Abstract
The X protein (HBX) of the hepatitis B virus (HBV) is not essential for the HBV life cycle in vitro but is important for productive infection in vivo. Our previous study suggests that interaction of HBX with the proteasome complex may underlie the pleiotropic functions of HBX. With the woodchuck model, we demonstrated that the X-deficient mutants of woodchuck hepatitis virus (WHV) are not completely replication defective, possibly behaving like attenuated viruses. In the present study, we analyzed the effects of the proteasome inhibitors on the replication of wild-type and X-negative HBV and WHV. Recombinant adenoviruses or baculoviruses expressing replicating HBV or WHV genomes have been developed as a robust and convenient system to study viral replication in tissue culture. In cells infected with either the recombinant adenovirus-HBV or baculovirus-WHV, the replication level of the X-negative construct was about 10% of that of the wild-type virus. In the presence of proteasome inhibitors, the replication of the wild-type virus was not affected, while the replication of the X-negative virus of either HBV or WHV was enhanced and restored to the wild-type level. Our data suggest that HBX affects hepadnavirus replication through a proteasome-dependent pathway.
Human hepatitis B virus (HBV) is a member of the Hepadnaviridae family, which includes the hepatitis viruses of the woodchuck, ground squirrel, tree squirrel, Pekin duck, and heron. HBV has a fourth open reading frame, termed the hepatitis B virus X (HBX) gene. The HBX gene is well conserved among the mammalian hepadnaviruses and codes for a 16.5-kDa protein. The protein can activate the transcription of a variety of viral and cellular genes (1, 7). Since HBX does not bind to DNA directly, its activity is thought to be mediated via protein-protein interaction. HBX has been shown to enhance transcription through AP-1 and AP-2 (2, 24) and to activate various signal transduction pathways (9, 11). Several recent studies have also identified possible cellular targets of HBX, including members of the CREB/ATF family (19), the TATA-binding protein (20), RNA polymerase subunit RPB5 (6), the UV-damaged DNA-binding protein (25), the replicative senescence p55sen (28), and the mitochondrial protein (31). HBX has also been shown to interact with p53 and inhibit its function (29, 30). Furthermore, X protein is necessary for the establishment of a productive infection in vivo (5, 37). Recent results have demonstrated that signaling through calcium may mediate a function of HBX in viral replication, and calcium chelator can inhibit viral replication by blocking the effect of HBX (4).
We have previously demonstrated that the proteasome complex is a cellular target of HBX (18, 34). We demonstrated that this interaction is functionally important in the pleiotropic effect of HBX (17). With the woodchuck model, we demonstrated that the X-deficient mutants of woodchuck hepatitis virus (WHV) are not completely replication defective, behaving like attenuated viruses (35).
Adenovirus and baculovirus vectors have been used for efficient transduction of foreign genes, especially in hepatocyte-derived cell lines. Recombinant adenovirus or baculovirus expressing hepadnavirus genome has recently been shown to be a robust and convenient system for studying HBV replication in tissue culture (10, 27). Such a system is superior to transfection of viral genomic DNA because it is more efficient and supports the full cycle of viral replication, including the production of covalently closed-circle DNA (cccDNA) (10, 27). In the present study, we constructed recombinant adenoviruses or baculoviruses expressing replicating HBV or WHV genomes with or without a functional X gene. Using these recombinant viruses, we determined the effects of proteasome inhibitors on the functions of the X protein in hepadnavirus replication and proved that proteasome inhibitors restored the replication defect of X-negative HBV and WHV.
MATERIALS AND METHODS
Plasmid construction.
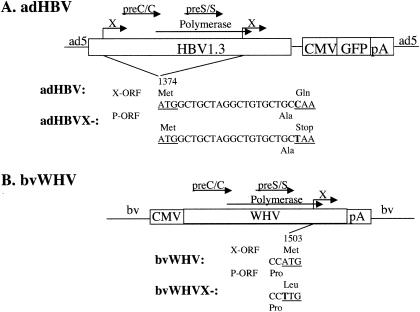
Recombinant adenovirus expressing the HBV genome was generated using the AdEasy system (16). A 1.3× genome of HBV DNA was cloned into an adenovirus vector to generate the adHBV recombinant virus, as previously described (27). For the construction of the HBV X mutant, a C-to-T mutation was introduced to create a premature stop codon of the X open reading frame at amino acid position 8 of the 5′ and 3′ terminal redundant region of the 1.3× genome (adHBVX−) (see Fig. 1A). To generate recombinant baculovirus expressing the WHV genome, the polyhedrin promoter of the baculovirus vector pFastBac (Bac-to-Bac; Gibco-BRL, Gaithersburg, Md.) was deleted, and a 1.2× full-length genome of an infectious WHV strain (13) driven by the cytomegalovirus promoter was cloned into the EcoRI sites of the promoterless pFastBac vector, resulting in the baculovirus-WHV wild type, bvWHV. The bvWHV X mutant was created by introducing an ATG-to-TTG mutation at the first translation initiation site of WHVX of bvWHV, resulting in bvWHVX− (see Fig. 1B).
FIG. 1.
Schematic diagram of adHBV and bvWHV constructs. (A) adHBV constructs. HBV1.3 represents the 1.3× genome of HBV. The X mutation and its approximate position are shown. (B) bvWHV constructs. WHV represents the 1.2× genome of WHV. The X mutation and its approximate position are shown. The nucleotide sequences are shown in the middle, and the amino acid sequences are shown at the top for the X open reading frame (X-ORF) and at the bottom for the overlapping pol open reading frame (P-ORF).
Cell cultures.
Sf9 insect cells were maintained in a flask at 28°C in Sf-900 II serum-free medium (Gibco-BRL). Recombinant baculoviruses were generated and amplified in Sf9 cells. The 293 cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) containing 10% fetal bovine serum in a humidified incubator (5% CO2) and used for generation and amplification of adenoviruses as described previously (27). The human hepatoma HepG2 or Huh7 cells were grown in Dulbecco's modified Eagle's medium (Gibco-BRL) containing 10% fetal bovine serum in a humidified incubator (5% CO2).
Proteasome inhibitors and calcium signaling reagents.
Proteasome-specific inhibitors lactacystin (Kamiya Biomedical Company, Seattle, Wash.), MG132 (Proscript, Inc., Cambridge, Mass.), and epoxomicin (Peptides Institute Inc., Osaka, Japan) and N-acetyl-l-leucinyl-l-methonal (ALLM; Sigma Chemical Co., St. Louis, Mo.), a potent calpain inhibitor that has only a very weak activity against the proteasome, were used. Calcium signaling reagents BAPTA-AM (cytosolic calcium chelator) and valinomycin (calcium mobilizing agent) were obtained from BIOMOL Research Laboratories Inc. (Plymouth Meeting, Pa.).
Infection of HepG2 or Huh7 cells.
HepG2 or Huh7 cells were infected with recombinant viruses at about 80% confluency at day 1 or 2 postseeding. The multiplicity of infection (MOI) was optimized. For bvWHV infection, HepG2 cells were infected (MOI of 50) for 2 h and washed twice with phosphate-buffered saline. After addition of fresh medium, different doses of proteasome inhibitors or calcium signaling reagents were added. The cells were harvested 3 days postinfection for the analyses of virus infections. For adHBV infection, HepG2 or Huh7 cells were infected at a MOI of 5 for 2 h, washed twice with phosphate-buffered saline, and maintained in fresh medium. After incubation overnight, different doses of proteasome inhibitors or calcium signaling reagents were added. The cells were harvested 1 day later for analysis of viral infection.
Viral nucleic acid analysis.
HepG2 cells were infected with recombinant adenovirus-HBV or baculovirus-WHV as described above. Three days after infection, the cells were harvested for viral RNA and DNA analyses. RNA was prepared by the guanidium isothiocyanate-acid-phenol method (TRIzol RNA isolation Kit; Invitrogen, Carlsbad, Calif.), analyzed by 1% formaldehyde agarose gel electrophoresis, and hybridized with 32P-labeled WHV- or HBV-specific probe. Viral replicative intermediates associated with intracellular core particles were isolated by ultracentrifugation of cell lysate through a 30% sucrose cushion and analyzed by Southern blot hybridization as described previously (15).
Viral antigen assays.
After bvWHV or adHBV infection, culture medium was analyzed for WHV surface antigen using solid-phase radioimmunoassay (35) or HBV surface antigen using AUSZYME (ABBOTT Laboratories, Abbott Park, Ill.) and for HBV e antigen using ETI-EBK PPLUS (DiaSorin Inc., Stillwater, Minn.).
RESULTS
Effect of X on viral replication in cells infected with recombinant viruses.
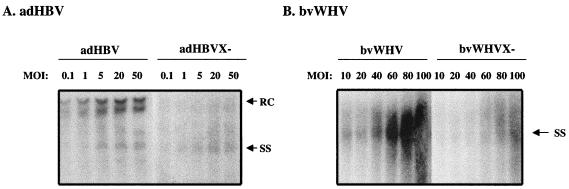
To study the effect of HBX on HBV replication, we generated an adenovirus (adHBV) expressing HBV and a recombinant baculovirus (bvWHV) expressing the WHV genome (Fig. 1) in HepG2 cells. HepG2 cells were infected with the recombinant viruses at MOIs ranging from 0.1 to 50 for adHBV or 10 to 100 for bvWHV. To evaluate viral replication, cytoplasmic core particles were prepared 3 days postinfection and the core-associated viral DNA was extracted for the analysis of replicative intermediates. Figure 2A shows HBV replication and Fig. 2B shows WHV replication in HepG2 cells. The X mutants replicated at about 10% of the wild-type HBV or WHV level, which is consistent with previous results (3, 35). Both wild-type and X mutants replicated at levels proportional to the MOI. Therefore, adHBV or bvWHV infection offers an efficient and dose-dependent replication of hepadnavirus genomes in the human hepatoma HepG2 cells.
FIG. 2.
Analysis of viral replication in HepG2 cells. HepG2 cells were infected with adHBV at MOIs ranging from 0.1 to 50 or bvWHBV at MOIs ranging from 10 to 100. The viral core particles were purified 3 days postinfection, and the viral DNA was extracted for Southern blot analyses. (A) Replication of adHBV; (B) replication of bvWHV. Results are representative of at least three independent experiments. RC, relaxed circular DNA; SS, single-stranded DNA.
Effect of proteasome inhibitors on hepadnavirus replication.
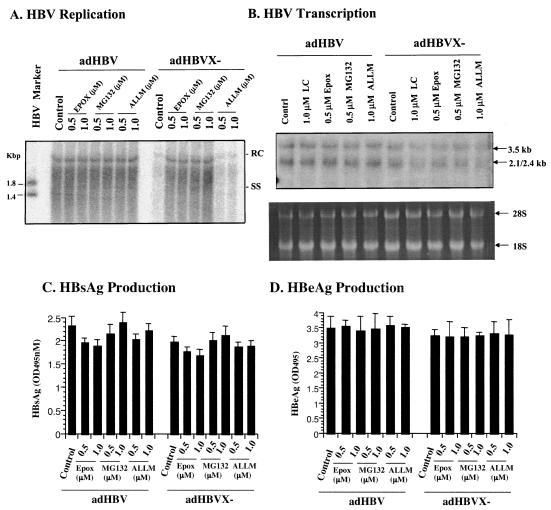
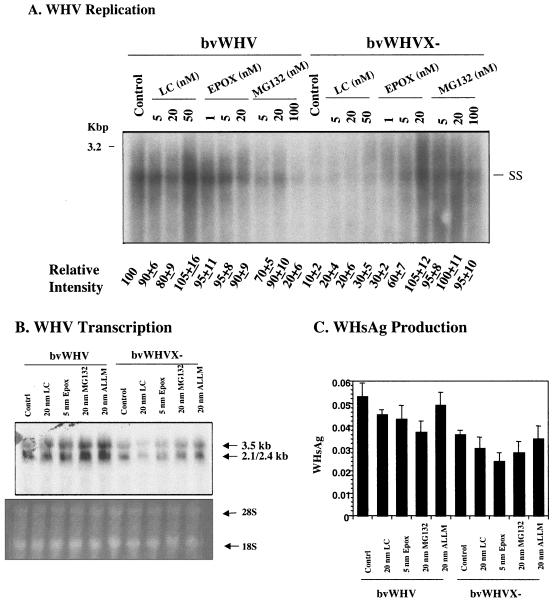
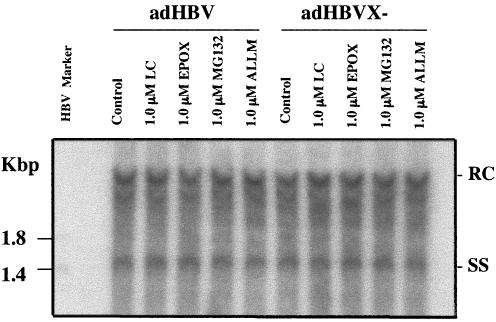
To test whether the proteasome pathway is involved in HBV replication, HepG2 cells were infected with adHBV or bvWHV, described above, and then treated with different doses of proteasome-specific inhibitors. The cells were also treated with ALLM, a calpain inhibitor with little effect on the proteasome activity, as a nonspecific control. After treatment, the cells were harvested and analyzed for viral gene expression and replication. In cells infected with recombinant viruses, the replication level of the X-negative virus was about 10% of that of the wild type. However, in the presence of proteasome inhibitors, the replication of the wild type was not affected, whereas the replication of the X-negative virus, either HBV (Fig. 3A) or WHV (Fig. 4A), was restored to the wild-type level. Treatment with ALLM had no effect on replication of either the wild type or the X-negative mutant for either HBV (Fig. 3A) or WHV (data not shown). To test whether the proteasome inhibitors have any effects on viral gene expression at the level of transcription and protein synthesis, total RNA from the infected cells was isolated and analyzed by Northern blotting, and the culture medium of the infected cells was assayed for viral antigens. There was little difference in viral transcription with or without proteasome inhibitors in cells infected with either wild-type or X-negative viruses (Fig. 3B for HBV and Fig. 4B for WHV). No significant differences were detected in viral antigen production (Fig. 3C and D for HBV and 4C for WHV). In the HBV and WHV experiments, presumably toxic effects of proteasome inhibitors were seen at the high concentration, contributing to the lower levels of replication, transcription, and/or protein production (Fig. 4). The proteasome inhibitors appeared to affect viral replication of only X-negative virus but not wild-type virus, suggesting that the proteasome pathway plays an important role in the function of HBX in viral replication.
FIG. 3.
Effects of proteasome inhibitors on HBV gene expression. HepG2 cells were infected with recombinant adHBV at a MOI of 5 and treated with two kinds of proteasome inhibitors, epoxomicin (EPOX) or MG132, at different doses (lactacystin was also tested for HBV transcription). As a control, cells were also treated with ALLM. The cells were harvested 2 days postinfection. (A) Effects on HBV replication. The core-associated DNA was extracted for the detection of replicative intermediates by Southern blotting with an HBV-specific probe. HBV Marker is the 3.2-kbp HBV genome digested with NcoI (1.8- and 1.4-kbp fragments). (B) Effects on HBV transcription. Total RNA was extracted for the detection of viral transcripts by Northern blotting. The 28S and 18S RNAs were shown as loading controls. (C and D) Effects on viral antigen production. The culture medium was tested for HBV surface antigen (HBsAg) (C) and HBV e antigen (HBeAg) (D). Results are representative of three independent experiments.
FIG. 4.
Effects of proteasome inhibitors on WHV gene expression. HepG2 cells were infected with recombinant bvWHV at a MOI of 50 and treated with three different proteasome inhibitors, lactacystin (LC), epoxomicin (EPOX), and MG132, at different doses. The cells were harvested for analysis 3 days postinfection. (A) Effects on WHV replication. The core-associated DNA was extracted for the detection of replicative intermediates by Southern blotting with WHV-specific probe. The signal intensities were quantitated and normalized to the control (control as 100), and means from three independent experiments are shown at the bottom. The WHV marker is full-length WHV DNA. (B) Effect on WHV transcription. Total RNA was isolated for the detection of viral transcripts by Northern blotting. The 28S and 18S RNAs were used as loading controls. (C) Effect on viral antigen production. Culture medium was assayed for WHV surface antigen (WHsAg). Results are representative of three independent experiments.
The HBV X gene is known to affect HBV replication in HepG2 cells but not Huh7 cells. We therefore infected Huh7 cells with AdHBV wild-type or X-negative viruses and examined the effect of proteasome inhibitors on HBV replication. No significant differences were detected in the levels of viral replication between wild-type and X-negative viruses (Fig. 5). Treatment with proteasome inhibitors (LC, EPOX, and MG132) or the calpin inhibitor ALLM had no effect on HBV replication of either wild-type or X-negative viruses (Fig. 5). Therefore, the proteasome inhibitors affect replication of X-negative HBV only in cells in which the X gene is functional.
FIG. 5.
Effects of proteasome inhibitors on HBV replication in Huh7 cells. Huh7 cells were infected with the recombinant adHBV wild-type or X-minus mutant at a MOI of 5 and treated with three kinds of proteasome inhibitors, lactacystin (LC), epoxomicin (EPOX), or MG132, at different doses. As a control, cells were also treated with ALLM. The cells were harvested for viral infection 2 days postinfection. The core-associated DNA was extracted for detection of replicative intermediates by Southern blotting with HBV-specific probe. HBV Marker is HBV DNA/NcoI fragments. Results are representative of three independent experiments.
Effect of calcium signaling on function of HBX in HBV replication.
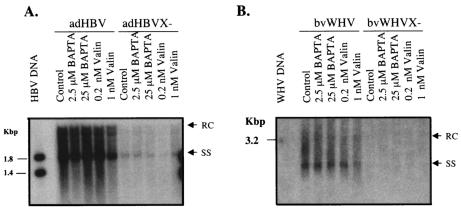
To examine whether the function of HBX in HBV replication is mediated by the calcium signaling pathway, as suggested by a recent publication (4), we tested the effects of various calcium signaling reagents on HBV replication using the recombinant virus systems. HepG2 cells were infected with adHBV or bvWHV and then treated with two calcium signaling reagents, BAPTA-AM (which chelates cytosolic calcium and inhibits calcium signaling) and valinomycin (which mobilizes intracellular calcium to activate the calcium signaling pathway). After 3 days of treatment, cytoplasmic viral core particles were isolated and the core-associated viral DNA was extracted for the analysis of replicative intermediates (Fig. 6). Treatment with either of the reagents appeared to have little effect on viral replication of either wild-type or X-negative viruses.
FIG. 6.
Effects of calcium signaling reagents on HBV replication. HepG2 cells were infected with adHBV at a MOI of 5 or with bvWHV at a MOI of 50. The cells were treated with BAPTA-AM (BAPTA) or valinomycin (Valin) for 3 days at two different doses as reported previously (4). The core-associated DNA was isolated for the detection of replicative intermediates by Southern blotting with HBV-specific (A) or WHV-specific (B) probe. Results are representative of three independent experiments. HBV marker is HBV DNA/NcoI fragments (A), and WHV marker is full-length WHV DNA (B).
DISCUSSION
The molecular mechanisms by which HBX attains its biological functions in HBV replication remain largely unresolved, despite compelling evidence that HBX is crucial for productive infection in vivo. Numerous studies have provided tantalizing evidence about the potential cellular targets and functions of HBX, but most of them have not been thoroughly tested in a relevant biological system nor rigorously examined with respect to viral replication. We have previously demonstrated the structural and functional interaction between HBX and the proteasome complex (17, 18, 34) and that the molecularly constructed HBX mutants deficient in interaction with proteasome in vitro behave as attenuated viruses in vivo (35). In this study we further demonstrated the biological importance of the proteasome complex in the functional pathway of HBX in the context of HBV replication.
Using specific proteasome inhibitors and two model systems that have been shown to be robust and reproducible in studying the complete replication cycle of HBV, including the generation of cccDNA, we showed that the reduced viral replication of the HBX-defective mutant can be restored in the presence of various proteasome-specific inhibitors. On the other hand, these inhibitors appear to have little effect on wild-type replication. This phenomenon is specific for the proteasome activities, because other protease inhibitors had no effect. HBV and WHV appear to behave the same way in response to the proteasome inhibitors. The doses of the proteasome inhibitors used in the study are low and have a minor or no toxic effect on the cells for the duration of treatment (with the exception of the higher dose of MG132). In addition, the specific effect of the proteasome inhibitors on X-negative but not wild-type viruses supports the notion that this observation is biologically relevant. The proteasome inhibitors had little effect on viral transcription and protein production. Therefore, the enhanced replication of the HBX mutant in the presence of the inhibitors appears to be operational at the posttranslational and most likely the assembly level. This is consistent with the functional effect of HBX on viral replication and the potential interaction point of the proteasome complex and HBX. HBX possibly inhibits the degradation of nucleocapsid by proteasome during the assembly process, as a mechanism of promoting viral replication and inhibiting the cellular antiviral pathway (26). In light of recent studies showing the importance of proteasome in the endogenous antiviral response to viral infection (22, 23), further experimentation is needed to prove our proposed mechanism of action by HBX.
Interestingly, interferons have been shown to be potent inhibitors of HBV replication either in vitro or in vivo (21, 32). One of the well-known interferon-activated targets is the proteasome complex, which, in addition to other changes, acquires enhanced proteolytic activities after treatment with interferons (8, 12). A recent study with HBV transgenic mice also pointed to a potentially critical role of proteasome activation in inhibiting HBV replication (32). All these intriguing observations provide additional support that HBX-mediated interaction with the proteasome may underlie an important aspect of HBV replication.
The doses of proteasome inhibitors used in this study are below the effective concentrations that are required to fully inhibit the proteasome activities in tissue culture, but they are within the inhibitory concentrations in standard in vitro proteasome assays using purified proteasome complexes (8, 17). It is possible that the multicatalytic functions of proteasome may be differentially sensitive to various inhibitors under diverse biological conditions. The lower dose with suboptimal inhibition of proteasome activities may be sufficient to effect the necessary changes for efficient viral replication in our model system. We could not use the fully effective doses because of the potential toxicity. Furthermore, in our previous publication, we were able to see effects of these concentrations of proteasome inhibitors on the HBX functions (17). There are newer generations of proteasome inhibitors that are less toxic and have been used in vivo with success (36). Perhaps these inhibitors can be used to study the function of HBX in the context of hepadnavirus replication in vivo.
A recent study demonstrated that HBX mediates its effect on HBV replication through activation of the calcium signaling pathway (4). Using the viral delivery systems, we also tested the effects of two compounds, one inhibiting intracellular calcium mobilization and the other functioning as a calcium ionophore to activate the calcium signal pathway, on viral replication. In this recent publication, the former compound, BAPTA-AM, inhibits wild-type HBV replication, and the latter compound, valinomycin, enhances replication of an HBX-negative mutant. However, BAPTA-AM was not tested on HBX mutant replication, nor was valinomycin tested on wild-type replication (4). Interesting, we saw little or no effect of these two compounds on replication of either the wild-type or HBX-negative viruses. The explanation of the contrasting results of the two studies is not clear, but it is possible that the difference could be due to the different experimental systems used in each study. In the study by Bouchard et al. (4), transfection of recircularized HBV genome was used, whereas we used the viral delivery system (either adenovirus or baculovirus) to transduce the viral genome. It is possible that the viral systems cause some untoward effects on HBV replication, so the functions of these compounds affecting the calcium signaling pathway are being masked with respect to viral replication. However, the viral delivery systems provide a more biological relevant model because of the generation of cccDNA, which is absent in the DNA transfection system. In addition, the observation that the proteasome inhibitors elicit a specific effect on the HBX-negative mutant but not the wild-type virus supports a physiological role of the proteasome in HBX-dependent HBV replication.
The ultimate proof of any pathways in the functions of HBX would have to be determined with an infectious and/or in vivo model. The recent description of a human hepatoma-derived cell line capable of supporting robust HBV infection and replication would be a valuable model with which to test these hypotheses (14). In addition, the availability of several HBV transgenic mouse lines capable of supporting either wild-type or HBX mutant replication (33) provides a rational approach for testing the pharmacologic effects of various compounds targeting these pathways.
Acknowledgments
We thank the members of the Liver Diseases Section of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, for helpful discussions.
REFERENCES
- 1.Aufiero, B., et al. 1990. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 9:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benn, J., et al. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum, H. E., Z.-S. Zhang, E. Galun, F. von Weizsacker, B. Garner, T. J. Liang, and J. R. Wands. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H.-S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheong, J., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgrove, R., et al. 1989. Transcriptional activation of homologous and heterologous genes by the hepatitis B virus X gene product in cells permissive for viral replication. J. Virol. 63:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coux, O., K. Tanaka, and A. L. Goldberg. 1996. Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65:801-847. [DOI] [PubMed] [Google Scholar]
- 9.Cross, J. C., P. Wen, and W. Rutter. 1993. Transactivation by hepatitis B virus X protein is promiscuous and dependent on mitogen-activated cellular serine/threonine kinases. Proc. Natl. Acad. Sci. USA 90:8078-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delaney, W. E., IV, and H. C. Isom. 1998. Hepatitis B virus replication in human HepG2 cells mediated by hepatitis B virus recombinant baculovirus. Hepatology 28:1134-1146. [DOI] [PubMed] [Google Scholar]
- 11.Doria, M., N. Klein, R. Lucito, and R. J. Schneider. 1995. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 14:4747-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaczynska, M., K. L. Rock, and A. L. Goldberg. 1993. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 365:264-267. [DOI] [PubMed] [Google Scholar]
- 13.Girones, M. R., et al. 1989. Complete nucleotide sequence of a molecular clone of woodchuck hepatitis virus that is infectious in the natural host. Proc. Natl. Acad. Sci. USA 86:1846-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gripon, P., S. Rumin, S. Urban, J. Le Seyec, D. Glaise, I. Cannie, C. Guyomard, J. Lucas, C. Trepo, and C. Guguen-Guillouzo. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 99:15655-15660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hasegawa, K., J. Huang, J. R. Wands, H. H Obata, and J. Liang. 1991. Association of hepatitis B viral precore mutations with fulminant hepatitis B in Japan. Virology 185:460-463. [DOI] [PubMed] [Google Scholar]
- 16.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, Z., Z. Zhang, E. Doo, O. Coux, A. L. Goldberg, and T. J. Liang. 1999. Hepatitis B virus X protein is both a substrate and a potential inhibitor of the proteasome complex. J. Virol. 73:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, J., et al. 1996. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maguire, H. F., et al. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 20.Qadri, I., et al. 1995. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc. Natl. Acad. Sci. USA 92:1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robek, M. D., S. F. Wieland, and F. V. Chisari. 2002. Inhibition of hepatitis B virus replication by interferon requires proteasome activity. J. Virol. 76:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santoro, M. G. 1996. Viral infection. In V. Feige, R. I. Morimoto, I. Yahara, and B. S. Polla (ed.), Stress inducible cellular responses. Birkhauser Verlag, Boston, Mass.
- 23.Schwartz, O., V. Marechal, B. Friguet, F. Arenzana-Seisdedos, and J. M. Heard. 1998. Antiviral activity of the proteasome on incoming human immunodeficiency virus type 1. J. Virol. 72:3845-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seto, E., et al. 1990. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature 344:72-74. [DOI] [PubMed] [Google Scholar]
- 25.Sitterlin, D., T. H. Lee, S. Prigent, P. Tiollais, J. S. Butel, and C. Transy. 1997. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J. Virol. 71:6194-6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sodeik, B. 2002. Unchain my heart, baby let me go—the entry and intracellular transport of HIV. J. Cell Biol. 159:393-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sprinzl, M. F., H. Oberwinkler, H. Schaller, and U. Protzer. 2001. Transfer of hepatitis B virus genome by adenovirus vectors into cultured cells and mice: crossing the species barrier. J. Virol. 75:5108-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, B. S., et al. 1998. Identification of a protein isolated from senescent human cells that binds to hepatitis B virus X antigen. Hepatology 27:228-239. [DOI] [PubMed] [Google Scholar]
- 29.Truant, R., et al. 1995. Direct interaction of the hepatitis B virus HBx protein with p53 leads to inhibition by HBx of p53 response element-directed transactivation. J. Virol. 69:1851-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, X. W., et al. 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 91:2230-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waris, G., K. W. Huh, and A. Siddiqui. 2001. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-κB via oxidative stress. Mol. Cell. Biol. 21:7721-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wieland, S. F., R. G. Vega, R. Muller, C. F. Evans, B. Hilbush, L. G. Guidotti, J. G. Sutcliffe, P. G. Schultz, and F. V. Chisari. 2003. Searching for interferon-induced genes that inhibit hepatitis B virus replication in transgenic mouse hepatocytes. J. Virol. 77:1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu, Z., T. S. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, Z., N. Torii, A. Furusaka, N. Malayaman, Z. Hu, and T. J. Liang. 2000. Structural and functional characterization of interaction between hepatitis B virus X protein and the proteasome complex. J. Biol. Chem. 275:15157-15165. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Z., N. Torii, Z. Hu, J. Jacob, and T. J. Liang. 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 108:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zollner, T. M., M. Podda, C. Pien, P. J. Elliott, R. Kaufmann, and W. H. Boehncke. 2002. Proteasome inhibition reduces superantigen-mediated T cell activation and the severity of psoriasis in a SCID-hu model. J. Clin. Investig. 109:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]