Abstract
The Saccharomyces cerevisiae [PSI+] prion is believed to be a self-propagating cytoplasmic amyloid. Earlier characterization of HSP70 (SSA1) mutations suggested that [PSI+] propagation is impaired by alterations that enhance Ssa1p's substrate binding. This impairment is overcome by second-site mutations in Ssa1p's conserved C-terminal motif (GPTVEEVD), which mediates interactions with tetratricopeptide repeat (TPR) cochaperones. Sti1p, a TPR cochaperone homolog of mammalian Hop1 (Hsp70/90 organizing protein), activates Ssa1p ATPase, which promotes substrate binding by Ssa1p. Here we find that in SSA1-21 cells depletion of Sti1p improved [PSI+] propagation, while excess Sti1p weakened it. In contrast, depletion of Fes1p, a nucleotide exchange factor for Ssa1p that facilitates substrate release, weakened [PSI+] propagation, while overproducing Fes1p improved it. Therefore, alterations of Hsp70 cochaperones that promote or prolong Hsp70 substrate binding impair [PSI+] propagation. We also find that the GPTVEEVD motif is important for physical interaction with Hsp40 (Ydj1p), another Hsp70 cochaperone that promotes substrate binding but is dispensable for viability. We further find that depleting Cpr7p, an Hsp90 TPR cochaperone and CyP-40 cyclophilin homolog, improved [PSI+] propagation in SSA1 mutants. Although Cpr7p and Sti1p are Hsp90 cochaperones, we provide evidence that Hsp90 is not involved in [PSI+] propagation, suggesting that Sti1p and Cpr7p functionally interact with Hsp70 independently of Hsp90.
Hsp70 is a universally conserved essential protein that acts in many cellular processes where proteins are incompletely folded, such as translation and membrane transport (7, 25). Additionally, Hsp70 expression is increased by stress, whereupon it protects cells by binding to partially unfolded proteins and preventing hydrophobic interactions that lead to aggregation. Proper functioning of Hsp70 in its diverse roles depends upon a finely tuned cycle of binding and release of the substrate that is regulated by ATP hydrolysis and nucleotide exchange.
Hsp70 has an amino-terminal ATPase domain that regulates the function of an adjacent substrate-binding domain (SBD). When ATP is bound, the SBD is “open” and rapidly binds and releases the substrate. Hydrolysis of ATP to ADP converts the SBD to a “closed” conformation, establishing a tight association with the substrate. Nucleotide exchange restores the ATP-bound state, returning the SBD to the open conformation and allowing release of the substrate.
Hsp70's ATPase activity is stimulated by substrate binding (41), indicating two-way communication between the domains, and is influenced by interactions with cochaperone proteins. Hsp40s, a conserved family of Hsp70 cochaperones that also bind partially folded proteins, stimulate Hsp70's ATPase activity. This stimulation is markedly elevated in the presence of the substrate, coupling ATP hydrolysis with substrate capture (23, 65). Return to the ATP-bound state is regulated by nucleotide exchange factors.
Function of eukaryotic Hsp70 is also regulated by cochaperones that contain tetratricopeptide repeat (TPR) motifs (52). Mammalian Hop1 (Hsp70/90-organizing protein) has two TPR regions that distinctly and simultaneously bind Hsp70 and Hsp90, forming a physical link between them (49). Hsp90 is another essential and stress-induced protein chaperone. Although its function during stress is unclear, Hsp90 has a well-defined role in assisting folding of steroid receptors and signaling kinases (48). Hsp70 acts in this process and has been shown to “accept” substrates from Hsp40 and “present” them to Hsp90 through an interaction facilitated by Hop1 (26). Other proteins involved in this pathway include CyP-40 cyclophilins, which are peptidyl-prolyl isomerases that have high affinity for immunosuppressants. Cyclophilins compete with Hop1 for binding to Hsp90 and displace Hop1 and Hsp70 from the complex.
The Hsp90 TPR cochaperone machinery is conserved in yeast and includes the Hop1 homolog Sti1p, which was shown recently to stimulate Ssa1p ATPase, and the CyP-40 cyclophilin homolog Cpr7p (8, 16, 45, 60). In addition to interacting with Hsp70 and Hsp90, these TPR proteins also interact with Hsp104, a chaperone that disaggregates protein aggregates in a reaction aided by Hsp70 and Hsp40 (1, 20). Hsp70 and Hsp90 have similar conserved amino acid residues at their extreme C termini that mediate interactions with TPR proteins and Hsp40 (18, 49). The C terminus of Hsp104 is similar but less conserved.
Yeast prions are proteins that misfold and form self-replicating aggregates, which are believed to propagate as amyloid (35, 63, 64). Ure2p and Sup35p, the protein determinants of the Saccharomyces cerevisiae prions [URE3] and [PSI+], respectively, rapidly and spontaneously form amyloid when purified (19, 32, 57). Additionally, fibrous structures of Ure2p have been detected in [URE3] but not [ure-o] cells (55), and the protease digestion pattern of Ure2p from [URE3] cells is identical to that of amyloid formed from purified Ure2p (40, 57).
In a manner analogous to mammalian amyloidoses, the aggregates act as “seed” to recruit and convert the soluble form of the protein into the same misfolded form as it joins the polymer. Yeast prions replicate in the cytoplasm and are infectious in that they are transmitted between cells during cell division and cell fusion. Sup35p (eRF3) encodes a translation release factor (56, 66). When [PSI+] is present, much Sup35 protein is aggregated and unavailable to function in translation termination, which causes a nonsense suppression phenotype. [PSI+] thus provides a simple yet powerful system for studying amyloid propagation in vivo.
In line with yeast prions being protein folding problems, altered abundance or function of a variety of protein chaperones can affect their propagation (9, 28, 33, 34, 42, 54). Most effects are observed upon overexpression of the chaperones, and mechanisms underlying the effects remain speculative. Among the chaperones Hsp104 is special in that both its lack or its overproduction can cause loss of [PSI+] (9). So an intermediate level of Hsp104's disaggregating activity appears to be critical for efficient [PSI+] propagation. Hsp104 plays an important role in maintaining prion seed number, presumably by breaking prion aggregates into more numerous self-propagating particles (9, 43, 47). The elimination of [PSI+] by overproduced Hsp104 is moderated somewhat by simultaneously overproducing Hsp70 Ssa1p (44). Our previous characterization of an Ssa1p mutant (Ssa1-21p) showed that it reduced generation of [PSI+] seeds (28). Despite impairing [PSI+] considerably, Ssa1-21p has little effect on cell growth under optimal or stressful conditions, suggesting that it interacts differently with amyloid than with other substrates.
We previously generated second-site mutations in Ssa1-21p that restored normal [PSI+] propagation to understand better how Hsp70 affects yeast prion propagation (27). Among others, several mutations were located in the conserved C-terminal motif (GPTVEEVD) known to be important for interactions with TPR cochaperones. Here, we used a candidate gene approach to identify relevant interacting proteins and found that depletion of specific TPR cochaperones improved [PSI+] propagation in SSA1-21 cells. Although the Hsp90 cochaperones Sti1p and Cpr7p had significant effects on [PSI+], we find evidence that Hsp90 is not involved in [PSI+] propagation, uncovering a functional interaction between these cochaperones and Hsp70 that appears independent of Hsp90. Our characterization of Hsp70 interactions with cochaperones reveals the altered Hsp70 function that antagonizes amyloid propagation and provides an explanation for effects seen in some earlier overexpression studies.
MATERIALS AND METHODS
Strains, media, growth conditions, and plasmids.
Yeast strains are listed in Table 1. Except for strains G658 through G663, which are transformants of G400-1C (MATa kar1-1 SUQ5 his3 leu2 lys2 trp1 ura3 ssa1::KanMX ssa2::HIS3 ssa3::TRP1 ssa4::ura3-1f/pRDW10 [27]), all are isogenic to 779-6A (MATα kar1-1 SUQ5 ade2-1 his3Δ202 leu2Δ1 trp1Δ63 ura3-52 [29]). Because Ssap function is essential, plasmids with SSA alleles can be maintained in G400-1C cells on nonselective media. Similarly, plasmids with HSC82 alleles can be maintained in hsc82Δ hsp82Δ cells without selection. SSA1 gene replacements were made as described previously (29) by transformation using alleles on BglII-SphI fragments from plasmids pJ126 and pJ127 (see below). These fragments contain the SSA coding region and 500 bp of 5′ and 3′ flanking DNA. The coding region of SSA2 was precisely replaced with HIS3 by transformation with DNA obtained by PCR-amplifying HIS3 using primers with 5′ homology to SSA2 untranslated DNA (3). Remaining genes were disrupted by transformation using KanMX cassettes (59). YPAD (excess adenine), 1/2YPD (limiting adenine), and synthetic media were as described previously (50, 51). Unless indicated otherwise, cells were grown at 30°C. Genetic methods were as described previously (22, 28). The presence or absence of [PSI+] was confirmed by both guanidine curing (below) and cytoduction, which is transmission of cytoplasm between strains through abortive mating (10, 28).
TABLE 1.
Yeast strains and growth rates
| Strain | Relevant genotypea
|
Generation time (min)b
|
||||
|---|---|---|---|---|---|---|
| SSA1 | SSA2 | CPR7 | STI1 | [psi−] | [PSI+] | |
| 1001 | + | + | + | + | 99 | 100 |
| 1002 | + | ssa2Δ | + | + | 100 | 109 |
| 1003 | + | + | cpr7Δ | + | 144 | 145 |
| 1004 | + | + | + | sti1Δ | 93 | 110 |
| 1005 | + | ssa2Δ | cpr7Δ | + | 117 | 122 |
| 1006 | + | ssa2Δ | + | sti1Δ | 108 | 111 |
| 1007 | + | + | cpr7Δ | sti1Δ | 175 | 151 |
| 1008 | + | ssa2Δ | cpr7Δ | sti1Δ | 192 | 167 |
| 1009 | SSA1636S | + | + | + | 108 | 106 |
| 1010 | SSA1636S | ssa2Δ | + | + | 108 | 111 |
| 1011 | ssa1Δ | + | + | + | 93 | 96 |
| 1012 | ssa1Δ | ssa2Δ | + | + | 141 | 173 |
| 1013 | SSA1-21 | + | + | + | 84 | 84 |
| 1014 | SSA1-21 | ssa2Δ | + | + | 102 | NA |
| 1015 | SSA1-21 | + | cpr7Δ | + | 127 | 125 |
| 1016 | SSA1-21 | + | + | sti1Δ | 90 | 108 |
| 1017 | SSA1-21 | ssa2Δ | cpr7Δ | + | 133 | NA |
| 1018 | SSA1-21 | ssa2Δ | + | sti1Δ | 111 | NA |
| 1019 | SSA1-21 | + | cpr7Δ | sti1Δ | 170 | 153 |
| 1020 | SSA1-21 | ssa2Δ | cpr7Δ | sti1Δ | 168 | 208 |
| 1021 | SSA1-21636S | + | + | + | 94 | 103 |
| 1022 | SSA1-21636S | ssa2Δ | + | + | 95 | 108 |
| 919 | fes1ΔSSA1 | 145 | 126 | |||
| 921 | fes1ΔSSA1-21 | 123 | NA | |||
| G612 | hsc82Δhsp82Δ/pHSC82(1-709) | ND | ND | |||
| G616 | hsc82Δhsp82Δ/pHSC82(1-704) | ND | ND | |||
| G658 | ssa1,2,3,4/pSSA1 | 90 | ND | |||
| G659 | ssa1,2,3,4/pSSA1-21 | 92 | NA | |||
| G660 | ssa1,2,3,4/pSSA1P636S | 105 | ND | |||
| G661 | ssa1,2,3,4/pSSA1-21P636S | 98 | ND | |||
| G662 | ssa1,2,3,4/pSSA1Δ8 | 136 | 165 | |||
| G663 | ssa1,2,3,4/pSSA1-21Δ8 | 265 | NA | |||
All except G658 through G663 are isogenic to wild-type strain 779-6A (see Materials and Methods). Various allele combinations are indicated under relevant genotype. G658 through G663 are G400-1C (see Materials and Methods [27]) with the indicated plasmids in place of pRDW10.
Growth rates were measured in liquid YPAD at 30°C. ND, not determined; NA, not applicable, since [PSI+] is unable to propagate in these strains.
The pRS series plasmids have been described (53). Plasmids pRDW10 (URA3) and pJ120 (LEU2) are single-copy vectors with SSA1 (27, 28). Plasmid pJ121 is pJ120 with L483W (the SSA1-21 mutation) in SSA1 (27). Plasmids pJ126 and pJ127 are pJ120 and pJ121, respectively, with a hisG::URA3::hisG cassette (2) inserted at an AatII site engineered 200 bp 3′ to the SSA1 termination codon. SSA1Δ8 and SSA1-21Δ8 are SSA1 and SSA1-21, respectively, which lack the eight C-terminal codons (GPTVEEVD) and were made by site-directed mutagenesis of pJ120 and pJ121. The URA3-based plasmid pHSSA1, a gift from E. Craig (University of Wisconsin, Madison; named pRSETB-SSA1 by her), has an amino-terminal His6-tagged SSA1 under control of the TEF2 promoter. Plasmids pHSSA1-21, pHSSA1Δ8, and pHSSA1-21Δ8 are pHSSA1 with the indicated allele in place of SSA1. Multicopy plasmids p423STI1 and p423FES1 are pRS423 with STI1 and FES1, respectively. The single-copy plasmid pL116 is pRS314 with the intact HSC82(1-709) gene and 500 bp of 5′ and 3′ flanking DNA, PCR amplified from strain 779-6A, on a BamHI fragment. Plasmid pL118 contains the truncated HSC82(1-704) allele lacking the five C-terminal codons (MEEVD).
Mutagenesis.
The SSA1-21 allele of SSA1 has a tryptophan in place of leucine at codon 483 (L483W). A screen for second-site suppressors of L483W was done as described previously (27). Briefly, plasmid pJ121 was randomly mutagenized with hydroxylamine and then used to transform strain G400-1C to leucine prototrophy. Transformant colonies were replicated onto plates lacking adenine (−ade) and containing 5-fluoro-orotic acid to simultaneously select for cells that propagated [PSI+] and had lost pRDW10, which requires that the mutant proteins both restore [PSI+] propagation and provide essential Ssa1p function. Site-directed mutagenesis on pJ126 and pJ127 using the QuikChange kit (Stratagene) and appropriate primers was done to create alleles for genomic gene replacements of SSA1.
Nonsense suppression (read-through) assays.
A dual-luciferase assay system was used (24). The system consists of bicistronic mRNAs encoding a translational fusion of Renilla and firefly luciferase genes with UGG or UAA at the sixth codon of the firefly gene (J. W. Harger and J. D. Dinman, unpublished data). Because the enzymes are expressed from the same mRNA and the product is a fusion protein, abundance of mRNA and protein are controlled for internally. Cells with plasmids pYDL505 (UGG) and pYDL506 (UAA) were grown in plasmid selective medium to an optical density at 600 nm (OD600) of 0.5 to 1. Cells in 1 ml of culture were centrifuged, washed, and then broken by agitation with glass beads in 0.3 ml of lysis buffer. Broken cells were centrifuged for 5 min at 8,000 rpm in a microcentrifuge (Sorvall; MC12) and luciferase activity of 5 ml of supernatant was assayed using the Promega dual-luciferase assay system in a Zylux FB15 luminometer.
Purification of His-tagged Ssa1p and Western blotting.
His6-tagged Ssa1p and Ssa1-21p were indistinguishable from the untagged proteins with respect to functions in growth and [PSI+] propagation (data not shown). Strain 1012 [psi−] transformants with the pHSSA1 series plasmids were transferred from solid medium lacking uracil to YPAD liquid. Cultures (OD600 = 1.0) were washed, suspended in lysis buffer (0.5× phosphate-buffered saline [pH 7.4], 50 mM KCl, 5 mM MgCl2, EDTA-free protease inhibitor cocktail tablets [Roche]), and broken by agitation with glass beads. Proteins were cross-linked for 60 min at 4°C with 1 mM dithiobis(succinimidylpropionate) (Sigma). The reaction was stopped with 5 mM lysine for 30 min at 4°C. Lysates (3 mg of protein) were diluted with equal volumes of binding buffer (1× phosphate buffer [pH 7.4], 10 mM imidazole, HisTrap kit [Amersham]) and incubated with nickel-charged resin for 1 h at 4°C. After five washes with binding buffer and then five washes with washing buffer (1× phosphate buffer [pH 7.4], 40 mM imidazole, HisTrap kit [Amersham]), resin-bound proteins were eluted with 120 μl of elution buffer (1× phosphate buffer [pH 7.4], 500 mM imidazole, HisTrap kit [Amersham]). Samples were boiled in 30 μl of 5× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer for 5 min, and 20-μl aliquots were subjected to Western analysis. Mouse anti-Hsp70 and rabbit anti-Hsp104 antibodies (SPA-822 and SPA-1040) were from Stressgen, rabbit anti-Ydj1p polyclonal antibodies were a gift from J. Brodsky (University of Pittsburgh), and rabbit anti-Sti1p and anti-Cpr7p polyclonal antibodies were generated for this study.
Guanidine curing of [PSI+].
[PSI+] cells grown on −ade plates at 25°C were used to inoculate liquid YPAD cultures, which were grown overnight at 30°C. These cultures were diluted in YPAD containing 3 mM guanidine-hydrochloride and maintained at an OD600 of ≤0.8 by dilution into fresh guanidine-containing medium. Samples were removed at each cell number doubling, measured as doubling of OD600, and spread onto three YPD plates at dilutions producing 300 to 500 colonies per plate. Entirely red colonies were scored as [psi−]. When guanidine curing was used as a confirmation of the presence of [PSI+], cells were grown to colonies on YPAD plates containing 3 mM guanidine and then assayed for the [PSI+] phenotype on YPD.
RESULTS
Impairment of [PSI+] prion propagation by Hsp70 mutation.
[PSI+] is a self-replicating aggregated form of Sup35p thought to be amyloid. Aggregation of Sup35p in [PSI+] cells causes nonsense suppression because much Sup35 protein is unavailable to function in translation termination. Mutants with the ade2-1 nonsense allele cannot grow without adenine and are red when grown on limiting amounts of adenine (e.g., on 1/2YPD) because of the accumulation of a pigmented substrate of Ade2p. Partial suppression of ade2-1 by [PSI+], which also requires the weakly UAA-suppressing tRNA SUQ5/SUP16, allows growth without adenine and eliminates the pigmentation (11). The relative strength of [PSI+] can be estimated from the degree of pigmentation and rate of growth without adenine, which reflect the degree of nonsense suppression caused by [PSI+] (13, 28). Another indicator of robustness of [PSI+] propagation is its mitotic stability, which typically correlates with nonsense suppression. Spontaneous mitotic loss of [PSI+] is seen as appearance of red, [psi−] colonies.
Figure 1 shows the various [PSI+] phenotypes of our strains, several of which were abnormal. To rule out the possibility that mutations were generating variants of [PSI+] that propagated atypically, all strains with abnormal [PSI+] phenotypes were used as cytoplasm donors in cytoduction crosses with wild-type [psi−] cells (see Materials and Methods). In all crosses, the wild-type recipients had a normal [PSI+] phenotype, indicating that the mutations weakened propagation of a normal form of [PSI+]. This test also confirms absence of [PSI+] in [psi−] cells. Where appropriate, we confirm that colony color reflected levels of nonsense suppression by quantifying stop codon read-through (Fig. 2).
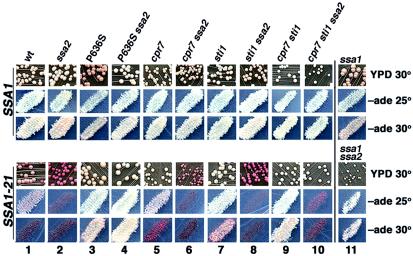
FIG. 1.
Effects of SSA1 and cochaperone mutations on [PSI+] phenotype. Cells were grown as colonies on YPD for 2 days at 30°C followed by 3 days at 25°C or as patches of cells replica plated onto −ade plates, which were incubated for 5 days at 25°C (−ade 25°) or for 3 days at the more stringent 30°C (−ade 30°). Representative areas of growth on plates are shown. Except for those in column 11, strains in upper panels have wild-type SSA1 and strains in lower panels have SSA1-21, as indicated. Strains in column 11 upper panels are ssa1Δ, and those in lower panels are ssa1Δ ssa2Δ. Relevant alleles of strains in each column are indicated; wt is wild type. Colors on YPD range from white to red, reflecting from most to least suppression of ade2-1 (see the text). The extent of ade2-1 suppression is also reflected as density of growth on −ade plates. Red cell color is enhanced on −ade plates compared with YPD plates, especially for nongrowing strains.
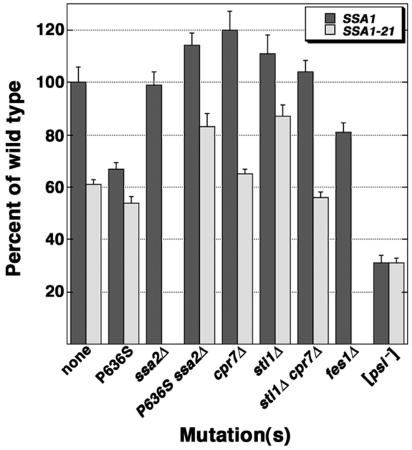
FIG. 2.
Quantified levels of [PSI+]-mediated nonsense suppression. Strains carry plasmids expressing a translational fusion of Renilla (upstream) and firefly (downstream) luciferases, with or without a termination codon (UAA) early in the firefly coding region. The ratio of activities of firefly to Renilla enzymes for the UAA construct versus the control construct [(firefly/Renilla)UAA/(firefly/Renilla)control] provides a measurement of UAA read-through. Read-through values for mutant strains are expressed as percentages relative to values for the wild-type strain. Assays were done in triplicate at least three times. SSA1-21 derivatives of ssa2Δ and fes1Δ strains are not represented, since they are unable to propagate [PSI+].
Our wild-type [PSI+] cells are white on YPD medium and grow well without adenine at both 25°C and the more stringent 30°C (Fig. 1, column 1, upper panels). [PSI+] is very stable in these cells, and we do not observe spontaneous appearance of red [psi−] colonies during routine handling. SSA1-21, an allele of the HSP70 gene SSA1 with the L483W mutation, causes frequent mitotic loss of [PSI+] and reduces nonsense suppression (Fig. 1, column 1, lower panels; Fig. 2). Effects of Ssa1-21p on [PSI+] are dominant over those of Ssa2p, which is another constitutively expressed cytosolic Hsp70 that is 97% identical to Ssa1p. When Ssa2p expression is abolished, Ssa1p abundance is elevated in a compensatory manner. While ssa2Δ cells have a normal [PSI+] phenotype, [PSI+] cannot propagate in SSA1-21 ssa2Δ cells (28) (Fig. 1, column 2).
C-terminal second-site suppressors of Ssa1-21p.
To understand better how Ssa1-21p function was altered, we previously isolated second-site mutations in Ssa1-21p that restored [PSI+] propagation in SSA1-21 ssa2Δ cells (27). Among others, three (P636S, E639K, and E640K) were in the extreme C terminus of Ssa1-21p. The conserved C-terminal octapeptide of Hsp70 (GPTVEEVD) mediates interactions with TPR-containing cochaperones and is necessary for optimal Hsp70 function (18, 49). Assuming that the three mutations similarly suppressed the impaired [PSI+] phenotype by altering Ssa1-21p interaction with TPR cochaperones, we chose P636S for further study.
Isogenic strains that contained chromosomal SSA1 and SSA1-21 alleles with or without the P636S substitution (SSA1-21P636S has both L483W and P636S) were constructed. An ssa1Δ strain and an isogenic ssa2Δ series were also tested. As originally isolated, SSA1-21P636S ssa2Δ cells had a normal [PSI+] phenotype (Fig. 1, compare column 4, lower panels, to column 1, upper panels). Therefore, the P636S substitution completely suppressed the [PSI+]-inhibitory effects of the Ssa1p L483W substitution in ssa2Δ cells.
The presence of Ssa2p in SSA1P636S or SSA1-21P636S cells reduced ability of [PSI+] to cause nonsense suppression (Fig. 1, compare columns 3 and 4; Fig. 2). The [PSI+] phenotype was also slightly weakened in ssa1Δ cells but was normal ssa2Δ cells (Fig. 1, columns 11 and 2, upper panels; Fig. 2). These results show that Ssa2p weakened [PSI+] in cells with compromised Ssa1p function and reveal a functional distinction between Ssa1p and Ssa2p with regard to [PSI+] propagation.
Depletion of TPR cochaperones restores [PSI+] propagation in SSA1-21 cells.
Since the extreme C terminus of Hsp70 is important for physical interaction with TPR motifs of cochaperones, we expected that the C-terminal suppressing mutations of Ssa1-21p were disrupting such interactions. Taking a candidate gene approach to identifying TPR cochaperones involved in the effects on [PSI+], we deleted separately the nonessential cyclophilin homologs CPR6 and CPR7 and HOP1 homologs SGT2 and STI1. Deleting CPR6 or SGT2 had no effect on growth or [PSI+] phenotype of wild-type or SSA1-21 strains. Deleting CPR7 or STI1 in wild-type cells also had no obvious effect. In contrast, deleting either of them in SSA1-21 cells reduced pigment accumulation and improved mitotic stability of [PSI+] (Fig. 1, lower panels, compare columns 5 and 7 to column 1). Deleting STI1 improved [PSI+] propagation better than deleting CPR7. On medium lacking adenine at the more stringent 30°C, SSA1-21 and SSA1-21 cpr7Δ cells were unable to form colonies but SSA1-21 sti1Δ cells grew well. As observed previously (16), deleting CPR7 reduced the growth rate (Table 1). These results show that Sti1p and Cpr7p were required for Ssa1-21p to have its full effects on impairing [PSI+].
Unlike the P636S mutation, deletion of STI1 or CPR7 did not restore [PSI+] propagation in SSA1-21 ssa2Δ cells. When both STI1 and CPR7 were deleted in SSA1-21 ssa2Δ cells, however, [PSI+] propagated stably (Fig. 1, lower panels, column 10). These [PSI+] cells did not grow without adenine, however, indicating that restoration of [PSI+] propagation was incomplete. Compared with the complete restoration of [PSI+] in SSA1-21 ssa2Δ cells by the P636S substitution in Ssa1-21p, this result suggests that other TPR cochaperones may affect [PSI+] propagation. One candidate is the essential Cns1p, which is functionally redundant with Cpr7p (14, 39).
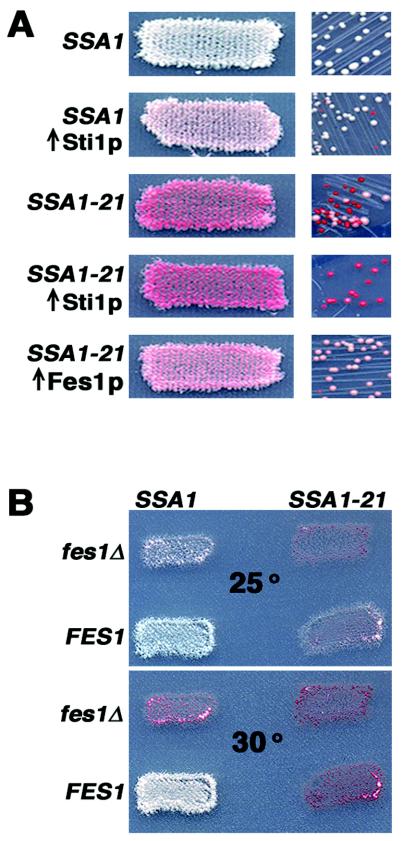
Overexpression of STI1 weakens [PSI+] and enhances SSA1-21 effects.
Since depleting Cpr7p and Sti1p improved [PSI+] propagation, we anticipated that increasing their abundance would have the opposite effect. In fact, others have shown that overproduction of Sti1p can weaken propagation of weak or hybrid forms of [PSI+] (33). We tested overproduction by transforming [PSI+] cells with high-copy-number plasmids carrying CPR7 and STI1. We also tested CNS1 because excess Cns1p restores normal growth to cpr7Δ cells (39). In wild-type cells, [PSI+] was not affected by excess Cpr7p or Cns1p, but overproduced Sti1p weakened [PSI+] propagation, increasing pigmentation and reducing mitotic stability of [PSI+] (Fig. 3A). Similarly, only excess Sti1p enhanced the impairment of [PSI+] propagation in SSA1-21 cells, increasing both pigment accumulation and frequency of mitotic loss of [PSI+] (Fig. 3A). In SSA1-21 cpr7Δ cells, overproduced Cns1p restored both growth rate and impairment of [PSI+] by Ssa1-21p (data not shown). Thus, in addition to being redundant with Cpr7p for growth, Cns1p also overlaps functionally with Cpr7p with regard to [PSI+] propagation through effects on Hsp70.
FIG. 3.
Opposing effects of Sti1p and Fes1p on [PSI+]. (A) Increased expression of Sti1p and Fes1p. Patches or colonies of [PSI+] SSA1 and SSA1-21 transformants of a high-copy-number plasmid, with or without the indicated genes, were grown on selection plates with limiting adenine for 2 days at 30°C. While [PSI+] can propagate in SSA1-21 cells overproducing Sti1p, the enhanced weakening of [PSI+] makes it difficult to distinguish its presence at the level of individual colonies. (B) Patches of SSA1 and SSA1-21 cells, having or lacking FES1 as indicated, were replica plated onto plates lacking adenine and incubated at the indicated temperature (given in degrees Celsius) for 2 days.
Deletion of FES1 weakens [PSI+] and enhances SSA1-21 effects.
While it is not known how the Hsp90 cochaperone Cpr7p affects Hsp70 function, Sti1p was recently shown to be an activator of Ssa1p ATPase (60) and thus promotes Ssa1p substrate binding. In contrast, the nucleotide exchange factor Fes1p accelerates release of ADP from Ssa1p (30), thereby facilitating Ssa1p's return to the open state and substrate release. Therefore, deleting Fes1p should prolong Ssa1p's closed state, which we predicted would adversely affect [PSI+], while excess Fes1p was expected to have the opposite effects. Indeed, we found that [PSI+] was unable to propagate in SSA1-21 cells lacking FES1 and propagated better in SSA1-21 cells that overproduced Fes1p (Fig. 3A and B). Moreover, deleting FES1 predictably weakened [PSI+] in wild-type cells, reducing both nonsense suppression and mitotic stability of [PSI+] (Figs. 2 and 3, data not shown).
TPR cochaperone deletion or P636S restores [PSI+] seed number in SSA1-21 cells.
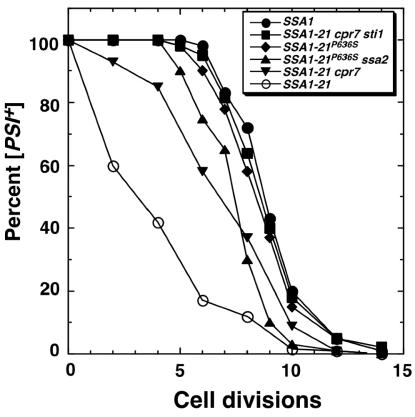
In order for [PSI+] to be maintained in a population, the number of transmissible prion particles, or seeds, must double, on average, in the time it takes cells to divide. SSA1-21 significantly reduces the average number of [PSI+] seeds per cell (28), which explains why it causes frequent loss of [PSI+]. We used guanidine curing of [PSI+] to determine if P636S and the cochaperone deletions restored [PSI+] seed generation in SSA1-21 cells. At millimolar concentrations, guanidine in growth media arrests replication of [PSI+] seeds, which then become diluted as cells divide, eventually giving rise to cells having lost [PSI+] (17). After adding guanidine to growing cultures, the rate of appearance of [psi−] cells provides an estimate of the average number of seeds per cell.
SSA1-21P636S cells had a near-wild-type curing profile, showing that the P636S substitution significantly restored seed number (Fig. 4). Deleting STI1 or CPR7 had no effect on [PSI+] curing in wild-type cells. In SSA1-21 cells, deleting STI1 restored the curing profile, similar to that of SSA1-21P636S cells, while deletion of CPR7 restored seed number less well (Fig. 4). When both CPR7 and STI1 were deleted in SSA1-21 cells, [PSI+] seed number was restored completely. These results show that Cpr7p and Sti1p affect Hsp70 function with respect to [PSI+] seed generation and are consistent with our other findings that altering Sti1p abundance had greater effects on [PSI+].
FIG. 4.
Guanidine curing of [PSI+]. The percentage of [PSI+] cells remaining in log-phase cultures was monitored as a function of cell divisions after addition of guanidine-hydrochloride to a final concentration of 3 mM.
Hsp70 C-terminal mutations variably affect interaction with cochaperones.
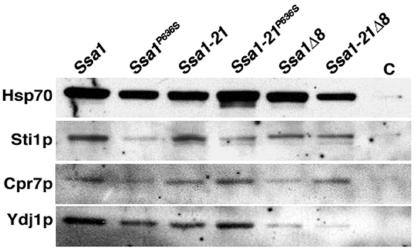
To assess the effects of the C-terminal mutations on cochaperone interactions, we compared abundance of proteins that copurified with Ssa1p. Amino-terminal His6-tagged versions of Ssa1p and Ssa1-21p were used to specifically isolate Ssa1p isoforms of Hsp70. Binding of Sti1p to Ssa1p and Ssa1-21p was reduced but not eliminated by both the P636S substitution and C-terminal deletion (Fig. 5). While these mutations modestly reduced the amount of Cpr7p associated with Ssa1p, they did not have a significant effect on the binding of Cpr7 to Ssa1-21p. Therefore, although the TPR interaction motif is important for binding of Sti1p and Cpr7p to Ssa1p, these cochaperones can bind Ssa1p at a different site. Nevertheless, since both point mutations and complete deletion of the C-terminal TPR interaction motif suppress Ssa1-21p impairment of [PSI+], this secondary interaction must have incomplete or no functionality with regard to effects on [PSI+]. Also, since more Cpr7p was associated with Ssa1-21p, the L483W substitution may enhance the secondary interaction with Cpr7.
FIG. 5.
Copurification of Hsp70-associated proteins. His6-tagged versions of Ssa1p and Ssa1-21p were purified on nickel affinity resin and subjected to Western analysis. Purified His6-tagged proteins are indicated at the top, and lane C is from a similar purification using lysates of cells expressing nontagged Ssa1p. The Hsp70 and Sti1p panels are from separate blots loaded with identical aliquots of the same samples. The Cpr7p and Ydj1p panels are from the Hsp70 and Sti1p blots, respectively, which were stripped and reprobed.
Deleting the C-terminal residues significantly reduced Hsp40 (Ydj1p) interaction, which agrees with earlier work showing that Hsp40 interaction with Hsp70 is reduced when Hsp70 lacks its four C-terminal residues (EEVD) (12). Hsp40s also interact with the ATPase domain of Hsp70 (31), and residual binding of Hsp40 to the deletion mutants is likely mediated by this interaction. The P636S mutation had little affect on binding of Hsp40 to Ssa1p or Ssa1-21p, suggesting that suppression of L483W effects by P636S is not due to reduced ability of Hsp40 to interact with Ssa1p.
Deleting Hsp90's TPR interaction motif has no effect on [PSI+].
Because Cpr7p and Sti1p are Hsp90 cochaperones, we anticipated that Hsp90 might be involved in the SSA1-21 effects on [PSI+]. If so, then disrupting the interaction between Hsp90 and TPR cochaperones would improve [PSI+] propagation in SSA1-21 cells. We tested this prediction by constructing strains that lacked both chromosomal HSP90 genes (HSC82 and HSP82) and expressed intact (Hsc82(1-709) or truncated (Hsc82(1-704) forms of Hsc82p. Expression of either Hsc82p or Hsp82, collectively referred to as Hsp90, is essential for viability. The Hsc82(1-704) protein lacks the C-terminal MEEVD amino acids, which mediate TPR interaction but are dispensable for viability (37).
The [PSI+] phenotypes of both wild-type and SSA1-21 cells were unchanged when the truncated form of Hsc82p was expressed in place of intact Hsc82p (data not shown). These results demonstrate that the Hsp90 TPR interaction motif is dispensable for [PSI+] propagation and suggest that Hsp70 can functionally interact with TPR cochaperones independently of Hsp90.
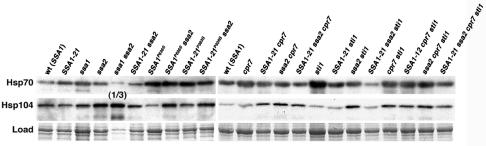
Altered Hsp70 and Hsp104 abundance does not correlate with effects on [PSI+].
Since altered abundance of Hsp70 and Hsp104 can have significant effects on [PSI+] propagation (9, 34, 44), we addressed the possibility that the SSA and cochaperone effects on [PSI+] were due to altered expression of Hsp70 and Hsp104 by comparing their abundance in our strains (Fig. 6). Although there was a complex pattern of Hsp70 and Hsp104 expression, there was no correlation between the levels of either of these proteins and [PSI+] phenotype. Also, there was no correlation between Hsp70 or Hsp104 abundance and ability to propagate [PSI+]. These results suggest that effects of SSA and cochaperone mutations on [PSI+] propagation were due to altered activity of individual proteins or complexes rather than altered abundance of Hsp70 and Hsp104.
FIG. 6.
Relative abundance of Hsp70 and Hsp104. Abundance of Hsp70 and Hsp104 was examined by Western analysis. Blots probed with anti-Hsp70 antibodies were stripped and reprobed with anti-Hsp104 antibodies. Representative portions of membranes, stained by amido black as a loading and transfer control, are shown (Load). Relevant alleles of strains are indicated at the top; wt is wild type. The ssa1Δ ssa2Δ sample was diluted 1:3, as indicated, because Hsp104 abundance in this strain is very high (compare amounts in the Load panel). The signal in the Hsp70 blot for this strain represents Ssa3p and Ssa4p, which are expressed when Ssa1p and Ssa2p both are absent.
Deletion of CPR7 correlated with elevated Hsp104 abundance, which is consistent with earlier data (15). Hsp104 was also elevated for most ssa2Δ strains but not for ssa1Δ strains, which reveals another functional distinction between Ssa1p and Ssa2p.
The Ssa1p C-terminal TPR interaction motif is dispensable for viability.
The SSA subfamily of cytosolic Hsp70 genes, which consists of four members (Ssa3p and Ssa4p are expressed only under nonoptimal growth conditions), are the only S. cerevisiae Hsp70s with the GPTVEEVD motif. Expression of at least one SSA gene is essential for growth (62). To determine the importance of the GPTVEEVD motif for essential Ssap function, plasmids carrying SSA1 and SSA1-21 alleles without it (SSA1Δ8 and SSA1-21Δ8, respectively) were expressed in a strain lacking all chromosomal SSA genes. Cells expressing intact SSA1 or SSA1-21 as the only SSA gene grew as well as the wild type (Table 1). Those expressing only SSA1Δ8 grew more slowly (Table 1). Thus, the TPR interaction motif was important but dispensable for essential Ssap function. [PSI+] propagated stably in this strain, but its presence slowed growth further. The SSA1-21Δ8 allele also supported growth, but only very weakly and if cells were [psi−] (Table 1). Because SSA1-21Δ8 did not support growth of [PSI+] cells, we could not test if the deletion overcame SSA1-21 effects on [PSI+] in the absence of other Ssap. However, as with the P636S substitution, the C-terminal deletion suppressed the effect of Ssa1-21p on [PSI+] in cells expressing Ssa2p (data not shown).
DISCUSSION
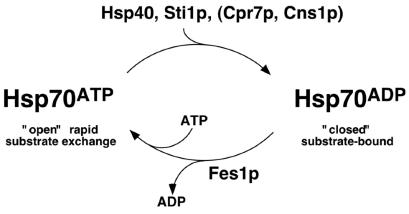
We identify new proteins whose altered abundance affects [PSI+] propagation and implies that amyloid propagation in vivo is impaired by alterations that promote conversion to, or stabilize, the substrate-bound state of Hsp70. Figure 7 depicts the Hsp70 reaction cycle and where these proteins act on it.
FIG. 7.
Regulation of Hsp70 (Ssa1p) reaction cycle by cochaperones. Hsp70 function is finely tuned by ATP hydrolysis and nucleotide exchange, which regulate substrate binding. It is known that Hsp40 (Ydj1p, Sis1p) and Sti1p stimulate ATP hydrolysis, promoting substrate binding, and that Fes1p accelerates ADP release and nucleotide exchange, promoting substrate release. Our data suggest that enhancing conversion of Hsp70 to the ADP-bound state or stabilizing this state impairs [PSI+] propagation. Our data also suggest that Cpr7p and Cns1p might promote or stabilize this step in the cycle.
Our earlier work showed that second-site mutations in Ssa1-21p that overcome the [PSI+]-impairing effect of the L483W substitution weaken substrate binding, which indicates that L483W enhances substrate binding of Ssa1p (27). Consistent with this interpretation, overproducing Sti1p, which promotes substrate binding by activating Ssa1p ATPase, also impaired [PSI+] propagation. Conversely, deletion of Sti1p restored [PSI+] propagation in SSA1-21 cells. As predicted and in contrast to the case with Sti1p, overproduction of Fes1p, which facilitates substrate release (30), counteracted the impairment of [PSI+] by Ssa1-21p, while depletion of Fes1p, which should prolong the substrate-bound state of Ssa1p, impaired [PSI+] in both wild-type and SSA1-21 cells. Thus, in addition to mutations in Hsp70 that alter its function directly, an imbalance of Hsp70 cochaperones can produce a similar [PSI+]-impairing effect by disrupting regulation of the Hsp70 reaction cycle.
Our results agree with previous data showing that excess Sti1p destabilizes some variants of [PSI+] (33) and suggest that this destabilization is mediated through effects on Ssa1p activity rather than altered expression of other Hsps as previously suggested. Hsp40s also stimulate Hsp70 ATPase, and in particular Ydj1p and Sis1p stimulate Ssa1p ATPase (38). Consistent with our conclusion, overproduction of the Hsp40 homologs Ydj1p, Sis1p, and Apj1p impairs propagation of yeast prions (33, 34, 42), and a suppressing mutation in Ssa1-21p described earlier is in a residue expected to disrupt Hsp40 interaction (27). Additionally, weakening of [PSI+] by Ydj1p overproduction is increased when Ssa1p is simultaneously elevated. Our data suggest that the previously described effects of Sti1p and Hsp40 overproduction on yeast prions can be attributed to stimulated Ssa1p ATPase.
Data presented here support our earlier interpretation that other mutations in Ssa1p that inhibit [PSI+] propagation also increase ATPase activity of Ssa1p. Aside from L483W, all of eight previously described Ssa1p mutations that impair [PSI+] are in the ATPase domain and cannot directly affect interaction with the substrate (27). The L483W substitution is within the substrate-binding domain but distant from the substrate-binding pocket, so its effects should also be indirect, perhaps altering interdomain communication. Together, effects of the Hsp70 and cochaperone mutations on [PSI+] suggest that the increase in ATPase is reflected in the reduced generation of amyloid seeds from preexisting material. Since cells expressing Ssa1-21p as the only essential Ssap are unable to maintain [PSI+] but grow as well as wild-type cells, an implication of our results is that it might be possible to therapeutically modulate ATPase activity of Hsp70 in other systems, directly or through cochaperones, in a way that would impair cytosolic amyloid propagation with minimal side effects.
How does promoting or prolonging substrate binding by Hsp70 impair [PSI+] propagation? In addition to the continued growth of polymers, propagation of yeast prions (infectious amyloid) requires the generation of new self-replicating seeds from preexisting material and efficient distribution of seeds between mother and daughter cells. Generation of yeast prion seeds requires Hsp104 protein disaggregating function, which may break the polymers into smaller, more numerous pieces that continue to propagate the structure and are more readily transmitted to daughter cells (4, 43, 47). Since SSA1-21 mutants have severalfold fewer seeds per cell, altered Hsp70 function can interfere with amyloid seeding. Because Ssa1-21p does not affect Hsp104's ability to provide thermotolerance (28) or to reactivate heat-denatured luciferase in vivo (G. Jung, unpublished data), its effect on [PSI+] might be direct. The simplest way to explain our data is that Ssa1-21p reduces generation of [PSI+] seeds by binding too avidly to Sup35p aggregates, which sterically restricts access to Hsp104 (27). If this explanation were true, then this altered function must affect disassembly or breakage of the presumed highly ordered amyloid fibers of yeast prions differently than disaggregation of amorphous, thermally denatured substrates.
A way to explain this difference is that amyloid is recognized differently as a substrate by Hsp70 and Hsp104. Since Hsp40 is thought to be able to present a substrate to Hsp70, and its substrate recognition overlaps that of Hsp70, Hsp40 also may contribute to the apparent substrate-specific effects on amyloid. In line with this idea, properties of a specific Hsp40 (Sis1p) have been suggested to enhance interaction of the yeast prion [PIN+]/[RNQ+] determinant Rnq1p with Hsp70 (36).
Another way that enhanced substrate binding by Hsp70 might impair [PSI+] is that Hsp70's ability to assist Hsp104 in resolubilizing protein from aggregates is altered. Although the mechanism of protein disaggregation by Hsp104 is unknown, Hsp70 and Hsp40 cooperate in this process, and together these three chaperones can resolubilize large protein aggregates in vitro (20). The altered function of the mutant Hsp70 may affect an aspect of a transient physical interaction between Hsp70 and Hsp104 or of their ability to act sequentially or simultaneously on a substrate. In the homologous bichaperone system of Escherichia coli, the Hsp104 homolog ClpB binds the substrate first, increasing exposure of surfaces for subsequent Hsp70 interaction (21, 61). If the reaction is similar in yeast, a substrate produced from Hsp104 interaction with amyloid may interact with Hsp70 in a way that allows it to retain the ability to propagate the self-replicating structure. By binding too avidly with this substrate, Ssa1-21p may interfere with the ability of the self-replicating conformation to be maintained. Such a scenario may explain why Ssa1-21p has a detrimental effect on amyloid without greatly affecting other cellular processes that require Hsp70 function.
Our data suggest that Cpr7p, whose effect on Hsp70's enzymatic function is unknown, may also act to enhance substrate binding by Ssa1p. Since more Cpr7p was associated with Ssa1-21p than with Ssa1p, and more Ssa1-21p is expected to be in the closed ADP-bound state, Cpr7p may preferentially bind the ADP-bound form of Hsp70. If so, then Cpr7p may be acting to stabilize the closed conformation rather than to induce conversion to this state. The additive effect of depleting both Cpr7p and Sti1p is consistent with this interpretation.
Deleting CPR6 or SGT2, other yeast cyclophilin and Hop1 homologs, respectively, had no effect on [PSI+] propagation in wild-type or SSA1-21 cells, showing that the effect of TPR cochaperones on [PSI+] was not a general one. Thus, with respect to [PSI+] there are clear functional differences in TPR cochaperone interactions with Ssa1p, which are likely rooted in differences in specificity or affinity. Such differences also may underlie functional distinctions between Ssa1p and Ssa2p regarding [PSI+] propagation (this study), the ability of excess Ssa1p but not Ssa2p to cure cells of the yeast [URE3] prion (50), or the specific requirement of Ssa2p for transport of certain proteins into prevacuolar (Vid) vesicles (6).
Our finding that deleting the TPR interaction motif (MEEVD) of Hsp90 had no effect on [PSI+] suggests that Hsp90 is not involved in Sti1p and Cpr7p effects on [PSI+] propagation. Despite wide interest in yeast prions and Hsp90 and the availability of many Hsp90 mutants, the only other experiments describing effects of Hsp90 on yeast prions were also negative, showing a lack of effect of overproduced Hsp90 on [PSI+] (44). Whether Hsp90 is required for [PSI+] propagation cannot be tested directly because it is essential for viability. However, we also found that [PSI+] propagation was unaffected in both wild-type and SSA1-21 cells by deletion of only HSC82, which reduces the overall abundance of Hsp90 to roughly 10% of wild-type levels (5), or when cells were treated with a wide range of concentrations of the Hsp90 inhibitors geldanamycin and radicicol (G. Jones, unpublished data). Together these observations suggest that Hsp90 function is not important for [PSI+] propagation. Our data therefore provide evidence suggesting that Sti1p and Cpr7p can functionally interact with Ssa1p independently of Hsp90. Cpr7 and CNS1 have been found together in complexes without Hsp90 (58), and our results may reveal effects of perturbation of such a complex.
Our findings illustrate the utility of yeast prions as a system for studying both amyloid propagation and protein chaperone function. In addition to advancing the understanding of how chaperones affect amyloid propagation in vivo, continued study with this unique system will provide new insights into the functions of the chaperone machinery in general.
Acknowledgments
We thank Andy Golden, Will Prinz, and Kevin O'Connell for helpful comments on the manuscript.
REFERENCES
- 1.Abbas-Terki, T., O. Donze, P. A. Briand, and D. Picard. 2001. Hsp104 interacts with Hsp90 cochaperones in respiring yeast. Mol. Cell. Biol. 21:7569-7575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borchsenius, A. S., R. D. Wegrzyn, G. P. Newnam, S. G. Inge-Vechtomov, and Y. O. Chernoff. 2001. Yeast prion protein derivative defective in aggregate shearing and production of new ′seeds.' EMBO J. 20:6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borkovich, K. A., F. W. Farrelly, D. B. Finkelstein, J. Taulien, and S. Lindquist. 1989. Hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol. Cell. Biol. 9:3919-3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, C. R., J. A. McCann, and H. L. Chiang. 2000. The heat shock protein Ssa2p is required for import of fructose-1,6-bisphosphatase into Vid vesicles. J. Cell Biol. 150:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 8.Chang, H. C., and S. Lindquist. 1994. Conservation of Hsp90 macromolecular complexes in Saccharomyces cerevisiae. J. Biol. Chem. 269:24983-24998. [PubMed] [Google Scholar]
- 9.Chernoff, Y. O., S. L. Lindquist, B. Ono, S. G. Inge-Vechtomov, and S. W. Liebman. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268:880-884. [DOI] [PubMed] [Google Scholar]
- 10.Conde, J., and G. R. Fink. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl. Acad. Sci. USA 73:3651-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, B. S. 1965. “Ψ” a cytoplasmic suppressor of super-suppressor in yeast. Heredity 20:505-521. [Google Scholar]
- 12.Demand, J., J. Luders, and J. Hohfeld. 1998. The carboxy-terminal domain of Hsc70 provides binding sites for a distinct set of chaperone cofactors. Mol. Cell. Biol. 18:2023-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derkatch, I. L., Y. O. Chernoff, V. V. Kushnirov, S. G. Inge-Vechtomov, and S. W. Liebman. 1996. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 144:1375-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolinski, K. J., M. E. Cardenas, and J. Heitman. 1998. CNS1 encodes an essential p60/Sti1 homolog in Saccharomyces cerevisiae that suppresses cyclophilin 40 mutations and interacts with Hsp90. Mol. Cell. Biol. 18:7344-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duina, A. A., H. M. Kalton, and R. F. Gaber. 1998. Requirement for Hsp90 and a CyP-40-type cyclophilin in negative regulation of the heat shock response. J. Biol. Chem. 273:18974-18978. [DOI] [PubMed] [Google Scholar]
- 16.Duina, A. A., J. A. Marsh, and R. F. Gaber. 1996. Identification of two CyP-40-like cyclophilins in Saccharomyces cerevisiae, one of which is required for normal growth. Yeast 12:943-952. [DOI] [PubMed] [Google Scholar]
- 17.Eaglestone, S. S., L. W. Ruddock, B. S. Cox, and M. F. Tuite. 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI+] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:240-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman, B. C., M. P. Myers, R. Schumacher, and R. I. Morimoto. 1995. Identification of a regulatory motif in Hsp70 that affects ATPase activity, substrate binding and interaction with HDJ-1. EMBO J. 14:2281-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glover, J. R., A. S. Kowal, E. C. Schirmer, M. M. Patino, J. J. Liu, and S. Lindquist. 1997. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell 89:811-819. [DOI] [PubMed] [Google Scholar]
- 20.Glover, J. R., and S. Lindquist. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94:73-82. [DOI] [PubMed] [Google Scholar]
- 21.Goloubinoff, P., A. Mogk, A. P. Zvi, T. Tomoyasu, and B. Bukau. 1999. Sequential mechanism of solubilization and refolding of stable protein aggregates by a bichaperone network. Proc. Natl. Acad. Sci. USA 96:13732-13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie, C., and G. R. Fink (ed.). 1991. Guide to yeast genetics and molecular biology, vol. 194. Academic Press, San Diego, Calif.
- 23.Han, W., and P. Christen. 2003. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J. Biol. Chem. 278:19038-19043. [DOI] [PubMed] [Google Scholar]
- 24.Harger, J. W., and J. D. Dinman. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA 9:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartl, F. U., and M. Hayer-Hartl. 2002. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295:1852-1858. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez, M. P., A. Chadli, and D. O. Toft. 2002. Hsp40 binding is the first step in the Hsp90 chaperoning pathway for the progesterone receptor. J. Biol. Chem. 277:11873-11881. [DOI] [PubMed] [Google Scholar]
- 27.Jones, G. W., and D. C. Masison. 2003. Saccharomyces cerevisiae Hsp70 mutations affect [PSI+] prion propagation and cell growth differently and implicate Hsp40 and tetratricopeptide repeat cochaperones in impairment of [PSI+]. Genetics 163:495-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung, G., G. Jones, R. D. Wegrzyn, and D. C. Masison. 2000. A role for cytosolic Hsp70 in yeast [PSI+] prion propagation and [PSI+] as a cellular stress. Genetics 156:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung, G., and D. C. Masison. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43:7-10. [DOI] [PubMed] [Google Scholar]
- 30.Kabani, M., J. M. Beckerich, and J. L. Brodsky. 2002. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22:4677-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley, W. L. 1999. Molecular chaperones: How J domains turn on Hsp70s. Curr. Biol. 9:R305-R308. [DOI] [PubMed] [Google Scholar]
- 32.King, C. Y., P. Tittmann, H. Gross, R. Gebert, M. Aebi, and K. Wuthrich. 1997. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc. Natl. Acad. Sci. USA 94:6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kryndushkin, D. S., V. N. Smirnov, M. D. Ter-Avanesyan, and V. V. Kushnirov. 2002. Increased expression of Hsp40 chaperones, transcriptional factors, and ribosomal protein Rpp0 can cure yeast prions. J. Biol. Chem. 277:23702-23708. [DOI] [PubMed] [Google Scholar]
- 34.Kushnirov, V. V., D. S. Kryndushkin, M. Boguta, V. N. Smirnov, and M. D. Ter-Avanesyan. 2000. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 10:1443-1446. [DOI] [PubMed] [Google Scholar]
- 35.Kushnirov, V. V., and M. D. Ter-Avanesyan. 1998. Structure and replication of yeast prions. Cell 94:13-16. [DOI] [PubMed] [Google Scholar]
- 36.Lopez, N., R. Aron, and E. A. Craig. 2003. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol. Biol. Cell 14:1172-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louvion, J. F., R. Warth, and D. Picard. 1996. Two eukaryote-specific regions of Hsp82 are dispensable for its viability and signal transduction functions in yeast. Proc. Natl. Acad. Sci. USA 93:13937-13942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273:27824-27830. [DOI] [PubMed] [Google Scholar]
- 39.Marsh, J. A., H. M. Kalton, and R. F. Gaber. 1998. Cns1 is an essential protein associated with the Hsp90 chaperone complex in Saccharomyces cerevisiae that can restore cyclophilin 40-dependent functions in Cpr7Δ cells. Mol. Cell. Biol. 18:7353-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masison, D. C., and R. B. Wickner. 1995. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science 270:93-95. [DOI] [PubMed] [Google Scholar]
- 41.Mayer, M. P., H. Schroder, S. Rudiger, K. Paal, T. Laufen, and B. Bukau. 2000. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7:586-593. [DOI] [PubMed] [Google Scholar]
- 42.Moriyama, H., H. K. Edskes, and R. B. Wickner. 2000. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol. 20:8916-8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ness, F., P. Ferreira, B. S. Cox, and M. F. Tuite. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22:5593-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Newnam, G. P., R. D. Wegrzyn, S. L. Lindquist, and Y. O. Chernoff. 1999. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol. 19:1325-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolet, C. M., and E. A. Craig. 1989. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol. Cell. Biol. 9:3638-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parsell, D. A., A. S. Kowal, M. A. Singer, and S. Lindquist. 1994. Protein disaggregation mediated by heat-shock protein Hsp104. Nature 372:475-478. [DOI] [PubMed] [Google Scholar]
- 47.Paushkin, S. V., V. V. Kushnirov, V. N. Smirnov, and M. D. Ter-Avanesyan. 1996. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 15:3127-3134. [PMC free article] [PubMed] [Google Scholar]
- 48.Pratt, W. B., and D. O. Toft. 2003. Regulation of signaling protein function and trafficking by the Hsp90/Hsp70-based chaperone machinery. Exp. Biol. Med. (Maywood) 228:111-133. [DOI] [PubMed] [Google Scholar]
- 49.Scheufler, C., A. Brinker, G. Bourenkov, S. Pegoraro, L. Moroder, H. Bartunik, F. U. Hartl, and I. Moarefi. 2000. Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101:199-210. [DOI] [PubMed] [Google Scholar]
- 50.Schwimmer, C., and D. C. Masison. 2002. Antagonistic interactions between yeast [PSI+] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 22:3590-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherman, F. 1994. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 52.Sikorski, R. S., M. S. Boguski, M. Goebl, and P. Hieter. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60:307-317. [DOI] [PubMed] [Google Scholar]
- 53.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sondheimer, N., N. Lopez, E. A. Craig, and S. Lindquist. 2001. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 20:2435-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speransky, V. V., K. L. Taylor, H. K. Edskes, R. B. Wickner, and A. C. Steven. 2001. Prion filament networks in [URE3] cells of Saccharomyces cerevisiae. J. Cell Biol. 153:1327-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stansfield, I., K. M. Jones, V. V. Kushnirov, A. R. Dagkesamanskaya, A. I. Poznyakovski, S. V. Paushkin, C. R. Nierras, B. S. Cox, M. D. Ter-Avanesyan, and M. F. Tuite. 1995. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 14:4365-4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor, K. L., N. Cheng, R. W. Williams, A. C. Steven, and R. B. Wickner. 1999. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science 283:1339-1343. [DOI] [PubMed] [Google Scholar]
- 58.Tesic, M., J. A. Marsh, S. B. Cullinan, and R. F. Gaber. 2003. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in Saccharomyces cerevisiae. J. Biol. Chem. 278:32692-32701. [DOI] [PubMed] [Google Scholar]
- 59.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 60.Wegele, H., M. Haslbeck, J. Reinstein, and J. Buchner. 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278:25970-25976. [DOI] [PubMed] [Google Scholar]
- 61.Weibezahn, J., C. Schlieker, B. Bukau, and A. Mogk. 2003. Characterization of a trap mutant of the AAA+ chaperone ClpB. J. Biol. Chem. 278:32608-32617. [DOI] [PubMed] [Google Scholar]
- 62.Werner-Washburne, M., D. E. Stone, and E. A. Craig. 1987. Complex interactions among members of an essential subfamily of Hsp70 genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 7:2568-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickner, R. B. 1994. Evidence for a prion analog in S. cerevisiae: the [URE3] non-Mendelian genetic element as an altered URE2 protein. Science 264:566-569. [DOI] [PubMed] [Google Scholar]
- 64.Wickner, R. B., K. L. Taylor, H. K. Edskes, M. L. Maddelein, H. Moriyama, and B. T. Roberts. 2000. Prions of yeast as heritable amyloidoses. J. Struct. Biol. 130:310-322. [DOI] [PubMed] [Google Scholar]
- 65.Wittung-Stafshede, P., J. Guidry, B. E. Horne, and S. J. Landry. 2003. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry 42:4937-4944. [DOI] [PubMed] [Google Scholar]
- 66.Zhouravleva, G., L. Frolova, X. Le Goff, R. Le Guellec, S. Inge-Vechtomov, L. Kisselev, and M. Philippe. 1995. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 14:4065-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]