This work reports that NLA mediates endocytosis and degradation of PHT1s via ubiquitination and functions cooperatively with PHO2 to regulate the abundance of PHT1s in different subcellular compartments. This finding uncovers a mechanism of modulation of Pi transport activity through the integration of miRNA-mediated posttranscriptional and ubiquitin-mediated posttranslational pathways.
Abstract
Members of the Arabidopsis thaliana PHOSPHATE TRANSPORTER1 (PHT1) family are key players in acquisition of Pi from the rhizosphere, and their regulation is indispensable for the maintenance of cellular Pi homeostasis. Here, we reveal posttranslational regulation of Pi transport through modulation of degradation of PHT1 proteins by the RING-type ubiquitin E3 ligase, NITROGEN LIMITATION ADAPTATION (NLA). Loss of function of NLA caused high Pi accumulation resulting from increases in the levels of several PHT1s at the protein rather than the transcript level. Evidence of decreased endocytosis and ubiquitination of PHT1s in nla mutants and interaction between NLA and PHT1s in the plasma membranes suggests that NLA directs the ubiquitination of plasma membrane–localized PHT1s, which triggers clathrin-dependent endocytosis followed by endosomal sorting to vacuoles. Furthermore, different subcellular localization of NLA and PHOSPHATE2 (PHO2; a ubiquitin E2 conjugase) and the synergistic effect of the accumulation of PHT1s and Pi in nla pho2 mutants suggest that they function independently but cooperatively to regulate PHT1 protein amounts. Intriguingly, NLA and PHO2 are the targets of two Pi starvation-induced microRNAs, miR827 and miR399, respectively. Therefore, our findings uncover modulation of Pi transport activity in response to Pi availability through the integration of a microRNA-mediated posttranscriptional pathway and a ubiquitin-mediated posttranslational regulatory pathway.
INTRODUCTION
P is an essential macroelement required for the biogenesis of biomembranes and genetic materials and is involved in energy transfer processes, regulation of enzymatic activities, and signal transduction (Marschner, 1995). In contrast with animals, plants are sessile and rely on acquisition of P from the rhizosphere. P is acquired by plants primarily in the form of Pi by Pi transporters (PHOSPHATE TRANSPORTER [PHT]) in the roots. However, Pi is not often readily available due to adsorption, precipitation with cations, or conversion into organic forms in the soil (Raghothama, 1999). In spite of fluctuations in externally available Pi, plants have to balance external Pi with internal needs to maintain cellular P homeostasis in order to coordinate growth, development, and reproduction. This is achieved by regulation of Pi uptake, mobilization, and partitioning to various organs (Chiou and Lin, 2011). Pi transporters located in the plasma membrane (PM) are the key players in regulating the initial uptake of Pi from the rhizosphere and subsequent Pi allocation inside plants (Chiou et al., 2001; Misson et al., 2004; Shin et al., 2004). Therefore, regulation of Pi transport activity provides the first line of protection against disturbance of cellular Pi homeostasis.
In Arabidopsis thaliana, members of the PHT1 family are responsible for direct Pi acquisition and distribution into different tissues (Nussaume et al., 2011). Among them, PHT1;1 and PHT1;4 are reported to be the major players in Pi acquisition under Pi-sufficient conditions (Shin et al., 2004). Many early studies emphasized the transcriptional regulation of PHT1 genes, which are upregulated by Pi deprivation (Muchhal et al., 1996; Muchhal and Raghothama, 1999; Karthikeyan et al., 2002; Misson et al., 2004; Shin et al., 2004). Posttranslational regulation of PHT1 proteins was not described until more recently. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1), an endoplasmic reticulum (ER)–localized protein, assists the trafficking of PHT1s to target to the PM (González et al., 2005; Bayle et al., 2011). Recently, we reported that PHO2 mediates the degradation of PHT1 proteins as well as PHOSPHATE1 (PHO1), which is involved in Pi loading into the xylem of roots, in the endomembrane (EM) system (Liu et al., 2012; Huang et al., 2013). PHO2, the target of Pi starvation-induced microRNA399 (miR399), encodes a ubiquitin E2 conjugase (Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006). Loss of function of PHO2 resulted in increased protein amounts of PHT1s and PHO1, leading to Pi toxicity (Hamburger et al., 2002; Liu et al., 2012; Huang et al., 2013). In addition, Bayle et al. (2011) observed that PHT1;1-GFP (for green fluorescent protein) is internalized from the PM and directed to vacuoles for degradation when external Pi is ample. These results suggest the existence of posttranslational regulation of PHT1s in the PM; however, the molecular mechanism underlying the endocytosis and subsequent degradation of PHT1s remains elusive.
Previously, we reported that microRNA827 (miR827) is induced by Pi deprivation, which posttranscriptionally represses its target gene NITROGEN LIMITATION ADAPTATION (NLA) (Hsieh et al., 2009). NLA was identified from a mutant that was less adaptive to nitrate limitation and showed an early senescence phenotype (Peng et al., 2007). Yaeno and Iba (2008) also showed that NLA functions as a negative regulator of the production of salicylic acid in the immune response. NLA encodes a protein containing an N-terminal SPX (for SYG1/PHO81/XPR1) domain, shown to be responsible for modulation of Pi sensing and uptake in yeast (Auesukaree et al., 2003; Giots et al., 2003; Hürlimann et al., 2007), and a C-terminal RING domain possessing putative ubiquitin E3 ligase activity. The N-terminal SPX domain and miR827-mediated regulation prompted us to study the role of NLA in regulating Pi homeostasis. Kant et al. (2011) reported that pht1;1 and phf1 act as suppressors of the nla mutant by reducing excessive Pi accumulation under nitrate-limited conditions. Although these observations hinted at a possible role for NLA in the regulation of Pi acquisition through modulation of downstream targets PHF1 and PHT1;1, direct evidence was lacking. In addition, NLA was previously reported to localize in the nucleus (Peng et al., 2007) where PHF1 and PHT1;1 are not present, leaving the mechanism by which NLA could regulate PHF1 and PHT1;1 a mystery.
In this study, we found that NLA is predominantly localized in the PM, where it interacts with PHT1s. In addition, we demonstrated that NLA-mediated ubiquitination of PHT1s triggers the endocytosis and vacuolar degradation of PHT1s. We further showed that NLA and PHO2 function cooperatively to regulate the abundance of PHT1s in different subcellular compartments. Together, these results illustrate an important mechanism through which plants modulate Pi transport activity in response to external Pi status via the integration of microRNA (miRNA)–mediated posttranscriptional and ubiquitin-mediated posttranslational regulatory pathways.
RESULTS
Mutation of NLA Causes an Increase in PHT1s at the Protein Rather Than the Transcript Level
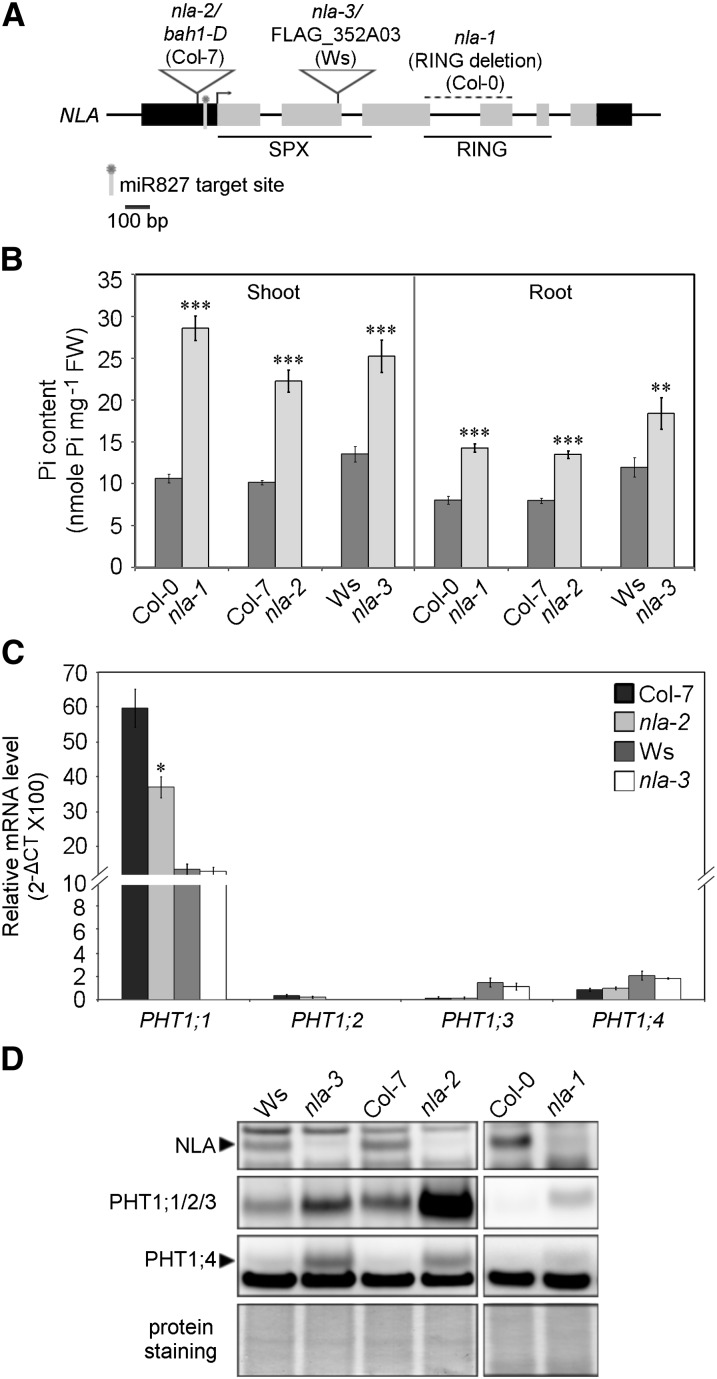
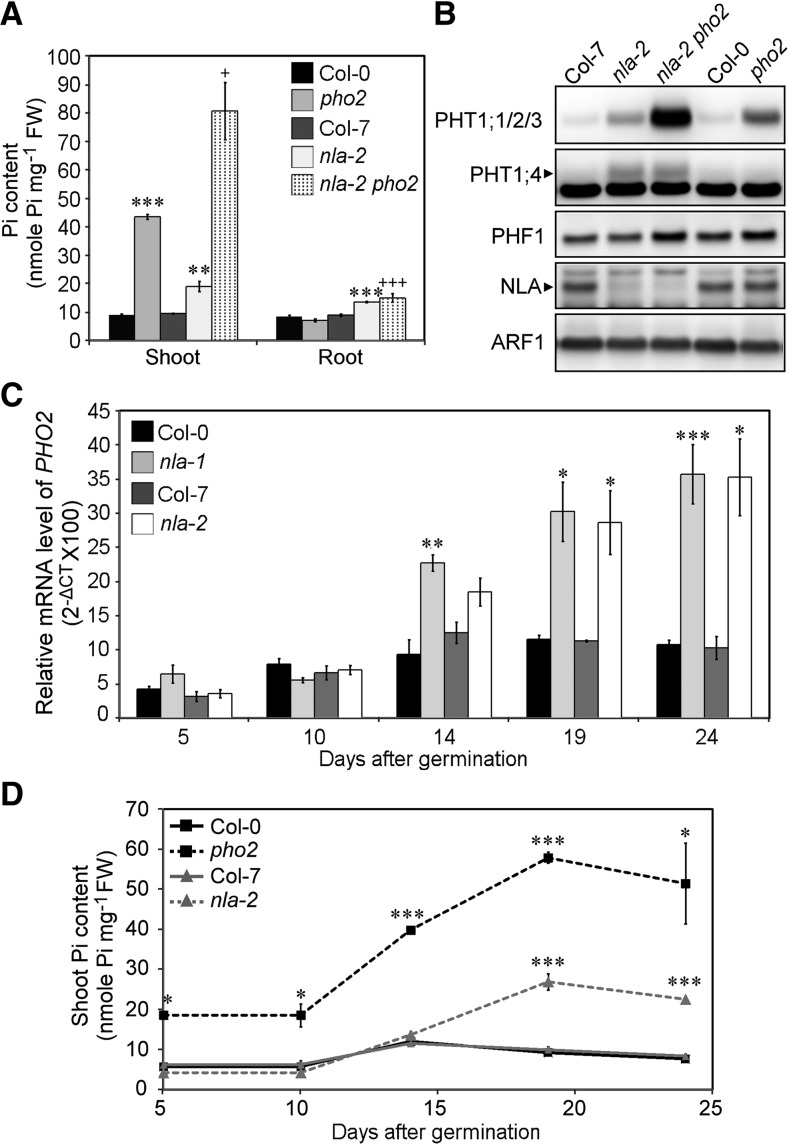
To reveal the function of NLA, we analyzed three allelic mutants, namely, nla-1, a RING-domain deletion mutant in the Columbia-0 background (Peng et al., 2007); bah1-D, a knockdown mutant in the Columbia-7 background (Yaeno and Iba, 2008) referred to hereafter as nla-2; and FLAG_352A03, a knockout mutant in the Wassilewskija background designated herein as nla-3 (Figure 1A). Under Pi-sufficient conditions, these three mutants contained around twofold higher content of Pi than the wild-type plants in both shoots and roots (Figure 1B); however, there were no visible morphological changes except for the slightly small size of nla-2 (see Supplemental Figure 1A online). Consistent with their high Pi accumulation, the nla mutants showed increased Pi acquisition activity relative to the wild type (see Supplemental Figure 1B online). We therefore inspected the expression of several PHT1s at both the transcript and protein levels by quantitative RT-PCR (qRT-PCR) and immunoblot analyses. While there were no increases in the mRNA levels of PHT1;1, PHT1;2, PHT1;3, and PHT1;4 (Figure 1C), their protein levels in all of the allelic mutants were increased notably compared with the wild type (Figure 1D; see Supplemental Figure 1C online). Under Pi deprivation conditions, PHT1 proteins were significantly upregulated, and no differences were observed between the wild type and nla mutants (see Supplemental Figure 1C online), suggesting that suppression of NLA by Pi starvation–induced miR827 may relieve its negative regulation of PHT1s. We also examined the expression of PHF1, but no differences between the wild type and mutants were observed (see Supplemental Figure 1C online). These results suggest that NLA modulates cellular Pi content via regulation of the protein abundance of PHT1s but not PHF1. It is known that loss of PHF1 function not only blocks the exit of PHT1 proteins from the ER but also decreases their stability, which in turn reduces Pi uptake activity (González et al., 2005; Huang et al., 2013). Suppression of the nla phenotype by PHF1 mutation as reported previously (Kant et al., 2011) is likely due to the reduction in PHT1 proteins.
Figure 1.
NLA Is Involved in the Regulation of Protein Abundance of PHT1s.
(A) Gene structure and mutation sites of NLA. Black bars, untranslated region; gray bars, coding region. Col, Columbia; Ws, Wassilewskija.
(B) Pi content of the wild type and nla mutants under Pi-sufficient conditions. n = 10; error bar indicates se. Student’s t test, mutants versus the wild type, **P < 0.01 and ***P < 0.005. FW, fresh weight.
(C) qRT-PCR analysis of PHT1;1, PHT1;2, PHT1;3, and PHT1;4 mRNA in the roots of the wild type and nla mutants. n = 3; error bar indicates se. Student’s t test, mutants versus the wild type, *P < 0.05. CT, cycle threshold.
(D) Immunoblot analysis of NLA, PHT1;1/2/3, and PHT1;4 proteins in the 19-d-old wild type and nla mutants under Pi-sufficient conditions.
NLA Is Important for Regulating the Abundance of PHT1s to Maintain Pi Homeostasis
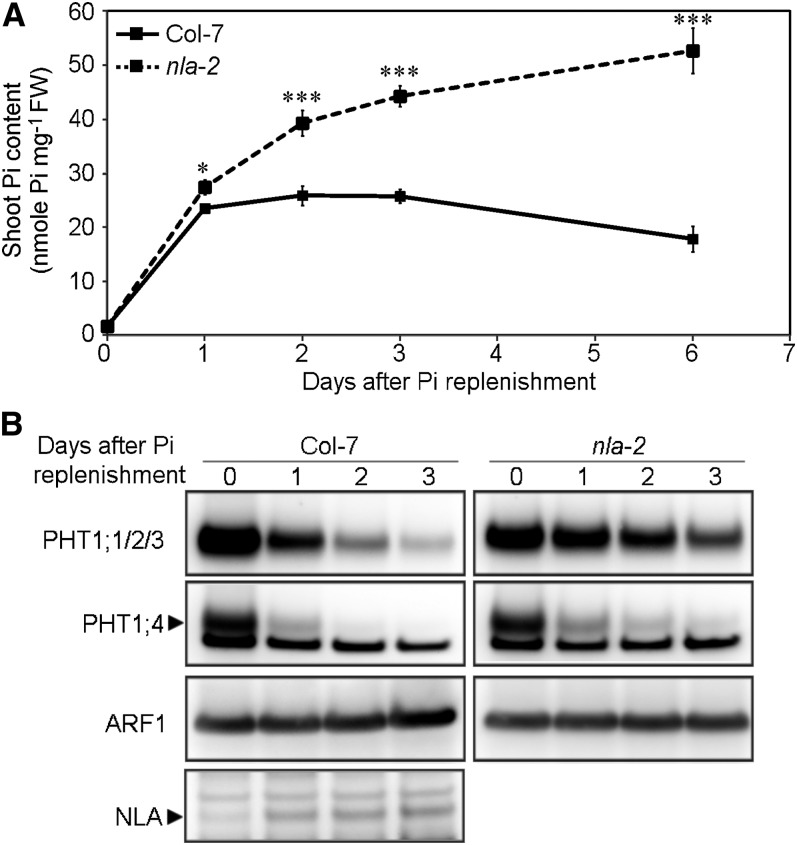
Because the expression of NLA is negatively regulated under Pi-deficient conditions, we next examined the responses of the wild type and nla-2 mutants after recovering from Pi deprivation when the repression of NLA was relieved to determine the effect of loss of function of NLA. Four-day Pi starvation lowered the Pi content in the shoots of the wild type and nla-2 mutants to a comparable level (the wild type, 1.61 ± 0.05; nla-2, 1.61 ± 0.10 nmol Pi mg−1 fresh weight). In the shoots of the wild type, Pi quickly accumulated 1 d after Pi replenishment, was maintained at a high level for 3 d, and then gradually declined (Figure 2A). By contrast, nla-2 mutants kept accumulating Pi after resupply, resulting in severe Pi toxicity as seen in necrosis at the edges of old leaves (see Supplemental Figure 2A online). In the wild type, primary miR827 transcripts were downregulated rapidly 1 d after Pi resupply (see Supplemental Figure 2C online), which was in line with the upregulation of the NLA proteins (Figure 2B). When the expression of PHT1;1, PHT1;2, PHT1;3, and PHT1;4 was examined, we found that their mRNAs were all reduced markedly 1 d after replenishment and maintained at a constant low level (see Supplemental Figure 2B online). The protein level of PHT1s in the wild type was also decreased 1 d after replenishment and decreased even further in the following days (Figure 2B). The mRNA and protein levels of PHT1s were not well correlated during Pi replenishment either in the wild type or nla-2 mutants, suggesting the existence of regulation in addition to the transcriptional regulation, likely protein degradation. Of note, nla-2 mutants showed much slower reduction in the protein levels of PHT1;1/2/3 (PHT1;1, PHT1;2, and PHT1;3) and PHT1;4 than the wild type (Figure 2B), despite there being no differences in their mRNA levels (see Supplemental Figure 2B online). These results indicate that NLA is necessary for maintenance of Pi homeostasis in response to fluctuations in external Pi supply and further support the role of NLA in regulating the protein levels of PHT1s. During Pi replenishment, the protein levels of PHT1s in the nla-2 mutants continued to gradually decrease, implying the involvement of an NLA-independent pathway (possibly mediated by PHO2; see below) or nonspecific proteolysis in the degradation of PHT1s.
Figure 2.
NLA-Mediated Regulation Is Important to Avoid Pi Overaccumulation.
(A) Shoot Pi content after Pi replenishment in the wild type and nla-2 mutants. n = 8; error bar indicates se. Student’s t test, mutants versus the wild type, *P < 0.05 and ***P < 0.005. FW, fresh weight.
(B) Immunoblot analysis of PHT1;1/2/3, PHT1;4, and NLA proteins in roots of the wild type and nla-2 mutants after Pi replenishment. The detection of ARF1 was used as a loading control.
NLA Is Predominantly Localized in the PM
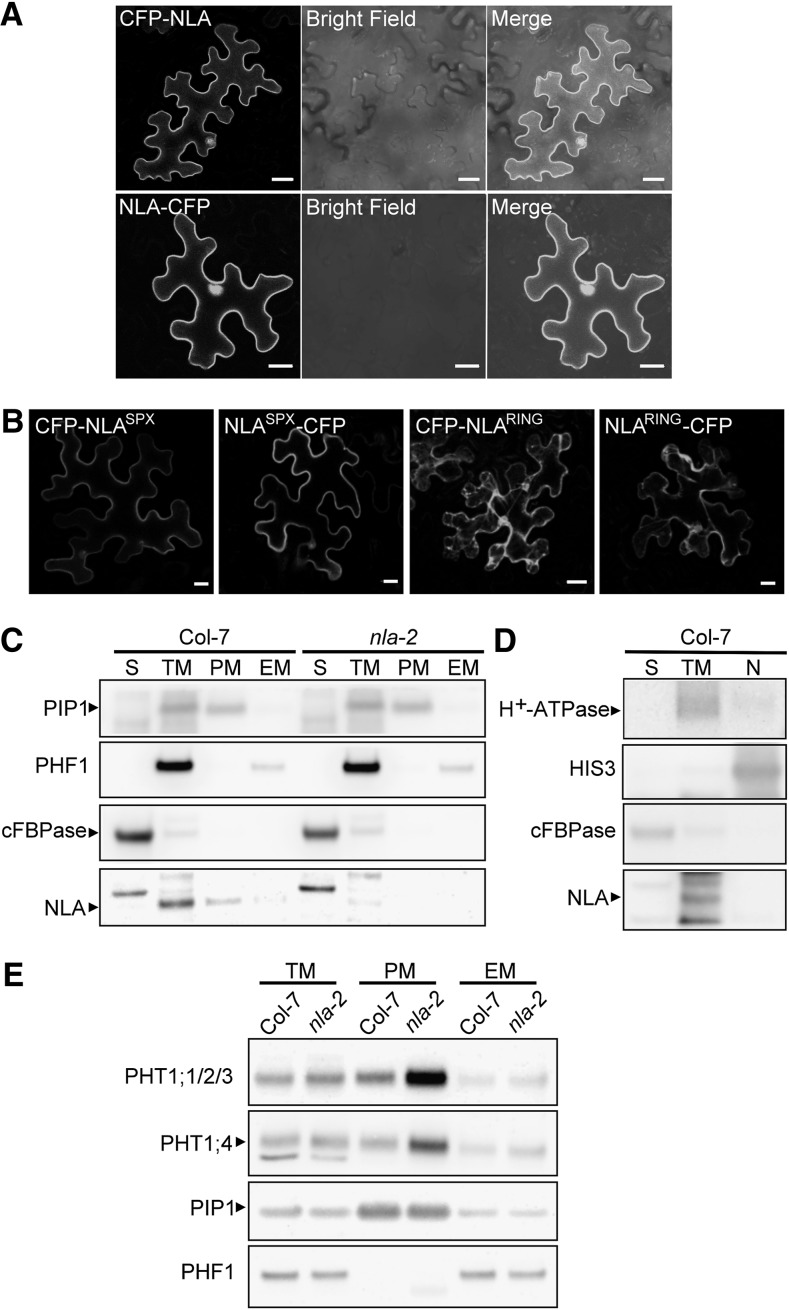
The C-terminal GFP-fused NLA protein was shown to localize in nuclear speckles (Peng et al., 2007), whereas the PHT1s function in the PM (Marmagne et al., 2004; Bayle et al., 2011). To investigate how NLA regulates the protein abundance of PHT1s, we reexamined the subcellular localization of NLA using N- or C-terminal tagging fluorescent protein in Arabidopsis mesophyll protoplasts and tobacco (Nicotiana benthamiana) leaves. In agreement with the previous observation in onion epidermal cells (Peng et al., 2007), NLA-GFP was detected only in the nuclei as a speckle-like structure in the protoplasts (see Supplemental Figure 3A online); however, NLA-CFP (for cyan fluorescent protein) was frequently observed in both nuclei and the PM in tobacco cells (Figure 3A). By contrast, both GFP-NLA and CFP-NLA were detected predominantly in the PM with occasional signals in nuclei regardless of which expression system was used (Figure 3A; see Supplemental Figure 3A online). The PM localization was confirmed by colocalization with FM4-64, a membrane-selective dye that binds to the outer leaflet of the PM (see Supplemental Figure 3B online). These results raise the possibility of dual localization of NLA in the PM and the nucleus or mistargeting of the fluorescent fusion proteins. To test which fusion protein is functional in vivo, a complementation test was performed by overexpressing GFP-NLA or NLA-GFP in nla-2 mutants. GFP-NLA was detected in the PM and the nucleus of root epidermal cells (see Supplemental Figure 3C online) and was able to reduce the shoot Pi content in nla-2 (see Supplemental Figure 3D online), suggesting that GFP-NLA is functional and PM-localized NLA is probably responsible for regulating Pi uptake. Unfortunately, we could not clearly define the function of NLA-GFP in vivo because its protein was undetectable.
Figure 3.
NLA Is Predominantly Localized in the PM.
(A) Subcellular localization of CFP-NLA and NLA-CFP in tobacco leaves. The “Merge” panel is an overlaid image of the CFP and bright field. Bars = 20 μm.
(B) Subcellular localization of truncated NLA proteins, NLASPX and NLARING, in tobacco leaves. Bars = 20 μm.
(C) and (D) Immunoblot analyses revealed predominant localization of NLA in the PM protein fraction rather than the EM, nuclear (N), or soluble protein (S) fractions of Arabidopsis roots. TM, total microsomal proteins. Col, Columbia.
(E) Immunoblot analysis of PHT1;1/2/3 and PHT1;4 in different membrane fractions of the wild type and nla-2 mutants.
Specific protein markers for different subcellular compartments were as follows: PIP1 and H+-ATPase, PM protein; PHF1, ER protein; cFBPase, cytosolic protein; and HIS3, nuclear protein.
Immunoblot analysis further showed the presence of native NLA proteins in microsomal fractions of the roots (see Supplemental Figure 3E online), specifically in the PM-enriched fraction, but not in the EM, soluble, or nuclear fractions (Figures 3C and 3D). Although we could not exclude the possibility of the presence of a small amount of NLA in the nucleus, our results clearly demonstrate that NLA is predominantly localized in the PM, hinting at a role for NLA in regulating PM-localized PHT1s. Interestingly, localization of NLA in the PM coincided with the increased level of PHT1s in the PM-enriched fraction of nla-2 mutants (Figure 3E).
We next dissected NLA protein into the N-terminal SPX domain (NLASPX) and C-terminal RING domain (NLARING) and examined their localizations in tobacco leaves by tagging fluorescent proteins at both termini. Interestingly, NLASPX was able to target to the PM and the nuclei similar to NLA, while NLARING was detected in the cytoplasm (Figure 3B). This suggests that the SPX domain–containing portion is sufficient to determine the localization of NLA.
It has been shown that PHT1;1 and PHT1;4 are expressed in the epidermis, cortex, and vasculature (Karthikeyan et al., 2002). To illustrate the cell types and tissues in which NLA-mediated regulation occurs, we then examined the expression pattern of NLA by fusing its promoter containing 5′ untranslated region (comprising the miR827 target site) with the β-glucuronidase (GUS) reporter gene. Under high Pi conditions, GUS staining was clearly observed in the root tips and vascular tissues of young roots. Interestingly, in the maturation zone proximal to the hypocotyls, only the cortex and epidermis were stained but not the vascular tissues (see Supplemental Figure 4A online). These observations suggest that NLA-mediated regulation occurs in different root cell types. Under Pi-deficient conditions, NLA is posttranscriptionally downregulated by miR827 (Hsieh et al., 2009), so the GUS gene was repressed (see Supplemental Figure 4A online). By contrast, the GUS activity driven by the MIR827 promoter was shown strongly in the Pi-starved roots. Similar to NLA expression pattern, the signal was restricted in the epidermis and cortex of the proximal part of the roots, but GUS staining was detected in all the cell types near the root tips (see Supplemental Figure 4B online). In shoots, the GUS activity driven by NLA or the miR827 promoter was broadly distributed, including in the vascular tissues and hydathodes (see Supplemental Figures 4C and 4D online), but the signals were weaker than those in roots, which is consistent with their transcript levels (Hsieh et al., 2009). The expression pattern of NLA under Pi-sufficient conditions mostly overlapped with that of MIR827 under Pi-deficient conditions, suggesting that miR827-mediated NLA repression occurs in these tissues.
NLA Directs the Degradation of PHT1s via Ubiquitination
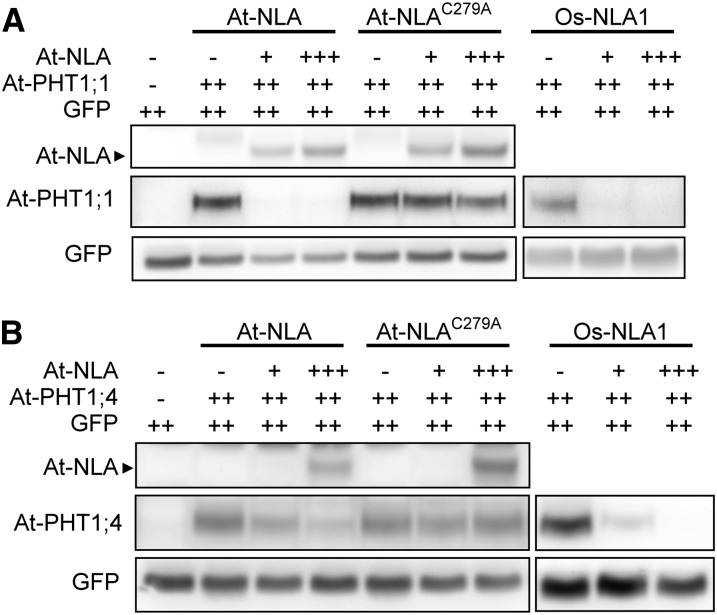
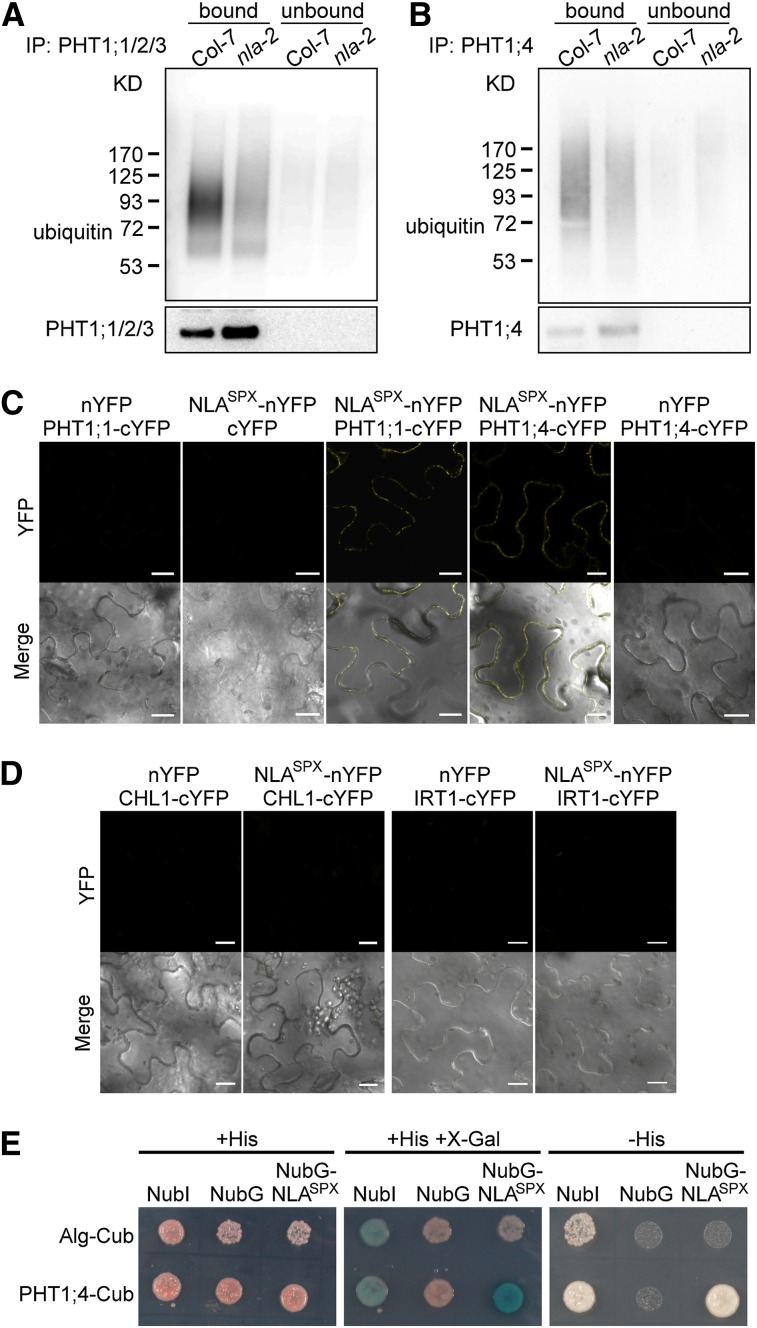
Because of the putative ubiquitin ligase activity of NLA and the increase of PHT1 proteins in nla mutants, we hypothesized that NLA mediates the degradation of PHT1 proteins via posttranslational ubiquitination. We mutated the conserved Cys at residue 279 in the RING domain of NLA (NLAC279A) to disrupt the RING domain and coexpressed NLA or NLAC279A with PHT1s in tobacco leaves. Immunoblot analyses showed that expression of NLA, but not of NLAC279A, resulted in the degradation of PHT1;1 and PHT1;4 (Figures 4A and 4B), suggesting that the RING domain of NLA is required for the degradation of PHT1s. Next, we examined the abundance of ubiquitin-conjugated PHT1s in the wild type and nla-2 mutants. Native PHT1;1/2/3 and PHT1;4 were immunoprecipitated using anti-PHT1;1/2/3 and anti-PHT1;4 antibodies, respectively, and ubiquitination was detected by immunoblotting using antiubiquitin antibody. We found that the ubiquitinated PHT1s in nla-2 mutants were notably decreased compared with the wild type, resulting in the increase of these PHT1s in the mutants (Figures 5A and 5B). These results indicate that NLA is involved in ubiquitin-mediated protein degradation of these Pi transporters. Although the ubiquitin ligase activity of NLA was not validated by in vitro ubiquitination assays (Stone et al., 2005), the previous report of NLA interacting with UBC8, a ubiquitin E2 conjugase (Peng et al., 2007), and our results suggest that NLA functions in vivo as a ubiquitin E3 ligase that is responsible for the ubiquitination of PHT1;1/2/3 and PHT1;4.
Figure 4.
The RING Domain of NLA Is Required for the Degradation of PHT1;1 and PHT1;4.
Coexpression of At-NLA, At-NLAC279A, or Os-NLA1 with At-PHT1;1 (A) or At-PHT1;4 (B) in tobacco leaves. GFP was used as the control for the infiltration event. Note that “+++” and “++” denotes fivefold and 2.5-fold increases in volume of infiltration relative to “+,” respectively.
Figure 5.
PHT1;1 and PHT1;4 Are the Ubiquitination Substrates of NLA.
(A) and (B) In vivo ubiquitination of PHT1;1/2/3 (A) and PHT1;4 (B) in the roots of the wild type and nla-2 mutants under Pi-sufficient conditions and after 1 d of Pi recovery, respectively. The corresponding antibodies were used for immunoprecipitation (IP) (bound). Immunoprecipitation without adding antibodies (unbound) was used as negative control. PHT1;1/2/3 and PHT1;4 protein levels of the immunoprecipitation products are shown in the bottom panels. Col, Columbia.
(C) and (D) BiFC analysis of the interaction between NLASPX and PHT1;1 or PHT1;4 (C) and NLASPX and CHL1 or IRT1 (D). Reconstituted fluorescence signals were detected in the PM of tobacco leaf cells when NLASPX-nYFP was coexpressed with PHT1;1- or PHT1;4-cYFP but not with CHL1-cYFP or IRT1-cYFP. Coexpression of nYFP or cYFP with the corresponding PHT1;1-cYFP or PHT1;4-cYFP or NLASPX-nYFP constructs was used as an additional control. The top panels are YFP images, and the bottom panels are overlaid images of YFP and bright field. Bars = 20 μm.
(E) The interaction between NLASPX and PHT1;4 by split-ubiquitin yeast two-hybrid analysis. NubI and NubG, the wild type and mutated N-terminal fragment of ubiquitin with isoleucine and glycine at position 13, respectively; Cub, C-terminal ubiquitin. Dolichyl-phosphate beta-glucosyltransferase (Alg)-Cub is the control that could interact with NubI but not with NubG and NubG-NLASPX.
In order to investigate whether PHT1;1 and PHT1;4 are the direct substrates of NLA, bimolecular fluorescence complementation (BiFC) was performed to examine the physical interaction between NLA and PHT1s in tobacco leaves. Reconstituted yellow fluorescent protein (YFP) signals from coexpression of NLAC279A and PHT1;4 were observed in the PM and colocalized with FM4-64 (see Supplemental Figures 5A and 5B online), but the interaction between NLA and PHT1;1 was inconclusive due to the strong background in the negative control. However, when we employed the N-terminal SPX domain region (NLASPX), reconstituted YFP signals in the PM were observed not only for NLASPX and PHT1;4, but also for NLASPX and PHT1;1 (Figure 5C), suggesting that NLA regulates PHT1;1 and PHT1;4 through interaction at its SPX domain in the PM. Next, we chose another two PM-localized nutrient transporters, a nitrate transporter (CHLORATE RESISTANT1 [CHL1]) and an iron transporter (IRON REGULATED TRANPSORTER1 [IRT1]) (Tsay et al., 1993; Eide et al., 1996; Vert et al., 2002) to test whether they can interact with NLASPX. As expected, no YFP signals were detected, supporting the specificity of the interaction between NLASPX and PHT1s (Figure 5D). In addition, we also examined the interaction using a split-ubiquitin yeast two-hybrid system. Consistent with the results of BiFC, NLASPX could interact with PHT1;4 in yeast (Figure 5E), but we could not determine the interaction between NLASPX and PHT1;1 because of lack of interaction in positive control for unknown reasons. Together, our results support the notion that NLA directs the ubiquitination of PHT1 proteins for degradation.
To identify the residues on PHT1;1 responsible for NLA-mediated degradation, we used a website to predict the potential ubiquitination sites (http://www.ubpred.org/). Three Lys residues located in the central loop (Lys-276 and Lys-279) and at the end (Lys-524) of PHT1;1 showed the highest scores. We therefore mutated all these Lys residues into Ala. When NLA was coexpressed with PHT1;1K524A, PHT1;1K276/279A, or PHT1;1K276/279/524A, all of the mutated forms of PHT1;1 were able to be degraded by NLA in tobacco leaves (see Supplemental Figure 6A online), suggesting that PHT1;1 might have other ubiquitination sites. In order to determine which region of PHT1;1 is sensitive to NLA-dependant degradation, we further dissected PHT1;1 into N-terminal (PHT1;1N1-279) and C-terminal (PHT1;1C234-524) portions, which contain the central loop and six transmembrane helixes at the N terminus and C terminus, respectively. When PHT1;1N1-279 or PHT1;1C234-524 was coexpressed with NLA in tobacco leaves, although the PHT1;1C234-524 signal was weak and inconclusive, PHT1;1N1-279 was sufficient for degradation by NLA (see Supplemental Figure 6B online). This suggests that N-terminal portion of PHT1;1 is able to be recognized by NLA as the substrate and targeted for ubiquitination and degradation pathway.
Ubiquitinated PHT1s Are Endocytosed from the PM and Trafficked to the Vacuole through the Endosomal Pathway
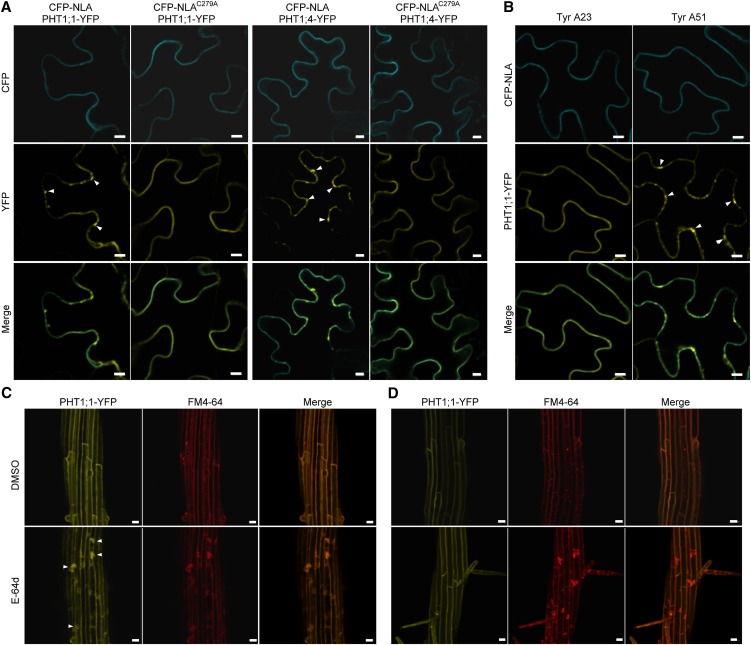
When PHT1;1-YFP or PHT1;4-YFP was coexpressed with CFP-NLA in tobacco leaves, we noticed that signals of PHT1;1 or PHT1;4 were not only found in the PM but also as a punctate structure adjacent to the PM, which was absent when PHT1;1 or PHT1;4 was coexpressed with CFP-NLAC279A (Figure 6A). To determine whether the punctate structure was the endocytosed PHT1s, Tyrphostin A23 (Tyr A23), an inhibitor of clathrin-dependent endocytosis (Banbury et al., 2003; Dhonukshe et al., 2007), or its ineffective analog, Tyrphostin A51 (Tyr A51), was applied. Tyr A23 but not Tyr A51 treatment inhibited the formation of the punctate structure, indicating that NLA promoted the endocytosis of PHT1;1 from the PM through a clathrin-dependent pathway (Figure 6B). We also introduced PHT1;1-YFP into the wild type and nla-2 mutants to examine the protein trafficking. The DMSO solvent had no effect on the PM localization of PHT1;1-YFP, whereas E-64d, which inhibits the fusion of the endosomes with the vacuole (Yamada et al., 2005), caused aggregation of PHT1;1-YFP in the wild type. The aggregated PHT1;1-YFP signals were colocalized with FM4-64 signals, representing the internalized PM (Figure 6C). However, aggregation signals of PHT1;1-YFP were rarely observed in nla-2 mutants (Figure 6D), very likely because of the reduction of endocytosis in PM-localized PHT1;1. This phenomenon could be consistently observed in several independent lines (see Supplemental Figure 7 online). These results indicate that NLA-mediated ubiquitination triggers clathrin-dependent endocytosis of PHT1s, which are subsequently sorted into the vacuolar degradation pathway through endosomes.
Figure 6.
NLA-Mediated Ubiquitination Triggers Clathrin-Dependent Endocytosis of PHT1;1 and PHT1;4 from the PM and Sorting into the Endosomal Trafficking Pathway.
(A) The punctate signals of YFP (arrowheads) were observed when PHT1;1-YFP or PHT1;4-YFP was coexpressed with NLA but not with NLAC279A in tobacco leaves. Bars = 10 μm.
(B) The punctate structure of PHT1;1-YFP caused by NLA was diminished by applying Tyr A23, an inhibitor of clathrin-dependent endocytosis, but not by its analog, Tyr A51. The signal was observed 24 h after the chemicals were applied. Bars = 10 μm.
(C) and (D) The effect of E-64d on the localization of PHT1;1-YFP in the wild type (C) and nla-2 mutant plants (D). The samples were observed 3 h after treatment with E-64d and FM4-64. DMSO was used as the negative control. The aggregation of PHT1;1-YFP in the wild type (C) is indicated by arrowheads. Bars = 20 μm.
Os-NLA1 Is Also Involved in the Degradation of Pi Transporters
In the rice (Oryza sativa) genome, there are two NLA homologs, Os-NLA1 and Os-NLA2, which share 61 and 42% amino acid identity with At-NLA, respectively (Lin et al., 2010; Secco et al., 2012). We then investigated whether Os-NLA possesses a similar function to At-NLA in the regulation of PHT1s. We chose Os-NLA1 for further characterization because it has higher similarity to At-NLA. First, we examined the subcellular localization of Os-NLA1 in tobacco leaves. Both CFP-OsNLA1 and OsNLA1-CFP were observed in the PM and nuclei, similar to At-NLA (see Supplemental Figures 8A and 8B online). Next, we coexpressed Os-NLA1 with At-PHT1;1 or At-PHT1;4 in tobacco leaves and found that both At-PHT1;1 and At-PHT1;4 were degraded by Os-NLA1 (Figures 4A and 4B), suggesting that Os-NLA1 plays a similar role as At-NLA in regulating the protein abundance of Pi transporters.
NLA and PHO2 Function Cooperatively to Regulate the Degradation of PHT1 Proteins
Because PHO2, a ubiquitin E2 conjugase, is also involved in regulating PHT1s through protein ubiquitination machinery (Huang et al., 2013), it is possible that NLA and PHO2 may function together in regulating PHT1s. We therefore examined their genetic interaction by investigating nla pho2 double mutants. Nineteen-day-old nla pho2 double mutants were smaller in size and showed more severe Pi toxicity symptoms, such as necrosis at leaf edges than the single mutants (see Supplemental Figure 9A online). Consistent with the phenotype, the Pi content of the double mutants was twofold to threefold higher than that of the single mutants (Figure 7A; see Supplemental Figure 9B online), and PHT1;1/2/3 proteins were also increased in the roots (Figure 7B; see Supplemental Figure 9C online). Intriguingly, the increased level of PHT1;4 was observed only in nla single and double mutants, but not in the wild type and pho2 mutants. By contrast, at the younger stage, PHT1;4 was only increased in pho2 mutants (see Supplemental Figure 9D online), suggesting that PHO2 and NLA are responsible for regulating the level of PHT1;4 at different growth stages. The level of PHF1 in nla pho2 double mutants was similar to that in pho2 mutants, suggesting that PHF1 is not regulated by NLA (Figure 7B; see Supplemental Figure 9C online). In addition, PHO2 is an ER/Golgi-localized protein (Liu et al., 2012), whereas NLA is predominantly localized in the PM (Figure 3C). The synergistic effects displayed in the double mutants and the different subcellular localizations suggest that NLA and PHO2 function independently in the regulation of the protein levels of PHT1s.
Figure 7.
NLA and PHO2 Function Cooperatively to Regulate the Degradation of PHT1 Proteins.
(A) and (B) The Pi content (A) and protein expression of PHT1;1/2/3, PHT1;4, and PHF1 (B) in nla-2 pho2 double and single mutants. n = 3; error bar indicates se; Student’s t test, single mutants versus the wild type, *P < 0.05, **P < 0.01, and ***P < 0.005; nla-2 pho2 versus pho2, +P < 0.05 and +++P < 0.005. Col, Columbia; FW, fresh weight. The detection of ARF1 was used as a loading control.
(C) and (D) qRT-PCR analysis of PHO2 mRNA in the root (C) and the shoot Pi content (D) of the wild type and nla mutants during plant growth. n = 3; error bar indicates se; Student’s t test, mutants versus the wild type, *P < 0.05, **P < 0.01, and ***P < 0.005. CT, cycle threshold.
It is of interest to note that excess Pi accumulated in nla mutants that were more than 14 days old, whereas Pi overaccumulated in pho2 seedlings of much younger ages (e.g., 5 d old) (Figure 7D; see Supplemental Figure 9G online), implying a temporal difference between the PHO2- and NLA-mediated regulatory pathways. In other words, PHO2-mediated regulation may start to function at an earlier growth stage, while NLA-mediated regulation may operate at a later stage. At later growth stages, PHO2 and NLA cooperate to regulate Pi transport. In fact, we noticed that PHO2 was upregulated in nla mutants grown to an older stage (Figure 7C), likely to compensate for the loss of NLA. By contrast, we did not observe any change in NLA expression in pho2 mutants (see Supplemental Figure 9E online), which could be because of the induction of miR827 in pho2 mutants through an unknown mechanism (see Supplemental Figure 9F online) counteracting the effect of the upregulation of NLA. Taken together, we conclude that the regulation of the degradation of PHT1s by PHO2 and NLA is differentiated both temporally and spatially.
DISCUSSION
Ubiquitination-Mediated Endocytosis as a Crucial and Conserved Mechanism for Regulation of the Abundance of PM Proteins
In this study, we provide multiple lines of evidence showing that NLA mediates the ubiquitination of PM-localized PHT1s, which triggers endocytosis and ultimately the degradation of PHT1s. This regulation is crucial to avoid the uptake of excess Pi especially when external Pi is ample. Pho84, a high-affinity Pi transporter in yeast, is internalized after phosphorylation and ubiquitination and is sorted into a vacuolar degradation pathway when Pi is resupplied (Lundh et al., 2009). Pho84 is composed of two bundles of six helical transmembrane segments connected by a long central loop (Lagerstedt et al., 2004). Interestingly, the deletion of the central loop abolished these posttranslational events, supporting the assertion that the ubiquitination and phosphorylation sites are gathered in this region (Peng et al., 2003; Lundh et al., 2009). The predicted structure of PHT1s is similar to that of Pho84 (Muchhal et al., 1996). However, when we mutated three Lys residues, two in the central loop and one in the end of PHT1;1, none of the resulting proteins were resistant to NLA-mediated degradation (see Supplemental Figure 6A online), suggesting that PHT1;1 might be ubiquitinated at other residues or at multiple sites for degradation. In plants, it has been shown that PIN-FORMED2 (PIN2) and IRT1 are recycled from the PM through ubiquitination on several residues, whereas monoubiquitination of REQUIRES HIGH BORON1 (BOR1) is sufficient for internalization (Barberon et al., 2011; Kasai et al., 2011; Leitner et al., 2012), suggesting that different PM proteins may require distinct ubiquitination modifications for internalization. Of note, we found that the N-terminal region of PHT1;1 was sufficient to be degraded by NLA when they were coexpressed in tobacco leaves (see Supplemental Figure 6B online), implying that the N-terminal portion of PHT1;1 might harbor the NLA-mediated ubiquitination sites. Further study will be required to pinpoint the ubiquitination sites and the type of ubiquitination of PHT1s.
In animals and yeast, it is known that selective internalization of proteins from the PM is a crucial and conserved mechanism that regulates cellular processes, such as the uptake of nutrients and reducing signal perception (Conner and Schmid, 2003; Dupré et al., 2004). Ubiquitin acts as a common sorting signal for the internalization of PM proteins. In plants, ubiquitin is also sufficient to lead artificial PM proteins into the endocytic pathway and sorting into the vacuole for degradation (Scheuring et al., 2012). As mentioned above, several PM-localized transporters, including IRT1, BOR1, and PIN2, have been shown to be internalized after posttranslational ubiquitination for constitutive recycling or turnover (Barberon et al., 2011; Kasai et al., 2011; Leitner et al., 2012), but the components of the ubiquitination machinery were unknown until very recently. IRT1 DEGRADATION FACTOR1, a RING-type ubiquitin E3 ligase, was identified to regulate the degradation of IRT1 via ubiquitination (Shin et al., 2013). Here, we provide insights into the regulation of PHT1 proteins, in which NLA functions as a ubiquitin E3 ligase catalyzing the process and triggering the endocytosis of PHT1;1 through a clathrin-dependent pathway. Various pharmacological treatments reported previously (Bayle et al., 2011) and our findings here (Figure 6; see Supplemental Figure 7 online) suggest that internalized PHT1;1 is sorted into the vacuolar degradation pathway through endosomes. Whether PHT1s are constitutively recycled between the PM and the trans-Golgi network and whether the endosome sorting passes through the trans-Golgi network as the transporters mentioned above require further investigation. It has been shown by yeast two-hybrid screening that UBC8 is the ubiquitin E2 conjugase that interacts with NLA (Peng et al., 2007). Furthermore, Dowil et al. (2011) reported that UBC8 and other members in subgroup VI of ubiquitin E2 conjugating enzymes can interact with membrane-anchored ubiquitin-fold proteins, which provide a docking site for ubiquitin conjugation in the PM. It will be interesting to see whether this subgroup of E2 enzymes function together with NLA to ubiquitinate PHT1 proteins at the PM.
In addition, the type of ubiquitin linkage is also important for determining the fate of the protein. Lys-63–linked ubiquitination of PM proteins has been implicated as a multivesicular body sorting signal in animals and yeasts (Galan and Haguenauer-Tsapis, 1997; Lauwers et al., 2009). A study of PIN2 suggested that a Lys-63–linked polyubiquitin chain could also be used as a vacuolar sorting signal to regulate PM protein turnover in plants (Leitner et al., 2012). Whether these PHT1s are modified by Lys-63–linked ubiquitin chains awaits further investigation.
As mentioned above, both phosphorylation and ubiquitination are required for internalization of yeast Pho84 from PM. Phosphoproteomic analyses have identified phosphorylation sites at the C terminus conserved among PHT1;1 to PHT1;4 (Nühse et al., 2004). Bayle et al. (2011) further pointed out the increase of phosphorylation at the C terminus of PHT1;1 and PHT1;4 under Pi-replete conditions compared with Pi-deprived conditions. It is worth examining whether these phosphorylation events are related to ubiquitination and internalization of PHT1s.
NLA-Mediated Regulation of PHT1s Is Conserved between Arabidopsis and Rice
Based on their similar subcellular localizations (see Supplemental Figure 8 online) and possession of the same ubiquitin ligase activity facilitating the degradation of PHT1 proteins (Figures 4A and 4B), we propose that NLA is functionally conserved between Arabidopsis and rice. Intriguingly, unlike At-NLA, Os-NLA1 is not the target of Os-miR827. However, microarray results show that its transcript is downregulated under Pi-deficient conditions (www.genevestigator.com), suggesting that Os-NLA1 is regulated by external Pi status through a different mechanism. The rice PHT1 family has 13 members (Paszkowski et al., 2002). Transcripts for Os-Pht1;1, which functions in Pi acquisition and distribution in rice roots, have been shown not to be responsive to Pi status (Sun et al., 2012). Os-Pht1;8 is also responsible for Pi acquisition. Its transcripts are abundant under Pi-sufficient conditions and are upregulated only 1.5-fold by Pi deficiency (Jia et al., 2011). Suppression of Os-Pht1;1 and Os-Pht1;8 reduces accumulation of Pi when Pi is ample (Jia et al., 2011; Sun et al., 2012), suggesting that these two transporters play an important role in Pi uptake under Pi-sufficient conditions. It will be interesting to see whether these two Pi transporters are the downstream targets of Os-NLA1 in rice.
The SPX Domain as a Protein–Protein Interaction Domain
It has been reported that the SPX domain is responsible for protein–protein interaction in yeast. Yeast Pho87 and Pho90, low-affinity Pi transporters, are negatively regulated by Spl2, which interacts with these transporters via their SPX domain (Hürlimann et al., 2009). Recently, Arabidopsis PHO1 was shown to interact with PHO2 through its SPX domain (Liu et al., 2012). SHORT HYPOCOTYL UNDER BLUE1, involved in transcriptional regulation of light signaling in Arabidopsis, was also reported to interact with a WRKY transcription factor, MINI3, via its SPX domain to trigger the seed development process (Kang et al., 2013). Here, we reported that NLASPX, which has no transmembrane domains, is sufficient for PM targeting and interacting with PHT1s (Figures 3B and 5C). These observations suggest that the association of NLA with the PM is through the interaction of its SPX domain with PM proteins and that the SPX domain carries an important regulatory role whose functions may depend on the interacting protein partners. There are 20 and 15 SPX domain–containing proteins in the Arabidopsis and rice genomes, respectively (Lin et al., 2010; Secco et al., 2012). Identification of the proteins that interact with the SPX domain of these proteins will help us reveal their functions.
Coordinated Regulation of PHO2 and NLA in Pi Transport
When studying the genetic interaction between PHO2 and NLA, we observed synergistic effects on Pi accumulation and protein expression of PHT1s in nla pho2 double mutants compared with the respective single mutants (Figure 7; see Supplemental Figure 9 online). We found that PHO2 is responsible for ubiquitination and degradation of PHT1s in the post-ER endocompartments (Huang et al., 2013), suggesting that PHO2 and NLA function in different subcellular compartments to regulate the protein levels of PHT1s. Kant et al. (2011) did not observe differences in Pi accumulation between nla pho2 double mutants and the respective single mutants after long-term nitrate limitation treatment. It is possible that single mutants had accumulated a very high level of Pi under such conditions and therefore were hardly distinguishable from the double mutants. Nevertheless, the interaction between nitrate and Pi is worth further study.
We also found that nla and pho2 mutants overaccumulated Pi and showed differential regulation of PHT1;4 at different growth stages, suggesting that PHO2 and NLA regulate Pi uptake coordinately in both a temporal and spatial manner (Figure 7; see Supplemental Figure 9 online). We observed that the increase of Pi in nla mutants coincided with the upregulation of PHO2 mRNA (Figure 7), indicating that these two pathways are coordinated to regulate Pi acquisition. Furthermore, NLA and PHO2 are tightly regulated by miR827 and miR399 in response to Pi status, demonstrating that crosstalk between these two miRNA regulatory pathways is important for Pi homeostasis.
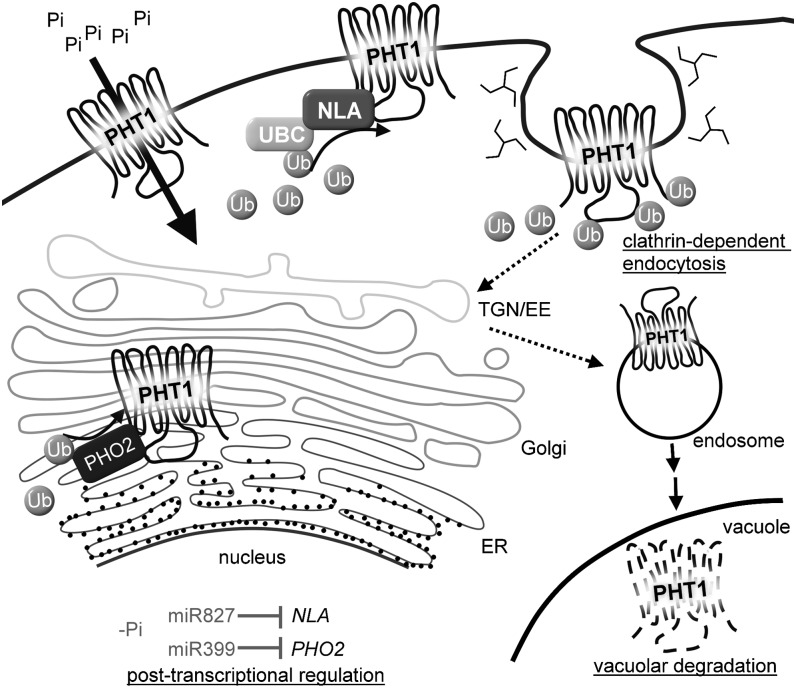
In conclusion, our study revealed a molecular mechanism through which Pi acquisition is modulated in response to external Pi availability, which involves interplay between two Pi starvation–induced miRNAs, miR399 and miR827, that comprises posttranscriptional and posttranslational regulation. In the model, we propose (Figure 8), under Pi-sufficient conditions, PHT1s are regulated by PHO2 and NLA in the EM system and the PM, respectively, via protein ubiquitination machinery. We have previously hypothesized that PHO2 might function as a chimeric E2-E3 enzyme (Liu et al., 2012) because of its direct interaction with PHO1 and its sequence similarity to a chimeric E2-E3 enzyme of human BRUCE/APOLLON (Hao et al., 2004). Therefore, PHO2 might possess E2 and E3 activities to regulate the abundance of PHT1s in the secretory pathway destined for the PM. On the other hand, NLA, a ubiquitin E3 ligase, might work with UBC8 to ubiquitinate PHT1s and regulate the abundance of PHT1s in the PM by endocytosis following protein degradation in the vacuole. When Pi is limited, both miR399 and miR827 are upregulated to reduce the mRNA levels of PHO2 and NLA, respectively, thereby alleviating the negative regulation of PHT1s at the protein level and reinforcing Pi uptake activity. In addition to Pi starvation responses, NLA is involved in the regulation of nitrate starvation responses (Peng et al., 2007) and defense responses (Yaeno and Iba, 2008), implying that it may have multiple substrates to modulate different stress responses. Identification of additional substrates will uncover the functions of NLA under biotic and abiotic stress conditions.
Figure 8.
A Working Model of the Regulation of Pi Uptake via MiRNA-Mediated Posttranscriptional and Ubiquitin-Mediated Posttranslational Regulation.
NLA and PHO2 function in the PM and EM system, respectively, to direct the degradation of PHT1s through protein ubiquitination machinery. Degradation of PHT1s is relieved by miR827- and miR399-mediated posttranscriptional cleavages on the transcripts NLA and PHO2, respectively, upon Pi deficiency. UBC designates a ubiquitin E2 conjugase that works together with NLA to ubiquitinate PHT1s in the PM.
METHODS
Plant Material and Growth Conditions
Seeds of the Arabidopsis thaliana wild type (Columbia-0 and Wassilewskija) and pho2 mutant (Delhaize and Randall, 1995) were obtained from the ABRC. Seeds of nla-1 and nla-1 pho2 were kindly provided by Steven J. Rothstein (University of Guelph, Canada) (Kant et al., 2011). Columbia-7 and bah1-D/nla-2 seeds were kindly provided by Koh Iba (Kyushu University, Japan) (Yaeno and Iba, 2008). nla-3 (FLAG_352A03) seeds were obtained from Institut National de la Recherche Agronomique–Versailles Genomic Resource Center, France. The nla-2 pho2 double mutant was generated by crossing nla-2 with the pho2 mutant. Seeds were germinated in half-strength Hoagland agar media containing 250 μM KH2PO4. For hydroponic culture, 10-d-old seedlings were transferred to half-strength Hoagland solution containing 250 μM KH2PO4 (+Pi). For Pi recovery, 14-d-old hydroponically cultured seedlings were transferred to Pi-free nutrient solution (−Pi) for 4 d and changed to full nutrient solution (+Pi) for 6 d. For high Pi treatment, 5-d-old seedlings were transferred to media containing 5 mM KH2PO4 (Hu et al., 2009) for 7 d.
Constructs and Gene Transformation
Genes of interest were amplified using the primer pairs listed in Supplemental Table 1 online, cloned into pCR8/GW/TOPO (Invitrogen) and then recombined into the desired Gateway destination vectors using the Gateway® LR Clonase™ II Enzyme Mix (Invitrogen). Except for Os-NLA1, all cloned genes were derived from Arabidopsis. The clone containing point mutated NLA or PHT1;1 was amplified using phosphorylated primers with the desired change. PHT1;1-YFP and PHT1;4-YFP were constructed by three-piece PCR to insert YFP at 10 amino acids upstream of the stop codon in PHT1s (Tian et al., 2004). Detailed information on constructs is provided in Supplemental Table 2 online.
The stable transgenic lines were generated using an Agrobacterium tumefaciens dipping procedure (Clough and Bent, 1998). Agrobacterium strain GV3101 was used for transformation.
Transformation of Arabidopsis Mesophyll Protoplasts
Transformation was conducted as described previously (Wu et al., 2009).
Agrobacterium-Mediated Infiltration of Tobacco Leaves
The Agrobacterium-mediated infiltration method was adopted from Bendahmane et al. (2000) with minor modification as described previously (Liu et al., 2012). For immunoblot analysis, samples were collected 3 d after infiltration for total or microsomal protein isolation and immunoblot analysis. To observe the fluorescence in tobacco (Nicotiana benthamiana) leaves, plants were grown for 3 to 4 d before observation. To stain the PM of epidermal cells, leaves were stained by 5 μM FM4-64 for 15 min before observation. For chemical treatment, the leaves that transiently expressed genes of interest for 3 d were infiltrated again with Tyr A23 or Tyr A51. After 24 h, leaves were collected for observation.
Chemical Treatment
Tyr A23 and Tyr A51 stocks (both 33 mM) were prepared in DMSO and diluted with water into 33 μM for use. E-64d stock (50 mM) was prepared in DMSO. Twelve-day-old seedlings were immersed in the half-strength Hoagland solution containing 250 μM KH2PO4, 5 μM FM4-64, and 50 μM E-64d or 0.1% DMSO for 3 h.
Fluorescence Microscopy
Fluorescent signals were observed under a Zeiss LSM 510 META NLO DuoScan and a Zeiss LSM 780 plus ELYRA S.1 with objective Plan-Apochromat ×20/0.8 mm and LD C-Apochomat ×40/1.1 W. Excitation/emission wavelengths were 458 nm/465 to 510 nm for CFP, 488 nm/495 to 520 nm for GFP, 514 nm/520 to 550 nm for YFP, and 561 nm/575 to 630 nm for FM4-64.
Measurement of Pi Contents and Pi Uptake Activity
Nineteen-day-old seedlings were used for Pi uptake assays. Pi contents and [33P]Pi uptake activity were determined as described previously (Chiou et al., 2006).
GUS Staining
GUS activity was detected by the modified method as described (Lin et al., 2005). The staining time for examining the expression pattern of NLA and MIR827 was 1 and 0.5 hour, respectively.
Protein Isolation and Immunoblot Analysis
Total proteins in tobacco leaves were isolated using SDS extraction buffer. The detailed methods of protein isolation and immunoblotting have been described previously (Liu et al., 2012). Sixty micrograms of total proteins of each sample were loaded for immunoblot analysis. GFP protein that was used as the control of infiltration event was detected by rabbit polyclonal GFP antibody (Abcam).
Membrane proteins were isolated from roots of hydroponically cultured 19-d-old Arabidopsis seedlings or high-Pi-treated 12-d-old Arabidopsis seedlings or infiltrated tobacco leaves. Tissues were ground in homogenization buffer containing 330 mM Suc, 50 mM Tris, pH7.5, 10 mM KCl, and 5 mM EDTA, with freshly added DTT, phenylmethanesulfonyl fluoride, and protease inhibitor cocktail (Roche). The homogenate was centrifuged at 4°C at 2000g for 15 min. The supernatant was saved and centrifuged at 4°C at 400,000g for 30 min. The supernatant was precipitated by acetone as the soluble protein fraction. The pellet was resuspended in the homogenization buffer without DTT as the total microsomal protein fraction for immunoblot or immunoprecipitation analyses. Fifteen to thirty micrograms of total microsomal protein and soluble protein fraction of each sample was loaded for immunoblot analysis. PM and EM protein-enriched fractions were isolated from total microsomal proteins by two-phase partitioning as described previously (Larsson et al., 1987). For each sample, 30 μg of soluble protein fraction, 25 μg of total microsomal protein fraction, and 8 μg of PM and EM were loaded to detect NLA; 10 μg of total microsomal protein fraction and EM, and 5 μg of PM was loaded to detect PHT1;1/2/3 and PHT1;4. The immunoprecipitation and detection of ubiquitination were performed as described previously (Kasai et al., 2011).
The method of nuclear protein isolation was modified from a previous protocol (Castle, 2003). Roots of 19-d-old Arabidopsis were ground using a homogenizer with homogenization buffer containing 300 mM Suc, 50 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2, and freshly added protease inhibitor cocktail (Roche). The homogenate was filtered through two layers of Miracloth and centrifuged at 4°C at 800g for 15 min. The pellet was saved as the crude nuclear fraction, and the supernatant was centrifuged at 4°C at 100,000g for 2 h to separate the microsomal protein fraction (pellet) from soluble fraction (supernatant). The pellets of nuclear and microsomal fractions were resuspended in homogenization buffer and sonicated. Proteins were precipitated by 100% trichloroacetic acid and resuspended in elution buffer (containing 60 mM Tris-HCl (pH8.5), 2% SDS, 2.5% glycerol, 0.13 mM EDTA, and protease inhibitor cocktail). Twenty micrograms of soluble protein, total microsomal protein, and nuclear fractions were loaded for immunoblot analysis.
The polyclonal antibodies for PHT1;1/2/3 (amino acid residues 266 to 285) and PHT1;4 (amino acid residues 265 to283) were raised against the central loop region (Liu et al., 2011; Huang et al., 2013). Because of high amino acid identity, the antibody against PHT1;1/2/3 recognizes all three proteins, PHT1;1, PHT1;2, and PHT1;3. The polyclonal antibodies for PHF1 (amino acid residues 382 to 398) and NLA (amino acid residues 300 to 318) were raised against the peptides at the C termini. The working concentrations of affinity-purified antibodies were 20 to 500 ng mL−1. Mouse monoclonal ubiquitin (P4D1) antibodies were purchased from Santa Cruz (1:5000). The polyclonal rabbit Histone 3 antibody was purchased from Abcam (1:1000). The polyclonal rabbit ARF1, H+-ATPase, PIP1, and cFBP1 antibodies were purchased from Agrisera (1:10000).
Split-Ubiquitin Yeast Two-Hybrid Assay
Split-ubiquitin yeast two-hybrid assay was performed following the manufacturer’s instructions provided with the DUALmembrane Kit (Dualsystems Biotech). The coding regions of PHT1;4 and NLASPX were cloned in frame into the vectors pTMBV4 and pDL-Nx, respectively. Yeast strain DSY-1 cells were cotransformed with these two constructs and plated onto synthetic medium lacking Leu and Trp. The protein–protein interactions was assessed by the growth of yeast colonies on synthetic medium lacking Leu, Trp, and His and the chloroform overlay β-galactosidase plate assay as described (Duttweiler, 1996). Dolichyl-phosphate beta-glucosyltransferase, a yeast ER-resident protein, was used as the control to confirm the interaction.
RNA Extraction, qRT-PCR, and Small RNA Gel Blot
Total RNA was isolated from 19-d-old Arabidopsis roots using RNAzol (Molecular Research Center). cDNAs were synthesized from 500 ng of total RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen) with oligo(dT) primer. qRT-PCR was performed using the Power SYBR Green PCR Master Mix kit (Applied Biosystems) on a 7300 Real-Time PCR system (Applied Biosystems) following the manufacturer’s instructions. UBQ10 was used as the internal control.
Total RNA for small RNA gel blots was isolated from 19-d-old Arabidopsis roots by TRIzol (Invitrogen). Ten micrograms total RNA was loaded. The methods used for the small RNA gel blots have been described previously (Hsieh et al., 2009).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: NLA, At1g02860; PHO2, At2g33770; PHT1;1, At5g43350; PHT1;2, At5g43370; PHT1;3, At5g43360; PHT1;4, At2g38940; and PHF1, At3g52190. The accession number of Os-NLA1 is Os07g0673200.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Mutation in NLA Causes Increased Pi Uptake Activity and PHT1 Protein Amounts.
Supplemental Figure 2. The Responses of the Wild Type and nla-2 Mutants after Recovery from Pi Deprivation.
Supplemental Figure 3. Subcellular Localization of NLA.
Supplemental Figure 4. Expression Pattern of NLA and MIR827.
Supplemental Figure 5. Analysis of Protein–Protein Interaction between NLA and PHT1;4.
Supplemental Figure 6. The N-Terminal Portion of PHT1;1 Is Sufficient for NLA-Mediated Degradation.
Supplemental Figure 7. NLA-Mediated Ubiquitination Triggers PHT1;1 Sorting into the Vacuole through the Endosomal Trafficking Pathway.
Supplemental Figure 8. Subcellular Localization of Os-NLA1.
Supplemental Figure 9. The Difference between NLA- and PHO2-Dependent Regulation of PHT1s.
Supplemental Table 1. Sequences of Primers Used in This Study.
Supplemental Table 2. Gene Constructs Used in This Study.
Supplementary Material
Acknowledgments
We thank Steven J. Rothstein and Koh Iba for kindly providing the seeds of nla-1 and nla-2, respectively. We thank Yi-Fang Tsay for kindly providing the plasmid pENTR/D-TOPO-CHL1. We thank Shu-Chen Shen from the Confocal Microscopic Core Facility at Academia Sinica for assistance in confocal microscopy images. We thank the members of the Plant Technology Core Laboratory at the Agricultural Biotechnology Research Center, Academia Sinica, for assistance in the sectioning of GUS staining tissues and protoplast transformation. This work was supported by Academia Sinica, Taiwan (Grant 98-CDA-L11), and the National Science Council of the Republic of China (Grants NSC100-2321-B-001-005 and NSC102-2321-B-001-047).
AUTHOR CONTRIBUTIONS
W.-Y.L., T.-K.H., and T.-J.C. designed the research. W.-Y.L. and T.-K.H. performed the experiments and analyzed the data. W.-Y.L. and T.-J.C. wrote the article.
Glossary
- PM
plasma membrane
- ER
endoplasmic reticulum
- miRNA
microRNA
- qRT-PCR
quantitative RT-PCR
- GFP
green fluorescent protein
- CFP
cyan fluorescent protein
- GUS
β-glucuronidase
- BiFC
bimolecular fluorescence complementation
- YFP
yellow fluorescent protein
- Tyr A23
Tyrphostin A23
- Tyr A51
Tyrphostin A51
- EM
endomembrane
References
- Auesukaree C., Homma T., Kaneko Y., Harashima S. (2003). Transcriptional regulation of phosphate-responsive genes in low-affinity phosphate-transporter-defective mutants in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306: 843–850 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S.I., Wu C.C., Huang Y.T., Su C.L., Chiou T.J. (2006). pho2, a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banbury D.N., Oakley J.D., Sessions R.B., Banting G. (2003). Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278: 12022–12028 [DOI] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conéjéro G., Curie C., Friml J., Vert G. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W.R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141: 988–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V., Arrighi J.F., Creff A., Nespoulous C., Vialaret J., Rossignol M., Gonzalez E., Paz-Ares J., Nussaume L. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendahmane A., Querci M., Kanyuka K., Baulcombe D.C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: Application to the Rx2 locus in potato. Plant J. 21: 73–81 [DOI] [PubMed] [Google Scholar]
- Castle, J.D. (2003). Purification of organelles from mammalian cells. Curr. Protoc. Immunol. (Hoboken: John Wiley & Sons), pp 56: 8.1B.1–8.1B.57. [DOI] [PubMed] [Google Scholar]
- Chiou T.J., Aung K., Lin S.I., Wu C.C., Chiang S.F., Su C.L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18: 412–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T.J., Lin S.I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Chiou T.J., Liu H., Harrison M.J. (2001). The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 25: 281–293 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conner S.D., Schmid S.L. (2003). Regulated portals of entry into the cell. Nature 422: 37–44 [DOI] [PubMed] [Google Scholar]
- Delhaize E., Randall P.J. (1995). Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 107: 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Aniento F., Hwang I., Robinson D.G., Mravec J., Stierhof Y.D., Friml J. (2007). Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17: 520–527 [DOI] [PubMed] [Google Scholar]
- Dowil R.T., Lu X., Saracco S.A., Vierstra R.D., Downes B.P. (2011). Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J. Biol. Chem. 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré S., Urban-Grimal D., Haguenauer-Tsapis R. (2004). Ubiquitin and endocytic internalization in yeast and animal cells. Biochim. Biophys. Acta 1695: 89–111 [DOI] [PubMed] [Google Scholar]
- Duttweiler H.M. (1996). A highly sensitive and non-lethal beta-galactosidase plate assay for yeast. Trends in Genetics. 12: 340–341 [DOI] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.M., Haguenauer-Tsapis R. (1997). Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giots F., Donaton M.C.V., Thevelein J.M. (2003). Inorganic phosphate is sensed by specific phosphate carriers and acts in concert with glucose as a nutrient signal for activation of the protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 47: 1163–1181 [DOI] [PubMed] [Google Scholar]
- González E., Solano R., Rubio V., Leyva A., Paz-Ares J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17: 3500–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y., Sekine K., Kawabata A., Nakamura H., Ishioka T., Ohata H., Katayama R., Hashimoto C., Zhang X., Noda T., Tsuruo T., Naito M. (2004). Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 6: 849–860 [DOI] [PubMed] [Google Scholar]
- Hsieh L.C., Lin S.I., Shih A.C., Chen J.W., Lin W.Y., Tseng C.Y., Li W.H., Chiou T.J. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151: 2120–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H.C., Wang Y.Y., Tsay Y.F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Huang T.-K., Han C.-L., Lin S.-I., Chen Y.-J., Tsai Y.-C., Chen Y.-R., Chen J.-W., Lin W.-Y., Chen P.-M., Liu T.-Y., Chen Y.-S., Chiou T.-J. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann H.C., Pinson B., Stadler-Waibel M., Zeeman S.C., Freimoser F.M. (2009). The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 10: 1003–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hürlimann H.C., Stadler-Waibel M., Werner T.P., Freimoser F.M. (2007). Pho91 Is a vacuolar phosphate transporter that regulates phosphate and polyphosphate metabolism in Saccharomyces cerevisiae. Mol. Biol. Cell 18: 4438–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Ren H., Gu M., Zhao J., Sun S., Zhang X., Chen J., Wu P., Xu G. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Li W., Zhou Y., Ni M. (2013). A WRKY transcription factor recruits the SYG1-like protein SHB1 to activate gene expression and seed cavity enlargement. PLoS Genet. 9: e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant S., Peng M., Rothstein S.J. (2011). Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A.S., Varadarajan D.K., Mukatira U.T., D’Urzo M.P., Damsz B., Raghothama K.G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai K., Takano J., Miwa K., Toyoda A., Fujiwara T. (2011). High boron-induced ubiquitination regulates vacuolar sorting of the BOR1 borate transporter in Arabidopsis thaliana. J. Biol. Chem. 286: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerstedt J.O., Voss J.C., Wieslander A., Persson B.L. (2004). Structural modeling of dual-affinity purified Pho84 phosphate transporter. FEBS Lett. 578: 262–268 [DOI] [PubMed] [Google Scholar]
- Larsson C., Widell, S., and Kjellbom, P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol. 148: 558–568 [Google Scholar]
- Lauwers E., Jacob C., André B. (2009). K63-linked ubiquitin chains as a specific signal for protein sorting into the multivesicular body pathway. J. Cell Biol. 185: 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner J., Petrášek J., Tomanov K., Retzer K., Pařezová M., Korbei B., Bachmair A., Zažímalová E., Luschnig C. (2012). Lysine63-linked ubiquitylation of PIN2 auxin carrier protein governs hormonally controlled adaptation of Arabidopsis root growth. Proc. Natl. Acad. Sci. USA 109: 8322–8327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.I., et al. (2010). Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51: 2119–2131 [DOI] [PubMed] [Google Scholar]
- Lin S.I., Wang J.G., Poon S.Y., Su C.L., Wang S.S., Chiou T.J. (2005). Differential regulation of FLOWERING LOCUS C expression by vernalization in cabbage and Arabidopsis. Plant Physiol. 137: 1037–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Aung K., Tseng C.Y., Chang T.Y., Chen Y.S., Chiou T.J. (2011). Vacuolar Ca2+/H+ transport activity is required for systemic phosphate homeostasis involving shoot-to-root signaling in Arabidopsis. Plant Physiol. 156: 1176–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Huang T.K., Tseng C.Y., Lai Y.S., Lin S.I., Lin W.Y., Chen J.W., Chiou T.J. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24: 2168–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundh F., Mouillon J.M., Samyn D., Stadler K., Popova Y., Lagerstedt J.O., Thevelein J.M., Persson B.L. (2009). Molecular mechanisms controlling phosphate-induced downregulation of the yeast Pho84 phosphate transporter. Biochemistry 48: 4497–4505 [DOI] [PubMed] [Google Scholar]
- Marmagne A., Rouet M.A., Ferro M., Rolland N., Alcon C., Joyard J., Garin J., Barbier-Brygoo H., Ephritikhine G. (2004). Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol. Cell. Proteomics 3: 675–691 [DOI] [PubMed] [Google Scholar]
- Marschner, H. (1995). Functions of mineral nutrients: Macronutrients. In Mineral Nutrition of Higher Plants, 2nd ed (London: Academic Press), pp 265–277, 537–594. [Google Scholar]
- Misson J., Thibaud M.-C., Bechtold N., Raghothama K., Nussaume L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55: 727–741 [DOI] [PubMed] [Google Scholar]
- Muchhal U.S., Pardo J.M., Raghothama K.G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 93: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchhal U.S., Raghothama K.G. (1999). Transcriptional regulation of plant phosphate transporters. Proc. Natl. Acad. Sci. USA 96: 5868–5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nühse T.S., Stensballe A., Jensen O.N., Peck S.C. (2004). Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16: 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., Nakanishi T.M., Thibaud M.C. (2011). Phosphate import in plants: Focus on the PHT1 transporters. Front. Plant Sci. 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paszkowski U., Kroken S., Roux C., Briggs S.P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 99: 13324–13329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S.P. (2003). A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Peng M., Hannam C., Gu H., Bi Y.M., Rothstein S.J. (2007). A mutation in NLA, which encodes a RING-type ubiquitin ligase, disrupts the adaptability of Arabidopsis to nitrogen limitation. Plant J. 50: 320–337 [DOI] [PubMed] [Google Scholar]
- Raghothama K.G. (1999). Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Scheuring D., Künzl F., Viotti C., Yan M.S., Jiang L., Schellmann S., Robinson D.G., Pimpl P. (2012). Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol. 12: 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D., Wang C., Arpat B.A., Wang Z., Poirier Y., Tyerman S.D., Wu P., Shou H., Whelan J. (2012). The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 193: 842–851 [DOI] [PubMed] [Google Scholar]
- Shin H., Shin H.S., Dewbre G.R., Harrison M.J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Shin L.J., Lo J.C., Chen G.H., Callis J., Fu H., Yeh K.C. (2013). IRT1 DEGRADATION FACTOR1, a RING E3 ubiquitin ligase, regulates the degradation of IRON-REGULATED TRANSPORTER1 in Arabidopsis. Plant Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Gu M., Cao Y., Huang X., Zhang X., Ai P., Zhao J., Fan X., Xu G. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G.-W., et al. (2004). High-throughput fluorescent tagging of full-length Arabidopsis gene products in planta. Plant Physiol. 135: 25–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsay Y.F., Schroeder J.I., Feldmann K.A., Crawford N.M. (1993). The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713 [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dédaldéchamp F., Gaymard F., Guerinot M.L., Briat J.F., Curie C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F.H., Shen S.C., Lee L.Y., Lee S.H., Chan M.T., Lin C.S. (2009). Tape-Arabidopsis Sandwich - A simpler Arabidopsis protoplast isolation method. Plant Methods 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno T., Iba K. (2008). BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 148: 1032–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Fuji K., Shimada T., Nishimura M., Hara-Nishimura I. (2005). Endosomal proteases facilitate the fusion of endosomes with vacuoles at the final step of the endocytotic pathway. Plant J. 41: 888–898 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.