Abstract
For all adeno-associated virus (AAV) serotypes, 60 monomers of the Vp1, Vp2, and Vp3 structural proteins assemble via an unknown mechanism to form an intact capsid. In an effort to better understand the properties of the capsid monomers and their role in viral entry and infection, we evaluated whether monomers from distinct serotypes can be mixed to form infectious particles with unique phenotypes. This transcapsidation approach consisted of the transfection of pairwise combinations of AAV serotype 1 to 5 helper plasmids to produce mosaic capsid recombinant AAV (rAAV). All ratios (19:1, 3:1, 1:1, 1:3, and 1:19) of these mixtures were able to replicate the green fluorescent protein transgene and to produce capsid proteins. A high-titer rAAV was obtained with mixtures that included either serotype 1, 2, or 3, whereas an rAAV of intermediate titer was obtained from serotype 5 mixtures. Only mixtures containing the AAV4 capsid exhibited reduced packaging capacity. The binding profiles of the mixed-virus preparations to either heparin sulfate (HS) or mucin agarose revealed that only AAV3-AAV5 mixtures at the 3:1 ratio exhibited duality in binding. All other mixtures displayed either an abrupt shift or a gradual alteration in the binding profile to the respective ligand upon increase of a capsid component that conferred either HS or mucin binding. The transduction of cell lines was used to further evaluate the phenotypes of these transcapsidated virions. Three transduction profiles were observed: (i) small to no change regardless of ratio, (ii) a gradual increase in transduction consistent with titration of a second capsid component, or (iii) an abrupt increase in transduction (threshold effect) dependent on the specific ratios used. Interestingly, an unexpected synergistic effect in transduction was observed when AAV1 helper constructs were combined with type 2 or type 3 recipient helpers. Further studies determined that at least two components contributed to this observed synergy: (i) heparin-mediated binding from AAV2 and (ii) an unidentified enhancement activity from AAV1 structural proteins. Using this procedure of mixing different AAV helper plasmids to generate “cross-dressed” AAV virions, we propose an additional means of classifying new AAV serotypes into subgroups based on functional approaches to analyze AAV capsid assembly, receptor-mediated binding, and virus trafficking. Exploitation of this approach in generating custom-designed AAV vectors should be of significant value to the field of gene therapy.
Adeno-associated virus (AAV), a member of the Parvoviridae family, is utilized as a virus vector for gene therapy. Eight different AAV serotypes have been described (6, 7, 12, 32, 43). Of these, AAV serotype 2 (AAV2) is the most studied and the only one used in gene therapy clinical trials to date (19, 21). The genomes of AAV serotypes are similar, consisting of two large open reading frames (ORFs) that are flanked by inverted terminal repeats (ITRs) (reviewed in references 24 and 25). The ITRs are the genetic elements responsible for the replication and packaging and the only cis elements required to generate recombinant AAV (rAAV). The left ORF encodes the four replication proteins (rep78, rep68, rep52, and rep40) responsible for site-specific integration, nicking activity, helicase activity, and regulation of AAV promoters.
The viral structural proteins (VP1, VP2, and VP3) that assemble into the virion shells are encoded by the right ORF. The first step in the viral infection process, cell attachment, is mediated by the interaction of specific regions on the capsid shell with cellular receptors and coreceptors (28, 36, 37). A pivotal contribution to the understanding of the molecular events that mediate attachment is the recent publication of the crystal structure of AAV2 (45). This information, in conjunction with mutational analyses of the AAV2 capsid, has allowed the identification of functional domains of the AAV2 capsid involved in binding to the primary cell surface receptor, heparan sulfate (HS) (23, 26, 31, 34, 42). Additional properties of viruses that can be attributed to the capsid include pathways for internalization and uncoating and immediate cellular responses to the capsid (4, 9, 10, 33). Unfortunately, these activities have yet to be mapped to specific regions of the capsid of any AAV serotype.
The driving force behind the manipulation of the AAV virion shell is to map and understand the contribution of the capsid amino acids, as well as to exploit this information to alter the tropism or transduction efficiency. Several studies have used a rational design method to insert or exchange small epitopes into the capsid shell predicted as a means of retargeting AAV and thereby altering the tropism (3, 13, 14, 27, 35, 42). These methods have met with various degrees of success. Others have proposed to utilize the preexisting differences of tropism between the serotypes (8, 30, 40). The serotypes of AAV exhibit specific phenotypes, as exemplified by their binding to particular cell surface receptors. For example, AAV2 and AAV3 bind to HS; however, their elution profiles are quite distinct, suggesting that these viruses interact differently (29). In contrast, AAV4 and AAV5 do not interact with HS but instead interact with sialic acid moieties, albeit via different linkages (22, 39). Furthermore, AAV1 does not appear to utilize HS, and its receptor and binding activity remain unknown. The recently described serotypes 7 and 8 have no known receptor, but since they closely resemble AAV1, except for the hypervariable region, they may share a common receptor (12). In an effort to further understand the infectious process of the various serotypes, investigators have swapped regions between serotypes within one subunit of the capsid, forming chimeric viruses homogenous in nature, by utilizing standard cloning techniques (18) or, in one case, by using a marker rescue approach (5).
Since more AAV serotypes are being isolated each year, the task of understanding serotype-specific infection with the long-term goal of developing more efficient and safer AAV vectors requires a focused effort on basic AAV virology. One approach to assist in classifying and understanding distinct phenotypes of specific AAV serotype and utilize this information to further evolve AAV as a viral vector is to generate rAAV with mixed or mosaic capsid shells (17). We undertook a comprehensive approach and mixed all combinations of available serotype plasmids (AAV1 to AAV5) at five ratios to control the final contribution of serotype-specific capsid components in the mature virion. Some of the properties of the two serotypes may be shared (17), some may be masked, and some may be dominant depending on the ratio of helper plasmids used. Most importantly, positive attributes of the two serotypes may be additive or synergistic in tests for viral transduction. Such possibilities were investigated both in vitro and in cell culture, and both expected and unexpected results were obtained at particular ratios of helper plasmids. Mixing different AAV helper plasmids to generate “cross-dressed” AAV virions can be applied to any and all AAV serotypes and should facilitate a basic understanding of AAV capsid assembly, receptor-mediated binding, and virus trafficking, attributes essential for vector development.
MATERIALS AND METHODS
Cells and plasmids.
The six cell lines utilized in the present study are available from the American Type Culture Collection. All were maintained at 37°C with 5% CO2 in their respective media, which was supplemented with 10% fetal bovine serum and penicillin-streptomycin. 293 HEK and HeLa cells were grown in Dulbecco modified Eagle medium (DMEM). CHOK1, CHO-pgsD, and CHO-pgsE, cells were grown in Ham F-12 medium. C2C12 cells were grown in minimal essential medium alpha.
The AAV serotype helper plasmids pXR1-5, as well as the plasmid pXX6-80, have been described elsewhere (29, 44). The H/N3761 and H2634 mutant AAV2 helper plasmids were described previously (31). Essentially, in the H/N3761 mutant, 12 bp encoding the amino acids AISP was inserted at position 3761 of the AAV2 genome. The H2634 mutant consisted of a 12-bp insertion encoding the amino acids GDIA at AAV2 nucleotide position 2634.
Recombinant virus production.
Recombinant AAV was produced by using a modified triple plasmid transfection protocol (15). A 15-cm dish of 293 HEK cells (ca. 2 × 107) was transfected by using Superfect reagent according to the manufacturer's instructions (Qiagen, Valencia, Calif.) with 7 μg of pTRCMVeGFP, 15 μg of XX6-80, and 10 μg of the serotype helper plasmid. To generate AAV virions containing capsid components of different serotypes, the amount of each helper plasmid transfected was varied depending on the ratio being assessed. For example, if AAV3 and AAV4 were mixed at a ratio of 19:1, then 9.5 μg of pXR3 and 0.5 μg of pXR4 were added to the transfection mixture.
The cells were harvested at 48 to 52 h posttransfection by scraping, and all subsequent manipulations were performed on ice unless otherwise noted. One-fifth of the cell lysate was set aside for Hirt DNA isolation (see below), while the remainder was subjected to low-speed centrifugation and resuspended in a solution of 3 μg of leupeptin/ml. The cell lysate was subjected to sonication under previously described conditions (31). A portion of the cell lysate was removed for the Western blot analyses described below. Benzonase (Sigma, St. Louis, Mo.) was added to the remainder at a final concentration of 1.2 U/μl, followed by incubation at 37°C for 30 to 60 min. Then, 0.58 g of cesium chloride was added to each milliliter of lysate, and the mixtures were centrifuged for 4 h at 100,000 rpm in a TLN 100 rotor in the Optima TLX ultracentrifuge (Beckman, Palo Alto, Calif.). Peak fractions from the gradient were determined by dot blot hybridization as previously described (31) by using a radiolabeled green fluorescent protein (GFP) probe.
Hirt assay.
DNA from the infected cell lysate was isolated as described by Hirt (20). Approximately 3 μg of DNA from each sample was digested with DpnI, fractionated on an agarose gel, transferred to a Nytran membrane, and probed with the GFP transgene.
Western blot analyses.
The protein concentration of transfected cell lysates was determined by the BCA protein assay (Pierce, Rockford, Ill.). Five micrograms of total protein was loaded in each lane, followed by separation by electrophoresis on a NuPage 10% polyacrylamide Bis-Tris gel (Invitrogen, Carlsbad, Calif.) and then transfer to a Hybond ECL membrane (Amersham Pharmacia Biotech, Piscataway, N.J.). The membrane was probed with either the B1 monoclonal antibody (41) specific for AAV capsid proteins or the Living Colors Anti-GFP monoclonal antibody (Clontech, Palo Alto, Calif.). The proteins were visualized by using a peroxidase-coupled secondary antibody and the Supersignal WestFemto kit (Pierce).
Electron microscopy.
Peak fractions of rAAV1/5 and rAAV2 were placed on a 400-mesh glow-discharged carbon grid by inverting on a 10-μl drop of virus. The grid was washed three times in 1× phosphate-buffered saline (PBS) for 1 min. The virus was then stained for 1 min with 2% phosphotungstic acid. The virus was visualized by using a Zeiss EM 910 electron microscope.
Heparin-binding assays.
Batch binding of rAAV to heparin agarose was performed as described previously (31). Briefly, equivalent particles of parental and mixed rAAV virions were applied to heparin agarose type 1 (H-6508; Sigma) in 1× PBS, allowed to bind for 1 h at room temperature, and centrifuged at low speed for 2 min, and supernatant (flowthrough) was then removed. Six washes of five bed-volumes of PBS-1 mM MgCl2 were performed, followed by a three-step elution of five bed-volumes of PBS-1 mM MgCl2 containing 0.5 M NaCl (step 1), 1.0 M NaCl (step 2), or 1.5 M NaCl (step 3). The numbers of rAAV particles present in the washes and the three-step elution were determined by dot blot hybridization.
Mucin-binding assays.
Mucin (Sigma) was linked to agarose as described by the manufacturer. Parental and mixed rAAV virion binding was determined in a manner analogous to that described above for heparin binding.
Flow cytometry.
For analysis of GFP expression, the cell lines pgsD, pgsE, CHOK1, C2C12, and HeLa (n = 5 × 105) were transduced in quadruplicate with 300 or 3,000 vector genomes (vg) of the parental or mixed rAAV/cell and were additionally infected with Ad dl309 at a multiplicity of infection (MOI) of 14. At 24 h posttransduction, cells were harvested and scored for GFP expression by using the FacSCAN 1 flow cytometer (Becton Dickinson, Franklin Lakes, N.J.). Some variation was observed between experiments possibly due to differences in gating, age of the adenovirus preparation, or even the passage number of the cell lines.
Heparin inhibition assay.
The ability of soluble HS to inhibit the binding of rAAV1 and rAAV2 to C2C12 cells was assayed as described previously (37). Briefly, rAAV1/2, rAAV1, or rAAV2 at 300 vg/cell was incubated in DMEM in the presence or absence of soluble HS at 30 μg/ml for 1 h at 37°C. Concurrently, C2C12 cells were incubated for 1 h in DMEM in the presence or absence of soluble HS. After the preincubation steps, the C2C12 cells were infected for 1 h with their respective rAAV preparations and additionally coinfected with adenovirus dl309 (Ad dl309) at an MOI of 14. At 24 h posttransduction, cells were harvested and scored for GFP expression as described above. Analogous methods were used to assess the ability of soluble HS to inhibit binding of rAAV3/5 to HeLa cells.
Neuraminidase assay.
The ability of neuraminidase treatment of HeLa cells to prevent infection of rAAV3/5 was assayed. Briefly, HeLa cells were pretreated for 1 h at 37°C with 10 U of neuraminidase (Sigma)/ml/h. After the preincubation step, HeLa cells were infected for 1 h with the respective rAAV preparation (3,000 vg/cell) and coinfected with Ad dl309 at an MOI of 14. At 24 h posttransduction, cells were harvested and scored for GFP expression as described above.
RESULTS
Design parameters of AAV serotype mixing.
The feasibility of mixing capsid proteins from different AAV serotypes was investigated by using the previously characterized AAV2 helper plasmid mutants H2634 and H/N3761 (31). Both helper plasmids produce noninfectious virions as a result of unique mutations in their capsid regions. Virus produced from the H/N3761 mutant contains all three capsid subunits (VP1, VP2, and VP3); however, this virus does not bind heparin, nor will it compete with wild-type AAV2 for transduction into cells. The H2634 mutant produces virus with only VP3 subunits that will bind heparin and has the ability to compete with wild-type AAV2 for transduction into cells but is not infectious. When these two helper plasmids are mixed at different ratios during the transfection stage of recombinant AAV2 production, infectious virus was obtained as judged by the ability to transduce HeLa cells (Table 1). These results indicated that the viable virions were not assembled homogeneously from the capsid subunits of one mutant but were instead composed of mixtures of subunits from each mutant. Importantly, these initial experiments indicated that advantageous phenotypes of each mutant could be built into a mosaic virus (Fig. 1) and further suggested that properties of different AAV serotypes might be able to be mixed in an analogous manner.
TABLE 1.
Complementation of AAV2 capsid mutant viruses via helper plasmid mixing
| Viral samplea | Ratiob or percentage | No. of particles (1011/ml) | Particle:TUc |
|---|---|---|---|
| H2634 | 100% | 1.24 | 0 |
| 9:1 | 0.79 | 3.6 × 104 | |
| H2634:H/N3761 | 4:1 | 1.01 | 1.4 × 104 |
| 7:3 | 1.47 | 1.0 × 104 | |
| 3:2 | 1.48 | 1.2 × 105 | |
| 1:1 | 1.02 | 1.9 × 105 | |
| 2:3 | 1.19 | 1.5 × 106 | |
| 3:7 | 2.48 | 0 | |
| 1:4 | 3.54 | 0 | |
| 1:9 | 1.40 | 0 | |
| H/N3761 | 100% | 2.77 | 0 |
Viral samples were described elsewhere (31).
Ratio refers to the mixing of helper plasmids during the transfection.
From each sample, 2.5 × 108 viral genomes were used to transduce 3 × 105 HeLa cells in triplicate. TU, transducing unit.
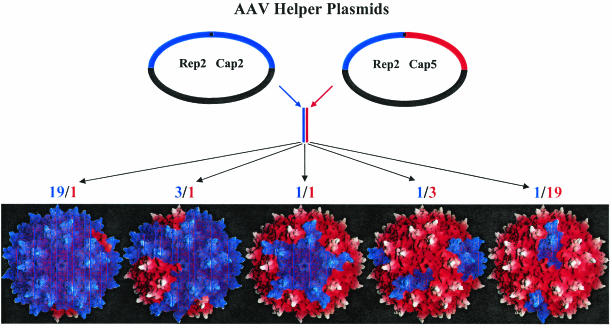
FIG. 1.
Plasmid mixing approach used to generate transcapsidated rAAV. Helper plasmid DNA containing the capsid gene from any two AAV serotypes (one is represented by a red capsid gene, and the other is represented by a blue capsid gene) was cotransfected at different ratios (19:1, 3:1, 1:1, 1:3, and 1:19) during the standard production scheme to generate rAAV particles. The topology maps of potential viral products are shown and colored according to the proportion of subunits from the ratio of helper plasmids in the transfection mixtures.
To investigate the possibility of serotype capsid mixing, helper plasmids containing the five currently available serotype capsid genes (AAV1 to AAV5) (29) were mixed pairwise at specific ratios of 19:1, 3:1, 1:1, 1:3, and 1:19 (Fig. 1). These helper plasmids have the same genetic backbone, efficiently replicate transgenes, produce replication and capsid proteins at nearly the same level, and package transgenes with AAV2 ITRs. As a result of these similarities and the number of DNA molecules used in the transfection (1.12 × 1012 in 10 μg), the number of molecules of each helper plasmid entering each cell should vary only slightly around the specific ratio. The resulting virions should be composed of subunits from each serotype used in the pairwise transfection mixture, and the ratio would determine the general composition of these virions (Fig. 1). For nomenclature purposes, the mixtures are reported as fractions in which the numerator represents the first parent plasmid and the denominator represents the second parent. Accordingly, the first number in the ratio corresponds to the first parent, and the second number in the ratio corresponds to second parent. For example, a mixed-virus preparation consisting of 25% AAV1 plasmid and 75% AAV2 plasmid would be referred to as AAV1/2 with a ratio of 1:3.
Physical characterization of mixed virions.
Control experiments were performed to examine the potential for recombination of the helper plasmid pairs. Hirt DNA was isolated at 48 h posttransfection and transformed into Escherichia coli, and individual plasmids were assessed to determine whether recombination between serotype plasmids occurred. After numerous colonies from separate transfections were screened by restriction digestion, no evidence was found for recombination (data not shown).
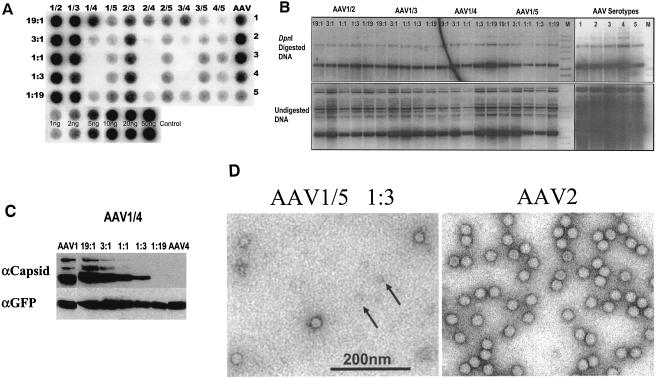
Dot blot analyses were performed on cesium gradient purified samples from the 50 AAV mixing combinations, as well as the five serotype control samples, to determine whether the resultant viruses could package the GFP transgene. A sample of the peak fraction from each mixture is shown (Fig. 2A), and the corresponding physical titers are presented in Table 2. Three general classes of capsid interactions were identified from this type of analyses. Class A mixtures consist of serotypes that mix well at all ratios examined and generate titers almost equivalent to their respective 100% controls. In these experiments, class A mixtures consisted of any AAV1, AAV2, or AAV3 combination. Class B mixtures consist of interactions of AAV5 with serotypes 1, 2, 3, and 4. The physical titers from all of these mixtures were of intermediate numbers and were similar to the AAV5 parental 100% control (see Table 2). Class C mixtures were composed of the serotypes (AAV1, AAV2, and AAV3) that do not mix very well with AAV4, as evidenced by the drastic reduction in physical titer of these mixtures outside of the 19:1 and 1:19 ratios.
FIG. 2.
(A) Dot blot of peak fractions from cesium gradient-purified AAV mixtures. 293 cells were transfected with AAV1 to AAV5 plasmids (AAV column) or combinations of the five serotypes (AAV serotype mixtures 1/2, 1/3, 1/4, 1/5, 2/3, 2/4, 2/5, 3/4, 3/5, and 4/5) at differing ratios (19:1, 3:1, 1:1, 1:3, and 1:19). The fractions on top of the blot indicate the AAV serotype helper plasmid contributors in the mixture and the ratios on the left side of the blot correspond to the relative amounts of all contributors in the mixture, respectively. Portions (2 μl) from the peak fraction of each cesium-purified virus preparation were applied to a GeneScreen Plus membrane and then probed with the GFP transgene that had been random primed. Duplicate DNA controls of the plasmid transgene are shown at the bottom of the blot. (B) Hirt DNA Southern blot analyses of AAV1/2, AAV1/3, AAV1/4, and AAV1/5 mixtures. 293 cells were transfected with either the AAV1 to AAV5 helper plasmids (AAV serotype lanes) or all ratios (19:1, 3:1, 1:1, 1:3, and 1:19) of the AAV1/2, AAV1/3, AAV1/4, and AAV1/5 helper plasmids to produce rAAV. Low-molecular-weight DNA was isolated 48 h posttransfection, and a portion was digested with DpnI. Then, 3 μg of DpnI-digested (top panel) or undigested (bottom panel) DNA was separated via a 1% agarose gel (M lane, DNA ladder). After transfer, the blot was probed with a random-primed GFP transgene. (C) Analyses of capsid protein composition from the AAV1/4 mixtures. Protein lysates from the transfection of either the AAV1 and AAV4 helper plasmids alone or mixed at different ratios (19:1, 3:1, 1:1, 1:3, and 1:19) were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis and transferred to a membrane. The blot was probed with either the B1 monoclonal antibody (top panel) directed against the carboxyl termini of the AAV2 VP3 protein or an anti-GFP antibody (bottom panel). (D) Transmission electron microscopy. Peak dot blot fractions of the AAV1/5 mixed virions (1:3 ratio) and rAAV2 were stained in 2% phosphotungstic acid and imaged by transmission electron microscopy. Arrows indicate potential fivefold capsid subunits.
TABLE 2.
Particle numbers for AAV pseudotypes and controlsa
| Ratio | No. of particles (1012)/ml for AAV serotype mixture:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1/2 | 1/3 | 1/4 | 1/5 | 2/3 | 2/4 | 2/5 | 3/4 | 3/5 | 4/5 | |
| 19:1 | 2.56 | 1.95 | 1.160 | 0.168 | 0.602 | 0.278 | 0.246 | 0.364 | 0.047 | 0.027 |
| 3:1 | 2.42 | 2.10 | 0.115 | 0.079 | 0.813 | 0.029 | 0.125 | 0.022 | 0.085 | 0.077 |
| 1:1 | 2.40 | 1.59 | 0.027 | 0.103 | 0.660 | 0.011 | 0.151 | 0.004 | 0.146 | 0.133 |
| 1:3 | 2.36 | 0.97 | 0.018 | 0.155 | 1.010 | 0.011 | 0.190 | 0.006 | 0.172 | 0.070 |
| 1:19 | 2.27 | 1.64 | 0.128 | 0.280 | 0.884 | 0.068 | 0.240 | 0.066 | 0.144 | 0.081 |
For unmixed AAVs 1, 2, 3, 4, and 5, the values were 0.858 × 1012, 1.840 × 1012, 1.760 × 1012, 0.836 × 1012, and 0.237 × 1012 particles/ml, respectively.
To eliminate the possibility that problems other than aberrant packaging of the transgene were responsible for the poor performance of the class C mixtures, additional physical properties of the mixed viruses or its corresponding transfection lysate were examined. The presence of replicating transgene DNA in all mixed and parental rAAV preparations was confirmed by analyses of Hirt DNA (Fig. 2B; AAV1/2, AAV1/3, AAV1/4, and AAV1/5 are shown). These results clearly demonstrate that replicating genomes were present at similar levels in transfection lysates of all mixed virus preparations, including those that produced low particle numbers.
Western blot analyses were performed on equivalent protein amounts from transfected cell lysates from each helper plasmid mixture ratio by using monoclonal antibodies against the AAV structural proteins or the GFP protein as a control (Fig. 2C, AAV1/4 mixtures are shown). The B1 monoclonal antibody recognizes the capsid proteins of all serotypes with the exception of AAV4 (29). Western blot analyses with this antibody on cell lysates from the AAV1/4 mixing experiments indicated that, as the contribution of AAV4 increases to 1:19, the ability of the antibody to recognize the capsid is lost. This absence in antibody recognition does not correlate with the loss of dot blot signal, which decreases as the contribution of AAV4 subunits increases toward the 1:1 ratio (Fig. 2A, see 1/4, 2/4, and 3/4 mixtures).
Electron microscopy was used to examine the physical particles of the three classes of mixed virions. Class A virions were indistinguishable from preparations of AAV2 virions at the resolution viewed, whereas no intact particles were observed from preparations of class C virions of intermediate ratio of 1:1 (data not shown). This further suggests that the lack of a dot blot signal in class C virus preparations does not represent an absence of subunits or packageable genomes but rather an inability to assemble capsids with the type and proportion of the capsid subunits present. Class B mixed viruses (AAV1/5 [1:3] shown as an example) were indistinguishable from AAV2 (Fig. 2D); however, smaller components were evident in these preparations (Fig. 2D, arrows) and may represent incompletely assembled or unassembled capsid components. These smaller entities were not observed in preparations of AAV2 (Fig. 2D, AAV2).
In vitro binding properties of mixed virus.
It has been shown that AAV2 and AAV3 bind heparin agarose (29, 37) and that both AAV4 and AAV5 bind mucin (1, 38). To determine whether the mixed virions exhibit binding properties similar to the parental virions or whether the binding to both heparin and mucin can be coupled through mixing of capsid components, the binding properties of each ratio of all mixtures except the 1/4, 2/4, and 3/4 mixed viruses that were deficient in generating sufficient test material were examined.
Equivalent particles of parental and all ratios of mixed virions were applied to either a heparin agarose or a mucin column, washed, and eluted in three steps of increasing salt. Samples from each column fraction were analyzed by dot blot analyses, and the corresponding percentages of bound and unbound virus are presented as pie charts (Fig. 3 and 4).
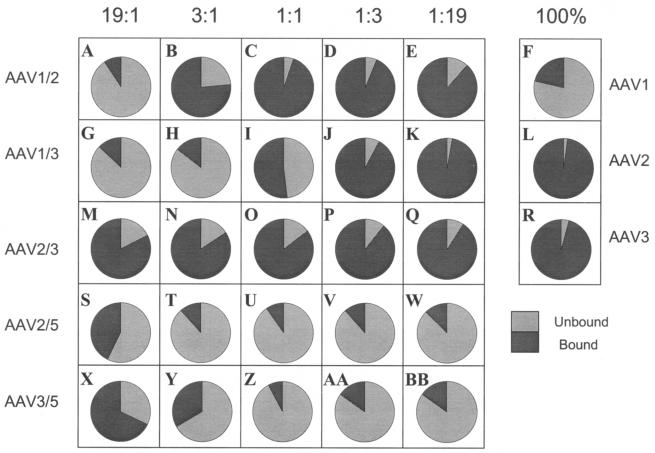
FIG. 3.
Heparin batch binding profiles of rAAV mixed virions. Totals of 1010 vg of each ratio (19:1, 3:1, 1:1, 1:3, or 1:19) of the rAAV1/2, rAAV1/3, rAAV2/3, rAAV2/5, and rAAV3/5 mixed virions or the rAAV1, rAAV2, or rAAV3 viral controls were applied to 50 μl of heparin agarose, washed, and eluted as described in Materials and Methods. Equal percentages of the total volume from each fraction were subjected to dot blot analyses and probed with the GFP transgene. The data are represented in pie charts and indicate the percent virus that bound (dark gray slices) or did not bind (light gray slices) to heparin.
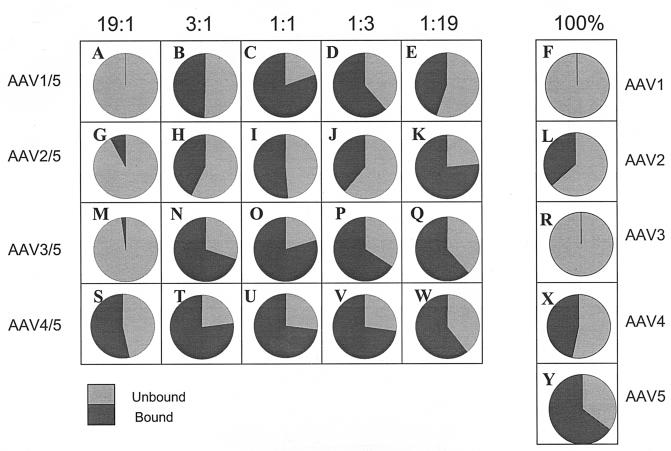
FIG. 4.
Mucin batch binding profiles of rAAV mixed virions. Totals of 109 vg of each ratio (19:1, 3:1, 1:1, 1:3, or 1:19) of the rAAV1/5, rAAV2/5, rAAV3/5, and rAAV4/5 mixed virions or the rAAV1 to rAAV5 viral controls were applied to 50 μl of mucin agarose, washed, and eluted as described in Materials and Methods. Equal percentages of the total volume from each fraction were subjected to dot blot analyses and probed with the GFP transgene. The data are represented in pie charts and indicate the percent virus that bound (dark gray slices) or did not bind (light gray slices) to mucin.
Class A mixed viruses interacted on the heparin column in two manners: either as an abrupt shift (Fig. 3A to E) or as a gradual change (Fig. 3G to K) in the heparin-binding profile upon increase of the second capsid component. Less than 10% of the AAV1/2 (19:1) mixture bound to heparin (Fig. 3A). Heparin binding abruptly changed to >75% at the 3:1 ratio (Fig. 3B). This finding is in contrast to AAV1/3 mixed viruses, with which there was a gradual change in heparin binding: <25% binding at the 3:1 ratio increasing to 50% at 1:1 ratio to >90% at the 1:3 ratio (Fig. 3H, I, and J). As expected, all ratios of the AAV2/3 mixtures bound heparin (Fig. 3M to Q).
Since neither AAV4 nor AAV5 bound heparin, the heparin-binding profiles were not determined for the AAV1/5 and AAV4/5 mixtures. In the case of the other class B interactions, the heparin-binding profiles of the AAV2/5 and AAV3/5 mixtures were essentially identical (Fig. 3S to BB); the 19:1 ratios were the only mixtures that significantly bound to heparin (Fig. 3S and X). Heparin-binding analyses of the class C mixes could not be determined because of the low particle numbers.
Mucin-binding experiments with class A parental or class A mixed virus did not result in significant mucin binding, as expected (data not shown). Interactions of class C mixtures on the mucin column could not be determined since these mixtures did not contain sufficient viral particles.
The mucin-binding profiles for the AAV1/5 and AAV3/5 mixtures resemble their respective parents in the extreme ratios (19:1 and 1:19; Fig. 4A, M, E, and Q). These mixtures at the 1:1 ratio bind to mucin more efficiently than AAV5 (Fig. 4C and O). AAV4/5 mixed viruses bound mucin as expected, since both parental serotypes individually bound mucin. Intermediate ratios of AAV4/5 (3:1, 1:1, and 1:3) appear to bind mucin better than either parent or the more extreme ratios 19:1 and 1:19 (Fig. 4S to W). Interestingly, only AAV3/5 (3:1) was able to bind both mucin and heparin agarose (Fig. 3Y and 4N). To determine whether individual particles bound both mucin and heparin agarose, a preparation of AAV3/5 (3:1) was used to first bind mucin. Virus eluted from this column was then used to bind heparin. The results from this experiment indicate that the AAV3/5 (3:1) preparation was able to bind both matrices at levels similar to those shown in Fig. 3Y and 4N. The control viruses (AAV3 and AAV5) were not able to successively bind the different matrices, indicating that the individual mosaic particles of AAV3/5 (3:1) were dual binders due to assembly of virus particles composed of mixed parental (types 3 and 5, respectively) capsid components (data not shown).
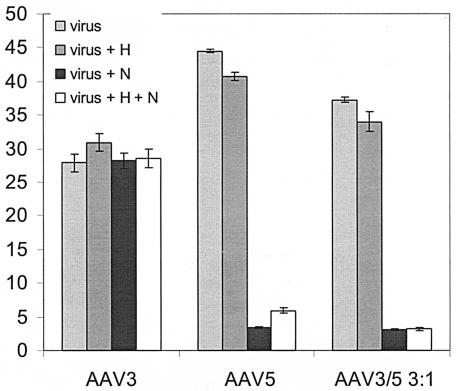
Both heparin competition and neuraminidase treatment of cells were used to assess whether this binding duality correlated with cell infection. Heparin pretreatment was unable to inhibit AAV3b (Fig. 5). This was unexpected since Handa et al. (16) previously demonstrated that AAV3a is inhibited by soluble heparin at these concentrations. The same concentration of heparin was able to inhibit the ability of rAAV2 to transduce C2C12 cells (see Fig. 7B). As expected, the ability of rAAV5 to transduce HeLa cells was greatly reduced upon pretreatment with neuraminidase, whereas the ability of rAAV3 to transduce these cells was unaffected (Fig. 5). More importantly, the AAV3/5 (3:1) mixture was reliant only on sialic acid for infectivity, as judged by the decrease in transduction of cells pretreated with neuraminidase (Fig. 5).
FIG. 5.
Neuraminidase and soluble heparan inhibition. HeLa cells, either untreated (virus) or pretreated with neuraminidase (N) and/or soluble heparin (H or H+N) for 1 h, were transduced with 3,000 vg of AAV3, AAV5, or AAV3/5 (3:1)/cell. The cells were either untreated or preincubated with soluble heparin for 1 h (H or H+N). After the completion of all preincubation steps, the respective virus was allowed to attach to cells and removed 1 h later, and fresh medium was applied. At 24 h posttransduction, cells were prepared for flow cytometry to measure the percent GFP expression. The graph indicates the percent GFP expression, with each bar representing an average of three separate infections, with the standard deviation indicated by an error bar. All transductions were performed in the presence of Ad dl309 at an MOI of 14.
FIG. 7.
(A) Transduction of C2C12 cells with mixtures of purified rAAV1 and rAAV2 virus. C2C12 cells were transduced with 300 vg of rAAV1, rAAV2, or both viruses mixed at the following ratios (19:1, 3:1, 1:1, 1:3, and 1:19)/cell. At 24 h posttransduction, cells were assayed for GFP expression via flow cytometry. The percent GFP expression is represented graphically. Each bar represents an average of three separate infections, with the standard deviation indicated by an error bar. All transductions were performed in the presence of Ad dl309 at an MOI of 14. (B) Heparin inhibition of rAAV1/2 mixed virions in C2C12 cells. C2C12 cells and rAAV1, rAAV2, and rAAV1/2 at the following ratios (19:1, 3:1, 1:1, 1:3, and 1:19) were preincubated for 1 h in the presence of 30 μg of heparin/ml. After preincubation, virus (300 vg/cells) was allowed to attach to cells and removed 1 h later, and fresh medium was applied. At 24 h posttransduction, the cells were prepared for flow cytometry to measure the percent GFP expression. The graph indicates the percent GFP expression, with each bar representing an average of three separate infections and with the standard deviation indicated by an error bar. All transductions were performed in the presence of Ad dl309 at an MOI of 14.
Transduction of different cell lines with AAV mixed virus.
Mixed virus preparations were analyzed for their ability to transduce human, mouse, and Chinese hamster cell lines. The CHO wild-type cells, K1, and derivatives with mutations in glycosaminoglycan biosynthesis (pgsD and pgsE cells) were selected since these cells demonstrate a biological phenomenon: heparin-dependent transduction of AAV2 (29, 37). HeLa cells are the standard line to titer rAAV, and C2C12 cells were used because they are known to bind AAV2 and yet are refractory to transduction (11). Either 300 or 3,000 vg of the mixed virus/cell was used in the transduction assays. All transduction assays were performed in the presence of adenovirus and flow cytometry was used to determine the percentage of cells expressing GFP.
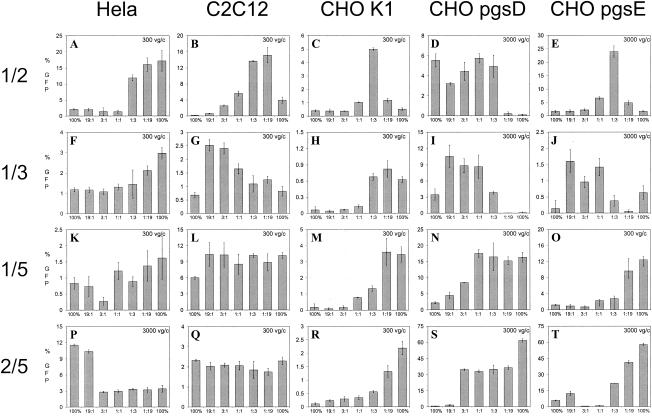
Four transduction profiles were apparent in these experiments: (i) a gradual change (titration), (ii) little or no change (plateau), (iii) abrupt changes (threshold), and (iv) synergistic changes as the amount of input plasmid increased from one parental serotype to the other (Fig. 6). The titration effect is seen in HeLa cells for the AAV1/3 (Fig. 6F) and in CHO K1 and pgsE cells for the AAV 2/5 mixtures (Fig. 6R and T). The plateau effect was observed in C2C12 cells for both the AAV1/5 and the AAV2/5 mixtures (Fig. 6L and Q). The threshold effect is clearly seen in HeLa cells infected with the AAV2/5 mixtures, for which the transduction efficiency decreased from 10 to 11% at the 19:1 ratio to 3% at 3:1 ratio (Fig. 6P). This pattern was also seen in HeLa cells with the AAV1/2 mixtures (Fig. 6A), in CHO K1 cells with the AAV1/3 (Fig. 6H) and 1/5 mixtures (Fig. 6M), in CHO pgsD cells with the AAV1/2 mixtures (Fig. 6D), and in CHO pgsE cells with the AAV1/5 mixtures (Fig. 6O). Finally, a synergistic pattern was observed in C2C12, CHOK1, and pgsE cells infected with vector from AAV1/2 mixtures (Fig. 6B, C, and E) and in C2C12, pgsD, and pgsE cells infected with vector from AAV1/3 mixtures (Fig. 6G, I, and J). This pattern resulted in some ratios of mosaic virus having a better transduction efficiency than either parental virus.
FIG. 6.
Transduction efficiency of reference cell lines with mixed rAAV. Cells (cell types are indicated on top) were transduced with all ratios (19:1, 3:1, 1:1, 1:3, and 1:19) of the rAAV1/3, rAAV1/2, rAAV1/5, or rAAV2/5 mixed virus and their corresponding parental controls (indicated as 100%). The vg/cell (vg/c) value used for each experiment is indicated in the upper right corner of each panel. At 24 h posttransduction, cells were assayed for GFP expression via flow cytometry. Percent GFP expression is represented graphically. Each bar represents an average of four separate infections, with the standard deviation indicated by an error bar. All transductions were performed in the presence of Ad dl309 at an MOI of 14.
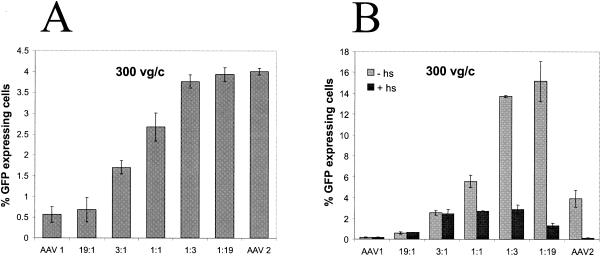
To test that the enhanced transduction observed from mosaic virus particles were not an artifact of mixing two parental serotypes virions, purified recombinant viruses from two serotypes (AAV1 and AAV2) were mixed at ratios identical to those used for the helper plasmids, and these mixtures were used to transduce C2C12 cells, with the same overall particle number (i.e., 300) that was used in the plasmid mixing experiments. A titration effect was observed in this instance as the ratio of the virus varied (Fig. 7A), in contrast to the synergistic effect observed (Fig. 6B) when rAAV1/2 mosaic viruses were used to transduce C2C12 cells. Based on in vitro binding studies and in vivo transduction assays, we probed known attributes of the parental virus to determine a mechanism for the synergism we observed from the mosaic virus particles.
Heparin inhibition of C2C12 cells transduced with AAV1/2 mixed viruses.
Heparin competition was used to determine whether the enhancement in transduction of C2C12 cells by AAV1/2 mixed viruses was due to HS-mediated binding. Recombinant AAV1/2 or the parental viruses were incubated in the presence or absence of soluble heparin for 1 h prior to transduction with 300 vg/cell. Soluble heparin reduced the enhanced transduction of 1:1, 1:3, and 1:19 ratios (Fig. 7B). However, the transduction of these mixtures was not completely abolished in the presence of HS-like AAV2 control (Fig. 7B). Instead, these mixtures plateau at levels identical to the 3:1 ratio, which was a modest enhancement over both parental viruses 1 and 2 in the presence of heparin. This suggests that not all of the enhancement was due to HS-mediated binding but rather related to other steps involved in vector transduction (e.g., coreceptor usage, trafficking, uncoating, or facilitation of second-strand synthesis [Fig. 8B ]).
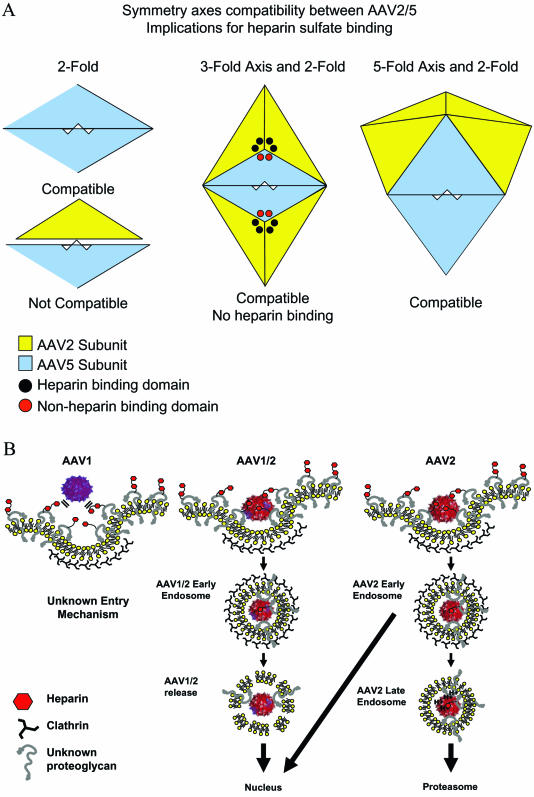
FIG. 8.
(A) Model of potential interactions between AAV2 and AAV5 capsid subunits. The triangular shapes represent individual subunits of AAV2 (yellow) or AAV5 (light blue). The heparin-binding domain of AAV2 is represented by black circles near the threefold axis of symmetry, whereas the corresponding non-heparin-binding domains of AAV5 are represented as red circles. AAV5 subunits can interact at the twofold axis of symmetry, whereas AAV2 and AAV5 subunits cannot form in a manner compatible with building intact virions (bottom left). The center depicts two threefold axes of symmetry consisting of a mosaic composed of AAV2 and AAV5 subunits. The twofold axis of symmetry is composed of interacting AAV5 subunits, which is consistent with virion formation. However, the ability of these threefold axes, composed of AAV2 andAAV5 subunits, to bind heparin is compromised. The interactions of AAV2 and AAV5 at the fivefold axis are also compatible with virion formation (right). (B) Model of AAV1/AAV2 mosaic virus interactions with C2C12 cells resulting in synergistic transduction. The topology map is used to depict the virion, with the blue subunits representing AAV1 and the red subunits representing AAV2. HS is represented as small hexagons attached to HS proteoglycans (HSPG) (gray question marks). AAV1 does not interact with C2C12 cells through HSPG, whereas AAV2 does (left panel). The AAV2 portion of AAV1/2 binds HSPG through the threefold axis of symmetry, and an endosome forms around the virion (middle panel). The trafficking of AAV1/2 through the cell may be controlled by the AAV1 portion of the virion by a route distinct from that used by AAV2. AAV2 internalizes through coated vesicles and yet traffics to late endosomes and proteosomes where a larger proportion is degraded (short arrow) and a portion is directed to the nucleus (long arrow) (right panel).
DISCUSSION
In the present study, AAV serotype helper plasmids mixed at various ratios were used to “cross-dress” AAV2 DNA templates into virion particles. Characterization of the virus particles for titer, receptor binding, and infectivity on specific cell lines determined at least three classes of AAV based on structural compatibility.
Previously, we grouped the AAV serotypes into three classes based on sequence homology (30). Here, we strengthen the definition of these classes and now propose that these classes be referred to as AAV subgroups A (AAV1, AAV2, and AAV3), B (AAV5), and C (AAV4) based on the nomenclature used for the human adenovirus serotypes. The importance of this distinction in classification is that the ever-increasing number of new AAV serotypes will primarily be grouped according to sequence homology and further refined functionally on the basis of capsid structural compatibility.
Critical to the interpretation of these experiments was the ability to confirm that mixed virus particles were generated. A number of controls were utilized to verify this, including the complementation and rescue of the mutants through plasmid mixing (Table 1) and analysis of Hirt DNA after it was cloned into E. coli to demonstrate recombination was not a factor for the generation of mixed viruses (data not shown). Western blot and Hirt analyses (Fig. 2B and C) indicate that the components necessary to form virions are present (i.e., AAV capsid proteins and monomer genomes) even when intact virions are not detected (i.e., subgroup A and C mixes). In addition, some mosaic virions bound to both HS and mucin columns, displaying the affinity of the respective parental virus to their putative receptor substrate (types 3 and 5, respectively). Further, to ensure transduction observations were the results of cross-dressed virions, preparations of premade serotype-specific virus were mixed at identical ratios, and distinct vector transduction profiles were generated compared to mosaic particles from AAV1/2 on C2C12 cells (compare Fig. 7A and B [minus heparin]). Together, these experiments confirmed the ability to generate mosaic or transcapsidated virus by mixing different ratios of helper plasmids.
Transcapsidation provides insight into AAV capsid structure.
Transcapsidation can be utilized as a method to functionally define structural relatedness among the serotypes without having to perform crystallographic studies on all new serotypes. For example, transcapsidation of subgroup A viruses (AAV1, AAV2, or AAV3) all resulted in efficient virion production (Fig. 2A), indicating that these three viruses are structurally related and that the AAV2 crystal structure (42) can be used with confidence when modeling changes in the AAV1 and AAV3 capsids.
In contrast, the viruses of subgroup A, when mixed with subgroup C (AAV4), inefficiently packaged genomes (Fig. 2A). Based on the crystal structure of AAV2, the capsid subunits interact at the two-, three-, and fivefold axes of symmetry to form stable virions (45). The inability to generate stable virus between subgroups A and C may reflect the failure of these structural subunits to interact at one or more of these axes of symmetry (Fig. 8A). Since this is a functional and not a homology test, this information could not be derived from the sequence only. To further support this hypothesis, a comparison of the AAV2 and AAV4 crystal structures revealed clashes between amino acids consistent with our proposed subgrouping (M. Agbandje-McKenna, unpublished data).
Transcapsidation between subgroups A and B generated reduced the viral titer at all ratios, suggesting an incompatibility of subunit interaction at only one axis (Fig. 8A). As an example, the subunits in AAV2/5 transcapsidated virions may form stable three- and fivefold axes but an unstable twofold axis. This would require identical subgroup pairing at the twofold axis to form stable virions (Fig. 8A). Based on this hypothesis, each pair of AAV5 subunits at the twofold axis would potentially reduce the affinity for heparin at adjacent threefold axes (Fig. 8A, threefold). The amino acids responsible for heparin binding of AAV2 are located near the peaks on the slope facing the threefold axis of symmetry (23, 26, 45). Xie et al. proposed that more than one threefold axis of symmetry may be necessary for physiological binding of heparin, and the juxtaposition of multiple sites may determine the strength of the interaction (45). In our mixed AAV2/5 virions, we observed a decrease in heparin binding (Fig. 3S to W), as well as decreased titers at all ratios, a finding consistent with this model. In addition, the physical characteristics of the AAV2/5 mixed virions were further supported by transduction experiments on appropriate cell lines showing that the binding experiments were relevant (Fig. 6; 2/5 HeLa [heparin +] and CHO pgsD [heparin −]). Another prediction based on this model would be an accumulation of partially assembled subunits, a by-product of this structural incompatibility. This prediction was supported by electron microscopic analysis, in which smaller structures were observed with the AAV1/5 mixed virions and also in subgroup A/B (Fig. 2D, arrows).
Transcapsidation provides insight into AAV binding.
The binding profiles of subgroup A/B mixtures on heparin and mucin matrices provide insight to the fundamental mechanism of viral binding. These mixtures bound mucin at all ratios except 19:1, at which 95% of the virion is composed of subgroup A subunits. These results indicate that AAV5 binds mucin through a few subunits. However, consistent with predictions from the AAV2 crystal structure, the corresponding heparin-binding profiles for subgroup A/B mixtures indicated that multiple subunits in juxtaposition were needed for heparin binding. This is evidenced by the loss of heparin binding for AAV2/5 at a ratio of 3:1 (Fig. 3T).
A biological consequence of the binding phenomenon is seen in the transduction data of AAV2/5 mixtures on HeLa cells, which have high levels of HS. As the composition of the virion changed from predominantly AAV2 at the 19:1 ratio to more AAV5 at the 3:1 ratio, the ability to transduce HeLa cells correspondingly dropped from ∼11 to 3% (Fig. 6P). In contrast, the ability of AAV2/5 to transduce an HS-deficient cell line (CHO pgsD), increased from 2 to >30% as the composition of the virion changed from type 2 to type 5 (Fig. 6S).
The binding profiles of group A/B mixed virus preparations to either heparin or mucin agarose revealed that AAV3/5 mixtures at the 3:1 ratio exhibited duality in binding. In an effort to determine whether this dual binding was of biological significance, HeLa cells were treated with neuraminidase, and the virus and cells were treated with soluble heparin (Fig. 5). The neuraminidase results demonstrated a significant dependence on sialic acid for transduction, correlating the mucin binding and transduction data. Interestingly, heparin inhibition was not observed for either the AAV3 control or the AAV3/5 mosaic particles (Fig. 5). This unexpected result may be attributed to amino acid differences between AAV3a, which is inhibited by heparin (16), and AAV3b, which was used in the present study and has never been assessed for heparin inhibition (32). A complete resolution of this observation will require mixed virions with AAV3a capsid components instead of AAV3b.
Transcapsidation can be used to produce new and novel AAV vectors.
A potential outcome from the transcapsidation experiments is that mixed viruses may exhibit a tropism distinct from either parent or that minor components may have significant influence on the properties of these cross-dressed virions. This is clearly seen in the unexpected enhancement in transduction of some cell lines when the AAV1 helper construct was combined with AAV2 or AAV3 helpers (Fig. 6B, C, E, G, I, and J). This synergy in transduction efficiency may result from sharing advantageous phenotypes from each of the parental serotypes. Alternatively, a unique property could result from a juxtaposition of subunits from different parents at a specific ratio or at a specific axis of symmetry.
A model to explain the enhanced transduction of AAV1/2 mixed virions is depicted in Fig. 8B and examines the viral entry and trafficking fates of the AAV1 parental, AAV2 parental, and AAV1/2 mixed virions. Transduction of C2C12 cells by AAV1 and AAV2 was relatively inefficient, in contrast to AAV1/2 mixtures at 1:3 and 1:19 ratios, at which transduction was >4-fold higher than for AAV2 and 20-fold higher than for AAV1 (Fig. 7B). The results of the heparin inhibition studies demonstrated AAV2 dependency on heparin to transduce C2C12 cells (11), whereas AAV1 was not affected (Fig. 7B). The AAV1/2 mixtures composed predominantly of AAV1 subunits were not sensitive to the presence of soluble heparin but became sensitive as the composition of the virions changed to include more AAV2 subunits. Interestingly, mosaic virions that were composed of more AAV 2 subunits demonstrated synergistic enhancement that correlated in part to ability of the viral particle to bind HS.
However, this was not the sole reason for enhanced transduction since, in the presence of HS, the transduction of C2C12 cells by AAV1/2 (1:1) or AAV1/2 (1:3) was still >5-fold higher than for either parent. It is possible that alternate receptors, coreceptors, or differences in trafficking between AAV1 and AAV2 are contributing factors (Fig. 8B).
The trafficking pathways of AAV2 and AAV5 have been shown to be different (2, 4, 9, 33). This raises the possibility that AAV1 not only uses a receptor different from that of AAV2 (Fig. 8B, AAV1) but may also use a different trafficking route. Once bound to the membrane by the heparin-binding contribution of AAV2, the AAV1/2 mosaic virions internalize into endosomes, where the small number of AAV1 subunits present in these mixtures may exert a dominant trafficking phenotype, and direct the AAV1/2 particles efficiently through the cytoplasm to the nucleus (Fig. 8B, AAV1/2). If this is the case, it would indicate a mechanism different from that proposed by Hauck and Xiao (17), who suggest that to gain a property the virion must be composed of a majority of that serotype structural component. In any case, additional studies should allow one to map the region(s) of AAV1 contributing the enhancement factor observed in our and their mosaic virions.
Conclusions and road map for new vector design.
The straightforward approach described here of mixing different AAV helper plasmids to generate cross-dressed AAV virions can be applied to all AAV serotypes and should allow for continued analyses of AAV capsid assembly, receptor-mediated binding, and virus trafficking. The transcapsidation of the AAV serotypes defines subgroups based on sequence homology, as well as structural relatedness. One advantage of this approach is that it represents a method to functionally define structural relatedness without having to perform crystallographic studies on all new serotypes. If this classification scheme is applied to the newly described serotypes AAV7 and AAV8, we would predict that by sequence homology they would fall into subgroup A. It remains to be determined whether this grouping would be maintained after structural compatibility experiments similar to the ones described here are performed. The outcome of these types of experiments may further define the addition of new subgroups (i.e., sequence homology to subgroup A but structural compatibility to subgroup C). It is possible that new and/or novel transduction profiles will be observed upon mixing serotype 7 or 8 with the members of subgroup A. Exploitation of this approach in generating custom-designed AAV vectors should be of significant value to the field of gene therapy.
Acknowledgments
We thank Mavis Agbandje-McKenna for her invaluable crystallography advice. We acknowledge the technical assistance of Angelique Camp in this study. We thank both Jennifer Giles and Josh Grieger for helpful discussion and critique of the manuscript.
This study was supported by NIH research grants 5P01GM059299 and 5P01HLL6973-01-02.
REFERENCES
- 1.Auricchio, A., E. O'Connor, M. Hildinger, and J. M. Wilson. 2001. A single-step affinity column for purification of serotype 5-based adeno-associated viral vectors. Mol. Ther. 4:372-374. [DOI] [PubMed] [Google Scholar]
- 2.Bantel-Schaal, U., B. Hub, and J. Kartenbeck. 2002. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J. Virol. 76:2340-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartlett, J. S., J. Kleinschmidt, R. C. Boucher, and R. J. Samulski. 1999. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab′γ)2 antibody. Nat. Biotechnol. 17:181-186. [DOI] [PubMed] [Google Scholar]
- 4.Bartlett, J. S., R. Wilcher, and R. J. Samulski. 2000. Infectious entry pathway of adeno-associated virus and adeno-associated virus vectors. J. Virol. 74:2777-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles, D. E., J. E. Rabinowitz, and R. J. Samulski. 2003. Marker rescue of adeno-associated virus (AAV) capsid mutants: a novel approach for chimeric AAV production. J. Virol. 77:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson, B. L., C. S. Stein, J. A. Heth, I. Martins, R. M. Kotin, T. A. Derksen, J. Zabner, A. Ghodsi, and J. A. Chiorini. 2000. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA 97:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douar, A. M., K. Poulard, D. Stockholm, and O. Danos. 2001. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 75:1824-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan, D., Q. Li, A. W. Kao, Y. Yue, J. E. Pessin, and J. F. Engelhardt. 1999. Dynamin is required for recombinant adeno-associated virus type 2 infection. J. Virol. 73:10371-10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan, D., Z. Yan, Y. Yue, W. Ding, and J. F. Engelhardt. 2001. Enhancement of muscle gene delivery with pseudotyped adeno-associated virus type 5 correlates with myoblast differentiation. J. Virol. 75:7662-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 99:11854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 5:1052-1056. [DOI] [PubMed] [Google Scholar]
- 14.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3:964-975. [DOI] [PubMed] [Google Scholar]
- 15.Haberman, R., G. K. Lux, T. McCown, and R. J. Samulski. 1999. Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration, p. 4.17.1-4.17.25. In J. Crawley, C. Gerfen, M. Rogawski, D. Sibley, P. Skolnick, and S. Wray (ed.), Current protocols in neuroscience, vol. 1. John Wiley & Sons, Inc., New York, N.Y.
- 16.Handa, A., S. Muramatsu, J. Qiu, H. Mizukami, and K. E. Brown. 2000. Adeno-associated virus (AAV)-3-based vectors transduce hematopoietic cells not susceptible to transduction with AAV-2-based vectors. J. Gen. Virol. 81:2077-2084. [DOI] [PubMed] [Google Scholar]
- 17.Hauck, B., L. Chen, and W. Xiao. 2003. Generation and characterization of chimeric recombinant AAV vectors. Mol. Ther. 7:419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hauck, B., and W. Xiao. 2003. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 77:2768-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.High, K. 2002. AAV-mediated gene transfer for hemophilia. Genet. Med. 4:56S-61S. [DOI] [PubMed] [Google Scholar]
- 20.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 21.Janson, C., S. McPhee, L. Bilaniuk, J. Haselgrove, M. Testaiuti, A. Freese, D. J. Wang, D. Shera, P. Hurh, J. Rupin, E. Saslow, O. Goldfarb, M. Goldberg, G. Larijani, W. Sharrar, L. Liouterman, A. Camp, E. Kolodny, J. Samulski, and P. Leone. 2002. Clinical protocol: gene therapy of Canavan disease: AAV-2 vector for neurosurgical delivery of aspartoacylase gene (ASPA) to the human brain. Hum. Gene Ther. 13:1391-1412. [DOI] [PubMed] [Google Scholar]
- 22.Kaludov, N., K. E. Brown, R. W. Walters, J. Zabner, and J. A. Chiorini. 2001. Adeno-associated virus serotype 4 (AAV4) and AAV5 both require sialic acid binding for hemagglutination and efficient transduction but differ in sialic acid linkage specificity. J. Virol. 75:6884-6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 77:11072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, C. M., Y. K. Lai, and P. E. Rakoczy. 2002. Adenovirus and adeno-associated virus vectors. DNA Cell Biol. 21:895-913. [DOI] [PubMed] [Google Scholar]
- 25.Muzyczka, N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97-129. [DOI] [PubMed] [Google Scholar]
- 26.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 77:6995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ponnazhagan, S., G. Mahendra, S. Kumar, J. A. Thompson, and M. Castillas, Jr. 2002. Conjugate-based targeting of recombinant adeno-associated virus type 2 vectors by using avidin-linked ligands. J. Virol. 76:12900-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qing, K., C. Mah, J. Hansen, S. Zhou, V. Dwarki, and A. Srivastava. 1999. Human fibroblast growth factor receptor 1 is a coreceptor for infection by adeno-associated virus 2. Nat. Med. 5:71-77. [DOI] [PubMed] [Google Scholar]
- 29.Rabinowitz, J. E., F. Rolling, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz, J. E., and R. J. Samulski. 2000. Building a better vector: the manipulation of AAV virions. Virology 278:301-308. [DOI] [PubMed] [Google Scholar]
- 31.Rabinowitz, J. E., W. Xiao, and R. J. Samulski. 1999. Insertional mutagenesis of AAV2 capsid and the production of recombinant virus. Virology 265:274-285. [DOI] [PubMed] [Google Scholar]
- 32.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanlioglu, S., P. K. Benson, J. Yang, E. M. Atkinson, T. Reynolds, and J. F. Engelhardt. 2000. Endocytosis and nuclear trafficking of adeno-associated virus type 2 are controlled by rac1 and phosphatidylinositol-3 kinase activation. J. Virol. 74:9184-9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi, W., G. S. Arnold, and J. S. Bartlett. 2001. Insertional mutagenesis of the adeno-associated virus type 2 (AAV2) capsid gene and generation of AAV2 vectors targeted to alternative cell-surface receptors. Hum. Gene Ther. 12:1697-1711. [DOI] [PubMed] [Google Scholar]
- 35.Shi, W., and J. S. Bartlett. 2003. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol. Ther. 7:515-525. [DOI] [PubMed] [Google Scholar]
- 36.Summerford, C., J. S. Bartlett, and R. J. Samulski. 1999. ανβ5 integrin: a coreceptor for adeno-associated virus type 2 infection. Nat. Med. 5:78-82. [DOI] [PubMed] [Google Scholar]
- 37.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 72:1438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walters, R. W., J. M. Pilewski, J. A. Chiorini, and J. Zabner. 2002. Secreted and transmembrane mucins inhibit gene transfer with AAV4 more efficiently than AAV5. J. Biol. Chem. 277:23709-23713. [DOI] [PubMed] [Google Scholar]
- 39.Walters, R. W., S. M. Yi, S. Keshavjee, K. E. Brown, M. J. Welsh, J. A. Chiorini, and J. Zabner. 2001. Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J. Biol. Chem. 276:20610-20616. [DOI] [PubMed] [Google Scholar]
- 40.Weber, M., J. Rabinowitz, N. Provost, H. Conrath, S. Folliot, D. Briot, Y. Cherel, P. Chenuaud, J. Samulski, P. Moullier, and F. Rolling. 2003. Recombinant adeno-associated virus serotype 4 mediates unique and exclusive long-term transduction of retinal pigmented epithelium in rat, dog, and nonhuman primate after subretinal delivery. Mol. Ther. 7:774-781. [DOI] [PubMed] [Google Scholar]
- 41.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 74:8635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie, Q., W. Bu, S. Bhatia, J. Hare, T. Somasundaram, A. Azzi, and M. S. Chapman. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. USA 99:10405-10410. [DOI] [PMC free article] [PubMed] [Google Scholar]