Abstract
Leukocyte immunoglobulin-like receptor (LILR) B3 and LILRA6 represent a pair of inhibitory/activating receptors with identical extracellular domains and unknown ligands. LILRB3 can mediate inhibitory signaling via immunoreceptor tyrosine-based inhibition motifs (ITIMs) in its cytoplasmic tail whereas LILRA6 can signal through association with an activating adaptor molecule, FcRγ, which bears a cytoplasmic tail with an immunoreceptor tyrosine-based activation motif (ITAM). The receptors are encoded by two highly polymorphic neighboring genes within the Leukocyte Receptor Complex (LRC) on human chromosome 19. Here we report that the two genes display similar levels of single nucleotide polymorphisms with the majority of polymorphic sites being identical. In addition, the LILRA6 gene exhibits copy number variation (CNV) whereas LILRB3 does not. A screen of healthy Caucasians indicated that 32% of the subjects possessed more than 2 copies of LILRA6, whereas 4% have only one copy of the gene per diploid genome. Analysis of mRNA expression in the major fractions of PBMCs showed that LILRA6 is primarily expressed in monocytes, similarly to LILRB3, and its expression level correlates with copy number of the gene. We suggest that the LILRA6 CNV may influence the level of the activating receptor on the cell surface, potentially affecting signaling upon LILRB3/A6 ligation.
Keywords: myeloid receptor, LILR, copy number variation
Introduction
LILRs are a family of molecules whose members are involved in the regulation of leukocyte activity (Anderson and Allen 2009). Genes encoding these receptors are clustered within the LRC on human chromosome 19q13. Based on their signaling capacities, LILRs are either inhibitory (LILRB) or activating (LILRA), with the exception of soluble LILRA3. The inhibitory LILRs possess ITIMs in their cytoplasmic tails, whereas the activating members lack intracellular signaling motifs but can associate with an ITAM bearing adaptor molecule, FcRγ, due to the presence of an arginine residue in their transmembrane domain close to the cell surface. Thus, ligation of LILRs can trigger signaling mediated by phosphorylation of ITIMs or ITAMs, similar to many other immune receptors (Daeron et al. 2008; Humphrey et al. 2005).
Despite structural similarity of their ectodomains, consisting of two or four immunoglobulin like domains, LILRs are diversified in ligand binding properties. The two best studied human LILRs, LILRB1 and LILRB2, bind human leucocyte antigen (HLA) class I molecules. Willcox et al. suggested categorizing human LILRs based on their potential to bind HLA class I (Willcox et al. 2003). Group 1 includes LILRB1, B2, A1, A2, and A3, which exhibit high conservation of the amino acid residues involved in class I binding. Besides LILRB1 and LILRB2, class I binding has been formally demonstrated for LILRA1 and LILRA3 in this group (Jones et al. 2011; Ryu et al. 2011). Group 2 members, LILRB3, B4, B5, A4, A5, A6, have poor conservation at the HLA class I contact sites and, therefore, are assumed to bind non-class I ligands. LILRA4 is the only molecule within this group that has an identified natural ligand, human bone marrow stromal antigen 2 (BST2), which is structurally unrelated to class I (Cao et al. 2009).
The group 2 proteins LILRB3 and LILRA6 represent a closely related inhibitory/activating pair that are structurally distant from other LILRs (Supplementary File 3 in (Sambrook et al. 2006)). These molecules are encoded by neighboring “head-to-tail” genes and possess indistinguishable ectodomains, implicating identical ligand binding properties for the two molecules. LILRB3 was first described in 1997 by Colonna et al. (Colonna et al. 1997) and Borges et al. (Borges et al. 1997), who named the new receptor ILT5 and LIR-3, respectively. LILRB3 was shown to display extensive polymorphism based on the identification of 15 variants in a pool of bone marrow cells from 51 donors (Colonna et al. 1997). LILRA6 demonstrated some level of polymorphism as well (Torkar et al. 2000). The high level of the LILRB3 variation was supported by data from hematopoietic stem cell transplant patients, who developed LILRB3-specific antibodies due to sequence disparities between donors and recipients in at least 5.4 % of cases (Pfistershammer et al. 2009).
Expression of LILRB3 was reported based on cell surface staining on monocytes, macrophages, dendritic cells (DCs), granulocytes, some T cells, basophils, eosinophils, and osteoclasts, but not on NK cells or B cells (Colonna et al. 1999; Mori et al. 2008; Sloane et al. 2004; Tedla et al. 2003). LILRB3 and LILRA6 expression was also demonstrated by RNA microarray analysis in monocytes, cultured macrophages, and osteoclasts (Mori et al. 2008). A recent study detected a high level of expression of LILRB3 in dermal CD14+ DCs, but not in Langerhans cells, by both RNA microarray and flow cytometry (Banchereau et al. 2012). Importantly, both antibody binding and RNA microarray detection used in these studies do not distinguish between LILRB3 and LILRA6 due to the structural similarity of the extracellular fragments used for their detection. Therefore, specific expression of LILRB3 vs. LILRA6 is unclear from the published data.
The physiological ligand for LILRB3/A6 remains unknown, but binding of Staphylococcus aureus to LILRB3 was detected when the molecule was expressed in NIH3T3 cells (Nakayama et al. 2007). As predicted, LILRB3 failed to bind HLA class I in a number of experiments (Allen et al. 2001; Colonna et al. 1998; Jones et al. 2011). LILRB3 was shown to be constitutively phosphorylated and associated with SH2 domain-containing tyrosine phosphatase 1 (SHP-1) in macrophages and osteoclasts derived in vitro (Mori et al. 2008). LILRB3 aggregation by antibody cross-linking resulted in inhibition of osteoclast development, similar to aggregation of LILRB1 and LILRB4. Further, antibody co-ligation of LILRB3 with the activating receptors LILRA2 or FcεRI down-regulated a basophil response to cross-linking of the activating receptors (Sloane et al. 2004). Recent work revealed that LILRB3 is upregulated on the surface of myeloid dendritic cells (mDCs) in elite HIV controllers and may contribute to a distinct mDC profile in this rare group of people (Huang et al. 2010). However, no studies questioned the role of LILRA6 specifically, although the antibodies used to evaluate LILRB3 protein expression and crosslink the receptor would also bind LILRA6.
We undertook a comprehensive genetic analysis of the LILRB3/A6 locus and investigated gene-specific expression. The data confirm the presence of high levels of non-synonymous variation in both genes. It was also apparent that this locus has been subjected to non-allelic homologous recombination (NAHR) over time, resulting in variable copy numbers of the activating LILRA6 gene, but maintenance of a single fixed copy of the inhibitory LILRB3 gene. We show that both genes are expressed in monocytes at the mRNA level, and their relative expression is influenced by the LILRA6 CNV, which may affect the net signaling upon LILRB3/A6 ligand engagement.
Material and Methods
Study subjects
Ten donors used for cDNA cloning were recruited at Massachusetts General Hospital (MGH) and gave written informed consent to participate, and the study was approved by the MGH Institutional Review Board. Healthy donors of European ancestry (N=228) were recruited at the Frederick National Laboratory. This study was approved by the protocol review office of the U.S. National Cancer Institute institutional review board. B cell lines from the CEPH (Centre d'Etude du Polymorphisme Humain) families were obtained from the Coriell Institute for Medical Research.
DNA extraction
DNA was extracted using the standard phenol/chloroform-based method.
Cloning of cDNA fragments
Total RNA was extracted from frozen PBMC using RiboPure RNAkit (Life Technologies) and reverse transcribed using SuperScript® III First-Strand Synthesis System (Life Technologies). The cDNA fragments were amplified using two pairs of primers, ILT2/ILT22 and ILT2/ILT26 (Table S1), and Platinum Taq High Fidelity (Life Technologies) with the following cycling conditions: 95°C - 15 sec, 64°C - 15 sec, 68°C - 4 min. The first pair of primers specifically amplified LILRB3 whereas the second pair amplified LILRA6 and LILRA2, which could be readily distinguished by sequence differences. The primers were designed using the alignment available in the dbLRC database, in which LILRA6 is erroneously labeled as LILRB6 (http://www.ncbi.nlm.nih.gov/gv/lrc). The fragments were cloned into TOPO-TA or Zero Blunt TOPO vector (Life Technologies). Plasmid DNA from multiple clones was extracted using the Agencourt CosMCPrep system (Beckman Coulter) in a 96-well format. The plasmids were sequenced using standard BigDye® Terminator v1.1 Cycle Sequencing Kit (Life Technologies). About 200 clones were analyzed for each gene. While we tried to minimize PCR artifacts by using a high fidelity enzyme and prolonged extension time, mutations and chimeras were still present in the sequenced clones. The artifacts were recognized based on their presence in only a single clone each, and true nucleotide sequences were obtained based on their occurrence in multiple clones, and partial verification at the genomic DNA level. The overall percentage of chimeric clones was 7% for LILRB3 and 20% for LILRA6, where the higher number of LILRA6 chimeras likely occurred due to co-amplification of LILRA2 in the same reaction. Genomic DNA fragments were amplified using two gene-specific pairs of primers: ILT5/ILT22 (LILRB3), ILT5/ILT26 (LILRA6) using Platinum Taq (Life Technologies) with the following cycling conditions: 95°C - 15 sec, 62°C - 15 sec, 68°C - 5 min. Partial sequencing of the corresponding genomic fragments was used for verification of allelic sequences from cDNA clones. The GenBank accession numbers for the LILRB3 and LILRA6 allelic sequences reported in this paper are KC918851- KC918876.
CNV genotyping
CNV genotyping was performed using RT2 SYBR Green qPCR Master Mixes with ROX (Qiagen) and the following pairs of primers at a concentration of 250 nM in triplicate: ILT62/63 (LILRB3), ILT55/56 (LILRA6), ZNF80F/R and GPR15F/R (Table S1). Amplification efficiencies were defined using serial dilutions of genomic DNA in yeast tRNA solutions as recommended by D’Haene et al. (D'Haene et al. 2010) and were in the range of 1.91–1.97. The cycling conditions in the ABI 7900 HT instrument were as follows: 10 min at 95°C, 1 cycle; 15 sec 95°C, 1 min 60°C, 40 cycles. The data was analyzed using qbasePLUS software (Biogazelle) with target-specific amplification efficiencies and two reference targets, ZNF80 and GPR15. The Taqman-based CNV genotyping was done using pre-designed assays, Hs04013741_cn (LILRB3), Hs04034020_cn (LILRA6), and the TERT reference assay according to manufacturer’s protocol in quadruplicate (Life Technologies). The Taqman CNV data was analyzed using the CopyCaller software v.1 (Life Technologies). Results were defined as highly confident based on the criteria recommended by the software: confidence value > 95% and Z score < 1.75.
Analysis of mRNA expression
PBMCs were isolated using a Ficoll technique and cell fractions were obtained by negative selection for CD8 and CD56, and positive selection for CD4, CD14, and CD19 using EasySep magnetic platform (StemCell Technologies). Total PBMCs were used for selection of all fractions with the exception of CD4 T cells, for which we used CD14 monocyte depleted PBMCs. Purity of selected fractions monitored by flow cytometry showed that >85% of cells expressed the selected marker with <1% CD14+ cells in non-monocyte fractions. For the analysis of fraction purity, antibodies to all of the selected CD antigens were obtained from BD Biosciences except anti-CD56 (Biolegend). Total RNA was extracted using RNeasy Plus Universal kit (Qiagen). RNA integrity and quantity was checked using the LabChip GX instrument (Perkin Elmer) and the quality score was > 9.0 for all samples. cDNA was synthesized using SuperScript VILO kit and random hexamers (Life Technologies). The primers and PCR conditions were the same as those used in the SYBR Green CNV genotyping method described above. The equimolar mix of the PCR fragments products was prepared by purification of the PCR products using QIAquick PCR purification kit (Qiagen) and quantification using the LabChip GX (Perkin Elmer). The mix was diluted in yeast tRNA solution (5 ng/ul) to ~ 10−7 ng/ul and used as a reference. The cDNA quantity was normalized to the total RNA amount used for the cDNA synthesis. Analyses were performed using the qbasePLUS (Biogazelle).
Results
LILRB3 and LILRA6 display high levels of allelic polymorphism
We amplified and cloned cDNA fragments of LILRB3 and LILRA6 from a group of ten donors. The LILRB3 fragment encompassed 97% of the coding sequence (nt 48–1898, NM_006864) and the LILRA6 fragment spanned the entire coding sequence of the gene (nt 61–1613, NM_024318). Among the ten individuals, we identified 13 unique alleles for each of the two genes, which we labeled numerically, LILRB3*01-13 and LILRA6*01-13 (Table 1, Figure S1). Four groups of alleles encoded identical proteins: LILRB3*05/10/13, LILRB3*03/12, LILRA6*01/13, and LILRA6*03/05/06/09. Thus, we isolated ten LILRB3 and nine LILRA6 unique amino acid sequences from ten people (Figure S2). Notably, most of the polymorphic sites in ectodomains were shared between the two proteins (20 out of 31 polymorphic aa) and more than half of the alleles did not reveal identical cDNA sequences in Genbank (Table S2). There were no identical LILRB3 genotypes among the ten donors, all of whom exhibited heterozygosity for this gene, and only two individuals possessed the same LILRA6 genotype (Table 1, P3 and P5, LILRA6*02/03). Surprisingly, we identified three distinct sequences of LILRA6 in three donors (Table 1; P6, P7, and P8) indicating that this gene exhibits CNV. Our further experiments (see below) confirmed the CNV in LILRA6, but not in LILRB3, which was consistent with the allelic genotypes.
Table 1.
Allelic genotypes determined by cDNA cloning and LILRA6 copy numbers in ten donors.
| Donor | LILRB3 alleles | LILRA6 alleles | LILRA6 CN |
|---|---|---|---|
| P1 | 03/05 | 01 | 2 |
| P2 | 01/02 | 01/02 | 2 |
| P3 | 03/04 | 02/03 | 3 |
| P4 | 09/10 | 04 | 1 |
| P5 | 03/11 | 02/03 | 3 |
| P6 | 02/05 | 02/05/12 | 3 |
| P7 | 01/03 | 01/03/06 | 3 |
| P8 | 06/07 | 07/10/11 | 3 |
| P9 | 08/13 | 01/08 | 2 |
| P10 | 12/13 | 09/13 | 2 |
Alignment of the alleles and corresponding amino acid sequences are shown in Figures 1S and 2S. Copy numbers (CN) were determined using two independent assays as shown in Figure 1.
While the overall majority of the sequenced clones for LILRB3 and LILRA6 were spliced similar to the reference sequences, NM_006864 and NM_024318, respectively, multiple splicing variants were detected for both genes. Interestingly, for one particular allele, LILRA6*10, we observed a deletion of exon 5 (297 bp) in all six clones sequenced, which results in the absence of the D3 domain in the corresponding protein (Figures S1–S2).
CNV in the LILRA6 locus
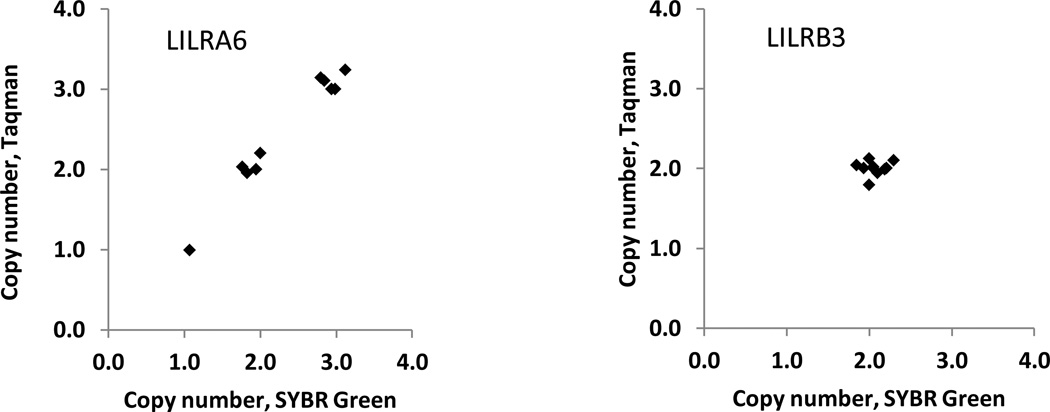
Using the same samples that were used for cDNA cloning, we tested for copy number polymorphism at the LILRB3/A6 locus by qPCR in two independent assays. The first assay employed gene-specific primers in SYBR Green qPCR and two reference genes, ZNF80 and GPR15, as recommended previously (D'Haene et al. 2010). The second method utilized commercial pre-designed Taqman primers and probes and the TERT gene as a reference. Distinct LILRB3/A6 areas were amplified in each assay: 3’untranslated region (3’UTR) fragments in the SYBR Green experiments, and intronic fragments in the Taqman PCRs (Table S1). The results obtained by the two assays were in good agreement and indicated that two copies of the LILRB3 gene were present in each sample, whereas the LILRA6 gene exhibited CNV, and its presence ranged from one to three copies per diploid genome (Table 1, Figure 1). Thus, within this small group of people, haplotypes exist in which LILRA6 is either present in multiple copies or completely absent.
Figure 1.
Copy numbers of LILRA6 and LILRB3 determined by two independent assays. The Sybr Green and Taqman PCRs targeted distinct fragments in each gene and demonstrated identical results (see Table 1).
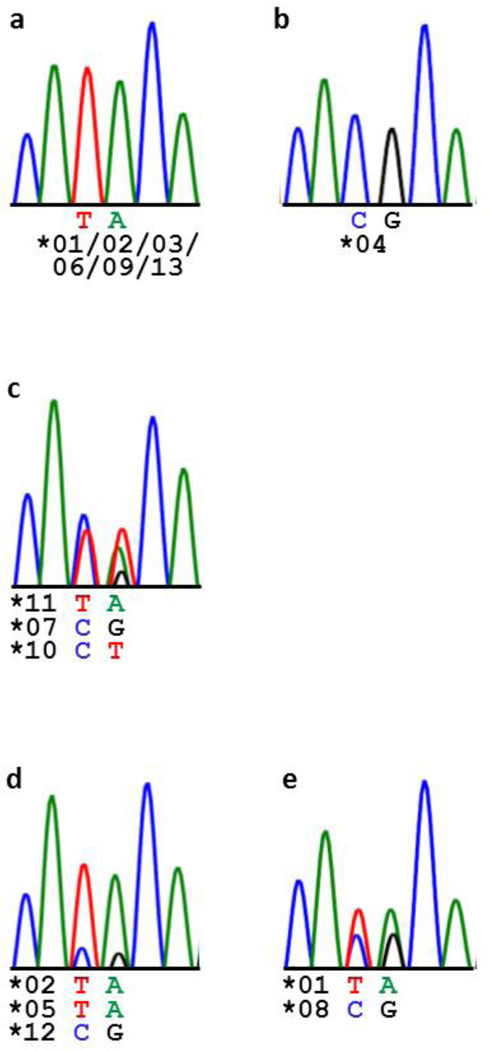
Sequencing genomic DNA also supported CNV of LILRA6. Figure 2 shows sequencing peak patterns at two neighboring polymorphic sites in exon 6 of LILRA6 (nt 1322–1323, NM_024318). Trimorphism was evident at position 1323 in donor P8 (Figure 2C), who had three copies of LILRA6 alleles with distinct nucleotides at this position, LILRA6*07/*10/*11 (G/T/A), as defined by the cDNA cloning (Figure S1). Further, comparison of the P6 and P9 patterns indicates a larger contribution of the “TA” variant in P6 as compared to P9 in agreement with their genotypes: P6 has two “TA” alleles and one “CG” allele whereas P9 has one “TA” allele and one “CG” allele (Figure 2D–E).
Figure 2.
Electrophoregrams of a polymorphic LILRA6 genomic fragment sequenced in donors P1, 2, 3, 5, 7, 10 (a), P4 (b), P8 (c), P6 (d), and P9 (e). Nucleotide variants at positions 1322–1323 (exon 6, NM_024318) and corresponding alleles are shown under the electrophoregrams. One representative picture is presented in A. Trimorphism in P8 (c) as well as comparison of P6 (d) and P9 (e) support CNV.
We genotyped the LILRA6 CNV in a cohort of 228 healthy Caucasians using the Taqman assay (Table 2). The copy numbers ranged between one and six. Approximately one third of all people had more than 2 copies of the gene, whereas ~4% possessed one copy. In contrast to LILRA6, LILRB3 did not exhibit CNV in 82 Caucasians tested.
Table 2.
LILRA6 copy number frequencies in a cohort of 228 healthy Caucasians.
| LILRA6 CN | N | % |
|---|---|---|
| 1 | 8 | 3.5 |
| 2 | 145 | 63.6 |
| 3 | 59 | 25.9 |
| 4 | 14 | 6.1 |
| 5 | 1 | 0.4 |
| 6 | 1 | 0.4 |
Copy numbers (CN) were determined with high confidence in 97% of the cohort. One donor with a single LILRA6 copy and two donors within each of the 2, 3, and 4 LILRA6 copy groups did not pass the high confidence criteria (see Material and Methods).
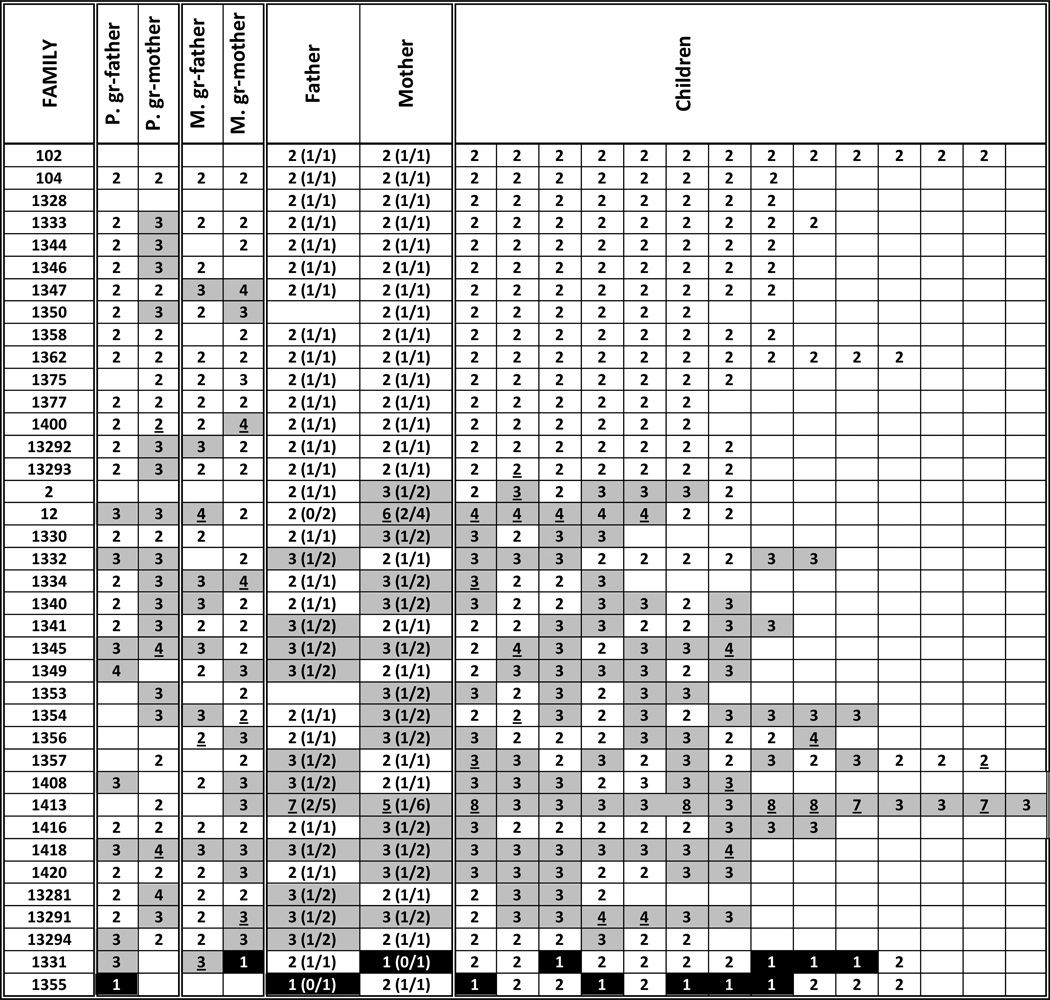
The LILRA6 CNV was also genotyped in 476 DNA samples representing 38 CEPH pedigrees (Figure 3). Children from 15 families had two copies of LILRA6, as did their parents (one family was missing a DNA sample from the father). In 21 families, however, at least one child had more than two copies, and except for one of these families, at least one parent also had more than two copies. The remaining two families contained multiple children and one parent with one copy of the gene. These data suggest that the LILRA6 CNV overall displays normal Mendelian inheritance.
Figure 3.
LILRA6 CNV in 38 CEPH pedigrees. Cells with copy numbers >2 are grey and with one copy are balck. Copy numbers that did not pass high confidence criteria are underlined (8% of all samples). Haplotypes based on segregation analysis are shown in parentheses. Families 2 and 12 – French; 102 and 104 – Venezuelan; others – Utah. Three groups of families are related: 102/104, 1344/1375, 1328/13281/13291/13292/13293/13294 (www.ccr.coriell.org; Figure S3). Blank cells in parents denote absence of DNA samples.
Expression of LILRA6 and LILRB3 mRNA in PBMCs
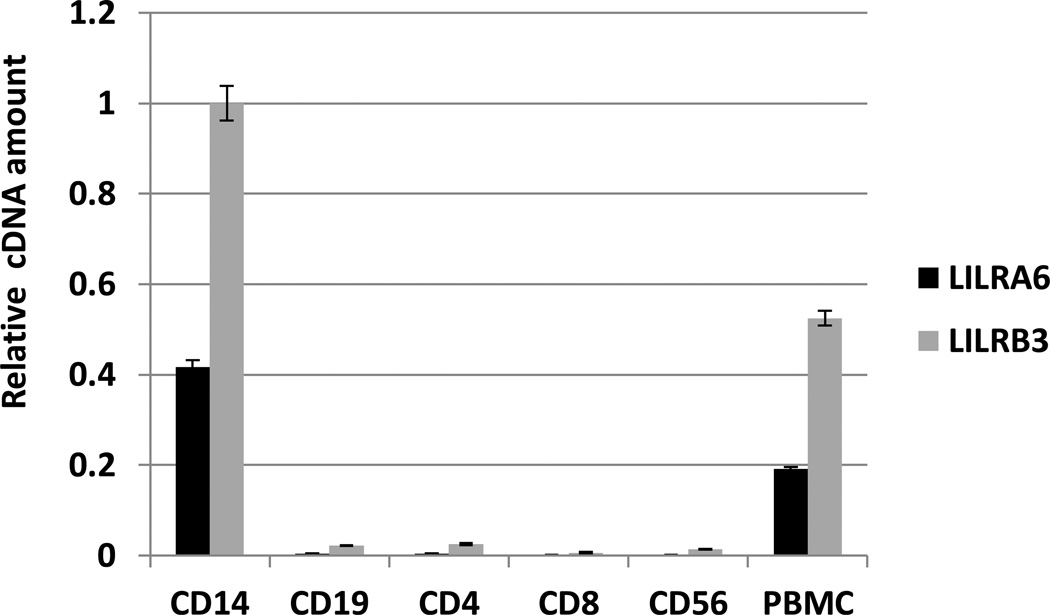
Expression of the LILRB3 and LILRA6 genes was estimated using qPCR in six major PBMCs fractions including monocytes, CD4+ T cells, CD8+ T cells, B cells and NK cells, which were isolated from a donor with 2 copies of LILRA6 (Figure 4). Both genes were expressed in monocytes, and at a very low level, if any, in other cell types. The amount of LILRA6 cDNA in monocytes appeared to be ~ 40 % the level of LILRB3 cDNA. Assuming the cDNA synthesis is equally efficient for the two genes, the activating gene is transcribed at a lower level than the inhibitory gene in monocytes of this donor.
Figure 4.
Analysis of LILRA6 and LILRB3 transcription in PBMC by SYBR Green qPCR. Cell fractions were isolated using EasySep selection and included monocytes (CD14), B cells (CD19), T cells (CD4 and CD8) and NK cells (CD56). Expression was quantified using equimolar mixture of LILRB3/A6 PCR fragments as reference and normalized to the total RNA input (measured in the LabChip DX).
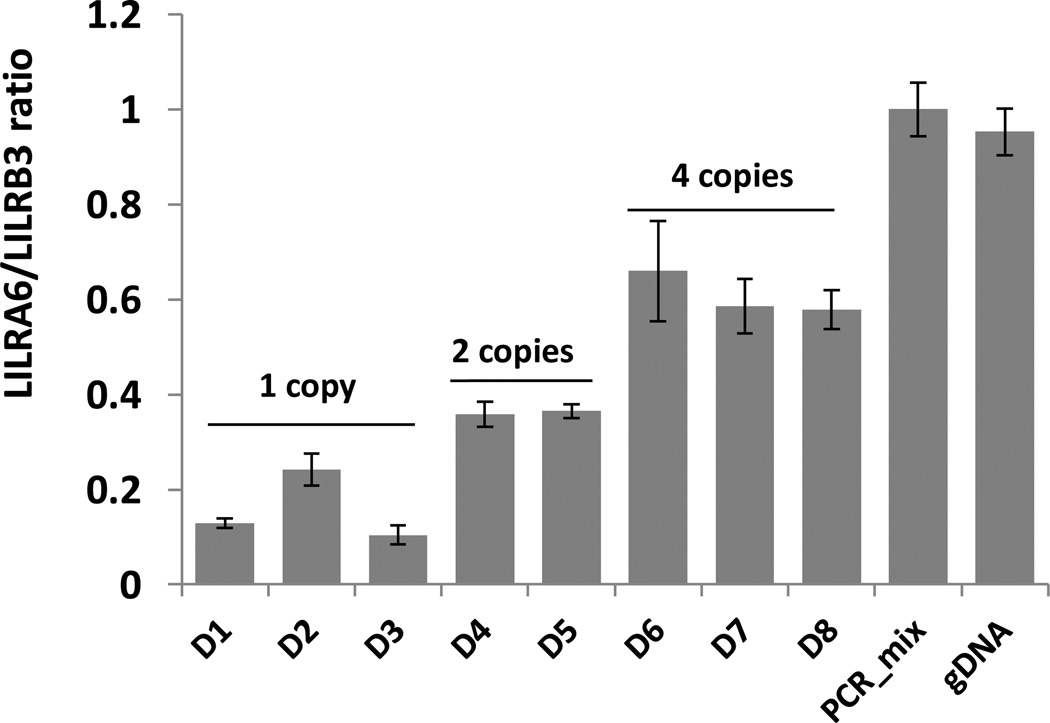
Next, we examined the influence of the CNV on relative expression of the two genes. PBMCs from eight donors with variable copy numbers were tested using qPCR. The results shown in Figure 5 indicate that the LILRA6/LILRB3 cDNA ratio correlates positively with gene copy number. In individuals with one, two and four LILRA6 copies, the averaged cDNA ratio was 0.2, 0.4 and 0.6, respectively. Thus, the LILRA6 CNV corresponds with the relative transcriptional level.
Figure 5.
Relative level of LILRA6/B3 mRNA expression correlates with the CNV genotype. Total RNA was isolated from PBMC of healthy donors and the corresponding cDNA was used in SYBR Green qPCR. The PCR_mix is an equimolar mixture of LILRB3 and LILRA6 PCR fragments. Genomic DNA from an individual with two copies of LILRA6 was used as a reference control (gDNA, LILRA6/LILRB3=1). All reactions were performed in triplicate.
Discussion
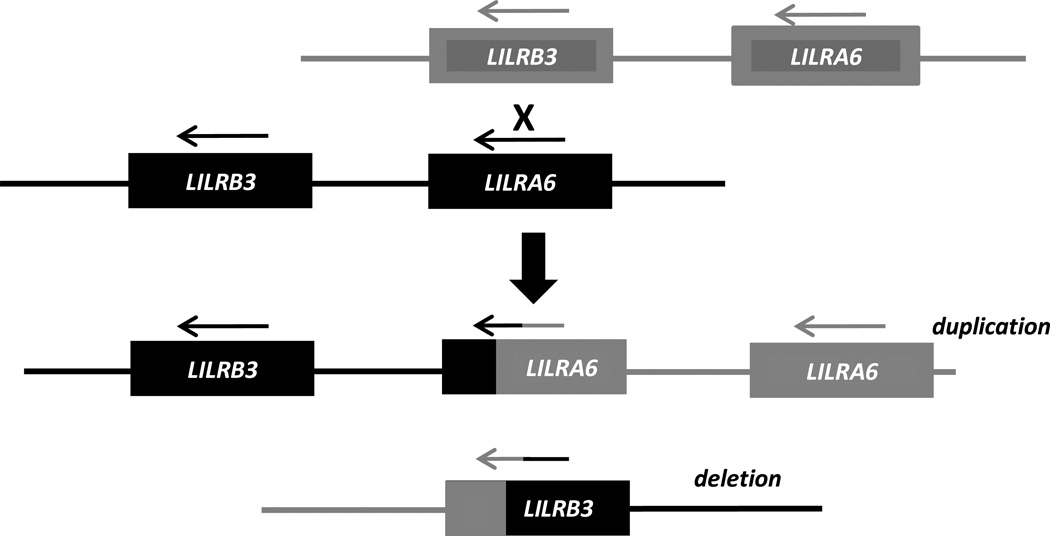
The exceptionally high level of the LILRB3 polymorphism has been known since its first identification (Colonna et al. 1997) and similar variation of its activating counterpart was expected (Torkar et al. 2000). Here we show that LILRA6 indeed exhibits comparable level of polymorphism to LILRB3 in exons encoding extracellular domains. Allelic variants isolated in ten individuals encoded ten LILRB3 and nine LILRA6 unique amino acid sequences. In addition, we demonstrate that LILRA6 can be present in variable copy numbers, which is not a characteristic of LILRB3. This polymorphism is likely to be a result of NAHR between LILRB3 and LILRA6 shown schematically in Figure 6. The configuration of the locus explains why only the activating counterpart is subject to duplications and deletions. The inhibitory gene is located downstream of the activating gene in a head-to-tail orientation and the receptors have type 1 topologies, where the extracellular domains are encoded by the 5‘ region of the gene. Inheritance of the 3’ region, which encodes the intracellular domain, determines whether a hybrid gene is activating or inhibitory. As exemplified in Figure 6, a crossover between in LILRB3 and LILRA6 results in two reciprocal chromosomes, one with a duplication and the other with a deletion of LILRA6. Importantly, any crossover between LILRB3 and LILRA6 will preserve the downstream location of LILRB3 relative to LILRA6 in head-to-tail orientation (as long as LILRA6 is not missing altogether on the haplotype). Thus, recombinant chromosomes will always maintain a single copy of the inhibitory form of the gene, while expanding/contracting activating forms of the gene. The structural identity of the ectodomains and sharing of polymorphic sites between LILRB3 and LILRA6 also points to the dynamic exchange of the genetic material between the receptors as a result of NAHR.
Figure 6.
Schematic representation of NAHR between chromosomes possessing one copy of each gene. In the duplicated haplotype, LILRB3 stays intact and the newly formed hybrid gene contains the 3’ end of LILRA6 and is therefore defined as LILRA6 due to its “activating” 3’ end. The reciprocal haplotype retains only a hybrid gene that has the 3’ end of LILRB3, which defines the gene as inhibitory. If the recombinant chromosomes shown in the scheme were involved in further NAHR, the two daughter recombinant chromosomes will always have a single copy of LILRB3, and LILRA6 will either eliminated altogether or it will be present as one copy or multiple tandem copies.
Distributions of the LILRA6 CNV in the CEPH pedigrees (Figure 3) indicate that the locus is fairly stable and the CNV is likely to be inherited in a Mendelian manner. Interestingly, the frequencies of the copy numbers in a cohort of 228 healthy Caucasians demonstrate a sizeable prevalence of duplications over deletions: 75 individuals have more than two copies (i.e. at least one duplicated haplotype) and 8 individuals have only one copy (i.e. one deleted haplotype; Table 2). Similarly, among CEPH family members, we identified 48 unrelated individuals with more than two copies, which included 42 grandparents and six parents, and only two unrelated members with one copy (see Figure S3 for relatedness between families). Segregation analysis provides a more stringent estimate of duplication prevalence, and by this method, we identified 28 duplicated vs three deleted haplotypes in 67 unrelated CEPH parents (Figures 3, S3). This dominance of duplications is not expected from NAHR, which theoretically results in either reciprocity (i.e. equal numbers of duplications and deletions) when interchromatid recombinations take place, as shown in Figure 6, or in deletions alone when intrachromatid recombinations occur (Gu et al. 2008). Thus, the skewed CNV distribution may be the result of selection pressure to preserve LILRA6 due to some functional benefit of this gene. Alternatively, the underlying mechanism for LILRA6 CNV may occur by a distinct, more complex process.
Previous reports on protein and mRNA expression did not distinguish between LILRB3 and LILRA6. We performed qPCR analysis, in which we used primers specifically targeting the 3’UTR of each gene. Our data indicated that both genes are primarily expressed in monocytes, and not in NK, T, and B cells, at the mRNA level. This is in line with previous data showing expression of LILRB3/A6 mostly in cells of myeloid origin (Banchereau et al. 2012; Borges et al. 1997; Colonna et al. 1999; Huang et al. 2010; Mori et al. 2008; Pfistershammer et al. 2009; Sloane et al. 2004; Tedla et al. 2003). We also demonstrate that there is a dosage effect of CNV on the LILRA6 mRNA level, and if mRNA level correlates to the protein level, then the CNV may affect receptor signaling. Our qPCR analyses indicate a lower level of LILRA6 transcripts compared with LILRB3 in cells at a steady state, even among individuals with four copies of LILRA6 (Figure 5). This ratio may change upon stimulation of the cells, although the ratio of inhibitory vs. activating transcripts does not necessarily predict signaling capacity since signaling is a complex function of multiple parameters.
Two activating LILRs, LILRA1 and LILRA3, have been demonstrated to bind to HLA class I, but with substantially lower affinity than do the inhibitory LILRB1 and LILRB2 (Jones et al. 2011; Ryu et al. 2011). This pattern is characteristic of many other paired receptors, such as KIR2DL1/KIR2DS1, SIRPα/SIRPγ and NKG2A/NKG2C (reviewed in (Kuroki et al. 2012)). The difference in affinities may reflect some common principle in ITIM/ITAM regulation of leukocytes, but we propose that it is the expression level and probably not the binding properties that differentiate LILRB3 and LILRA6. Binding of these LILRs to their putative ligand(s) may be influenced by polymorphisms in the molecules, but these polymorphisms are common to the inhibitory and activating forms. Thus, similarity in ligand binding capacity of LILRB3 and LILIRA6 is a unique concept as compared to other paired receptors. It will be interesting to investigate the distinct regulatory mechanisms of LILRB3 and LILRA6 expression under normal conditions and after stimulation with cytokines or exposure to pathogens.
Understanding the physiological relevance of the genetic polymorphism of the LILRB3/A6 locus described herein will require further investigation at the protein level. Our findings will be helpful in interpreting functional data, particularly upon identification of a ligand.
Supplementary Material
Acknowledgments
This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, Frederick National Lab, Center for Cancer Research. We would like to thank Colm O’huigin, Daniel McVicar, and Ludmila Lyakh for helpful discussions and the BSP CCR Genetics Core for technical assistance.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen RL, Raine T, Haude A, Trowsdale J, Wilson MJ. Leukocyte receptor complex-encoded immunomodulatory receptors show differing specificity for alternative HLA-B27 structures. J Immunol. 2001;167:5543–5547. doi: 10.4049/jimmunol.167.10.5543. [DOI] [PubMed] [Google Scholar]
- Anderson KJ, Allen RL. Regulation of T-cell immunity by leucocyte immunoglobulin-like receptors: innate immune receptors for self on antigen-presenting cells. Immunology. 2009;127:8–17. doi: 10.1111/j.1365-2567.2009.03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Zurawski S, Thompson-Snipes L, Blanck JP, Clayton S, Munk A, Cao Y, Wang Z, Khandelwal S, Hu J, McCoy WHt, Palucka KA, Reiter Y, Fremont DH, Zurawski G, Colonna M, Shaw AS, Klechevsky E. Immunoglobulin-like transcript receptors on human dermal CD14+ dendritic cells act as a CD8-antagonist to control cytotoxic T cell priming. Proc Natl Acad Sci U S A. 2012;109:18885–18890. doi: 10.1073/pnas.1205785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges L, Hsu ML, Fanger N, Kubin M, Cosman D. A family of human lymphoid and myeloid Ig-like receptors, some of which bind to MHC class I molecules. J Immunol. 1997;159:5192–5196. [PubMed] [Google Scholar]
- Cao W, Bover L, Cho M, Wen X, Hanabuchi S, Bao M, Rosen DB, Wang YH, Shaw JL, Du Q, Li C, Arai N, Yao Z, Lanier LL, Liu YJ. Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J Exp Med. 2009;206:1603–1614. doi: 10.1084/jem.20090547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Nakajima H, Navarro F, Lopez-Botet M. A novel family of Ig-like receptors for HLA class I molecules that modulate function of lymphoid and myeloid cells. J Leukoc Biol. 1999;66:375–381. doi: 10.1002/jlb.66.3.375. [DOI] [PubMed] [Google Scholar]
- Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, Lopez-Botet M. A common inhibitory receptor for major histocompatibility complex class I molecules on human lymphoid and myelomonocytic cells. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonna M, Samaridis J, Cella M, Angman L, Allen RL, O'Callaghan CA, Dunbar R, Ogg GS, Cerundolo V, Rolink A. Human myelomonocytic cells express an inhibitory receptor for classical and nonclassical MHC class I molecules. J Immunol. 1998;160:3096–3100. [PubMed] [Google Scholar]
- D'Haene B, Vandesompele J, Hellemans J. Accurate and objective copy number profiling using real-time quantitative PCR. Methods. 2010;50:262–270. doi: 10.1016/j.ymeth.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Daeron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- Gu W, Zhang F, Lupski JR. Mechanisms for human genomic rearrangements. Pathogenetics. 2008;1:4. doi: 10.1186/1755-8417-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Burke PS, Cung TD, Pereyra F, Toth I, Walker BD, Borges L, Lichterfeld M, Yu XG. Leukocyte immunoglobulin-like receptors maintain unique antigen-presenting properties of circulating myeloid dendritic cells in HIV-1-infected elite controllers. J Virol. 2010;84:9463–9471. doi: 10.1128/JVI.01009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey MB, Lanier LL, Nakamura MC. Role of ITAM-containing adapter proteins and their receptors in the immune system and bone. Immunol Rev. 2005;208:50–65. doi: 10.1111/j.0105-2896.2005.00325.x. [DOI] [PubMed] [Google Scholar]
- Jones DC, Kosmoliaptsis V, Apps R, Lapaque N, Smith I, Kono A, Chang C, Boyle LH, Taylor CJ, Trowsdale J, Allen RL. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol. 2011;186:2990–2297. doi: 10.4049/jimmunol.1003078. [DOI] [PubMed] [Google Scholar]
- Kuroki K, Furukawa A, Maenaka K. Molecular recognition of paired receptors in the immune system. Front Microbiol. 2012;3:429. doi: 10.3389/fmicb.2012.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori Y, Tsuji S, Inui M, Sakamoto Y, Endo S, Ito Y, Fujimura S, Koga T, Nakamura A, Takayanagi H, Itoi E, Takai T. Inhibitory immunoglobulin-like receptors LILRB and PIR-B negatively regulate osteoclast development. J Immunol. 2008;181:4742–4751. doi: 10.4049/jimmunol.181.7.4742. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Underhill DM, Petersen TW, Li B, Kitamura T, Takai T, Aderem A. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178:4250–4259. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- Pfistershammer K, Lawitschka A, Klauser C, Leitner J, Weigl R, Heemskerk MH, Pickl WF, Majdic O, Bohmig GA, Fischer GF, Greinix HT, Steinberger P. Allogeneic disparities in immunoglobulin-like transcript 5 induce potent antibody responses in hematopoietic stem cell transplant recipients. Blood. 2009;114:2323–2332. doi: 10.1182/blood-2008-10-183814. [DOI] [PubMed] [Google Scholar]
- Ryu M, Chen Y, Qi J, Liu J, Fan Z, Nam G, Shi Y, Cheng H, Gao GF. LILRA3 binds both classical and non-classical HLA class I molecules but with reduced affinities compared to LILRB1/LILRB2: structural evidence. PLoS One. 2011;6:e19245. doi: 10.1371/journal.pone.0019245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook JG, Bashirova A, Andersen H, Piatak M, Vernikos GS, Coggill P, Lifson JD, Carrington M, Beck S. Identification of the ancestral killer immunoglobulin-like receptor gene in primates. BMC Genomics. 2006;7:209. doi: 10.1186/1471-2164-7-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloane DE, Tedla N, Awoniyi M, Macglashan DW, Jr., Borges L, Austen KF, Arm JP. Leukocyte immunoglobulin-like receptors: novel innate receptors for human basophil activation and inhibition. Blood. 2004;104:2832–2839. doi: 10.1182/blood-2004-01-0268. [DOI] [PubMed] [Google Scholar]
- Tedla N, Bandeira-Melo C, Tassinari P, Sloane DE, Samplaski M, Cosman D, Borges L, Weller PF, Arm JP. Activation of human eosinophils through leukocyte immunoglobulin-like receptor 7. Proc Natl Acad Sci U S A. 2003;100:1174–1179. doi: 10.1073/pnas.0337567100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torkar M, Haude A, Milne S, Beck S, Trowsdale J, Wilson MJ. Arrangement of the ILT gene cluster: a common null allele of the ILT6 gene results from a 6.7-kbp deletion. Eur J Immunol. 2000;30:3655–3662. doi: 10.1002/1521-4141(200012)30:12<3655::AID-IMMU3655>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.