contrast, this regulatory circuitry is disrupted by constitutive or aberrant activation of mTORC1, as occurs during nutrient excess or in our genetic model. As a consequence, induction of M2 polarization by the synergistic interactions of the Akt and JAK/STAT pathways is impaired (Fig. 6a,b). Conversely, elevated mTORC1 activity and consequent downregulation of Akt signaling may facilitate increased responses to LPS stimulation (Fig. 6b). Finally, we note that such “hard-wiring” of mTORC1-Akt signaling to cytokine signaling could have relevance to other immunological contexts, given that many cytokines that regulate immune cell function engage the Akt pathway (and presumably also mTORC1).
In conclusion, our study highlights a key role of the mTORC1 pathway in control of macrophage polarization. Such control is likely to be of particular relevance for adipose tissue macrophage (ATMs). In the lean state, ATMs with a M2 phenotype maintain an anti-inflammatory environment and adipocyte insulin sensitivity, while in obesity, ATMs with a M1 phenotype produce inflammatory cytokines and promote insulin resistance and metabolic dysfunction. We propose that nutrient sensing by mTORC1 may directly regulate the pathophysiological switch of ATMs during obesity, extending the current model for regulation of the pathophysiological M2 to M1 switch 62. Indeed, given that macrophages are critical orchestrators of diverse physiological responses, regulation of macrophage activation by the mTOR pathway may have profound consequences in many settings, including helminth infection, inflammation, allergy, and tissue repair.
Methods
BMDMs
Briefly, femurs were removed from mice after CO2 euthanasia, and cells were liberated using a mortar and pestle. For macrophage differentiation, bone marrow derived cells were plated in petri dishes with 1640 RPMI media (10% FCS, Penicillin/Streptomycin, 2mM L-Glutamine) supplemented with MCSF-containing L929 cell supernatant for seven days. MCSF differentiated macrophages were harvested and plated in tissue culture dishes for subsequent experiments. For M1-like activation, 0.5-0.7×106 BMDMs were plated in 12-well tissue culture dishes and treated with 10ng/ml LPS (Invivogen). For M2(a) polarization, cells were treated with 10ng/ml IL-4 (Peprotech). For M2b polarization, cells were treated with 10ng/ml IL-10 (Peprotech). For M2b polarization, cells were treated simultaneously with 10ng/ml LPS (Invivogen) and either unopsonized sheep red blood cells (SRBC) (Lampire biological laboratories) or SRBCs opsonized with 1:400 anti-SRBC IgG (cat# 55806 MP Biomedicals).
Mice
To generate mice with targeted deletion of Tsc1 in myeloid lineage cells, mice with flanking loxP Tsc1 alleles (Tsc1fl/fl) were crossed to LysozymeM Cre transgenic mice, both on a B6 background 38, 63. Cre-recombinase activity results in deletion of exons 17 and 18 of Tsc1, generating a null allele 64. Male mice aged 12 weeks were used for in vivo chitin administration and male mice aged 6–8 weeks were used for in vivo IL-4 administration. For Rictor deletion in vivo, tamoxifen (VWR) was suspended in sunflower seed oil (VWR) at 10mg/ml, and 200μl/25g body weight was injected into 10-week old Rictorfl/fl and Rictorfl/fl UbiquitinC- CreERT2 mice once daily for 7 days 65. Bone-marrow from such mice were used to generate Rictor-deficient macrophages. Mice were maintained at Harvard Medical School and Massachusetts Institute of Technology, and all procedures were performed in accordance with the guidelines set forth by the Institutional Animal Care and Use Committees at each institution.
Immunoblotting
For protein sample preparation, cells were washed twice with cold PBS following stimulation and lysed in 1% NP-40 buffer with EDTA-free protease inhibitor tablets (Roche Diagnostics) and phosphatase inhibitors. Protein concentration in lysates was determined using the Bradford method. Equal amounts of protein were loaded onto SDS-PAGE gels and subsequently transferred to PVDF membranes for immunoblotting with primary antibodies as indicated. Full blots of all immunoblots shown in the main article are included in Supplementary Fig. S9.
Antibodies and Reagents
Primary antibodies were purchased from Cell Signaling (all at 1:1000 dilution in 5% BSA), except for the following: α- Tubulin (Sigma, 1:5000), β-Actin (Sigma,1:2000), Flag-M2 (Sigma, 1:1000), and PPARγ (Santa Cruz E-8, 1:1000) and PPARδ (Santa Cruz, 1:250). For flow cytometry, antibodies were used to CD11b-PE (BD Biosciences) and F4/80-FITC (BioLegend). Inhibitors were used as follows: MK2206 1μM (Selleck), Aktviii 10μM (Enzo), Rapamycin 20nM (LC Laboratories), Troglitazone 1μM (Cayman) and GW501516 100 μM (Enzo).
Arginase Assays
Arginase assay was described previously 66. Briefly, 0.5×106 cells/well in 12-well plates were stimulated with IL-4 for 12–48h. Cells were lysed in 0.1% TritonX-100 lysis buffer with protease inhibitors. Lysates with equal amounts of protein were incubated with 500mM L-Arginine for 45 minutes at 37°C, followed by acid stop solution. The degradation of L-arginine to urea was measured by adding 9% isonitrosopropiophenone in 100% ethanol. Absorbance was read at 540nm in a microplate reader. All samples were read in triplicate.
Fatty Acid Oxidation
BMDMs were plated 0.7×106 cells/well in 12-well tissue culture dishes in complete RPMI and stimulated with IL-4 for 36h. After stimulation, cells were washed with PBS and loaded with low glucose DMEM + 2% fatty acid-free BSA (Lampire Biologicals) for 30 minutes at 37°C. After 30 minutes, cells were washed with PBS and given 3H-labeled palmitic acid (2μCi/well, MP Biomedicals) in low glucose DMEM, with 2% fatty acid-free BSA and 0.2mM unlabeled oleic acid (Sigma). After 4h, 100μl of media was collected and the isolation of 3H2O was performed using trichloroacetic acid followed by chloroform-methanol extraction. The water-soluble fraction was collected in 5ml of EcoLume (MP Biomedicals) scintillation fluid and counted for 5 minutes using a Beckman LS6500 scintillation counter. Cells were lysed in 500μl of 0.1N NaOH and total protein was determined using the Bradford method. Background 3H was subtracted from the CPM (counts per minute) value and all samples were normalized to mg of total protein. All samples were performed in duplicate.
Myr-Akt-Transduction
To make retrovirus particles, 293T cells were co-transfected with pBabe empty vector (EV) Puro or pBabe Puro Myr Flag Human-Akt1 purchased from Addgene (plasmid 15294) along with pCL-Eco (Imgenex #10045P) using Lipofectamine 2000 (Invitrogen). Transfected 293T media was changed the next day and placed at 32C. Viral supernatant was collected on day 2 and day 3 post-transfection. Fresh bone marrow was plated on the same day as transfection above and transduced with media containing viral supernatant (50% viral supernatant containing pBabe EV or pBabe Puro Myr Flag Human-Akt1, 40% RPMI complete media, and 10% CMG-media) on day 2 and day 3 and selected using 4 μg/ml puromycin on days 4–7.
ELISA
Cytokine concentration was determined using for IL-10, TNFα, and IL-6 using ELISA kits purchased from BioLegend. Briefly, experimental supernatants were collected and centrifuged at 3000 g/5min. Supernatants were analyzed in duplicate per manufacturers protocol.
Chromatin Immunoprecipitation
For STAT6 ChIP, BMDMS (5×106) were plated in 6cm tissue culture plates and stimulated for 2h with 10ng/ml IL-4. Cells were subsequently fixed with 1% formaldehyde for 10 min at room temperature. Formaldehyde was quenched with glycine. After collecting cells, lysis was performed using 500μl of SDS buffer (1% SDS, 10mM EDTA, 50mM Tris-Cl pH 8) plus protease and phosphatase inhibitors. Cells were subsequently sonicated for 3 min on ice with 1 sec pulses using a Misonix 4000 sonicator to shear chromatin. Following sonication, samples were diluted to 3ml with ChIP Dilution Buffer (0.01% SDS, 1.1% TritonX-100, 1.2mM EDTA, 16.7mM Tris-Cl pH 8, 167mM NaCl) and precleared for 1h at 4°C with Protein A salmon sperm/agarose beads (Millipore). Precleared chromatin was split into 400μl aliquots for IP with either 5μg of STAT-6 (M-20 ChIP grade, Santa Cruz) or for no antibody control overnight at 4°C. IPs were incubated with Protein A salmon sperm/agarose beads (Millipore) the following day for 3h at 4°C. After 3h, beads were spun down and ~10% of chromatin was taken for input and processed in parallel to IP samples. Antibody-bead complexes were then washed with low salt (0.1%SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-Cl pH 8, 150mM NaCl), high salt (0.1%SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris-Cl pH 8, 500mM NaCl), and lithium chloride (0.25M LiCl, 1% deoxycholic acid, 1%NP-40, 1mM EDTA, 10mM Tris-Cl pH 8) buffers followed by two washes with TE buffer. Chromatin-antibody complexes were eluted with elution buffer (1%SDS + 0.1M sodium bicarbonate) and crosslinks were reversed using sodium chloride and incubation at 60°C for 4h. Samples were then incubated 1h with Proteinase K (Roche) at 60°C. DNA was purified using PCR purification columns (Qiagen) and used for quantitative PCR with primers generated to the Arg1 promoter region. Fold enrichment was calculated as ChIP signals divided by no antibody control and normalized to input.
Quantitative PCR
To measure gene expression in BMDMs and PECs, RNA was isolated using RNA Bee (Tel-Test) per manufacturers protocol and used to make cDNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A Bio-Rad C1000 Thermocycler was used to analyze the samples under the following conditions: 95°C (5 min), 50 cycles of 95°C (10 s), 60°C (10 s), and 72°C (20 s). Reaction mixture consisted of 4 μl cDNA, 1.5 μl 3 μM primers for each gene used in the study (F+R), 2 μl H2O, and 7.5 μl 2× SYBR green (Bio-Rad). Samples from BMDMs were normalized to hypoxanthine phosphoribosyltransferase (HPRT) and samples from PECs were normalized to the macrophage marker CD68. Data was analyzed by means of the CFX Manger Software (Bio-Rad) using the delta/delta CT method. Sequences for all qPCR primers are shown in Supplementary Table S1.
Chitin Administration
Chitin (Sigma) was prepared as previously described 48. Briefly, chitin was washed 3 times with PBS and then sonicated (Misonix Sonicator 4000) for 30 minutes on ice. The dissolved chitin was filtered and diluted with PBS to 4 μg/ml. 800ng chitin dissolved in 200 μl PBS of was injected intraperitoneally and PECs were collected 48 hours post injection.
IL-4 Complex Administration
Long acting IL-4 complex (IL-4c) was prepared as previously described 47. Briefly, IL-4 (Peprotech) was suspended at a concentration of 500 μg/ml and mixed with anti-mouse IL-4 (BioXcell clone 11b11) at a molar ratio of 2:1 (weight 1:5) and incubated 1–2 minutes at room temperature. IL-4c was suspended in normal saline to a concentration of 25μg/ml IL-4 and 125 ug/ml of 11b11. Each mouse was injected intraperitoneally with 200μl of IL-4c (5 μg IL-4 and 25 μg 11b11) on day 0 and 2, and PECs were collected on day 4.
Dual Luciferase Assays
BMDMs were electroporated using Amaxa nucleofector and mouse macrophage nucleofector kit (Lonza) with PPAR-Firefly luciferase plasmid (C.H. Lee, Harvard School of Public Health) or STAT6-Firefly luciferase (purchased from Addgene-plasmid #35554) along with Renilla-Luciferase plasmid as a transfection control. The PPAR-Firefly luciferase plasmid consists of 3 copies of the Acox1 PPAR response element 67 upstream of an SV40 minimal promoter. BMDMs were treated with IL-4 and/or troglitazone 4 hours post electroporation for 24 hours. Cell lysates were collected and analyzed using the Promega Dual-Luciferase Reporter Assay System.
Statistical Analysis
Statistical analysis was carried out using Prism (Graph Pad) software. The student’s t-test was used to determine statistical significance, defined as p<0.05.
Supplementary Material
Acknowledgments
This project was supported by a NIH grant R01AI102964 (to T.H.). A.J.C. is a recipient of a Ford Foundation Predoctoral Fellowship. B.D.M was supported by a NIH grant R01- CA122617 and I.B.-S. by a LAM Foundation Fellowship. D.M.S. was funded in part by a Julie Martin Mid-Career Award in Aging Research from the American Federation of Aging Research (AFAR) and is an Investigator of the Howard Hughes Medical Institute. D.W.L. is supported by a K99/R00 award from the NIH/NIA (1K99AG041765-01A1). We also thank C.H. Lee for critical reading of the manuscript and S.H. Liu for technical advice.
Footnotes
Author contributions
V.B. and A.J.C designed and performed experiments, analyzed data, and wrote the paper. T.H. supervised the project, including experimental design and data analysis, and edited the paper. I.B.-H. and D. W. L. contributed technical expertise. D.M.S. and B.D.M. provided reagents and mice.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Odegaard JI, et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007;447:1116–1120. doi: 10.1038/nature05894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang K, et al. Adipocyte-derived Th2 cytokines and myeloid PPARdelta regulate macrophage polarization and insulin sensitivity. Cell Metab. 2008;7:485–495. doi: 10.1016/j.cmet.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Bossche J, et al. Pivotal Advance: Arginase-1-independent polyamine production stimulates the expression of IL-4-induced alternatively activated macrophage markers while inhibiting LPS-induced expression of inflammatory genes. J Leukoc Biol. 2012;91:685–699. doi: 10.1189/jlb.0911453. [DOI] [PubMed] [Google Scholar]
- 7.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer T, et al. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 2003;112:645–657. doi: 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc Natl Acad Sci U S A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem Soc Trans. 2009;37:217–222. doi: 10.1042/BST0370217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong-Kopera H, et al. TSC1 stabilizes TSC2 by inhibiting the interaction between TSC2 and the HERC1 ubiquitin ligase. J Biol Chem. 2006;281:8313–8316. doi: 10.1074/jbc.C500451200. [DOI] [PubMed] [Google Scholar]
- 17.Pan H, O’Brien TF, Zhang P, Zhong XP. The role of tuberous sclerosis complex 1 in regulating innate immunity. J Immunol. 2012;188:3658–3666. doi: 10.4049/jimmunol.1102187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 20.Filardy AA, et al. Proinflammatory clearance of apoptotic neutrophils induces an IL- 12(low)IL-10(high) regulatory phenotype in macrophages. J Immunol. 2010;185:2044–2050. doi: 10.4049/jimmunol.1000017. [DOI] [PubMed] [Google Scholar]
- 21.Martinez FO, Sica A, Mantovani A, Locati M. Macrophage activation and polarization. Front Biosci. 2008;13:453–461. doi: 10.2741/2692. [DOI] [PubMed] [Google Scholar]
- 22.Szanto A, et al. STAT6 transcription factor is a facilitator of the nuclear receptor PPARgamma-regulated gene expression in macrophages and dendritic cells. Immunity. 2010;33:699–712. doi: 10.1016/j.immuni.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acacia de Sa Pinheiro A, et al. IL-4 induces a wide-spectrum intracellular signaling cascade in CD8+ T cells. J Leukoc Biol. 2007;81:1102–1110. doi: 10.1189/jlb.0906583. [DOI] [PubMed] [Google Scholar]
- 24.Wang IM, Lin H, Goldman SJ, Kobayashi M. STAT-1 is activated by IL-4 and IL-13 in multiple cell types. Mol Immunol. 2004;41:873–884. doi: 10.1016/j.molimm.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 25.Bhattacharjee A, Shukla M, Yakubenko VP, Mulya A, Kundu S, Cathcart MK. IL-4 and IL-13 employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free Radic Biol Med. 2013;54:1–16. doi: 10.1016/j.freeradbiomed.2012.10.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tontonoz P, et al. Adipocyte-specific transcription factor ARF6 is a heterodimeric complex of two nuclear hormone receptors, PPAR gamma and RXR alpha. Nucleic Acids Res. 1994;22:5628–5634. doi: 10.1093/nar/22.25.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–252. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 28.Reilly SM, Lee CH. PPAR delta as a therapeutic target in metabolic disease. FEBS Lett. 2008;582:26–31. doi: 10.1016/j.febslet.2007.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurster AL, Withers DJ, Uchida T, White MF, Grusby MJ. Stat6 and IRS-2 Cooperate in Interleukin 4 (IL-4)-Induced Proliferation and Differentiation but Are Dispensable for IL-4-Dependent Rescue from Apoptosis. Mol Cell Biol. 2002;22:117–126. doi: 10.1128/MCB.22.1.117-126.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heller NM, et al. Type I IL-4Rs selectively activate IRS-2 to induce target gene expression in macrophages. Sci Signal. 2008;1:ra17. doi: 10.1126/scisignal.1164795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang X, Yu S, Tanyi JL, Lu Y, Woodgett JR, Mills GB. Convergence of multiple signaling cascades at glycogen synthase kinase 3: Edg receptor-mediated phosphorylation and inactivation by lysophosphatidic acid through a protein kinase C-dependent intracellular pathway. Mol Cell Biol. 2002;22:2099–2110. doi: 10.1128/MCB.22.7.2099-2110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 34.Eldar-Finkelman H, Seger R, Vandenheede JR, Krebs EG. Inactivation of glycogen synthase kinase-3 by epidermal growth factor is mediated by mitogen-activated protein kinase/p90 ribosomal protein S6 kinase signaling pathway in NIH/3T3 cells. J Biol Chem. 1995;270:987–990. doi: 10.1074/jbc.270.3.987. [DOI] [PubMed] [Google Scholar]
- 35.Zhang HH, Lipovsky AI, Dibble CC, Sahin M, Manning BD. S6K1 regulates GSK3 under conditions of mTOR-dependent feedback inhibition of Akt. Mol Cell. 2006;24:185–197. doi: 10.1016/j.molcel.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrington LS, et al. The TSC1–2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol. 2004;166:213–223. doi: 10.1083/jcb.200403069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 38.Yecies JL, et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 2011;14:21–32. doi: 10.1016/j.cmet.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fritsche L, et al. Insulin-induced serine phosphorylation of IRS-2 via ERK1/2 and mTOR: studies on the function of Ser675 and Ser907. Am J Physiol Endocrinol Metab. 2011;300:E824–836. doi: 10.1152/ajpendo.00409.2010. [DOI] [PubMed] [Google Scholar]
- 40.Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends Biochem Sci. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 41.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332:1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu PP, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holt LJ, Siddle K. Grb10 and Grb14: enigmatic regulators of insulin action--and more? Biochem J. 2005;388:393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick KR, et al. Grb10 inhibits insulin-stimulated insulin receptor substrate (IRS)- phosphatidylinositol 3-kinase/Akt signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor. J Biol Chem. 2003;278:8460–8467. doi: 10.1074/jbc.M208518200. [DOI] [PubMed] [Google Scholar]
- 45.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mora A, Komander D, van Aalten DM, Alessi DR. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol. 2004;15:161–170. doi: 10.1016/j.semcdb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 47.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 49.Reese TA, et al. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature. 2007;447:92–96. doi: 10.1038/nature05746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weichhart T, et al. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565–577. doi: 10.1016/j.immuni.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, et al. Macrophage-induced tumor angiogenesis is regulated by the TSC2- mTOR pathway. Cancer Res. 2012;72:1363–1372. doi: 10.1158/0008-5472.CAN-11-2684. [DOI] [PubMed] [Google Scholar]
- 52.O’Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 53.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11:889–896. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 54.Wills-Karp M, Finkelman FD. Untangling the complex web of IL-4- and IL-13- mediated signaling pathways. Sci Signal. 2008;1:pe55. doi: 10.1126/scisignal.1.51.pe55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat Rev Mol Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 56.Luyendyk JP, Schabbauer GA, Tencati M, Holscher T, Pawlinski R, Mackman N. Genetic analysis of the role of the PI3K-Akt pathway in lipopolysaccharide-induced cytokine and tissue factor gene expression in monocytes/macrophages. J Immunol. 2008;180:4218–4226. doi: 10.4049/jimmunol.180.6.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan W, et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arranz A, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proc Natl Acad Sci U S A. 2012;109:9517–9522. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guertin DA, et al. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 2006;11:859–871. doi: 10.1016/j.devcel.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Jacinto E, et al. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 2006;127:125–137. doi: 10.1016/j.cell.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 61.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chawla A, Nguyen KD, Goh YP. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clausen BE. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Research. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 64.Kwiatkowski DJ, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 65.Lamming DW, et al. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science. 2012;335:1638–1643. doi: 10.1126/science.1215135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sinha P, Clements VK, Ostrand-Rosenberg S. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol. 2005;174:636–645. doi: 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- 67.Liu S, et al. Role of peroxisome proliferator-activated receptor {delta}/{beta} in hepatic metabolic regulation. J Biol Chem. 2011;286:1237–1247. doi: 10.1074/jbc.M110.138115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.