Drought dramatically affects plant reproductive development and crop yield, but previous research has primarily focused on vegetative development. This work uses a scheme that allows plant growth with limited water to study plant responses to drought during reproductive development. The authors examine floral developmental defects and identify key genes affecting this process.
Abstract
Drought dramatically affects plant growth and crop yield, but previous studies primarily examined responses to drought during vegetative development. Here, to study responses to drought during reproductive development, we grew Arabidopsis thaliana plants with limited water, under conditions that allowed the plants to initiate and complete reproduction. Drought treatment from just after the onset of flowering to seed maturation caused an early arrest of floral development and sterility. After acclimation, plants showed reduced fertility that persisted throughout reproductive development. Floral defects included abnormal anther development, lower pollen viability, reduced filament elongation, ovule abortion, and failure of flowers to open. Drought also caused differential expression of 4153 genes, including flowering time genes FLOWERING LOCUS T, SUPPRESSOR OF OVEREXPRESSION OF CO1, and LEAFY, genes regulating anther and pistil development, and stress-related transcription factors. Mutant phenotypes of hypersensitivity to drought and fewer differentially expressed genes suggest that DEHYDRATION RESPONSE ELEMENT B1A may have an important function in drought response in flowers. A more severe filament elongation defect under drought in myb21 plants demonstrated that appropriate stamen development requires MYB DOMAIN PROTEIN 21 under drought conditions. Our study reveals a regulatory cascade in reproductive responses and acclimation under drought.
INTRODUCTION
Abiotic stresses such as drought, salinity, and high and low temperatures negatively affect plant growth and development, causing cellular water deficit, cell membrane injury, loss of enzyme activities, and other defects and resulting in severe reductions of crop yields (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). Drought also enhances the damage caused by other stresses (Farooq et al., 2009). Genes for acclimation to water deficit help to minimize loss of plant productivity during drought. Such genes have been identified and analyzed in many studies, especially during vegetative development (Kawasaki et al., 2001; Zhu, 2001; Breshears et al., 2005; Schröter et al., 2005; Chaves et al., 2009). For example, one of the plant responses to drought is to increase the expression levels of genes involved in osmolyte synthesis and metabolism, thereby enhancing drought tolerance (Kreps et al., 2002; Yancey, 2005).
Plant hormone pathways and many gene families, including PYRABACTIN RESISTANCE1 (PYR1), PROTEIN PHOSPHATASE2C (PP2C), SNF1-RELATED PROTEIN KINASE (SnRK), DEHYDRATION RESPONSE ELEMENT B (DREB), NO APICAL MERISTEM/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR/CUPSHAPED COTYLEDON (NAC) and HEAT SHOCK TRANSCRIPTION FACTOR (HSF) families, have important functions in response to different environmental stresses (Zhu, 2002; Ooka et al., 2003; Busch et al., 2005; Park et al., 2009; Klingler et al., 2010). The hormone abscisic acid (ABA) plays key roles in response and acclimation to biotic and abiotic stresses during vegetative development (Fujita et al., 2006; Hirayama and Shinozaki, 2007). ABA is synthesized in response to drought and acts at least in part by binding to ABA receptors of the PYR/PYR1-LIKE (PYL)/REGULATORY COMPONENTS OF ABA RECEPTORS (RCAR) family in Arabidopsis thaliana (Fujii et al., 2009; Melcher et al., 2009). The ABA-PYR/PYL/RCAR complexes bind to and negatively regulate PP2Cs. The binding of PYR/PYL/RCAR to PP2Cs releases and facilitates the phosphorylation of SnRK2s, which then induce downstream responses, including the expression of transcription factors (Park et al., 2009; Umezawa et al., 2009; Cutler et al., 2010).
ABA induces the expression of several genes encoding transcription factors (Shinozaki et al., 2003). Among the encoded proteins are the ABA-RESPONSIVE ELEMENT-BINDING PROTEIN (AREB)/ABA-RESPONSIVE ELEMENT BINDING FACTOR (ABF) proteins, which bind to the ABA-RESPONSIVE ELEMENT (ABRE) cis-acting element regulating RESPONSIVE TO DESSICATION29B (RD29B) expression (Uno et al., 2000). In addition, an ABA-independent pathway is also important for abiotic stress responses in plant; this pathway includes members of the DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN (DREB) family. For example, DREB2, also known as C-repeat binding factor, can activate via the DEHYDRATION-RESPONSIVE ELEMENT (DRE)/C-REPEAT (CRT) cis-acting element in the RD29A promoter to induce expression in response to drought, high salinity, or cold (Liu et al., 1998). In addition, DREB1D, DREB1E, and DREB1F can be induced by osmotic stress such as drought and high salinity (Haake et al., 2002; Magome et al., 2004). The functions of these genes during reproductive development are not known.
Flower development requires both early organ identity genes and later genes for organogenesis and tissue formation (Ma, 2005). In Arabidopsis, APETALA1 (AP1) and APETALA2 (AP2) are required for the A function of the ABC model for organ identity, APETALA3 (AP3) and PISTILLATA are needed for the B function, and AGAMOUS (AG) is essential for the C function (Ma, 2005). Other genes important for anther development include SPL/NZZ (Schiefthaler et al., 1999; Yang et al., 1999; Ito et al., 2004), EXCESS MICROSPOROCYTES1/EXTRA SPOROGENOUS CELLS (EMS1/EXS) (Canales et al., 2002; Zhao et al., 2002), TAPETUM DETERMINANT1 (TPD1) (Yang et al., 2003, 2005), MALE STERILITY1 (MS1) (Wilson et al., 2001), ABORTED MICROSPORES (AMS) (Sorensen et al., 2003), and DYSFUNCTIONAL TAPETUM1 (DYT1) (Zhang et al., 2006). In addition, SHOOT MERISTEMLESS (STM) (Long et al., 1996), SHATTERPROOF (SHP) (Liljegren et al., 2000; Pinyopich et al., 2003), WUSCHEL (WUS) (Mayer et al., 1998; Lohmann et al., 2001), and SEPALLATA (SEP) (Pelaz et al., 2000; Honma and Goto, 2001) are necessary for female development.
Plant vegetative growth is greatly affected by environmental changes, including drought, especially during seed germination and seedling development (Shinozaki and Yamaguchi-Shinozaki, 2007). In contrast to the many studies on the effect of drought on vegetative development, there have been few reports about the effect of drought on reproductive growth. Crops such as wheat (Triticum aestivum and rice (Oryza sativa) show partial male sterility under water deficit, leading to significant reduction of grain production (Sheoran and Saini, 1996; Lalonde et al., 1997; Saini, 1997). Maize (Zea mays) exhibits embryo abortion soon after pollination in response to drought stress (Andersen et al., 2002), and a transcriptomic analysis of fertilized maize ovaries under water deficit revealed that many events, such as ABA signaling and senescence, were activated by drought (Kakumanu et al., 2012). Furthermore, Arabidopsis exhibits female reproductive organ abortion because of salt stress (Park et al., 2004; Sun et al., 2004). Another study uncovered altered expression of genes encoding 65 transcription factors in response to salt stress, including 6 ETHYLENE RESPONSE FACTOR (ERF)/AP2-family members and 11 NAC proteins (Sun et al., 2005). However, these studies did not characterize reproductive development.
To investigate the effect of drought on flower and fruit development in Arabidopsis, we devised a scheme of controlled drought stress that allows the plant to survive and reproduce. Plants treated with this scheme, with maintenance of 30 to 35% soil moisture, showed reduced productivity, similar to crops experiencing water deficits in the field. Morphological analysis demonstrated that drought affected several aspects of flower development, producing effects that include fewer flowers formed, decreased elongation of filaments, and delayed anther development and dehiscence. We also used microarray (Affymetrix ATH1 GeneChip) analysis to examine transcriptomic changes under drought, identifying thousands of genes that potentially mediate drought responses in the flower, including genes encoding transcription factors that likely play crucial regulatory roles. In particular, we found that DREB1A acts as an early drought response regulator in the Arabidopsis flower, and MYB DOMAIN PROTEIN21 (MYB21) is important for filament elongation under drought especially in the drought recovery process. Our analysis revealed distinct phases of responses at both the morphological and transcriptomic levels, supporting a regulatory cascade for reproductive acclimation to drought.
RESULTS
Sustained Water Stress Reveals Severe Effects on Growth and Water Content
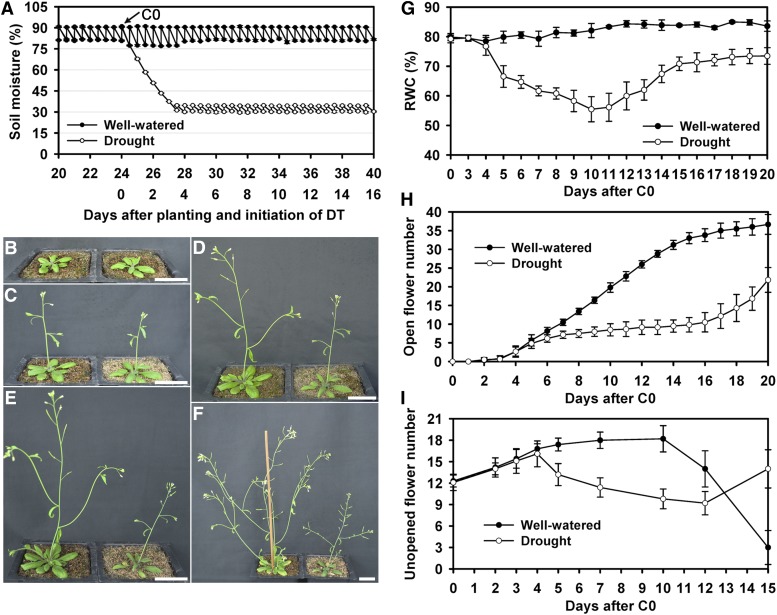
To allow Arabidopsis plants to survive and produce flowers and siliques under constant water deficit, we tested and optimized a scheme that both presented severe drought stress to the plants and permitted their continued growth and reproduction (see Methods for details). Briefly, wild-type Arabidopsis (Col-0) seeds were sown one per pot, and allowed to grow under well-watered (WW) conditions (80 to 90% soil moisture) for approximately 24 d, until they bolted, with approximately eight to nine true leaves and a main inflorescence stem (referred to as the main stem hereafter) of 1 cm (Figure 1). At this point (defined as control day 0, C0), half of the plants continued under WW, whereas the other half (drought-treated [DT]) was not watered for approximately 80 h, allowing the soil moisture to decrease to 35% at treatment day 3 (T3). Soil moisture was then maintained between 30% and 35% by daily addition of the appropriate amount of water as determined by lysimetry (Figure 1A) until tissues were harvested or seeds became mature. According to the definition of principal growth stages of Arabidopsis (Boyes et al., 2001), the drought treatment started from growth stage 5.90 to growth stage 9.70.
Figure 1.
Soil Water Moisture and Plant Growth during the Treatment Period.
(A) Soil moisture at d 20 to 40 from when the seeds were planted; the arrow points to control day 0 (C0) immediately before drought treatment. The data points and error bars represent the mean and se (n = 5).
(B) to (F) WW plants (left) and DT plants (right) at C0 (B), C3/T3 (C), C5/T5 (D), C7/T7 (E), and C10/T10 (F).
(G) RWC over the course of the treatment. The values for DT plants on days 5 to 20 are significantly different from those of the wild type (P < 0.01 by a Student’s t test.). The data points and error bars represent the mean and se (n = 9). Bar = 3 cm.
(H) The cumulative numbers of opened flowers on the main stem of WW plants and DT plants during the 20-d drought treatment. The DT plants differ significantly from the WW plants on days 6 to 20 (P < 0.01 by a Student’s t test.). The data points and error bars represent the mean and se (n = 6).
(I) The number of floral buds (from stages 8 to 12) on the main inflorescence stem, with significant difference between the DT and WW plants on days 5 to 12 (P < 0.01 by a Student’s t test.). The data points and error bars represent the mean and se (n = 6).
[See online article for color version of this figure.]
We observed that water deficit affected vegetative organ development (Figure 1; see Supplemental Figure 1 online). The inflorescences of WW (at C3) and DT (T3) plants had three to four opened flowers, but the main stem was shorter in DT plants than WW plants (Figure 1C). One day later (C4 and T4), the WW and DT plants had similar numbers of open flowers (five to six); however, the DT plants clearly had shorter main stems and branches than those of the WW plants (see Supplemental Figure 1G online). At C5, new rosette leaves formed on the WW plants, but not on the T5 DT plants (Figure 1D). The WW plants continued to generate new flowers, resulting in a total of seven to eight open flowers, whereas the DT inflorescences had fewer open flowers (six to seven; see below for more details). DT plants had more wilted leaves and reduced height than WW plants (Figure 1D). Two days later (T7), leaves of DT plants were obviously wilted, and the main stem was not upright (Figure 1E). Moreover, inflorescence development appeared to be arrested, with fewer newly opened flowers, in contrast to the WW plants, which had more newly open flowers (Figure 1E). At T8 to 9, the DT plants had no newly opened flowers (see Supplemental Figures 1I and 1J online). At T10, DT plants had only a few siliques and these were reduced in length, whereas the WW plants formed more branches with healthy siliques containing seeds (Figure 1F).
Relative water content (RWC) (Smart, 1974) was measured in the leaf as an indicator of plant water status. The leaf RWC of DT plants decreased from 76.67 ± 3.05% (at T4) to 66.47 ± 3.68% (T5), then 61.67 ± 1.65% (T7), and finally 55.47 ± 4.25% (T10), as the water availability decreased, similar to other reports (Gigon et al., 2004; Nilson and Assmann, 2010). In contrast, the WW plants had leaf RWC of approximately 80% throughout this period. We also found that the leaf RWC of DT plants gradually recovered to approximately 70% from T10 to T20 (Figure 1G). Therefore, this scheme of 30 to 35% soil moisture caused severe effects on plant growth yet still allowed reproductive development, and was used to investigate flower development under drought stress.
Drought Postponed and Reduced Arabidopsis Floral Maturation
Because drought appeared to affect flower maturation (production of open flowers), we examined the inflorescences over a period of 20 d (Figure 1; see Supplemental Figures 1K to 1X online) and found that the number of mature flowers formed on the main stem (the main stem was assayed unless otherwise noted) in the DT plants was significantly reduced (21.8 ± 3.3; n = 6) compared with the WW plants (36.7 ± 2.7; n = 6), with a P value of 6.51×10−6 (Student’s t test). The reduction was especially severe from T7 to T13, with zero to one newly opened flower each day (Figure 1H). In contrast, WW plants had steady production of two to four newly opened flowers every day from C4 to C13 (Figure 1H). The results suggested that the rapid expansion/elongation of floral organs during floral stages 8 to 12 (Smyth et al., 1990) might be particularly dependent on adequate water availability. In addition, the number of unopened floral buds at stages 8 to 12 increased from C0 to C3/T3 in both WW and DT plants, respectively (Figure 1I). Although the number of floral buds remained nearly constant in the WW plants for approximately10 d, the number dramatically decreased in DT plants (Figure 1I), with only about the half of the normal number at T10, indicating retardation of new bud formation and growth. The number of floral buds subsequently recovered in the DT plants by T15 (Figure 1I).
In short, in WW Arabidopsis plants, the rate of flower maturation increased after flowering and then reached a steady state. However, in DT plants, the oldest buds were able to mature (Figure 1I; see Supplemental Figures 1T to 1V online), but then the plants coped by reducing the rate of new bud formation and growth at stages 8 to 12, nearly stopping flower maturation for several days, before recovering and resuming flower development at a reduced steady state rate. These results suggest that Arabidopsis is able to respond to reduced water availability by developing and reproducing at a decreased rate, consistent with the reduction and subsequent partial recovery of RWC described in the previous section.
Flower Development Was First Arrested and Then Showed Partial Recovery
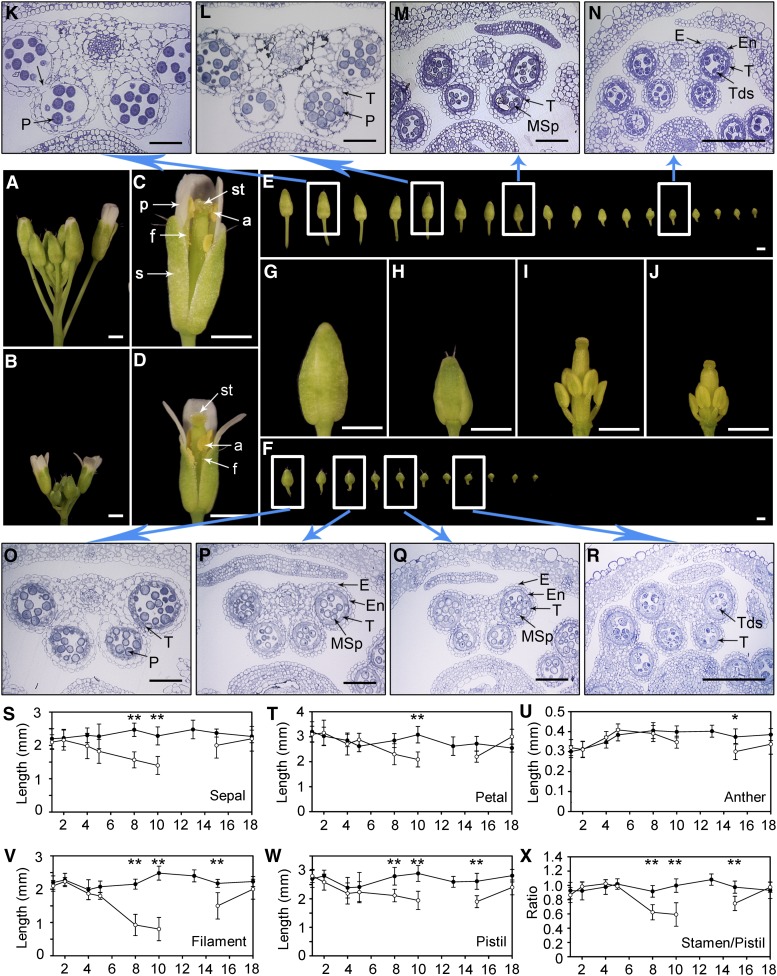
Next we examined flower development in WW (C10) (Figure 2A) and DT inflorescences (T10) (Figure 2B) at higher resolution. Open flowers of WW plants (Figure 2C) had long pistils and stamens of similar height, presumably allowing facile delivery of pollen, but open flowers from DT plants (Figure 2D) had underdeveloped filaments too short to reach the top of the pistil, which would impede pollen delivery. One or more nearly open floral buds with white petals just emerging from under the sepals were seen in WW plants (Figure 2A), but not in DT plants (Figure 2B); the unopened DT buds were much smaller than those in the WW plants, suggesting that they were immature (Figures 2E and 2F). WW plants had approximately 18 buds at stages 8 to 12, but the DT plants had only ∼11 buds at these stages (Figures 2E and 2F); most of the WW buds were longer and wider, with longer pedicels, than those from the DT plants. Dissected oldest buds from WW and DT plants (Figures 2G to 2J) revealed that in the DT bud, the sizes for all floral organs were reduced, particularly the filament length.
Figure 2.
Drought Affects Development of Reproductive Organs.
(A) An example of WW inflorescences at C10, with one opened flower and many floral buds.
(B) At T10, growth of DT inflorescences was retarded and flower buds were much smaller than those of WW inflorescences.
(C) The newest opened flower (one petal was removed) from the WW inflorescence of (A). The filaments were sufficiently long for pollen grains to be deposited onto the stigma.
(D) The newest opened flower (one petal was removed) from the DT inflorescence of (B). The filaments were apparently too short to deliver the pollen to the stigma.
(E) Floral buds from the WW inflorescence, shown from the oldest (left, floral stage 12) to the youngest (right, stage 8).
(F) Floral buds from a DT plant at stage 12 to stage 8.
(G) A stage 12 floral bud from a WW inflorescence.
(H) A stage 12 floral bud from a DT inflorescence.
(I) Sepals and petals were removed from the floral bud in (G).
(J) Sepals and petals were removed from the floral bud in (H).
(K) to (N) Anther cross-sections of a C10 WW inflorescence from the second floral bud (K), fifth bud (L), eighth bud (M), and fourteenth bud (N).
(O) to (R) Anther sections of a T10 DT inflorescence from the first bud (O), third bud (P), fifth bud (Q), and eighth bud (R).
(S) to (W) Lengths of sepals (S), petals (T), anthers (U), filaments (V), and pistils (W). The data points and error bars represent the mean and se (n = 6).
(X) The ratio lengths of stamens to pistils. *Significantly different (P < 0.05). **Highly significant (P < 0.01) by a Student’s t test. The data points and error bars represent the mean and se (n = 6).
Bar in (A) to (J) = 1 mm; bar in (K) to (R) = 50 µm.
An examination of anther sections of floral buds of WW (C10) plants showed that, in a stage 12 bud (just before opening), the anther lacked the tapetum as a result of degeneration and had begun to dehisce (Figure 2K, arrow). The fifth oldest bud had anthers with a degenerating tapetum surrounding mature pollen (Figure 2L). The eighth bud had smaller anthers with a thick tapetum and immature vacuolated pollen (Figure 2M). In the anther of the fourteenth bud, the tapetum surrounds tetrads with microspores produced from meiosis (Figure 2N). In the DT plants, the anther showed delayed development (Figures 2O to 2R), still having a thick tapetum surrounding the developing pollen grains in the oldest bud (Figure 2O), resembling an anther found in a much younger WW bud (Figure 2M). In the third and fifth DT buds, the pollen grains had large vacuoles, typical of stage 8 anthers (Figures 2P and 2Q). The anther of the eighth bud contained tetrads (Figure 2R), indicating the completion of meiosis at stage 7. These observations indicate that anther development in younger floral buds of DT plants was arrested at floral stages 9 to 10, with nearly mature pollen grains, possibly preserving the pollen for future release.
We then measured floral organ lengths of newly opened flowers (Figures 2S to 2X). Although the lengths of sepals (Figure 2S), petals (Figure 2T), and pistils (Figure 2W) were reduced slightly in the DT flowers, the length of filaments was decreased dramatically (Figure 2V), whereas the length of the anthers was close to that in the WW flowers (Figure 2U). The reduction in filament length was detectable at T5 and most severe at T10. From T10 to T14, the DT plants usually had no newly opened flowers, thus no data for organ lengths are available (Figures 2S to 2W). Subsequently, from T15 to T18, the flower organs showed recovery to near normal lengths. In self-pollinating Arabidopsis, pollination is facilitated because it has similar pistil and stamen lengths; in addition, the filament makes up most of the stamen length (Figures 2U and 2V). Because the length reduction is more severe for the filament than the pistil, the ratio of stamen to pistil is much less in DT flowers than in WW flowers, especially from T8 to T10 (Figure 2X). From T15 to T18, this ratio returned to near 1.0 (Figure 2X), which would allow pollination and subsequent seed production.
As a test for reproductive fitness, we examined pollen viability using Alexander’s stain (Johnson-Brousseau and McCormick, 2004) and found that the pollen from stage 12 WW floral buds stained a deep pink color, indicative of viability (see Supplemental Figure 2 online). Although the pollen from DT plants after ≤5 d of treatment displayed a similar color, the pollen from T7 and T9 flower buds had a lighter pink color (see Supplemental Figure 2 online), suggesting that the cytoplasmic activity might be slightly reduced. We also examined in vitro pollen germination and found that mature pollen grains (n > 500) from the WW plants had higher germination rates (67.2 ± 12.1%) than those from DT plants (38.6 ± 15.6%) after 7 d of drought treatment (see Supplemental Figure 2 online). The results suggest that pollen viability was reduced under long-term severe drought condition, although the in vitro assay provided sufficient water for pollen germination.
Productivity Shows a Biphasic Response to Controlled Drought
The observed negative effect of drought on flower development suggested that total ultimate yield would be reduced. To examine this aspect, the plants were allowed continue to develop until silique growth was complete, at approximately C35/T55. The WW plants first generated one to two nearly sterile siliques (as is normal under our conditions) and then produced more than 35 siliques (see Supplemental Figure 3A online), each containing 45 to 50 seeds (Figure 3V). However, the DT plants exhibited three different silique phenotypes (see Supplemental Figure 3B online). After the same initial one to two sterile siliques, the next two to three siliques were fertile but contained fewer seeds; these were from the nearly mature flowers that were able to open at the beginning of drought treatment when the soil moisture level decreased from 75% to 40% (Figure 1A). The next group of four to six siliques was sterile or nearly sterile, derived from the maturing floral buds when the plants first encountered severe drought at 30 to 35% soil moisture; these floral buds showed the most severe reduction of organ size. The next set of ∼20 siliques was fertile with reduced seed set of ∼25 seeds, gradually decreasing toward the top of the inflorescence (see Supplemental Figure 3B online). Therefore, the biphasic response of a short-term arrest (an acute phase) followed by a longer-term recovery at a reduced level (prolonged phase) observed for floral organ sizes is also manifest in terms of mature seed production. It is possible that an acute phase is needed for the DT plants to reprogram reproductive development to enter the prolonged phase; this prolonged response phase then uses the limited water resource to support both flower and silique development, albeit with fewer flowers and seeds per silique.
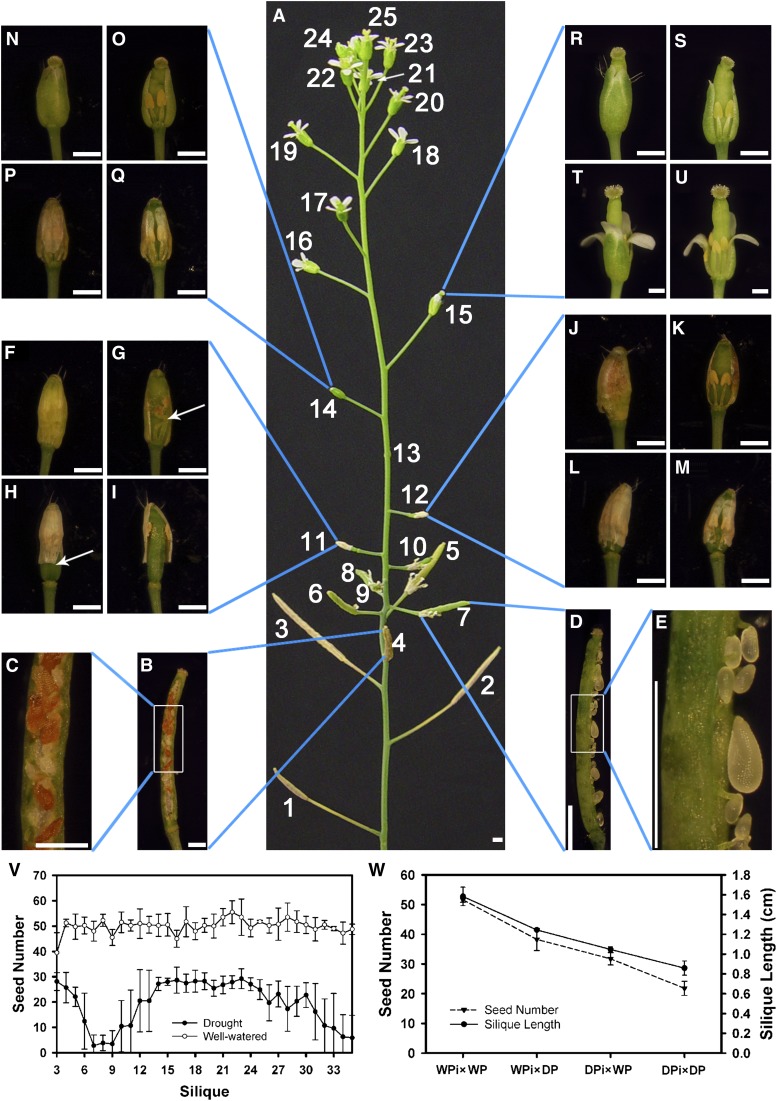
Figure 3.
Flower Development after 20 d of Drought Treatment.
(A) The main inflorescence stem of a DT plant photographed at T20. The fruits and flowers are labeled from the oldest to the youngest (B) to (G), (J), (K), (N), (O), (R), and (S) were from (A). (H), (I), (L), (M), (P), (Q), (T), and (U) were from another inflorescence at the same positions as indicated by the blue lines.
(B) The fourth fruit from (A) after dissection.
(C) A portion of the fourth fruit with higher magnification.
(D) The seventh fruit with fresh ovules and seeds shown.
(E) A portion of (D) with higher magnification, showing normal and small ovules.
(F) The eleventh bud.
(G) The eleventh bud after dissection, showing reduced filament length (arrow).
(H) The eleventh bud after maturation, showing the detachment at the base of sepals that were not separated at the top.
(I) The dissected bud of (H), showing detached short stamens.
(J) The twelfth bud.
(K) The dissected twelfth bud with short filaments.
(L) The twelfth bud after maturation with sepals detached at the base.
(M) The dissected bud from (L).
(N) The fourteenth bud.
(O) The dissected fourteenth bud.
(P) The fourteenth bud after maturation.
(Q) The dissected bud from (P).
(R) The fifteenth bud.
(S) The dissected fifteenth bud.
(T) The fifteenth bud after maturation.
(U) The bud from (T) with two sepals and a petal removed.
(V) The comparison of seed count per silique on the main stem from WW and DT plants. P < 0.01 by a Student’s t test. The data points and error bars represent the mean and se (n = 5).
(W) The average seed number and silique length of different crossing plants: WW plants (WPi×WP), WW pistil pollinated with DT pollen (WPi×DP), DT pistil pollinated with WW pollen (DPi×WP), and DT plants (DPi×DP). The data points and error bars represent the mean and se (n = 3).
Bar in (A) to (T) = 1 mm.
To illustrate the biphasic response, we examined plants that had been treated for 20 d (T20), when they had produced a number of siliques and were still producing flowers (Figure 3A). At this time, the first 10 flowers had passed the time of pollination and exhibited different degrees of fertility, consistent with what we observed for older plants as described above. In particular, from the third to the ninth silique, the siliques were progressively shorter (Figure 3A). In the fourth silique, there were shrunken seeds and white aborted ovules (Figures 3B and 3C), suggesting that pollen, ovule, and/or seed development were affected. In the seventh silique, only a few seeds formed and most ovules appeared aborted (Figures 3D and 3E), indicating a greater effect of drought. Other siliques (fifth, sixth, eighth, and ninth) were also sterile or nearly sterile, illustrating a component of the acute response.
The eleventh flower bud, the first unopened bud on the stem, had anthers with normal appearance and elongated filaments that were bent, suggesting that the sepals had not opened and so the internal organs could not extend further (Figures 3F and 3G). The sepals of similar buds still adhered to each other 5 d (T25) later, even though they had separated from the base of the flower bud (Figures 3H and 3I). Similar sepal phenotypes were also observed for the twelfth (Figures 3J to 3M) and the thirteenth flowers. In the fourteenth flower, the pistil protruded out of the sepals, but the filaments were short (Figures 3N and 3O), presumably resulting in ineffective pollination (Figures 3P and 3Q). In the fifteenth flower, the pistil was even more obviously longer than the sepals (Figures 3R and 3S), leading to possible failure in pollination (Figures 3T and 3U). Strikingly, T20 plants had recovered and entered the prolonged phase of drought response, with a burst of blossom, illustrated here by the plant in Figure 3A, with 10 mature flowers (numbers 16 to 25) that had just opened within 2 d, consistent with the earlier observation that floral organ sizes recovered to close to normal sizes by T15 (Figures 2S to 2X). These and later flowers then led to the subsequent generation of relatively fertile siliques, as shown in Supplemental Figure 3B online.
To test the effect of drought on female reproductive development, we performed reciprocal crosses between WW and DT plants, with additional crosses as controls (Figure 3W; see Supplemental Figures 3B and 3C online), and determined the silique length and seed number 2 weeks later. The results indicated that drought also reduced female fertility in addition to negative effects on pollen development, although there was likely also an effect of drought on the plants (other than the female reproductive organs).
Overview and Analysis of Gene Expression in Flower under Drought Stress
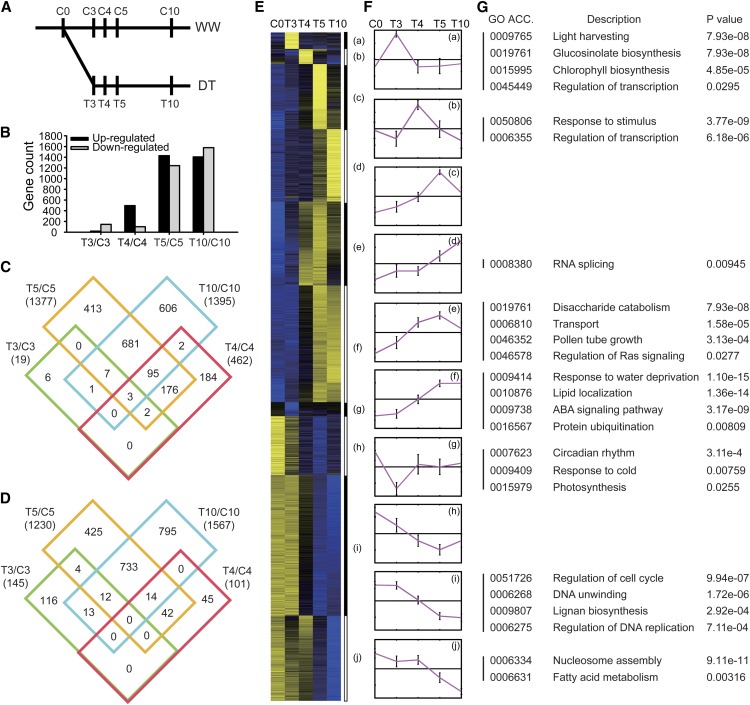
The dramatic reprogramming of reproductive growth in response to drought suggested likely changes in gene expression. To characterize the transcriptomes of unopened floral buds in response to drought, we analyzed RNA samples from WW (C0, C3, C4, C5, and C10) or DT plants (T3, T4, T5, and T10). The expression data were highly reproducible (Pearson correlation coefficients >0.985) (Figure 4A; see Supplemental Figure 4 online). Using q-values of less than 0.01 for significantly different expression and at least twofold change, we identified a total of 4153 genes that were differentially expressed between the WW and DT samples on the corresponding days (T3 versus C3, T4 versus C4, etc) (see Supplemental Data Set 1 online). The number of differentially expressed genes increased moderately from 164 on T3 (19 upregulated and 145 downregulated) to 563 on T4 (462 upregulated and 101 downregulated), then dramatically to 2607 genes on T5 (1377 upregulated and 1230 downregulated), and finally increased further only slightly to 2962 on T10 (1395 upregulated and 1567 downregulated) (Figure 4B; see Supplemental Table 1 online). Among the 19 genes upregulated at T3, the majority (13) also increased on T4, T5, and/or T10. Similarly, the majority (278 among 482) of the upregulated genes on T4 were also higher at T5 and/or T10 (Figure 4C). Therefore, most of the early induced genes tended to remain induced. By contrast, the majority (116 among 145) of downregulated genes on T3 were not persistently reduced at T4, T5, or T10 (Figure 4D), suggesting that the decreased expression of these genes was a transient response. Among the genes that were upregulated (1395) or downregulated (1567) on T10, 786 and 759, respectively, were also increased on T5 (Figures 4C and 4D), indicating that many of the transcriptomic changes had occurred by day 5.
Figure 4.
Analysis of Transcriptomes from Inflorescences of WW and DT Plants.
(A) Samples were collected at C0, C3, C4, C5, and C10 for WW plants and T3, T4, T5, and T10 for DT plants.
(B) The numbers of the differentially expressed genes for T3/C3, T4/C4, T5/C5, and T10/C10.
(C) Venn diagram analysis of all upregulated genes of each group.
(D) Venn diagram analysis of all downregulated genes of each group.
(E) K-means clustering of differentially expressed genes. Expression distribution of genes activated or repressed 3 d after drought treatment. The number indicates log2 ratio of the fold change between the DT groups with the control group before start of the drought treatment (C0). Yellow color represents genes that have higher expression levels after drought stress and blue indicates reduced expression.
(F) The pink lines represent the mean expression profiles for each cluster. The y-axis is in log2 scale and the x-axis shows log2 (T0/C0), log2 (T3/C0), log2 (T4/C0), log2 (T5/C0), and log2 (T10/C0) in sequence.
(G) The GO enrichment analysis of genes in each cluster, with P values.
Coexpression Pattern of Drought-Responsive Genes in Flowers
To further examine the floral transcriptomic response to drought, we applied hierarchical clustering to all 4153 genes differentially expressed between WW and DT floral buds, resulting in 10 large clusters. Using the K-means (K = 10) approach in the hierarchical clusters, genes were assigned to 1 of 10 clusters, which were then visualized with heat map and centroid views (Figures 4E and 4F), revealing general patterns of transcriptomic profiles during the drought treatment.
The first four clusters (Figures 4Fa to 4Fd) contained genes with drought-induced significant increase in expression that peaked on days 3, 4, 5, and 10, respectively, suggesting that their functions might be particularly important in a transient fashion. Clusters 5 and 6 (Figures 4Fe and 4Ff) contained genes with high levels of expression at two time points, suggesting more sustained functions, with the genes in cluster 5 acting earlier than those in cluster 6. In contrast, the genes from clusters 7 to 10 (Figures 4Fg to 4Fj) exhibited various patterns of drought-induced decrease in expression. The genes in cluster 7 (Figure 4Fg) showed a transient decrease in expression on day 3, whereas those in cluster 8 (Figure 4Fh) had persistent reduction from T4 to T10. The expression of genes in clusters 9 (Figure 4Fi) and 10 (Figure 4Fj) showed reduced expression at T5, with further reduction on T10 for genes in cluster 10.
Functional Prediction of Different Drought-Responsive Gene Clusters
To gain further insight into the changing transcriptomic landscapes in the DT flowers, we performed gene ontology (GO) annotation enrichment analysis using singular enrichment analysis provided by agriGO (Du et al., 2010). In both clusters 1 and 2, genes involved in transcriptional regulation were enriched (Figure 4G), suggesting that changes in transcriptional regulation were one of the early responses to drought stress. On the other hand, in cluster 4, the genes that function in RNA splicing were enriched, suggesting a potential increase in the level of protein diversity attributable to alternative splicing in the response at day 10. In addition, genes for glucosinolate biosynthesis were enriched in cluster 1, suggesting a possible function in drought response; this is consistent with a function of glucosinolates in the regulation by ABA of stomatal opening (Zhao et al., 2008) and is possibly related to the role of glucosinolates as regulators of biotic response (Hopkins et al., 2009).
Among the genes enriched in cluster 5 (e) are genes related to pollen tube growth, particularly GTP binding signal proteins and disaccharide catabolism (Figure 4G). For example, RHO-RELATED PROTEIN FROM PLANTS 1 (ROP1) and ROP-INTERACTIVE CRIB MOTIF-CONTAINING PROTEIN 6 (RIC6) function in the polarized growth of pollen tubes (Wu et al., 2001; Hwang et al., 2005). The genes encoding disaccharide enzymes such as β-galactosidase 7 (BGAL-7), BGAL-11, and BGAL-13, which play a role in pollen development and fertilization (Jakobsen et al., 2005; Wang et al., 2008), also showed elevated expression, suggesting that enhanced activity of these gene products is needed to promote pollen development and function under drought stress. Genes related to disaccharide catabolism (7.93e-08) and transport (1.58e-05) are highly enriched, which suggests the possible regulation of transport of sugars and other osmolytes in response to drought. At a later time in cluster 6 (Figure 4Ff), genes responsive to water deprivation and ABA-mediated signaling pathways were highly enriched. Among them are regulators of ABA signaling, including ABI1, ABI2, ABI5, ABF1, ABF3, and ABF4, and genes encoding PP2CA, SNRK2.3, and SNRK2.6 (OST1) proteins; these genes are known to function in ABA-mediated responses to abiotic stresses (Luan, 2003; Cutler et al., 2010; Yoshida et al., 2010). In addition, genes functioning in the ABA-independent pathway, such as those coding for DREB1B, DREB2A, DREB2B, RD20, and RD22 (Bernoux et al., 2008; Matsukura et al., 2010), were enriched (Figure 4G).
Among the downregulated clusters, genes related to photosynthesis and circadian rhythm were enriched in cluster 7 (g) (Figure 4G), suggesting that these functions were reduced transiently as an early drought response. In cluster 9 (i), the genes for cell cycle regulation and DNA replication were enriched (Figure 4G), including those coding for cyclins and cyclin-dependent kinases. In cluster 10 (j), genes for nucleosome subunits were highly enriched (Figure 4G), including those for histone H2A, histone H2B, histone H3, and histone H4. The reduced expression of the genes in clusters 9 and 10 suggests that cell proliferation was reduced soon after the drought treatment, consistent with the observed morphological changes.
Early Induction of ABA and Jasmonic Acid Signaling Genes
In addition to the analysis of GO categories and enrichment, we also examined the early regulation of drought response. From the phenotypic analysis, we found that after the onset of reproductive development, both the WW and DT plants continued to grow, with increased rate of floral maturation in the WW plants (Figure 1), suggesting that floral gene expression might change during this period. Indeed, 517 genes were induced (twofold change with a q-value less than 0.01) at C3 compared with C0. To identify genes that specifically responded to drought, we compared the T3 data with those from either C0 or C3 (see Supplemental Figure 5C online). A total of 98 genes were upregulated in T3 compared with both C0 and C3, such as DREB1A, ABF, NAC DOMAIN CONTAINING PROTEIN19, RD20, and RD29A, which are known as stress-responsive regulators involved in ABA-independent and ABA-dependent pathways (Jiang et al., 2009; Lata and Prasad, 2011) and jasmonate response (Bu et al., 2008). Furthermore, genes for putative negative regulators of ABA signaling, PP2CA, HAI1, and HAI2, were also induced (Table 1), suggesting the importance of a fine balance of positive and negative regulation in the floral response to drought. Therefore, the early floral transcriptomic responses to drought included components that were previously identified for the vegetative phase (Urano et al., 2010; Bhaskara et al., 2012). A gene encoding a key enzyme for methyl jasmonate (MeJA) synthesis, jasmonic acid carboxyl methyltransferase (JMT) (Seo et al., 2001), was also induced at T3. AT5G63380, which is involved in jasmonic acid (JA) biosynthesis (Schneider et al., 2005), was also upregulated. JA signaling is important not only for biotic stress but also abiotic stress and development; in addition, the crosstalk between ABA and JA signaling contributes to the balance between growth and defense (Lackman et al., 2011).
Table 1. Drought-Related Genes Showing Early Response in Flowers at T3.
| AGI | Description | T3/C0 | q-Value |
|---|---|---|---|
| ABA-responsive positive regulator | |||
| AT1G52890 | NAC DOMAIN CONTAINING PROTEIN19 | 1.26 | 5.42E-05 |
| AT4G34000 | ABA RESPONSIVE ELEMENTS-BINDING FACTOR3 | 1.67 | 4.27E-08 |
| AT5G52310 | RESPONSIVE TO DESSICATION29A | 2.09 | 1.25E-07 |
| AT2G33380 | RESPONSIVE TO DESSICATION20 | 1.00 | 1.74E-07 |
| AT5G66400 | RESPONSIVE TO ABA18 | 1.36 | 2.97E-05 |
| AT1G69260 | ABI FIVE BINDING PROTEIN | 1.48 | 1.08E-07 |
| AT5G13630 | H SUBUNIT OF MG-CHELATASE | 1.20 | 1.18E-08 |
| ABA-responsive negative regulator | |||
| AT3G11410 | PROTEIN PHOSPHATASE 2CA | 1.09 | 1.89E-05 |
| AT5G59220 | HIGHLY ABA-INDUCED PP2C GENE 1 | 1.01 | 2.37E-05 |
| AT1G07430 | HIGHLY ABA-INDUCED PP2C GENE 2 | 1.49 | 1.83E-07 |
| ABA-independent regulator | |||
| AT4G25480 | DEHYDRATION RESPONSE ELEMENT B1A | 1.45 | 8.39E-08 |
| MeJA biosynthesis enzyme | |||
| AT1G19640 | JASMONIC ACID CARBOXYL METHYLTRANSFERASE | 1.01 | 3.97E-05 |
These genes showed an early response under drought treatment in flowers (98 genes, in the a, b, and e groups of Supplemental Figure 5C, are significantly induced at T3 compared with C0 or C3 with at least twofold difference and q-values < 0.01, but not in the group of genes induced at C3 compared with C0; the latter genes likely reflect changes in inflorescence development independent of drought). The fold changes (T3/C0) are calculated from the average expression values of DT samples (T3) to those of the WW control (C0).
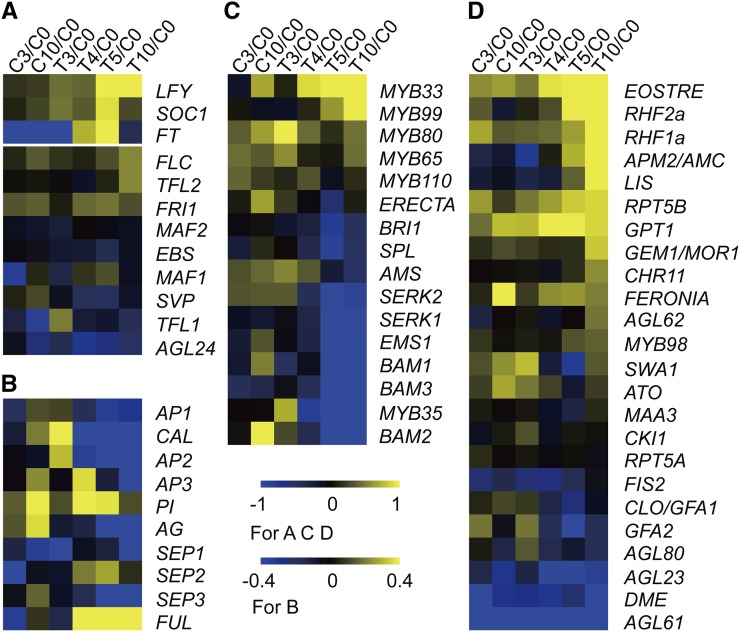
The Expression of Floral Genes Was Affected by Drought
To determine whether flowering genes were affected by drought, we examined the expression of positive regulators such as LFY, SOC1, and FT and negative regulators FLC, TFL1, TFL2, AGL24, MAF1, MAF2, and FRI1 (Putterill et al., 2004; Bernier and Périlleux, 2005; Jung and Müller, 2009). We found that LFY, SOC1, and FT were upregulated by drought after 4 or 5 d of treatment. In contrast, the negative regulators were not obviously induced under drought (Figure 5A; see Supplemental Figure 6A online). From C3 to C10 in the flowers of WW plants, the expression of the ABC function genes controlling floral organ identity (Ma, 2005) increased slightly or was maintained at similar levels. However, their expression increased slightly during early drought treatment and then decreased with further drought (Figure 5B; see Supplemental Figure 6B online). These results suggest complex drought effects on floral regulatory genes. Because the ABC genes also function during floral organ development, their expression at T3 to T5 might have supported the completion of bud development, whereas their reduced expression levels at T10 are consistent with the transient developmental arrest of floral buds. The increase of LFY, SOC1, FT, and FUL expression at T4 and T5 might be needed for the recovery of floral development shown in Figures 2 and 3.
Figure 5.
The Expression of Floral Genes Was Affected by Drought.
(A) Expression heat maps for relative expression of flowering time related genes.
(B) Expression heat maps for relative expression of ABC model genes.
(C) Expression heat maps for relative expression of anther development related genes.
(D) Expression heat maps for relative expression of pistil development related genes.
The relative expression levels (log2) above 0 represent upregulation, whereas those below 0 represent downregulation.
Anther development requires a number of genes, including SPL, AMS, EMS1, DYT1, MYB33, MYB35, MYB65, MYB80, MYB99, MYB110, BAM1, BAM2, BAM3, and ERECTA (Zhao et al., 2002; Hord et al., 2006; Zhang et al., 2006; Wijeratne et al., 2007; Hord et al., 2008). A majority of these genes were repressed by drought stress (Figure 5C; see Supplemental Figure 6C online), providing an explanation for the observed retardation of anther development under drought. However, some genes for pistil development were induced by drought, such as AGL80, EOSTRE, RHF1a, RHF2a, and GPT1 (Niewiadomski et al., 2005; Portereiko et al., 2006; Pagnussat et al., 2007; Liu et al., 2008). The expression levels of other pistil genes were maintained at nearly normal levels (Figure 5D; see Supplemental Figure 6D online). These results are consistent with the observation that drought stress did not affect the development of the pistil as severely as it did anther development.
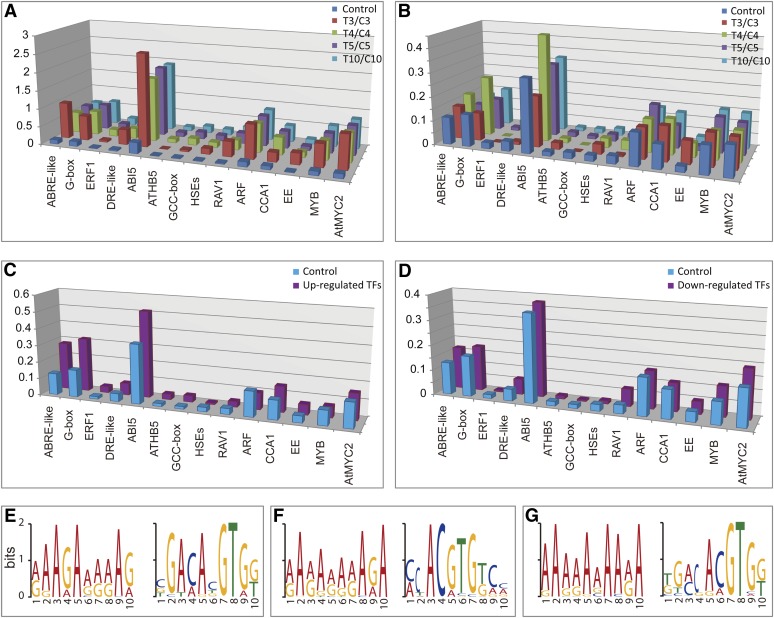
Cis-Regulatory Element Analysis of the Differentially Expressed Genes
The changes of gene expression under drought, especially those encoding transcription factors, suggested that transcriptional regulation is an important mechanism of floral response to drought. To obtain further evidence for a drought-responsive transcriptional network, we analyzed cis-regulatory elements (CREs) for the putative promoter regions of the differentially expressed genes. First, we examined the CREs within 500 bp of upstream transcription start site (TSS), a region that is commonly analyzed as putative promoter region and has a high frequency of CREs distribution (Wang et al., 2011). We found that many CREs were overrepresented in the 500 bp upstream of the 2176 upregulated genes but not in the downregulated genes (Figures 6A and 6B; see Supplemental Data Set 2 online), suggesting that these CREs might mediate drought-responsive transcriptional regulation. Among the promoter regions of 214 upregulated transcription factors, the G-box (ABRE or other similar ones) (P = 1.54E-04) and DC3 PROMOTER-BINDING FACTOR (P = 3.16E-03) were overrepresented (Figure 6C; see Supplemental Data Set 3 online); however, no CREs were found to show a greater frequency in the promoter regions of downregulated transcription factors than upregulated transcription factors (Figure 6D; see Supplemental Data Set 3 online). For comparison, we also examined regions 1000 bp and 3000 bp upstream of the TSS, and found that only the G-box or the related ABRE and ABRE-like binding sites were overrepresented in the drought-induced genes compared with promoter regions of all annotated genes (see Supplemental Figure 7 and Supplemental Data Set 2 online), suggesting that G-box and ABRE-like sites might be particularly important in mediating drought-induced transcriptional regulation in the Arabidopsis flower, similar to the finding from responses in vegetative tissue (Dolferus et al., 1994; Freeling et al., 2007).
Figure 6.
CRE Analysis of Differentially Expressed Genes and Prediction of New CREs.
(A) CRE analysis within the 500 bp promoter region of all upregulated genes.
(B) CRE analysis within the 500 bp promoter region of all downregulated genes.
(C) CRE analysis within the 500 bp promoter region of all upregulated transcription factors.
(D) CRE analysis within the 500 bp promoter region of all downregulated genes encoding transcription factors.
(E) Sequence logo of CRE prediction for the differentially expressed genes at T4.
(F) Sequence logo of CRE prediction for the differentially expressed genes at T5.
(G) Sequence logo of CRE prediction for the differentially expressed genes at T10.
To obtain evidence for other possible CREs, we predicted putative CREs in the putative promoters of differentially expressed transcription factor genes at T3, T4, T5, or T10 using Multiple Em for Motif Elicitation (MEME) software (Figures 6E to 6G). We found that the motif RRRRRRRRRR (R = A/G) was at high levels in the promoters of upregulated genes for transcription factors and was located close to the TSS, suggesting that this motif might be a novel transcriptional enhancer. The G-box (CACGTG) and G-box–like binding sites were also detected at high levels in promoter regions of drought-induced transcription factor genes, supporting an important role of these motifs in drought response during flower development.
A Potential Early Role for DREB1A in Drought Response in Flower
Among the genes induced by drought, 19 were increased by at least twofold at T3 compared with C3 (see Supplemental Table 2 online); only one of these genes, DREB1A, encodes a transcription factor, suggesting a key role in the early response to drought stress in the flower. DREB1A is one of the 65 Arabidopsis DREB transcription factors in the ERF superfamily (Lata and Prasad, 2011), and is a known regulator of cold response (Liu et al., 1998; Shinwari et al., 1998). Other DREB genes that play important roles in drought and cold responses (Shinozaki and Yamaguchi-Shinozaki, 2007) were not significantly induced in the flower at T3 (see Supplemental Figure 8 and Supplemental Table 3 online).
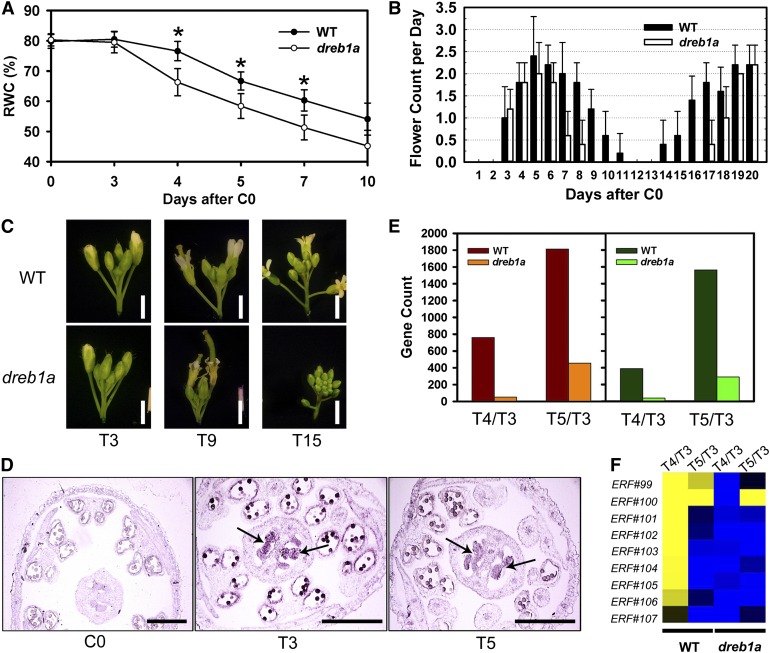
The dreb1a T-DNA mutant (CS872453) (see Supplemental Figure 8 online) showed an early wilting phenotype and was hypersensitive to water deprivation with significantly lower leaf RWC than the wild type from T4 to T7 of the drought treatment (Figure 7A; see Supplemental Figure 9 online). In the prolonged drought response, the dreb1a mutant showed a late recovery phenotype from the extreme environment (Figure 7B). At T3, both wild type and MT had healthy inflorescences with newly open flowers. However, at T9, flower opening was already arrested in dreb1a but not in wild type. At T15, wild-type plants recovered from the drought response with opening of new flowers, but in dreb1a floral buds remained closed (Figure 7C; see Supplemental Figure 10 online). Viability of dreb1a pollen was similar to that from wild-type plants as determined by in vitro germination assay (see Supplemental Figure 9 online), suggesting that it might not be critical for pollen development under drought. In situ hybridization showed that DREB1A was induced at relatively high level in the pistil after 3 d of drought treatment (Figure 7D). Therefore, among DREB genes, DREB1A likely plays an important and specific role in very early response to drought stress in the Arabidopsis flower.
Figure 7.
Phenotypes and Gene Expression of the dreb1a Mutant.
(A) Leaf RWC of dreb1a compared with the wild type. Asterisks indicate values that are significantly different with P < 0.01 by a Student’s t test. The data points and error bars represent the mean and se (n = 6). WT, Wild type.
(B) Number of newly opened flowers each day during the DT from C0 to T20. The data points and error bars represent the mean and se (n = 8). WT, Wild type.
(C) Phenotype of the dreb1a inflorescence compared with that of the wild type under drought. WT, Wild type.
(D) DREB1A was not expressed in the wild type under WW condition at day 0 but was expressed at a high level in the wild-type ovule under DT at days 3 and 5 (arrows).
(E) Numbers of differentially expressed genes at T4/T3 and T5/T3 in dreb1a and the wild type. Warm colors indicate upregulated genes and cool colors indicate downregulated genes. WT, Wild type.
(F) A heat map of IXa and IXb subfamilies of ERF genes. WT, Wild type.
Bar in (C) = 2 mm; bar in (D) = 200 µm.
We also performed microarray experiments with the ATH1 gene chips using mRNAs of the dreb1a flowers at T3, T4, and T5 and found that dramatically fewer genes were differentially expressed due to drought than those in the wild type (Figure 7E). At T4, only 51 genes were upregulated compared with T3 in dreb1a, in contrast to 760 in wild type. At T5 in dreb1a, there were only 454 genes upregulated compared with T3, whereas the wild type had many more (1813) genes significantly induced by drought. We also found that there were many fewer genes repressed compared with the wild type, with 39 genes at T4/T3 and 290 genes at T5/T3. Furthermore, genes involved in response to ABA signaling, water deprivation, and salt stress were highly enriched among genes induced by drought at T3 in the wild type but not induced by drought in dreb1a at T3 (see Supplemental Figure 8 online). These results suggest that DREB1A is a key regulator of drought response in Arabidopsis flowers.
To gain a better understanding about the function of DREB1A in the context of other members of the ERF subfamily of genes encoding AP2-domain proteins, we analyzed the expression patterns of all ERF genes and found that the genes in IXa and IXb subgroups (Nakano et al., 2006) showed drought-affected expression changes in a different manner in dreb1a from those observed in wild-type plants under drought (Figure 7F). In the wild type, this group of genes was transiently and highly induced by drought at T4, but interestingly, there was no induction found in the dreb1a mutant. Among these, ERF99 to ERF107, also named AtERF1, AtERF2, AtERF5, AtERF6, and AtERF13 (Nakano et al., 2006), are known to have specific binding activity for GCC boxes and function in the regulation of biotic stress responses by ethylene and JA (Fujimoto et al., 2000; Oñate-Sánchez and Singh, 2002; Moffat et al., 2012). Our results suggest that these genes also potentially function in response to water deprivation during flower development.
MYB21 Functions in Promoting Fertility under Drought
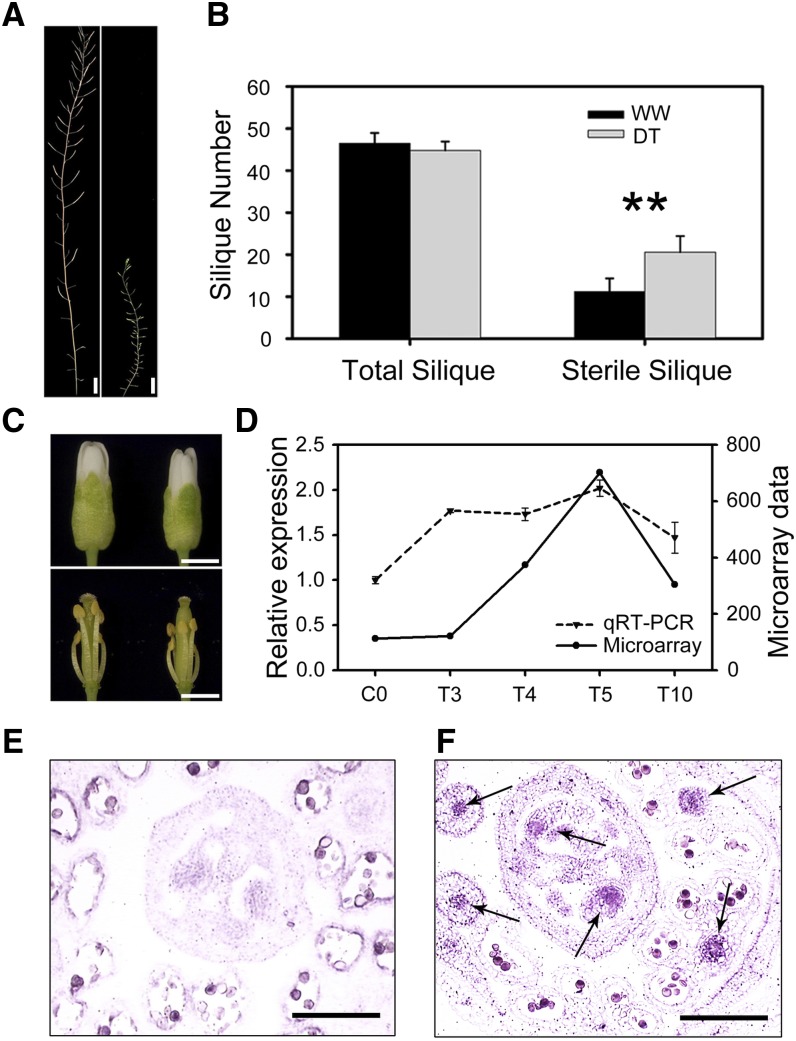
MYB21 is known to regulate filament elongation and anther dehiscence (Mandaokar et al., 2006). The expression of MYB21 increased at T4, suggesting that MYB21 is involved in the drought response in the flower. Although the knockout mutant myb21-2 (SALK_042711C) under WW conditions exhibited some defects in stamen development and reduced fertility of early siliques, the DT myb21-2 mutant showed an even more severe phenotype than WW plants (Figure 8A), with nearly twice as many sterile siliques (Figure 8B) compared with the wild-type plants. These observations indicate that MYB21 is important for the fertility that was maintained in the wild type under drought. In addition, we compared the floral buds from both the wild type and the myb21 mutant under drought and found that when the wild type had partially recovered, the mutant still showed the defect in filament elongation, resulting in a more severe pollination defect in the mutant than in the wild type (Figure 8C). Both microarray and quantitative RT-PCR showed induction of MYB21 in inflorescence under our drought assay (Figure 8D); in situ hybridization results showed MYB21 was highly expressed in ovule and filament in wild type after 5 d of drought treatment but not in WW plants (Figures 8E and 8F). Therefore, MYB21, through promotion of filament elongation, is likely an important component of the partial recovery after the severe drought response.
Figure 8.
Molecular and Morphological Analyses of myb21 Mutants in Response to Severe Drought Stress.
(A) myb21 main stem under the WW condition (left) and under DT (right).
(B) Total fruit number and the sterile silique number on the main stem under the WW condition and DT. Asterisks indicate values that are significantly different with P < 0.01 by a Student’s t test. The data points and error bars represent the mean and se (n = 6).
(C) Wild type (top left) and myb21 (top right) stage 13 floral buds after 20-d drought treatment and the phenotypes after removing sepals and petals (bottom panel).
(D) Quantitative RT-PCR and microarray results of MYB21 in the wild type under drought at days 0, 3, 4, 5, and 10. Mean and sd of two biological replicates are presented in each case.
(E) In situ hybridization results showed MYB21 expression was at a very low level in the wild type under WW conditions at day 5.
(F) MYB21 expression was at a high level in wild type ovule and filament under drought treatment at day 5 (arrows).
Bar in (A) = 1 cm; bar in (C) = 2 mm; bar in (F) = 200 µm.
DISCUSSION
Plant responses to different environmental stresses have been studied extensively, revealing that responses to drought during vegetative development include changes in cellular metabolism and in growth and yield (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). The acclimation to drought includes induction of plant defensive genes, repression of plant developmental genes, enhancement of stomata closure, inhibition of cell proliferation and differentiation, and generation of reactive oxygen species (Xiong et al., 2002; Xu et al., 2010). However, our understanding of how plant reproductive tissues respond to such stresses remains very limited. In this study, using a scheme for soil water deficit (Figure 1) to simulate environmental drought stress, we investigated how flower development was altered in response to drought and how the floral transcriptome was reprogrammed during the drought treatment. We found that flower number and size were both reduced, floral organ lengths, especially the ratio of stamen to pistil length, were less than normal, and anther and pollen development were affected, resulting in a decrease in seed production. In maize, Boyer et al. also found that flower development was sensitive to water limitation especially during the early phases; ovaries and pollen were affected due to inhibition of invertase activity and sugar transport at low water potential (Boyer and Westgate, 2004; McLaughlin and Boyer, 2004; Mäkelä et al., 2005). At the transcriptome level, we also found that the number of genes differentially expressed during early drought treatment was relatively small, increasing dramatically soon afterward, providing a molecular explanation for the morphological changes in the flower under drought.
General Properties of Plant Response to Drought in Reproductive Development
From our observation, 5 d of severe drought treatment resulted in the reduction of the leaf RWC to below 70% (Figure 1C), which has been shown to be a critical point for wilting for many crop species (Sengupta and Majumder, 2009). Plants respond to low RWC by reducing their growth, and in this study, the floral organs in DT plants were smaller than those in WW plants. Simultaneously, the number of floral buds (stages 8 to 12) decreased under drought (Figure 2), suggesting that initiation of floral buds was sensitive to water deficit. This supports the idea that stressful environments could limit cell division in the meristem and the transport of nutrients to different plant tissues (Nilcolas et al., 1985). On the other hand, the nearly mature floral buds (stages 11 and 12) could open during very early drought treatment, suggesting that the plant devoted limited water resources to completing flower development. At the same time, younger buds initially failed to mature, with unopened sepals that inhibited the extension of internal organs (Figure 3). Therefore, one possible strategy by which plants may cope with recent drought stress is to sacrifice some young floral buds and to reduce new bud formation, thereby allowing the use of the limited water and nutrients to support the continued development of older flowers and immature seeds.
After the plants had experienced persistent low water availability, flower development improved compared with the initial severe response, with resumption of floral maturation and less reduction of floral organ lengths. These flowers would later produce siliques with reduced seed number compared with the siliques from WW plants. Therefore, the plant had switched to a prolonged response with less extreme phenotypes and sustained, albeit reduced, seed production. Plants may regulate seed abortion, thus providing availability of the limited water and nutrients for the development of fewer seeds, rather than evenly distributing the nutrients to all young seeds with the potential consequence of the underdevelopment of all seeds. This suggests that drought can induce, as part of a long-term acclimation, physiological and developmental changes of the source sink balance such that limited yield is protected under extreme conditions. This is consistent with a previous report that plants use a series of mechanisms to regulate vegetative growth and reproduction to maximize plant fitness (Liu et al., 2003). Plants can adjust the expenditure of maternal resources at different stages of plant development, which controls the flower number and further development of reproductive cells (Lloyd, 1980). In wheat, anther sink strength is also important for maintenance of pollen fertility and grain number under water stress, and such reprogramming also agrees with models for plant acclimation to stressful conditions that alter plant fertility by adjusting resource distribution (Sun et al., 2004; Ji et al., 2010).
Preferentially Expressed Genes Reveal Their Roles in Drought Response
The GO enrichment for the upregulated (2176) and downregulated (2199) genes suggests the crucial roles of those enriched groups involved in response to drought. Among the upregulated genes, two enriched groups were annotated as stress responsive and reproductive development related (see Supplemental Figure 5A). In particular, the highly enriched genes included those predicted to respond to stimuli, especially the hormone ABA and water deprivation, suggesting that they are important for drought response in the developing flower. In addition, genes annotated for pollen tube growth and postembryonic development were also enriched, suggesting an enhancement of these functions under drought. The enriched pollen tube-related genes, such as the symporters SHAKER POLLEN INWARK K+ CHANNEL and PEPTIDE TRANSPORTER 5, are known or predicted to function in transporting ions, protons, and peptides (Mouline et al., 2002; Hammes et al., 2010), highlighting the potential importance of osmotic adjustment, signaling transduction and starvation response in the flower under drought.
Furthermore, cell wall-modifying enzyme pectinesterases (PMEs) and their inhibitors (PMEIs), such as VGD1 and PMEI2, are important for pollen tube growth (Röckel et al., 2008). Pectins are important structural determinants of the cell wall in dicots and their esterified forms are associated with active cell expansion or growth such as pollen tube tip growth. PMEs de-esterify pectins and mainly exist in the lateral regions of the pollen tube, whereas PMEIs can promote pollen tube tip growth (Röckel et al., 2008). The balance of these functions modulates pollen tube cell wall stability and cell expansion (Bosch et al., 2005; Jiang et al., 2005). The enrichment of these genes among drought-induced genes suggests that elevated levels of such activities are needed to modulate cell wall integrity and cellular growth when there is greater osmotic stress and there are limited resources for growth.
Among the downregulated genes (see Supplemental Figure 5B), the enriched categories included key regulators of the cell cycle and DNA replication such as cyclins and cyclin-dependent kinases and regulators of microtubule-associated motors. These results are consistent with the finding that drought stress can inhibit the normal cell cycle process, thereby reducing plant cell division and differentiation (Setter and Flannigan, 2001). Other enriched groups include biosynthetic genes for JA and glucosinolate, which promote defense against disease and herbivore attack (Adie et al., 2007; Clay et al., 2009), suggesting that the energy costly defense responses were repressed by drought. JA also enhances filament elongation (Song et al., 2011); the reduction of expression of JA biosynthetic genes provides an explanation for the decreased filament length observed in DT flowers. The expression of genes encoding lipid transfer proteins was also reduced under drought stress, consistent with previous observation that lipid transfer protein genes are differentially regulated by various abiotic stresses (Wu et al., 2004; Oshino et al., 2007; Choi et al., 2008).
The Strategy of Reproductive Development under Drought
Our morphological study showed that stamen growth and development was highly sensitive to drought (Figure 2). From the transcriptomics results, we also found that many genes important for anther development showed reduced expression under drought, providing a link between gene expression and phenotype (Figure 5). The pistil and the subsequent fruit support ovule development and seed formation, respectively. In addition, the numbers of ovules and seeds are small, whereas pollen grains are numerous and in excess; therefore, partially sacrificing pollen development could reduce the need for resources while not greatly decreasing fertility. We found that pistils had higher drought tolerance than stamens, providing a means for the species to survive extreme environments, consistent with the induction of many pistil-related genes under drought and their possible role in the formation of a healthy pistil under stress. These results suggest a possible strategy by which plants protect yield against extreme environments by providing better protection of female development than male development.
During the later stages of normal flower development, the filament elongates dramatically for efficient delivery of the pollen to the pistil, ensuring subsequent fertilization. We found that after 8 d of drought treatment, filament length of DT plants was obviously shortened compared with WW plants, whereas pistil length was altered to a lesser extent (Figure 2). In addition, anther dehiscence was slightly delayed in the DT plants compared with WW plants. These findings are consistent with observations in rice (Liu et al., 2006). The shortened filament and delayed anther dehiscence greatly reduced pollen delivery and pollen availability to the pistil, thereby decreasing pollination. Subsequently, DT flowers exhibited recovery, with resumption of organ elongation, although to a lesser extent than in WW plants. Arabidopsis MYB family members MYB21, MYB24, and MYB108 are positive regulators of filament elongation and anther dehiscence via the JA and gibberellic acid (GA) signaling pathways (Mandaokar et al., 2006; Cheng et al., 2009; Mandaokar and Browse, 2009). We found that MYB21 was induced in the flower by drought, suggesting that MYB21 contributes to the recovery in the DT flower. Indeed, the myb21-2 mutant flowers showed a delay in recovery of filament elongation under drought treatment compared with wild-type plants. Although MYB21 is an important regulator in JA and GA signaling pathways (Cheng et al., 2009), its role in drought response is novel. Because ABA is a key hormone in drought responses, it will be important to investigate whether MYB21 serves to integrate different hormonal pathways, particularly the possible crosstalk between JA/GA and ABA in drought response, as hormone crosstalk has been shown to be important for biotic stresses (Bari and Jones, 2009). The induction of the expression of COI1, TIR1, and GID1A/B/C, genes encoding the JA/IAA/GA receptors, under drought suggests their crosstalk and cofunction in response to drought (Griffiths et al., 2006; Katsir et al., 2008; Calderón Villalobos et al., 2012); however, genes for brassinosteroid signaling receptors, BRI1 and BAK1 (Russinova et al., 2004), were not induced in our study (see Supplemental Figure 11 online). More experimental work is needed to further elucidate their relationship and crosstalk in response to drought.
Transcriptome Analysis Suggests the Mechanism of Plant Response to Drought Stress
We investigated floral transcriptomic changes under drought stress to probe gene activities during flower development affected by drought treatment, to obtain clues regarding gene regulation in response to drought in the Arabidopsis flower. Among upregulated genes, those encoding transcription factors were highly enriched, indicating that they play important roles in regulating downstream genes in response to drought. Surprisingly, only one transcription factor, DREB1A, was found from the T3/C3 comparison, which revealed 19 genes upregulated by at least twofold (see Supplemental Table 2 online). This suggests that DREB1A might be a key regulator of drought early response in Arabidopsis flowers. Previous studies showed that DREB1A is mainly involved in cold response (Maruyama et al., 2004; Ito et al., 2006); however, our results showed that DREB1A probably also plays an important role in drought response in flowers. Many drought-responsive genes detected here are key regulators in ABA signaling, including DREB1A, ABFs, NAC DOMAIN CONTAINING PROTEIN19, RD20, RD29A, and 3 PP2Cs (Table 1) (Kasuga et al., 2004; Bu et al., 2008; Xue et al., 2008; Yoshida et al., 2010).
Flowering time is influenced by environmental factors, including photoperiod and temperature, and requires the functions of many genes (Bernier and Périlleux, 2005). Although a recent study reported that FT is induced by drought and accelerates the floral transition (Riboni et al., 2013), how drought and osmotic stresses affect flowering was still largely unknown. Our results also indicated that FT could be induced after 4 to 5 d of drought treatment. In addition, other positive regulators of flowering, such as LFY, SOC1, were induced (Figure 5). At this point, plants have already sensed the stress signal and begun regulating reproductive growth, such as acceleration of flower differentiation and seed maturation. The induction also could be an indication that these genes have important functions under drought conditions to shorten the life cycle and promote survival under extreme environments, and is consistent with the findings that plants often flower earlier when grown under unfavorable conditions (Verslues and Juenger, 2011). The promotion of reproductive growth at the early phase of drought would not persist at the late phase because of the continuing limitation of water. Accordingly, the transcript levels of FT and SOC1 were reduced at day 10 compared with day 5, suggesting that a prolonged response might decrease the level of positive flowering regulators, thereby reducing the number of flowers under long-term severe drought.
Gene Regulatory Network for Plant Development under Drought Stress
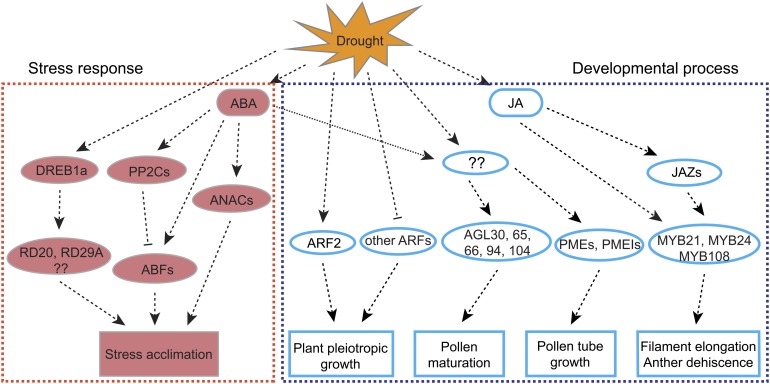
Previous studies have demonstrated that phytohormones coordinate plant growth and development under different environments (Wolters and Jürgens, 2009). ABA functions in many plant developmental processes and stress responses, including seed dormancy and osmotic stress. JA has multiple functions, including in plant defense and reproduction (Avanci et al., 2010; Robert-Seilaniantz et al., 2011). Our transcriptomic data also support a model that includes important roles of ABA and JA (Figure 9). For stress response (red boxes in Figure 9), the PP2C and NAC families have essential functions in the ABA-dependent pathways, and the DREB family is involved in an ABA-independent pathway for response to abiotic stresses (Ooka et al., 2003; Xue et al., 2008; Matsukura et al., 2010). From our transcriptome analysis, those gene family members showed significant induction in expression by drought in flowers, suggesting the possibility of similar roles in both reproductive and vegetative tissues.
Figure 9.
A Proposed Regulatory Network of Drought Response in Arabidopsis Flowers.
Ovals represent genes with known functions in either stress response or developmental process; boxes show different proposed functions. Interactions are represented by dashed lines with arrows representing positive regulatory relationships based on past studies. Interactions with hammer-ended dashed lines represent inhibition. Red symbols indicate stress related, whereas cyan symbols indicate developmental regulation.
[See online article for color version of this figure.]
Another group of genes regulating pollen maturation and tube growth in Arabidopsis are the Mδ subgroup of the MADS box family (Parenicová et al., 2003; Adamczyk and Fernandez, 2009) (Figure 9). The induction of those genes suggests that pollen maturation and tube growth could be accelerated by drought stress as a strategy for plants to survive drought stress and minimize loss of reproductive yield. During fertilization, pollen tube elongation and maturation require the polar accumulation of PMEs and PMEIs, which are key modifiers of pectin, a major cell wall component at the growing pollen tube tip (Bosch et al., 2005; Röckel et al., 2008) (Figure 9). The enrichment of genes for PMEs and PMEIs among upregulated genes under drought suggests that pollen tube growth is very sensitive to water deficit and is subject to enhanced control by a group of cell wall–related genes. As discussed above, several MYB genes are important for filament elongation and dehiscence, both affecting pollen delivery (Figure 9), as supported by both transcriptomic data and mutant phenotypes.
The proposed regulatory network (Figure 9) provides only a brief outline for the regulation of the two aspects of response to drought stress in the Arabidopsis flower: physiological response and developmental acclimation. Much further work is needed to identify additional components and to demonstrate the interactions among the components. Insights gained from such studies will likely be helpful in efforts to improve crop yield under drought and other environmental stresses.
METHODS
Plant Material and Drought Treatments
Morphological characterization and expression profiling under different water conditions were performed with the Arabidopsis thaliana ecotype Col-0. Seeds were directly sown into pots with no drainage holes containing 100 g soil mixture of dry Metro-Mix 360 soil (Sun Gro Horticulture Canada) and dry Turface profile greens grade (Profile Products) in a 3:2 ratio by volume, with a water-holding capacity of 90 g (defined as 90% soil moisture, also indicated as 90% field capacity). Seeds were stratified in a cold room for 2 d at 4°C in the dark. After stratification, the pots with seeds were transferred into a Conviron growth chamber (Conviron), which was set at our standard conditions (22°C, 16/8 h day/night photoperiod, ∼300 μmol m−2 s−1 photon flux, 60% humidity) until just after flowering (bolting) when half of the plants were subjected to drought stress. A few seeds were planted in each pot and then extra seedlings in the pot were removed, leaving only one plant per pot with similar growth status prior to drought treatment.
For the drought experiment, two treatments (WW, 90% soil moisture or field capacity; DT, 35% soil moisture or field capacity) were performed. Pots were arranged according to a randomized design and their positions were changed daily. Plants were WW until after just after bolting (the main stem was ∼1 cm high) when drought treatment was started by withholding water (defined as day 0 for drought treatment, C0, principal growth stage 5.80-5.90) (Boyes et al., 2001) and the humidity of the growth chamber was changed from 60% to 30% to accelerate the soil water evaporation. For the DT plants, the relative soil moisture content decreased sharply (Figure 1A); after ∼80 h, the relative soil moisture content was near 35% of the soil water-holding capacity (defined as day 3 for drought treatment). At this point, the chamber humidity was changed back to 60%. The soil water condition was then maintained (Figure 1A) by daily watering (pots were weighed and watered twice per day) until almost all of the fruits were mature and ready to harvest (∼50 d from C0). For WW plants (control), 90% of the soil moisture was maintained until tissue harvest or after seed maturation (principal growth stage 9.70) (Boyes et al., 2001).
For the tissue sampling, only unopened flower samples were collected at days 0, 3, 4, 5, and 10 of water stress treatment from both WW and DT plants (indicated as C0, C3, C4, C5, and C10 and T3, T4, T5, and T10), totaling nine samples. Two biological replicates were collected for each sample, and each replicate consisted of 18 inflorescences from 18 plants. Flower samples were frozen in liquid nitrogen right after sampling and stored at −80°C.
RWC of Leaf
RWC is the percentage of the water content in a given tissue at sampling as related to the water content at full turgor (Yamasaki and Dillenburg, 1999). To estimate RWC, leaves were collected, wrapped immediately in foodservice foil, and stored in the dark. The leaf pieces were weighed to determine fresh weight (FW). The leaves were then immersed in distilled water overnight, before weighing to obtain the turgid weight (TW). Then the leaves were dried in an oven for 24 h before weighing for the dry weight (DW), with RWC = [(FW − DW)/(TW − DW)] (Sharp et al., 1990).
Pollen Viability and Germination Assay
To determine pollen viability, Alexander’s stain was used. The stain contains acid fuchsin, which stains cytoplasm and mitochondria (Alexander, 1969). Pollen grains with high viability and cell activity are purple or red with a green outline, grains with low viability or cell activity are pink or light red, and aborted pollen grains are green inside and outside. First, the inflorescences with unopened flower buds were fixed with Carnoy’s fixative (absolute alcohol:chloroform:glacial acetic acid = 6:3:1, vol) for 2 h at room temperature. The fixative was later discarded and staining solution was added to stain the plant tissues at 50°C for 12 h. Before observation, the samples were washed with 25% glycerol for 8 h at room temperature. Tissues were observed using a Nikon Eclipse E400 microscope, and all images were prepared using Adobe Systems Photoshop 9.0 software.
Pollination
For pollination experiments, stamens were removed under a dissecting microscope from stage 12 flowers and the dissected plants were returned to the growth chamber and allowed to recover for ∼8 h. Pollen grains from anthers of mature flowers were immediately applied to fully cover the stigma of the emasculated flowers.
Size Measurements of Reproductive Organs
To measure the lengths of reproductive organs, we dissected the flower buds into the separate parts sepals, petals, stamens, and pistils under a dissecting scope (SMZ-U; Nikon). Parts were photographed at a fixed magnification using a camera system (Optronics Digital Microscope Cameras). Thereafter, the images were measured using ImageJ software (http://rsbweb.nih.gov/ij/). The Student's t test was used for comparison of means. P-values < 0.05 and 0.01 were considered statistically significant and highly statistically significant, respectively.
RNA Isolation and Microarray Analysis
Total RNA was isolated from unopened flower buds using RNeasy columns according to the manufacturer’s instructions (Qiagen). Arabidopsis whole-genome ATH1 arrays (Affymetrix) were used for this project, with more than 22,500 probe sets representing ∼24,000 genes. Labeling and hybridization of RNA and data collection were performed at the Genomics Core Facility at Pennsylvania State University. Two biological replicates were performed using independent tissue samples for each control at C0, C3, C4, C5, and C10 and drought treatment at T3, T4, T5, and T10.
Microarray datasets (*.cel files) were analyzed using R software (http://www.r-project.org). First, the bioconductor packages affy, AnnotationDbi, limma, and qvalue were applied to compare signals between WW plants and DT plants. The robust multiarray average method was used for normalization. We compared DT and WW samples at each time point and also compared all DT samples with the control C0. Pearson correlation coefficients were determined between replicates. The genes with a q-value (false discovery rate) greater than 0.01 were screened out, and the differentially expressed genes were then further selected and analyzed. Genome-wide Arabidopsis gene annotation was performed based on the The Arabidopsis Information Resource 10 (TAIR10) GO data (GOSlim_Plants) and the bulk download tool at the TAIR website was also used for downloading small reference sets. The file (affy_ATH1_array_elements-2010-12-20) was used to identify unique probes and filter out crosshybridization probes (ftp://ftp.Arabidopsis.org/home/tair/Microarrays/Affymetrix/). GO enrichment analysis was performed using agriGO, a publicly accessible analysis tool and database (http://bioinfo.cau.edu.cn/agriGO). The Z-score value was used and further converted to a P value to infer statistical significance (Du et al., 2010). Cluster analysis for gene categorization was performed with the TIGR Multi-Experiment Viewer (http://www.tm4.org/mev/). Microarray data were validated by quantitative RT-PCR of 26 genes. Detailed primer information is listed in Supplemental Table 6 online.
Promoter Motif Analysis
Three types of putative promoter sequence (500 bp, 1 kb, and 3 kb sequences upstream of the start codon) of the genes on the ATH1 microarray were downloaded from TAIR. We obtained 101 cis binding sites from AGRIS (Palaniswamy et al., 2006) that were then analyzed by perl. The number of each binding site was counted. We then compared the mean in DT samples against the means of each cis-element site in the whole genome. The P value was calculated using the Student’s t test and obtained according to occurrences of these CREs in DT gene groups and in all genes. The motifs in drought upregulated genes are more significant than the motifs in downregulated genes, so we set 1.0E-10 and 1.0E-05 as the significance thresholds for the motifs, respectively (Wang et al., 2011). Identification of new conserved motifs was accomplished with MEME software (version 4.9.0; http://meme.nbcr.net/meme/cgi-bin/meme.cgi), a program designed to identify frequent motifs in biopolymers (Bailey and Elkan, 1994).
RNA in Situ Hybridization
Nonradioactive RNA in situ hybridization was performed as previously described (Hord et al., 2006). Arabidopsis wild-type inflorescences were collected at days 0, 3, 4, and 5 of drought treatment and fixed in FAA (3.5% formaldehyde, 5% acetic acid, and 50% ethanol alcohol) fixative. The inflorescence samples were vacuumed twice in the fixative each for 10 min, with fixation for at least 2 h at room temperature. Samples were then dehydrated and embedded in paraffin, and 8 µm-thick sections were prepared and mounted onto slides. After dewaxing of all slides with Histo-Clear, the slides were dehydrated and baked. The antisense and sense RNAs of DREB1A and MYB21 were labeled with digoxigenin by in vitro transcription of linearized pGEM-T-DREB1A and pGEM-T-MYB21, which carry fragments of the DREB1A cDNA amplified with gene-specific primers oMC7398 (5′-CTCAGATTATTATTTCCATTTTTAGTA) and oMC7399 (5′-GAAACTGAATCAATTTAATTTACACT), and MYB21 cDNA amplified with gene-specific primers oMC7323 (5′-CTCGTTGTAGAGGTTTGGGG) and oMC7324 (5′-TATGACCCCTTCAACAATTTTAT). Antidigoxigenin antibodies coupled with alkaline phosphatase and nitro blue tetrazolium were used to detect the hybridization signal.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers DREB1A (AT4G25480), MYB21 (AT3G27810), ROP1 (AT3G51300), RIC6 (AT2G20430), ABI1 (AT4G26080), ABI2 (AT5G57050), ABI3 (AT3G24650), ABI4 (AT2G40220), ABI5 (AT2G36270), ABF1 (AT1G49720), ABF3 (AT4G34000), ABF4 (AT3G19290), PP2CA (AT3G11410), SNRK2.3 (AT5G66880), SNRK2.6 (AT4G33950), DREB1B (AT4G25490), DREB2A (AT5G05410), DREB2B (AT3G11020), RD20 (AT2G33380), RD22 (AT5G25610), RD29A (AT5G52310), CO (AT5G15840), TFL1 (AT5G03840), TFL2 (AT5G17690), MAF2 (AT5G65050), FUL (AT5G60910), ERECTA (AT2G26330), MYB24 (AT5G40350), and MYB108 (AT3G06490). Other accession numbers can be found in Supplemental Table 4 online. The wild-type drought project microarray data are available from Gene Expression Omnibus (GEO) at National Center for Biotechnology Information (NCBI) with the series entry GSE40998. Access to these data is provided at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hoxdowwwocaobmandacc=GSE40998. The dreb1a (CS872453) microarray data are available from GEO at NCBI with the series entry GSE43724. Access to these data is provided at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=fdazviywgskayzuandacc=GSE43724.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypic Comparison of WW Plants and DT Plants.
Supplemental Figure 2. Alexander Staining of the Anthers and in Vitro Germination of Pollen from WW Plants and DT Plants.
Supplemental Figure 3. Silique Growth Was Affected by Drought Stress.
Supplemental Figure 4. Correlation between Two Biological Replicates at Each Time Point.
Supplemental Figure 5. Venn Diagram and Enrichment Analysis of Differentially Expressed Genes.
Supplemental Figure 6. Quantitative PCR Verification of Microarray Data.
Supplemental Figure 7. cis-Regulatory Element Analysis of 1 kb and 3 kb Promoters of Differentially Expressed Genes.
Supplemental Figure 8. The dreb1a T-DNA Mutant Information and DREB Genes Expression Pattern under Drought Stress in Flower.
Supplemental Figure 9. Phenotypes of dreb1a Compared with Wild Type under Early Drought Stress.
Supplemental Figure 10. Phenotypes of dreb1a Compared with Wild Type under Prolonged Stress.
Supplemental Figure 11. Changes in Expression Patterns of Genes for IAA, GA, JA, and brassinosteroid Pathways under Drought Stress.
Supplemental Table 1. The Number of Genes with Expression Levels above the 50 Threshold (log2 value > 5.64) in Each Dataset and the Number of Genes Differentially Expressed in Each Dataset Compared with Multiple Controls.
Supplemental Table 2. GO Annotation of Genes Differentially Expressed at T3 versus C3.
Supplemental Table 3. The Expression of DREB Genes in Arabidopsis Flower under Drought Stress.
Supplemental Table 4. Primers for Quantitative PCR.
Supplemental Data Set 1. The List of 4153 Genes that Were Differentially Expressed in Any of the Four Datasets.
Supplemental Data Set 2. Statistical Analysis of the cis-Regulatory Element within 500 bp, 1 kb, and 3kb Promoter Regions of Differentially Expressed Genes.
Supplemental Data Set 3. Statistical Analysis of the cis-Regulatory Element within 500 bp, 1 kb, and 3kb Promoters Region of Differentially Expressed Transcription Factors.
Supplementary Material
Acknowledgments
We thank Haikun Zhang and Craig Praul of the Genomics Core Facility for help in performing microarray hybridizations. We also thank Jiong Wang and Yi Hu for help with plant care. This work was supported by a grant from the US Department of Energy (DE-FG02-02ER15332 to H.M.) and funds from Rijk Zwaan as well the Biology Department and the Huck Institutes of the Life Sciences at the Pennsylvania State University.
AUTHOR CONTRIBUTIONS
Z.S. and H.M. designed the research. Z.S., X.M., H.G., N.L.S., and B.G. performed different aspects of the research. Z.S., H.M., and S.M.A wrote the article.
References
- Adamczyk B.J., Fernandez D.E. (2009). MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol. 149: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie B.A., Pérez-Pérez J., Pérez-Pérez M.M., Godoy M., Sánchez-Serrano J.J., Schmelz E.A., Solano R. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19: 1665–1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Andersen M.N., Asch F., Wu Y., Jensen C.R., Naested H., Mogensen V.O., Koch K.E. (2002). Soluble invertase expression is an early target of drought stress during the critical, abortion-sensitive phase of young ovary development in maize. Plant Physiol. 130: 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanci N.C., Luche D.D., Goldman G.H., Goldman M.H. (2010). Jasmonates are phytohormones with multiple functions, including plant defense and reproduction. Genet. Mol. Res. 9: 484–505 [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 28–36 [PubMed] [Google Scholar]
- Bari R., Jones J.D. (2009). Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488 [DOI] [PubMed] [Google Scholar]
- Bernier G., Périlleux C. (2005). A physiological overview of the genetics of flowering time control. Plant Biotechnol. J. 3: 3–16 [DOI] [PubMed] [Google Scholar]
- Bernoux M., Timmers T., Jauneau A., Brière C., de Wit P.J., Marco Y., Deslandes L. (2008). RD19, an Arabidopsis cysteine protease required for RRS1-R-mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 20: 2252–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskara G.B., Nguyen T.T., Verslues P.E. (2012). Unique drought resistance functions of the highly ABA-induced clade A protein phosphatase 2Cs. Plant Physiol. 160: 379–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch M., Cheung A.Y., Hepler P.K. (2005). Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol. 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J.S., Westgate M.E. (2004). Grain yields with limited water. J. Exp. Bot. 55: 2385–2394 [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. (2001). Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breshears D.D., et al. (2005). Regional vegetation die-off in response to global-change-type drought. Proc. Natl. Acad. Sci. USA 102: 15144–15148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Jiang H., Li C.B., Zhai Q., Zhang J., Wu X., Sun J., Xie Q., Li C. (2008). Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res. 18: 756–767 [DOI] [PubMed] [Google Scholar]
- Busch W., Wunderlich M., Schöffl F. (2005). Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J. 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Calderón Villalobos L.I., et al. (2012). A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 8: 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C., Bhatt A.M., Scott R., Dickinson H. (2002). EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Curr. Biol. 12: 1718–1727 [DOI] [PubMed] [Google Scholar]
- Chaves M.M., Flexas J., Pinheiro C. (2009). Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. (Lond.) 103: 551–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., Song S., Xiao L., Soo H.M., Cheng Z., Xie D., Peng J. (2009). Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5: e1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi A.M., Lee S.B., Cho S.H., Hwang I., Hur C.G., Suh M.C. (2008). Isolation and characterization of multiple abundant lipid transfer protein isoforms in developing sesame (Sesamum indicum L.) seeds. Plant Physiol. Biochem. 46: 127–139 [DOI] [PubMed] [Google Scholar]
- Clay N.K., Adio A.M., Denoux C., Jander G., Ausubel F.M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. (2010). Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Dolferus R., Jacobs M., Peacock W.J., Dennis E.S. (1994). Differential interactions of promoter elements in stress responses of the Arabidopsis Adh gene. Plant Physiol. 105: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64– 70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooq M., Wahid A., Kobayashi N., D., F., and Basra, S.M.A. (2009). Plant drought stress: Effects, mechanisms and management. Sustainable Agric. 29: 185–212 [Google Scholar]
- Freeling M., Rapaka L., Lyons E., Pedersen B., Thomas B.C. (2007). G-boxes, bigfoot genes, and environmental response: Characterization of intragenomic conserved noncoding sequences in Arabidopsis. Plant Cell 19: 1441–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Chinnusamy V., Rodrigues A., Rubio S., Antoni R., Park S.Y., Cutler S.R., Sheen J., Rodriguez P.L., Zhu J.K. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto S.Y., Ohta M., Usui A., Shinshi H., Ohme-Takagi M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Fujita Y., Noutoshi Y., Takahashi F., Narusaka Y., Yamaguchi-Shinozaki K., Shinozaki K. (2006). Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol. 9: 436–442 [DOI] [PubMed] [Google Scholar]
- Gigon A., Matos A.R., Laffray D., Zuily-Fodil Y., Pham-Thi A.T. (2004). Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot. (Lond.) 94: 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths J., Murase K., Rieu I., Zentella R., Zhang Z.L., Powers S.J., Gong F., Phillips A.L., Hedden P., Sun T.P., Thomas S.G. (2006). Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haake V., Cook D., Riechmann J.L., Pineda O., Thomashow M.F., Zhang J.Z. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 130: 639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes U.Z., Meier S., Dietrich D., Ward J.M., Rentsch D. (2010). Functional properties of the Arabidopsis peptide transporters AtPTR1 and AtPTR5. J. Biol. Chem. 285: 39710–39717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2007). Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12: 343–351 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409: 525–529 [DOI] [PubMed] [Google Scholar]
- Hopkins R.J., van Dam N.M., van Loon J.J. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54: 57–83 [DOI] [PubMed] [Google Scholar]
- Hord C.L., Chen C., Deyoung B.J., Clark S.E., Ma H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18: 1667–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord C.L., Sun Y.J., Pillitteri L.J., Torii K.U., Wang H., Zhang S., Ma H. (2008). Regulation of Arabidopsis early anther development by the mitogen-activated protein kinases, MPK3 and MPK6, and the ERECTA and related receptor-like kinases. Mol. Plant 1: 645–658 [DOI] [PubMed] [Google Scholar]
- Hwang J.U., Gu Y., Lee Y.J., Yang Z. (2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16: 5385–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Wellmer F., Yu H., Das P., Ito N., Alves-Ferreira M., Riechmann J.L., Meyerowitz E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430: 356–360 [DOI] [PubMed] [Google Scholar]
- Ito Y., Katsura K., Maruyama K., Taji T., Kobayashi M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2006). Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 47: 141–153 [DOI] [PubMed] [Google Scholar]
- Jakobsen M.K., Poulsen L.R., Schulz A., Fleurat-Lessard P., Møller A., Husted S., Schiøtt M., Amtmann A., Palmgren M.G. (2005). Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a type V P-type ATPase. Genes Dev. 19: 2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X., Shiran B., Wan J., Lewis D.C., Jenkins C.L., Condon A.G., Richards R.A., Dolferus R. (2010). Importance of pre-anthesis anther sink strength for maintenance of grain number during reproductive stage water stress in wheat. Plant Cell Environ. 33: 926–942 [DOI] [PubMed] [Google Scholar]
- Jiang H., Li H., Bu Q., Li C. (2009). The RHA2a-interacting proteins ANAC019 and ANAC055 may play a dual role in regulating ABA response and jasmonate response. Plant Signal. Behav. 4: 464–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Yang S.L., Xie L.F., Puah C.S., Zhang X.Q., Yang W.C., Sundaresan V., Ye D. (2005). VANGUARD1 encodes a pectin methylesterase that enhances pollen tube growth in the Arabidopsis style and transmitting tract. Plant Cell 17: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Brousseau S.A., McCormick S. (2004). A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 39: 761–775 [DOI] [PubMed] [Google Scholar]
- Jung C., Müller A.E. (2009). Flowering time control and applications in plant breeding. Trends Plant Sci. 14: 563–573 [DOI] [PubMed] [Google Scholar]
- Kakumanu A., Ambavaram M.M., Klumas C., Krishnan A., Batlang U., Myers E., Grene R., Pereira A. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 160: 846–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Miura S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45: 346–350 [DOI] [PubMed] [Google Scholar]
- Katsir L., Schilmiller A.L., Staswick P.E., He S.Y., Howe G.A. (2008). COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. USA 105: 7100–7105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki S., Borchert C., Deyholos M., Wang H., Brazille S., Kawai K., Galbraith D., Bohnert H.J. (2001). Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 13: 889–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler J.P., Batelli G., Zhu J.K. (2010). ABA receptors: The START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61: 3199–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps J.A., Wu Y., Chang H.S., Zhu T., Wang X., Harper J.F. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackman P., et al. (2011). Jasmonate signaling involves the abscisic acid receptor PYL4 to regulate metabolic reprogramming in Arabidopsis and tobacco. Proc. Natl. Acad. Sci. USA 108: 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde S., Beebe D.U., Saini H.S. (1997). Early signs of disruption of wheat anther development associated with the induction of male sterility by meiotic-stage water deficit. Sex. Plant Reprod. 10: 40–48 [Google Scholar]
- Lata C., Prasad M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62: 4731–4748 [DOI] [PubMed] [Google Scholar]
- Liljegren S.J., Ditta G.S., Eshed Y., Savidge B., Bowman J.L., Yanofsky M.F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404: 766–770 [DOI] [PubMed] [Google Scholar]
- Liu F.L., Andersen M.N., Jensen C.R. (2003). Loss of pod set caused by drought stress is associated with water status and ABA content of reproductive structures in soybean. Funct. Plant Biol. 30: 271–280 [DOI] [PubMed] [Google Scholar]
- Liu J., et al. (2008). Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. Plant Cell 20: 1538–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.X., Liao D.Q., Oane R., Estenor L., Yang X.E., Li Z.C., Bennett J. (2006). Genetic variation in the sensitivity of anther dehiscence to drought stress in rice. Field Crops Res. 97: 87–100 [Google Scholar]
- Liu Q., Kasuga M., Sakuma Y., Abe H., Miura S., Yamaguchi-Shinozaki K., Shinozaki K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd D.G. (1980). Sexual strategies in plants. I. An hypothesis of serial adjustment of maternal investment during one reproductive session. New Phytol. 86: 69–79 [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803 [DOI] [PubMed] [Google Scholar]
- Long J.A., Moan E.I., Medford J.I., Barton M.K. (1996). A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379: 66–69 [DOI] [PubMed] [Google Scholar]
- Luan S. (2003). Protein phosphatases in plants. Annu. Rev. Plant Biol. 54: 63–92 [DOI] [PubMed] [Google Scholar]
- Ma H. (2005). Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu. Rev. Plant Biol. 56: 393–434 [DOI] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2004). dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 37: 720–729 [DOI] [PubMed] [Google Scholar]
- Mäkelä P., McLaughlin J.E., Boyer J.S. (2005). Imaging and quantifying carbohydrate transport to the developing ovaries of maize. Ann. Bot. (Lond.) 96: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Browse J. (2009). MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiol. 149: 851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A., Thines B., Shin B., Lange B.M., Choi G., Koo Y.J., Yoo Y.J., Choi Y.D., Choi G., Browse J. (2006). Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. Plant J. 46: 984–1008 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sakuma Y., Kasuga M., Ito Y., Seki M., Goda H., Shimada Y., Yoshida S., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 38: 982–993 [DOI] [PubMed] [Google Scholar]
- Matsukura S., Mizoi J., Yoshida T., Todaka D., Ito Y., Maruyama K., Shinozaki K., Yamaguchi-Shinozaki K. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283: 185–196 [DOI] [PubMed] [Google Scholar]
- Mayer K.F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95: 805–815 [DOI] [PubMed] [Google Scholar]
- McLaughlin J.E., Boyer J.S. (2004). Sugar-responsive gene expression, invertase activity, and senescence in aborting maize ovaries at low water potentials. Ann. Bot. (Lond.) 94: 675–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K., et al. (2009). A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat C.S., Ingle R.A., Wathugala D.L., Saunders N.J., Knight H., Knight M.R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7: e35995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouline K., Véry A.A., Gaymard F., Boucherez J., Pilot G., Devic M., Bouchez D., Thibaud J.B., Sentenac H. (2002). Pollen tube development and competitive ability are impaired by disruption of a Shaker K(+) channel in Arabidopsis. Genes Dev. 16: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P., Knappe S., Geimer S., Fischer K., Schulz B., Unte U.S., Rosso M.G., Ache P., Flügge U.I., Schneider A. (2005). The Arabidopsis plastidic glucose 6-phosphate/phosphate translocator GPT1 is essential for pollen maturation and embryo sac development. Plant Cell 17: 760–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M.E., Gleadow R.M., Dalling M.J. (1985). Effect of post-anthesis drought on cell division and starch accumulation in developing wheat grains. Ann. Bot. (Lond.) 55: 433–444 [Google Scholar]
- Nilson S.E., Assmann S.M. (2010). Heterotrimeric G proteins regulate reproductive trait plasticity in response to water availability. New Phytol. 185: 734–746 [DOI] [PubMed] [Google Scholar]
- Oñate-Sánchez L., Singh K.B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128: 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooka H., et al. (2003). Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Oshino T., Abiko M., Saito R., Ichiishi E., Endo M., Kawagishi-Kobayashi M., Higashitani A. (2007). Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol. Genet. Genomics 278: 31–42 [DOI] [PubMed] [Google Scholar]
- Pagnussat G.C., Yu H.J., Sundaresan V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19: 3578–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniswamy S.K., James S., Sun H., Lamb R.S., Davuluri R.V., Grotewold E. (2006). AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L., de Folter S., Kieffer M., Horner D.S., Favalli C., Busscher J., Cook H.E., Ingram R.M., Kater M.M., Davies B., Angenent G.C., Colombo L. (2003). Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.O., Hwang S., Hauser B.A. (2004). The phenotype of Arabidopsis ovule mutants mimics the morphology of primitive seed plants. Proc. Biol. Sci. 271: 311–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelaz S., Ditta G.S., Baumann E., Wisman E., Yanofsky M.F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405: 200–203 [DOI] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424: 85–88 [DOI] [PubMed] [Google Scholar]
- Portereiko M.F., Lloyd A., Steffen J.G., Punwani J.A., Otsuga D., Drews G.N. (2006). AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J., Laurie R., Macknight R. (2004). It’s time to flower: The genetic control of flowering time. Bioessays 26: 363–373 [DOI] [PubMed] [Google Scholar]
- Riboni M., Galbiati M., Tonelli C., Conti L. (2013). GIGANTEA enables drought escape response via abscisic acid-dependent activation of the florigens and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS. Plant Physiol. 162: 1706–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M., Jones J.D. (2011). Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49: 317–343 [DOI] [PubMed] [Google Scholar]
- Röckel N., Wolf S., Kost B., Rausch T., Greiner S. (2008). Elaborate spatial patterning of cell-wall PME and PMEI at the pollen tube tip involves PMEI endocytosis, and reflects the distribution of esterified and de-esterified pectins. Plant J. 53: 133–143 [DOI] [PubMed] [Google Scholar]
- Russinova E., Borst J.W., Kwaaitaal M., Caño-Delgado A., Yin Y., Chory J., de Vries S.C. (2004). Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 16: 3216–3229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H.S. (1997). Effects of water stress on male gametophyte development in plants. Sex. Plant Reprod. 10: 67–73 [Google Scholar]
- Schiefthaler U., Balasubramanian S., Sieber P., Chevalier D., Wisman E., Schneitz K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96: 11664–11669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Kienow L., Schmelzer E., Colby T., Bartsch M., Miersch O., Wasternack C., Kombrink E., Stuible H.P. (2005). A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 280: 13962–13972 [DOI] [PubMed] [Google Scholar]
- Schröter D., et al. (2005). Ecosystem service supply and vulnerability to global change in Europe. Science 310: 1333–1337 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Majumder A.L. (2009). Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta 229: 911–929 [DOI] [PubMed] [Google Scholar]
- Seo H.S., Song J.T., Cheong J.J., Lee Y.H., Lee Y.W., Hwang I., Lee J.S., Choi Y.D. (2001). Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter T.L., Flannigan B.A. (2001). Water deficit inhibits cell division and expression of transcripts involved in cell proliferation and endoreduplication in maize endosperm. J. Exp. Bot. 52: 1401–1408 [DOI] [PubMed] [Google Scholar]
- Sharp R.E., Hsiao T.C., Silk W.K. (1990). Growth of the Maize primary root at low water potentials: II. Role of growth and deposition of hexose and potassium in osmotic adjustment. Plant Physiol. 93: 1337–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheoran I.S., Saini H.S. (1996). Drought-induced male sterility in rice: Changes in carbohydrate levels and enzyme activityies associated with the inhibition of starch accumulation in pollen. Sex. Plant Reprod. 9: 161–169 [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58: 221–227 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Yamaguchi-Shinozaki K., Seki M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Shinwari Z.K., Nakashima K., Miura S., Kasuga M., Seki M., Yamaguchi-Shinozaki K., Shinozaki K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Smart R.E. (1974). Rapid estimates of relative water content. Plant Physiol. 53: 258–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S., Qi T., Huang H., Ren Q., Wu D., Chang C., Peng W., Liu Y., Peng J., Xie D. (2011). The Jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect Jasmonate-regulated stamen development in Arabidopsis. Plant Cell 23: 1000–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen A.M., Kröber S., Unte U.S., Huijser P., Dekker K., Saedler H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33: 413–423 [DOI] [PubMed] [Google Scholar]
- Sun K., Hunt K., Hauser B.A. (2004). Ovule abortion in Arabidopsis triggered by stress. Plant Physiol. 135: 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K., Cui Y., Hauser B.A. (2005). Environmental stress alters genes expression and induces ovule abortion: Reactive oxygen species appear as ovules commit to abort. Planta 222: 632–642 [DOI] [PubMed] [Google Scholar]
- Umezawa T., Sugiyama N., Mizoguchi M., Hayashi S., Myouga F., Yamaguchi-Shinozaki K., Ishihama Y., Hirayama T., Shinozaki K. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y., Furihata T., Abe H., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano K., Kurihara Y., Seki M., Shinozaki K. (2010). ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Verslues P.E., Juenger T.E. (2011). Drought, metabolites, and Arabidopsis natural variation: A promising combination for understanding adaptation to water-limited environments. Curr. Opin. Plant Biol. 14: 240–245 [DOI] [PubMed] [Google Scholar]
- Wang R.S., Pandey S., Li S., Gookin T.E., Zhao Z., Albert R., Assmann S.M. (2011). Common and unique elements of the ABA-regulated transcriptome of Arabidopsis guard cells. BMC Genomics 12: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W.Z., Song L.F., Zou J.J., Su Z., Wu W.H. (2008). Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol. 148: 1201–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne A.J., Zhang W., Sun Y., Liu W., Albert R., Zheng Z., Oppenheimer D.G., Zhao D., Ma H. (2007). Differential gene expression in Arabidopsis wild-type and mutant anthers: Insights into anther cell differentiation and regulatory networks. Plant J. 52: 14–29 [DOI] [PubMed] [Google Scholar]
- Wilson Z.A., Morroll S.M., Dawson J., Swarup R., Tighe P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28: 27–39 [DOI] [PubMed] [Google Scholar]
- Wolters H., Jürgens G. (2009). Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat. Rev. Genet. 10: 305–317 [DOI] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S., Yang Z. (2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Robertson A.J., Liu X., Zheng P., Wilen R.W., Nesbitt N.T., Gusta L.V. (2004). A lipid transfer protein gene BG-14 is differentially regulated by abiotic stress, ABA, anisomycin, and sphingosine in bromegrass (Bromus inermis). J. Plant Physiol. 161: 449–458 [DOI] [PubMed] [Google Scholar]
- Xiong L., Schumaker K.S., Zhu J.K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14 (Suppl): S165–S183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Zhou G., Shimizu H. (2010). Plant responses to drought and rewatering. Plant Signal. Behav. 5: 649–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Wang D., Zhang S., Ehlting J., Ni F., Jakab S., Zheng C., Zhong Y. (2008). Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics 9: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K., Shinozaki K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yamasaki S., Dillenburg L.R. (1999). Measurements of leaf relative water content in Araucaria angustifolia. Rev. Bras. Fisiol. Vegetal 11: 69–75 [Google Scholar]
- Yancey P.H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 208: 2819–2830 [DOI] [PubMed] [Google Scholar]
- Yang S.L., Xie L.F., Mao H.Z., Puah C.S., Yang W.C., Jiang L., Sundaresan V., Ye D. (2003). Tapetum determinant1 is required for cell specialization in the Arabidopsis anther. Plant Cell 15: 2792–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.L., Jiang L., Puah C.S., Xie L.F., Zhang X.Q., Chen L.Q., Yang W.C., Ye D. (2005). Overexpression of TAPETUM DETERMINANT1 alters the cell fates in the Arabidopsis carpel and tapetum via genetic interaction with excess microsporocytes1/extra sporogenous cells. Plant Physiol. 139: 186–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W.C., Ye D., Xu J., Sundaresan V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13: 2108–2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zhang W., Sun Y., Timofejeva L., Chen C., Grossniklaus U., Ma H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133: 3085–3095 [DOI] [PubMed] [Google Scholar]
- Zhao D.Z., Wang G.F., Speal B., Ma H. (2002). The excess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16: 2021–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Zhang W., Stanley B.A., Assmann S.M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2001). Plant salt tolerance. Trends Plant Sci. 6: 66–71 [DOI] [PubMed] [Google Scholar]
- Zhu J.K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53: 247–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.