Abstract
Hypoxia-inducible factor 1α (HIF-1α) controls the cellular responses to hypoxia, activating transcription of a range of genes involved in adaptive processes such as increasing glycolysis and promoting angiogenesis. However, paradoxically, HIF-1α also participates in hypoxic cell death. Several gene products, such as BNip3, RTP801, and Noxa, were identified as HIF-1α-responsive proapoptotic proteins, but the complicated hypoxic cell death pathways could not be completely explained by the few known genes. Moreover, molecules linking the proapoptotic signals of HIF-1α directly to mitochondrial permeability transition are missing. In this work, we report the identification of an HIF-1α-responsive proapoptotic molecule, HGTD-P. Its expression was directly regulated by HIF-1α through a hypoxia-responsive element on the HGTD-P promoter region. When overexpressed, HGTD-P was localized to mitochondria and facilitated apoptotic cell death via typical mitochondrial apoptotic cascades, including permeability transition, cytochrome c release, and caspase 9 activation. In the process of permeability transition induction, the death-inducing domain of HGTD-P physically interacted with the voltage-dependent anion channel. In addition, suppression of HGTD-P expression by small interfering RNA or antisense oligonucleotides protected against hypoxic cell death. Taken together, our data indicate that HGTD-P is a new HIF-1α-responsive proapoptotic molecule that activates mitochondrial apoptotic cascades.
Hypoxia is the most common cellular stress, with important pathological implications in many disease processes, including cerebral ischemia and myocardial infarction (15). Cells in hypoxia express a variety of adaptive or death gene products to satisfy altered metabolic demands or to remove irreversibly damaged cells (4). Adaptive genes allow increased O2 delivery to the peripheral tissues through vasodilation and angiogenesis, facilitate ATP synthesis through the glycolytic pathway, or reduce proliferative rates (12, 29, 30). In addition, antiapoptotic Bcl-2 family proteins prevent hypoxic cell death by stabilization of mitochondria or inhibition of caspase activation (28). However, in the case of severe hypoxic damage beyond the cell's adaptive capability, death-promoting genes are expressed, resulting in necrosis or apoptosis (4).
Hypoxia-inducible factor 1α (HIF-1α) is known to be a master transactivator in hypoxia, which is induced, stabilized, and translocated to the nucleus to regulate the transcription of a variety of genes involved in adaptive responses such as increased O2 delivery and angiogenesis (5, 31). While HIF-1α participates largely in adaptive responses to hypoxia, paradoxically it also mediates hypoxic cell death via the interaction with p53 or modulation of its effector expression (3, 14, 17). Although several proapoptotic genes induced by HIF-1α have been reported (3, 17, 37), the hypoxic cell death pathway might be too complicated to be explained by the few known genes.
To better understand the molecular mechanisms underlying hypoxic cell death, we have been attempting to identify novel genes that mediate hypoxic cell death by subtraction suppression hybridization. In this process, the HGTD-P gene, which was initially cloned as a novel gene expressed in human dendritic cells (GenBank accession no. AF201944), was identified, but it has not been functionally characterized yet. In this work, we show that HGTD-P was a death-inducing effector molecule downstream of HIF-1α. In addition, HGTD-P facilitated cell death by induction of the mitochondrial permeability transition (PT) via interaction with the voltage-dependent anion channel (VDAC), a component of the permeability transition pores (PTP). Suppression of endogenous HGTD-P expression with short interfering RNA (SiRNA) or antisense oligonucleotides rescued cells from hypoxic assaults by stabilizing mitochondria. HGTD-P thus transmits apoptotic signals which are sensed by HIF-1α to the mitochondrial PT. In vivo characterization of HGTD-P in knockout mice is in progress.
MATERIALS AND METHODS
Subtractive hybridization.
Subtractive hybridization was performed with a PCR-Select cDNA subtraction kit (Clontech) according to the manufacturer's protocol. Briefly, subtractive cDNA library was prepared with polyadenylated RNAs purified from SK-N-MC cells incubated under normoxic (20% O2) (driver) and hypoxic (0.5% O2) (tester) conditions for 4 h. The tester cDNAs were digested with RsaI and linked with adaptor oligonucleotides. The tester cDNA was hybridized with excess driver cDNA digested with RsaI (the first hybridization), and subtracted single-stranded cDNA was converted to double-stranded DNA by a second hybridization with another adaptor-linked first hybridization. The subtracted cDNA was amplified by PCR with the primer set located in the adaptors and inserted into the TA vector (Clontech).
Cell culture, hypoxic conditions, and cell death analysis.
PC-3 prostatic cancer cells were cultured in RPMI medium containing 10% fetal bovine serum under 5% CO2. Primary cortical neuronal cells were prepared from a postnatal day 1 BALB/c mouse as described (1). On day 4 of culture, cytosine arabinoside was added at a final concentration of 10 μM in order to prevent nonneuronal cell proliferation, and neurons were used on day 7. For hypoxic conditions, cells in medium gassed with 95% N2-5% CO2 were transferred to a hypoxic chamber equipped with an air lock and continuously gassed with 0.5% O2-5% CO2, with the balance being nitrogen. Apoptotic cells exhibiting altered DNA morphology following 4′,6′-diamidino-2-phenylindole (DAPI) staining, were enumerated manually under a fluorescence microscope.
Generation of anti-HGTD-P polyclonal antibody.
Anti-HGTD-P polyclonal antibody L16 was produced by immunizing a rabbit with keyhole limpet hemocyanin-conjugated amino acid residues 33 to 48 (DLKRINGFCTKPQESP) of HGTD-P. Antibody was purified from the immune serum by affinity chromatography.
Semiquantitative reverse transcription-PCR analysis.
We converted 1 μg of total RNA extracted from normoxia- or hypoxia-stimulated cells to cDNA. The reverse transcription-PCR exponential phase was determined on 22 to 32 cycles to allow semiquantitative comparisons. The PCR regimen for HGTD-P involved an initial denaturation step of 94°C for 5 min, followed by 32 cycles at 94°C for 30 s, 56°C for 45 s, and 72°C for 45 s on a GeneAmp PCR system 9700 (Perkin-Elmer).
Luciferase assay.
A 2-kb fragment of the human HGTD-P promoter was amplified by PCR with primers F0 (5′-GGTACCCGAGGCAAGAGGATTATTT-3′) and R0 (5′-AAGCTTCGACGCGGTGTACAGGCC-3′), which introduced KpnI and HindIII sites, respectively. This fragment was cloned in frame with the translation start codon of the luciferase reporter gene in pGL2-Basic (Promega) to generate the HGTD-P-Luc reporter construct. pGL2-Luc-1 and pGL2-Luc-2 were constructed with F1 (5′-GGTACCGAAGCTGGAAACTGAGTTCA-3′) and F2 primer (5′-GGTACCTCCTCCTGGAACTGGGCCAG), respectively, and the R0 primer. HGTD-P-Luc-3, HGTD-P-Luc-4, and HGTD-P-Luc-5 were constructed with primers F0 and R3 (5′-AAGCTTCATGTTAAAAGGGTTGGGAC-3′), F0 and R4 (5′-AAGCTTTGCCATTCCTCTCCTACTTT-3′), and F5 (5′-GGTACCGAAGCTGGAAACTGAGTTCA) and R5 (5′-AAGCTTCATGTTAAAAGGGTTGGGAC), respectively.
The hypoxia-responsive element mutant form of pGL2-Luc-2 (pGL2-Luc-2M) was generated by changing the core sequence of the hypoxia-responsive element (CGTG) to GCAC by the splicing overlap extension method (26). The expression vector carrying HIF-1α (pcDNA-HIF-1α) was cotransfected with luciferase reporter constructs by the calcium phosphate precipitation method. Luciferase activity was measured on samples containing equivalent amounts of protein with a luminometer (Victor) and luciferase assay reagents (Promega).
Plasmid construction and transient transfection.
DNAs encoding human HGTD-P, HIF-1α, Bcl-2, and Bax were amplified by PCR from a human kidney cDNA library (Clontech) and subcloned into the pCDNA3.1 or the TA vector tagged with His or V5 (Clontech). Various deletion mutants of HGTD-P were constructed by the splicing overlap extension method (26) and cloned into the TA vector. Cells were transiently transfected with Transfast reagent (Promega) unless specified otherwise.
Determination of mitochondrial membrane potential.
Transfected or untransfected PC-3 cells were cultured under the indicated conditions, and then Mitotracker Red CMXRos (Molecular Probes, Inc.) was added to the medium at a final concentration of 400 nM. After 20 min of incubation, the cells were harvested and washed three times with phosphate-buffered saline (PBS). The fluorescence intensity was measured by flow cytometry (FACSCalibur; Becton Dickinson) with the CellQuest program.
Caspase assays.
Caspase 3 and 8 activity was measured with the Colorimetric caspase 3 and caspase 8 assay kits (Calbiochem), respectively, according to the manufacturer's instructions. Caspase 9 activity was measured with the colorimetric N-acetyl-Leu-Glu-His-Asp-p-nitroanilide (Ac-LEHD-pNA) substrate in the presence or absence of the caspase 9 inhibitor Ac-LEHD-CHO. Briefly, 50 μl of cell lysate was obtained from 106 cells in lysis buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 0.1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate [CHAPS], 1 mM dithiothreitol, 0.1 mM EDTA), incubated for 5 min on ice, and then centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were incubated with Ac-LEHD-pNA substrate in the presence or absence of the caspase 9 inhibitor. Absorbance at 405 nm was determined ≈1 h following initiation of the reaction. Activity was expressed as the change over the control value after correction for the baseline value (protein and buffer without colorimetric substrate).
Immunocytochemistry.
PC-3 cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.1% Triton X-100. After blocking with 3% bovine serum albumin in PBS, the cells were incubated with anti-HSP60 or anti-V5 antibody for 1.5 h at room temperature. Afterwards, the cells were stained with secondary antibody conjugated with fluorescein isothiocyanate or Texas Red and viewed under a confocal microscope (META 510, Zeiss).
Immunoprecipitation and immunoblotting.
PC-3 cells (3 × 106/10-cm culture dish) were transiently transfected with 1 μg of each plasmid and Transfast reagent (Promega) and harvested after 48 h of transfection. Solubilized extracts in lysis buffer were precleared, and the resultant supernatants were incubated with primary antibody (3 μg) at 4°C for 2 h. Immunoprecipitates were collected by incubating protein G-Sepharose for 1 h, followed by centrifugation at 4°C. After being washed three times, the immunoprecipitates and immunodepleted final supernatant were subjected to Western blotting. The proteins were separated on sodium dodecyl sulfate-12% polyacrylamide gels and transferred to nitrocellulose membranes. The blots were sequentially incubated with primary antibody and secondary antibody conjugated with horseradish peroxidase, followed by enhanced chemiluminescence-based detection (Amersham Pharmacia Biotech).
Suppression of endogenous HGTD-P expression.
For inhibition of endogenous HGTD-P expression in PC-3 cells, an SiRNA mammalian expression vector (pRNA-U6.1-HGTD-P) was constructed. Following the search for candidate target SiRNA sequence, a 70-bp SiRNA with sense (5′-AGAACAAGATGCGAGTGAA-3′), loop (5′-TTCAAGAGA-3′), and antisense (5′-TTCACTCGCATCTTGTTCT-3′) sequences was inserted into the pRNA-U6.1/Neo GenScript SiRNA expression vector (GenScript). The selected sequence was subjected to a BLAST search to ensure that the human genome sequence other than HGTD-P was not targeted. pRNA-U6.1-HGTD-P was transfected into PC-3 cells with Transfast reagent, and stably transfected cells were selected with neomycin.
To suppress expression of the endogenous HGTD-P in mouse neuronal cells, sense (5′-CTCGCCCACCCCGACACCTC-3′) and antisense (5′-CTCCACAGCCCCACCCGCTC-3′) oligonucleotides were delivered by the streptolysin O technique, described elsewhere (38). Briefly, after being washed in serum-free Dulbecco's modified Eagle's medium, 106 day 7 cells were exposed to dithiothreitol-activated streptolysin O at 20 μmol/liter of sense or antisense oligonucleotide in serum-free Dulbecco's modified Eagle's medium. After incubation for 10 min at 37°C, serum-containing medium was used to reseal the cells. Suppression of HGTD-P expression was validated by Western blotting.
RESULTS
Hypoxia induces transcription and translation of HGTD-P.
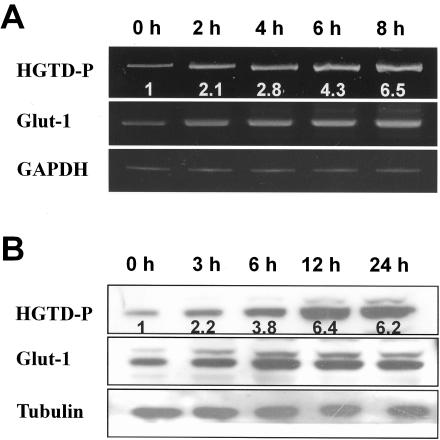
Subtractive hybridization was performed to identify proapoptotic genes induced by hypoxia in SK-N-MC neuroblastoma cells. Among the putative hypoxia-regulated genes detected by subtractive hybridization, we focused on HGTD-P. HGTD-P was initially identified as a novel gene expressed in human dendritic cells (GenBank accession no. AF201944) and as estradiol-induced gene 5 (E2IG5; GenBank accession no. AF191020) (6). To confirm the results of subtractive hybridization, reverse transcription-PCR analysis was performed with mRNA extracted from hypoxia-exposed PC-3 cells. As shown in Fig. 1A, an increase in HGTD-P transcripts was initially found at 2 h of hypoxia, with pronounced induction, up to sixfold, around 8 h of stimulation. Furthermore, the time course of HGTD-P induction paralleled that of Glut-1, a well-characterized hypoxia-inducible gene (44). Maximal induction was seen around 12 h after hypoxia (Fig. 1B). Upregulation of HGTD-P transcription and translation in hypoxia was seen in a variety of human and mouse cells, including neuroblastomas, leukemias, and breast cancers (data not shown), indicating that our findings were not cell type specific.
FIG. 1.
Hypoxia induces transcriptional and translational upregulation of HGTD-P. PC-3 cells in degassed medium were exposed to hypoxic conditions (0.5% O2) for the indicated periods. (A) cDNA was synthesized from total RNAs extracted from PC-3 cells exposed to hypoxia for the indicated time periods and subjected to reverse transcription-PCR analysis; 32, 30, and 26 cycles of amplification were performed for HGTD-P, Glut-1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively. Mean increases in HGTD-P mRNA expression (HGTD-P/GAPDH) compared with that of normoxic cells are presented. (B) We subjected 30 μg of cell lysates extracted from hypoxia-exposed cells for the indicated time periods to Western blot analysis with polyclonal anti-HGTD-P (L16), anti-Glut-1, or antitubulin antibody. The mean increases in HGTD-P protein expression (HGTD-P/tubulin) compared with that of normoxic cells are presented.
HGTD-P is downstream of HIF-1α.
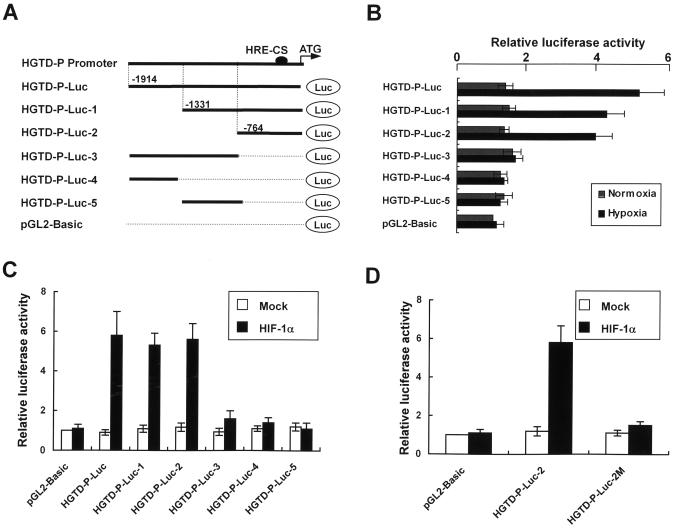
Hypoxia-responsive induction of HGTD-P led us to determine the promoter region of HGTD-P that responded to hypoxia. Promoter assays were performed with various luciferase reporter plasmids under normoxic and hypoxic conditions, as shown in Fig. 2A. Luciferase activities were increased 5-, 4.1-, and 3.8-fold in HGTD-P-Luc, HGTD-P-Luc-1, and HGTD-P-Luc-2, respectively, under hypoxic conditions, while no significant increases were noted under normoxia (Fig. 2B). In contrast, significant enzyme activity was not observed in HGTD-P-Luc-3-, HGTD-P-Luc-4-, HGTD-P-Luc-5, or pGL2-Basic-transfected cells (Fig. 2B). Our results suggest that a 764-bp fragment of the promoter region beginning from the initiation ATG responds to hypoxia.
FIG. 2.
HGTD-P promoter responds to hypoxia via HIF-1α. (A) HGTD-P promoter and various luciferase reporter constructs. The hypoxia-responsive element site (solid circle) is shown. Numbering refers to the region of the HGTD-P promoter inserted into the parental pGL2-Basic vector relative to the ATG translation initiation site at +1. (B) PC-3 cells were transiently transfected with 1 μg of the indicated luciferase reporter plasmids. After 24 h of transfection, the cells were exposed to hypoxia for 4 h or cultured in normoxia, and then luciferase activities were determined. For each construct tested, increases in luciferase activities (hypoxia versus normoxia) are presented. The luciferase activity of the cells transfected with pGL2-Basic in normoxia was arbitrarily set at 1. (C) PC-3 cells were transiently cotransfected with 1 μg of the indicated luciferase reporter plasmids and pcDNA-HIF-1α or mock vector and then cultured in normoxia. After 24 h of cotransfection, luciferase activities were determined. The luciferase activity of the cells cotransfected with pGL2-Basic and mock vector was arbitrarily set at 1. (D) A mutant form of HGTD-P-Luc-2 (HGTD-P-Luc-2M) was constructed by changing the core sequence of the hypoxia-responsive element (CGTG) to GCAC. PC-3 cells were cotransfected with the indicated luciferase reporter plasmid and pcDNA-HIF-1α or mock vector. Luciferase activities were determined as described above.
HIF-1α is known to be a major transcription regulator under hypoxic conditions. HIF-1α regulates the transcription of a variety of genes by binding to a hypoxia-responsive element. Thus, in order to determine whether HGTD-P upregulation by hypoxia was driven through an HIF-1α-dependent pathway, PC-3 cells were cotransfected with pcDNA-HIF-1α and the indicated luciferase plasmid. Cotransfection of HGTD-P-Luc, HGTD-P-Luc-1, or HGTD-P-Luc-2 with pcDNA-HIF-1α increased luciferase activity under normoxic conditions (Fig. 2C), suggestive of HIF-1α-dependent transcriptional upregulation. This result led us to search for a hypoxia-responsive element consensus motif in the 764-bp fragment, (T,G,C) (A,G) CGTG (C,G,A) (G,T,C) (G,T,C) (G,T,C), which allowed no more than single-base mismatch outside the CGTG core sequence, as proposed by Wenger and Gassmann (43). A single hypoxia-responsive element motif fitting Wenger's consensus located 75 bp upstream of the translation start codon was found. We constructed a site-directed mutant of the hypoxia-responsive element motif (HGTD-P-Luc-2M), changing 5′-GGCGTG-3′ to 5′-GGGCAC-3′. This mutation of the hypoxia-responsive element core sequence abolished the increase in luciferase activity in cells cotransfected with pcDNA-HIF-1α and HGTD-P-Luc-2 (Fig. 2D). Collectively, our data demonstrate that HGTD-P is an HIF-1α-dependent gene that responds to hypoxia via a hypoxia-responsive element motif in its promoter region.
HGTD-P is a mitochondrion-targeted protein with two TMs.
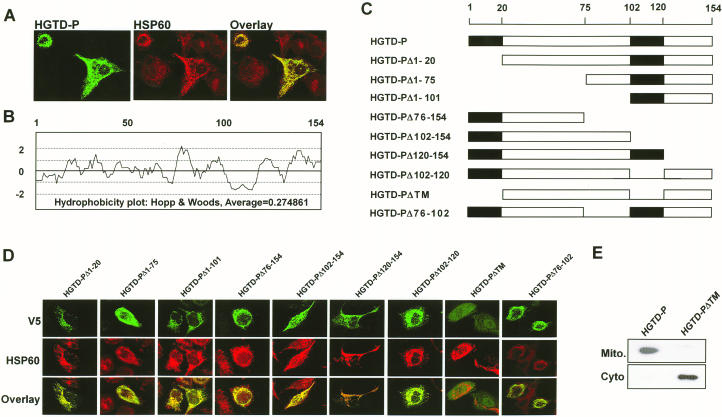
To determine the subcellular localization of human HGTD-P, we constructed an expression construct encoding the full-length HGTD-P protein with a carboxy-terminal V5/His tag (pcDNA-HGTD-P). Immunofluorescence staining with anti-V5 antibody following transient transfection of pcDNA-HGTD-P to PC-3 cells showed a punctate staining pattern suggestive of mitochondrial localization (Fig. 3A, left panel). Comparison of the staining pattern with that of HSP60 (Fig. 3A, middle panel), a component of mitochondrial matrix protein (36), showed complete colocalization (Fig. 3A, right panel), indicative of mitochondrial targeting of HGTD-P. Mitochondrial localization of HGTD-P was further confirmed by Western analysis after cell fractionation (Fig. 3E).
FIG. 3.
HGTD-P is a mitochondrion-localized protein. (A) PC-3 cells were transiently transfected with pcDNA-HGTD-P. At 24 h after transfection, the cells were double stained with mouse anti-V5 antibody conjugated with fluorescein isothiocyanate (HGTD-P) and rabbit anti-HSP60 antibody conjugated with Texas Red (HSP60). The stained images were combined to compare the staining patterns of the proteins (overlay), and their coincidence is indicated by yellow. (B) Hydrophobicity plot of HGTD-P determined with the DNASIS program. (C) Schematic representation of HGTD-P deletion mutants. (D) PC-3 cells were transiently transfected with 1 μg of the indicated deletion mutants. After 24 h, the cells were double stained with anti-V5 and anti-HSP60 antibodies and then visualized with fluorescein isothiocyanate (V5)- and Texas Red (HSP60)-conjugated antibodies. The stained images were combined to compare the staining patterns of the proteins (overlay), and their coincidence is indicated by the conversion of green (fluorescein isothiocyanate) and red (Texas Red stain) to yellow. (E) PC-3 cells were transiently transfected with 1 μg of HGTD-P or HGTD-PΔTM. After 24 h of transfection, cells were harvested, and Western blot analysis with anti-V5 antibody was performed after cellular fractionation. Mito., mitochondrial fraction; Cyto., cytoplasmic fraction.
We next analyzed the hydrophobicity of human HGTD-P to map the regions responsible for mitochondrial localization with the DNASIS program. HGTD-P appears to have two putative transmembrane domains (TMs), one from amino acids 1 to 20 and the other from amino acids 102 to 120 (Fig. 3B). To elucidate the roles of the putative TMs and other regions in mitochondrial targeting, we generated a series of deletion mutants of HGTD-P (Fig. 3C), each encompassing a putative TM or other region. PC-3 cells were transiently transfected with the mutant plasmids, followed by double staining with anti-V5 and anti-HSP60 antibodies. HGTD-PΔTM, in which both putative TMs were deleted, failed to show mitochondrial localization (Fig. 3D and E), while all other mutants were targeted to the mitochondria (Fig. 3D). These results indicate that the presence of at least one TM is sufficient for mitochondrial localization of HGTD-P.
HGTD-P is a proapoptotic molecule.
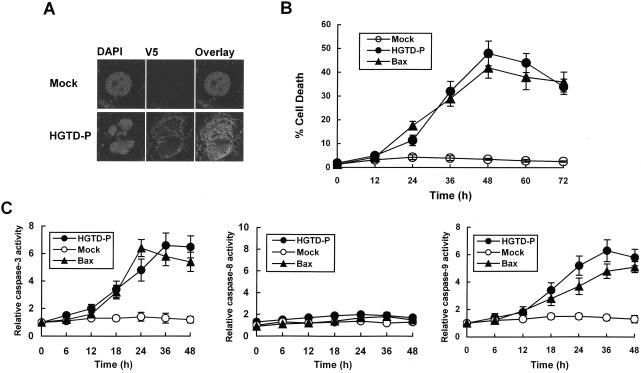
HIF-1α induces transactivation of target genes that participate in the adaptive response or cell death. We have shown above that HGTD-P is an HIF-1α-dependent mitochondrial protein. To characterize the effects of HGTD-P on cell viability, PC-3 cells were transiently transfected with pcDNA-HGTD-P or mock vector and double stained with anti-V5 antibody and DAPI. Apoptotic cells with condensed chromatin patterns or fragmented nuclei among the HGTD-P-expressing cells were counted under the fluorescence microscope (Fig. 4A). Over time, progressively more HGTD-P-expressing cells became apoptotic, starting around 24 h and reaching a peak by 48 h (Fig. 4B). Transfections with the same amount of Bax construct were performed to compare the apoptotic potencies of HGTD-P and Bax (Fig. 4B and C). Caspase 3 and 9 activities were increased in line with cell death, while the increase in caspase 8 activity was insignificant (Fig. 4C). Significant caspase activities were not noted in mock-transfected cells. Our results show that HGTD-P is a proapoptotic molecule, inducing cell death via caspase 9- and 3-dependent pathways.
FIG. 4.
HGTD-P induces apoptotic cell death with caspase activation. PC-3 cells were transfected with 1 μg of mock vector, pcDNA-HGTD-P, or pcDNA-Bax. (A) After 48 h of transfection, cells were harvested, double stained with DAPI and anti-V5 antibody, and visualized with a confocal microscope. (B) Transfected cells with fragmented nuclei or chromatin condensation were counted at the indicated time points, and percent cell death was deetermined. (C) Cells were harvested at the indicated time points after transfection, and caspase assays were performed as described in the text. The caspase activities of mock-transfected cells were arbitrarily set at 1. The increase in caspase activity is presented.
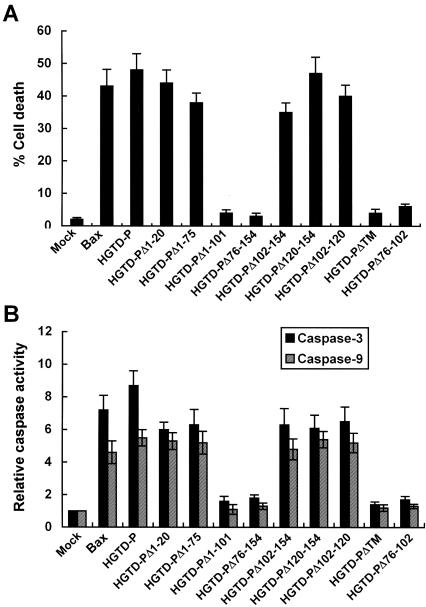
Region encompassing amino acid residues 76 to 102 is essential for apoptogenic activity of HGTD-P.
Our data above showed that the presence of at least one putative TM was essential for mitochondrial localization of HGTD-P. To investigate the possible relationship between mitochondrial localization and proapoptotic activity of HGTD-P and to identify domains essential for its activity, various mutant plasmids were transfected into PC-3 cells, and cell death assays were performed. As shown in Fig. 5A, HGTD-P, HGTD-PΔ1-20, HGTD-PΔ1-75, HGTD-PΔ102-154, HGTD-PΔ120-154, and HGTD-PΔ102-120 facilitated apoptotic cell death, while HGTD-PΔ1-101, HGTD-PΔ76-154, HGTD-PΔTM, and HGTD-PΔ76-102 lost killing activity. Furthermore, the results of the caspase 3 and caspase 9 assays were in line with those of the cell death assays (Fig. 5B). Bax was used for comparison of apoptotic potency with HGTD-P (Fig. 5A and B). Our results indicate that 27 amino acid residues between amino acids 76 and 102 from the N terminus (death-inducing domain [DID]) were essential for proapoptotic activity of HGTD-P. In addition, mitochondrial targeting of HGTD-P via a TM is indispensable for optimal activity of HGTD-P, since HGTD-PΔTM, which contains the DID but not a TM (Fig. 3C and D), lost apoptogenic activity (Fig. 5).
FIG. 5.
Death-inducing domain (DID) is crucial for the apoptotic activity of HGTD-P. PC-3 cells were transiently transfected with 1 μg of mock, pcDNA-Bax, pcDNA-HGTD-P, or the indicated mutant plasmid and then harvested after 48 h of transfection. (A) Following double staining with DAPI and anti-V5 antibody, transfected cells with fragmented nuclei or chromatin condensation were counted under a fluorescence microscope. (B) Caspase 3 and caspase 9 assays. The caspase activities of mock-transfected cells were arbitrarily set at 1. The increase in caspase activity is presented.
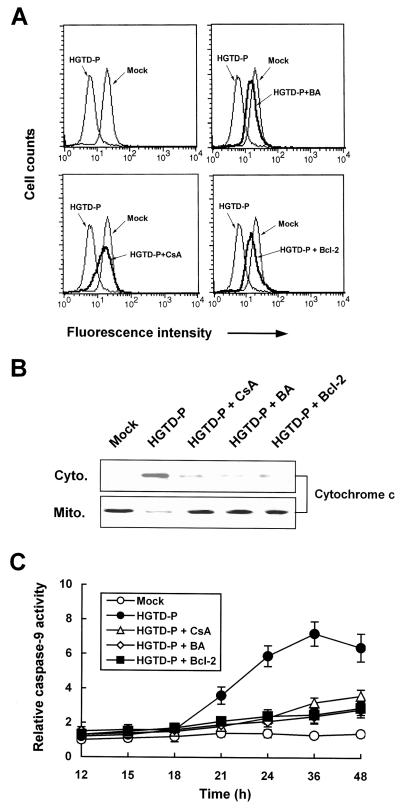
Induction of permeability transition by HGTD-P.
To investigate the molecular pathways of HGTD-P that induce cell death, we initially examined mitochondrial apoptotic cascades because HGTD-P was targeted to mitochondria and overexpression of HGTD-P induced caspase 9 activity (Fig. 3A and E and Fig. 4C). PC-3 cells were transiently transfected with pcDNA-HGTD-P, and then mitochondrial transmembrane potential (MTP, Δψm), a hallmark of PT, was determined by assessing changes in the fluorescence intensity of potential-sensitive Mitotracker Red CMXRos accumulated in mitochondria. A decrease in MTP was noted in HGTD-P-transfected cells after 16 h of transfection (Fig. 6A, left upper panel). We further examined whether loss of Δψm could be inhibited by PT inhibitors such as cyclosporine and bonkrekic acid or Bcl-2 overexpression. PT inhibitors and Bcl-2 overexpression suppressed MTP loss initiated by HGTD-P transfection (Fig. 6A). In parallel with PT induction, PT inhibitor- or Bcl-2-sensitive cytochrome c release (Fig. 6B) and caspase 9 activation (Fig. 6C) were also detected after HGTD-P transfection. Our data indicate that HGTD-P induced PT, with resultant cytochrome c release and caspase 9 activation.
FIG. 6.
HGTD-P induces mitochondrial PT with resultant cytochrome c release and caspase 9 activation. PC-3 cells were transiently transfected with 1 μg of mock vector or pcDNA-HGTD-P in the presence or absence of 20 μM cyclosporine (CsA) or 100 μM bonkrekic acid (BA) or cotransfected with 1 μg of pcDNA-HGTD-P and 1 μg of pcDNA-Bcl-2. (A) After 24 h of transfection, cells were incubated with 400 nM potential-sensitive Mitotracker Red CMXRos, and then MTP was determined by flow cytometry. (B) After 36 h of transfection, cells were harvested, fractionated, and subjected to Western blotting for cytochrome c. Cyto., cytosolic compartment; Mito., mitochondrial compartment. (C) Cells were harvested at the indicated time points, and the caspase 9 assay was performed. The caspase activities of mock-transfected cells were arbitrarily set at 1. The increase in caspase activity is presented.
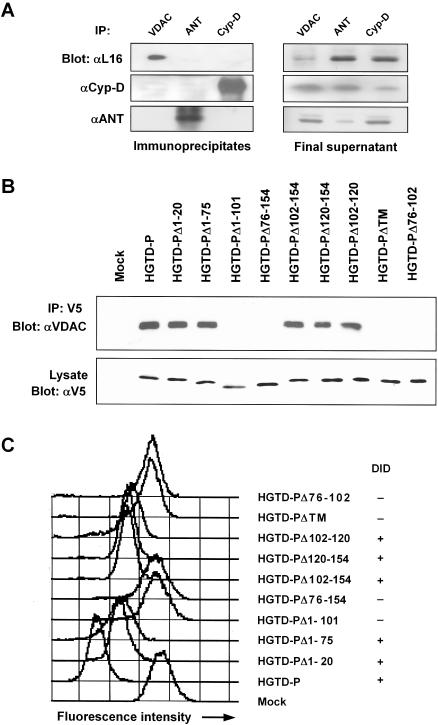
HGTD-P interacts with the VDAC to induce PT.
The cause of mitochondrial PT is the opening of a nonspecific pore known as the mitochondrial permeability transition pore (PTP), which is a protein aggregate composed of cyclophilin D, VDAC, and adenine nucleotide translocator (ANT) at the contact sites between the inner and outer mitochondrial membrane (48). The data shown above indicated that induction of PT and mitochondrial localization were indispensable for cell death triggered by HGTD-P. Thus, we investigated whether HGTD-P interacts with a component(s) of the PTP. Cell lysates were immunoprecipitated with anti-VDAC, -ANT, or -cyclophilin D antibody and then immunoblotted with anti-HGTD-P antibody L16. HGTD-P was coprecipitated with VDAC but not with ANT or cyclophilin D (Fig. 7A, left upper panel). Western blots for HGTD-P, cyclophilin D, and ANT from immunoprecipitates (Fig. 7A, left middle and lower panels) or immunodepleted final supernatants (Fig. 7A, right panels) supported the specificity of immunoprecipitation.
FIG. 7.
HGTD-P interacts with VDAC. (A) Cell lysates from PC-3 cells were immunoprecipitated (IP) with goat anti-VDAC, -ANT, or -cyclophilin D antibody and immunoblotted with anti-HGTD-P (L16) (αL16), anti-cyclophilin D, or anti-ANT antibody (left panels). The immunodepleted final supernatant was subjected to Western blotting with anti-HGTD-P (L16), anti-cyclophilin D, or anti-ANT antibody (right panels). (B) Lysates from cells transfected with the indicated plasmids were immunoprecipitated with anti-V5 antibody and immunoblotted with anti-VDAC antibody (upper panel). Total-cell lysates were analyzed by immunoblotting with anti-V5 antibody (lower panel). (C) PC-3 cells were transfected with the indicated plasmids, and MTP was determined with Mitotracker Red CMXRos by flow cytometry. The presence or absence of the DID in each plasmid is presented as + and −, respectively.
We next examined whether DID played any role in the coprecipitation of HGTD-P with VDAC, since DID was essential for cell death. PC-3 cells were transiently transfected with a wild-type or mutant form of HGTD-P, immunoprecipitated with anti-V5 antibody, and blotted with anti-VDAC antibody. In agreement with the results of the cell death and caspase assays (Fig. 5), HGTD-P, HGTD-PΔ1-20, HGTD-PΔ1-75, HGTD-PΔ102-154, HGTD-PΔ120-154, and HGTD-PΔ102-120 were coprecipitated with VDAC, while HGTD-PΔ1-101, HGTD-PΔ76-154, HGTD-PΔTM, and HGTD-PΔ76-102 were not (Fig. 7B). These results indicate that the DID is essential for binding of HGTD-P to VDAC.
To further elucidate the relationship between HGTD-P binding to VDAC and capacity for PT induction, we evaluated alteration of the MTP in PC-3 cells transfected with the mutant plasmids. In parallel with the immunoprecipitation results, loss of Δψm was noted in HGTD-P-, HGTD-PΔ1-20-, HGTD-PΔ1-75-, HGTD-PΔ102-154-, HGTD-PΔ120-154-, and HGTD-PΔ102-120-transfected cells, but not in HGTD-PΔ1-101-, HGTD-PΔ76-154-, HGTD-PΔTM-, or HGTD-PΔ76-102-transfected cells (Fig. 7C). Collectively, our data suggest that HGTD-P induces PT by binding to VDAC and that DID is essential for it.
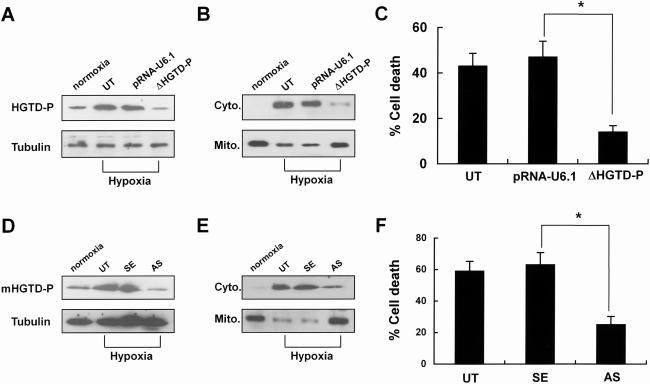
Suppression of HGTD-P expression attenuates cell death from hypoxic injury.
We showed that HGTD-P was downstream of HIF-1α and had proapoptotic activity. To assess the role of HGTD-P in cell death after hypoxic assaults, we determined the effects of suppressing endogenous HGTD-P expression on mitochondrial catastrophe and hypoxic cell death. We constructed a stable PC-3 cell line transfected with the pRNA-U6.1-HGTD-P SiRNA mammalian expression vector (ΔHGTD-P) or empty pRNA-U6.1 (pRNA-U6.1). Suppression of HGTD-P expression was evaluated by Western blotting (Fig. 8A). Untransfected PC-3 cells, PC-3 cells transfected with empty pRNA-U6.1, and the HGTD-P SiRNA mammalian expression vector were cultured in medium containing 10% fetal bovine serum under hypoxic conditions for 6 h and subjected to Western blotting for cytochrome c release or the cell death assay. As shown in Fig. 8B, suppression of HGTD-P expression inhibited hypoxia-induced cytochrome c release. In line with the mitochondrial parameters, hypoxia-induced cell death was also suppressed (Fig. 8C).
FIG. 8.
Suppression of HGTD-P rescues cells from the hypoxic assault. (A) PC-3 cells were untransfected (UT) or stably transfected with empty pRNA-U6.1 vector (mock) or pRNA-U6.1-HGTD-P (ΔHGTD-P). Cells were exposed to hypoxia for 6 h in medium containing 10% fetal bovine serum, and then the expression levels of HGTD-P were determined by Western blotting with anti-HGTD-P (L16) antibody. Equal loading of proteins was confirmed by Western blotting for tubulin. (B) PC-3 cells were exposed to hypoxia for 6 h and subjected to Western blotting for cytochrome c after cell fractionation. Cyto., cytoplasmic compartment; Mito., mitochondrial compartment. (C) PC-3 cells were exposed to hypoxia for 6 h, and then cell death assays were performed by counting apoptotic cells after DAPI staining. *, P < 0.05. (D) Mouse primary cortical neurons were untreated (UT) or treated with sense (SE) or antisense (AS) oligonucleotides and then exposed to hypoxia for 4 h in medium containing 10% fetal bovine serum. The expression levels of HGTD-P were determined by Western blotting with anti-HGTD-P (L16) antibody. Equal loading of proteins was confirmed by Western blotting for tubulin. (E) Mouse neuronal cells were exposed to hypoxia for 4 h and subjected to Western blotting for cytochrome c after cell fractionation. Cyto., cytoplasmic compartment; Mito., mitochondrial compartment. (F) Mouse neuronal cells were exposed to hypoxia for 4 h, and then cell death assays were performed by counting apoptotic cells after DAPI staining. *, P < 0.05. HGTD-P expression under normoxic conditions is shown for comparison (A, B, D, and E).
Cellular responses to hypoxic stimuli could differ depending on cell type or cellular status. To investigate the role of HGTD-P in the hypoxic cell death of postmitotic cells, mouse primary cortical neurons were employed. Suppression of HGTD-P expression by antisense oligonucleotide (Fig. 8D) reduced hypoxia-induced cytochrome c release and cell death rates of primary neuronal cells (Fig. 8E and F). Together, our data indicate that HGTD-P is a mediator of hypoxic cell death via mitochondrial cascades in both proliferating and postmitotic cells.
DISCUSSION
Hypoxia and ischemia participate in numerous pathological processes such as ischemic stroke, myocardial infarction, and chronic degenerative disorders (5). When exposed to hypoxia, cells transactivate a variety of genes to adapt to altered metabolic status or to remove irreversibly damaged cells (4). Among the transcription factors that respond to hypoxia, HIF-1α and p53 appear to be key modulators, because they transactivate numerous genes responsible for increased oxygen delivery, cell cycle arrest, and apoptosis (21, 25, 46). Although survival genes and their molecular pathways that protect against cell death in response to hypoxia are rather well documented, the death-promoting genes induced by hypoxia are less clear. Here we report the identification of HGTD-P as a new HIF-1α-responsive proapoptotic molecule.
Our initial experiments by subtraction hybridization, reverse transcription-PCR, and Western blot analysis strongly suggested the hypoxia-inducible properties of HGTD-P. Luciferase reporter assays with mutant plasmids and a cotransfection assay with HIF-1α clearly demonstrated that HGTD-P lies downstream of HIF-1α. The majority of genes downstream of HIF-1α participate in survival signaling pathways against hypoxic assault, increasing oxygen delivery or reducing metabolic demands for adaptation to hypoxic conditions (11, 13, 32). Paradoxically, HIF-1α serves as a prodeath gene as well (15, 25). Based on the findings presented in this study, HGTD-P is a death-inducing gene that responds to hypoxia via HIF-1α.
To date, several molecules that serve as downstream effectors of HIF-1α (3, 17, 35) or cooperate with HIF-1α to mediate hypoxic cell death (7, 14) have been found. BNip3 is a well-characterized proapoptotic mediator that responds to HIF-1α (3, 18, 27). As they are likely HIF-1α target genes, the mRNA levels of HGTD-P and BNip3 were increased shortly after hypoxia (Fig. 1A) (2). In contrast, accumulation of HGTD-P protein started at 3 h of hypoxia, while that of BNip3 was delayed to 4 days of hypoxic environment (Fig. 1B) (2). This finding may suggest that HGTD-P is an effector in the early stages of hypoxia, while BNip3 responds to chronic hypoxia. This finding may have clinical relevance for the selection of therapeutic targets in disorders caused by hypoxia.
There are also substantial differences in the molecular pathways of cell death. Hypoxia may induce apoptotic or necrotic cell death, depending on the type and status of the cells and the severity of oxygen deprivation (9). Whatever types of cell death may be triggered by hypoxia, mitochondria are in the central arena of hypoxic signals. This is thought to occur by several mechanisms: (i) disruption of electron transport and oxidative phosphorylation, leading to energy depletion, (ii) alteration of redox status, resulting in generation of reactive oxygen species, and (iii) induction of PT with resultant release of proapoptotic molecules such as cytochrome c into the cytoplasm (19, 40). Although the molecular mechanisms of cell death in energetic catastrophe, which is frequently encountered in acute severe oxygen deprivation or reactive oxygen species-induced cell death, are rather well documented, the molecule(s) directly responsible for PT induction in response to hypoxia has not been clearly identified yet. BNip3 induces necrosis-like cell death without the involvement of cytochrome c release or caspase activation, although the precise mechanisms are still controversial (8, 16, 41). RTP801, a recently identified HIF-1α-responsive gene, seems to participate not only in a protective pathway against hypoxic injury but also in proapoptotic effects, depending on cell context (35).
The death-inducing activity of RTP801 appears to be associated with reactive oxygen species, although details of the intracellular signaling pathways affected by RTP801 overexpression have not been determined yet. In contrast, HGTD-P triggers apoptosis via typical mitochondrial pathways, including PT induction, cytochrome c release, and sequential caspase 9 and 3 activations. Recently we reported that the BH3-only Bcl-2 family protein Noxa is an HIF-1α-responsive proapoptotic effector molecule that mediates hypoxic cell death (17). Although both HGTD-P and Noxa promoters responded to HIF-1α via the hypoxia-responsive element consensus motif, the molecular mechanisms of cell death are different in that Noxa-induced cell death occurs via the reactive oxygen species-dependent pathway. It is not clear at this point whether reactive oxygen species participate in HGTD-P-mediated cell death. The diversity of cell death pathways induced by HIF-1α-responsive effector molecules, including BNip3, RTP801, Noxa, and HGTD-P, may suggest that complicated signaling networks of cell death mechanisms are induced by HIF-1α. Whether these molecules transmit HIF-1α death signals individually or act in concert to induce cell death remains to be elucidated.
Mitochondrial catastrophic processes induced by HGTD-P were significantly blocked by conventional PT inhibitors (bonkrekic acid and cyclosporine) and Bcl-2 overexpression. Antiapoptotic Bcl-2 family proteins such as Bcl-2 and Bcl-XL are known to act at multiple levels of apoptotic pathways: (i) the sequestration of Apaf-1 (23), (ii) prevention of the release of cytochrome c and MTP loss (22, 45), and (iii) physical interaction with proapoptotic proteins (26). Bcl-2/Bcl-XL forms heterodimers with HGTD-P to suppress the proapoptotic activity of HGTD-P, which may be one mechanism whereby Bcl-2/Bcl-XL protects cells against hypoxic injury (M.-J. Lee, J.-Y. Kim, and J.-H. Park, unpublished data). HIF-1α-driven cell death may occur in a p53-dependent manner (42). HIF-1α stabilizes p53, which upregulates and translocates Bax to mitochondria, thereby releasing cytochrome c in response to hypoxia (7, 14, 20). The results presented in this report categorize HGTD-P as a new HIF-1α-dependent gene, linking its hypoxic signals to mitochondria.
Subcellular localization and cell death assays with wild-type and mutant forms of HGTD-P showed that HGTD-P is targeted to the mitochondria via two TMs and that mitochondrial localization of HGTD-P is indispensable for cell death induction, since deletion of both TMs (HGTD-PΔTM) caused the loss of death-inducing activity as well as mitochondrial localization. However, it seems that mitochondrial targeting per se is not sufficient for the functions of HGTD-P, because the DID deletion mutant possessing both TMs (HGTD-PΔ102-120) was targeted to the mitochondria without inducing cell death. Instead, it is speculated that the TMs of HGTD-P may allow a physical association with its target molecule, VDAC. Immunoprecipitation analysis indicated that the interaction of HGTD-P with VDAC via the DID is crucial for PT induction.
VDAC participates in apoptosis by releasing apoptogenic factors such as cytochrome c (10, 47) from the mitochondria to the cytosol to activate the apoptotic cascade, although the detailed molecular mechanisms of how VDAC forms a protein-conducting channel for the passage of cytochrome c are still controversial (34, 39). Several pro- or antiapoptotic proteins, including Bcl-2 family proteins and kinases, have been reported to interact physically with VDAC to modulate its function (2, 24, 33), suggesting that VDAC might also play a distinct role independent of channel formation. However, it is not clear at present whether HGTD-P participates directly in channel formation in association with VDAC or modulates its channel-forming activity.
The suppression of endogenous HGTD-P expression by SiRNA or antisense oligonucleotides stabilized mitochondria and rescued cells from hypoxic injury, indicating that HGTD-P is a candidate mediator linking the complex hypoxic signals of HIF-1α to the mitochondrial catastrophe and cell death. However, it has not yet been determined whether hypoxia-induced HGTD-P expression has any relevance in an ischemic animal model or clinical hypoxia-induced disorders. It will be interesting to study the physiological significance of HGTD-P in transgenic and knockout animals.
In conclusion, we have shown that HGTD-P is a proapoptotic effector of HIF-1α that directly links the complicated network of proapoptotic signals from HIF-1α to the mitochondrial PTP.
Acknowledgments
This work was supported by a grant (R01-2001-000-00200-0) from the Basic Research Program of the Korea Science & Engineering Foundation. This work was also funded by the Korea Science & Engineering Foundation through the Biomedical Research Center for Reactive Oxygen Species at Kyung Hee University (R13-2002-020-01002-0).
REFERENCES
- 1.Almeida, A., and J. M. Medina. 1998. A rapid method for the isolation of metabolically active mitochondria from rat neurons and astrocytes in primary culture. Brain Res. Protoc. 2:209-214. [DOI] [PubMed] [Google Scholar]
- 2.Brdiczka, D., P. Kaldis, and T. Wallimann. 1994. In vitro complex formation between the octamer of mitochondrial creatine kinase and porin. J. Biol. Chem. 269:27640-27644. [PubMed] [Google Scholar]
- 3.Bruick, R. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA 97:9082-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunn, H. F., and R. O. Poyton. 1996. Oxygen sensing and molecular adaptation to hypoxia. Physiol. Rev. 76:839-885. [DOI] [PubMed] [Google Scholar]
- 5.Carmeliet, P., Y. Dor, J. M. Herbert, D. Fukumura, K. Brusselmans, M. Dewerchin, M. Neeman, F. Bono, R. Abramovitch, P. Maxwell, C. J. Koch, P. Ratcliffe, L. Moons, R. K. Jain, D. Collen, E. Keshert, and E. Keshet. 1998. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 394:485-490. [DOI] [PubMed] [Google Scholar]
- 6.Charpentier, A. H., A. K. Bednarek, R. L. Daniel, K. A. Hawkins, K. J. Laflin, S. Gaddis, M. C. MacLeod, and C. M. Aldaz. 2000. Effects of estrogen on global gene expression: identification of novel targets of estrogen action. Cancer Res. 60:5977-5983. [PubMed] [Google Scholar]
- 7.Chen, D., M. Li, J. Luo, and W. Gu. 2003. Direct interactions between HIF-1 alpha and Mdm2 modulate p53 function. J. Biol. Chem. 278:13595-13598. [DOI] [PubMed] [Google Scholar]
- 8.Chen, G., R. Ray, D. Dubik, L. F. Shi, J. Cizeau, R. C. Bleackley, S. Saxena, R. D. Gietz, and A. H. Greenberg. 1997. The E1B 19K/Bcl-2-binding protein Nip3 is a dimeric mitochondrial protein that activates apoptosis. J. Exp. Med. 186:1975-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, D. W. 1996. Ischemia-induced neuronal apoptosis. Curr. Opin. Neurobiol. 6:667-672. [DOI] [PubMed] [Google Scholar]
- 10.Crompton, M. 1999. The mitochondrial permeability transition pore and its role in cell death. Biochem. J. 341:233-249. [PMC free article] [PubMed] [Google Scholar]
- 11.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner, L. B., Q. Li, M. S. Park, W. M. Flanagan, G. L. Semenza, and C. V. Dang. 2001. Hypoxia inhibits G1/S transition through regulation of p27 expression. J. Biol. Chem. 276:7919-7926. [DOI] [PubMed] [Google Scholar]
- 13.Goda, N., H. E. Ryan, B. Khadivi, W. McNulty, R. C. Rickert, and R. S. Johnson. 2003. Hypoxia-inducible factor 1α is essential for cell cycle arrest during hypoxia. Mol. Cell. Biol. 23:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halterman, M. W., C. C. Miller, and H. J. Federoff. 1999. Hypoxia-inducible factor-1α mediates hypoxia-induced delayed neuronal death that involves p53. J. Neurosci. 19:6818-6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halterman, M. W., and H. J. Federoff. 1999. HIF-1α and p53 promote hypoxia-induced delayed neuronal death in models of CNS ischemia. Exp. Neurol. 159:65-72. [DOI] [PubMed] [Google Scholar]
- 16.Kim, J.-Y., J.-J. Cho. J. Ha, and J.-H. Park. 2002. The carboxy terminal C-tail of BNip3 is crucial in induction of mitochondrial permeability transition in isolated mitochondria. Arch. Biochem. Biophys. 398:147-152. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J.-Y., H.-J. Ahn, J.-H. Ryu, K. Suk, and J.-H. Park. 2004. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1. J. Exp. Med. 199:113-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubasiak, L. A., O. M. Hernandez, N. H. Bishopric, and K. A. Webster. 2002. Hypoxia and acidosis activate cardiac myocyte death through the Bcl-2 family protein BNIP3. Proc. Natl. Acad. Sci. USA 99:12825-12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malhotra, R., Z. Lin, C. Vincenz, and F. C. Brosius III. 2001. Am. J. Physiol. Cell Physiol. 281:C1596-C1603. [DOI] [PubMed] [Google Scholar]
- 20.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 21.Moll, U. M., and A. Zaika. 2001. Nuclear and mitochondrial apoptotic pathways of p53. FEBS Lett. 493:65-69. [DOI] [PubMed] [Google Scholar]
- 22.Narita, M., S. Shimizu, T. Ito, T. Chittenden, R. I. Lutz, H. Matsuda, and Y. Tsujimoto. 1998. Bax interacts with the permeability transition pore to induce permeability transition and cytochrome c release in isolated mitochondria. Proc. Natl. Acad. Sci. USA 95:14681-14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan, G., K. O'Rourke, and V. M. Dixit. 1998. Caspase-9, Bcl-XL, and Apaf-1 form a ternary complex. J. Biol. Chem. 273:5841-5845. [DOI] [PubMed] [Google Scholar]
- 24.Pastorino, J. G., N. Shulga, and J. B. Hoek. 2002. Mitochondrial binding of hexokinase II inhibits Bax-induced cytochrome c release and apoptosis. J. Biol. Chem. 277:7610-7618. [DOI] [PubMed] [Google Scholar]
- 25.Piret, J. P., D. Mottet, M. Raes, and C. Michiels. 2002. Is HIF-1alpha a pro- or an anti-apoptotic protein? Biochem. Pharmacol. 64:889-892. [DOI] [PubMed] [Google Scholar]
- 26.Ray, R., G. Chen, C. Vande Velde, J. Cizeau, J.-H. Park, J. C. Reed, R. D. Gietz, and A. H. Greenberg. 2000. BNIP3 heterodimerizes with Bcl-2/Bcl-X(L) and induces cell death independent of a Bcl-2 homology 3 (BH3) domain at both mitochondrial and nonmitochondrial sites. J. Biol. Chem. 275:1439-1448. [DOI] [PubMed] [Google Scholar]
- 27.Regula, K. M., K. Ens, and L. A. Kirshenbaum. 2002. Inducible expression of BNIP3 provokes mitochondrial defects and hypoxia-mediated cell death of ventricular myocytes. Circ. Res. 91:226-231. [DOI] [PubMed] [Google Scholar]
- 28.Saikumar, P., Z. Dong, J. M. Weinberg, and M. A. Venkatachalam. 1998. Mechanisms of cell death in hypoxia/reoxygenation injury. Oncogene 17:3341-3349. [DOI] [PubMed] [Google Scholar]
- 29.Salnikow, K., T. Kluz, M. Costa, D. Piquemal, Z. N. Demidenko, K. Xie, and M. V. Blagosklonny. 2002. The regulation of hypoxic genes by calcium involves c-Jun/AP-1, which cooperates with hypoxia-inducible factor 1 in response to hypoxia. Mol. Cell. Biol. 22:1734-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semenza, G. L. 1999. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu. Rev. Cell Dev. Biol. 15:551-578. [DOI] [PubMed] [Google Scholar]
- 31.Semenza, G. L. 2000. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J. Appl. Physiol. 88:1474-1480. [DOI] [PubMed] [Google Scholar]
- 32.Semenza, G. L., P. H. Roth, H. M. Fang, and G. L. Wang. 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269:23757-23763. [PubMed] [Google Scholar]
- 33.Shi, Y., J. Chen, C. Weng, R. Chen, Y. Zheng, Q. Chen, and H. Tang. 2003. Identification of the protein-protein contact site and interaction mode of human VDAC1 with Bcl-2 family proteins. Biochem. Biophys. Res. Commun. 305:989-996. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, S., and Y. Tsujimoto. 2000. Proapoptotic BH3-only Bcl-2 family members induce cytochrome c release, but not mitochondrial membrane potential loss, and do not directly modulate voltage-dependent anion channel activity. Proc. Natl. Acad. Sci. USA 97:577-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shoshani, T., A. Faerman, I. Mett, E. Zelin, T. Tenne, S. Gorodin, Y. Moshel, S. Elbaz, A. Budanov, A. Chajut, H. Kalinski, I. Kamer, A. Rozen, O. Mor, E. Keshet, D. Leshkowitz, P. Einat, R. Skaliter, and E. Feinstein. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22:2283-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soltys, B. J., and R. S. Gupta. 1992. Interrelationships of endoplasmic reticulum, mitochondria, intermediate filaments, and microtubules—a quadruple fluorescence labeling study. Biochem. Cell Biol. 70:1174-1186. [DOI] [PubMed] [Google Scholar]
- 37.Sowter, H. M., P. J. Ratcliffe, P. Watson, A. H. Greenberg, and A. L. Harris. 2001. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 61:6669-6673. [PubMed] [Google Scholar]
- 38.Spiller, D. G., R. V. Giles, J. Grzybowski, D. M. Tidd, and R. E. Clark. 1998. Improving the intracellular delivery and molecular efficacy of antisense oligonucleotides in chronic myeloid leukemia cells: a comparison of streptolysin-O permeabilization, electroporation, and lipophilic conjugation. Blood 91:4738-4746. [PubMed] [Google Scholar]
- 39.Sugiyama, T., S. Shimizu, Y. Matsuoka, Y. Yoneda, and Y. Tsujimoto. 2002. Activation of mitochondrial voltage-dependent anion channel by apro-apoptotic BH3-only protein Bim. Oncogene 21:4944-4956. [DOI] [PubMed] [Google Scholar]
- 40.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, J. Mangion, E. Jacotot, P. Costantini, M. Loeffler, N. Larochette, D. R. Goodlett, R. Aebersold, D. P. Siderovski, J. M. Penninger, and G. Kroemer. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441-446. [DOI] [PubMed] [Google Scholar]
- 41.Vande Velde, C., J. Cizeau, D. Dubik, J. Alimonti, T. Brown, S. Israels, R. Hakem, and A. H. Greenberg. 2000. BNIP3 and genetic control of necrosis-like cell death through the mitochondrial permeability transition pore. Mol. Cell. Biol. 20:5454-5468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 406:307-310. [DOI] [PubMed] [Google Scholar]
- 43.Wenger, R. H., and M. Gassmann. 1997. Oxygen and the hypoxia-inducible factor-1. Biol. Chem. 378:609-616. [PubMed] [Google Scholar]
- 44.Wood, S. M., M. S. Wiesener, K. M. Yeates, N. Okada, C. W. Pugh, P. H. Maxwell, and P. J. Ratcliffe. 1998. Selection and analysis of a mutant cell line defective in the hypoxia-inducible factor-1 alpha-subunit (HIF-1alpha). Characterization of hif-1alpha-dependent and -independent hypoxia-inducible gene expression. J. Biol. Chem. 273:8360-8368. [DOI] [PubMed] [Google Scholar]
- 45.Yang, J., X. Liu, K. Bhalla, C. N. Kim, A. M. Ibrado, J. Cai, T. I. Peng, D. P. Jones, and X. Wang. 1997. Prevention of apoptosis by Bcl-2: release of cytochrome c from mitochondria blocked. Science 275:1129-1132. [DOI] [PubMed] [Google Scholar]
- 46.Zaman, K., H. Ryu, D. Hall, K. O'Donovan, K. I. Lin, M. P. Miller, J. C. Marquis, J. M. Baraban, G. L. Semenza, and R. R. Ratan. 1999. Protection from oxidative stress-induced apoptosis in cortical neuronal cultures by iron chelators is associated with enhanced DNA binding of hypoxia-inducible factor-1 and ATF-1/CREB and increased expression of glycolytic enzymes, p21waf1/cip1, and erythropoietin. J. Neurosci. 19:9821-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zamzami, N., and G. Kroemer. 2001. The mitochondrion in apoptosis: how Pandora's box opens. Nat. Rev. Mol. Cell. Biol. 2:67-71. [DOI] [PubMed] [Google Scholar]
- 48.Zoratti, M., and I. Szabo. 1995. The mitochondrial permeability transition. Biochim. Biophys. Acta 1241:139-176. [DOI] [PubMed] [Google Scholar]