The authors report the structure of ten plastid genomes from hemi- and holoparasitic plants and their closest nonparasitic relative. Structural plastome reconfiguration is associated with obligate parasitism. The extent of genome reduction after the loss of photosynthesis is governed by dispensable genes’ proximity to essential genes and the position in operons or by alternative gene function.
Abstract
Nonphotosynthetic plants possess strongly reconfigured plastomes attributable to convergent losses of photosynthesis and housekeeping genes, making them excellent systems for studying genome evolution under relaxed selective pressures. We report the complete plastomes of 10 photosynthetic and nonphotosynthetic parasites plus their nonparasitic sister from the broomrape family (Orobanchaceae). By reconstructing the history of gene losses and genome reconfigurations, we find that the establishment of obligate parasitism triggers the relaxation of selective constraints. Partly because of independent losses of one inverted repeat region, Orobanchaceae plastomes vary 3.5-fold in size, with 45 kb in American squawroot (Conopholis americana) representing the smallest plastome reported from land plants. Of the 42 to 74 retained unique genes, only 16 protein genes, 15 tRNAs, and four rRNAs are commonly found. Several holoparasites retain ATP synthase genes with intact open reading frames, suggesting a prolonged function in these plants. The loss of photosynthesis alters the chromosomal architecture in that recombinogenic factors accumulate, fostering large-scale chromosomal rearrangements as functional reduction proceeds. The retention of DNA fragments is strongly influenced by both their proximity to genes under selection and the co-occurrence with those in operons, indicating complex constraints beyond gene function that determine the evolutionary survival time of plastid regions in nonphotosynthetic plants.
INTRODUCTION
Photosynthesis is the primary function of plastids (chloroplasts). The genome retained in the plastid organelle of land plants (the plastome) therefore encodes numerous structural proteins required for photosynthesis as well as ribosomal proteins and structural RNAs (Palmer, 1985; Wicke et al., 2011). Because of the selective pressure on photosynthesis-related elements, plastid chromosomes in most land plants are conserved in terms of structure, gene content, and nucleotide substitution rates (Raubeson and Jansen, 2005). Typically, the plastome comprises two single copy regions (large single-copy region [LSC] and small single-copy region [SSC]) that are separated by two virtually identical large inverted repeats (IRs). The latter play an important role in stabilizing plastid genome structure (Maréchal and Brisson, 2010). Other factors contributing to structural conservation across plastomes are the predominantly uniparental inheritance of plastids (Bock, 2007; Zhang and Sodmergen, 2010) and the suppression of potentially mutagenic repeats, such as small dispersed and simple sequence repeats and repetitive elements larger than 50 bp (Raubeson et al., 2007).
Highly diverged plastid chromosomes are found in nonphotosynthetic plants (dePamphilis and Palmer, 1990; dePamphilis, 1995; Nickrent et al., 1997; Krause, 2011), which parasitize other flowering plants (parasitic plants sensu stricto) or more rarely mycorrhizal fungi (myco-heterotrophs). Hemiparasites still carry out photosynthesis to some extent, while holoparasites almost completely rely on a host for water as well as inorganic and organic nutrients. In nonphotosynthetic plants, photosynthesis-associated genes are no longer required, may become pseudogenes, and are eventually deleted, resulting in a functional and physical reduction of the plastid genome (Wolfe et al., 1992a, 1992b; Delavault et al., 1996; Funk et al., 2007; McNeal et al., 2007; Wickett et al., 2008; Delannoy et al., 2011; Logacheva et al., 2011). The extent and speed of plastome reduction both appear to be lineage specific. For instance, in the broomrape family (Orobanchaceae), the gene encoding the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco; rbcL) is retained and expressed in species of Lathraea (toothwort) and Harveya, whereas in species of Hyobanche as well as in the broomrapes Orobanche and Phelipanche, it is only retained as a pseudogene or lost completely (Delavault et al., 1995; Wolfe and dePamphilis, 1997; Lusson et al., 1998; Leebens-Mack and dePamphilis, 2002; Randle and Wolfe, 2005; Young and dePamphilis, 2005; Leebens-Mack and dePamphilis, 2007). Relaxed functional constraints as a result of the loss of photosynthesis also affect plastid-encoded housekeeping genes, implying that gene function alone is insufficient to explain the patterns of gene loss (Wimpee et al., 1991; Wolfe et al., 1992b; Colwell, 1994; Funk et al., 2007; McNeal et al., 2007; Delannoy et al., 2011).
Understanding course, tempo, and mechanisms of plastome evolution after the loss of photosynthesis requires comparative analyses of closely related nonparasitic (autotrophic) and parasitic species with different degrees of trophic specialization. The only family fulfilling this requirement is the broomrape family, Orobanchaceae (Westwood et al., 2010). It includes a single autotrophic lineage, the genus Lindenbergia, with about a dozen species, which is the sister group to a large clade of ∼2000 hemi- and holoparasitic species (Bennett and Mathews, 2006; McNeal et al., 2013). Within Orobanchaceae, loss of photosynthesis has occurred at least three times independently (Bennett and Mathews, 2006; McNeal et al., 2013), once thereof in the clade that includes beechdrops (Epifagus virginiana), the first parasitic plant with a fully sequenced plastome (dePamphilis and Palmer, 1990; Wolfe et al., 1992b).
Here, we compare the complete plastome sequences of 11 Orobanchaceae, including the nonparasitic Lindenbergia philippensis, one obligate hemiparasite from the earliest diverging parasitic branch of the family, and nine holoparasites from an exclusively nonphotosynthetic clade (Bennett and Mathews, 2006; McNeal et al., 2013). Our sampling represents a single loss of photosynthesis and allows us to infer the modes and mechanisms of plastome reduction after the transition to holoparasitism. To this end, we used rigorous statistical testing to identify factors governing the pattern of functional and physical genome reduction. Specifically, we investigated the frequency and relative order of structural rearrangements and gene losses and tested whether gene loss is governed by (1) gene function, (2) proximity to dispensable genes, (3) colocalization with conserved genes within the same operon, (4) gene length, or (5) strandedness. We also evaluated the interrelation of gene loss and the accumulation of mutagenic elements and structural reconfigurations as well as the shifts in nucleotide composition under relaxed selection.
RESULTS
Diversity, Size, Gene Content, and Structure of Plastomes
We sequenced and examined the plastid chromosomes of an autotrophic species, one photosynthetic obligate parasite, and nine nonphotosynthetic parasites from the broomrape family, Orobanchaceae (Figure 1; see Supplemental Table 1 online). Table 1 and Figure 1 provide an overview of the physical properties and gene content of the 11 plastomes; more information regarding single genes is provided as Supplemental Data Set 1 online, in Supplemental Methods 1 online, and in Supplemental References 1 online.
Figure 1.
Physical Maps of the Plastid Chromosomes of Photosynthetic and Nonphotosynthetic Members of Orobanchaceae.
All genes are colored according to functional complexes. Pseudogenes are indicated by Ψ. Brackets on top indicate different lifestyles.
Table 1. Overview of Physical Properties of Plastid Chromosomes in Nonparasitic and Parasitic Orobanchaceae.
| Taxon | Size (bp) | Gene Content (Protein-Codinga/tRNAa/rRNAb) | Protein-Coding Regions (%) | Noncoding Regions (%) | Structural RNAs (%) | GC (%)c | GC Protein Coding (%)d | GC Noncoding (%)d | GC Structural RNAs (%)d |
|---|---|---|---|---|---|---|---|---|---|
| Lindenbergia | 155,103 | 79/30/4 | 51.07 | 41.29 | 7.64 | 37.80 | 38.16 | 33.42 | 54.57 |
| Schwalbea | 160,911 | 74/30/4 | 50.82 | 41.81 | 7.36 | 38.08 | 38.82 | 33.73 | 54.31 |
| Epifagus | 70,028 | 21/17/4 | 43.07 | 41.58 | 15.35 | 36.01 | 34.72 | 29.84 | 50.20 |
| Conopholis | 45,673 | 21/18/4e | 49.29 | 37.86 | 12.84 | 33.92 | 35.66 | 28.88 | 52.45 |
| Cistanche | 94,380 | 27/26/4 | 33.40 | 54.36 | 12.24 | 36.57 | 36.21 | 32.66 | 53.44 |
| Boulardia | 80,361 | 26/21/4 | 42.12 | 44.15 | 13.74 | 35.76 | 35.72 | 29.84 | 53.82 |
| O. crenata | 87,529 | 32/27/4 | 39.88 | 36.92 | 13.21 | 35.19 | 35.37 | 29.84 | 53.39 |
| O. gracilis | 65,535 | 28/25/4e,f | 49.42 | 36.09 | 14.50 | 34.51 | 34.79 | 28.24 | 52.95 |
| Myzorrhiza | 120,840 | 42/28f/4f | 36.85 | 53.44 | 9.71 | 36.69 | 37.19 | 32.17 | 53.65 |
| P. purpurea | 62,891 | 28/22/4e,g | 50.15 | 39.74 | 10.11 | 31.09 | 34.62 | 24.95 | 51.66 |
| P. ramosa | 61,709 | 28/22/4e,g | 43.50 | 46.43 | 10.07 | 32.04 | 34.91 | 26.77 | 52.12 |
Number of unique genes.
Unless mentioned otherwise, rDNA is present in two sets.
GC content of the entire plastid chromosome.
GC content of unique plastome segments (IR region removed where present).
IR loss/loss of one rDNA operon.
One rRNA operon may be nonfunctional.
Partial duplication of rrn16.
The plastid chromosome of the autotrophic Lindenbergia philippensis is 155,103 kb in length and resembles that of most eudicots in terms of gene order and coding capacity. Apart from small (<100 bp) shifts of the IR junctions into the large and small single-copy regions, the L. philippensis plastome is colinear with that of tobacco (Nicotiana tabacum). Different from tobacco, however, the gene encoding the plastid translation initiation factor 1 (infA) is intact and potentially functional in L. philippensis. Plastid chromosomes are 160,911 kb large in the hemiparasitic American chaffseed (Schwalbea americana) and range between 120,840 and 45,673 kb in the holoparasites (Figure 1), with American squawroot (Conopholis americana) having the smallest plastid chromosome reported from land plants so far. Factors responsible for size variation include (partial) gene deletions and shifts of IR boundaries up to the complete loss of one IR (Table 1, Figure 1).
No genes have been physically lost in S. americana, but several genes are pseudogenes (and hence are functionally lost). Flanking the breakpoint of an inversion in the LSC region, the accD gene coding for the β-subunit of the acetyl-CoA carboxylase for lipid biosynthesis appears to be truncated at the 5′ end, lacking an unambiguously identifiable translation start codon in frame. Compared with N. tabacum and L. philippensis, the remainder of the reading frame shows extreme sequence divergence, including large indels and several premature stop codons. Functional losses in S. americana also affect genes for the plastid NADH dehydrogenase complex (ndh genes). The gene ndhF is split into three fragments, parts of which are relocated from the SSC into the IR; ndhA lacks parts of its intron and one exon. NdhD and ndhG contain several indels and multiple premature stop codons; ndhG additionally lacks a translation start site.
Among the 113 unique genes present in the nonparasitic L. philippensis, we identified a minimum common set of 16 protein-coding genes and 19 structural RNA genes that appear to be essential in the holoparasites (Figure 1, Table 2). The protein-coding genes include 13 genes encoding ribosomal protein subunits and three genes functioning in other housekeeping pathways, such as RNA maturation (matK) and preprotein import (ycf1; Kikuchi et al., 2013). The precise role of the conserved ycf2 gene is still unknown, but protein domain analysis (Wolfe, 1994) indicates a housekeeping role rather than a metabolic function. The genes accD and clpP, encoding the proteolytic subunit of the ATP-dependent caseinolytic protease, have intact reading frames in most parasites. In Phelipanche, however, the accD gene is highly diverged, with long insertions or deletions (indels), and appears to lack a standard start codon. ClpP experienced independent intron losses in Orobanche, Phelipanche, and California broomrape (Myzorrhiza californica). Additionally, in Phelipanche ramosa, a partial duplication resulted in aberrant copies of clpP, which we therefore classify as a pseudogene until further experimental evidence of its functionality is available. The minimum set of retained structural RNA genes encompasses 15 unique tRNAs and four rRNAs. An insertion in the stem of a tRNA gene (trnSGGA) in C. americana may indicate its pseudogenization, which, in consequence, would reduce the number of essential RNA genes in Orobanchaceae to 18.
Table 2. List of Essential Genes in 10 Parasitic Orobanchaceae.
| Gene Class | Gene IDs | Remarks |
|---|---|---|
| Protein genesa | matK, rpl16, rpl2, rpl20, rpl33, rpl36, rps11, rps12, rps14, rps18, rps2, rps4, rps7, rps8, ycf1, ycf2 | In Phelipanche, ycf1 is fragmented and shows strong sequence divergence and therefore may be a pseudogene. |
| tRNAb | trnDGUC, trnEUUC, trnfMCAU, trnHGUG, trnICAU, trnLCAA, trnLUAG, trnMCAU, trnNGUU, trnPUGG, trnQUUG, trnSGCU, trnSUGA, trnWCCA, trnYGUA | trnICAU exists in two divergent copies in M. californica. |
| rRNA | rrn16, rrn23, rrn4.5, rrn5 | In Phelipanche, one rRNA gene set is deleted and rrn16 is partially duplicated in the LSC; one copy of rrn16 is deleted from the O. gracilis plastome. |
accD and clpP await experimental verification of functionality in Phelipanche and S. americana.
trnSUGA may be a pseudogene in C. americana.
The nine holoparasites vary greatly in the number and type of retained genes. Except for the plastid-encoded polymerase (PEP) genes (rpo), housekeeping genes (50S ribosomal protein genes [rpl], 30S ribosomal protein genes [rps], and structural RNAs) are commonly conserved, and only a few have been lost or pseudogenized (e.g., rpl14, rpl23, rps15, rps16, trnAUGC, and trnGUCC) in the plastomes of some, but not all, holoparasites. In a few cases (rps7, 8, 12, and 18), unambiguous identification of the translation start and stop codon requires experimental verification (see Supplemental Data Set 1 online). Genes related to photosynthesis are mostly pseudogenized (photosystem I genes, psaA, B, and C; photosystem II genes, psbA, C, K, and Z) or absent (psaI, J, and K; psbB, D, E, F, I, J, L, M, and T, most ndh and pet genes, encoding subunits of the cytochrome b6/f complex) from the holoparasite plastomes. In Myzorrhiza californica, however, several photosynthesis genes (e.g., petG and psbF, I, J, K, and L) are retained. The rbcL genes is retained as an intact reading frame in M. californica, whereas it resides as a pseudogene in the plastomes of E. virginiana, C. americana, Cistanche phelypaea, Boulardia latisquama, and Orobanche crenata; rbcL was deleted from the plastomes in O. gracilis and Phelipanche. In M. californica and Phelipanche, all genes encoding subunits of the thylakoid ATP synthase complex (atp genes) are conserved with apparently intact reading frames (see Supplemental Data Set 1 online); some atp genes are also retained in Orobanche. Reading frames for both ycf1 and ycf2 are present in all nine holoparasites. In Phelipanche, the ycf1 gene is highly divergent and appears to be 5′-truncated, lacking an unambiguously identifiable translation start. Sequencing errors resulting from frequent repeats and long homopolymers cannot be ruled out for the ycf1 region, even after Sanger resequencing; ycf1 was therefore excluded from subsequent analyses.
In both Phelipanche species and, to a smaller extent, in O. gracilis, introns are reduced in number, although the coding sequences of the genes that normally contain them are intact. In Phelipanche, the genes rps12, clpP, rpl2, and trnKUUU/matK all lack their normal group IIA introns; however, trnLUAA, rpl16, and atpF retained their group I and group IIB intron, respectively. In O. gracilis, both clpP introns (group IIA and IIB) as well as the trnGUCC intron (group IIB) are absent. Long (>10 bp) poly(A) stretches are found near the coding sequences of clpP, rpl2, and 3′rps12 in Phelipanche as well as clpP in O. gracilis. In Phelipanche, a large fragment of the 16S rRNA gene (rrn16), bordered by a long stretch of noncoding DNA (ΨaccD-Ψrrn16 spacer: 2.6 to 2.8 kb) that shows no significant similarity to known plastid DNA regions replaces the rbcL gene between atpB and accD.
Relative to the outgroup L. philippensis, IR expansions occurred in the hemiparasite S. americana as a result of the relocation of ΨndhA/ΨndhF fragments and in the holoparasite C. phelypaea where the IRs expand into the ΨpsbA-trnK-matK region. Whereas O. crenata shows no structural modifications, the IRs in the O. gracilis plastome encompass only about two-thirds of the rDNA operon and a few tRNA genes. In P. purpurea, IR regions are even shorter, extending only over the ycf2 gene. In C. americana and P. ramosa, one IR copy is lost completely (Figure 1).
Inversions of the LSC often coincide with modifications of the IR regions (Figure 1; see Supplemental Figure 1 online). This is the case in the hemiparasite S. americana and in the holoparasites Cistanche, Orobanche, and Phelipanche. In S. americana, the accD-rbcL region is inverted relative to L. philippensis, and the inferred ancestral gene order, and this breakpoint may have affected the accD reading frame (see above). In O. gracilis, the fragment between the pseudogene of a photosystem assembly factor (Ψycf3) and trnSGCU in the LSC is inverted and coincides with the deletion of another tRNA gene (trnGUCC). Gene order is most extensively reconfigured in P. purpurea (Figure 1), in which the rpl32-trnLUAG region (normally located in the SSC) has been duplicated by relocation into the ycf2-rps7 segment. These inversions apparently happened in the common ancestor of the two Phelipanche species and were followed by further independent inversion events and the loss of the IR from the P. ramosa plastome. Inversions in the other holoparasite species occurred at least three times independently (see Supplemental Figure 1 online).
Plastid Repetitive DNA
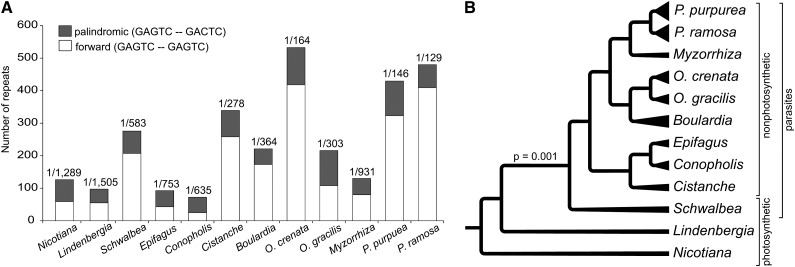
As in Nicotiana, DNA repeats account for ∼17.7% of the Lindenbergia plastome amounting to about one repetitive element per ∼1.5 kb (Figure 2A). Repeat density is much higher in the parasites, with an average of one repeat per ∼0.75 kb in E. virginiana and up to one repeat per ∼0.13 kb in Phelipanche (Figure 2A). Reverse complement repeats between 20 and 50 bp as well as repeats longer than 100 bp are mainly responsible for the increase in repeat DNA in the parasites (see Supplemental Figure 2 online). In the hemiparasite S. americana, the holoparasites B. latisquama, M. californica, and Orobanche slight accumulations of repeats occur around the IR-SSC junctions and near the center of the LSC (see Supplemental Figure 3 online). By contrast, repeats are dispersed nearly uniformly in the autotrophic L. philippensis.
Figure 2.
Repeat DNA in Plastomes of Orobanchaceae.
(A) Proportions of different repeat types in plastomes of 11 Orobanchaceae species and tobacco. Numbers above individual repeat columns indicate the repeat density, where, for example, 1/500 means that one repeat occurs every 500 bp. The direction is given for each repeat type.
(B) Evolution of repeats in Orobanchaceae. The strong evolutionary trend of increasing numbers of repetitive DNA in plastid genomes after the transition to heterotrophy is shown by the P value from an LRT evaluating constant-variance random walk versus directional random walk models to explain repeat variation. The number of repeats is illustrated by differently sized triangles at the tip of each terminal branch. Brackets indicate different lifestyles.
With N. tabacum included, there is a significant trend toward increased repeat densities in the plastomes of the hemi- and holoparasitic species (likelihood ratio test [LRT] of a constant-variance random walk versus a directional random walk model: P = 0.001; Table 3, Figure 2B); repeat density and the degree of parasitism are positively correlated (covariance versus no covariance among traits: LRT P = 0.003). A test for the mode of trait evolution in parasites (gradual versus punctuated evolution as measured by the branch-length parameter κ) revealed that changes in repeat density are consistent with a punctuated mode of evolution (LRT Pκ = 0.349; Table 3), implying that selection against repeats has been relaxed already in the common ancestor of hemi- and holoparasites. Repeat density is positively correlated with the extent of physical genome reduction (constant-variance random walk versus directional random walk model: LRT P < 0.001; Table 3). Our data suggest a slight, yet only statistically marginally significant (Pκ = 0.079) trend of increasing repeat density as genome reduction proceeds. A covariance versus no-covariance analysis among the two traits reveals no statistical support (LRT P = 0.371; Table 3), implying that relative genome reduction and repeat density are not correlated per se.
Table 3. Results of Phylogenetic Analyses Evaluating Evolution of Selected Plastome Traits in Orobanchaceae.
| Constant-Variance Random Walk (H0) versus Directional Random Walk Model (H1) of Trait Evolution | Gradual (H0) versus Punctuated (H1) Trait Evolutiona | Covariance (H0) versus No Covariance (H1) among Traitsa | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Traits Tested | -lnL H0b | -lnL H1 | LRT | P Value | -lnL H1 | LRT | P Value | -lnL H1 | LRT | P Value |
| Parasitism and repeat density | 89.654 | 83.138 | 13.032 | 0.001 | 83.577 | 0.878 | 0.349 | 87.978 | 8.802 | 0.003 |
| Relative genome reduction and repeat density | 140.136 | 131.557 | 17.158 | <0.001 | 130.016 | 3.083 | 0.079 | 131.957 | 0.800 | 0.371 |
| Parasitism and GC % | 28.563 | 20.185 | 16.756 | <0.001 | 20.304 | 0.237 | 0.626 | 20.389 | 0.406 | 0.524 |
| Parasitism and coding GC % | 25.285 | 15.825 | 18.920 | <0.001 | 11.729 | 8.192 | 0.004 | 15.990 | 0.331 | 0.565 |
| Parasitism and noncoding GC % | 32.073 | 22.708 | 18.729 | <0.001 | 22.786 | 0.156 | 0.692 | 23.145 | 0.874 | 0.350 |
| Loss of photosynthesis and GC % | 26.408 | 19.695 | 13.426 | 0.001 | 16.675 | 6.040 | 0.014 | 20.049 | 0.708 | 0.400 |
| Loss of photosynthesis and coding GC % | 20.260 | 13.016 | 14.490 | <0.001 | 9.444 | 7.144 | 0.008 | 15.651 | 5.269 | 0.022 |
| Loss of photosynthesis and noncoding GC % | 30.146 | 22.663 | 14.966 | <0.001 | 19.117 | 7.092 | 0.008 | 22.805 | 0.284 | 0.594 |
H0 is the best supported model from the test constant-variance random walk versus directional random walk model.
lnL, log likelihood.
Factors Shaping Segmental Deletions
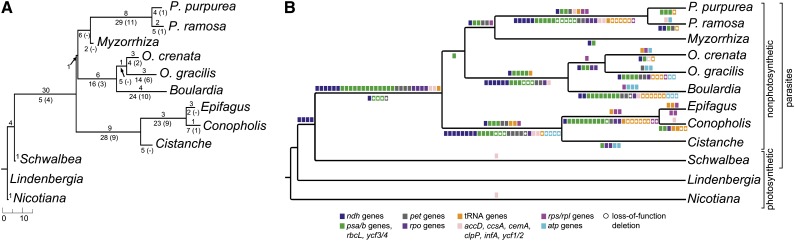
We reconstructed the ancestral set of protein-coding genes, rRNA genes, and tRNA genes of the nine holoparasites using maximum likelihood and an unconstrained model allowing for different rates of state changes. The loss of photosynthesis coincides with the pseudogenization of 31 out of 49 plastid genes for photosystems and photosynthetic electron transport (Figure 3; see Supplemental Data Set 2 and Supplemental Figure 4 online). Our inference suggests that these genes were not immediately deleted, with the possible exceptions of ndhA, petB, and psbT/N/H. Four ndh genes were likely functionally lost already after the transition to an obligate hemiparasitic lifestyle (Figure 3). All genes for the PEP and the tRNAs AlaUGC became pseudogenes during or shortly after the transition to holoparasitism (i.e., before the diversification of the extant lineages; Figure 3B; see Supplemental Data Sets 2 and 3 online). By contrast, most ribosomal protein genes (e.g., rpl14, 23, 32; rps3, 15, 16, and 19), tRNA isoacceptors (e.g., IleAUC, LeuUAA, LysUUU, and PheGAA), and infA, accD, clpP, and rbcL represent independent and repeated functional losses, occurring at a later stage of holoparasitic evolution. A replacement by cytosolic subunits is likely in species with an apparently nonfunctional plastid copy of the accD and/or the clpP gene, the products of which are involved in nonphotosynthetic pathways.
Figure 3.
Series of Functional and Physical Losses of Plastid Genes.
Graphical summary of the number of gene losses (A) and the losses of functional classes of genes (B) based on the reconstruction of plastid gene contents at ancestral nodes in Orobanchaceae. Pseudogenization is illustrated above the branches, whereas gene deletion is shown below the branches. In (A), an arrow indicates the correct placement of a value, and in (B), dots mark loss-of-function deletions (i.e., those without pseudogenization at any of the parent nodes).
Visual inspection of extant and reconstructed ancestral LSC regions (Figure 4) suggests that retention of dispensable DNA may depend on the localization in an operon-like transcription unit (see Supplemental Table 2 online) or relate to the proximity of essential genes (i.e., genes present in all taxa studied here; see Supplemental Data Set 4 online). Deletion of dispensable DNA may be affected by the length of the dispensable region (shorter regions will be lost less frequently than longer ones; Lohan and Wolfe, 1998) or by strandedness. The latter was shown to play a significant role in plastome evolution with respect to large-scale genome reconfigurations, localized gene losses, and relatively high amounts of repetitive DNA (Cui et al., 2006), features also observed in Orobanchaceae plastomes. We used multiple regression analyses and 12 models of different complexity (with one to four predictor variables) to investigate the influence of these physical properties on the survival time of nonessential DNA fragments. The model with the two predictor variables distance to essential genes and localization within an operon outperformed all other tested hypotheses, including those with only one of these two predictors (Akaike information criterion [AIC] of the best-fit model: 359.3; AIC of the distance-only model: 363.3; AIC of the operon-only model: 366.7; see Supplemental Table 3 online). The best-fit model is statistically reliable (overall F-test: F, 8.115, P < 0.001), and both variables collectively have a significant impact on survival time (Student’s t test: t = −3.098 with P = 0.003 for distance and t = −2.447 with P = 0.017 for operon). Therefore, essential genes in combination with the organization of genes in operon-like transcription units seem to provide protection from rapid deletion of dispensable gene regions.
Figure 4.
Evolution of Plastomes Based on Likelihood Reconstructions of Ancestral Gene Contents and the Rearrangement History.
The reconstructed plastid LSCs plus their adjacent regions are shown at all nodes of the Orobanchaceae phylogeny (thick gray lines), except for the ancestor of Phelipanche and Myzorrhiza because it differs from M. californica only by having not yet lost rpoC1, petN, psaA, petL, and ndhH. Pseudogenes are indicated by gray gene boxes and gene names with a Ψ-prefix, whereas deletions are indicated by black arrowheads and the corresponding gene name(s). Gene labels with an asterisk indicate intron losses. Blue pennons indicate the breakpoints of an inversion (I1 to I5); the range and orientation of these inversions are denoted in both the ancestral and the derived plastid genome, and the genes flanking inversion breakpoints are labeled accordingly. For simplicity, SSC and IR are not shown here.
Nucleotide Compositional Bias and Codon Usage
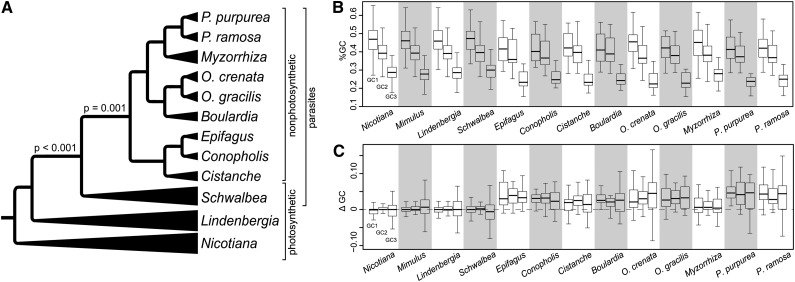
Functional genome reduction corresponds to variation in the GC content of the plastid chromosomes in Orobanchaceae (Table 1, Figure 5). Compared with the autotrophs N. tabacum and L. philippensis as well as to the hemiparasite S. americana, the nine holoparasites show a 2 to 6% lower total GC content (Table 1). In protein-coding regions, GC reduction amounts to 3 to 4%, while in structural RNAs it remains nearly unaltered (Table 1). The IR-lacking holoparasitic species possess a relatively low GC content in their noncoding regions, with only 25% GC in Phelipanche and 28% in C. americana, compared with >30% in the other holoparasites (Table 1). The GC poor plastomes also appear to accumulate inversions as well as structural mutations around the IR (Figures 1 and 4, Table 1). LRTs (Table 3) show that reduction of the GC content is directional and coincides with the transition to heterotrophy (constant-variance random walk versus directional random walk model LRT P < 0.001, Pκ = 0.626; Figure 5A) and, moreover, that it increases gradually in holoparasites (LRT P = 0.001, Pκ = 0.014; Figure 5A).
Figure 5.
Evolution of GC Content in Plastomes of Orobanchaceae.
(A) A strong evolutionary trend of reduction in total GC content occurs with the transition from an autotrophic to a parasitic lifestyle and continues in nonphotosynthetic lineages. P values from LRTs at key nodes of lifestyle changes evaluate constant-variance random walk versus directional random walk models to explain GC variation in parasites and nonparasites.
(B) GC content at different codon positions of intact plastid protein-coding genes for nine nonphotosynthetic and four photosynthetic plants.
(C) Variation of GC content at different codon positions in coding regions of parasites and nonparasites assessed as the difference (ΔGC) to a reference genome (Aucuba japonica, Garryaceae). In (B) and (C), a line inside each box designates the median across 31 conserved plastid genes; the whisker ends are at the 5th and 95th percentiles.
In L. philippensis and S. americana, the median GC content differs among codon positions with the first position having the highest GC content; these differences almost disappear in the holoparasites (Figures 5B and 5C). By contrast, the holoparasites show a marked AT richness (LRT P = 0.001, Pκ = 0.014; Table 3, Figure 5A), which affects the first and second positions such that the GC content of the first codon position converges to that of the second (Figures 5B and 5C).
To investigate whether the compositional bias depends on the functional class of a gene, we evaluated differences in base pair distribution in 31 matching genes from the 10 hemi- and holoparasitic Orobanchaceae and 10 nonparasitic asterid relatives (including L. philippensis), using a series of alpha error-corrected Wilcoxon tests. The set of 31 genes corresponds to the commonly retained genes plus the atp genes that are potentially intact in some of the holoparasites (see Supplemental Data Set 1 online). Results show that codon positions in the parasites show a general shift toward AT (see Supplemental Tables 4 to 6 online): Fourteen of the 31 genes tested contain significantly higher amounts of AT at both the first and the second codon sites; 13 more genes exhibit significant changes of nucleotide composition in either the first or the second codon position. Among the latter are three atp genes (atpE, H, and I), accD, and clpP (see Supplemental Table 5 online; Figure 5C). Regarding GC content, only four genes (rps14, rpl22, 23, and 32) did not differ between parasites and nonparasites.
Most parasitic Orobanchaceae utilize similar codons as nonparasites (see Supplemental Table 6 online), irrespective of GC content and regardless of the number of preserved tRNA genes (see Supplemental Figures 5 and 6 online). However, under- or overuses of some codons are observed in B. latisquama (AlaUGC), E. virginiana (SerUGA and IleUGU), C. americana (SerACU), and C. phelypaea (ProUGG) compared with the other taxa. The difference in usage of the two preferred codons for the Val tRNA rises from <2 to >5% in M. californica, O. gracilis, and Phelipanche (see Supplemental Table 6 online). All changes in codon usage are apparently not related to the number and type of retained tRNA isoacceptors.
DISCUSSION
Evolution of Chromosomal Size and Architecture after the Loss of Photosynthesis
A main finding of this study is the uneven extent to which plastomes in a clade of nine closely related holoparasites have diverged from the ancestral plastome of autotrophic Orobanchaceae, represented here by L. philippensis. The plastome of the latter is colinear with that of related autotrophic angiosperms, while structural changes have occurred in the obligate hemiparasite S. americana. This is similar to the situation in other hemiparasitic flowering plants, such as Asian witchweed (Striga asiatica; also belonging to Orobanchaceae) and Cuscuta (dodder) species (Downie and Palmer, 1992; dePamphilis et al., 1997; Funk et al., 2007; McNeal et al., 2007). Apparently, structural maintenance relaxes with reduced functional constraints on photosynthesis resulting in the progressive nonfunctionalization of the plastome. This likely leads to the accumulation of mutagenic factors as observed here, including microsatellites, long homopolymer stretches, forward or palindromic repeats of any length, and a low GC content (Table 1, Figure 2; see Supplemental Figures 2 and 3 online). Those elements increase intramolecular and illegitimate recombination (Ogihara et al., 1988; Fejes et al., 1990; Sears et al., 1996; Müller et al., 1999; Gray et al., 2009; Maréchal and Brisson, 2010), which in turn may cause the deletion of dispensable plastome fragments.
Our statistical tests revealed that the shift toward AT richness in coding and noncoding regions is associated with the transition to a holoparasitic lifestyle. With the exception of some tRNAs in the Epifagus/Conopholis/Cistanche clade (Wolfe et al., 1992a), the shift in AT richness has no immediate and notable effect on codon usage (Figure 5; see Supplemental Figures 5 and 6 online). This is probably attributable to extensive superwobbling, where a tRNA species with an unmodified uridine reads all nucleotides in the third codon position, but at the expense of translation efficiency (Rogalski et al., 2008). However, alternative hypotheses, such as random drift, cannot be excluded because of the scarce sampling of completely sequenced plastid genomes in lamiid nonparasites. Extreme AT richness also characterizes the remnant plastid (apicoplast) genomes of parasitic Apicomplexa, such as Plasmodium and Toxoplasma (Sato, 2011). Thus, a gradually increasing nucleotide compositional bias may be a general feature in the process of physical plastome reduction in secondarily heterotrophic organisms as long as organelle genomes maintain a residual coding capacity.
Large inversions and modifications around the IR as observed in Phelipanche, O. gracilis, C. americana (see Supplemental Figure 1 online), the earth orchid Rhizanthella (Delannoy et al., 2011), and some Cuscuta species (Funk et al., 2007; McNeal et al., 2007) presumably represent recent and idiosyncratic chromosomal reconfigurations. The trigger for these genomic rearrangements likely is the relaxed selective pressure and the progressive nonfunctionalization of the plastome. However, the same argument cannot be made for large-scale rearrangements in plastomes of nonparasites (Palmer and Thompson, 1982; Palmer et al., 1987; Perry and Wolfe, 2002; Raubeson and Jansen, 2005; Haberle et al., 2008; Guisinger et al., 2011).
Unlike in the Epifagus lineage, several group IIA introns are absent from the plastomes of Phelipanche species (in rps12, clpP, rpl2, and trnKUUU/matK) and O. gracilis (in clpP). The same ones have also been lost in Cuscuta species (McNeal et al., 2009), which lack the plastid encoded splicing factor MatK as well. Except for the clpP intron, all of these introns associate with MatK (Liere and Link, 1995; Zoschke et al., 2010). As in Cuscuta species, the concerted loss of IIA introns may indicate an impaired splicing mechanism attributable to reduced activity or malfunction of MatK or other nuclear-encoded splicing factors, although matK losses are not known from Orobanchaceae so far. Long poly(A) stretches near the rpl2, rps12, and clpP genes in Phelipanche and O. gracilis suggest multiple cDNA recombination events responsible for the intron losses. The occurrence of unknown DNA in place of the plastid rbcL gene in Phelipanche implies either rapid sequence divergence or recombination with DNAs from other cellular genome compartments (e.g., by DNA repair–related recombination events).
Evolutionary Sequence of Gene Loss
Of the 113 unique plastid genes found in L. philippensis, only 16 protein-coding genes and 19 structural RNAs (tRNAs and rRNAs) are retained in all nine holoparasites. Deletion of photosynthesis genes is almost complete in E. virginiana and C. americana (Figures 1 and 3; see Supplemental Data Set 1 online), both of which have also lost notably more housekeeping genes than the remaining parasites, thus approaching the inferred minimum set of genes in Orobanchaceae. Clearly, pseudogenization and loss of plastid genes (usually preceded by pseudogenization: Figures 3 and 4) proceeded at a highly lineage-specific tempo. We inferred that changes to the plastid gene content are not all initiated by the loss of photosynthesis, but already accompany the transition to a parasitic lifestyle as seen in the obligate hemiparasite S. americana. This resembles findings in the cryptically photosynthetic Cuscuta (Krause, 2011; Braukmann et al., 2013) and in photosynthetic obligate myco-heterotrophic Ericaceae (Braukmann and Stefanović, 2012). The thylakoid NADH-dehydrogenase complex represents the earliest functional loss during the transition to heterotrophy (Figures 3 and 4; see Supplemental Figure 4 online), and all heterotrophic plants studied so far show pseudogenization or deletion of ndh genes (Wickett et al., 2008; Delannoy et al., 2011; Logacheva et al., 2011; Braukmann and Stefanović, 2012; Braukmann et al., 2013). This is also the case for the holoparasite Lathraea (Delavault et al., 1996), which belongs to another clade of predominantly hemiparasitic Orobanchaceae, distant from Schwalbea. Because of the lack of data for facultative hemiparasites (Schwalbea is an obligate hemiparasite), it is not possible to determine whether the loss of ndh genes is associated with the single transition to heterotrophy in general (as reconstructed here) or with the shift from facultative to obligate parasitism, the latter having occurred multiple times in Orobanchaceae (Bennett and Mathews, 2006). Loss of the plastid Ndh complex is thought not to affect the fitness of heterotrophs (Krause, 2011) because it is nonessential for cell survival under nonstress conditions (Peltier and Cournac, 2002).
Unlike the other housekeeping genes, the PEP genes had already been functionally lost in the last common ancestor of the nine holoparasites studied here (Figure 3; see Supplemental Figure 4 online), as well as in the unrelated holoparasitic Orobanchaceae Lathraea (Lusson et al., 1998). PEP genes are lost early in Cuscuta species, too (Krause et al., 2003; Revill et al., 2005; Funk et al., 2007; McNeal et al., 2007). In autotrophs, PEP transcription is important for the efficient functioning of the photosynthesis light reaction complexes and other photosynthesis-related subunits (Hajdukiewicz et al., 1997), although malfunction or loss of PEP can be compensated for by a nuclear-encoded polymerase in parasites and nonparasites (Lusson et al., 1998; Legen et al., 2002; Krause et al., 2003; Berg et al., 2004). Relaxed pressure on the rapid assembly of photosynthesis complexes as a result of a dark vegetative phase during host-root attachment may render rpo gene function dispensable at early developmental stages in obligate heterotrophic Orobanchaceae.
In contrast with the common and early loss of ndh genes and PEP, tempo and order of nonfunctionalization of the plastid-encoded genes for photosystems (psa and psb) and for the photosynthetic electron transport (pet and atp) are lineage specific (Figures 3 and 4; see Supplemental Data Set 2 online). An exception to the extensive loss of photosynthesis-related genes is the ATP synthase complex. In Phelipanche and M. californica, all six atp genes are conserved and potentially functional, resembling findings in the heterotrophic green alga Prototheca wickerhamii (Knauf and Hachtel, 2002), the myco-heterotrophic liverwort Aneura mirabilis (Wickett et al., 2008), and Cuscuta species (Funk et al., 2007; McNeal et al., 2007). A reduced subset of atp genes exists in the two Orobanche species and in Cistanche deserticola, which, unlike C. phelypaea, has retained pseudogenes of atpA, atpB, atpF, and atpE (Li et al., 2013). Prolonged functional conservation of genes after the loss of photosynthesis may indicate involvement in alternative pathways, as is the case for rbcL, the large subunit of Rubisco (Wolfe and dePamphilis, 1997; Lusson et al., 1998; Randle and Wolfe, 2005; Wicke et al., 2011). Besides its function during photosynthetic carbon fixation, Rubisco contributes to Ser and Gly synthesis in the C2 pathway (Tolbert, 1997) and to a glycolysis bypassing reaction (Schwender et al., 2004). Similarly, atp gene retention suggests a prolonged functionality of the thylakoid ATP synthase complex because of either an involvement in ATP synthesis from a source other than the photosynthetic proton gradient or the requirement of ATP hydrolysis in plastids of at least some holoparasites. Maintenance of functional constraints on some genes of the photosynthesis apparatus during the initial period of the holoparasitic life is also supported by the small extent of plastome reduction in M. californica.
In Orobanchaceae, housekeeping genes are lost idiosyncratically after the transition to holoparasitism (Figure 3; see Supplemental Data Sets 2 and 3 online), and losses affect structural RNAs as well as essential ribosomal protein genes (rps3, 15, 16, and 19 and rpl14, 22, 23, and 32) (Fleischmann et al., 2011). Deletion of ribosomal proteins has also been reported from other nonphotosynthetic angiosperms (Delannoy et al., 2011; Logacheva et al., 2011; Wicke et al., 2011), holoparasitic algae (de Koning and Keeling, 2006), and the apicoplast of apicomplexan parasites (Sato, 2011). Expression and import of nuclear-encoded elements for the translation apparatus are maintained (Wolfe et al., 1992a; Ems et al., 1995; de Koning and Keeling, 2004; Jackson et al., 2011), which suggests that plastid ribosomes are still functionally assembled even in parasites with highly reduced plastomes. The uncorrelated and independent losses of ribosomal proteins and tRNAs, which occur comparably late during plastome reduction, may thus point to an increased (functional) transfer of plastid DNA to the nuclear genomes in parasites.
Factors Affecting Gene Retention
In spite of the species-specific deletion of dispensable gene regions after the transition to a nonphotosynthetic lifestyle, several factors seem to determine retention or loss of genes (Figure 4). Using multiple regression analyses, we were able to show that the survival of nonessential regions (as pseudogenes or potentially still functional genes) is best explained by both their distance to an essential element and the colocalization in an operon (see Supplemental Table 3 online). By contrast, gene length and strandedness play no statistically significant role for gene retention in Orobanchaceae. A neighboring-gene effect, yet in conjunction with gene size, has also been suggested in a previous analysis simulating the loss of tRNAs from the Epifagus plastome (Lohan and Wolfe, 1998). Evidently, plastid operons and essential genes collectively provide a degree of protection from rapid deletion and thus reduce the speed with which dispensable regions are deleted from the plastome. A similar protection may emanate from nongenic essential elements, such as transcription promoting, terminating, and processing sequences as well as replication origins. As the regression models testing the operon effect and the neighboring gene effect did not fully take phylogenetic relationships into account, our results should be viewed with some caution.
The long-term retention of atp genes as intact open reading frames in some holoparasitic lineages remains puzzling, as this cannot be explained sufficiently by aspects of large-scale chromosomal evolution as discussed above. Therefore, the tempo and sequence of nonfunctionalization of plastid genes may also be affected by an as yet unknown function beyond photosynthesis and by the rate of functional gene transfer from the plastid to the nucleus.
METHODS
Taxon Sampling and Sequencing Approaches
The complete plastomes were sequenced from one autotrophic species (Lindenbergia philippensis), one photosynthetic obligate parasite (American chaffseed [Schwalbea americana]), and eight nonphotosynthetic parasitic species of Orobanchaceae (Boulardia latisquama, Cistanche phelypaea, Conopholis americana, California broomrape [Myzorrhiza californica], Orobanche crenata, Orobanche gracilis, Phelipanche purpurea, and Phelipanche ramosa). The data set was complemented with the plastid DNA sequence of beechdrops (Epifagus virginiana) (Wolfe et al., 1992b) from the National Center for Biotechnology Information (NCBI) GenBank (NC_001568). Voucher information and sequencing approaches are summarized in Supplemental Table 1 online. Sequences of the complete plastomes have been determined using either fosmid libraries (modified after McNeal et al., 2006) or shotgun-pyrosequencing from total genomic DNA. A detailed description of the experimental procedures is provided in Supplemental Methods 2 online.
Sequence Assembly, Finishing, and Contig Verification
Shotgun Sanger and pyrosequencing data from fosmid clones were quality trimmed, and the vector sequences were removed prior to assembly with SeqMan I (DNA Star) or Geneious 5.6. Contaminated sequences were removed by BLASTing against custom contaminant databases. Subsequently, a BLAST search was conducted against custom plastid genes and genome databases as well as plastid protein databases employing the NCBI local BLAST package. BLAST hits were considered with E-values < 10e−25 for BLASTn and 10e−10 for BLASTx, respectively, and a minimal contiguous alignment length of 100 bp. Postassembly of contigs was performed manually using the Phylogenetic Data Editor (available at http://www.phyde.de) and Geneious 5.6, respectively. Gaps and uncertain regions were closed and verified by PCR amplification and Sanger sequencing. PCR with custom primers was used to verify overlapping regions between different fosmids as well as junctions between single-copy and the IR regions.
Plastid genomes sequenced from total genomic DNA were reconstructed from two independent assemblies: 454 raw reads were extracted from the sff file using a third-party script provided along with the MIRA assembler software (Chevreux et al., 1999). Adapters plus the adjacent 10 bp were clipped off, and quality trimmed reads were assembled de novo under the accurate assembly mode with MIRA 3 (Chevreux et al., 1999). Contigs were BLASTed against custom plastid databases as described above. Positive matches were extracted and postassembled gene by gene with high stringency in SeqMan I. Contigs were preannotated using DOGMA (Wyman et al., 2004) and then overlapped manually. Contig joins and uncertain regions including microsatellite-like fragments were PCR verified and confirmed by Sanger sequencing. A second parallel approach involved sorting and extracting 454 reads of a potential plastid origin using the NCBI BLAST suite and subsequent assembly of plastid-like reads with CAP3 (identity cutoff 97%, minimal overlap 90 bp, and maximum gap length 5 bp). CAP3 contigs were preannotated using DOGMA, manually joined, and checked for incongruences with MIRA contigs. Cases of incongruence (mainly satellite regions) were verified by PCR and Sanger sequencing. Annotation of finished plastid chromosome sequences was performed using DOGMA with some manual refinement. Reference-assisted assembly of Phelipanche plastomes was performed in MIRA 3 using P. purpurea fosmids (covering IR and LSC) as backbone sequences.
Comparative Plastome Analysis
Descriptive features of the plastid genomes were obtained with SeqState 1.4.1 (Müller, 2005). Unless noted otherwise, tobacco (Nicotiana tabacum; NC_001879) (Shinozaki et al., 1986), Mimulus guttatus (Mimulus Genome Project, Department of Energy Joint Genome Institute; http://www.phytozome.net/mimulus), and L. philippensis were used as core autotrophic groups for analysis of sequence data.
Self-self dot plots using Geneious 5.6 were analyzed for an overview of repeat distribution, and REPuter (Kurtz et al., 2001) was employed to quantify all forward and reverse complement (i.e., palindromic) repeats longer than 20 bp applying a Hamming distance of 3; repeats with an E-value > 0.1 were not considered.
The program CodonW (http://codonw.sourceforge.net/) was employed for the analyses of codon usage and base pair composition in plastid coding regions. Statistical analyses and hypothesis testing were performed employing custom R scripts (http://www.R-project.org).
Pairwise Wilcoxon tests with sequential alpha-error correction were employed to evaluate differences in A, T, C, and G distribution, total GC content, and GC content at different codon positions. To evaluate differences among gene-specific GC at different codon positions, unpaired Wilcoxon tests were performed between parasites and an equally sized set of closely related autotrophic taxa: snapdragon (Antirrhinum majus; GQ996966 to GQ997048) (Moore et al., 2010), Atropa belladonna (NC_004561) (Schmitz-Linneweber et al., 2002), coffee (Coffea arabica; NC_008535) (Samson et al., 2007), Jasminum nudiflorum (NC_008407) (Lee et al., 2007), Nerium oleander (GQ997630 to GQ997712) (Moore et al., 2010), Olea europaea (NC_013707) (Mariotti et al., 2010), tomato (Solanum lycopersicum; DQ347959) (Daniell et al., 2006), tobacco (Nicotiana tabacum), M. guttatus, L. philippensis, and Aucuba japonica (GQ997049 to GQ997131) (Moore et al., 2010). Aucuba, sister to the remainder lamiid species, was also used as a reference to build gene-wise distances for box plots of variation in GC content between photosynthetic taxa and Orobanchaceae. Because the genes infA and rps12 were missing in the original Olea plastid genome annotation, we BLAST searched and extracted them from the genome reference sequence (localizations of infA in refseq: c82247 to 82480; localization of rps12-exons in refseq: c72298 to 72411, c99915 to 99941, and c100477 to 100708).
Reconstruction of the Ancestral Gene Content and the Rearrangement History
For tree-based analyses, phylogenetic relationships were inferred based on a concatenated data set of all plastid ribosomal protein genes, using tobacco and M. guttatus as outgroups. Species-specific absence of a marker gene was treated as an indel event and included as binary coded data. Using PAUP 4.0b, a maximum likelihood (ML) tree was reconstructed using the GTR+G+I model selected by the AIC in ModelTest 3.7 (Posada and Crandall, 1998). We used four rate categories for the gamma distribution describing rate heterogeneity across sites; the gamma shape parameter, proportion of invariable sites, nucleotide frequencies, and substitution rates of the GTR model were estimated via ML. One thousand bootstrap replicates were run with the same settings. In addition, two runs of one million generations each were run in MrBayes 3.2 using default priors. Each run consisted of one cold and seven heated chains (temperature set to 0.2), sampling every 100th generation. The initial 10% of trees was discarded as the burn-in fraction. Results of both inferences were congruent and highly supported.
Based on the consensus tree, we reconstructed the ancestral genome structure for all Orobanchaceae nodes for 30 tRNAs, four RNAs, and 79 protein-coding genes (excluding conserved but cryptic open reading frames,such as ycf15 and ycf62), using the ML approach implemented in BayesTraits (Pagel et al., 2004). Based on a multistate matrix where genes were encoded as functionally present, pseudogenized, or deleted, state changes and probabilities at internal nodes were computed with an unconstrained six-parameter model. Fifty likelihood attempts were run on the consensus tree using the most-recent-common-ancestor mode.
The history of large-scale rearrangements among plastomes was traced using the Bayesian approach implemented in Badger 1.02/barphlye (Larget et al., 2005; Darling et al., 2008). The evolution of plastomes in nonphotosynthetic plants is mainly driven by gene loss, but current algorithms and software cannot cope with this particular scenario. Consequently, the raw gene data could not be used as a primary input for Badger because of dissimilar gene contents. To circumvent this problem, we determined the maximum amount of locally colinear blocks among all sequenced Orobanchaceae genomes with progressive Mauve 2.3.1 (Darling et al., 2010) using a seed weight of 21 and a custom gap open penalty of −200 to account for (relatively) small gaps. Based on this alignment, a permutation matrix was constructed and transformed as input for Badger, which was run in the MC3 (for Metropolis-Coupled Markov chain Monte Carlo) mode with eight parallel chains of one million cycles each, sampling every tenth tree, and ancestral permutations, which were then evaluated using the barphlye scripts.
Statistical Analyses
BayesTraits was employed to evaluate evolutionary trends in traits such as GC content and repeats. Specifically, we tested whether the change in a trait is explained significantly better by a directional random walk model than by a constant-variance random walk model; an LRT was used to assess the significance of the difference between models. We also tested the mode of trait evolution (gradual versus punctuated) by constraining the BayesTraits-specific parameter kappa (κ) to κ = 0 to measure the effect of branch lengths upon trait changes (Pagel, 1994); significance was again assessed by a LRT. Correlation of traits was further tested by constraining the covariance to zero and then carrying out a LRT on the constrained and unconstrained model. Treegraph II (Stöver and Müller, 2010) was employed to visualize the results.
Gene retention in relation to being part of an operon (operon effect), proximity to conserved genes (neighboring-gene effect), gene length, and/or strandedness was tested by fitting multiple linear regression models (in R, including the fRegression package) that differed in the combinations of predictor variables. The overall fit of the models was compared employing the AIC. To account for the nonindependence of data resulting from phylogenetic relatedness in the correlation tests, we grouped plastid genes according to their survival time. Deletion of a gene was coded/scored as 0 and presence (irrespective of functionality) was coded as 1. Thus, a conserved gene could score a maximum of 9, indicating highest survival of a genic fragment over the tree; rapidly lost genic fragments (i.e., genes lost in all parasites) scored 0. We computed the shortest distance to a gene’s closest conserved neighbor (downstream or upstream) in the last common photosynthetic ancestor deduced from L. philippensis (see Supplemental Data Set 4 online), with conserved being defined as universally present in all investigated taxa. For testing the operon effect we encoded genes in a binary matrix according to their localization in a transcription unit (see Supplemental Table 2 online). Gene length and strandedness were parsed from the L. philippensis plastome annotation (see Supplemental Data Set 4 online).
Accession Numbers
All newly sequenced plastid genomes have been deposited at the European Nucleotide Archive (see Supplemental Table 1 online), and all data sets from whole-genome shotgun pyrosequencing are deposited at the Sequence Read Archive under accession number SRA047928.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Locally Colinear Blocks Inferred with Progressive Mauve.
Supplemental Figure 2. Proportion and Length of Repetitive DNA in Plastid Genomes of Orobanchaceae.
Supplemental Figure 3. Self-Self Dot Plots of Plastid Genomes of Orobanchaceae and Nicotiana.
Supplemental Figure 4. Graphical Summary of Plastid Gene Losses Inferred by Ancestral State Reconstruction.
Supplemental Figure 5. Results of Coalescence Analyses of Codon Usage for Intact Plastid Genes.
Supplemental Figure 6. Results of Coalescence Analyses of Codon Usage for 59 Degenerated Plastid Codons.
Supplemental Table 1. Plant Material Used for Plastome Sequencing.
Supplemental Table 2. Organization of Transcription Units in Angiosperm Plastid Genomes.
Supplemental Table 3. Detailed Results from Multiple Regression Analyses.
Supplemental Table 4. Results of Pairwise Wilcoxon Tests (P Values) Evaluating Differences in GC Content and Nucleotide Composition of Coding Regions between Nonparasites and Parasites.
Supplemental Table 5. Results of Unpaired Wilcoxon Tests (P Values) Evaluating Differences in GC Content of Coding Regions between Nonparasites and Parasites.
Supplemental Table 6. Codon Usage in Photosynthetic and Nonphotosynthetic Orobanchaceae.
Supplemental Data Set 1. Detailed Overview of the Gene Content of 11 Photosynthetic and Nonphotosynthetic Orobanchaceae.
Supplemental Data Set 2. Ancestral State Reconstructions for 79 Plastid Protein-Coding Genes.
Supplemental Data Set 3. Ancestral State Reconstructions for 30 Plastid tRNA Genes and Four Plastid rRNA Genes.
Supplemental Data Set 4. Proximity of Nonessential Plastid Genes to Conserved Genic Elements in Photosynthetic and Nonphotosynthetic Orobanchaceae Ancestors.
Supplemental Methods 1. Details Regarding Gene- and Species-Specific Annotation.
Supplemental Methods 2. Details of Experimental Procedures Used for Plastid Genome Sequencing.
Supplemental References 1. References Cited in the Supplemental Data.
Supplementary Material
Acknowledgments
We thank Monika Ballmann, Karola Maul (University of Bonn, Germany), Thomas Münster (Max Planck Institute for Plant Breeding, Cologne, Germany), Paula Ralph, Lena Landherr (Penn State University, Pennsylvania), and Christian Gülly (Medical University Graz, Austria) for technical support, Klaus Bahr, Wolfram Lobin (Botanical Garden Bonn), Barbara Ditsch (Botanical Garden Dresden, Germany), Mats Hjertson (Uppsala University, Sweden), and Kay Kirkman (Joseph W. Jones Ecological Research Center, Georgia) for plant material, and two anonymous reviewers for valuable comments and suggestions on an earlier version of this article. Financial support came from the Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung [FWF] Grant 19404 to G.M.S.), the German Science Foundation (Deutsche Forschungsgemeinschaft [DFG] Grants MU2875/2 to K.F.M. and RE603/9-1 to S.S.R.), and the U.S. National Science Foundation (Grants DEB-0120709 and DBI-0701748 to C.W.D.).
AUTHOR CONTRIBUTIONS
S.W., G.M.S., and C.W.D. designed the research. S.W. performed research and analyzed the data. G.M.S., C.W.D., K.F.M., D.Q., N.J.W., and Y.Z. assisted with experiments and/or contributed to data analyses. S.S.R. contributed large-scale data. S.W. and G.M.S. wrote the article. C.W.D., K.F.M., D.Q., N.J.W., and S.S.R. critically revised the article.
Glossary
- LSC
large single-copy region
- SSC
small single-copy region
- IR
inverted repeat
- Rubisco
ribulose-1,5-bisphosphate carboxylase/oxygenase
- LRT
likelihood ratio test
- Pκ
branch length parameter κ
- AIC
Akaike information criterion
- PEP
plastid-encoded polymerase
- NCBI
National Center for Biotechnology Information
- ML
maximum likelihood
References
- Bennett J.R., Mathews S. (2006). Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am. J. Bot. 93: 1039–1051 [DOI] [PubMed] [Google Scholar]
- Berg S., Krause K., Krupinska K. (2004). The rbcL genes of two Cuscuta species, C. gronovii and C. subinclusa, are transcribed by the nuclear-encoded plastid RNA polymerase (NEP). Planta 219: 541–546 [DOI] [PubMed] [Google Scholar]
- Bock, R. (2007). Structure, function, and inheritance of plastid genomes. In Cell and Molecular Biology of Plastids, Topics in Current Genetics, R. Bock, ed (Berlin and Heidelberg: Springer), pp. 29–63. [Google Scholar]
- Braukmann T., Kuzmina M., Stefanović S. (2013). Plastid genome evolution across the genus Cuscuta (Convolvulaceae): Two clades within subgenus Grammica exhibit extensive gene loss. J. Exp. Bot. 64: 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann T., Stefanović S. (2012). Plastid genome evolution in mycoheterotrophic Ericaceae. Plant Mol. Biol. 79: 5–20 [DOI] [PubMed] [Google Scholar]
- Chevreux, B., Wetter, T., and Suhai, S. (1999). Genome sequence assembly using trace signals and additional sequence information. In German Conference on Bioinformatics, R. Giegerich, R. Hofestädt, T. Lengauer, W. Mewes, D. Schomburg, M. Vingron and E. Wingender, eds (Braunschweig-Bielefeld, Germany:GBF-Braunschweig and University of Bielefeld), pp. 45–56. [Google Scholar]
- Colwell, A.E. (1994). Genome Evolution in a Nonphotosynthetic Plant, Conopholis americana PhD dissertation (St. Louis, WA: Washington University). [Google Scholar]
- Cui L., Leebens-Mack J., Wang L.S., Tang J., Rymarquis L., Stern D.B., dePamphilis C.W. (2006). Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evol. Biol. 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H., Lee S.-B., Grevich J., Saski C., Quesada-Vargas T., Guda C., Tomkins J., Jansen R.K. (2006). Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor. Appl. Genet. 112: 1503–1518 [DOI] [PubMed] [Google Scholar]
- Darling A.E., Mau B., Perna N.T. (2010). progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A.E., Miklós I., Ragan M.A. (2008). Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 4: e1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A.P., Keeling P.J. (2004). Nucleus-encoded genes for plastid-targeted proteins in Helicosporidium: Functional diversity of a cryptic plastid in a parasitic alga. Eukaryot. Cell 3: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A.P., Keeling P.J. (2006). The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E., Fujii S., Colas des Francs-Small C., Brundrett M., Small I. (2011). Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol. Biol. Evol. 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delavault P.M., Russo N.M., Lusson N.A., Thalouarn P. (1996). Organization of the reduced plastid genome of Lathraea clandestina, an achlorophyllous parasitic plant. Physiol. Plant. 96: 674–682 [Google Scholar]
- Delavault P.M., Sakanyan V., Thalouarn P. (1995). Divergent evolution of two plastid genes, rbcL and atpB, in a non-photosynthetic parasitic plant. Plant Mol. Biol. 29: 1071–1079 [DOI] [PubMed] [Google Scholar]
- dePamphilis, C.W. (1995). Genes and Genomes. In Parasitic Plants, M.C. Press and J.D. Graves, eds (London: Chapman and Hall), pp. 177–205. [Google Scholar]
- dePamphilis C.W., Palmer J.D. (1990). Loss of photosynthetic and chlororespiratory genes from the plastid genome of a parasitic flowering plant. Nature 348: 337–339 [DOI] [PubMed] [Google Scholar]
- Downie S.R., Palmer J.D. (1992). Restriction site mapping of the chloroplast DNA inverted repeat - A molecular phylogeny of the Asteridae. Ann. Mo. Bot. Gard. 79: 266–283 [Google Scholar]
- Ems S.C., Morden C.W., Dixon C.K., Wolfe K.H., dePamphilis C.W., Palmer J.D. (1995). Transcription, splicing and editing of plastid RNAs in the nonphotosynthetic plant Epifagus virginiana. Plant Mol. Biol. 29: 721–733 [DOI] [PubMed] [Google Scholar]
- Fejes E., Engler D., Maliga P. (1990). Extensive homologous chloroplast DNA recombination in the pt14 Nicotiana somatic hybrid. Theor. Appl. Genet. 79: 28–32 [DOI] [PubMed] [Google Scholar]
- Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schöttler M.A., Bock R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk H.T., Berg S., Krupinska K., Maier U.G., Krause K. (2007). Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B.N., Ahner B.A., Hanson M.R. (2009). Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res. 18: 559–572 [DOI] [PubMed] [Google Scholar]
- Guisinger M.M., Kuehl J.V., Boore J.L., Jansen R.K. (2011). Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: Rearrangements, repeats, and codon usage. Mol. Biol. Evol. 28: 583–600 [DOI] [PubMed] [Google Scholar]
- Haberle R.C., Fourcade H.M., Boore J.L., Jansen R.K. (2008). Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 66: 350–361 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P.T.J., Allison L.A., Maliga P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K.E., Habib S., Frugier M., Hoen R., Khan S., Pham J.S., Ribas de Pouplana L., Royo M., Santos M.A., Sharma A., Ralph S.A. (2011). Protein translation in Plasmodium parasites. Trends Parasitol. 27: 467–476 [DOI] [PubMed] [Google Scholar]
- Kikuchi S., Bédard J., Hirano M., Hirabayashi Y., Oishi M., Imai M., Takase M., Ide T., Nakai M. (2013). Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Knauf U., Hachtel W. (2002). The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol. Genet. Genomics 267: 492–497 [DOI] [PubMed] [Google Scholar]
- Krause K. (2011). Piecing together the puzzle of parasitic plant plastome evolution. Planta 234: 647–656 [DOI] [PubMed] [Google Scholar]
- Krause K., Berg S., Krupinska K. (2003). Plastid transcription in the holoparasitic plant genus Cuscuta: Parallel loss of the rrn16 PEP-promoter and of the rpoA and rpoB genes coding for the plastid-encoded RNA polymerase. Planta 216: 815–823 [DOI] [PubMed] [Google Scholar]
- Kurtz S., Choudhuri J.V., Ohlebusch E., Schleiermacher C., Stoye J., Giegerich R. (2001). REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29: 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larget B., Kadane J.B., Simon D.L. (2005). A Bayesian approach to the estimation of ancestral genome arrangements. Mol. Phylogenet. Evol. 36: 214–223 [DOI] [PubMed] [Google Scholar]
- Lee H.-L., Jansen R.K., Chumley T.W., Kim K.-J. (2007). Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol. Biol. Evol. 24: 1161–1180 [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J.H., dePamphilis C.W. (2002). Power analysis of tests for loss of selective constraint in cave crayfish and nonphotosynthetic plant lineages. Mol. Biol. Evol. 19: 1292–1302 [DOI] [PubMed] [Google Scholar]
- Legen J., Kemp S., Krause K., Profanter B., Herrmann R.G., Maier R.M. (2002). Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Li X., Zhang T.-C., Qiao Q., Ren Z., Zhao J., Yonezawa T., Hasegawa M., Crabbe M.J.C., Li J., Zhong Y. (2013). Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 8: e58747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K., Link G. (1995). RNA-binding activity of the matK protein encoded by the chloroplast trnK intron from mustard (Sinapis alba). Nucleic Acids Res. 23: 917–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logacheva M.D., Schelkunov M.I., Penin A.A. (2011). Sequencing and analysis of plastid genome in mycoheterotrophic orchid Neottia nidus-avis. Genome Biol. Evol. 3: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohan A.J., Wolfe K.H. (1998). A subset of conserved tRNA genes in plastid DNA of nongreen plants. Genetics 150: 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusson N.A., Delavault P.M., Thalouarn P.A. (1998). The rbcL gene from the non-photosynthetic parasite Lathraea clandestina is not transcribed by a plastid-encoded RNA polymerase. Curr. Genet. 34: 212–215 [DOI] [PubMed] [Google Scholar]
- Maréchal A., Brisson N. (2010). Recombination and the maintenance of plant organelle genome stability. New Phytol. 186: 299–317 [DOI] [PubMed] [Google Scholar]
- Mariotti R., Cultrera N.G., Díez C.M., Baldoni L., Rubini A. (2010). Identification of new polymorphic regions and differentiation of cultivated olives (Olea europaea L.) through plastome sequence comparison. BMC Plant Biol. 10: 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J.R., Bennett J.R., Wolfe A.D., Mathews S. (2013). Phylogeny and origins of holoparasitism in Orobanchaceae. Am. J. Bot. 100: 971–983 [DOI] [PubMed] [Google Scholar]
- McNeal J.R., Kuehl J.V., Boore J.L., de Pamphilis C.W. (2007). Complete plastid genome sequences suggest strong selection for retention of photosynthetic genes in the parasitic plant genus Cuscuta. BMC Plant Biol. 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J.R., Kuehl J.V., Boore J.L., Leebens-Mack J.H., dePamphilis C.W. (2009). Parallel loss of plastid introns and their maturase in the genus Cuscuta. PLoS ONE 4: e5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J.R., Leebens-Mack J.H., Arumuganathan K., Kuehl J.V., Boore J.L., DePamphilis C.W. (2006). Using partial genomic fosmid libraries for sequencing complete organellar genomes. Biotechniques 41: 69–73 [DOI] [PubMed] [Google Scholar]
- Moore M.J., Soltis P.S., Bell C.D., Burleigh J.G., Soltis D.E. (2010). Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 107: 4623–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A.E., Kamisugi Y., Grüneberg R., Niedenhof I., Hörold R.J., Meyer P. (1999). Palindromic sequences and A+T-rich DNA elements promote illegitimate recombination in Nicotiana tabacum. J. Mol. Biol. 291: 29–46 [DOI] [PubMed] [Google Scholar]
- Müller K.F. (2005). SeqState: Primer design and sequence statistics for phylogenetic DNA datasets. Appl. Bioinformatics 4: 65–69 [DOI] [PubMed] [Google Scholar]
- Nickrent D.L., Ouyang Y., Duff R.J., dePamphilis C.W. (1997). Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol. Biol. 34: 717–729 [DOI] [PubMed] [Google Scholar]
- Ogihara Y., Terachi T., Sasakuma T. (1988). Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc. Natl. Acad. Sci. USA 85: 8573–8577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. (1994). Detecting correlated evolution on phylogenies: A general method for the comparative analysis of discrete characters. Proc. Biol. Sci. 255: 37–45 [Google Scholar]
- Pagel M., Meade A., Barker D. (2004). Bayesian estimation of ancestral character states on phylogenies. Syst. Biol. 53: 673–684 [DOI] [PubMed] [Google Scholar]
- Palmer J.D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19: 325–354 [DOI] [PubMed] [Google Scholar]
- Palmer J.D., Osorio B., Aldrich J., Thompson W.F. (1987). Chloroplast DNA evolution among legumes: Loss of a large inverted repeat occurred prior to other sequence rearrangements. Curr. Genet. 11: 275–286 [Google Scholar]
- Palmer J.D., Thompson W.F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29: 537–550 [DOI] [PubMed] [Google Scholar]
- Peltier G., Cournac L. (2002). Chlororespiration. Annu. Rev. Plant Biol. 53: 523–550 [DOI] [PubMed] [Google Scholar]
- Perry A.S., Wolfe K.H. (2002). Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J. Mol. Evol. 55: 501–508 [DOI] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. (1998). MODELTEST: Testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Randle C.P., Wolfe A.D. (2005). The evolution and expression of RBCL in holoparasitic sister-genera Harveya and Hyobanche (Orobanchaceae). Am. J. Bot. 92: 1575–1585 [DOI] [PubMed] [Google Scholar]
- Raubeson, L.A., and Jansen, R.K. (2005). Chloroplast genomes of plants. In Diversity and Evolution of Plant-Genotypic and Phenotypic Variation in Higher Plants, R.J. Henry, ed (Wallingford, UK: CABI Publishing), pp. 45–68. [Google Scholar]
- Raubeson L.A., Peery R., Chumley T.W., Dziubek C., Fourcade H.M., Boore J.L., Jansen R.K. (2007). Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics 8: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revill M.J.W., Stanley S., Hibberd J.M. (2005). Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J. Exp. Bot. 56: 2477–2486 [DOI] [PubMed] [Google Scholar]
- Rogalski M., Karcher D., Bock R. (2008). Superwobbling facilitates translation with reduced tRNA sets. Nat. Struct. Mol. Biol. 15: 192–198 [DOI] [PubMed] [Google Scholar]
- Samson N., Bausher M.G., Lee S.-B., Jansen R.K., Daniell H. (2007). The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: Organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plant Biotechnol. J. 5: 339–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S. (2011). The apicomplexan plastid and its evolution. Cell. Mol. Life Sci. 68: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Regel R., Du T.G., Hupfer H., Herrmann R.G., Maier R.M. (2002). The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: The role of RNA editing in generating divergence in the process of plant speciation. Mol. Biol. Evol. 19: 1602–1612 [DOI] [PubMed] [Google Scholar]
- Schwender J., Goffman F., Ohlrogge J.B., Shachar-Hill Y. (2004). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782 [DOI] [PubMed] [Google Scholar]
- Sears B.B., Stoike L.L., Chiu W.L. (1996). Proliferation of direct repeats near the Oenothera chloroplast DNA origin of replication. Mol. Biol. Evol. 13: 850–863 [DOI] [PubMed] [Google Scholar]
- Shinozaki K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5: 2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöver B.C., Müller K.F. (2010). TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert N.E. (1997). The C2 oxidative photosynthetic carbon cycle. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 1–25 [DOI] [PubMed] [Google Scholar]
- Westwood J.H., Yoder J.I., Timko M.P., dePamphilis C.W. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15: 227–235 [DOI] [PubMed] [Google Scholar]
- Wicke S., Schneeweiss G.M., dePamphilis C.W., Müller K.F., Quandt D. (2011). The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 76: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickett N.J., Zhang Y., Hansen S.K., Roper J.M., Kuehl J.V., Plock S.A., Wolf P.G., DePamphilis C.W., Boore J.L., Goffinet B. (2008). Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol. Biol. Evol. 25: 393–401 [DOI] [PubMed] [Google Scholar]
- Wimpee C.F., Wrobel R.L., Garvin D.K. (1991). A divergent plastid genome in Conopholis americana, an achlorophyllous parasitic plant. Plant Mol. Biol. 17: 161–166 [DOI] [PubMed] [Google Scholar]
- Wolfe A.D., dePamphilis C.W. (1997). Alternate paths of evolution for the photosynthetic gene rbcL in four nonphotosynthetic species of Orobanche. Plant Mol. Biol. 33: 965–977 [DOI] [PubMed] [Google Scholar]
- Wolfe K.H. (1994). Similarity between putative ATP-binding sites in land plant plastid ORF2280 proteins and the FtsH/CDC48 family of ATPases. Curr. Genet. 25: 379–383 [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Morden C.W., Ems S.C., Palmer J.D. (1992a). Rapid evolution of the plastid translational apparatus in a nonphotosynthetic plant: Loss or accelerated sequence evolution of tRNA and ribosomal protein genes. J. Mol. Evol. 35: 304–317 [DOI] [PubMed] [Google Scholar]
- Wolfe K.H., Morden C.W., Palmer J.D. (1992b). Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc. Natl. Acad. Sci. USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman S.K., Jansen R.K., Boore J.L. (2004). Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255 [DOI] [PubMed] [Google Scholar]
- Young N.D., dePamphilis C.W. (2005). Rate variation in parasitic plants: Correlated and uncorrelated patterns among plastid genes of different function. BMC Evol. Biol. 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Sodmergen (2010). Why does biparental plastid inheritance revive in angiosperms? J. Plant Res. 123: 201–206 [DOI] [PubMed] [Google Scholar]
- Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. (2010). An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.