This work demonstrates that plants integrate the efficiency of photosynthesis over a period of days and transduce that information into a daily rate of gibberellin synthesis. This enables a plant to match its growth rate to its environment without unnecessary short-term fluctuations.
Abstract
A plant’s eventual size depends on the integration of its genetic program with environmental cues, which vary on a daily basis. Both efficient carbon metabolism and the plant hormone gibberellin are required to guarantee optimal plant growth. Yet, little is known about the interplay between carbon metabolism and gibberellins that modulates plant growth. Here, we show that sugar starvation in Arabidopsis thaliana arising from inefficient starch metabolism at night strongly reduces the expression of ent-kaurene synthase, a key regulatory enzyme for gibberellin synthesis, the following day. Our results demonstrate that plants integrate the efficiency of photosynthesis over a period of days, which is transduced into a daily rate of gibberellin biosynthesis. This enables a plant to grow to a size that is compatible with its environment.
INTRODUCTION
Phenotypic plasticity is the capacity of an organism to change its phenotype to adapt to environmental changes (Nicotra et al., 2010). Plants are sessile organisms that cannot escape a fluctuating environment that affects metabolic processes such as photosynthesis (Wiese et al., 2007; Graf et al., 2010; Alter et al., 2012) and, consequently, their phenotypes. Seminal studies have highlighted that internal signals such as hormones also have a massive impact on plant growth and development (Yamaguchi, 2008; Davies, 2010). Plant hormone–driven growth is also of primary importance in practical applications. The introduction of stem-dwarfing traits into cereal crops (i.e., wheat [Triticum aestivum] and rice [Oryza sativa]) improved plant architecture and allowed greater allocation of resources to the grain. This was a key step in the spectacular increases in food production obtained since the 1960s and known as the Green Revolution (Hedden, 2003; Sakamoto and Matsuoka, 2004). Identification of the genes responsible for dwarfism in cereal crops showed that most genes relate to regulating the physiology of gibberellins (GAs), a class of plant hormones (Peng et al., 1999; Hedden, 2003).
GA biosynthesis is mostly controlled by a homeostatic mechanism based on the negative feedback regulation of GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), both involved in the synthesis of bioactive GAs, and activation of the GA 2-oxidases, which convert bioactive GAs to inactive forms (Yamaguchi, 2008). Arabidopsis thaliana mutants that are defective in either GA synthesis or signaling are dwarf (Yamaguchi, 2008; Hedden and Thomas, 2012), whereas overexpression of the GA biosynthesis enzyme GA20ox results in increased leaf size (Gonzalez et al., 2010), indicating that this trait is strongly influenced by GAs.
Besides plant hormones, such as GAs, growth inevitably depends on the efficiency of the plant’s primary metabolism. Plants are exposed to the alternation between night and day and variable light levels. During the day, photosynthesis provides sugars that are partitioned among growth and metabolic processes, and storage as transitory starch in the leaves. During the night, starch is degraded through a circadian clock–regulated pathway (Graf et al., 2010) to fuel continued growth (Wiese et al., 2007). The synthesis of transitory starch is of great importance for growth and Arabidopsis mutants defective in either starch synthesis or degradation are dwarf (Caspar et al., 1985, 1991; Lin et al., 1988a, 1988b; Yu et al., 2001; Schneider et al., 2002; Niittylä et al., 2004; Stitt and Zeeman, 2012).
Nutrient sensing and hormonal signaling are interconnected in plants (Krouk et al., 2011), but little is known about whether or how the growth-promoting roles of photosynthesis and plant hormones are integrated. Exposure to elevated carbon dioxide (CO2) enhances growth and results in a significant increase in sugar and starch content in Arabidopsis leaves (Teng et al., 2006). Remarkably, exposure to higher CO2 levels also increases the GA content, suggesting a link between the effects of higher CO2 on photosynthesis, starch, and the production of GAs (Teng et al., 2006). However, this view was recently challenged by evidence suggesting that growth induced by exposure to elevated CO2 is at least partly uncoupled from the effect of GAs (Ribeiro et al., 2012).
Here, we show that carbohydrates produced by photosynthesis modulate the synthesis of GAs and that it is also through this mechanism that plant size is determined. We found that in order to modulate hormone-driven growth, plants exploit the metabolism of starch at night as a reliable measure of how efficient photosynthesis was the previous day. The synthesis of GAs and growth was shown to be dampened when sugar starvation resulted from inefficient metabolism of starch at night. Our results demonstrate that plants integrate the efficiency of photosynthesis over a period of days and transduce that information into a daily rate of GA synthesis. This enables a plant to grow to a size that is compatible with the environment where it lives.
RESULTS
The Dwarf Phenotype of Starch Mutants Can Be Reverted by Exogenous GAs
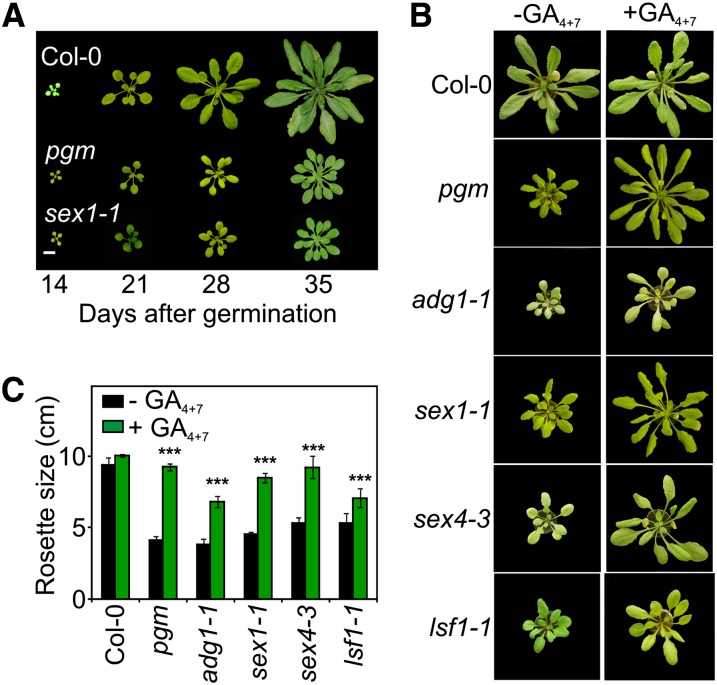
Arabidopsis growth during the day mostly relies on sugars produced by photosynthesis, while growth at night is fueled by the mobilization of transitory leaf starch (Wiese et al., 2007; Graf et al., 2010). Arabidopsis mutants (Caspar et al., 1985, 1991; Lin et al., 1988a, 1988b; Yu et al., 2001; Stitt and Zeeman, 2012) defective in either starch synthesis (phosphoglucomutase [pgm] and ADP glucose phosphorylase1 [adg1-1]) or degradation (starch excess1 [sex1-1], starch excess4 [sex4-3], like sex four [lsf1-1]) are dwarf, and flowering is delayed (Figures 1A and 1B; see Supplemental Figures 1A to 1D and Supplemental Table 1 online). Similar traits have also been observed in mutants defective in GA synthesis or signaling (Wilson et al., 1992; Graf et al., 2010; Hedden and Thomas, 2012). This prompted us to test whether the dwarfism of starch mutants could be reverted by exogenously applying GAs. Unexpectedly, GA-treated starch mutants responded by growing to a size that, in several cases, was close to the wild type (Figures 1B and 1C). Exogenous GAs supplied to pgm and sex1-1 increased fresh weight, but not dry weight (see Supplemental Figures 1E and 1F online), indicating that GAs boosted leaf elongation alone, without rescuing the defective carbon metabolism in the mutants. This conclusion was supported by the measurement of photosynthetic rate, which was unaffected by the GA treatment (see Supplemental Figure 2 online).
Figure 1.
GAs Rescue the Dwarf Phenotype of Starch Mutants to a Large Extent.
(A) Representative growth phenotypes of wild-type (Col-0), pgm, and sex1-1 plants from 14 to 35 d after germination. Bar = 1.5 cm.
(B) Effect of GA4+7 application on growth of wild-type (Col-0), pgm, adg1-1, sex1-1, sex4-3, and lsf1-1 plants.
(C) Rosette size of Col-0 and mutants in absence (−GA4+7) or presence (+GA4+7) of 10 µM GA4+7. Data are mean values of 10 replicates ± sd (two-way analysis of variance; ***P < 0.001).
We then wanted to know whether leaf expansion boosted by exogenous GAs (Figures 1B and 1C) had an impact on carbon allocation, and we found that GA treatment had a negligible effect on the wild type (see Supplemental Figure 3A online). However, in pgm, GAs significantly reduced Glc and Suc levels (see Supplemental Figure 3A online), suggesting a higher use of these carbohydrates to fuel GA-induced leaf expansion during the day (Wiese et al., 2007). In sex1-1, GA treatment caused a reduction in the level of starch that was only slightly compensated for by an increase in sucrose and fructose (see Supplemental Figure 3A online), indicating a diversion of photosynthates to fuel GA-dependent growth and not starch synthesis. We found that the GA effect on carbohydrate partitioning enhanced the expression of four starvation-related genes (see Supplemental Figure 3B online; Gonzali et al., 2006; Usadel et al., 2008). This thus indicates that both pgm and sex1-1 suffered from severe starvation when treated with GAs, as a result of the increased growth rates that apparently exceeded the amount of carbohydrates available.
GA Synthesis Is Dampened in Mutants Defective in Starch Synthesis or Degradation
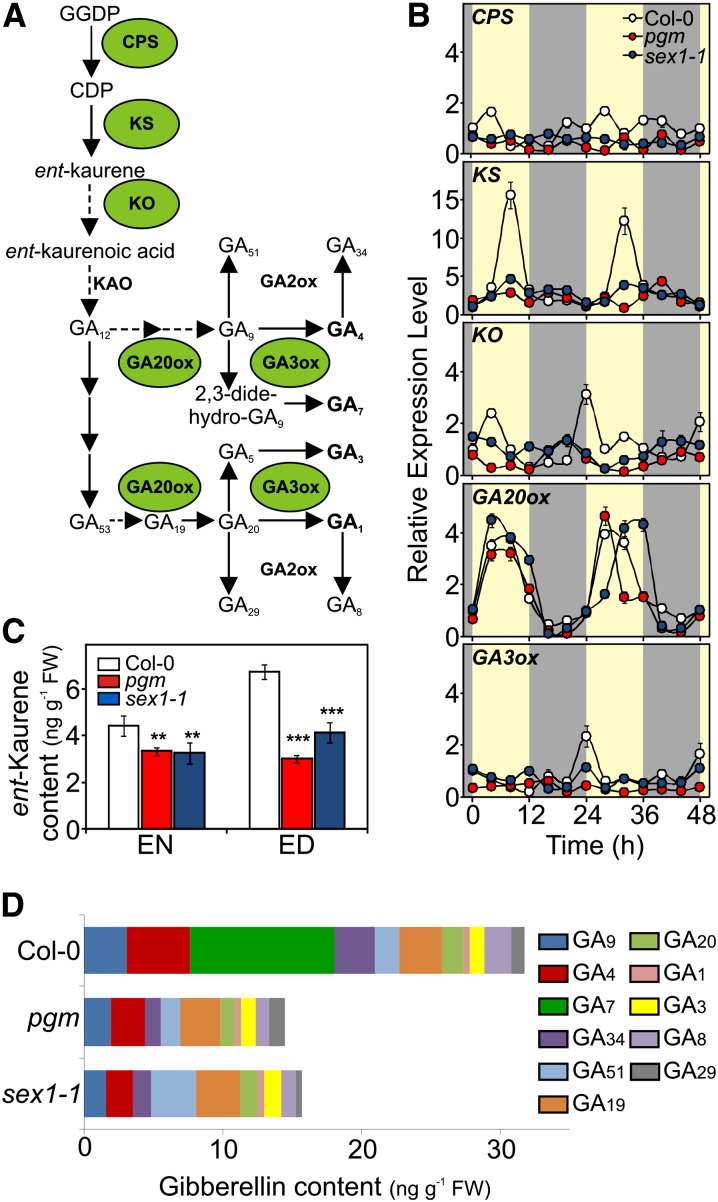
The ability of GAs to rescue the dwarfism of starch mutants suggested that they may be defective in the synthesis of these hormones, a hypothesis that was confirmed by the experiments described here. GAs are synthesized from geranylgeranyl diphosphate via a multistep process (Figure 2A) (Olszewski et al., 2002; Yamaguchi, 2006; Graf et al., 2010; Hedden and Thomas, 2012) regulated by both developmental and environmental stimuli (Hedden and Thomas, 2012). Reductions in the expression of copalyl diphosphate synthase and ent-kaurene oxidase were observed in pgm and sex1-1, while the diurnal expression of GA20ox1 (At4g25420), which is under circadian control (Hisamatsu et al., 2005), was unaffected. GA3ox1 (At1g15550) expression was also largely unaffected (Figure 2B). The expression level of other GA20ox and GA3ox genes was not significantly changed compared with the wild type (see Supplemental Figures 4A and 4B online).
Figure 2.
Alterations in Starch Metabolism Affect GA Biosynthesis.
(A) Pathways of GA synthesis in Arabidopsis. The enzymes involved are ent-copalyl diphosphate (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA20-oxidase (GA20ox), GA3-oxidase (GA3ox), and GA2-oxidase (GA2ox).
(B) Diurnal changes in the relative expression levels of genes involved in the GA biosynthetic pathway in Col-0, pgm, and sex1-1 plants grown under a 12-h-light/12-h-dark photoperiod. Expression levels are expressed as relative units assuming as unitary the value of the wild type (Col-0) at the beginning of the day (0 h). Yellow background, day; gray background, night.
(C) Endogenous level of ent-kaurene (ng g−1 fresh weight [FW]) in Col-0, pgm, and sex1-1 at the end of night (8 am) and day (8 pm). Asterisks indicate significant differences from the wild type (two-way analysis of variance; **P < 0.01 and ***P < 0.001).
(D) Level of GAs (ng g−1 fresh weight) in Col-0, pgm, and sex1-1 at the end of day (8 pm). Values are means (±sd) of three independent replicates.
The expression of ent-kaurene synthase (KS) during the diurnal cycle had a distinct pattern, which peaked in the afternoon. Strikingly, this peak was absent in both pgm and sex1-1 (Figure 2B). The increased expression of KS in the afternoon correlates well with higher levels of ent-kaurene in the wild type at the end of the day (Figure 2C), and, remarkably, the level of GAs also was higher at the end of the day (see Supplemental Figure 5 and Supplemental Table 2 online). Lower kaurene content, on the other hand, was detected in pgm and sex1-1, which was consistent with their lower KS expression (Figure 2C). Profiling of GA levels revealed a reduced metabolic flow to biologically active GAs (Yamaguchi, 2008; Hedden and Thomas, 2012), especially GA4 and GA7, in both pgm and sex1-1 (Figure 2D; see Supplemental Table 3 online). Whereas the GA4 level significantly increased at the end of day in the wild type (see Supplemental Figures 5 and 6 online), this was not the case in pgm and sex1-1 (see Supplemental Figure 6 online).
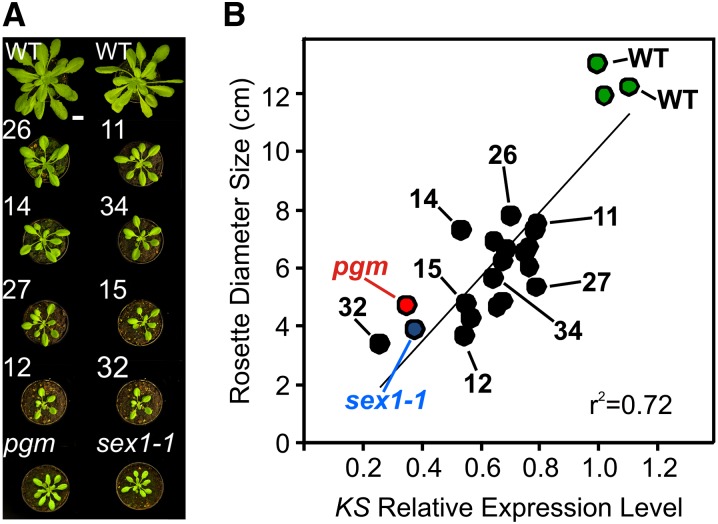
Fleet et al. (2003) showed that plants are able to maintain GA homeostasis and normal morphology even when KS expression is greatly increased. In order to verify if a decreased expression of KS similar to that observed in starch mutants would result in reduced plant size, KS RNA interference (RNAi) lines (hereafter, KSi) were studied. All of the KSi lines showed a dwarf phenotype, in some cases similar to that of pgm and sex1-1 (Figure 3A; see Supplemental Figure 7A online). A significant reduction in KS expression was observed in the KSi plants (see Supplemental Figure 7B online), correlating with the rosette diameter (Figure 3B), thus indicating that decreased KS expression to the level observed in pgm and sex1-1 results in a comparable reduction in rosette size.
Figure 3.
Reduction in KS Expression Level in RNAi Knockdown Lines Results in Decreased Rosette Diameter.
(A) Representative growth phenotypes of 4-week-old Col-0 (WT) and selected KSi lines and pgm and sex1-1 plants. Bar = 1.5 cm.
(B) Correlation between the rosette diameter (y axis) and KS expression levels (x axis) of wild-type Col-0 (green symbols), KSi knockdown lines (closed symbols), pgm (red symbol), and sex1-1 (blue symbol) mutants. For pictures and transcript levels of the complete set of plants, see Supplemental Figure 7 online.
Sugar Starvation at Night Influences KS Expression and Growth
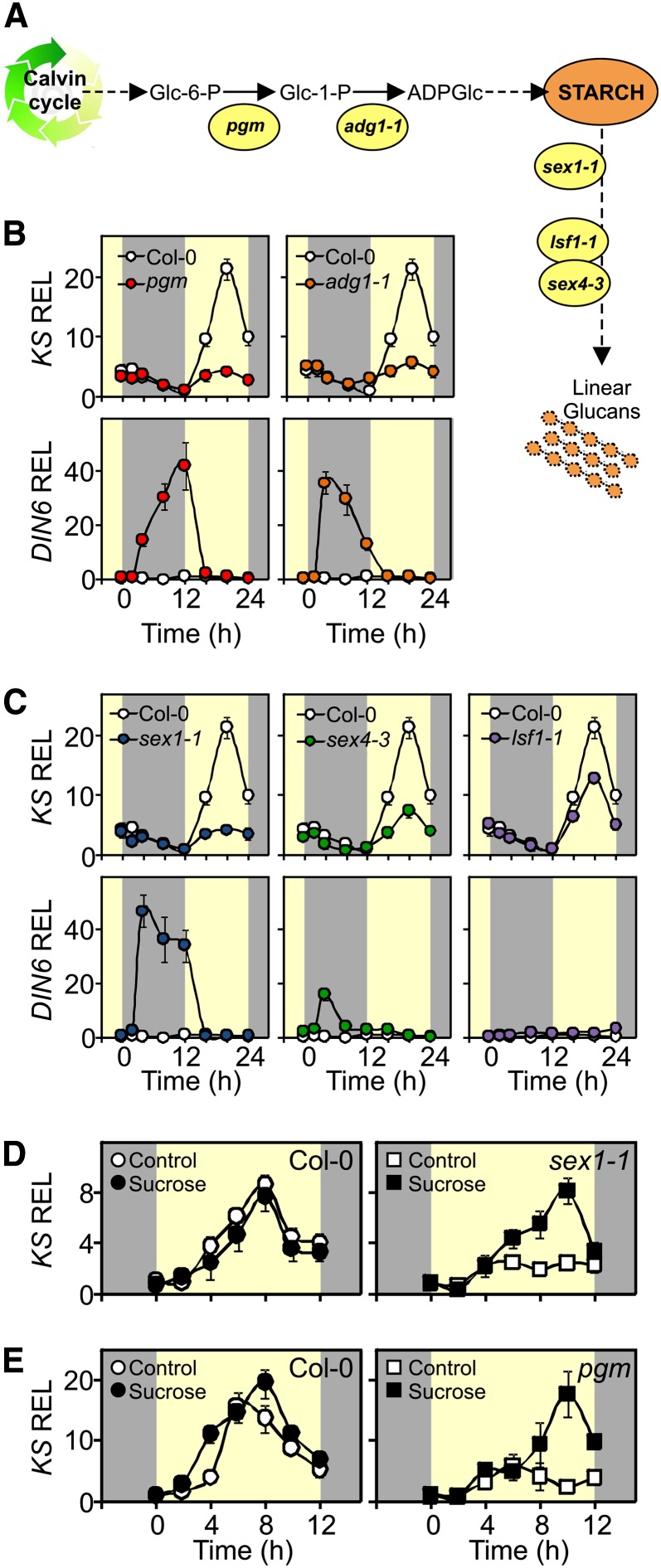
Our next question was whether the lower GA level observed in starch mutants was related to the altered nighttime carbohydrate metabolism, which has been shown to be typical for these mutants (Streb and Zeeman, 2012). The starchless mutants pgm and adg1-1 (Figure 4A) showed expression of the typical marker for sugar starvation (dark inducible6 [DIN6]; Gonzali et al., 2006; Usadel et al., 2008) during the night (Figure 4B) and negligible expression of KS the following day. Sugar starvation at night was confirmed by other starvation-induced markers, such as At1g76410 and trehalose 6-P synthase (see Supplemental Figures 8A and 8B online). Starch degradation downstream of SEX1 involves SEX4 and LSF1 (Figure 4A). Both sex1-1 and sex4-3 show reduced sugar content at night (Caspar et al., 1991; Zeeman and ap Rees, 1999), induction of the sugar starvation marker at night, and reduced KS expression the next day (Figure 4C). In lsf1-1, which has the least severe starch-excess phenotype of the mutants studied (Comparot-Moss et al., 2010), KS expression was the closest to the wild-type expression (Figure 4C). In order to gain direct evidence linking night starvation to the regulation of KS, we sprayed Suc on the leaves of pgm and sex1-1 during the night. Interestingly, the Suc treatment restored the peak of KS expression the following day (Figures 4D and 4E). The same result was obtained using the adg1-1 mutant (see Supplemental Figure 9 online). This further supports the idea that sugar starvation at night triggers the downregulation of KS during the following light period.
Figure 4.
Relationship between Night Starvation and Expression of KS during the Following Light Period.
(A) Pathway of starch synthesis and degradation in Arabidopsis leaves. The enzymatic steps affected by each mutation are indicated.
(B) and (C) Comparisons of the expression of KS and the starvation reporter gene (DIN6) in Col-0 and starch mutants analyzed in a 12-h-light/12-h-dark photoperiod (gray/yellow background corresponds to night/day). The relative expression level (REL) at the end of night of the wild type (Col-0) was set to 1.
(D) and (E) Transcript levels of KS in Col-0, sex1-1 (D), and pgm (E) mutants treated with 1% Suc starting 13 h before dawn (closed symbols). Control plants (open symbols) were sprayed with water. The relative expression level at the end of night of the wild type (Col-0) was set to 1.
Values are means (±sd) of three replicates from a single experiment. An independent experiment gave comparable results.
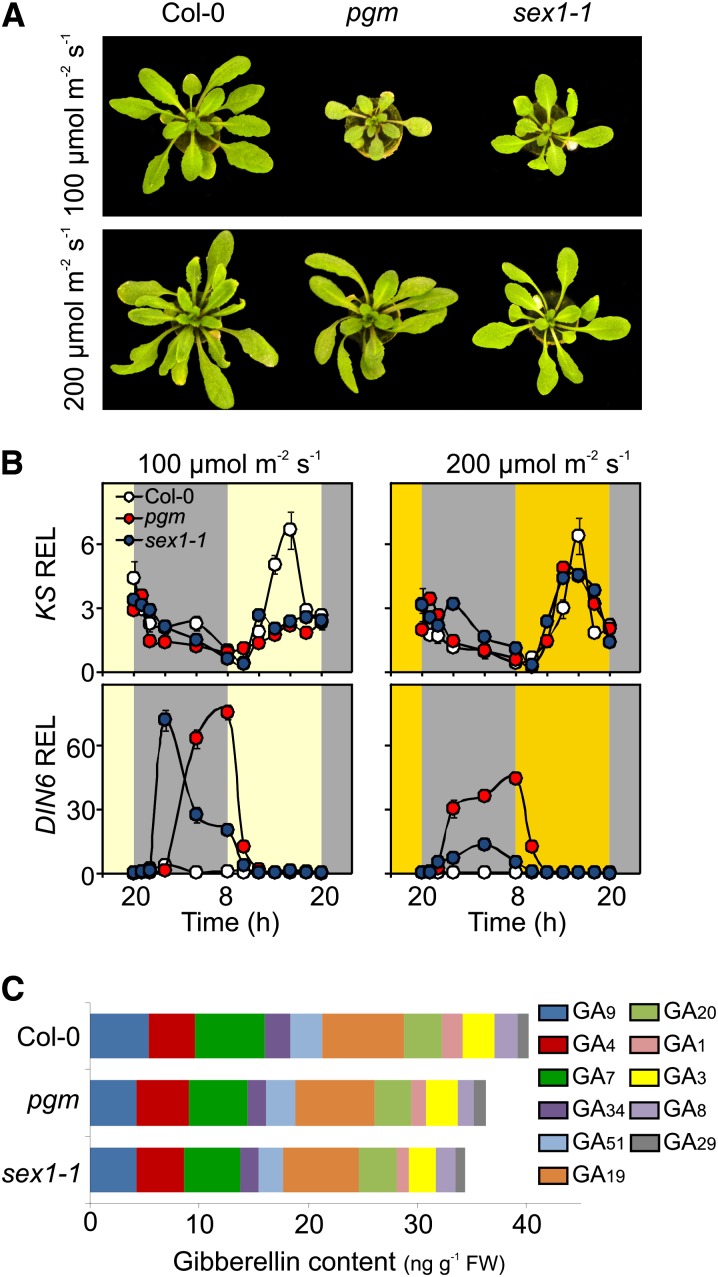
When grown at twice the normal light intensity, both pgm and sex1-1 reached a size close to that of the wild type (Figure 5A). Although the amount of carbohydrates was unchanged in pgm and sex1-1 (see Supplemental Figure 10A online), the level of starvation was lower in these plants when grown at 200 μmol m2 s−1 (Figure 5B; see Supplemental Figure 10B online), and the afternoon peak of KS expression was present (Figure 5B). Since we can exclude that the metabolism of carbohydrates at night can be improved in pgm and sex1-1 by growing these mutants at higher light intensity, it is tempting to speculate that autophagy at night is responsible for the lower starvation at night observed in plants grown at 200 μmol m2 s−1. Indeed, autophagy ameliorates the night energy status of pgm (Izumi et al., 2013). Consistent with the reduced dwarfism in pgm and sex1-1 plants grown at 200 μmol m2 s−1 (Figure 5A), no significant differences were found in the GA content of wild-type, pgm, and sex1-1 plants at the end of the day (Figure 5C), and only a moderately lower GA level was detected in pgm and sex1-1 at the end of the night (see Supplemental Figures 11A and 11B online).
Figure 5.
Higher Light Intensity Reduces Starvation Symptoms at Night and Restores KS Expression.
(A) Phenotype of Col-0, pgm, and sex1-1 plants at 100 or 200 µmol m−2 s−1 light intensity.
(B) Transcript levels of KS and DIN6 in Col-0, pgm, and sex1-1 plants at 100 or 200 µmol m−2 s−1 light intensity. The relative expression level (REL) at the end of night (hour 8) of the wild type (Col-0) was set to 1.
(C) Level of GAs (ng g−1 fresh weight [FW]) in Col-0, pgm, and sex1-1 grown at 200 µmol m−2 s−1 light intensity at the end of day (8 pm).
Values are means (±sd) of three independent replicates.
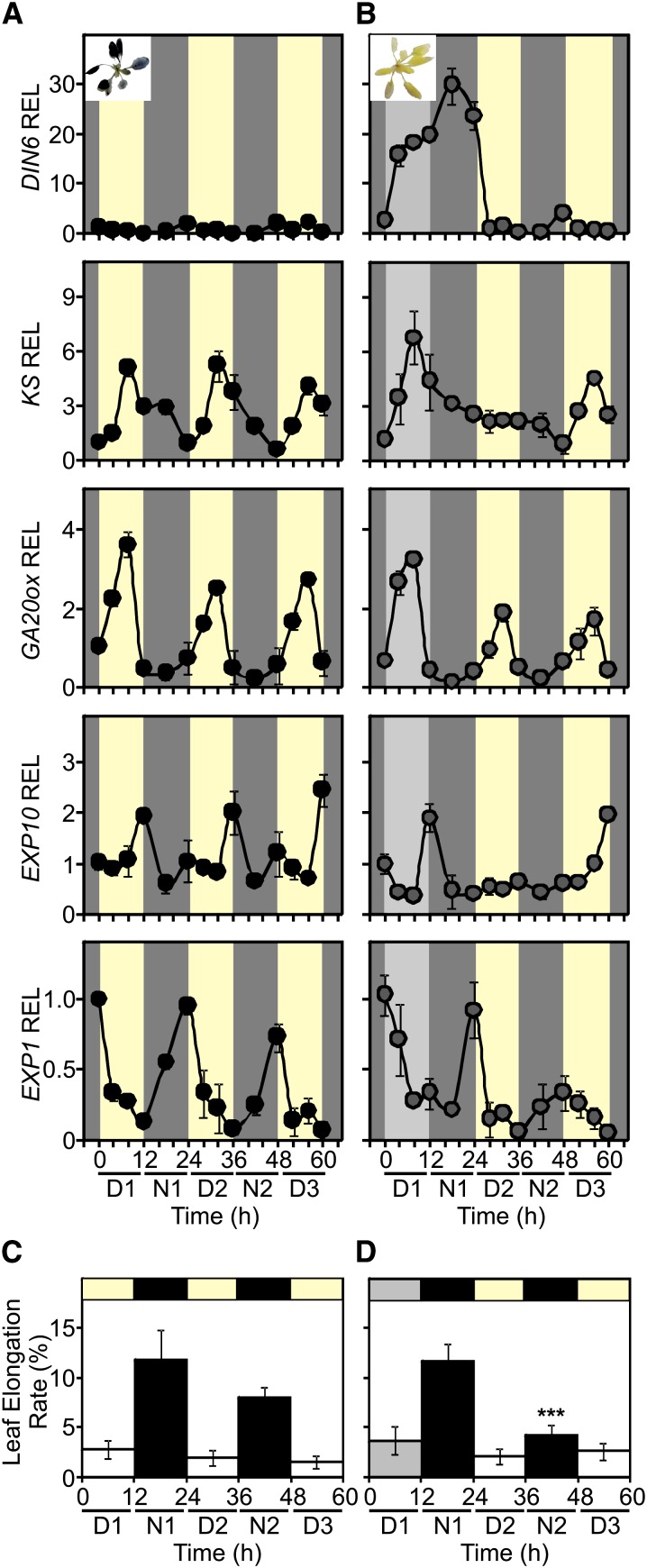
So, was the altered KS expression pattern unique to starch mutants, or might it also be observed in wild-type plants experiencing night starvation? To examine this, we kept a batch of wild-type plants under normal day/night conditions (Figure 6A), while a second batch of plants was exposed to low light for 1 d, followed by two additional days under normal day/night conditions (Figure 6B). The low light treatment resulted in low starch content at the end of the first day, together with high expression of the sugar starvation marker (Figure 6B; see Supplemental Figures 12A and 12B online). Over the next 2 d, starch synthesis resumed and the expression of the marker for sugar starvation was consequently low (Figure 6B). In line with what we observed in starch mutants, while KS expression was unaffected during the day under low light, the afternoon peak of KS expression was lost during the second day but resumed the third day (Figure 6B). The expression pattern of GA20ox, on the other hand, was unaffected by the low light over the 3 d (Figure 6B), which is in agreement with the absence of GA20ox downregulation in starch mutants (Figure 2B). Remarkably, leaf elongation at night (Figures 6C and 6D) was reduced during the second night in low-light-treated plants. Collectively, the results indicate that (1) low starch synthesis during the day was unable to modulate KS expression on the same day, (2) abnormal starch metabolism during the night, leading to sugar starvation, affected the expression of KS the following day, and (3) only after one night with normal starch metabolism was the KS afternoon peak of expression restored.
Figure 6.
KS Expression Is Adjusted to Unexpected Changes in Light Intensity during the Previous Day.
(A) Col-0 plants grown in a 12-h-light/12-h-dark photoperiod at 100 µmol m−2 s−1 light intensity were used as controls. D, day; N, night.
(B) Another group of plants was subjected to a 12-h treatment at low-light intensity (10 µmol m−2 s−1; light-gray background) and then transferred to normal light intensity (yellow background). DIN6, KS, GA20ox, EXP10, and EXP1 expression analysis over 3 d was performed. Expression in the control set of plants (A) at time 0 was set to 1. Pictures show Lugol’s staining of a representative plant at the end of the first day.
(C) and (D) Leaf elongation under the experimental conditions described in (A) and (B), respectively.
Values are means (±sd) of three replicates from two independent experiments.
The next step was to explore whether the low expression of KS the day after a night starvation event would affect the expression of GA-regulated genes. We found that EXP10, an expansin that modulates leaf growth (Cho and Cosgrove, 2000), was upregulated by GAs (see Supplemental Figure 13A online) and displayed a diurnal expression peak at the end of the day (Figure 6A). Although EXP10 was not expressed when the KS expression peak was abolished following a night starvation event (Figure 6B), interestingly it was restored the next day. Similar behavior was observed for EXP1, a GA-induced expansin (see Supplemental Figure 13B online) expressed in stomatal guard cells (Zhang et al., 2011), whose expression rose rapidly each night (Figure 6A). In plants that were exposed to low light for just 1 d, EXP1 expression was severely dampened the night immediately following the period when there had been no KS expression peak (Figure 6B). This indicates that the consequences of low-light treatment are delayed in terms of their impact on KS expression, as was to be expected.
DISCUSSION
Plant growth depends on sugars produced by photosynthesis. These sugars provide cells with energy through respiration along with the metabolic intermediates used to build up biomass. Mineral nutrients are also required to sustain growth (Sinclair, 1992), providing structural elements for macromolecule synthesis, such as proteins and nucleic acids. Both sugars and mineral nutrients also act as signaling molecules that modulate plant development (Rolland et al., 2006; Schachtman and Shin, 2007). In addition, plant growth is driven by hormones and, regardless of the efficiency of photosynthesis and mineral nutrition, plants that are deficient in growth-promoting hormones show dwarfism or other severe developmental defects (Davies, 2010). However, how the plant integrates signaling from carbohydrates and nutrients with the hormonal regulation of plant growth is still largely unknown.
The amount of starch a plant can accumulate in the leaves is a direct consequence of how efficient photosynthesis is in the overall growth environment. Our results indicate that nighttime sugar starvation, arising from defective starch metabolism, orchestrates GA metabolism in plants to ensure that growth-related processes proceed with an adequate carbon supply. Our experiments revealed the existence of a clear diurnal regulation of KS (Figure 2B), resulting in enhanced ent-kaurene and GA synthesis at the end of the day (Figures 2C and 2D; see Supplemental Figures 5 and 6 online). In line with this, the rate of auxin biosynthesis at the end of the day has also been shown to be twice that at the beginning of the day (Sairanen et al., 2012). Sugars upregulate auxin biosynthesis (Sairanen et al., 2012), indicating that the synthesis of growth hormones depends on the availability of sugars in order to be able to coordinate growth. Attempting hormone-driven growth at a rate exceeding carbon availability is maladaptive and results in severe starvation symptoms (see Supplemental Figure 3B online). It is thus tempting to speculate that plants reset hormone-driven growth to a pace that is compatible with their carbon resources when photosynthesis is unable to meet the requirements for a normal growth rate.
A low amount of starch at night or the inability to use it results in reduced GA synthesis (Figure 2). The signaling event(s) modulating the synthesis of GAs originate during the previous night and are based on the level of starvation experienced by the plant. The fact that KS expression inversely correlates with the expression of starvation-induced genes but not with sugar content itself indicates that the signaling mechanism regulating KS expression relies on the overall nighttime carbon starvation, which is the result of both starch availability and autophagy, as recently demonstrated (Izumi et al., 2013). This starvation-dependent repression of GA-triggered growth is in line with the model proposed by Stitt and Zeeman (2012) for the coordination of starch breakdown and growth at night, extending the mechanisms linking starch to growth to hormonal regulation. Furthermore, Bläsing et al. (2005) highlighted the importance of responses to sugar starvation rather than to high levels in diurnal gene regulation in the light. Lunn et al. (2006) demonstrated that a greater reserve of starch is accumulated when its supply was insufficient during the previous night, in agreement with the idea of a day-by-day adjustment of metabolism to avoid an imbalance between starch availability and its use to fuel growth. If at night starch is unable to meet the requirements for growth, such as in genotypes that are defective in starch metabolism (Figure 4) or after a day under low light (Figure 6), sugar starvation can be severe and the daily peak of KS expression is abolished, thus reducing the GA level and leaf elongation.
The availability and daily fluctuation in sugar levels are likely to contribute to the array of signals required to ensure the rhythmic growth of plants (Todd et al., 2008). The circadian clock controls starch degradation (Graf et al., 2010) and regulates the transcription of GA receptors, resulting in a higher stability of DELLA proteins during the day and a higher GA sensitivity at night (Arana et al., 2011). This, together with higher GA levels at the end of the day (see Supplemental Figure 5 online) guarantees that growth takes place predominantly at night (peaking at dusk) (Wiese et al., 2007; Todd et al., 2008). At night, carbohydrates are available only if an adequate level of transitory starch was produced the previous day. Indeed, the growth phase of the starch-free1 mutant (which is basically equivalent to pgm) was shifted compared with the wild type, with reduced nocturnal and increased afternoon growth activity (Wiese et al., 2007). This indicates that sugar availability, which is very high in the afternoon but very low at night in lsf1-1 and pgm, overrides the hormonal-dependent temporal growth pattern. We explain this by the starvation-dependent reduction of GA synthesis in pgm (Figure 2D), which prevents an otherwise prevailing growth-promoting period at night.
The evidence that sugar availability affects both auxin (Sairanen et al., 2012) and GA biosynthesis (as outlined in this work) suggests that carbon allocation is key to ensuring a growth pattern that is suited to the plant’s environment. This also implies the need to ensure an adequate balance between root and shoot growth. Nutrients such as nitrate and phosphate influence root architecture. Nitrate induces root branching by modifying auxin distribution (Krouk et al., 2010), which is likely to result in the allocation of more sugars to the root system. Likewise, plants that suffer phosphorus starvation respond by simultaneously reducing shoot growth, through a reduction in bioactive GA level, and increasing root proliferation (Jiang et al., 2007). Further work is required to examine the extent of the regulatory network connecting shoot/root growth and fluctuating environmental conditions through a carbohydrate-dependent signaling pathway that modulates hormonal responses.
We have provided evidence for a model that describes plant growth (Figure 7) based on the efficiency of starch metabolism at night, taken as an integrator of photosynthesis activity from the previous day. A low rate of starch metabolism at night signals unfavorable growth conditions to which the next day’s GA syntheses adapt. This enables diurnal changes in carbon availability at night and growth responses to be integrated, over several days, in order to reach the most suitable growth rate for the environment inhabited by the plant, without excessive sensitivity to short-term environmental fluctuations.
Figure 7.
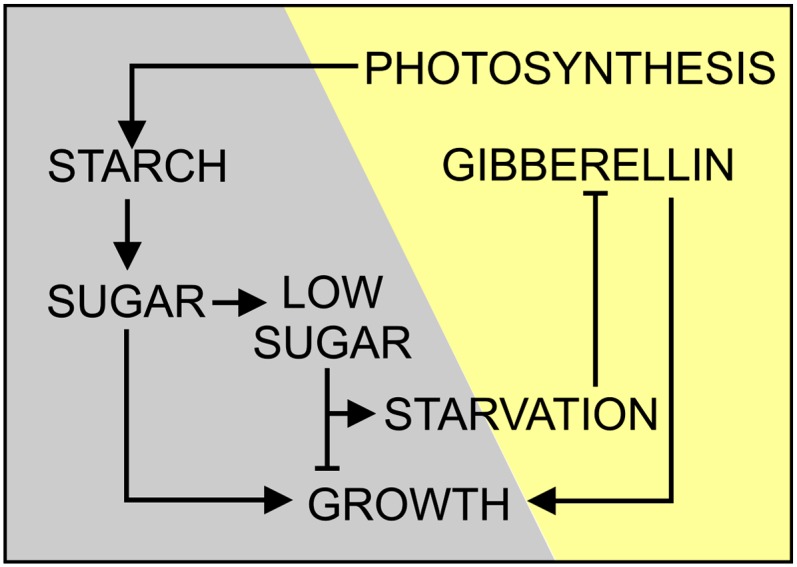
Model Depicting the Regulation of Growth by Sugar Starvation and GAs.
Growth at night requires both the presence of transitory starch and the synthesis of GAs. Should photosynthesis be unable to provide enough starch to support growth at night, a starvation signal will be generated that downregulates the synthesis of GAs the next day to balance the hormone-driven growth with the lower carbon availability.
METHODS
Plant Material
Experiments were performed using Arabidopsis thaliana accession Columbia-0 (Col-0) and its mutants, pgm (Caspar et al., 1985), sex1-1 (Caspar et al., 1991; Yu et al., 2001; Ritte et al., 2002), adg1-1 (Lin et al., 1988a, 1988b; Wang et al., 1998), sex4-3 (Zeeman and ap Rees, 1999; Kötting et al., 2009), and lsf1-1 (Comparot-Moss et al., 2010). Col-0 (N1093) seeds and seeds of the mutated lines pgm (N210), adg1-1 (N3094), and sex1-1 (N3093) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk/home.html). The other mutants were kindly provided by Martin Steup (Institute of Biochemistry and Biology, University of Potsdam, Germany). In most experiments, plants were grown using a hydroponic system (Gibeaut et al., 1997). Seeds were stratified at 4°C in the dark for 48 h and germinated at 22°C day/18°C night with a 12-h photoperiod. The quantum irradiance was 100 µmol photons m−2 s−1, unless otherwise stated. Measurements of the size of rosettes and leaf elongation were taken using a caliber measure tool. Leaf (blade and petiole) elongation rate (%) was calculated by measuring the same leaves (intermediate length) at 8 am and 8 pm. The percentage of elongation rate is calculated by setting at 100 the starting length (e.g., 8 am) and subtracting the length at the end point (e.g., at 8 pm; expressed as a percentage of that of the starting point). A total of 55 leaves were monitored for each of the two experimental conditions described in Figures 6C and 6D.
Total RNA Extraction and Quantitative PCR
RNA extraction, removal of genomic DNA, cDNA synthesis, and quantitative RT-PCR analyses were performed as described previously (Paparelli et al., 2012). GAPDH and 40SrRNA were used as reference genes. Three replicates for each experiment were used, and the average expression value was calculated. For a list of the primers used and designed using QuantPrime (http://quantprime.mpimp-golm.mpg.de/; Arvidsson et al., 2008), see Supplemental Table 4 online.
Application of Exogenous GA
The effect of GAs on starch mutants and Col-0 plants was tested by applying 5 μL of a solution of 10 µM GA4+7 onto the shoot apex. The GA4+7 was dissolved with drops of ethanol in distilled water. Control plants were treated with an equal volume of ethanol. The treatment was started 3 weeks after germination and repeated every 3 d. After 2 weeks of treatment, rosette size was measured and transcript levels were analyzed.
Screening of KSi Transgenic Lines
RNAi knockdown mutants using the pAGRIKOLA system (Hilson et al., 2004) were used. Col-0 transformants with the pAGRIKOLA construct for At1g79460 (KS) were obtained from the Nottingham Arabidopsis Stock Centre (N291353 or CATMA1a68545). This set of seeds is a mix of wild-type Col-0, heterozygotes, and homozygotes for the RNAi construct. To select plants that contain the RNAi construct, the specific gene sequence tag fragment (324 bp) was amplified by PCR on genomic DNA with primers for pAGRIKOLA (for a list of the primers used, see Supplemental Table 5 online). The KS expression in the wild type and in the positive transgenic lines was measured by quantitative RT-PCR analyses, collecting samples at the time of KS afternoon expression peak.
Quantitative Analysis of Endogenous GA
Approximately 3 to 5 g of 30-d-old Col-0, pgm, and sex1-1 rosettes, collected at the end of the light phase, were extracted and purified as previously described (Mariotti et al., 2011). The material was homogenized in cold 80% (v/v) methanol using a mortar and pestle. Fifty nanograms of deuterated GAs ([17,17-2H2]-GA9, [17,17-2H2]-GA4, [17,17-2H2]-GA34, [17,17-2H2]-GA7, [17,17-2H2]-GA19, [17,17-2H2]-GA20, [17,17-2H2]-GA29, [17,17-2H2]-GA1, [17,17-2H2]-GA8, [17,17-2H2]-GA3, and [17,17-2H2]-GA5) were added as internal standards to account for purification losses. Methanol was evaporated under a vacuum at 35°C, and the aqueous phase was partitioned against ethyl acetate, after adjusting the pH to 2.8. The extracts were dried and resuspended in 0.3 to 0.5 mL of distilled water with 0.01% acetic acid and 10% methanol. HPLC analysis was performed with a Kontron instrument equipped with a UV absorbance detector operating at 214 nm, while the gas chromatography–tandem mass spectrometry analysis was performed on a Saturn 2200 quadruple ion trap mass spectrometer coupled to a CP-3800 gas chromatograph (Varian Analytical Instruments) equipped with a MEGA snc (http://www.mega.mi.it) 1MS capillary column (30 m × 0.25-mm i.d. and 0.25-µm film thickness) as described (Fambrini et al., 2011). GAs were identified by comparing the full mass spectra with those of the authentic compounds. Quantification was performed with reference to standard plots of concentration ratios versus ion ratios, obtained by analyzing known mixtures of unlabeled and labeled GAs.
Analysis of Endogenous ent-Kaurene
Approximately 3 to 5 g of 30-d-old plants, collected at the end of the light and dark phases, were homogenized in cold 80% (v/v) methanol. One hundred nanograms of deuterated kaurene ([17,17-2H2]-kaurene) was added as an internal standard. Methanol was evaporated under N2 flow, and the aqueous phase was partitioned against hexane. The extracts were dried under N2 and resuspended in 0.3 to 0.5 mL of water with 0.01% (v/v) acetic acid and 90% (v/v) acetonitrile. HPLC analysis was performed as described above. The fraction corresponding to the elution volume of standard kaurene was analyzed with gas chromatography–tandem mass spectrometry as previously outlined (Fambrini et al., 2011).
Extraction and Analysis of Carbohydrates
Four-week-old rosettes with a fresh weight of 300 to 500 mg were harvested rapidly into liquid N2. Frozen samples were homogenized then extracted in perchloric acid, as previously described (Paparelli et al., 2012). Glc, Suc, Fru, and starch were measured enzymatically on the neutralized supernatant (soluble sugars) (Guglielminetti et al., 1995) and the insoluble pellet (starch) (Critchley et al., 2001).
Qualitative Analysis of Leaf Starch Content by Lugol Staining
Rosettes of individual plants were harvested at the end of the light period and boiled in 50 mL 80% (v/v) ethanol. Decolored plants were stained with a fresh iodine solution (I2/KI [5 g KI and 0.5 g I2 in 500 mL of distilled water]) for 5 min, destained in water for 1 to 2 h, and photographed immediately.
Photosynthetic Gas Exchange Measurement
Gas exchange of whole rosettes was measured using a custom-built multichamber system connected in parallel to an infrared gas analyzer (Licor7000). Individual plants were introduced into each of the eight chambers, and after an adaptation period of 2 d, the gas exchange was measured for a complete 12-h-light/12-h-dark cycle. During the measurement, air with 380 ppm CO2 and a relative humidity of 65% was channeled through the system with a flow rate of 200 µmol s−1. Four plants treated with GAs and four controls (each of the genotypes used, without GA treatment) were measured in each run. Each chamber was measured consecutively for 6 min, and the average value was taken for the light and dark periods. At the end of the light period, a photograph was taken to calculate the projected leaf area of each plant. Based on the projected leaf area, photosynthetic and respiration rates were calculated with the ΔCO2 and Δwater values gained from the gas exchange system.
Treatment with Suc
The effect of Suc on sex1-1 and Col-0 30-d-old plants was tested by spraying a solution of 1% Suc (dissolved in water) directly on leaves twice (at 7 pm and at midnight) during the night before the sampling. Control plants were treated with an equal volume of water. Sampling was done 13 h after the beginning of the Suc treatment, starting at dawn, and every 2 h.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At4g02780 (copalyl diphosphate synthase), At1g79460 (KS), At5g25900 (ent-kaurene oxidase), At1g15550 (GA3ox1), At1g80340 (GA3ox2), At4g21690 (GA3ox3), At1g80330 (GA3ox4), At4g25420 (GA20ox1), At5g51810 (GA20ox2), At5g07200 (GA20ox3), At1g60980 (GA20ox4), At1g44090 (GA20ox5), At3g47340 (DIN6), At1g70290 (TPS8), At1g76410, At3g59940, At1g26770 (EXP10), and At1g69530 (EXP1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Phenotypes of pgm and sex1-1 Mutants Grown in a 12-h-Light/12-h-Dark Photoperiod.
Supplemental Figure 2. Effect of GA Treatment on Photosynthesis, Respiration, and Transpiration Rates in Leaves of Col-0, pgm, and sex1-1 Plants.
Supplemental Figure 3. Exogenous GA Treatment Disrupts the Normal Expression Pattern of Starvation Genes.
Supplemental Figure 4. Diurnal Changes in the Relative Expression Levels of Genes Encoding Enzymes Involved in the Late Steps of GA Biosynthesis (GA20oxs and GA3oxs) in Col-0, pgm, and sex1-1 Plants Grown under a 12-h-Light/12-h-Dark Photoperiod.
Supplemental Figure 5. Level of GAs (ng g−1 FW) in Col-0 at the End of Night (8 am) and End of Day (8 pm).
Supplemental Figure 6. Comparison of GA4 Content (ng g−1 FW) in Col-0, pgm, and sex1-1 at the End of Night (8 am) and End of Day (8 pm).
Supplemental Figure 7. Screening of KSi Lines Grown in a 12-h-Light/12-h-Dark Photoperiod.
Supplemental Figure 8. Altered Expression of Sugar Starvation Marker Genes in Starch Mutants.
Supplemental Figure 9. Suc Supplementation in the adg1-1 Mutant the Night before Restores the Afternoon KS Expression Peak the Day After.
Supplemental Figure 10. Effect of Light Intensity on Soluble Sugar Levels and on the Expression of Sugar Starvation Marker Genes in Col-0, pgm, and sex1-1 Plants.
Supplemental Figure 11. Effect of Light Intensity on the GA Content in Col-0, pgm, and sex1-1 Plants.
Supplemental Figure 12. Expression of Sugar Starvation Marker Genes in Plants Exposed to Low-Light Intensity for 1 d.
Supplemental Figure 13. Expression of EXP10 and EXP1 Is Modulated by GAs.
Supplemental Table 1. List and Description of the Mutants Used in This Work.
Supplemental Table 2. Levels of GAs (ng g−1 FW) at the End of Night and at the End of Day in Wild-Type Plants Grown at 100 µmol m−2 s−1 Irradiance.
Supplemental Table 3. Levels of GAs (ng g−1 FW) at the End of Day in Col-0, pgm, and sex1-1 Mutants.
Supplemental Table 4. Primers Used for Gene Expression Analysis Using Real-Time Quantitative RT-PCR.
Supplemental Table 5. Primers Used to Test the Insertion in the AGRIKOLA RNAi Lines.
Supplementary Material
Acknowledgments
We thank Alison M. Smith (John Innes Centre, UK), Martin Steup (University of Potsdam, Germany), Elena Loreti (Consiglio Nazionale delle Ricerche, Italy), and Francesco Licausi (Scuola Superiore Sant’Anna, Italy) for invaluable discussions. S.C.Z. and K.K. gratefully acknowledge the support of the SystemsX.ch initiative, Plant Growth in a Changing Environment. This research was financially supported by Scuola Superiore Sant’Anna.
AUTHOR CONTRIBUTIONS
E.P., S.G., J.T.v.D., S.C.Z., and P.P. designed the experiments. E.P., S.P., and G.N. performed the experiments. L.M. and N.C. carried out the GA analysis. K.K. measured the photosynthetic gas exchange. E.P. and P.P. wrote the article. All the authors discussed and commented on the content of the article.
Glossary
- GA
gibberellin
- CO2
carbon dioxide
- RNAi
RNA interference
- Col-0
Columbia-0
References
- Alter P., Dreissen A., Luo F.-L., Matsubara S. (2012). Acclimatory responses of Arabidopsis to fluctuating light environment: Comparison of different sunfleck regimes and accessions. Photosynth. Res. 113: 221–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arana M.V., Marín-de la Rosa N., Maloof J.N., Blázquez M.A., Alabadí D. (2011). Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 108: 9292–9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvidsson S., Kwasniewski M., Riano-Pachon D.M., Müller-Röber B. (2008). Quant-Prime – A flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9: 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bläsing O.E., Gibon Y., Günther M., Höhne M., Morcuende R., Osuna D., Thimm O., Usadel B., Scheible W.R., Stitt M. (2005). Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T., Huber S.C., Somerville C.R. (1985). Alterations in growth, photosynthesis and respiration in a starch deficient mutant of Arabidopsis thaliana (L.) Heynh deficient in chloroplast phosphoglucomutase. Plant Physiol. 79: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar T., Lin T.-P., Kakefuda G., Benbow L., Preiss J., Somerville C.R. (1991). Mutants of Arabidopsis with altered regulation of starch degradation. Plant Physiol. 95: 1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.-T., Cosgrove D.J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97: 9783–9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comparot-Moss S., et al. (2010). A putative phosphatase, LSF1, is required for normal starch turnover in Arabidopsis leaves. Plant Physiol. 152: 685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley J.H., Zeeman S.C., Takaha T., Smith A.M., Smith S.M. (2001). A critical role for disproportionating enzyme in starch breakdown is revealed by a knock-out mutation in Arabidopsis. Plant J. 26: 89–100 [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (2010). Plant Hormones: Biosynthesis, Signal Transduction, Action! 3rd ed. (New York: Springer). [Google Scholar]
- Fambrini M., Mariotti L., Parlanti S., Picciarelli P., Salvini M., Ceccarelli N., Pugliesi C. (2011). The extreme dwarf phenotype of the GA-sensitive mutant of sunflower, dwarf2, is generated by a deletion in the ent-kaurenoic acid oxidase1 (HaKAO1) gene sequence. Plant Mol. Biol. 75: 431–450 [DOI] [PubMed] [Google Scholar]
- Fleet C.M., Yamaguchi S., Hanada A., Kawaide H., David C.J., Kamiya Y., Sun T.P. (2003). Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 132: 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut D.M., Hulett J., Cramer G.R., Seemann J.R. (1997). Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiol. 115: 317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., et al. (2010). Increased leaf size: Different means to an end. Plant Physiol. 153: 1261–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzali S., Loreti E., Solfanelli C., Novi G., Alpi A., Perata P. (2006). Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J. Plant Res. 119: 115–123 [DOI] [PubMed] [Google Scholar]
- Graf A., Schlereth A., Stitt M., Smith A.M. (2010). Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl. Acad. Sci. USA 107: 9458–9463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielminetti L., Perata P., Alpi A. (1995). Effect of anoxia on carbohydrate metabolism in rice seedlings. Plant Physiol. 108: 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2003). The genes of the green revolution. Trends Genet. 19: 5–9 [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25 [DOI] [PubMed] [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14 (10B): 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamatsu T., King R.W., Helliwell C.A., Koshioka M. (2005). The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 138: 1106–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi M., Hidema J., Makino A., Ishida H. (2013). Autophagy contributes to nighttime energy availability for growth in Arabidopsis. Plant Physiol. 161: 1682–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Gao X., Liao L., Harberd N.P., Fu X. (2007). Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 145: 1460–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kötting O., Santelia D., Edner C., Eicke S., Marthaler T., Gentry M.S., Comparot-Moss S., Chen J., Smith A.M., Steup M., Ritte G., Zeeman S.C. (2009). STARCH-EXCESS4 is a laforin-like Phosphoglucan phosphatase required for starch degradation in Arabidopsis thaliana. Plant Cell 21: 334–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 18: 927–937 [DOI] [PubMed] [Google Scholar]
- Krouk G., Ruffel S., Gutiérrez R.A., Gojon A., Crawford N.M., Coruzzi G.M., Lacombe B. (2011). A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 16: 178–182 [DOI] [PubMed] [Google Scholar]
- Lin T.P., Caspar T., Somerville C.R., Preiss J. (1988a). Isolation and characterization of a starchless mutant of Arabidopsis thaliana (L.) Heynh lacking ADPglucose pyrophosphorylase activity. Plant Physiol. 86: 1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.P., Caspar T., Somerville C.R., Preiss J. (1988b). A starch deficient mutant of Arabidopsis thaliana with low ADPglucose pyrophosphorylase activity lacks one of the two subunit of the enzyme. Plant Physiol. 88: 1175–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn J.E., Feil R., Hendriks J.H., Gibon Y., Morcuende R., Osuna D., Scheible W.R., Carillo P., Hajirezaei M.R., Stitt M. (2006). Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem. J. 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti L., Picciarelli P., Lombardi L., Ceccarelli N. (2011). Fruit-set and early fruit growth in tomato are associated with increases in indoleacetic acid, cytokinin, and bioactive gibberellin contents. J. Plant Growth Regul. 30: 405–415 [Google Scholar]
- Michael T.P., Breton G., Hazen S.P., Priest H., Mockler T.C., Kay S.A., Chory J. (2008). A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol. 6: e225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra A.B., Atkin O.K., Bonser S.P., Davidson A.M., Finnegan E.J., Mathesius U., Poot P., Purugganan M.D., Richards C.L., Valladares F., van Kleunen M. (2010). Plant phenotypic plasticity in a changing climate. Trends Plant Sci. 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Niittylä T., Messerli G., Trevisan M., Chen J., Smith A.M., Zeeman S.C. (2004). A previously unknown maltose transporter essential for starch degradation in leaves. Science 303: 87–89 [DOI] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: Biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparelli E., et al. (2012). Misexpression of a chloroplast aspartyl protease leads to severe growth defects and alters carbohydrate metabolism in Arabidopsis. Plant Physiol. 160: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., et al. (1999). ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261 [DOI] [PubMed] [Google Scholar]
- Ribeiro D.M., Araújo W.L., Fernie A.R., Schippers J.H.M., Mueller-Roeber B. (2012). Action of gibberellins on growth and metabolism of Arabidopsis plants associated with high concentration of carbon dioxide. Plant Physiol. 160: 1781–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritte G., Lloyd J.R., Eckermann N., Rottmann A., Kossmann J., Steup M. (2002). The starch-related R1 protein is an alpha-glucan, water dikinase. Proc. Natl. Acad. Sci. USA 99: 7166–7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland F., Baena-Gonzalez E., Sheen J. (2006). Sugar sensing and signaling in plants: Conserved and novel mechanisms. Annu. Rev. Plant Biol. 57: 675–709 [DOI] [PubMed] [Google Scholar]
- Sairanen I., Novák O., Pěnčík A., Ikeda Y., Jones B., Sandberg G., Ljung K. (2012). Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T.K., Matsuoka M. (2004). Generating high-yielding varieties by genetic manipulation of plant architecture. Curr. Opin. Biotechnol. 15: 144–147 [DOI] [PubMed] [Google Scholar]
- Schachtman D.P., Shin R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58: 47–69 [DOI] [PubMed] [Google Scholar]
- Schneider A., Häusler R.E., Kolukisaoglu U., Kunze R., van der Graaff E., Schwacke R., Catoni E., Desimone M., Flügge U.I. (2002). An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J. 32: 685–699 [DOI] [PubMed] [Google Scholar]
- Sinclair T.R. (1992). Mineral nutrition and plant growth response to climate change. J. Exp. Bot. 43: 1141–1146 [Google Scholar]
- Stitt M., Zeeman S.C. (2012). Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 15: 282–292 [DOI] [PubMed] [Google Scholar]
- Streb, S., and Zeeman, S.C. (2012). Starch metabolism in Arabidopsis. In The Arabidopsis Book 9:e0160, /10.1199/tab.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng N., Wang J., Chen T., Wu X., Wang Y., Lin J. (2006). Elevated CO2 induces physiological, biochemical and structural changes in leaves of Arabidopsis thaliana. New Phytol. 172: 92–103 [DOI] [PubMed] [Google Scholar]
- Usadel B., Bläsing O.E., Gibon Y., Retzlaff K., Höhne M., Günther M., Stitt M. (2008). Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol. 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., Lue W.L., Yu T.S., Long J.H., Wang C.N., Eimert K., Chen J. (1998). Characterization of ADG1, an Arabidopsis locus encoding for ADPG pyrophosphorylase small subunit, demonstrates that the presence of the small subunit is required for large subunit stability. Plant J. 13: 63–70 [DOI] [PubMed] [Google Scholar]
- Wiese A., Christ M.M., Virnich O., Schurr U., Walter A. (2007). Spatio-temporal leaf growth patterns of Arabidopsis thaliana and evidence for sugar control of the diel leaf growth cycle. New Phytol. 174: 752–761 [DOI] [PubMed] [Google Scholar]
- Wilson R.N., Heckman J.W., Somerville C.R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2006). Gibberellin biosynthesis in Arabidopsis. Phytochem. Rev. 5: 39–47 [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yu T.S., et al. (2001). The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell 13: 1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeeman S.C., ap Rees T. (1999). Changes in carbohydrate metabolism and assimilate export in starch-excess mutants of Arabidopsis. Plant Cell Environ. 22: 1445–1453 [Google Scholar]
- Zhang X.Q., Wei P.C., Xiong Y.M., Yang Y., Chen J., Wang X.C. (2011). Overexpression of the Arabidopsis α-expansin gene AtEXPA1 accelerates stomatal opening by decreasing the volumetric elastic modulus. Plant Cell Rep. 30: 27–36 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.