Abstract
We identified in the transcriptome analysis of patients with alcoholic hepatitis (AH) osteopontin (OPN) as one of the most up-regulated genes. Here, we used a translational approach to investigate its pathogenic role. OPN hepatic gene expression was quantified in patients with AH and other liver diseases. OPN protein expression and processing were assessed by immmunohistochemistry, Western blotting and ELISA. OPN gene polymorphisms were evaluated in patients with alcoholic liver disease. The role of OPN was evaluated in OPN−/− mice with alcohol-induced liver injury. OPN biological actions were studied in human hepatic stellate cells and in precision-cut liver slices. Hepatic expression and serum levels of OPN were markedly increased in AH compared to normal livers and other types of chronic liver diseases and correlated with short-term survival. Serum levels of OPN also correlated with hepatic expression and disease severity. OPN was mainly expressed in areas with inflammation and fibrosis. Two proteases that process OPN (thrombin and MMP-7) and cleaved-OPN were increased in livers with AH. Patients with AH had a tendency of a lower frequency of the CC genotype of the +1239C SNP of the OPN gene compared to patients with alcohol abuse without liver disease. Importantly, OPN−/− mice were protected against alcohol-induced liver injury and showed decreased expression of inflammatory cytokines. Finally, OPN was induced by LPS and stimulated inflammatory actions in hepatic stellate cells.
Conclusion
Human and experimental data suggest a role for OPN in the pathogenesis of AH. Further studies should evaluate OPN as a potential therapeutic target.
Keywords: alcoholic liver disease, translational research, cytokines, liver fibrosis
Alcoholic liver disease (ALD) is a major cause of advanced liver disease worldwide. Despite its relevance as an etiological factor of liver cirrhosis and liver-related mortality, there are not major advances in the treatment of this disease(1). In particular, there are no targeted therapies that attenuate liver injury and fibrosis in patients with alcohol abuse. The development of such therapies is hampered by the poor knowledge of disease mechanisms in humans. One of the scenarios in which new therapies is urgently needed is alcoholic hepatitis (AH), a clinical condition characterized by inflammation, massive hepatic steatosis, pericellular fibrosis and severe hepatocellular damage(2). The development of episodes of AH heavily impacts the progression and severity of ALD. In its severe cases, AH carries a high short-term mortality (20–30% of mortality at 3 months). Patients with severe AH are prompted to develop liver failure, portal hypertension and severe bacterial infections. Unfortunately, current therapies (i.e. corticosteroids) fail in many patients and the attempts to develop targeted therapies such as the use of tumor necrosis factor α (TNFα) blocking agents have not been successful (3). Because rodents are resistant to develop advanced stages of ALD, human studies are required to identify new targets for therapy. In our laboratory, we previously found that CXC chemokines are markedly overexpressed in patients with AH and their hepatic expression correlate with disease severity(4,5). Moreover, we recently performed a functional analysis of a high-throughput transcriptome study, which revealed that genes encoding extracellular matrix proteins are differentially expressed in livers with AH(6). Among these proteins, osteopontin (OPN) was the most up-regulated one. Because OPN is a highly active molecule that can act as a true cytokine, the current study was aimed at specifically investigating the role of OPN in AH as a potential target for therapy.
OPN or secreted phosphoprotein 1 (SPP1) is an extracellular matrix protein that can also act as a neutrophil-attracting chemokine. It is synthesized by multiple tissues and secreted into body fluids. In target cells, OPN binds to a high number of integrins and CD44 to promote profibrogenic and inflammatory actions(7). Importantly, OPN undergoes a high number of post-translational modifications including serine-phosphorylation, O-glycosylation and proteolytic processing(8). Importantly, the proteolytic actions of proteases such as thrombin or matrix metalloproteinase 7 (MMP-7) yield a more active form of OPN(9).
There is growing evidence that OPN plays a major role in the wound healing response to acute and chronic injury in many organs including the lung and the kidney(10,11). In addition, recent studies indicate that OPN participates in the pathogenesis of hepatic steatosis, inflammation and the resulting fibrosis in different types of liver diseases(12,13). In particular, OPN seems to play a major role in the pathogenesis of nonalcoholic steatohepatitis, a condition that resembles the histological findings of ALD including steatosis and neutrophil infiltration. The mechanisms by which OPN regulates steatohepatitis include Hedgehog signaling pathway stimulation(14). At the cellular level, OPN is highly secreted by hepatic stellate cells (HSC) to induce cell cytokine release and collagen synthesis(15).
To investigate the role of OPN in AH, we performed a translational study using different approaches including gene expression, protein content and DNA polymorphisms analysis in patients with AH as well as experimental studies in mice lacking OPN, in precision-cut liver slices and in cultured liver cells.
Materials and Methods
Selection of Patients with Alcoholic Hepatitis and other Liver Diseases and Control Livers
Patients admitted to the Liver Unit, Hospital Clínic of Barcelona with clinical, analytical and histological features of AH from 2007 to 2010 were prospectively included in the study (n=48). Inclusion criteria were described previously(4,5,16). Patients with hepatocellular carcinoma or any other potential cause of liver disease were excluded from the study. Liver biopsy was obtained using a transjugular approach and portal pressure gradient was measured. All patients received nutritional support as well as psychological support for achieving alcohol abstinence.
Three control groups were also included. In all cases, a liver biopsy was available. First, patients with hepatitis C virus (HCV)-induced liver disease (genotype 1) (n=28). No patient had previous antiviral therapy. Second, patients with compensated cirrhosis due to HCV or past-history of alcohol abuse (in all cases patients were abstinent for at least 6 months) (n=29). Third, patients with morbid obesity who underwent a laparoscopic liver biopsy during bariatric surgery and had criteria of nonalcoholic steatohepatitis (NASH) (n=47), according to Kleiner’s criteria(17). Finally, fragments of normal liver tissue were obtained from optimal cadaveric liver donors (n=3) or resection of liver metastases (n=4) as described in detail previously(6).
All liver specimens were analyzed by an expert pathologist and a part of the biopsy was submerged into a RNA stabilization solution (RNAlater). The protocol was approved by the Ethics Committee of the Hospital Clinic and all patients gave informed consent.
Selection of Patients for Genetic Studies
Patients were included from the University Hospital of Salamanca (Spain) and Amiens University Hospital (France). Patients with compensated alcoholic liver cirrhosis (n=102), alcoholic patients without liver disease (n=196) and patients with biopsy-proven AH (n=100) were included(18). Patients with cirrhosis had either histological diagnosis or clinical, analytical and imaging signs of compensated cirrhosis(19). Alcoholic patients without ALD had no clinical evidences of liver disease, normal liver blood tests and normal liver ultrasonography. Finally, patients with AH were diagnosed following the same criteria used in the current study. All patients were negative for viral hepatitis and other known causes of chronic liver disease. In addition, sex-matched healthy volunteers (n=152) were included in the study as negative controls. All of them consumed less than 10 g of ethanol per day and none had a story of alcohol abuse or alcohol dependence and neither did their first or second-degree relatives. The study was approved by both Ethical Committees and all patients gave informed consent.
Real-Time Polymerase Chain Reaction Analysis
RNA isolation was performed on liver biopsies from patients with AH (n=44), HCV (n=23), NASH (n=31) and compensated cirrhosis (n=12) and from fragments of normal livers (n=7). Quantitative real-time PCR (qPCR) was performed as described in Supplementary Methods.
OPN serum levels analysis
Serum samples were obtained from peripheral and suprahepatic blood and stored at −80°C.OPN serum levels were measured in patients with AH (n=26), HCV (n=22), NASH (n=23) and compensated alcoholic cirrhosis (n=17) and from healthy volunteers (n=5) using the Quantikine Human Osteopontin Immunoassay Kit (R&D Systems, Minneapolis, MN).
Immunohistochemistry
Paraffin-embedded sections were incubated with the primary antibody anti-human OPN (Abcam, Cambridge, UK) following the immunohistochemistry protocol described in the Supplementary methods.
Western blotting studies
Processed OPN was analyzed in serum proteins from patients with AH (n=16), compensated alcoholic cirrhosis (n=16) and healthy volunteers (n=7) using a specific antibody to detect MMP-cleaved OPN by Western blotting (Abcam, Cat. N° ab8448). Western blotting studies were performed in human and cellular samples as described in Supplementary methods.
OPN single nucleotide polymorphisms (SNP) analysis
Genomic DNA extraction was performed in nucleated peripheral blood using standard methods and stored at −20°C. Samples were mixed on PCR plates for simultaneous analysis in a blind fashion. Four SNPs of the SPP1 gene (Pubmed references: rs28357094, rs9138, rs11730582 and rs2728127) were analyzed in the selected population. Genotyping was carried out using specific TaqMan® SNP Genotyping Assays (Applied Biosystems) and PCR reactions were read in a sequence detection system in an ABI PRISM 7000 instrument (Applied Biosystems-Life Technologies Corporation, Carlsbad, CA).
Studies in experimental alcoholic liver disease in mice
Male OPN-deficient (OPN−/−) mice were generated by Dr. Gao’s laboratory (NIH, Bethesda, MD) and ethanol was administered following the chronic-binge model as described previously(20). Briefly, OPN−/− and WT mice were divided into two groups (n=10). Ethanol groups were fed with a liquid diet containing 5% ethanol for 10 days, whereas control group received a pair-fed diet. After this period, ethanol groups were gavaged with single doses of ethanol (5g/Kg body weight, 20% ethanol), whereas control group were gavaged with isocaloric dextrin maltose. Mice were then sacrificed 9 hours after the gavage and sample collection was performed. All animal experiments were approved by the NIAAA Animal Care and Use Committee.
Precision-cut liver slices from mice and cell cultures
Two-hundred and fifty µm high-precision cut slices were obtained from freshly isolated mice livers using a vibratome instrument VT1000S (Leica Microsystems, Wetzlar, Germany). The optimized cutting procedure and the incubation conditions were followed as described previously(6). Human primary HSC were isolated from healthy fragments of liver donors following the Nycodenz gradient protocol. Additionally, three different cell lines were used in a subset of experiments, including LX-2 cells (a kind gift from Scott L. Friedman, Mount Sinai Hospital, New York, NY), a human HSC line, RAW264 cells, a mouse macrophage line and EL4, a mouse T type lymphoma line. Both liver slices and cells were led to stabilize and further treated with lipopolysaccharide (LPS), recombinant human or mouse TNFα, recombinant human or mouse transforming growth factor β (TGFβ), recombinant human OPN (rhOPN), recombinant human Interleukin 1-β (IL-1β) and recombinant human platelet-derived growth factor (PDGF), all purchased from Sigma-Aldrich, St. Louis, MO.
Statistical Analysis
Results of quantitative variables are expressed as mean ± SE unless otherwise specified. The differences between groups were analyzed using non-parametric tests (Mann-Whitney U test) for continuous variables and χ2 for categorical variables. Correlations between variables were evaluated using Spearman’s rho. Cox regression analysis was employed to identify variables associated with mortality. Survival curves by the Kaplan-Meier method were created and compared using the log-rank test. Regarding the SNP analysis, allele and genotype frequencies of patients were compared by means of χ2 test or Fisher’s exact test when necessary and a logistic regression analysis was employed to adjust for potential confounders. A Bonferroni correction for multiple testing was applied to SNP analysis, by which only those comparisons that generated a P value < 0.05/n (n= total number of comparisons) were considered significant. Statistical analysis was performed using SPSS version 14.0 for Windows (SPSS Inc., Chicago, IL).
Results
General Characteristics of Patients with AH
Forty-eight patients were included in the study with clinical, analytical and histological characteristics of AH. Overall ninety-day mortality was 20%. Seventy-nine percent (n=38) of patients had severe AH at admission, as defined as an ABIC score >6.71(16). Patients were predominantly male (63%) and the mean age was 49 years. The majority of patients had severe sinusoidal portal hypertension (mean HVPG: 19.5±0.8 mmHg). The main epidemiological, clinical and analytical characteristics of patients are shown in Table 1.
Table 1.
Clinical, Analytical and Hepatic Hemodynamic Characteristics of Patients with Alcoholic Hepatitis (AH) (n=48).
| Variables | Mean ± SE or % |
|---|---|
| Age (years) | 49 ± 1.12 |
| Male (%) | 63 |
| BMI (kg/m2) | 25.69 ± 1.04 |
| Alcohol intake (gr/d) | 107.29 ± 5.17 |
| 90-day mortality (%) | 20 |
| Analytical and hemodynamic parameters | |
| Glucose (mg/dl) | 109 ± 5 |
| Creatinine (mg/dl) | 0.83 ± 0.06 |
| AST (U/L) | 157 ± 12 |
| ALT (U/L) | 61 ± 5 |
| GGT (U/L) | 588 ± 81 |
| Bilirubin (mg/dl) | 12.7 ± 1.27 |
| Albumin (mg/dl) | 26.2 ± 0.58 |
| Platelet count (x109/L) | 13.2 ± 1.36 |
| Leucocyte count (x109/L) | 9.9 ± 0.78 |
| INR | 1.87 ± 0.11 |
| HVPG (mmHg) | 19.49 ± 0.84 |
| Cirrhosis (%) | 62 |
| Scoring systems | |
| Maddrey’s DF | 60.48 ± 6.27 |
| MELD score | 22 ± 0.9 |
| ABIC score | 7.63 ± 0.19 |
The mean length of the biopsy specimens was 6.0±0.6 millimeters, the mean number of fragments was 5.0±0.4 and the mean number of portal spaces was 6±1. The majority of patients had moderate or marked steatosis (69%) of diffuse distribution, marked hepatocyte ballooning (61%), marked necroinflammation (37%) and cirrhosis (62%).
OPN is Overexpressed in Patients with AH
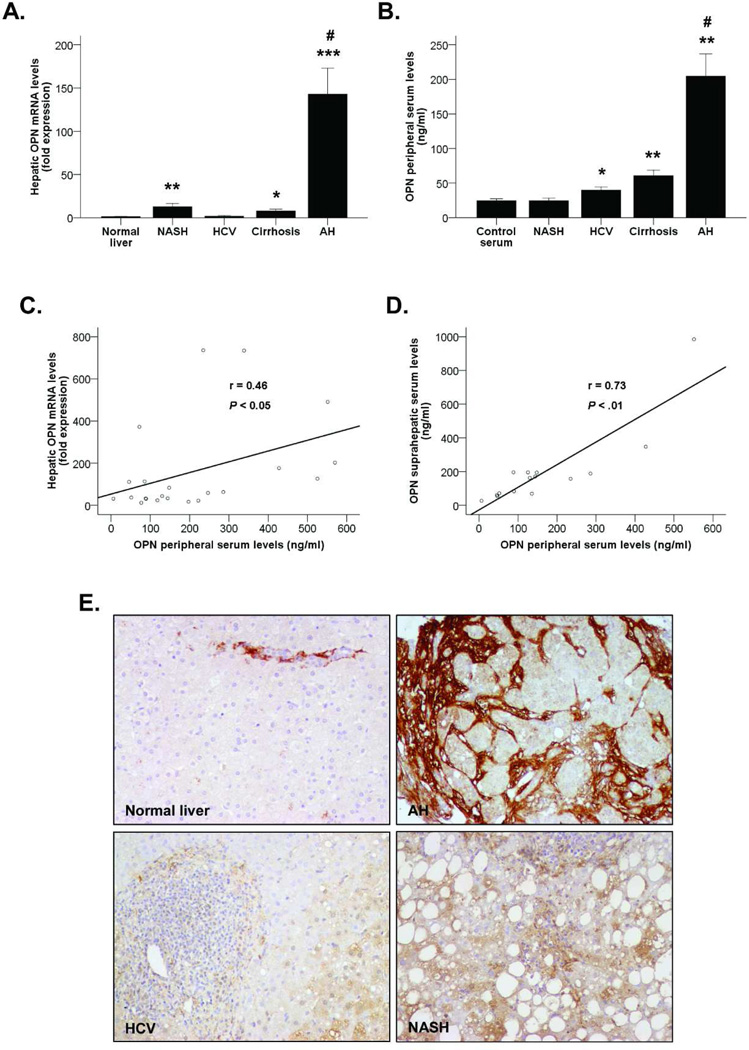
We previously detected by microarray analysis that OPN was extremely up-regulated in AH(6). Confirmatory gene expression analysis revealed that OPN was almost 150-times overexpressed in patients with AH compared to normal livers (P<0.001, Figure 1A). Among other types of liver diseases, patients with NASH -but not with HCV- had a moderate increase in hepatic OPN expression (P<0.01). Importantly, OPN was up-regulated in liver with compensated cirrhosis (P<0.05), but at a lesser extent than in patients with AH (P<0.001).
Figure 1.
(A) OPN hepatic gene expression in patients with AH (n=44), NASH (n=31), HCV (n=23) and compensated cirrhosis (n=12) compared to normal livers (n=7) (*P<0.05, **P<0.01, ***P<0.001 vs. normal livers, #P<0.001 vs. other groups). (B) OPN levels in peripheral serum of patients with AH (N=26), NASH (n=23), HCV (n=22) and compensated cirrhosis (n=17) and healthy controls (n=5) (*P<0.01, **P<0.001 vs. controls, #P<0.001 vs. other groups). (C) Correlation between OPN hepatic gene expression and OPN levels in peripheral serum of patients with AH (P<0.05). (D) Correlation between OPN levels in suprahepatic serum and OPN levels in peripheral serum of patients with AH (P<0.01). (E) Immunohistochemistry of paraffin sections from liver biopsies of patients with AH, HCV and NASH and fragments of normal livers stained with the anti-OPN antibody (200X magnification).
Next, we found that OPN serum levels were much higher in patients with AH compared to patients with other liver diseases and healthy controls (P<0.001). Patients with both HCV and compensated cirrhosis also had increased OPN levels compared to healthy controls (P<0.01 and P<0.001, respectively, Figure 1B). The gender of the patients did not influence either the hepatic expression or the serum levels of OPN. Importantly, OPN hepatic mRNA and serum levels correlated in patients with AH (P<0.05, Figure 1C). Furthermore, suprahepatic serum levels paralleled also with OPN measured in peripheral veins (P<0.01) (Figure 1D). Taken together, these results indicate that OPN is markedly overexpressed in AH and could be useful as a disease biomarker. Moreover, the damaged liver seems to be a source of circulating OPN in patients with AH.
Finally, OPN protein expression and deposition in liver tissue was assessed by immunohistochemistry. The results revealed a prominent OPN staining mainly focused in fibrotic areas of livers of patients with AH, while it was barely detected in normal livers and in patients with NASH and HCV (Figure 1E). These findings strongly support the hypothesis that OPN is overexpressed in patients with AH, both at the RNA and protein levels.
OPN Hepatic Expression and Serum Levels Correlate with Disease Severity in Patients with AH
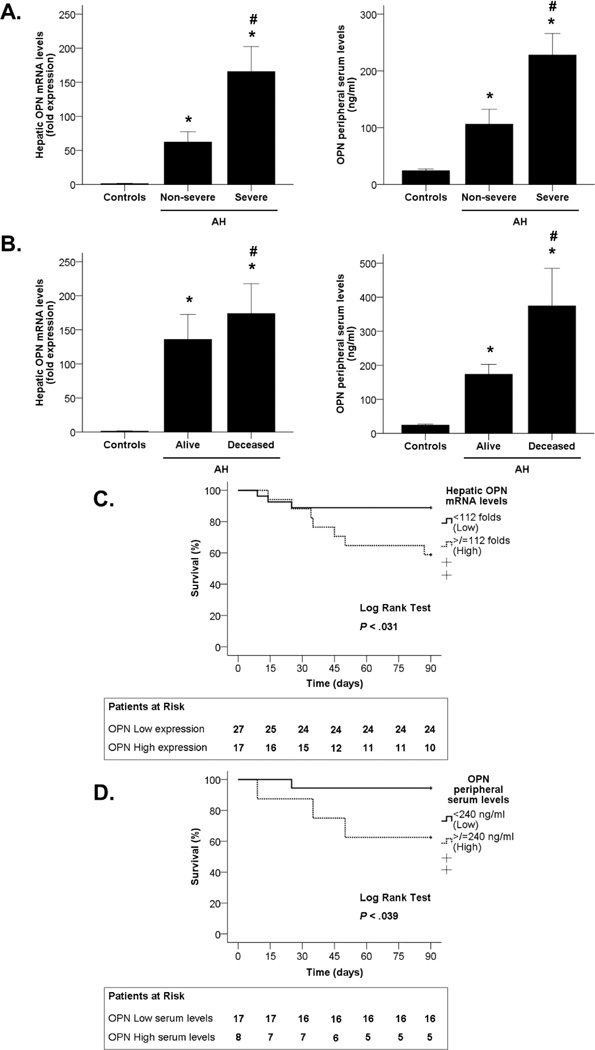
We next explored whether OPN gene and serum levels correlate with disease severity, which was assessed using the ABIC score(16). Patients were stratified into non-severe AH (ABIC <6.71; n=10) and severe AH (ABIC >6.71; n=38). OPN hepatic gene expression and serum levels were significantly higher in patients with severe AH than in those with non-severe AH (P<0.05 in both, Figure 2A).
Figure 2.
(A) Association between OPN hepatic gene expression and OPN levels in peripheral serum with disease severity (measured by the ABIC score) in patients with AH and control groups (*P<0.05). (B) Association between OPN hepatic gene expression and OPN levels in peripheral serum with 90-day mortality in patients with AH and control groups (*P<0.05). (C) Kaplan-Meier curve showing 90-day mortality according to OPN hepatic gene expression in patients with AH (P<0.05). (D) Kaplan-Meier curve showing 90-day mortality according to OPN levels in peripheral serum in patients with AH ( P <0.05).
Furthermore, short-term mortality was assessed at 90 days. OPN mRNA and serum levels were higher in patients who did not survive (P<0.05 in both, Figure 2B). Additionally, a Kaplan-Meier curve approach was used for determining if both hepatic OPN expression and OPN serum levels are good parameters for the prediction of patient’s short-term mortality. Receiver-operating curve (ROC) analysis was used to identify a cut-off value of 112-fold expression to define patients with low and high hepatic OPN gene expression and a cut-off value of 240 ng/ml to define patients that had low or high circulating OPN levels (P<0.05 in both, Supplementary Figure 1). The Kaplan-Meier curves showed that both parameters predict mortality in patients with AH (P<0.05), as the group of patients that had increased OPN hepatic expression and serum levels also had increased number of deaths during the 90-day period (Figures 2C and 2D). These results strongly indicate that OPN hepatic expression predicts disease severity in patients with AH.
We finally investigated whether OPN expression correlates with the expression of other neutrophil-attractive chemokines involved in the pathogenesis of AH, such as the CXC family(4,5). We found that OPN gene expression correlated with hepatic gene expression of different CXC chemokines (P<0.05, P<0.001), specially interleukin 8 (Supplementary Figure 2).
OPN Post-Transcriptional Modifications in AH
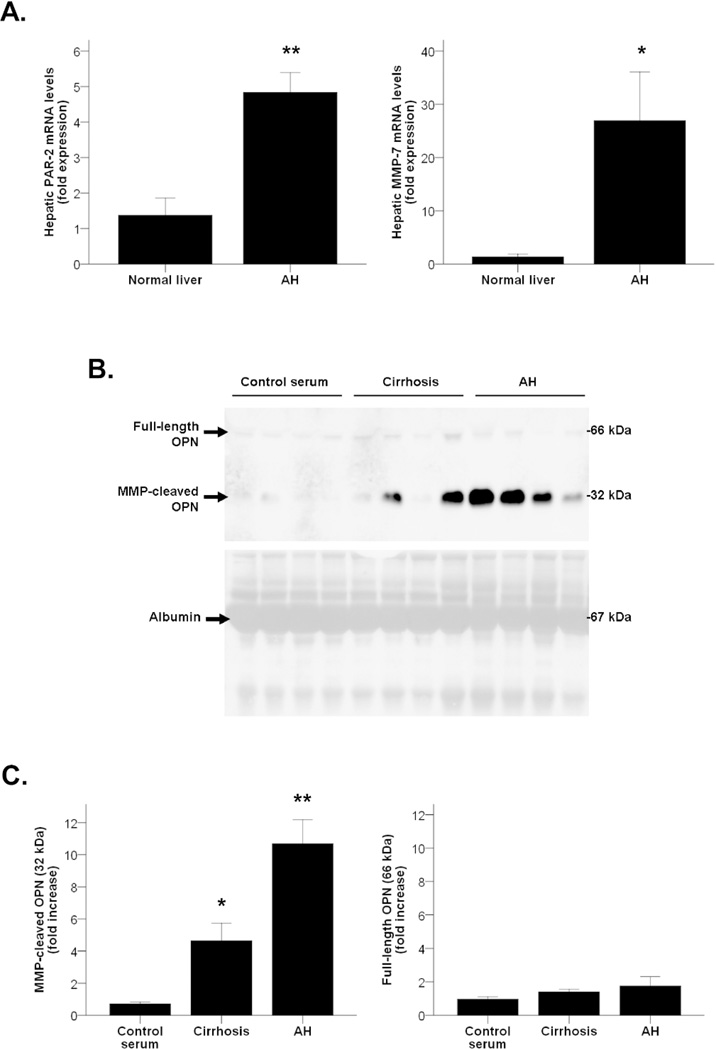
As thrombin (PAR-2) and MMP-7 are known to process OPN and amplify its pathogenic activity(9), we then explored whether the resulting cleaved-OPN is increased in AH. We first found by qPCR analysis that both thrombin and MMP-7 were overexpressed in patients with AH compared to normal livers (P<0.001 and P<0.05, respectively, Figure 3A). Furthermore, because the amount of proteins obtained in a transjugular biopsy is highly limited and Western blotting analysis could not be performed, the analysis performed on serum proteins revealed that patients with AH had increased levels of MMP-cleaved fragments of OPN -but not full-length fragments- in serum, compared to healthy controls (P<0.001). Furthermore, patients with compensated alcoholic cirrhosis had more cleaved OPN than healthy controls in serum (P<0.05), but at a lesser extent than patients with AH (Figures 3B and 3C). Interestingly, these results suggest that the 10-fold increase of OPN serum levels found in patients with AH compared with healthy controls as measured by ELISA (Figure 1B) is mainly due to the increased content of the MMP-cleaved form of OPN.
Figure 3.
(A) PAR-2 (thrombin) and MMP-7 hepatic gene expression in patients with AH compared to normal livers (*P<0.05, **P<0.001). (B) Representative Western blot of peripheral serum samples of patients with AH, cirrhosis and control serums (n=4) using the anti-OPN antibody and representative Ponceau staining of the whole membrane showing serum albumin. Ponceau staining serves to prove an equal sample loading to the membrane. (C) Quantification of full-length and MMP-cleaved OPN isoforms in peripheral serum of patients with AH, compensated cirrhosis and healthy controls (*P<0.05, **P<0.001).
OPN Single Gene Polymorphisms and AH
To gain more insight in the potential functional role of OPN, we next investigated whether OPN gene variations predispose patients with alcohol abuse to develop AH. A SNP analysis of the OPN gene was performed in a cohort of alcoholic patients with different degrees of ALD, including patients with alcohol abuse without liver disease (n=196), patients with compensated alcoholic liver cirrhosis (n=102) and patients with AH (n=100). The main characteristics of the patients with AH are shown in Supplementary Table 1. Furthermore, a group of healthy controls (n=152) were added in the study. Four SNPs were analyzed, including three within the promoter region (−1748 A/G, −443 T/C and −66 T/G) and one within the 3’ UTR (+1239 A/C). All these SNPs were associated with pathological features in different diseases(21–23).
The results of the SNP analysis are shown in Table 2. Patients with compensated cirrhosis did not show changes in the genotype frequencies of the four SNP analyzed compared with alcoholics without liver disease and healthy controls. Patients with AH did not show different genotype frequencies of the −443 T/C and −66 T/G SNPs but had a lower frequency of the CC genotype of the 3’ UTR +1239 A/C SNP compared with alcoholics without liver disease and healthy controls (P<0.05). This result, however, was non-significant when multiple correction testing was aplied for the 16 comparisons performed in Table 2 (alfa = 0,003). Importantly, although there were no differences in the frequencies of the −1748 A/G SNP among alcoholics with or without liver disease, we found that the presence of the AA genotype in patients with AH was associated with increased mortality at 1, 3 and 6 months. Specifically, patients with AH and the AA genotype of this SNP had a 75% mortality at 6 months whereas patients with AH and AG or GG genotype had a 50% mortality (P<0.05). Importantly, the presence of AA genotype remained an independent predictor factor of 6-month mortality after adjustment by age, sex and MELD score (P = 0.046, OR=2.65 (1.02–6.9)). This result may suggest that the presence of genetic variances in the OPN gene could modulate the prognosis of patients with ALD.
Table 2.
Genotyping frequencies of OPN gene polymorphisms.
| SNP ID | Genotype | Cirrhosis vs. AWLD | Cirrhosis vs. controls | AH vs. AWLD | AH vs. controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cirrhosis | AWLD | P | Cirrhosis | Controls | P | AH | AWLD | P | AH | Controls | P | ||
| rs2728127−1748 A/G | AA | 47 (46.1) | 95 (48.7) | 47 (46.1) | 72 (47.7) | 46 (50.0) | 95 (48.7) | 46 (50.0) | 72 (47.7) | ||||
| AG | 48 (47.1) | 81 (41.5) | 0.548 | 48 (47.1) | 61 (40.4) | 0.327 | 40 (43.5) | 81 (41.5) | 0.633 | 40 (43.5) | 61 (40.4) | 0.390 | |
| GG | 7 (6.9) | 19 (9.7) | 7 (6.9) | 18 (11.9) | 6 (6.5) | 19 (9.7) | 6 (6.5) | 18 (11.9) | |||||
| rs11730582−443 T/C | TT | 26 (25.5) | 56 (28.6) | 26 (25.5) | 32 (21.1) | 21 (21.6) | 56 (28.6) | 21 (21.6) | 32 (21.1) | ||||
| TC | 53 (52.0) | 84 (42.9) | 0.307 | 53 (52.0) | 83 (54.6) | 0.709 | 49 (50.5) | 84 (42.9) | 0.362 | 49 (50.5) | 83 (54.6) | 0.787 | |
| CC | 23 (22.5) | 56 (28.6) | 23 (22.5) | 37 (24.3) | 27 (27.8) | 56 (28.6) | 27 (27.8) | 37 (24.3) | |||||
| rs28357094−66 T/G | TT | 57 (55.9) | 117(60.3) | 57 (55.9) | 89 (58.6) | 62 (62.0) | 117(60.3) | 62 (62.0) | 89 (58.6) | ||||
| TG | 42 (41.2) | 68 (35.1) | 0.503 | 42 (41.2) | 55 (36.2) | 0.540 | 31 (31.0) | 68 (35.1) | 0.598 | 31 (31.0) | 55 (36.2) | 0.598 | |
| GG | 3 (2.9) | 9 (4.6) | 3 (2.9) | 8 (5.3) | 7 (7.0) | 9 (4.6) | 7 (7.0) | 8 (5.3) | |||||
| rs9138+1239 A/C | AA | 50 (49.0) | 83 (42.8) | 50 (49.0) | 79 (53.0) | 23 (44.2) | 83 (42.8) | 23 (44.2) | 79 (53.0) | ||||
| AC | 45 (44.1) | 91 (46.9) | 0.458 | 45 (44.1) | 55 (36.9) | 0.430 | 29 (55.8) | 91 (46.9) | 0.049 | 29 (55.8) | 55 (36.9) | 0.011 | |
| CC | 7 (6.9) | 20 (10.3) | 7 (6.9) | 15 (10.1) | 0 (0) | 20 (10.3) | 0 (0) | 15 (10.1) | |||||
Data are presented as absolute frequencies (%). Genotyped patients for each SNP do not equal the total number of subjects listed in each category since some SNPs could not be genotyped for technical reasons in some patients. AWLD: alcoholic patients without liver disease. AH: alcoholic hepatitis.
Attenuated Susceptibility to Alcohol-induced Liver Injury in Mice Lacking OPN
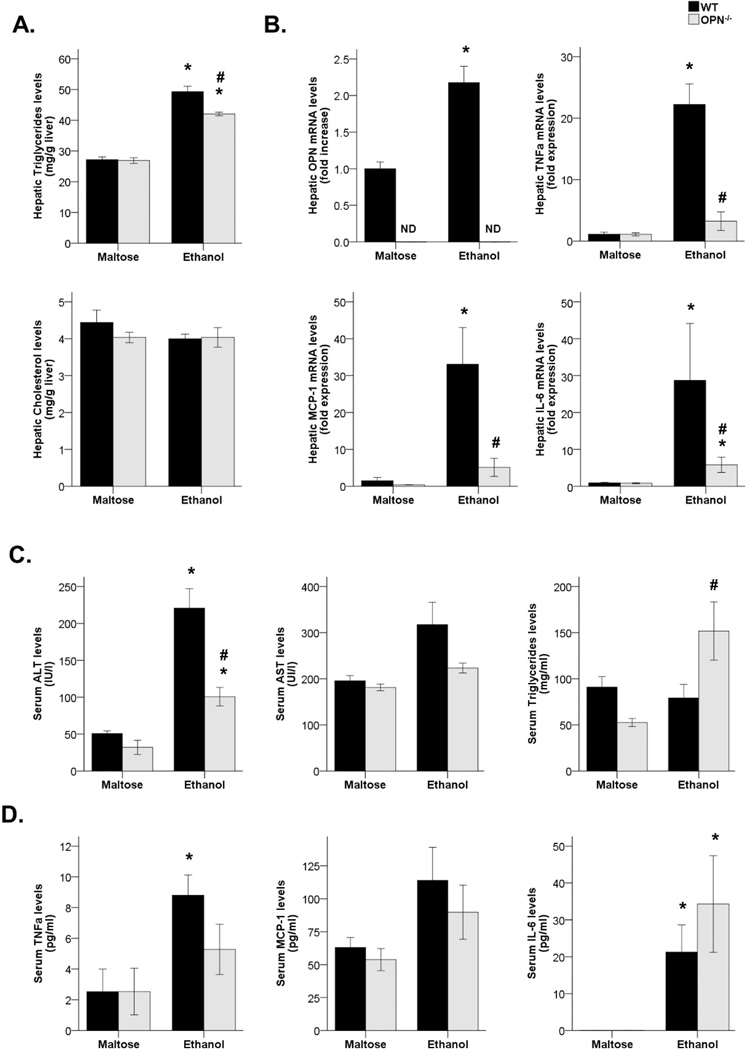
We next explored whether OPN plays a pathogenic role in an experimental model of ALD. WT and OPN−/− maltose-fed groups did not show differences on both hepatic and serum parameters. In contrast, WT ethanol-fed mice had higher hepatic triglyceride levels compared with WT-fed mice (P<0.05), but they were reduced in OPN−/− ethanol-fed mice (P<0.05). Hepatic cholesterol levels did not change between groups (Figure 4A). OPN hepatic gene expression was increased after ethanol feeding in WT mice (P<0.05, Figure 4B). Furthermore, liver inflammation was quantified by analyzing hepatic gene expression of pro-inflammatory cytokines such as TNFα, MCP-1 and IL-6. All three genes were highly increased in WT ethanol-fed mice compared with their maltose-fed group (P<0.05), whereas their expression was almost suppressed on OPN−/− ethanol-fed group (P<0.05, Figure 4B).
Figure 4.
(A) Triglyceride and cholesterol hepatic content in WT and OPN-deficient mice fed with maltose or ethanol diet (*P<0.05 vs. WT maltose-fed mice, #P<0.05 vs. WT ethanol-fed mice). (B) OPN, TNFα, MCP-1 and IL-6 hepatic gene expression in WT and OPN-deficient mice fed with maltose or ethanol diet (*P<0.05 vs. WT maltose-fed mice, #P<0.05 vs. WT ethanol-fed mice; ND, Non-detected). (C) Serum ALT, AST and triglyceride levels of WT and OPN-deficient mice fed with maltose or ethanol diet (*P<0.01 vs. WT maltose-fed mice, #P<0.05 vs. WT ethanol-fed mice). (D) Serum TNFα, MCP-1 and IL-6 levels of WT and OPN-deficient mice fed with maltose or ethanol diet (*P<0.05 vs. WT maltose-fed mice).
Regarding serum parameters, ALT serum levels were highly increased in WT ethanol-fed mice compared with their maltose-fed group (P<0.01), but were reduced in OPN−/− ethanol-fed mice (P<0.01). However, no differences were observed in serum AST, and serum triglycerides were increased in OPN−/− ethanol-fed mice (P<0.05, Figure 4C). In addition, no differences were noted in the cytokine serum profile between the WT and the OPN−/− ethanol-fed groups (Figure 4D).
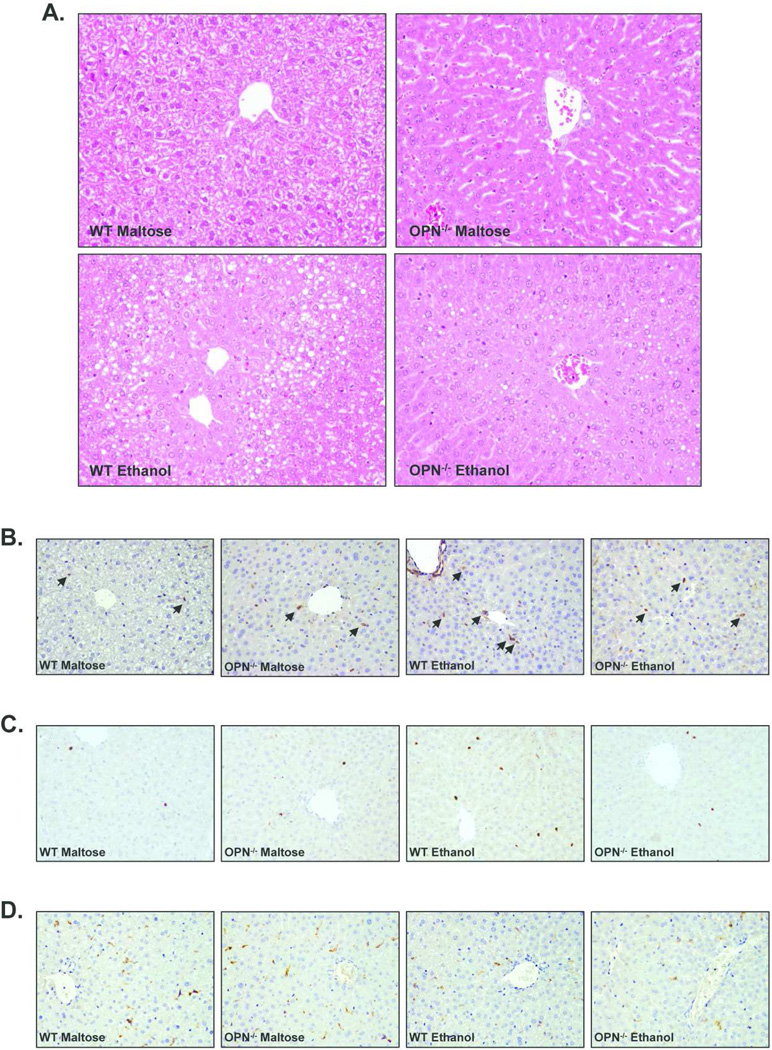
Hematoxylin and eosin staining of paraffin liver sections showed that ethanol administration increased the degree of microsteatosis and hepatocellular injury in WT mice. However, OPN−/− ethanol-fed mice had reduced steatosis and the hepatic parenchyma was more conserved than the WT ethanol-fed mice (Figure 5A). Immunohistochemical studies performed for the assessment of hepatic inflammation, injury and immune cell infiltration revealed that ethanol administration increased the number of p65-positive cells in WT and OPN−/− mice compared to the maltose-fed groups, but was slightly lower in OPN−/− mice (Figure 5B). Furthermore, WT ethanol-fed mice also had an increased number of infiltrating neutrophils as showed by myeloperoxidase (MPO) staining, that was partially attenuated in OPN−/− ethanol-fed mice (Figure 5C). However, macrophage infiltration tested by F4/80 staining remained unaltered after ethanol administration in both WT and in OPN−/− mice (Figure 5D).
Figure 5.
(A) Hematoxylin and eosin staining of paraffin liver sections from WT and OPN-deficient (OPN−/−) mice (200X magnification). (B, C and D) Immunohistrochemistry studies of paraffin liver sections from WT and OPN-deficient (OPN−/−) mice stained with anti-p65 antibody (B), with anti-MPO antibody (C) and with anti-F4/80 antibody (D) (200X magnification). In some cases, positive cells are marked with arrows.
Taken together, these results suggest that OPN mediates, at least in part, the inflammatory response to ethanol, as showed by decreased hepatic expression of pro-inflammatory cytokines, decreased NFkB activation of inflammatory cells and decreased neutrophil infiltration in OPN−/− ethanol-fed mice. As a result, mice deficient in OPN gene are partially, but not completely, protected against alcohol-induced liver injury.
Mechanisms Underlying Increased OPN Synthesis and Pathogenic Effects
We next analyzed the production of OPN in different in vitro approaches including primary human HSC, in mouse precision-cut liver slices and in RAW264 and EL4 immune cell lines. Among different mediators of liver injury, we focused on LPS, a pathogenic factor in ALD (24) known to regulate OPN production and secretion (25,26).
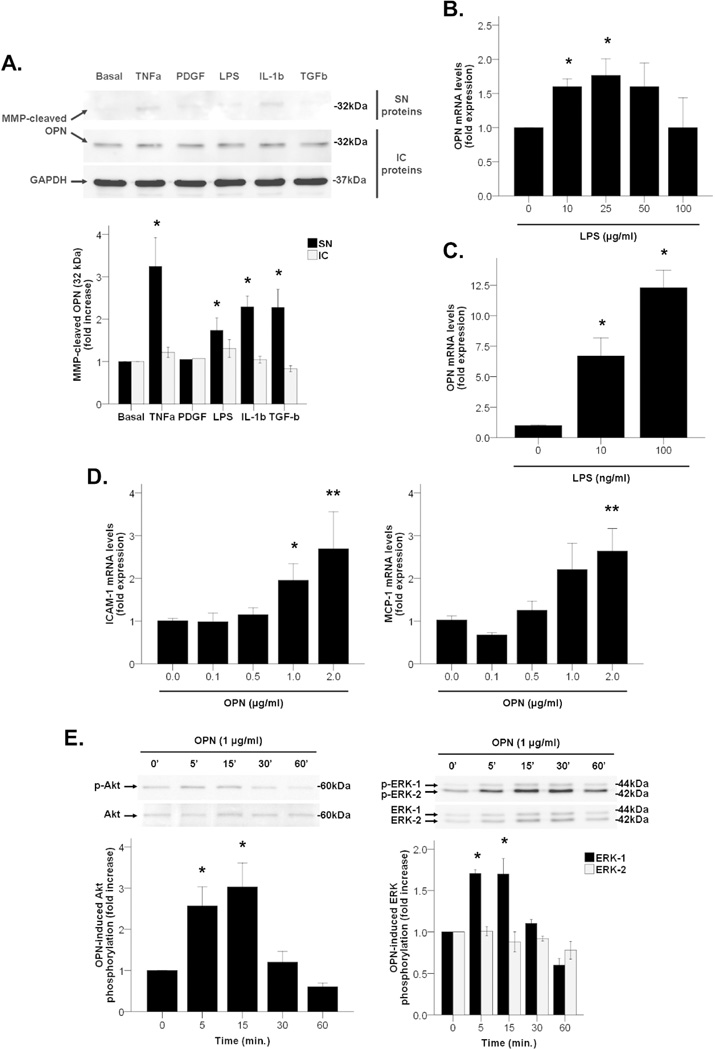
Western blotting studies showed that MMP-cleaved fragments of OPN were detected both intracellularly and extracellularly in activated primary HSC (Figure 6A, Western blot panel). OPN was highly produced under basal (untreated) conditions and intracellular synthesis remained unaltered after incubation with LPS or other key inflammatory mediators of the pathogenesis of AH including TNFα, PDGF, IL-1β and TGFβ. However, there was a marked increase on the secretion of MMP-cleaved fragments of OPN in supernatants of HSC treated with TNFα, LPS, IL-1β and TGFβ (P<0.05, Figure 6A). These results suggest that activated HSC could be a source of processed OPN after liver injury.
Figure 6.
(A) OPN protein expression and secretion to cell supernatant in HSC incubated with mediators of liver injury up to 48 hours (*P<0.05 vs. basal). (B) OPN gene expression in mouse liver slices incubated with LPS up to 48 hours (*P<0.05 vs. LPS 0 µg/ml). (C) OPN gene expression in RAW264 cells incubated with LPS up to 24 hours (*P<0.05 vs. LPS 0 ng/ml). (D) MCP-1 and ICAM-1 gene expression in HSC incubated with recombinant OPN up to 24 hours (*P<0.05 vs. OPN 0 µg/ml, **P<0.01 vs. rhOPN 0 µg/ml). (E) Activation of intracellular signaling pathways in HSC incubated with recombinant OPN up to different short times (*P<0.05 vs. time 0 min.).
Furthermore, mouse precision cut-liver slices incubated with LPS had increased gene expression of OPN at 48 hours (P<0.05, Figure 6B), but not after incubation with TNFα and TGFβ (data not shown). Similarly, LPS increased OPN gene expression in RAW264 cells -a macrophage cell line- in a dose-dependent manner (P<0.05, Figure 6C), but not in EL4 cells -a T cell line- (data not shown). These results suggest that in addition to activated HSC, the hepatic parenchyma and the resident and infiltrating macrophages could be also a source of OPN in a damaged liver.
We next explored the biological effects of recombinant OPN in primary HSC and LX-2 cells. In HSC, OPN induced gene expression of cell adhesion molecules such as ICAM-1 and pro-inflammatory cytokines such as MCP-1 in a dose-dependent manner (P<0.05, Figure 6D). In addition, OPN induced Akt and ERK-1, but not ERK-2, phosphorylation in LX-2 cells at short incubation times (P<0.05, Figure 6E), whereas neither JNK nor p38 signaling pathways were induced (data not shown). Taken together, these results suggest that OPN may participate in HSC activation inducing pro-inflammatory effects.
Discussion
The current study provides evidence that OPN is overexpressed in AH and may play a pathogenic role in this severe clinical condition. This assumption is based on two main results: first, OPN hepatic gene expression paralleled with disease severity in patients with well-characterized AH and second, mice lacking OPN are resistant to develop ethanol-induced liver damage. Our results are in accordance with previous experimental studies showing that OPN plays a role in liver injury and fibrogenesis in different experimental models of acute and chronic liver damage (13–15). Moreover, our results confirm a recent study showing that OPN is overexpressed in NAFLD and could play a pathogenic role (14). Because NAFLD share many histological features with AH including fat accumulation, PMN infiltration and hepatocellular damage, it is conceivable that OPN is a major pathogenic factor in steatohepatitis, regardless of the causative agent. It is important to stress, however, that AH represents the most severe form of ALD. Whether OPN is also overexpressed in mild forms of ALD is unknown and it deserves further investigation.
The most relevant result of this study is that OPN is markedly overexpressed in patients with AH and that the extent of hepatic expression correlates with disease survival. In fact, we recently identified OPN as the second most up-regulated gene in a recent study analyzing the whole transcriptome in patients with AH(6). The functional analysis of this microarray study revealed that ECM-integrin receptor interaction is differentially regulated in patients with AH, reinforcing a potential pathogenic role for OPN. Histological studies revealed that OPN was mainly expressed in areas with myofibroblast accumulation and active fibrogenesis. However, this study did not specifically address the exact cell source of OPN in ALD, so further studies are needed for assessing this question. In fact, OPN could be produced by other cell types in the liver including hepatocytes, biliary cells or leucocytes. Because fibrogenic mediators such as thrombin and MMP-7 are capable to cleave OPN to yield more active peptides(9), we also studied whether their expression parallel OPN. We provide evidence that both proteases are markedly overexpressed in AH and that cleaved OPN fragments are increased in these patients. This finding could be relevant, since preventing OPN cleavage and/or interfering cleaved fragments of OPN represent potential sites for therapeutic intervention. Future experimental studies should assess this hypothesis.
Another relevant result of our study is that OPN serum levels are particularly elevated in AH. It is well known that OPN can serve as a serum marker of advanced cirrhosis since its serum levels correlated with the degree of fibrosis in different human diseases such a chronic hepatitis C and NAFLD (14,15). Our study confirmed this hypothesis, since OPN serum levels were found increased in compensated cirrhosis compared to controls. Interestingly, OPN levels were much more elevated in patients with AH, suggesting that the accelerated fibrosis and inflammation in these patients results in an increased release of OPN. This hypothesis is supported by the finding that serum and suprahepatic OPN levels correlate with hepatic gene expression. Further studies should be performed to explore whether serum OPN, together with other proteins, can be used a serum markers of AH. This is a clinically relevant issue since many centers do not perform transjugular biopsies and the development of serum markers of AH would be of great clinical interest.
Because increased OPN hepatic expression could be a consequence rather that a mediator in AH, two different functional studies were employed. First, we challenged mice lacking OPN to ethanol-induced liver injury. OPN-deficient mice developed less severe liver injury and pro-inflammatory gene expression than wild-type mice. These results indicate that OPN is no merely a structural ECM protein produced by myofibroblasts that passively distorts the liver architecture, but that it actually participates in the inflammatory response to injury. Our in vitro studies showing that OPN induces pro-inflammatory effects in primary HSC support this assumption. However, we found that the reduced hepatic expression of pro-inflammatory cytokines and the hepatic content of triglycerides found in OPN-deficient mice after ethanol feeding did not correlate with their increased serum levels. There are some possible explanations to address this question. First, alcohol do not only induces liver inflammation but also a systemic inflammatory response and inflammatory changes in other organs such as the heart, the kidney and the nervous system. In addition, it also increases fatty acid mobilization from extrahepatic sources including visceral adipose tissue. Therefore, circulating levels of cytokines do not necessarily correlate with intrahepatic expression. We previously demonstrated that intrahepatic expression of TNF, a key circulating cytokine in alcohol-induced organ damage, was not increased in the livers from patients with AH (4). Moreover, inflammatory cytokines frequently act in an autocrine and paracrine manner and locally regulate parenchymal function and the wound healing response to injury. In this line, previous studies from our lab demonstrated that intrahepatic, rather than systemic expression of cytokines, predict disease outcome in patients with AH (4–6). Our second functional approach was to explore whether gene variations of OPN modulate the response to alcohol abuse and predisposes patients to develop severe forms of AH. We found that patients with AH had a lower frequency of the CC genotype of the 3’ UTR +1239 A/C SNP compared with alcoholics without liver disease. However, this result did not reach statistical significance after correction for multiple testing due to the relative small sample size. Larger studies in the setting of international networks are needed to elucidate whether OPN gene variations influence in the individual susceptibility to develop an episode of AH among heavy drinkers. Interestingly, the presence of the AA genotype of the −1748 A/G SNP in patients with AH was associated with increased mortality at 1, 3 and 6 months. The presence of AA genotype remained a predictor factor of 6-month mortality after adjustment by age, sex and the MELD score. Although this result suggests that the genetic variations of OPN could influence the clinical severity of AH, it should be considered exploratory and larger epidemiological studies have to be performed to confirm this finding. Also, mutagenic analysis should determine the influence of these SNPs in the expression of OPN by liver cells, its affinity to its receptors and its susceptibility to undergo post-transcriptional regulation.
Finally, we explored in cultured HSC and immune cells and in precision-cut liver slices the potential mechanisms implicated in the increased OPN expression in livers with AH. Two results were particularly relevant. First, TNFα, a known mediator of ALD, did not induce any expression of OPN. In contrast, LPS stimulated dose-dependently OPN expression. These results suggest that OPN could mediate, at least in part, the inflammatory and fibrogenic effects of increased LPS in the liver. Future experimental studies should test this hypothesis. In addition, we found that HSC and infiltrating inflammatory cells could be an important source of OPN in a damaged liver. This assumption is reinforced by a previous study in our lab that compared gene expression profiles of freshly-isolated HSC from cirrhotic and normal livers by microarray analysis. This study showed that OPN was 16-fold up-regulated in HSC activated in cirrhotic livers compared to normal livers (27).
In summary, our study demonstrated that OPN is markedly overexpressed in patients with AH, especially in those with more severe disease, and that OPN may play a role in experimental ethanol-induced liver injury in mice. Collectively, these results suggest that OPN may be an appealing target for therapy in these patients. There are, however, several issues that should be addressed in future studies. First, the precise cellular source of OPN in AH is unknown. Second, it is unclear whether OPN also pays a role in hepatic fibrogenesis in patients with AH. The results of two recent studies suggest that hepatic expression of OPN correlates with the degree of hepatic fibrosis in patients with alcoholic and non-alcoholic steatohepatitis (28,29). Also, OPN is expressed in areas of myofibrobalsts accumulation and exerts fibrogenic properties in cultured HSC (14,15). Although we did not find a direct correlation between the degree of fibrosis and the expression of OPN, these recent studies strongly suggest that OPN could play a fibrogenic role in patients with steatohepatitis, regardless of its cause. And most importantly, future preclinical studies should explore whether targeting OPN is safe and effective as a therapeutic option in patients with AH.
Supplementary Material
Acknowledgments
The authors thank Cristina Millán, Montserrat Moreno and Elena Juez for their excellent technical support, the personnel of the Liver Hemodynamic Unit in obtaining the liver samples, and Rosa Miquel from the Pathology Unit for the histological analysis of the liver samples. This work was performed in Centre Esther Koplowitz (CEK).
Grant support: This work was supported by grants from the Instituto de Salud Carlos III (FIS PI080237 and FIS PS09/01164 to Dr. Bataller and Dr. Caballería, respectively). S. Affò received a grant from IDIBAPS. Dr. Sancho-Bru received grants from Instituto Carlos III, Miguel Servet (CP11/00071) and from European Commission within its FP7 Cooperation Programme and the European Cosmetics Association (COLIPA) HeMiBio - HEALTH-F5-2010-266777. J. Altamirano received a grant from Fundación Banco Bilbao Vizcaya Argentaria.
Abbreviations
- ALD
Alcoholic liver disease
- ALT
alanine aminotransferase
- AH
alcoholic hepatitis
- AST
aspartate aminotransferase
- AUROC
area under the receiver operating curve
- ECM
extracellular matrix
- ERK
extracellular signal-regulated kinase
- GGT
gamma-glutamyl transpeptidase
- HCV
hepatitis C virus
- HSC
hepatic stellate cell
- ICAM-1
intercellular cell adhesion molecule 1
- IL-6
interleukin 6
- IL-1β
interleukin 1β
- INR
international normalized ratio
- JNK
c-Jun N-terminal kinase
- LPS
lipopolysaccharide
- MCP-1
monocyte chemotactic protein 1
- MMP
matrix metalloproteinase
- MPO
myeloperoxidase
- NASH
nonalcoholic steatohepatitis
- OPN
osteopontin
- PDGF
platelet-derived growth factor
- SNP
single nucleotide polymorphism
- SPP1
secreted phosphoprotein 1
- TGFβ
transforming growth factor β
- TNFα
tumor necrosis factor α
Footnotes
Conflict of interest statement: We declare that we have no conflict of interest.
Contributor Information
Oriol Morales-Ibanez, Email: oriol.mibanez@gmail.com.
Marlene Domínguez, Email: dominguezgarcia1@hotmail.com.
Sung H. Ki, Email: shki@chosun.ac.kr.
Miguel Marcos, Email: migmarmar10@hotmail.com.
Javier F. Chaves, Email: felipe.chaves@uv.es.
Eric Nguyen-Khac, Email: nguyen-khac.eric@chu-amiens.fr.
Hakim Houchi, Email: hakim.houchi@upicardie.fr.
Silvia Affò, Email: silviaaffo@gmail.com.
Pau Sancho-Bru, Email: psancho@clinic.ub.es.
José Altamirano, Email: jtaltami@clinic.ub.es.
Javier Michelena, Email: jmichele@clinic.ub.es.
Juan Carlos García-Pagán, Email: jcgarcia@clinic.ub.es.
Juan G. Abraldes, Email: jgon@clinic.ub.es.
Vicente Arroyo, Email: varroyo@clinic.ub.es.
Juan Caballería, Email: caballer@clinic.ub.es.
Francisco-Javier Laso, Email: laso@usal.es.
Bin Gao, Email: bgao@mail.nih.gov.
Ramón Bataller, Email: ramon_bataller@med.unc.edu.
Reference List
- 1.Altamirano J, Bataller R. Alcoholic liver disease: pathogenesis and new targets for therapy. Nat Rev Gastroneterol Hepatol. 2011;8:491–501. doi: 10.1038/nrgastro.2011.134. [DOI] [PubMed] [Google Scholar]
- 2.Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141:1572–1585. doi: 10.1053/j.gastro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boetticher NC, Peine CJ, Kwo P, Abrams GA, Patel T, Aqel B, Boardman L, et al. A randomized, double-blinded, placebo-controlled multicenter trial of etanercept in the treatment of alcoholic hepatitis. Gastroenterology. 2008;135:1953–1960. doi: 10.1053/j.gastro.2008.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colmenero J, Bataller R, Sancho-Bru P, Bellot P, Miquel R, Moreno M, Jares P, et al. Hepatic expression of candidate genes in patients with alcoholic hepatitis: correlation with disease severity. Gastroenterology. 2007;132:687–697. doi: 10.1053/j.gastro.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 5.Dominguez M, Miquel R, Colmenero J, Moreno M, García-Pagán JC, Bosch J, Arroyo V, et al. Hepatic expression of CXC chemokines predicts portal hypertension and survival in patients with alcoholic hepatitis. Gastroenterology. 2009;136:1639–1650. doi: 10.1053/j.gastro.2009.01.056. [DOI] [PubMed] [Google Scholar]
- 6.Affò S, Dominguez M, Lozano JJ, Sancho-Bru P, Rodrigo-Torres D, Morales-Ibanez O, Moreno M, et al. Transcriptome analysis identifies TNF Superfamily Receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang KX, Denhardt DT. Osteopontin: role in immune regulation and stress responses. Cytokine Growth Factor Rev. 2008;19:333–345. doi: 10.1016/j.cytogfr.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Anborgh PH, Mutrie JC, Tuck AB. Pre- and post-translational regulation of osteopontin in cancer. J Cell Comun Signal. 2011;5:111–122. doi: 10.1007/s12079-011-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kazanecki CC, Uzwiak DJ, Denhardt DT. Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J Cell Biochem. 2007;102:912–924. doi: 10.1002/jcb.21558. [DOI] [PubMed] [Google Scholar]
- 10.Sabo-Attwood T, Ramos-Nino ME, Eugenia-Ariza M, MacPherson MB, Butnor KJ, Vacek PC, McGee SP, et al. Osteopontin modulates inflammation, mucin production, and gene expression signatures after inhalation of asbestos in a murine model of fibrosis. Am J Pathol. 2011;178:1975–1985. doi: 10.1016/j.ajpath.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolak T, Kim H, Ren Y, Kim J, Vaziri ND, Nicholas SB. Osteopontin modulates angiotensin II-induced inflammation, oxidative stress, and fibrosis of the kidney. Kidney Int. 2009;76:32–43. doi: 10.1038/ki.2009.90. [DOI] [PubMed] [Google Scholar]
- 12.Seth D, Gorrell MD, Cordoba S, McCaughan GW, Haber PS. Intrahepatic gene expression in human alcoholic hepatitis. J Hepatol. 2006;45:306–320. doi: 10.1016/j.jhep.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Kiefer FW, Zeyda M, Gollinger K, Pfau B, Neuhofer A, Weichhart T, Säemann MD, et al. Neutralization of osteopontin inhibits obesity-induced inflammation and insulin resistance. Diabetes. 2010;59:935–946. doi: 10.2337/db09-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Syn WK, Choi SS, Liaskou E, Karaca GF, Agboola KM, Oo YH, Mi Z, et al. Osteopontin is induced by hedgehog pathway activation and promotes fibrosis progression in nonalcoholic steatohepatitis. Hepatology. 2011;53:106–115. doi: 10.1002/hep.23998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urtasun R, Lopategi A, George J, Leung TM, Lu Y, Wang X, Ge X, et al. Osteopontin, an oxidant stress-sensitive cytokine, up-regulates collagen-i via integrin α(V) β(3) engagement and PI3K-pAkt-NFκB signaling. Hepatology. 2012;55:594–608. doi: 10.1002/hep.24701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez M, Rincón D, Abraldes JG, Miquel R, Colmenero J, Bellot P, García-Pagán JC, et al. A new scoring system for prognostic stratification of patients with alcoholic hepatitis. Am J Gastroenterol. 2008;103:2747–2456. doi: 10.1111/j.1572-0241.2008.02104.x. [DOI] [PubMed] [Google Scholar]
- 17.Kleiner DE, Brunt EM, Van NM, Behling C, Contos MJ, Cummings OW, Ferrell LD, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen-Khac E, Thevenot T, Piquet MA, Benferhat S, Goria O, Chatelain D, Tramier B, et al. Glucocorticoids plus N-acetylcysteine in severe alcoholic hepatitis. N Engl J Med. 2011;365:1781–1789. doi: 10.1056/NEJMoa1101214. [DOI] [PubMed] [Google Scholar]
- 19.Marcos M, Pastor I, González-Sarmiento R, Laso FJ. A functional polymorphism of the NFKB1 gene increases the risk for alcoholic liver cirrhosis in patients with alcohol dependence. Alcohol Clin Exp Res. 2009;33:1857–1862. doi: 10.1111/j.1530-0277.2009.01023.x. [DOI] [PubMed] [Google Scholar]
- 20.Ki SH, Park O, Zheng M, Morales-Ibanez O, Kolls JK, Bataller R, Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu YW, Tu HF, Wang IK, Wu CH, Chang KW, Liu TY, Kao SY. The implication of osteopontin (OPN) expression and genetic polymorphisms of OPN promoter in oral carcinogenesis. Oral Oncol. 2010;46:302–306. doi: 10.1016/j.oraloncology.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 22.D’Alfonso S, Barizzone N, Giordano M, Chiocchetti A, Magnani C, Castelli L, Indelicato M, et al. Two single-nucleotide polymorphisms in the 5' and 3' ends of the osteopontin gene contribute to susceptibility to systemic lupus erythematosus. Arthritis Rheum. 2005;52:539–547. doi: 10.1002/art.20808. [DOI] [PubMed] [Google Scholar]
- 23.Liu CC, Huang SP, Tsai LY, Wu WJ, Juo SH, Chou YH, Huang CH, et al. The impact of osteopontin promoter polymorphisms on the risk of calcium urolithiasis. Clin Chim Acta. 2010;411:739–743. doi: 10.1016/j.cca.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Mandrekar P, Szabo G. Signalling pathways in alcohol-induced liver inflammation. J Hepatol. 2009;50:1258–1266. doi: 10.1016/j.jhep.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo H, Cai CQ, Schroeder RA, Kuo PC. Osteopontin is a negative feedback regulator of nitric oxide synthesis in murine macrophages. J Immunol. 2001;166:1079–1086. doi: 10.4049/jimmunol.166.2.1079. [DOI] [PubMed] [Google Scholar]
- 26.Guo H, Mi Z, Bowles DE, Bhattacharya SD, Kuo PC. Osteopontin and protein kinase C regulate PDLIM2 activation and STAT1 ubiquitination in LPS-treated murine macrophages. J Biol Chem. 2010;285:37787–37796. doi: 10.1074/jbc.M110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Sancho-Bru P, Bataller R, Gasull X, Colmenero J, Khurdayan V, Gual A, Nicolás JM, et al. Genomic and functional characterization of stellate cells isolated from human cirrhotic livers. J Hepatol. 2005;43:272–282. doi: 10.1016/j.jhep.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 28.Patouraux S, Bonnafous S, Voican CS, Anty R, Saint-Paul MC, Rosenthal-Allieri MA, Agostini H, et al. The osteopontin level in liver, adipose tissue and serum is correlated with fibrosis in patients with alcoholic liver disease. PLoS One. 2012;7:e35612. doi: 10.1371/journal.pone.0035612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Syn WK, Agboola KM, Swiderska M, Michelotti GA, Liaskou E, Pang H, et al. NKT-associated hedgehog and osteopontin drive fibrogenesis in non-alcoholic fatty liver disease. Gut. 2012;61:1323–1329. doi: 10.1136/gutjnl-2011-301857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.