NOPPERABO1 encodes a PUB-ARM–type E3 ubiquitin ligase localized at the plasma membrane that functions as a positive factor for schizogenous intercellular space formation in the liverwort Marchantia polymorpha. A significant T-DNA–tagged mutant in the emerging model liverwort, which occupies a critical position in the evolution of land plants, is reported.
Abstract
The vast majority of land plants develop gas-exchange tissues with intercellular spaces (ICSs) connected directly to the air. Although the developmental processes of ICS have been described in detail at the morphological and ultrastructural level in diverse land plants, little is known about the molecular mechanism responsible for ICS formation. The liverwort Marchantia polymorpha develops a multilayered tissue with a large ICS (air chamber), whose formation is initiated at selected positions of epidermal cells. We isolated a mutant of M. polymorpha showing impaired air-chamber formation, nopperabo1 (nop1), from T-DNA–tagged lines. In nop1 plants, no ICS was formed; consequently, a single-layered epidermis developed on the dorsal side of the thallus. The causal gene NOP1 encodes a Plant U-box (PUB) E3 ubiquitin ligase carrying tandem ARMADILLO (ARM) repeats in the C terminus. An in vitro ubiquitination assay indicated that the NOP1 protein possesses E3 ubiquitin ligase activity in a U-box–dependent manner. Confocal microscopy and biochemical analysis showed that NOP1 was localized to the plasma membrane. Our investigation demonstrated the essential role of the PUB-ARM–type ubiquitin ligase in ICS formation in M. polymorpha, which sheds light on the molecular mechanism of schizogenous ICS formation in land plants.
INTRODUCTION
All living organisms exchange gases to sustain vital activities. For single-celled organisms, this can be accomplished by direct contact of the cells with the environment. However, for large multicellular organisms, gas exchange is accomplished through systems specialized for circulation and gas uptake. For example, animals have a closed or open circulatory system pumped by a heart, and gas exchange with the environment occurs through lungs or gills. By contrast, plants lack such a system for circulation and gas exchange. Instead, many plants develop intercellular spaces (ICSs), connected to the external environment via small pores, stomata, or air pores, which allow for efficient gas exchange (Raven, 1996; Jackson and Armstrong, 1999; Evans, 2004). Development of the gas exchange system is critical in plants, as gaseous carbon dioxide is the only substrate for carbon fixation in photosynthesis. Spongy mesophyll in the leaves of vascular plants is a typical example of tissue containing ICSs connected to the external environment via stomata. ICSs are extensively developed in stems and roots of wetland plants that grow in hypoxic soils (Seago et al., 2005; Joshi and Kumar, 2012). ICSs also are present in bryophytes (e.g., stomata in the sporophyte of mosses and hornworts and air chambers in the gametophyte of some liverworts; Renzaglia et al., 2000). Fossil records of stomata and ICSs in early land plants found in the 400-million-year-old Rhynie chert, that is, an early Devonian deposit preserving fossils of plants and animals at the cellular level, indicate that presence of ICSs is one of the most conservative characters in embryophytes (Edwards et al., 1998). Throughout the evolution of land plants, internalization of the gas exchange surface by ICSs is implicated to be one of the critical developments for adaptation to terrestrial environments, together with development of the cuticle, stomata, and the water conduction system (Raven, 2002).
In the most cases, the developmental origin of ICSs in mesophyll is schizogenous, in which development results in cell wall separation between adjacent cells in the middle lamella, rather than lysigenous, in which cell death creates the gas space (Sachs et al., 1882; Dale, 1988). Although the ICS developmental process has been described in detail in a number of species at the histological level, little is known about the underlying molecular mechanism (Jackson and Armstrong, 1999; Evans, 2004).
The Marchantiales, a group of liverworts that includes Marchantia polymorpha, develop specialized organs, termed air chambers, on the dorsal surface of the gametophyte plant body, which is termed the thallus (Smith, 1955). The air chambers in M. polymorpha contain a large ICS containing photosynthetic filaments developed from the subepidermis and connected with the external environment by means of air pores formed in the epidermis at the center of the chamber (Barnes and Land, 1907; Apostolakos et al., 1982). The morphology of the air pores and filaments in the air chambers are analogous to stomata and spongy mesophyll in leaves of vascular plants and are believed to facilitate gas exchange during photosynthesis, transpiration, and respiration, especially under waterlogged conditions (Schönherr and Ziegler, 1975; Green and Snelgar, 1982). The development of air pores and air chambers in Marchantiales has been studied since the nineteenth century (Barnes and Land, 1907). The developmental process was elucidated on the basis of a series of histological observations on Marchantia paleacea (Apostolakos et al., 1982; Apostolakos and Galatis, 1985). According to detailed observations by Apostolakos, the ontogeny of the air pores and air chambers of Marchantia begins with the schizogenous development of protodermal ICSs, termed initial apertures, between the anticlinal walls of protodermal cells of the thallus apex (Apostolakos et al., 1982). An air chamber results from broadening of the base of the initial apertures by coordination of the rate of cell division and growth of the protodermal and subprotodermal cells surrounding the initial aperture (Apostolakos et al., 1982). Despite a number of histological studies over the course of the last century focusing on the morphology and development of air pores and air chambers in Marchantiales, the molecular mechanism responsible is unclear.
M. polymorpha has a long history as an experimental organism. There is renewed interest in the potential of M. polymorpha as a developmental model organism because of its critical evolutionary position (Bowman et al., 2007) and the availability of molecular genetic tools, including transformation techniques (Chiyoda et al., 2008; Ishizaki et al., 2008; Kubota et al., 2013) and gene-targeting strategies (Ishizaki et al., 2013).
In this study, we isolated a mutant, called nopperabo1 (nop1), of the liverwort M. polymorpha that shows impaired formation of air chambers by T-DNA tagging. Consistent with previous descriptions, schizogenous ICSs were observed at an early stage of air-pore and air-chamber development in the wild type, but no such structure was observed in nop1. Molecular and genetic characterization enabled identification of the causal gene NOP1, which encodes an E3 ubiquitin ligase localized on the plasma membrane. This represents an example of successful forward genetics by T-DNA tagging in bryophytes. Potential roles of the plasma membrane–localized E3 ligase NOP1 in the regulation of ICS formation in M. polymorpha are discussed.
RESULTS
Isolation and Characterization of nop1 in M. polymorpha
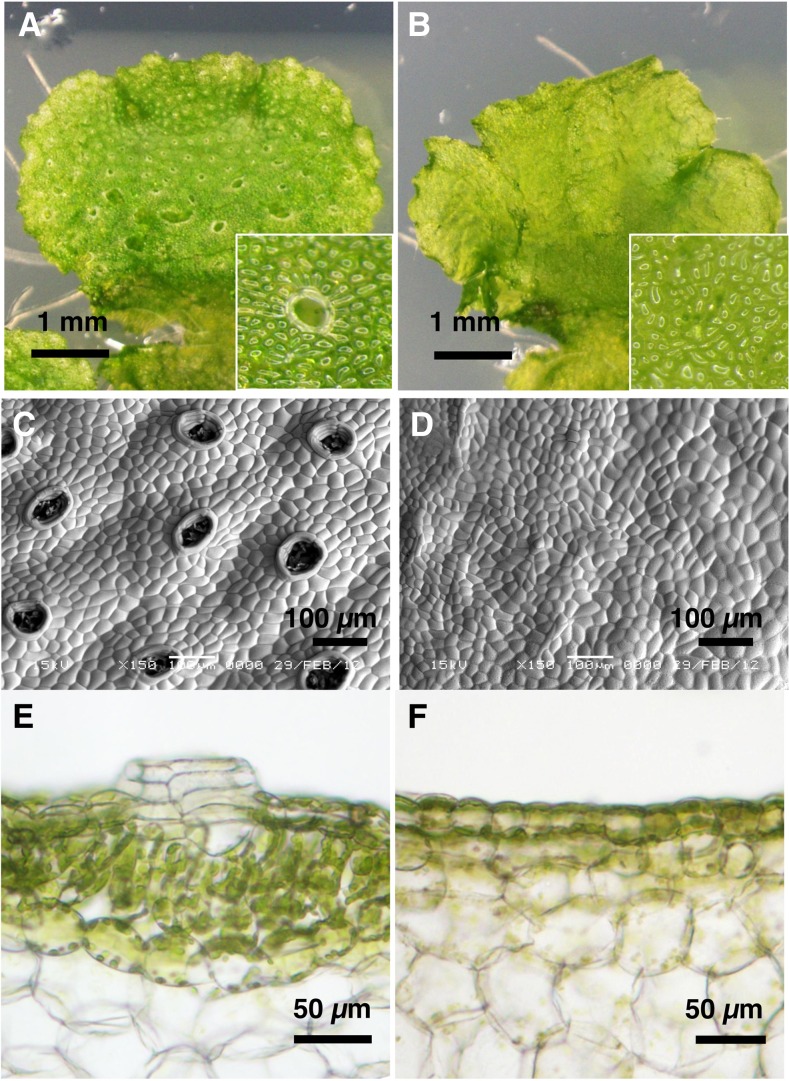
A mutant of M. polymorpha with impaired air-chamber formation was identified during the screening of 10,000 T-DNA–tagged lines for morphological phenotypes of the thallus. We named the mutant nop1, after a famous monster in Japanese folklore whose face has no eyes, nose, or mouth. Numerous air pores, stomata-like structures associated with each air chamber, were distributed regularly on the entire dorsal surface of the wild-type thallus (Figures 1A and 1C), whereas no air pores were observed on the dorsal surface of the nop1 thallus (Figures 1B and 1D). Transverse sections of the thallus showed that well-defined air chamber structures formed on the dorsal side of the thallus in wild-type plants (Figure 1E), whereas a single epidermal layer, distinguished by the presence of well-developed chloroplasts, was observed on the dorsal side of the thallus in nop1 plants (Figure 1F). No distinct impairment of the growth rate of nop1 thalli compared with that of wild-type thalli was observed when cultured on agar medium under continuous white fluorescent light at 22°C.
Figure 1.
Air-Chamber Phenotype of nop1 Mutant.
(A) and (B) Ten-day-old thalli grown from gemmae of the wild type (A) and nop1 (B) and magnified images (insets).
(C) and (D) Scanning electron micrographs of wild-type (C) and nop1 (D) thalli.
(E) and (F) Transverse sections (150-µm thick) from agar-embedded 10-d-old thalli of the wild type (E) and nop1 (F).
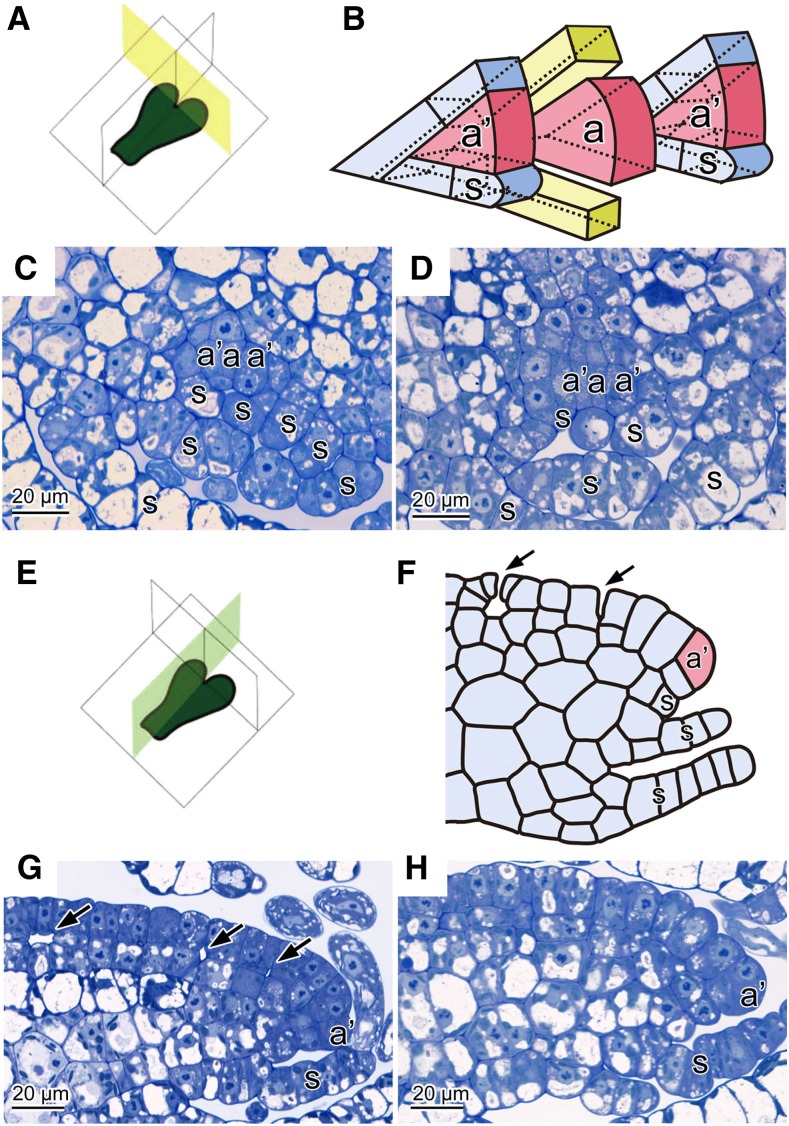
To investigate the phenotype of nop1 in more detail, the initial stage of dorsal tissue development was observed in the wild type and nop1 using a series of histological sections (Figure 2). The transverse section at the apical notch area of wild-type plants showed a single apical cell (a in Figure 2) with four planes of sectioning: one dorsal, one ventral, and two laterals (Figures 2A to 2C). In general, the apical cell and lateral derivatives (subapical cells) of marchantialian liverworts have the same shape and segmentation pattern (Figure 2B) (Crandall-Stotler, 1981; Shimamura, 2012). The ventral derivatives of subapical cells develop into rows of leaf-like ventral scales (s in Figure 2) (Crandall-Stotler, 1981; Shimamura, 2012). In the transverse sections, there were no obvious differences in the shape of apical cells, the segmentation pattern around the apical cell, and the tissue differentiation pattern on the ventral side between the wild type and nop1 (Figures 2C and 2D).
Figure 2.
Histological Observation of Thalli of the Wild Type and nop1 Mutant.
(A) Diagram showing transverse planes in which thallus sections were cut.
(B) Schematic illustration of the pattern of dorsiventral division of the apical cell and subapical cells of M. polymorpha.
(C) and (D) Transverse sections of wild-type (C) and nop1 (D) thalli.
(E) Diagram showing vertical longitudinal planes in which thallus sections were cut.
(F) Schematic illustration of a typical vertical longitudinal section around the subapical cell of wild-type M. polymorpha.
(G) and (H) Vertical longitudinal sections of wild-type (G) and nop1 (H) thalli.
Arrows indicate the initiation of ICSs in the wild type ([F] and [G]). a, apical cell; a’, subapical cell; s, ventral scale.
ICSs are formed between the dorsal derivatives of an apical and subapical cells (Figures 2E and 2F) (Crandall-Stotler, 1981; Apostolakos et al., 1982; Shimamura, 2012). A vertical longitudinal section of the thallus wing in the apical region of the wild type is shown in Figures 2G and 2F. Along the margin of the thallus wing were wedge-shaped subapical cells (a’ in Figure 2) that have meristematic activity and that give rise to derivative cells on both dorsal and ventral sides (Crandall-Stotler, 1981; Shimamura, 2012). The derivative cells on the dorsal side further divided periclinally and generated the protodermal and subprotodermal cell layers. ICSs first appeared between the anticlinal walls of protodermal cells as an initial aperture (Figures 2F and 2G). Through the course of cell divisions and growth of the protodermal and subprotodermal cells surrounding the initial aperture, the base of the initial aperture broadened and the primary air chamber was formed (arrows in Figures 2F and 2G). In vertical longitudinal sections of the same region in nop1, although wedge-shaped subapical cells and periclinal cell divisions to form protodermal and subprotodermal cell layers were observed, no ICS was formed among the protodermal and subprotodermal cells (Figure 2H). No indication of ICS formation was observed in serial sections in various orientations in the apical region. These observations indicated that in nop1, the initial stage of air-pore and air-chamber development was defective; that is, ICS formation was schizogenous.
Molecular Characterization of nop1 Mutant
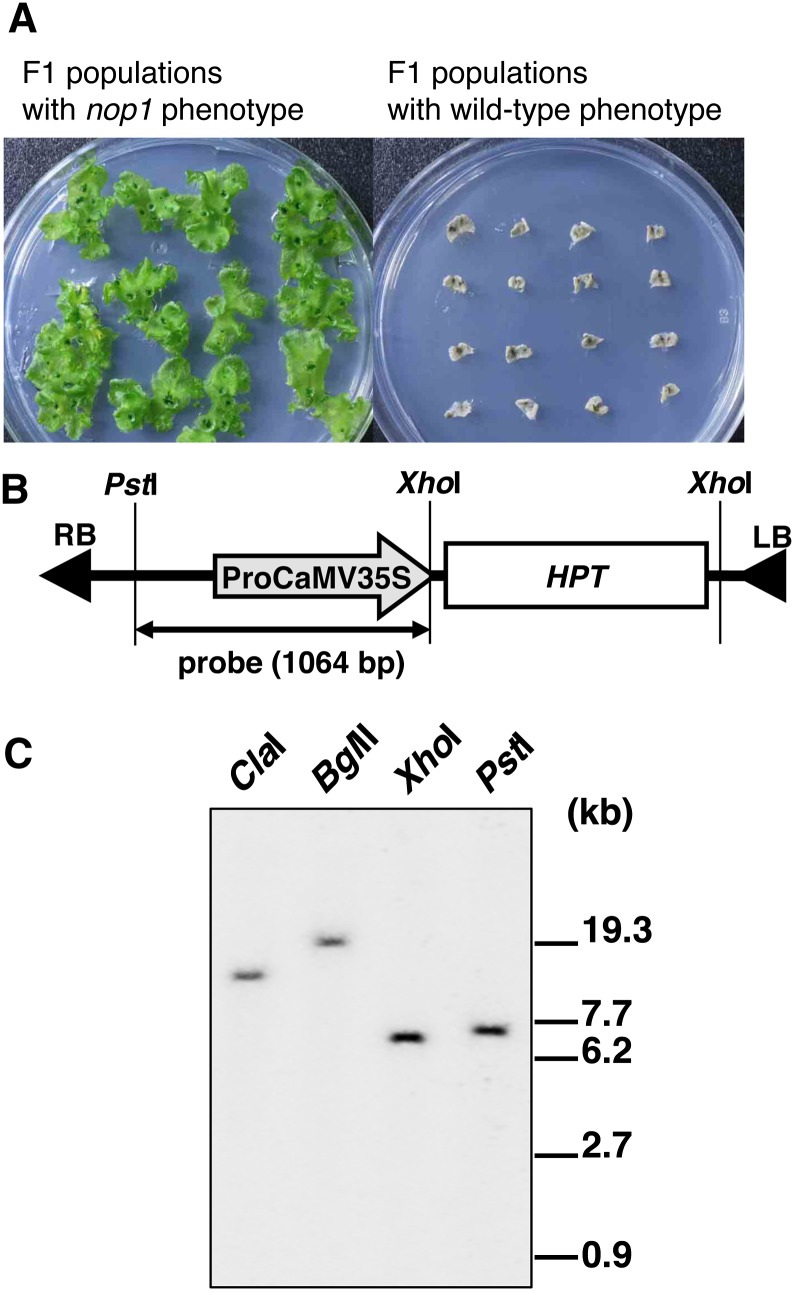
Segregation in an F1 population generated by a cross between nop1 and the wild-type accession Takaragaike-1 (Tak-1) was 123:135 (nop1:NOP1), which showed a good fit to the expected 1:1 ratio as indicated by the χ2 test. The nop1 phenotype in 123 F1 progeny was completely linked with hygromycin resistance (Figure 3A).
Figure 3.
T-DNA Insertion in the nop1 Genome.
(A) Segregation of the nop1 phenotype with the hygromycin resistance marker. F1 individuals obtained from a cross between nop1 and Tak-1 showed nop1 and wild-type phenotypes in the ratio of 1:1. F1 individuals that showed the nop1 phenotype survived (left), whereas F1 individuals that showed the wild-type phenotype (right) died after transfer to medium containing hygromycin.
(B) Schematic representation of the T-DNA region in pCAMBIA1300. RB, right border; LB, left border; ProCaMV35S, cauliflower mosaic virus 35S promoter; HPT, hygromycin phosphotransferase.
(C) Genomic DNA gel blot analysis of nop1. DNA extracted from nop1 plants was digested with ClaI, BglII, XhoI, and PstI, fractionated by electrophoresis, and allowed to hybridize to the cauliflower mosaic virus 35S promoter probe shown in (B).
To identify the causal gene of nop1, a genomic DNA gel blot analysis of the mutant was performed. Four restriction enzymes (ClaI, BglII, XhoI, and PstI) were used to digest genomic DNA isolated from nop1 thalli. The cauliflower mosaic virus 35S promoter fragment was used as the probe for hybridization (Figure 3B). DNA gel blot analysis with the four enzyme digests displayed single bands (Figure 3C). These results suggested that a single T-DNA insertion is tightly linked to nop1.
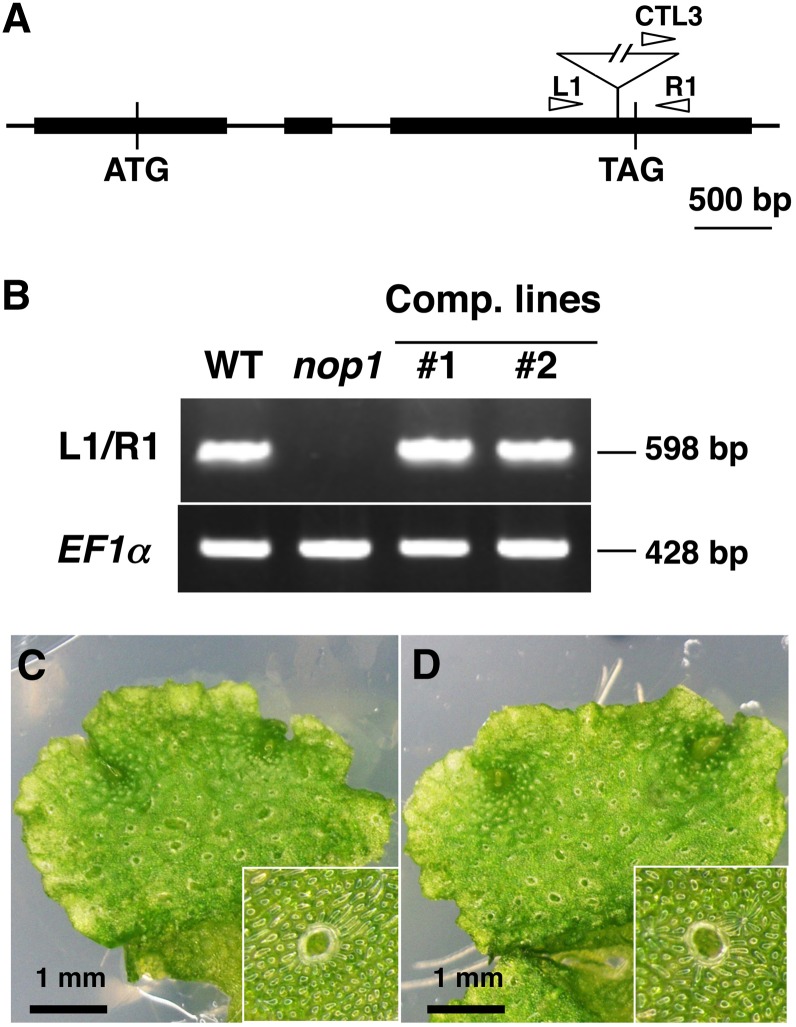
The junction sequences between the M. polymorpha genome and T-DNA were amplified from nop1 by thermal asymmetric interlaced–PCR (TAIL-PCR), and nucleotide sequences were determined. The T-DNA appeared to be inserted within the gene-coding sequence (Figure 4A). The site of T-DNA insertion was confirmed by sequencing of genomic PCR products amplified from nop1 (see Supplemental Figure 1 online). RT-PCR using a primer pair designed to span the T-DNA insertion site (Figure 4A) successfully amplified a cDNA fragment of expected size in the wild type, whereas no amplification was observed in nop1 plants using the same primer set (Figure 4B). The M. polymorpha ELONGATION FACTOR1-α gene (EF1α) was amplified in all samples at similar levels, which demonstrated the integrity of the RNA preparation. These results confirmed that transcripts spanning the T-DNA insertion site are absent in the nop1 mutant.
Figure 4.
Identification of the Causal Gene for nop1.
(A) Gene organization of NOP1. Arrow heads represent the positions of primers used for genotyping of wild-type and transgenic lines. Solid rectangles indicate exons, and an open triangle indicates the position of the T-DNA insertion in nop1.
(B) RT-PCR analysis of total RNA from the wild type (WT), nop1, and complemented lines, with the primer sets indicated on the left. The positions of each primer in the NOP1 genomic locus are indicated in (A).
(C) and (D) Phenotype of complemented lines #1 (C) and #2 (D) and magnified images (insets).
In order to further confirm that the nop1 phenotype was caused by this T-DNA insertion, we conducted a complementation experiment. The genomic DNA from 5.7 kb upstream of the putative start codon to 0.2 kb downstream of the 3′ untranslated region was introduced into the nop1 mutant (see Supplemental Figure 1 online), and two such lines restored the expression of this gene (Figure 4B) together with air-chamber formation (Figures 4C and 4D). These results indicated that the nop1 phenotype was caused by disruption of the introduced gene, which we named NOP1.
NOP1 Encodes a Plant U-Box E3 Ubiquitin Ligase
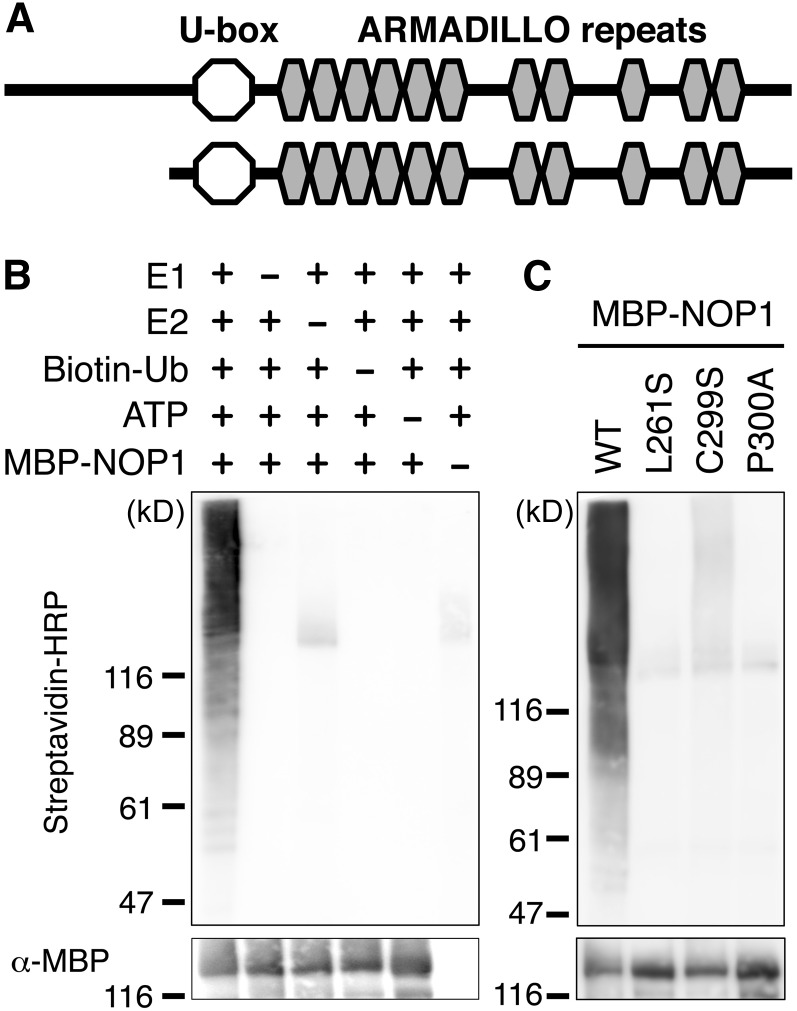
Sequences of NOP1 cDNAs that had been identified from our EST databases revealed that the deduced amino acid sequence of NOP1 was related to the plant U-box (PUB) protein family (Figure 5A; see Supplemental Figure 2 online). The U-box domain is structurally similar to the RING finger motif, and U-box–containing proteins are proposed to function as E3 ubiquitin ligases that catalyze the transfer of ubiquitin from the ubiquitin-conjugating enzyme (E2) to the target for ubiquitination (Aravind and Koonin, 2000; Ohi et al., 2003). In addition to the U-box, PUB proteins carry other predicted domains, such as tandem ARMADILLO (ARM) repeats, Ser/Thr kinase, WD40 repeats, tetratricopeptide repeats, or a peptidyl-prolyl isomerase domain. The PUB-ARM family has a plant-specific domain organization (Samuel et al., 2006; Yee and Goring, 2009). Domain structure analysis showed that the U-box at the N terminus of NOP1 was highly conserved and followed by a region containing at least 11 ARM repeats (Figure 5A; see Supplemental Figure 2 online), which are presumably important for protein–protein interactions (Samuel et al., 2006; Tewari et al., 2010). A subset of PUB-ARM proteins in Arabidopsis thaliana, such as At PUB18 and At PUB19 (Mudgil et al., 2004), contain the conserved domain with unknown function at the N-terminal end of the U-box (UND). Although there are six potential in-frame start codons upstream of the U-box–coding region, even the most upstream start codon did not allow coding of the UND. The amino acid sequence of NOP1 showed a high overall similarity, including the U-box domain followed by the ARM repeats, to homologs from bryophytes to angiosperms (see Supplemental Figure 2 online). A phylogenetic analysis suggested that NOP1 belongs to a subgroup of PUB-ARMs with an extended C-terminal ARM repeat region containing up to 12 ARM repeats but no UND (see Supplemental Figure 3 online). In summary, the NOP1 structure is typical of a PUB-ARM protein.
Figure 5.
In Vitro Ubiquitination Assay of the NOP1 Protein.
(A) Schematic diagram of the NOP1 protein structure. Modular architecture of NOP1 was analyzed using SMART software (Letunic et al., 2012). Top panel: Structure of the possible longest NOP1 protein. Bottom panel: Region of NOP1 protein used for in vitro ubiquitination assay.
(B) The MBP-NOP1 protein was incubated with or without E1, E2, biotin-Ub, and ATP. Ubiquitination in a heterogenous collection of higher molecular mass proteins was detected with HRP–bound streptavidin (top). Recombinant MBP-NOP1 was detected with anti-MBP antibody (bottom).
(C) MBP-NOP1 and a series of U-box mutant MBP-NOP1 proteins were incubated with E1, E2, biotin-Ub, and ATP for 120 min and subjected to protein gel blot analysis as in (B). WT, the wild type.
The presence of the U-box domain suggested that NOP1 is an E3 ubiquitin ligase and functions in protein ubiquitination. To determine whether NOP1 is a functional E3 ubiquitin ligase, we performed an in vitro ubiquitination assay using the protein containing the U-box domain and the ARM repeats of NOP1 purified from Escherichia coli as a C-terminal fusion with maltose binding protein (MBP) (Figure 5). Ubiquitination assays were performed using human E1 and E2 enzymes and biotinylated ubiquitin (biotin-Ub). Ubiquitinated proteins were detected using horseradish peroxidase (HRP)–bound streptavidin, and recombinant MBP-NOP1 was detected using an anti-MBP antibody. A polyubiquitination tail was detected when E1, E2, biotin-Ub, and ATP were present (Figure 5B). The E3 ligase activity increased in an incubation time–dependent manner (see Supplemental Figure 4 online) and was not observed in the absence of any of the essential components of the reaction (Figure 5B). Thus, NOP1 functions as an E3 ubiquitin ligase in vitro.
We then investigated if the U-box domain is essential for the E3 ligase activity of NOP1. The U-box domain of E3 ligases is predicted to be involved in the interaction with the E2 ubiquitin-conjugating enzyme, and several of its amino acids are reported to be essential for this interaction (Pringa et al., 2001; Ohi et al., 2003). The highly conserved Leu-261, Cys-299, and Pro-300 (see Supplemental Figure 2 online) were mutated to Ser, Ser, and Ala, respectively, in the NOP1 protein. These mutated proteins were expressed as MBP fusions, and the E3 ligase activity was assayed. Ubiquitin-ligase activity was greatly reduced in these mutant NOP1 proteins (Figure 5C). Therefore, we concluded that an intact U-box domain was essential for NOP1 E3 ligase activity.
Plasma Membrane Localization of NOP1
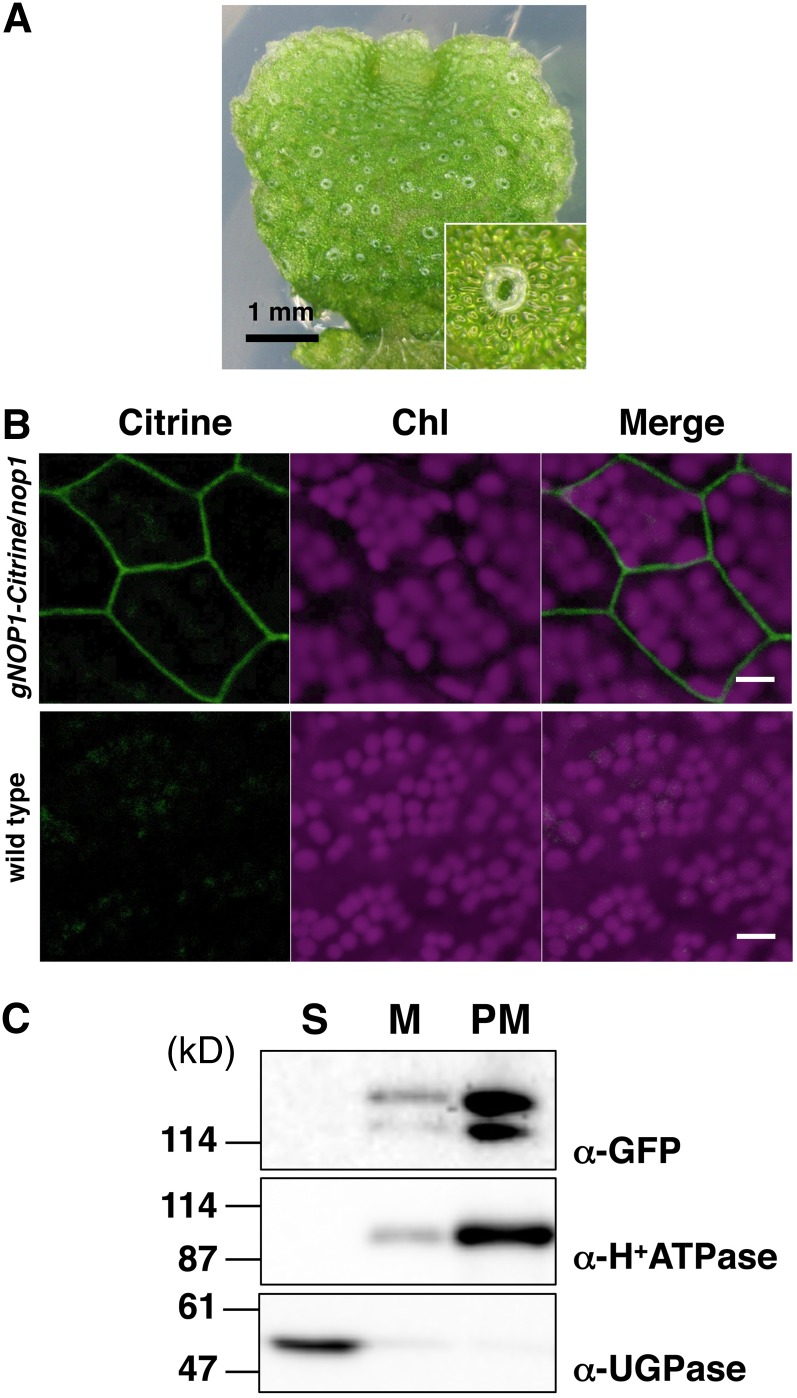
To investigate the expression specificity and the subcellular localization of NOP1, we generated nop1 plants expressing NOP1 translationally fused to the Citrine reporter protein under the control of the NOP1 promoter (hereafter designated as gNOP1-Citrine/nop1). The impaired air-chamber phenotype of nop1 was completely rescued by gNOP1-Citrine/nop1 (Figure 6A), which indicated that NOP1-Citrine is functional. Fluorescence microscopy analysis of this line showed that the fluorescence from the NOP1-Citrine fusion protein was distributed widely throughout the thallus, including in the apical regions where the development of air chambers is initiated (see Supplemental Figure 5 online). Under higher magnification, it was observed that NOP1-Citrine was mainly localized to the plasma membrane and was absent from the nucleus and the cytosol (Figure 6B). The subcellular localization of NOP1-Citrine was further investigated by subcellular fractionation using the plants expressing NOP1-Citrine. Soluble, total membrane, and plasma membrane fractions were prepared from thalli of M. polymorpha expressing NOP1-Citrine (Figure 6C). Immunoblot analysis demonstrated strong enrichment of the plasma membrane H+-ATPase (Maudoux et al., 2000) in the plasma membrane fraction compared with that of the total membrane fraction, whereas this protein was below the detection limit in the soluble fraction, in which cytosolic UDP-Glc pyrophosphorylase (UGPase) was enriched, which indicated successful fractionation. Immunoblot analyses with an anti–green fluorescent protein (GFP) antibody specifically detected the expressed NOP1-Citrine fusion proteins at 142 and 116 kD (Figure 6C); these proteins may represent the translational products from the most upstream start codon and the one closest to the U-box, respectively. Alternatively, the 142-kD band might correspond to a NOP1-Citrine protein consisting of the 116-kD product with posttranslational modifications. A similar partitioning pattern in the three fractions for NOP1-Citrine and the H+-ATPase marker (Figure 6C) indicated that NOP1-Citrine was localized to the plasma membrane. These data suggested that the NOP1 protein predominantly localized to the plasma membrane could be functional.
Figure 6.
Plasma Membrane Localization of NOP1.
(A) Phenotype of gNOP1-Citrine/nop1 #1 and a magnified image (inset). Introduction of the NOP1 gene fused with Citrine in the C terminus rescued the impaired air-chamber phenotype of nop1.
(B) Confocal microscopy of thallus cells. The panels show Citrine fluorescence, chlorophyll (Chl) autofluorescence, and merged images from left to right, respectively. Thallus cells of the gNOP1-Citrine/nop1 #1 (top panels) and the wild type (bottom panels) were observed. Bar = 10 µm.
(C) Protein gel blot analysis of the soluble fraction (S), total membrane fraction (M), and plasma membrane fraction (PM) from gNOP1-Citrine/nop1 #1. Fusion proteins were detected with an anti-GFP antibody. H+-ATPase and UGPase were chosen as subcellular markers for the plasma membrane–bound protein and for the cytosolic protein, respectively.
DISCUSSION
Development of schizogenous ICSs has been reported in many plants since the nineteenth century (Sachs et al., 1882). However, little is known about the responsible molecular mechanism (Evans, 2004). Here, we identified the NOP1 gene as an essential factor for ICS formation in the liverwort M. polymorpha. Histological examination of air-pore and air-chamber development in Marchantiales indicates that the ICS is of schizogenous origin (Apostolakos et al., 1982). Our histological observations confirmed that schizogenous ICSs are formed in the initial stage of air-pore and air-chamber development in wild-type M. polymorpha (Figure 2). In nop1, a periclinal cell division followed by anticlinal cell divisions, which generate protodermal and subprotodermal cell layers, occur normally in the apical region of nop1 as in the wild type. However, there was no indication for the presence of an initial aperture, which later enlarged into a subepidermal ICS in the air chamber connected to the external environment via the air pore (Figure 2). These observations indicated the critical role played by NOP1 in the formation of schizogenous ICS.
NOP1 encodes a protein that shows high overall similarity to the PUB-ARM protein family (see Supplemental Figure 2 online), which has a U-box domain implicated in E3 ubiquitin ligases (Yee and Goring, 2009). Several of the PUB-ARM proteins show E3 ubiquitin ligase activity in vitro (Stone et al., 2003; Mudgil et al., 2004; Zeng et al., 2004). Correspondingly, we demonstrated the U-box–dependent E3 ubiquitin ligase activity of NOP1 in vitro (Figure 5). Furthermore, confocal microscopy and biochemical analyses showed localization of NOP1 to the plasma membrane (Figure 6). These results demonstrated that NOP1 is a PUB-ARM–type E3 ubiquitin ligase localized to the plasma membrane. It should be noted that no transmembrane helix was predicted to be present in NOP1. Interestingly, a close Arabidopsis homolog of NOP1, PUB44/SAUL (see Supplemental Figure 3 online), associates with the plasma membrane via the C terminus ARM repeats, and the subcellular localization was linked to its in vivo function (Drechsel et al., 2011). The ARM repeat domain is a highly conserved right-handed superhelix of α-helices involved in protein–protein interactions (Samuel et al., 2006; Tewari et al., 2010). The plasma membrane association of NOP1 might also be dependent on the C terminus ARM repeats via interaction with the plasma membrane–localized regulatory protein and critical for the regulatory mechanism of ICS formation. Recently, Salt et al. (2011) suggested a complex regulatory mechanism for subcellular localization of PUB44/SAUL. Under a variety of conditions, such as treatment with abscisic acid, methyl jasmonate, and auxin, as well as coexpression with receptor kinases, the localization of PUB44/SAUL was observed to change to the cytoplasm, plasma membrane, or nucleus in tobacco (Nicotiana tabacum) Bright Yellow-2 cells (Salt et al., 2011). Though the regulatory mechanism for the subcellular localization of PUB44/SAUL has not been elucidated, those observations suggest the involvement of various modes of action and the potential involvement of PUB44/SAUL in plasma membrane–localized receptor kinase signaling.
At the onset of ICS formation, the external periclinal walls of the protodermal cells bulge locally and form small surface cavities at the site where the ICS will open (Apostolakos and Galatis, 1985). Ultrastructural examination of air-pore and air-chamber formation in M. paleacea revealed that the anticlinal cell wall below the cavity thickens, and such cell wall remodeling below the cavity expands gradually down to the subprotodermal cell layer, which is accompanied by displacement of microtubule systems (Apostolakos and Galatis, 1985). The initiation of ICS is restricted to a subset of anticlinal cell walls between three or more protodermal cells around the apical notch region (Apostolakos et al., 1982). To determine the position of ICS formation, cells must communicate for perception of their relative position within the dorsal side of the apical notch region to regulate cell wall remodeling for ICS initiation. In Arabidopsis, cell separation during floral organ abscission after pollination and lateral organ emergence are regulated by the peptide INFLORESCENCE DEFICIENT IN ABSCISSION, which signals through the Leu-rich repeat receptor-like kinases HAESA and HAESA-LIKE2, and the process is involved in the regulation of cell wall remodeling (Stenvik et al., 2008; Kumpf et al., 2013). Increasing evidence implicates the involvement of PUB-ARM proteins in various plant receptor-like kinase signaling pathways, and the ARM domains mediate binding to the kinase domains (Gu et al., 1998; Kim et al., 2003; Samuel et al., 2006, 2008; Mbengue et al., 2010; Lu et al., 2011). For example, the PUB-ARM proteins PUB12 and PUB13 attenuate the activity of the Arabidopsis flagellin-sensing receptor 2 by ubiquitination and subsequent degradation (Lu et al., 2011). Similarly, NOP1 may participate in a plasma membrane–localized receptor-like kinase signaling pathway that regulates ICS formation of air chambers via ubiquitination and degradation of target protein(s). Given that NOP1 is expressed widely throughout the thallus (see Supplemental Figure 5 online), there are likely to be target proteins and regulatory factors of NOP1 that function as key factors regulating ICS formation in M. polymorpha. Further investigations of NOP1-mediated air-chamber development in M. polymorpha should shed light on the regulatory mechanism of ICS formation and its evolution in land plants.
The development of ICSs is considered to be a critical step in the evolution of land plants (Renzaglia et al., 2000; Raven, 2002). The ICSs in air chambers must also be significant for members of the Marchantiales in ever-changing terrestrial environments, which are sometimes dry and sometimes waterlogged, since bryophytes are generally associated with wet habitats. There were no clear differences in thallus growth between nop1 and the wild type when cultured on agar medium under continuous white light irradiation at optimum temperature. The humidity was maintained at a high level, but the thalli were never submerged in water in these culture conditions. The significance of ICSs in the growth of M. polymorpha is difficult to detect under such laboratory conditions. Further research on the growth rate and photosynthetic properties of the wild type and nop1 under various conditions (e.g., dry and waterlogged conditions) should be conducted to define the functional significance of ICSs. The use of the air-chamber-less mutant nop1 should provide an ideal experimental system to clarify the physiological significance of ICSs and their evolutionary contribution to plants in various environments.
METHODS
Plant Material and Growth Conditions
Male and female accessions of Marchantia polymorpha, Tak-1 and Tak-2, respectively (Ishizaki et al., 2008), were maintained asexually. F1 spores generated by crossing Tak-2 and Tak-1 were used for transformation. Formation of sexual organs was induced by far-red irradiation as described previously (Chiyoda et al., 2008). Mature sporangia were collected 3 to 4 weeks after crossing, air-dried for 7 to 10 d, and stored at −80°C until use.
Plants were cultured using half-strength Gamborg’s B5 medium (Gamborg et al., 1968) containing 1% agar under 50 to 60 µmol photons m−2 s−1 continuous white fluorescent light at 22°C unless otherwise defined.
Generation and Screening of T-DNA–Tagged Lines of M. polymorpha
Agrobacterium tumefaciens strain GV2260 transformed by the pCAMBIA1300 binary vector was used for preparation of T-DNA–tagged lines. Transformation of M. polymorpha was performed by cocultivation of sporelings with Agrobacterium as described previously (Ishizaki et al., 2008). Hygromycin-resistant transformants were first selected on half-strength B5 agar medium containing 1% agar, 10 mg L−1 hygromycin (Wako Pure Chemical Industries), and 100 mg L−1 cefotaxime (Claforan; Sanofi-Aventis). After growth of hygromycin-resistant transformants to 3 to 5 mm diameter, independent T1 lines were transferred to fresh agar plates containing half-strength B5 medium containing 1% agar and 100 mg L−1 cefotaxime, grown for 3 to 4 weeks, and screened for abnormal thallus morphology by visual examination of phenotypes. Isogenic lines (the G1 generation) were established from a T1 line of morphological phenotypes using gemmae that arose asexually from single initial cells as described previously (Ishizaki et al., 2012). Plants grown from gemmae of G1 lines (the G2 generation) were used for further experiments.
Phenotype Analysis and Histology
Three-week-old thalli developed from gemmae were dissected into small pieces and transferred to fixative solution containing 4% glutaraldehyde in 0.05 M phosphate buffer, pH 7.0, and evacuated with a water aspirator until the specimens sank. After fixation for 3 h at room temperature, the samples were rinsed in 0.05 M phosphate buffer and postfixed in 1% buffered osmium tetroxide at room temperature for 2 h. The samples were dehydrated in a graded ethanol series and embedded in Quetol 651 plastic resin (Nissin EM) using a graded series of propylene oxide and resin. Polymerization of the resin was performed at 60°C for 24 h. Semithin sections (0.5- to 1-μm thickness) for light microscopy were obtained with an Ultramicrotome (Ultracut R; Leica Microsystems) using glass knives and stained with toluidine blue O.
For scanning electron microscopy, 10-d-old thalli were frozen in liquid nitrogen and observed with a JSM-6060LVS scanning electron microscope (JOEL).
DNA Gel Blot Analysis
Total DNA was extracted from ∼5 g of fresh weight of tissue using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980) with modifications as described previously (Ishizaki et al., 2013). Two micrograms of genomic DNA was digested overnight with ClaI, BglII, XhoI, and PstI at 37°C. The DNA was fractionated by electrophoresis in a 0.8% (w/v) agarose gel and blotted onto a positively charged nylon membrane Biodyne A (PALL). The 1064-bp PstI-XhoI fragment excised from pCAMBIA1300 was used as a probe to detect the cauliflower mosaic virus 35S promoter in the T-DNA of pCAMBIA1300. The blotted membranes were hybridized in Church hybridization buffer (Church and Gilbert, 1984) at 65°C with a probe labeled with [α-32P]deoxycytidine 5′-triphosphate by the Random Primer Labeling Kit Version 2 (Takara Bio). Washing and analyses of blots were performed as described previously (Chiyoda et al., 2008).
TAIL-PCR and Sequencing of TAIL-PCR Products
TAIL-PCR amplification was performed in accordance with the method of Liu et al. (1995) with minor modifications using KOD FX DNA polymerase (TOYOBO) and the PCR protocol listed in Supplemental Table 1 online. Primary PCR was performed using ∼40 ng of genomic DNA as a template. The following nested primers for T-DNA were designed: CTL1, CTL2, and CTL3 for the left border and CTR1, CTR2, and CTR3 for the right border of T-DNA in pCAMBIA1300. Each of these primers was paired with one of the three arbitrary degenerate primers AD1, AD2, and AD3. TAIL-PCR products of tertiary reactions were analyzed using 1% agarose gel electrophoresis. Distinct bands were excised from agarose gels and purified using the QIAquick gel extraction kit (Qiagen). Purified PCR products were sequenced using the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems). All primers used in this study are listed in Supplemental Table 2 online.
Complementation of nop1
For complementation, the binary plasmid containing the NOP1 genomic fragment from 5.7 kb upstream of the putative start codon to 0.2 kb downstream of the 3′ untranslated region and a chlorosulfuron resistance marker gene (Ishizaki et al., 2013) was used. The plasmid was transformed into regenerating thalli of nop1 as described previously (Kubota et al., 2013). Selection was conducted at a concentration of 0.5 μM chlorosulfuron (DuPont).
Genotyping
Small pieces (3 × 3 mm) of thalli were taken from individual plants and crushed with a micropestle in 100 μL of buffer containing 100 mM Tris-HCl, pH 9.5, 1 M KCl, and 10 mM EDTA. Sterilized water (400 µL) was added to each tube, and 1 μL of aliquot of the extract was used as a template for PCR using KOD FX Neo DNA polymerase (Toyobo). To confirm the T-DNA insertion position in the mutant and the success of complementation, 1 μL from the extract was used for PCR with the primers L1, R1, and CTL3. Each primer position and primer combinations used for genotyping of both types are shown in Figure 4.
Expression Analysis by RT-PCR
Total RNA was isolated using Qiagen RNeasy (Qiagen). First-strand cDNA was synthesized from 1 µg of total RNA with ReverTra Ace reverse transcriptase (Toyobo) and oligo(dT) primer. PCR amplification of the NOP1 cDNA-specific sequence was performed using primers L1 and R1. PCR amplification of the cDNA encoding the EF1α gene with primers MpEF1-L and MpEF1-R served as a control. These reactions were performed using the GeneAmp PCR system 9700 (Applied Biosystems).
Phylogenetic Analysis
U-box domains of NOP1 and 41 Arabidopsis thaliana PUB-ARM proteins were detected using SMART (Schultz et al., 1998) (http://smart.embl-heidelberg.de). Multiple sequence alignments of the U-box domains including the U-box domain of Saccharomyces cerevisiae PRP19 were constructed using the MUSCLE program (Edgar, 2004) implemented in Geneious software version 6.1.6 (Biomatters; http://www.geneious.com/) with default parameters. The obtained alignment (see Supplemental Data Set 1 online) was used to estimate the unrooted maximum likelihood phylogenetic tree with the PhyML program version 2.1.0 (Guindon and Gascuel, 2003) implemented in the Geneious software using the LG model and four categories of rate substitution. Tree topology, branch length, and substitution rates were optimized, and the tree topology was searched using the nearest-neighbor interchange method. Bootstrap values were computed from 2000 trials.
Recombinant Protein Expression and Purification
The NOP1 fragment was amplified by PCR using the primers CDS-MBP-L and CDS-MBP-R and cloned into pMAL-c2 (New England Biolabs) using the InFusion HD cloning kit (Takara Bio), which generated the plasmid pMAL-NOP1 for expression of the MBP-NOP1 fusion protein in bacterial cells. pMAL-NOP1 derivatives each carrying a mutation of Leu-261 to Ser (L261S), Cys-299 to Ser (C299S), or Pro-300 to Ala (P300A) were generated by PCR using the primer pairs L261S-F/L261S-R, C299S-F/C299S-R, and P300A-F/P300A-R, respectively. Wild-type and mutated constructs were transformed into Escherichia coli strain Rosetta 2 (DE3; Merck Millipore). To express proteins, overnight cultures were diluted 1:200 in a total volume of 100 mL. Induction of proteins was performed using isopropyl-β-d-thiogalactopyranoside for 12 h at 18°C. Following induction, cells were harvested by centrifugation at 4°C and 4000g for 15 min, resuspended in 5 mL of sonication buffer (150 mM NaCl, 20 mM Tris-HCl, and 10% glycerol, pH 7.4) to which a protease inhibitor cocktail (Complete tablet; Roche Diagnostics) and 0.1 mg mL−1 of lysozyme had been added, and sonicated. After centrifugation (4°C and 14,000g for 10 min) of the obtained cell lysates, recombinant proteins in the supernatants were purified by affinity chromatography using amylose resin (New England Biolabs) and used for the in vitro ubiquitination assay.
Ubiquitination Assays
Reactions were performed in a total volume of 50 μL using components of the Ubiquitinylation kit (Enzo Life Sciences). Each reaction contained 0.1 µM human E1, 2.5 µM E2 (UbcH5b, His6 tagged), 2.5 µM biotin-Ub, 1× ubiquitination buffer, 1 mM DTT, 1 unit of inorganic pyrophosphatase (Sigma-Aldrich), and 3 µg of recombinant MBP-NOP1 protein as indicated. The reactions were incubated for 2 h at 30°C and stopped by adding 50 μL 2× nonreducing loading buffer. Protein samples were resolved on 6% SDS-PAGE gels followed by protein gel blot analysis using a 1:4000 dilution of HRP–labeled streptavidin (High Sensitivity Streptavidin-HRP; Thermo Fisher Scientific) and a 1:10,000 dilution of monoclonal anti-MBP antibody (Clone MBP-17; Sigma-Aldrich).
Subcellular Localization Analysis Using Confocal Microscopy
In order to generate the gNOP1-Citrine construct, the promoter region and the coding region without the stop codon of NOP1 were amplified by genomic PCR using the primer sets PRO-L/PRO-R2 and gCDSL/gCDSR-ns, respectively, and cloned into pENTR/D-TOPO (Life Technologies) to generate pMM001 and pMM002, respectively. The 4.4-kb AvrII-AscI DNA fragment of pMM002 was subcloned into the AvrII and AscI sites of pMM001 to generate the plasmid pMM003, which contained the NOP1 genomic fragment from 5.7 kb upstream of the putative start codon to the coding region up to the stop codon. The resultant NOP1 cassette was subcloned into pMpGWB307 using LR Clonase II (Life Technologies) in accordance with the manufacturer’s protocol, which generated fusion of the NOP1 gene with Citrine at the C terminus. The binary plasmid was transformed into regenerating thalli of nop1 by the method described previously (Kubota et al., 2013). Selection was conducted with 0.5 µM chlorosulfuron (Dupont).
Fluorescence derived from Citrine and chloroplasts was monitored in 14-d-old thalli by confocal laser scanning microscopy (FV1000; Olympus) using 515-nm laser light for excitation, and detection windows ranged from 525 to 565 nm for Citrine and from 650 to 750 nm for chloroplast autofluorescence.
Membrane Fractionation
For fractionation, thallus tissues were homogenized with Polytron (Kinematica) in ∼3 volumes of the tissue weight of 500 mM Suc, 20 mM EDTA, 20 mM EGTA, 50 mM sodium fluoride, 1% (w/v) polyvinylpyrrolidone, 20 µM p-amidinophenylmethanesulfonyl fluoride, 2 mM DTT, 10% (w/v) glycerol, and 50 mM Tris-MES, pH 8.0. The homogenates were filtered and centrifuged at 5000g for 15 min. The supernatants were centrifuged at 200,000g for 60 min, and then the precipitates were suspended in 0.33 M Suc, 2 mM DTT, and 10 mM potassium phosphate buffer, pH 7.8 (Suc-KP buffer) and used as the total membrane fraction. The plasma membrane fraction was purified from the total membrane fraction by the aqueous polymer two-phase partition method (Yoshida et al., 1983; Santoni, 2007) as follows. The total membrane fraction was mixed with a separation solution (3.0 g of polyethylene glycol-3350 [Sigma-Aldrich], 3.0 g of Dextran 500 [SERVA], 28 mL of Suc-KP buffer, and 4.8 mL of 0.3 M NaCl in Suc-KP buffer), vigorously agitated at 4°C for 5 min, and then centrifuged at 5000g for 10 min. The resulting supernatant fractions were diluted with 0.3 M Suc, 10 mM boric acid, 9 mM KCl, 5 mM EDTA, 5 mM EGTA, 50 mM NaF, 2 mM DTT, and 10 mM Tris-MES, pH 8.3, and then centrifuged at 200,000g for 60 min. The precipitates were suspended in the same buffer and stored at −80°C as the plasma membrane fraction. Protein concentration was measured with a protein assay solution (Bio-Rad). Proteins were separated on 10% SDS-PAGE gels, transferred onto polyvinylidene difluoride membranes, and detected with antibodies as indicated in each figure. The antibodies were diluted as follows: 1:5000 for anti-GFP (Invitrogen), 1:10,000 for anti-UGPase (AgriSera), and anti-H+-ATPase (AgriSera), in combination with a 1:10,000 dilution of anti-rabbit IgG HRP–conjugated second antibody (GE Healthcare). Signals were detected using the ECL Plus Western Blotting Detection System (GE Healthcare) and the ImageQuant LAS-4010 digital imaging system (GE Healthcare).
Accession Numbers
Sequence data from this article can be found in the DDBJ/GenBank/EMBL databases under the following accession numbers: AB830886 (mRNA of NOP1) and AB830887 (the genomic locus of NOP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Molecular Characterization of the T-DNA Insertion Site of nop1.
Supplemental Figure 2. Amino Acid Sequence Alignment of NOP1 and Seven Other PUB-ARM Proteins.
Supplemental Figure 3. Phylogeny of NOP1 and Arabidopsis PUB-ARM Proteins.
Supplemental Figure 4. Time-Dependent Assay of in Vitro Ubiquitination of NOP1 Protein.
Supplemental Figure 5. Expression of the NOP1 Gene in Developing Thallus.
Supplemental Table 1. Cycle Settings Used for TAIL-PCR.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Alignment of the U-Box domains of NOP1 and 41 Arabidopsis PUB-ARM Proteins Used for the Phylogenetic Analysis Shown in Supplemental Figure 3.
Supplementary Material
Acknowledgments
We thank Makoto Shirakawa, Miho Takemura, and Katsuyuki T. Yamato for technical advice and discussions. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research on Priority Area (23012025 to T.K.) and for Scientific Research on Innovative Area (25113009 to T.K. and 25119711 and 25114510 to K.I.), the Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (C) (24580140 to K.I. and 24570048 to R.N.), for Challenging Exploratory Research (24658095 to T.K.), and for Japan Society for the Promotion of Science Fellows (25-5776 to M.M.), and by the Asahi Glass Foundation and SUNTORY Foundation for Life Sciences (to K.I.).
AUTHOR CONTRIBUTIONS
K.I. and T.K. designed the research. K.I., M.M., M.S., A.M., and R.N. performed the experiments. K.I., M.M., M.S., R.N., and T.K. analyzed the data. K.I., M.S., R.N., and T.K. wrote the article.
Glossary
- ICS
intercellular space
- TAIL-PCR
thermal asymmetric interlaced–PCR
- UND
N-terminal end of the U-box
- UGPase
UDP-Glc pyrophosphorylase
- GFP
green fluorescent protein
References
- Apostolakos P., Galatis B. (1985). Studies on the development of the air pores and air chambers of Marchantia paleacea. II. Ultrastructure of the initial aperture formation with particular reference to cortical microtubule organizing centres. Can. J. Bot. 63: 744–756 [Google Scholar]
- Apostolakos P., Galatis B., Mitrakos K. (1982). Studies on the development of the air pores and air chambers of Marchantia paleacea I. light microscopy. Ann. Bot. (Lond.) 49: 377–396 [Google Scholar]
- Aravind L., Koonin E.V. (2000). The U box is a modified RING finger - A common domain in ubiquitination. Curr. Biol. 10: R132–R134 [DOI] [PubMed] [Google Scholar]
- Barnes C.R., Land W.J.G. (1907). Bryological papers. I. The origin of air chambers. Bot. Gaz. 44: 197–213 [Google Scholar]
- Bowman J.L., Floyd S.K., Sakakibara K. (2007). Green genes-comparative genomics of the green branch of life. Cell 129: 229–234 [DOI] [PubMed] [Google Scholar]
- Chiyoda S., Ishizaki K., Kataoka H., Yamato K.T., Kohchi T. (2008). Direct transformation of the liverwort Marchantia polymorpha L. by particle bombardment using immature thalli developing from spores. Plant Cell Rep. 27: 1467–1473 [DOI] [PubMed] [Google Scholar]
- Church G.M., Gilbert W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crandall-Stotler, B. (1981). Morphology/anatomy of hepatics and anthocerotes. In Advances in Bryology, W. Schultze-Motel, ed (Vaduz, Lichtenstein: J. Cramer), pp. 315–398. [Google Scholar]
- Dale J.E. (1988). The control of leaf expansion. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39: 267–295 [Google Scholar]
- Drechsel G., Bergler J., Wippel K., Sauer N., Vogelmann K., Hoth S. (2011). C-terminal armadillo repeats are essential and sufficient for association of the plant U-box armadillo E3 ubiquitin ligase SAUL1 with the plasma membrane. J. Exp. Bot. 62: 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D., Kerp H., Hass H. (1998). Stomata in early land plants: An anatomical and ecophysiological approach. J. Exp. Bot. 49: 255–278 [Google Scholar]
- Evans D.E. (2004). Aerenchyma formation. New Phytol. 161: 35–49 [Google Scholar]
- Gamborg O.L., Miller R.A., Ojima K. (1968). Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Green T.G.A., Snelgar W.P. (1982). A comparison of photosynthesis in two thalloid liverworts. Oecologia 54: 275–280 [DOI] [PubMed] [Google Scholar]
- Gu T., Mazzurco M., Sulaman W., Matias D.D., Goring D.R. (1998). Binding of an arm repeat protein to the kinase domain of the S-locus receptor kinase. Proc. Natl. Acad. Sci. USA 95: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. (2003). A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Chiyoda S., Yamato K.T., Kohchi T. (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49: 1084–1091 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Nonomura M., Kato H., Yamato K.T., Kohchi T. (2012). Visualization of auxin-mediated transcriptional activation using a common auxin-responsive reporter system in the liverwort Marchantia polymorpha. J. Plant Res. 125: 643–651 [DOI] [PubMed] [Google Scholar]
- Ishizaki K., Johzuka-Hisatomi Y., Ishida S., Iida S., Kohchi T. (2013). Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci. Rep. 3: 1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M.B., Armstrong W. (1999). Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1: 274–287 [Google Scholar]
- Joshi R., Kumar P. (2012). Lysigenous aerenchyma formation involves non-apoptotic programmed cell death in rice (Oryza sativa L.) roots. Physiol. Mol. Biol. Plants 18: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Cho H.S., Kim D.M., Lee J.H., Pai H.S. (2003). CHRK1, a chitinase-related receptor-like kinase, interacts with NtPUB4, an armadillo repeat protein, in tobacco. Biochim. Biophys. Acta 1651: 50–59 [DOI] [PubMed] [Google Scholar]
- Kubota A., Ishizaki K., Hosaka M., Kohchi T. (2013). Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Biochem. 77: 167–172 [DOI] [PubMed] [Google Scholar]
- Kumpf R.P., Shi C.L., Larrieu A., Stø I.M., Butenko M.A., Péret B., Riiser E.S., Bennett M.J., Aalen R.B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. (2012). SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 40 (Database issue): D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.G., Mitsukawa N., Oosumi T., Whittier R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8: 457–463 [DOI] [PubMed] [Google Scholar]
- Lu D., Lin W., Gao X., Wu S., Cheng C., Avila J., Heese A., Devarenne T.P., He P., Shan L. (2011). Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. (2000). A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 275: 17762–17770 [DOI] [PubMed] [Google Scholar]
- Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22: 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgil Y., Shiu S.H., Stone S.L., Salt J.N., Goring D.R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-box E3 ubiquitin ligase family. Plant Physiol. 134: 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M.G., Thompson W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8: 4321–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M.D., Vander Kooi C.W., Rosenberg J.A., Chazin W.J., Gould K.L. (2003). Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 10: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringa E., Martinez-Noel G., Muller U., Harbers K. (2001). Interaction of the ring finger-related U-box motif of a nuclear dot protein with ubiquitin-conjugating enzymes. J. Biol. Chem. 276: 19617–19623 [DOI] [PubMed] [Google Scholar]
- Raven J.A. (1996). Into the voids: The distribution, function, development and maintenance of gas spaces in plants. Ann. Bot. (Lond.) 78: 137–142 [Google Scholar]
- Raven J.A. (2002). Selection pressures on stomatal evolution. New Phytol. 153: 371–386 [DOI] [PubMed] [Google Scholar]
- Renzaglia K.S., Duff R.J.T., Nickrent D.L., Garbary D.J. (2000). Vegetative and reproductive innovations of early land plants: Implications for a unified phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355: 769–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, J., Bennett, A.W., Vines, S.H., and Thiselton-Dyer, W.T. (1882). Text-book of Botany: Morphological and Physiological. (Oxford: Clarendon Press). [Google Scholar]
- Salt J.N., Yoshioka K., Moeder W., Goring D.R. (2011). Altered germination and subcellular localization patterns for PUB44/SAUL1 in response to stress and phytohormone treatments. PLoS ONE 6: e21321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel M.A., Salt J.N., Shiu S.H., Goring D.R. (2006). Multifunctional arm repeat domains in plants. Int. Rev. Cytol. 253: 1–26 [DOI] [PubMed] [Google Scholar]
- Samuel M.A., Mudgil Y., Salt J.N., Delmas F., Ramachandran S., Chilelli A., Goring D.R. (2008). Interactions between the S-domain receptor kinases and AtPUB-ARM E3 ubiquitin ligases suggest a conserved signaling pathway in Arabidopsis. Plant Physiol. 147: 2084–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoni, V. (2007). Plant plasma membrane protein extraction and solubilization for proteomic analysis. In Plant Proteomics, H. Thiellement, M. Zivy, C. Damerval, and V. Méchin, eds (New York: Humana Press), pp. 93–109. [DOI] [PubMed] [Google Scholar]
- Schönherr J., Ziegler H. (1975). Hydrophobic cuticular ledges prevent water entering the air pores of liverwort thalli. Planta 124: 51–60 [DOI] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P., Ponting C.P. (1998). SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seago J.L., Jr, Marsh L.C., Stevens K.J., Soukup A., Votrubová O., Enstone D.E. (2005). A re-examination of the root cortex in wetland flowering plants with respect to aerenchyma. Ann. Bot. (Lond.) 96: 565–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M. (2012). An introduction to the taxonomy and morphology of Marchantia polymorpha. BSJ-Review 3: 84–113 [Google Scholar]
- Smith, G.M. (1955). Cryptogamic Botany: Bryophytes and Pteridophytes, Vol. II. (New York, Toronto, London: McGraw-Hill Book Company). [Google Scholar]
- Stenvik G.E., Tandstad N.M., Guo Y., Shi C.L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Anderson E.M., Mullen R.T., Goring D.R. (2003). ARC1 is an E3 ubiquitin ligase and promotes the ubiquitination of proteins during the rejection of self-incompatible Brassica pollen. Plant Cell 15: 885–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari R., Bailes E., Bunting K.A., Coates J.C. (2010). Armadillo-repeat protein functions: Questions for little creatures. Trends Cell Biol. 20: 470–481 [DOI] [PubMed] [Google Scholar]
- Yee D., Goring D.R. (2009). The diversity of plant U-box E3 ubiquitin ligases: From upstream activators to downstream target substrates. J. Exp. Bot. 60: 1109–1121 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Uemura M., Niki T., Sakai A., Gusta L.V. (1983). Partition of membrane particles in aqueous two-polymer phase system and its practical use for purification of plasma membranes from plants. Plant Physiol. 72: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.