Abstract
Mammalian muscle cell differentiation is a complex process of multiple steps for which many of the factors involved have not yet been defined. In a screen to identify the regulators of myogenic cell fusion, we found that the G-protein coupled receptor 56 (GPR56) gene was transiently upregulated during the early fusion of human myoblasts. Human mutations in GPR56 cause the disease bilateral frontoparietal polymicrogyria (BFPP), however the consequences of receptor dysfunction on muscle development have not been explored. Using knockout mice, we defined the role of GPR56 in skeletal muscle. GPR56−/− myoblasts have decreased fusion and smaller myotube sizes in culture. In addition, loss of GPR56 expression in muscle cells results in decreases or delays in the expression of MyoD, myogenin, and NFATc2. Our data suggest that these abnormalities result from decreased GPR56-mediated SRE and NFAT signaling. Despite these changes, no overt differences in phenotype were identified in the muscle of GPR56 knockout mice, which presented only a mild but statistically significant elevation of serum creatine kinase (CK) compared to wildtype. In agreement with these findings, clinical data from 13 BFPP patients revealed mild serum CK increase in only 2 patients. In summary, targeted disruption of GPR56 in mice results in myoblast abnormalities. The absence of a severe muscle phenotype in GPR56 knockout mice and human patients suggests that other factors may compensate for the lack of this GPCR during muscle development and that the motor delay observed in these patients is likely not due to primary muscle abnormalities.
Keywords: GPR56, myoblast, skeletal muscle, dystroglycanopathies, SRE
Introduction
During muscle development, muscle progenitor cells in the somites migrate out to the limb buds and undergo two waves of myogenesis to form mature muscle [1]. This process of differentiation proceeds through several steps: progenitor cell proliferation and migration, the commitment to differentiation, myoblast-myoblast adhesion, and the fusion of cells to form syncytial myofibers [2]. These steps, largely recapitulated during adult muscle regeneration in vivo and myoblast differentiation in vitro, are regulated by the coordination of many factors. In particular, the “master” transcription factors of the basic helix-loop-helix (bHLH) family direct myogenic differentiation in a sequential manner [3]. Precursor cells in the somites are specified to the myogenic lineage through the expression of the bHLH factor, myogenic factor 5 (Myf5) [4]. Myogenic differentiation 1 (MyoD) is expressed shortly after and also specifies myogenic precursor cells [5]. Although normally expressed in different cells [6, 7], the loss of one can result in a compensatory upregulation of the other [8, 9]. After migration to the limb buds and exit from the cell cycle, the expression of myogenin induces myoblasts to differentiate [10]. Myogenin promotes the expression of factors that lead to cell-cell adherence and fusion, resulting in the formation of multinucleated myofibers.
Some of the cell-surface effectors of muscle cell differentiation and fusion have also been identified. Cell-cell adhesion molecules such as NCAM, N-cadherin, M-cadherin, ADAM12, and VCAM-1/VLA-4 are involved in cell-cell adhesion of muscle cells [2]. Other proteins have also been implicated as important for fusion, but their specific roles remain unclear. Part of the difficulty in identifying the role of cell surface proteins is that many of them act cooperatively and/or in parallel, thus complementing each other’s function [11]. A complete understanding of the molecular regulation of muscle development awaits a more complete characterization of these molecules as well as the identification of other as yet unknown factors that are involved.
A previous study of ours suggested that GPR56 expression is upregulated during the early differentiation of myoblasts [12]. Pull-down assays demonstrated that GPR56 localizes to a tetraspanin microdomain specified by the tetraspanins CD81 and CD9, which are associated with the Gαq/11 subunit [13]. CD81 and CD9 have each been implicated as partners that promote the fusion of myoblasts [14]. GPR56 expression has been associated with the migration and adhesion of neural progenitor cells, gliomas, and melanoma cells [15–21], processes that are also important for the differentiation of myoblasts into multinucleated myotubes.
GPR56 belongs to the adhesion subfamily of G-protein coupled receptors (aGPCRs), which are characterized by their large extracellular N-terminal structure and a GPCR proteolytic site, a GPS motif [22]. The GPS site is auto-catalytically cleaved during protein translation through the action of a GAIN (GPCR-autoproteolysis inducing) domain that encompasses both the GPS motif and regions N-terminal to it [23]. The resulting extracellular N-terminal fragment and membrane-bound C-terminal fragment then re-associate with each other non-covalently at the cell surface [24–26]. Recessive mutations in GPR56 that result in the loss of GPR56 protein at the surface of the cell, particularly those that disturb the cleavage of the GPS domain, cause the rare neurodevelopmental disease bilateral frontoparietal polymicrogyria (BFPP) [OMIM #606854] [27, 28]. Patients with BFPP are characterized by mental retardation, motor developmental delays, seizures, and defects in the brainstem and cerebellum [29–32, 27, 33]. The defects in the brainstem and cerebellum manifest through the development of polymicrogyria, which are aberrantly small convolutions on the brain surface [34]. Patients with the muscle dystroglycanopathies, particularly Muscle-Eye-Brain disease and Walker-Warburg Syndrome also display these “cobblestone” brain abnormalities [34, 35] in addition to developing severe muscular dystrophies [36–38]. Given the overlap in brain abnormalities seen in these diseases, there has been speculation regarding the potential role of GPR56 in skeletal muscle.
To address this question, we have completed studies using GPR56 knockout mice, silencing RNA in a differentiating myoblast cell line (i.e. in C2C12 cells), and luciferase assays to explore receptor-mediated signaling. We found that GPR56 is transiently upregulated in myocytes (differentiated myoblasts that have not yet fused) and nascent myotubes. This increase follows the induction of MyoD expression and is concurrent with myogenin expression. Loss of GPR56 results in decreased myoblast fusion in culture. Our data suggest that this abnormality may reflect decreased receptor-mediated SRF and NFAT signaling. Despite the disruption of GPR56-mediated pathways, no differences in myofiber size or fiber type specification were detected in GPR56 knockout mice. In a study of regenerating muscle, GPR56 KO mice showed delays in the expression of MyoD and myogenin. Analysis of serum creatine kinase levels revealed a mild but statistically significant increase in GPR56 knockout mice compared to wildtype. This finding was consistent with the clinical data obtained from 13 BFPP patients, where only 2 exhibited elevated serum CK levels. In summary, our data suggest that GPR56 supports the activation of SRE and NFAT signaling which in turn promote myoblast differentiation but not myofiber hypertrophy or fiber type specification. The lack of a muscle phenotype in GPR56 knockout mice suggests that other factors may compensate for the lack of GPR56 during muscle development. Therefore, the motor delay observed in BFPP patients is not likely caused by a primary muscle defect.
Results
GPR56 is transiently expressed during muscle cell differentiation
To define the timing of GPR56 expression, primary mouse myoblasts were isolated and induced to differentiate by serum withdrawal. At various times throughout differentiation, mRNA and protein lysates were collected (Figure 1A–C). Both GPR56 mRNA (Figure 1B) and protein (Figure 1C) are transiently induced during the early fusion of primary mouse myoblasts (D1, D2) and quickly downregulated during later fusion stages. GPR56 protein expression in myoblast cultures follows the onset of MyoD expression and is concomitant with myogenin expression (Figure 1C), suggesting that GPR56 is present in post-mitotic myocytes committed to fusion.
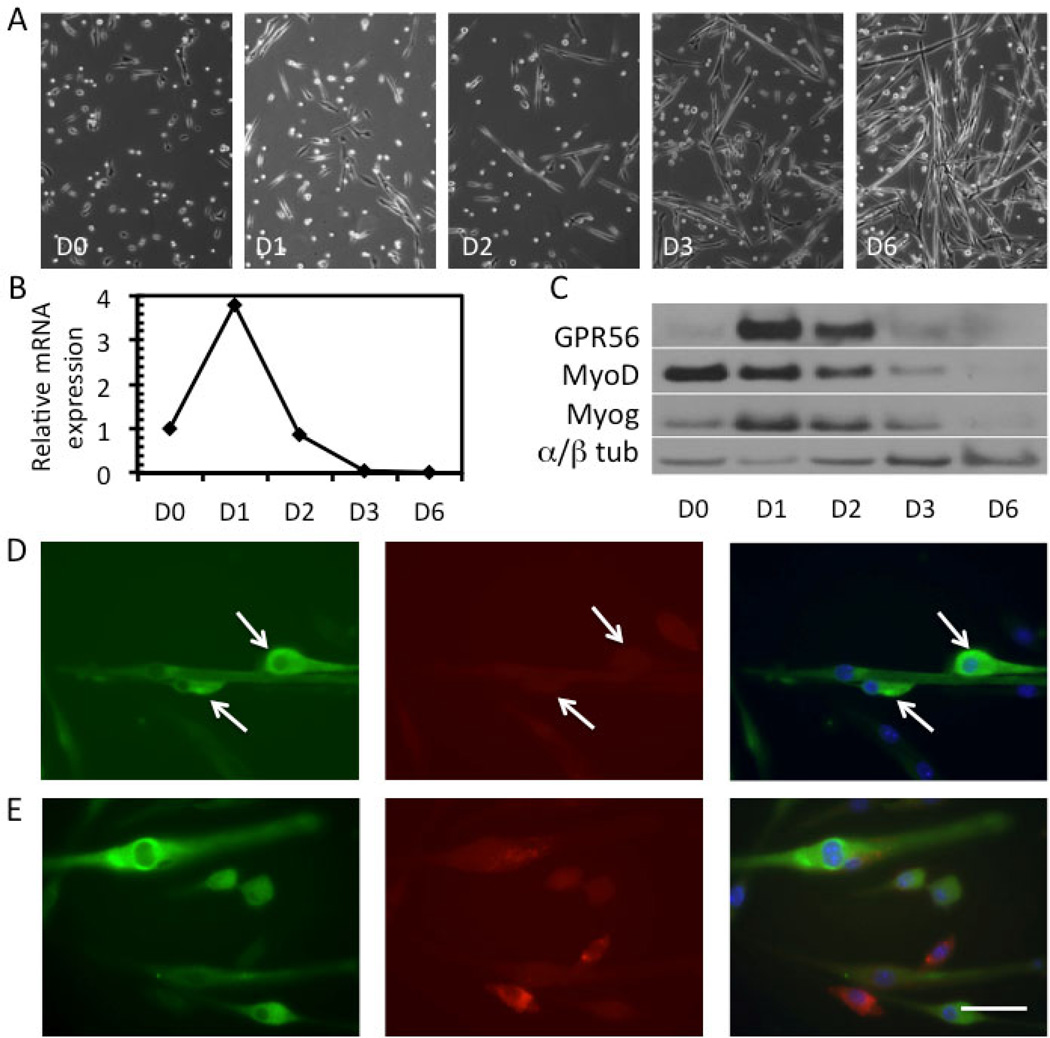
Figure 1. GPR56 is transiently expressed in the early differentiation phase of mouse myoblasts.
A. Phase images of mouse myoblasts induced to differentiate over the course of 6 days (D0 – D6) illustrating the degree of myotube formation. B. Upregulation of GPR56 mRNA expression at D1 by qRT-PCR in primary mouse myoblasts which then rapidly decreases. C. Protein expression of GPR56, MyoD, myogenin, and α/β-tubulin (loading control) in myoblasts at D0 to D6, as assessed by Western blot. GPR56 protein expression peaks at D1 and rapidly decreases by D3, where little expression remains. D–E. GPR56 (green) and caveolin-1 (red) staining in differentiating primary mouse myoblasts at D1. DAPI (blue) was used to stain nuclei. Arrows point to GPR56+ cells that are positioned closely and elongated, suggesting that the cells are readying for fusion or fusing. Scale bar = 50 µm.
The myoblast cultures at D1 and D2, where GPR56 expression is the highest, contain a mixture of proliferating and quiescent myoblasts, committed myocytes, and early myotubes with few nuclei. To determine which of these cells were expressing GPR56, we performed immunofluorescence staining on myoblasts at D1 (Figure 1D–E, Figure S1). GPR56 was detected in mononuclear cells, some of which were in close association with myotubes (Figure 1D, arrows). There was also slight immunoreactivity in myotubes, particularly surrounding the nuclei. The cultures were co-stained with caveolin-1, which is expressed in myoblasts but not myocytes [39], to better distinguish whether the GPR56-positive mononuclear cells were in myoblasts or myocytes (Figure 1D, E). The cells did not co-express GPR56 and caveolin-1, suggesting that GPR56 is only expressed in differentiating myocytes.
Loss of GPR56 results in decreased myoblast fusion through decreases in SRE and NFAT activity
To determine the role of GPR56 during myoblast differentiation, GPR56 was silenced using two separate shRNA constructs in the mouse C2C12 myoblast cell line (Figure 2A). Both GPR56 shRNA constructs efficiently silenced GPR56 mRNA and protein expression (Figure 2B, C). MyoD expression did not seem dramatically altered in GPR56-silenced C2C12 cultures, while myogenin expression was decreased, particularly with shRNA construct 3. Myoblast fusion and myotube size were significantly decreased in the GPR56-silenced C2C12 cells at day 5 following induction of differentiation (Figure 2D–F).
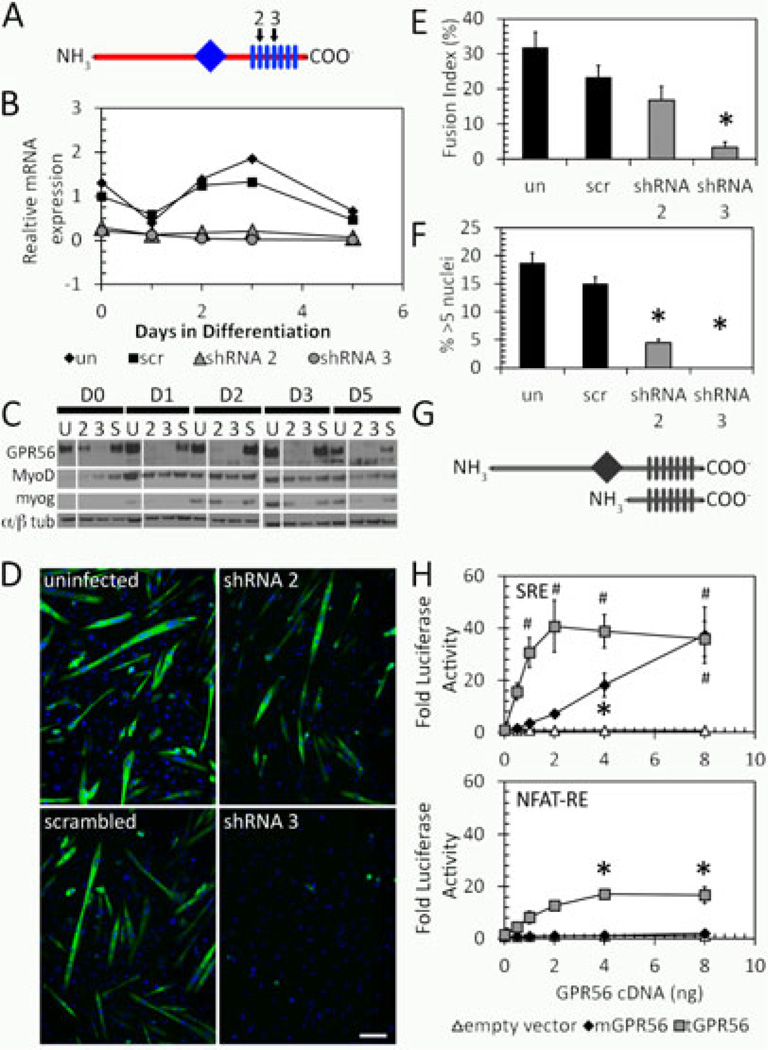
Figure 2. GPR56 is involved in directing myoblast fusion through SRF and NFAT pathways.
A. Schematic diagram showing the location of GPR56 shRNA constructs (2, 3, black arrows) against GPR56 transmembrane domains (rectangles) 2 and 4. The diamond indicates the G-proteolytic site. B. GPR56 mRNA expression by RT-qPCR in silenced C2C12s. Both shRNA2 and 3 effectively silenced the expression of GPR56. C. Western blot of GPR56, MyoD, and myogenin proteins in silenced C2C12 cells. D. Myosin Heavy Chain staining in GPR56-silenced cultures shows decreased myotube formation in GPR56 shRNA2 and 3 silenced cells. Scale bar = 50 µm. E. Fusion is decreased in GPR56-silenced cells at day 5 following differentiation. Un, uninfected; scr, scrambled oligo. * p < 0.01. F. Myotube size is decreased in GPR56-silenced cells at day 5 following differentiation. *p < 0.01. un, uninfected; scr, scrambled oligo. G. Schematic showing full-length and truncated GPR56. Diamond = G proteolytic site. Rectangles = transmembrane domains. H. Luciferase reporter assays in HEK293 cells of full-length (mGPR56, black diamond) or truncated (tGPR56, gray squares) GPR56 with luciferase reporter constructs driven by serum response element (SRE) or NFAT response element (NFAT-RE). GPR56 induces signaling from both SRE and NFAT-RE. * p<0.05. # p<0.001. n=3.
Multiple transcriptional pathways are involved in the early differentiation of myoblasts into myocytes and small myotubes in vitro. In particular, evidence that GPR56 downstream signaling includes the activation of the serum response element (SRE) and nuclear factor of activated T-cells response element (NFAT-RE) in gene promoters has been reported [40]. The SRE DNA element is functionally equivalent to the CArG box in myogenic cells [41], to which the serum response factor (SRF) binds. To confirm that GPR56 activates the SRE and NFAT-RE, we used vectors expressing full-length GPR56 (mGPR56) and a constitutively active variant, which includes an N-terminal GPR56 truncation (tGPR56) [26]. We assessed how expression of these constructs activated either an SRE or NFAT-RE luciferase reporter gene (Figure 2G). Truncated GPR56 activated both the SRE and NFAT reporters whereas the full-length receptor activated only the SRE luciferase construct (Figure 2H).
Primary myoblasts from GPR56-knockout mice exhibit decreased differentiation and fusion
To extend our findings to an in vivo model, primary myoblasts were isolated from littermate wildtype and GPR56 knockout mice. Myoblasts were FACS-sorted from dissociated limb and back muscles [42], differentiated for 5 days, and analyzed for their fusion competence (Figure 3A–D). Knockout myoblasts exhibited a decreased ability to fuse, as measured by their fusion index at days 2 and 5 in differentiation media (Figure 3A, B). Quantification of the myotube size demonstrates that knockout cells also form smaller myotubes at D2 (Figure 3C), whereas by D5 the myotube size is not significantly different between knockout and wildtype cultures. The ability of the knockout myotubes to grow to sizes similar to those of wildtype myotubes by D5 suggests that GPR56 plays a role only in the early stages of myoblast fusion.
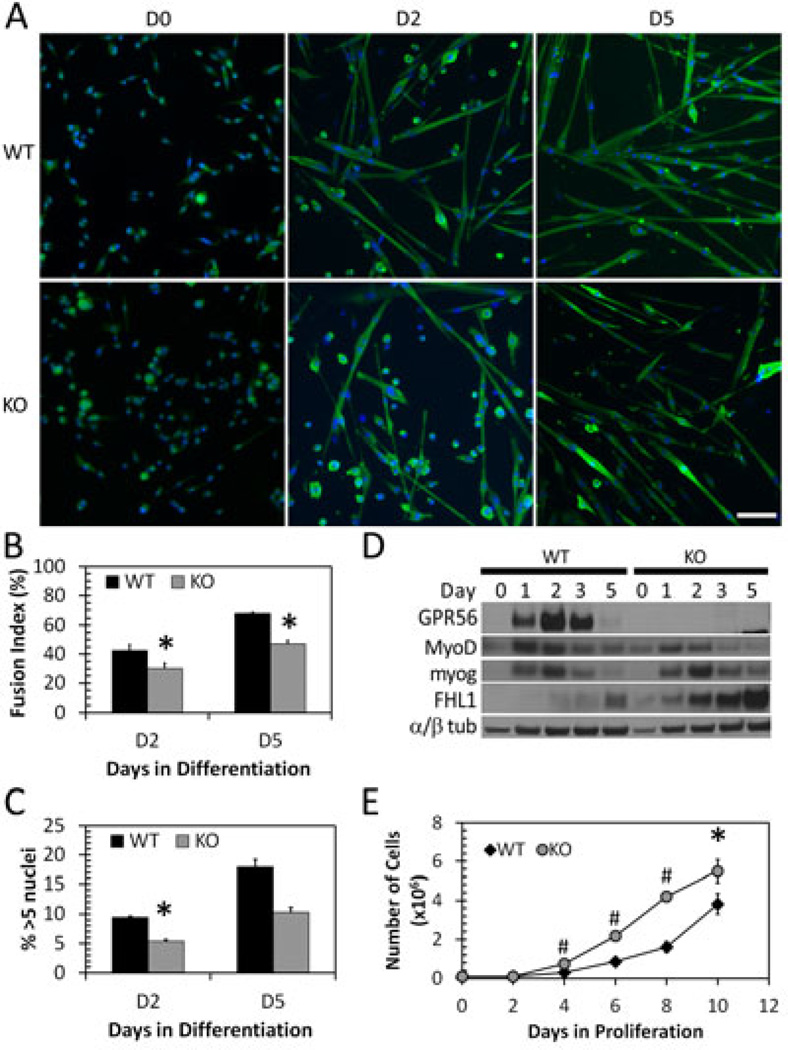
Figure 3. GPR56 KO myoblasts fuse less and have decreased MyoD expression.
A. WT and GPR56 KO mouse myoblasts undergoing differentiation at D0, D2, and D5. Scale bar = 50 µm. Green, Desmin (D0) or myosin heavy chain (D2, D5). Blue, nuclei. B. Fusion index in WT and KO differentiating mouse myoblasts. GPR56 KO myoblasts have decreased fusion at D2 and D5.* p < 0.05, n = 4. C. Overall myotube size as measured by percentage of myotubes with greater than 5 nuclei in WT and KO differentiating cultures. * p <0.05. D. Protein expression by Western blot of GPR56, MyoD, myogenin, and α/β-tubulin in differentiating myoblasts at D0-D5. GPR56 KO myoblasts show decreased MyoD expression at days 3 and 5, and increased FHL1 expression. E.GPR56 KO myoblasts (gray circles) proliferate more that WT myoblasts (black diamonds). (*p = <0.05, # = p < 0.001, n = 4 trials).
To more precisely define GPR56 function in relation to myogenic differentiation, we looked at the protein expression of myogenic transcription factors (Figure 3D). Decreases in signaling to the SRF transcription factor could result in reduced expression of the downstream transcription factors MyoD and myogenin, which regulate differentiation [43–45]. Indeed, MyoD expression appeared decreased at days 3 and 5 in the differentiating knockout compared with wildtype myoblasts at similar time points, whereas there was no change in the early marker of differentiation, myogenin. To confirm whether NFAT signaling was altered, we examined the expression of the transcriptional co-activator, FHL1, which supports NFATc1 and NFATc2 in promoting myoblast fusion and myotube growth after the onset of myogenin expression [46–48]. FHL1 was upregulated in the differentiating knockout myoblast cultures (Figure 3D).
Additionally, GPR56 knockout primary myoblasts proliferated significantly faster than wildtype myoblasts over the course of 10 days (Figure 3E). These data support the conclusion that loss of GPR56 results in less efficient commitment of myoblasts to differentiation, which manifests as decreased fusion ability in vitro.
GPR56 knockout muscle is morphologically normal
Mutations in GPR56 result in the human genetic disease, BFPP. Patients with BFPP share similarities in brain pathology with dystroglycanopathy patients (Figure 4A) whom also exhibit severe muscle defects, particularly muscular dystrophy [34]. Some patients with BFPP have a motor delay or early muscle hypotonia resulting in the consideration of congenital myopathy or muscular dystrophy as a diagnosis [33, 49]. These parallels suggest that BFPP patients may also have a specific muscle defect. A review of serum creatine kinase (CK) levels in 13 BFPP patients revealed that 2 patients had slightly elevated values (Figure 4B). Given that muscle biopsies for these patients were not available, muscle from GPR56 knockout mice were analyzed. The gastrocnemius and tibialis anterior muscles of 1- to 3-month old wildtype and knockout mice were examined after H&E staining (Figure 4C). They revealed no signs of myopathy or dystrophy, such as fibrosis, necrosis, or increased fiber size heterogeneity. To determine whether the in vitro myoblast fusion defect translated to decreased myofiber size in vivo, the myofiber sizes of knockout versus wildtype mouse gastrocnemius muscle were quantified. This analysis revealed no significant difference (Figure 4D). However, we found a slight but statistically significant increase in the serum CK levels of GPR56 knockout mice compared to wildtype mice (Figure 4E, n=11–12 mice/group; p=0.012).
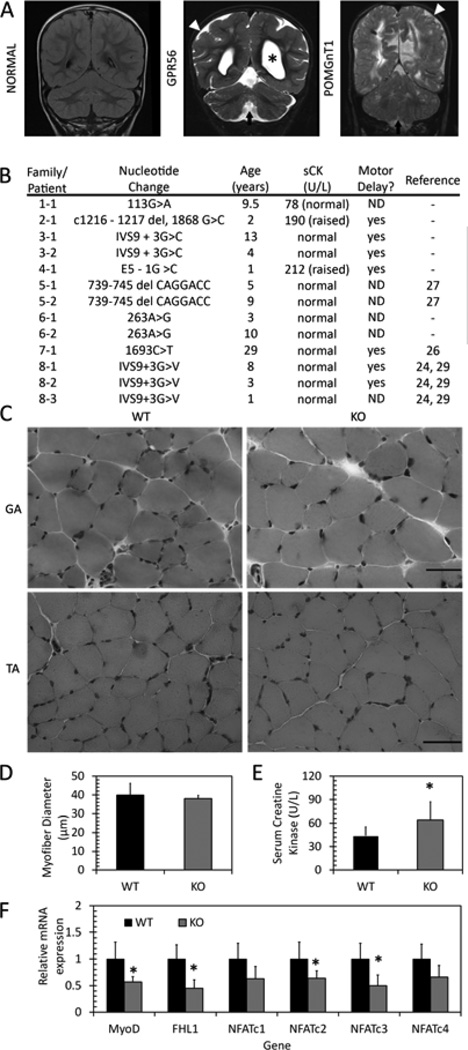
Figure 4. Muscle phenotypes in BFPP patients and GPR56 knockout mice.
A. Representative coronal flair MRI image from an unaffected individual (NORMAL) and coronal T2 images from individuals with confirmed mutations in GPR56 and POMGnT1. Patients exhibit enlarged ventricles (asterisks), presence of diffused cortical abnormalities (white arrowhead) and presence of cerebellar abnormalities, including a small vermis in the GPR56 patient (arrow). B. Serum creatine kinase levels and motor developmental delays in patients with BFPP. ND: Not determined. C. H&E staining of one-month-old gastrocnemius (top, GA) and tibialis anterior (bottom, TA) muscles shows no difference between wildtype and knockout muscle. Scale bars = 50 µm. D. Myofiber diameter in TA muscle shows no difference between WT and KO. E. Serum CK levels in WT and KO mice shows slightly elevated serum CK levels in knockout mice. *p=0.012, n = 11–12. F. mRNA expression in WT and KO gastrocnemius muscle. Expression of MyoD, FHL1, NFATc2, and NFATc3 are decreased in KO muscle. * p<0.05, n = 6.
The GPR56-downstream signaling pathways that were altered in differentiating myogenic cells were also analyzed for their expression in the muscle. MyoD, a transcriptional target of SRF signaling during myoblast commitment and differentiation [43, 50, 44], has decreased mRNA expression in GPR56 knockout muscle compared to wildtype muscle (Figure 4F); this decrease is in agreement with the MyoD protein expression data in differentiating muscle cells (Figure 3D). Similarly, the expression of NFAT family members that are involved in commitment and early myoblast fusion are also affected in GPR56 knockout mice. NFATc3 activity supports MyoD-directed myogenesis [51, 52], whereas NFATc2 is activated in early myotubes [46]. The mRNA expression of both NFATc2 and NFATc3 were significantly downregulated in GPR56 knockout muscle (Figure 4F). In addition, FHL1, whose transcription is also under the control of SRE [53], exhibited decreased expression in knockout mouse muscle (Figure 4F). These decreases in mRNA expression suggest that both SRE and NFAT signaling pathways are altered in GPR56 knockout mouse muscle.
The fusion defects seen in vitro do not translate to defective muscle regeneration in vivo
To determine if the decreased fusion and myotube size seen in vitro translated into reduced myofiber size during muscle regeneration, the tibialis anterior muscles of wildtype and knockout mice were injured by injection with cardiotoxin (Figure 5). GPR56 mRNA expression in regenerating wildtype muscle is transiently increased and peaks at day 4 after cardiotoxin injection (Figure 5B). Morphologically, there was no gross defect in the timing or extent of regeneration in knockout compared to wildtype muscle (Figure 5A). There was also no significant difference in myofiber diameter four days after cardiotoxin injection between knockout and wildtype muscle (Figure 5C). At six and eighteen days, there were no differences in myofiber diameter (Figure 5C).
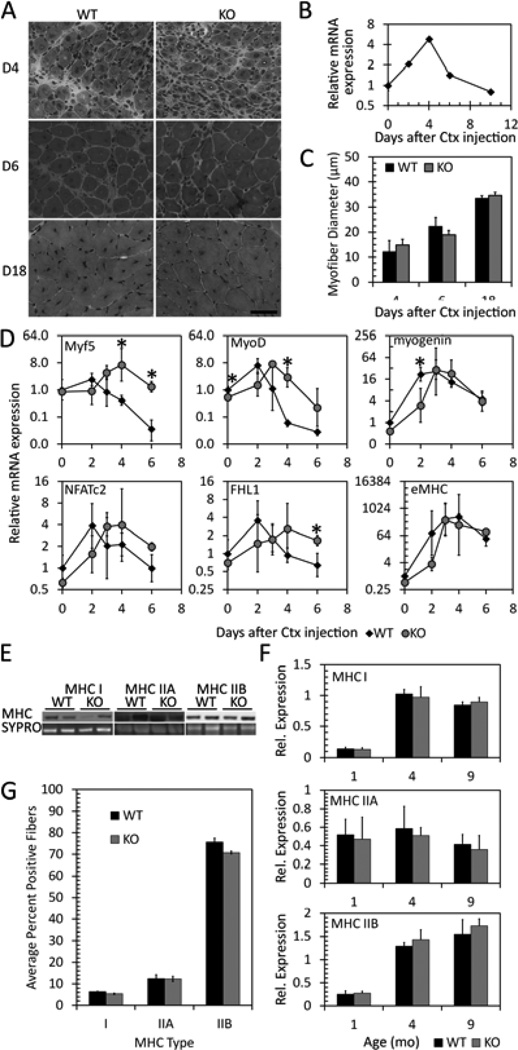
Figure 5. Loss of GPR56 affects the expression of myogenic transcription factors during regeneration but does not affect myofiber size.
A. H&E staining of GPR56 WT and KO gastrocnemius muscle at days 4, 6, and 18 after cardiotoxin injury. KO morphology and timing does not look different from WT. B. mRNA expression of GPR56 by RT-qPCR shows transient upregulation of GPR56 during regeneration. C. Myofiber diameter in cardiotoxin-injured WT and GPR56 KO gastrocnemius muscle shows no difference in diameter between WT and KO. D. mRNA expression by RT-qPCR of various genes in WT (black diamond) and GPR56 KO (gray circle) cardiotoxin-injured muscle. Myf5, MyoD, and myogenin are delayed in expression in KO muscle. * p<0.05, n = 3. E. Sample Western blots of myosin heavy chain protein expression. F. Quantification of the amount of MHC I, IIA, or IIB protein expression by Western blot in WT and KO gastrocnemius muscle in mice of various ages shows no difference in the amount of MHC isoforms between WT and KO. G. Quantification of the % of positive MHC I, IIA, or IIB fiber types in KO versus WT muscles, based on immunofluorescence staining in 4 littermate pairs.
We then examined the expression of critical myogenic transcription factors in the regenerating wildtype and knockout muscles. In knockout mice, the peak in Myf5 and MyoD expression were delayed (Figure 5D). The timing of later stages of myofiber nuclear accretion as indicated by the expression of NFATc2, FHL1, and embryonic myosin heavy chain, however, seem to match the timing in wildtype mice. Overall, although some of the molecular determinants of myoblast differentiation were significantly different or delayed in expression in the knockout muscle compared to wildtype, muscle regeneration did not seem affected.
The NFAT signaling pathways activated during myoblast commitment and differentiation are also used to regulate fiber type differentiation [54, 55]. Because the loss of GPR56 in knockout mice affected these pathways during muscle cell differentiation, we examined whether myofiber type specification was altered in the gastrocnemius muscle at various ages. No differences in protein expression between wildtype and GPR56 knockout mouse muscles were detected by Western blotting using antibodies specific to myosin heavy chains (MHC) type I, IIA, and IIB (Figure 5E, F). We also used immunofluorescence to manually count the individual myofibers and ensure that the lack of differences in MHC expression detected by Western blot was not due to a lack of sensitivity (Figure S2, Figure 5G). We isolated the gastrocnemius muscle from one-month-old littermates, stained sequential sections with antibodies against MHC I, IIA, or IIB together with anti-laminin to outline the myofibers, and quantified the proportion of positive myofibers for each MHC type. No differences in the proportion of fiber types were found between wildtype and knockout muscle (Figure 5G). Thus, although GPR56 signals through the SRF and NFAT pathways, it does not affect the NFAT-directed specification of fiber types.
Discussion
Mutations in GPR56 result in the human disease BFPP [27, 33, 56]. The similarities in brain phenotype between BFPP and the dystroglycanopathies have resulted in their classification as similar diseases [34, 49]. Both of these diseases are caused through a loss of binding between cell membrane proteins and the extracellular matrix (α-dystroglycan to laminin [57, 58] and GPR56 to collagen III [21]), which suggests a shared disease mechanism. We previously found that GPR56 is upregulated in differentiating human fetal muscle cells [12], suggesting that its loss could affect muscle cell differentiation and result in a muscle phenotype. Thus, we investigated GPR56’s role in the skeletal muscle using the GPR56 knockout mouse in conjunction with cell-based assays.
Our in vitro studies delineate a role for GPR56 in the commitment of myoblasts to early differentiation. GPR56 localizes to myocytes and nascent myotubes, while the luciferase assays link GPR56 expression with the activation of promoters containing the SRF DNA-binding elements. These data together with the previous findings demonstrating that GPR56 activates RhoA [17, 21, 26] draw a possible pathway for GPR56 at the cell surface to activate RhoA signaling and subsequently activate the SRF-mediated transcription of target genes in the nuclei of myoblasts committed to differentiation.
Our findings that full-length and truncated mouse GPR56 can activate luciferase driven by SRE agree with a previous study showing that different human isoforms of GPR56 were able to activate SRE-driven luciferase to varying degrees [40]. In myoblasts, the SRF transcription factor binds an SRE DNA element within the MyoD promoter to promote the transcription of MyoD during proliferation and differentiation [43, 50, 44]. The activation of SRF transcriptional activity, in turn, is dependent on RhoA [50]. The upregulation of MyoD expression, in correlation with a switch in SRF phosphorylation [59], induces proliferating myoblasts to exit the cell cycle [5]. MyoD then switches to a differentiation program to prepare the cells for fusion. The inhibition of SRF expression or activity has been shown to lead to decreased MyoD [50] and myogenin expression [60]. In support of these possible signaling links between GPR56, SRF, and MyoD, we saw a decrease in MyoD mRNA expression in GPR56 knockout muscle, as well as decreases in MyoD and myogenin expression in differentiating GPR56 knockout and GPR56-silenced C2C12 myoblasts, respectively. Satellite cells lacking MyoD continue to proliferate and inefficiently express myogenin when induced to differentiate [61, 62]; in our studies, GPR56 knockout myoblasts showed increased proliferation rates compared to wildtype myoblasts. These data support the conclusion that GPR56 might signal through the SRF to promote transcription of the myogenic regulators of myoblast commitment to differentiation (i.e., MyoD).
This transcriptional program of myogenic differentiation is aided by many cofactors, including NFATc3. NFATc3 potentiates the ability of MyoD to activate the myogenic differentiation program [51]. NFATc2 is then activated in the nascent myotubes to promote further fusion [46]. Our data demonstrates that loss of GPR56 resulted in significantly decreased NFATc3 and NFATc2 expression. In our luciferase assays, constitutively active GPR56 stimulated NFAT transcription to a lesser degree than SRE transcription. One of the target genes of the SRF is FHL1 [53, 45], a co-activator of NFAT transcription in the muscle [48]. Increased FHL1 expression was seen in GPR56 knockout myoblasts, but not in GPR56 silenced C2C12 cells. This inconsistency suggests that GPR56 activation of NFAT-RE is not direct, but instead through the induction of FHL1 transcription following activation of the SRF.
Clearly, the in vitro loss of GPR56 negatively affected the ability of myoblasts to efficiently differentiate and fuse, but these effects did not alter muscle development or regeneration in vivo. The initial examination of GPR56 knockout muscle histology found no gross changes in muscle histology or myofiber size. We did, however, find a small but statistically significant increase in the serum CK levels, which indicates the presence of myofiber membrane damage, as seen in muscular dystrophy [63, 64]. However, in the case of GPR56 knockout mice, the degree of serum CK elevation was much smaller than what is seen in a mouse model of the dystroglycanopathies [65]. That the increase was slight, albeit significant, is in agreement with other studies that did not detect raised serum CK levels in BFPP patients [49] and with our findings of two patients with only slightly elevated serum CK. Therefore, the loss of GPR56 in human muscle did not lead to a severe phenotype in this tissue.
Following acute injury, GPR56 knockout muscle regenerated normally although the expression of Myf5 and MyoD were delayed. The overall mild phenotype in vivo suggests that GPR56 could be one of many factors that promote commitment and differentiation and that the redundancy in function with other genes can compensate for its loss. Myf5 is known to be able to compensate for loss of MyoD, [8, 9, 6] and its delayed but emphatic increase during regeneration may act as a compensatory mechanism for the less efficient expression of MyoD in GPR56 knockout muscle. This hypothesis is supported by our in vitro findings that, despite a significantly decreased ability of GPR56 knockout myoblasts to fuse at early time points, the myotube sizes were not different from wildtype myotube sizes at later time points (D6). In addition, the fusion defect was more prominent in the GPR56-silenced C2C12 cells, as opposed to the primary GPR56 knockout myoblasts. Because the knockout myoblasts develop in a GPR56-null environment and were isolated from postnatal muscle, it is possible that they adapted their differentiation mechanisms to compensate for the loss of GPR56. These compensatory mechanisms would not have had time to develop in GPR56-silenced C2C12 cells and could be one reason for why the phenotype of silenced cells was more severe than in primary knockout cells.
In skeletal muscle, four major myofiber types can be classified by their myosin heavy chain contractile abilities [66]. Type I fibers have slow myosin heavy chains, whereas Types IIA, IIX, and IIB have fast myosin heavy chains. Intrinsic and extrinsic factors lead to the specification, maintenance, and switching of a particular fiber type [67]. In vivo RNA silencing of different NFATs shows that fiber type specification is influenced and dictated by different combinations of four isoforms of NFAT (c1 through c4) [55]. All four NFAT isoforms studied play roles in maintaining the Type I fiber type, whereas only NFATc2 through NFATc4 are required for Type IIA and Type IIX fibers, and only NFATc4 for Type IIB fibers. In our study, despite decreases in NFATc2 and NFATc3 expression in GPR56 knockout mice, no differences in fiber type specification were seen in GPR56 knockout mouse muscle. These findings agree with reports that knockouts of the NFAT family members have little to no effect on fiber type specification [46, 52] and support our conclusion that GPR56 signaling through NFAT is likely to be important only during the early stages of fusion.
Despite a clear role for GPR56 in myoblast commitment and differentiation, there are several reasons why its loss may not severely impact muscle function in BFPP patients and GPR56-null mice. GPR56 expression is transient and restricted to early differentiation in both myoblasts in culture and regenerating muscle in vivo. It has been suggested that cell-surface molecules that have roles in myoblast differentiation exhibit less severe phenotypes when individually knocked-out in mice due to redundancy in their functions [11]. Although the functions of α-dystroglycan and GPR56 in brain development seem similar, the relative importance of their ligands may differ in the brain versus skeletal muscle. A clear role for the importance of α-dystroglycan binding to laminin has been established in both tissues [68–70]. While laminin is a major component of the mature muscle basal lamina [71], GPR56’s ligand collagen III is expressed only transiently during development [72]. Similarly, α-dystroglycan is an integral component of the dystrophin-associated protein complex, which is expressed in both developing and mature muscle, whereas GPR56 is expressed transiently. Thus, while in the brain α-dystroglycan and GPR56 serve similar functions in aiding neuronal cell migration, in skeletal muscle their pattern of expression and downstream signaling pathways are clearly distinct. These studies demonstrate that while loss of GPR56 function in myogenic cells leads to decreased ability to fuse due to altered signaling through SRF, skeletal muscle development overall is not affected.
Materials and Methods
Human Subjects
Clinical details on patients and families affected by GPR56 mutations and other neurological conditions were obtained with written consent as part of their participation in research studies at Boston Children’s Hospital and Beth Israel Deaconess Medical Center. The studies were approved by the Internal Review Board at each participating institution and performed in accordance with the ethical standards covering human subjects research.
Animals
GPR56 knockout mice (B6N.129S5-Gpr56tm1Lex/Mmcd) were generated by Genentech/Lexicon Genetics. The absence of GPR56 protein was previously verified [18]. Animals were euthanized by CO2 asphyxiation and the appropriate muscles dissected. Unless otherwise stated, tissue was collected from one-month-old male animals.
For evaluation of serum creatine kinase levels, mice between 5 and 10 months of age were nicked in the tail vein and 200 µL blood was collected. Blood was allowed to coagulate for 1 hour at room temperature and then centrifuged at 16,100 g in a table-top microfuge for 1.5 min. Twenty microliters of plasma serum were used in the CK-NAC (UV-Rate) CK test (Stanbio #0910, Stanbio Laboratory, Boerne, TX, USA) to determine serum CK levels. For each blood sample, two measurements were taken and averaged. A total of 11 WT and 12 KO animals were analyzed. Data were analyzed for statistical significance using the Student’s t-test (unpaired, homoscedastic) in Excel with significance at p<0.05. The error bars represent the standard deviation between samples.
For cardiotoxin injections, mice (n = 5) were anesthetized with isoflurane and a total of 15 µL of 0.5 µg/mL cardiotoxin (Sigma #C9759-5MG, Sigma-Aldrich, St. Louis, MO, USA) in PBS was injected into three different sites in the right TA with a Hamilton syringe (26 G needle). Mice were sacrificed at 2, 3, 4, 6, and 18 days after cardiotoxin injury. Left (uninjured) TA muscles were used as uninjured (D0) controls.
For tissue section analyses, tissue was snap-frozen by embedding in OCT (Tissue-Tek #4583, VWR, Arlington Heights, IL, USA) in liquid nitrogen-cooled isopentane. For mRNA and protein analyses, tissue was snap-frozen in liquid nitrogen. All animals were handled in accordance with protocols approved by the Institutional Animal Care and Use Committee at Boston Children’s Hospital.
Hematoxylin and Eosin (H&E) staining and determination of myofiber diameter
For myofiber diameter analysis, ten micron sections of TA muscle tissue were taken from the approximate belly of the muscle. For cardiotoxin-injured muscle, sections were taken from the injured area. Tissue sections were stained as previously described [73] and imaged using a Nikon E1000 microscope with a SPOT Insight Color 3.2.0 camera using SPOT 4.5.9.9 software (Spot Imaging Solutions, Diagnostic Instruments Inc, Sterling Heights, MI, USA.).
Fiber diameter was measured using ImageJ 1.4r (NIH) and the plug-in “Measure and Label.java.” For cardiotoxin-injured muscle, only fibers with centrally-located nuclei were measured. For each mouse, fiber diameter measurements within 2–3 fields were averaged. Approximately 140 fibers were counted per field, totaling 280–420 fibers per mouse. Each data bar represents the measurements of 3–5 mice. Error bars represent the standard deviation between the measurements for the mice. Significance was determined using an unpaired Student’s t-test.
Determination of fiber type proportions by IHC
Sequential 10 µm muscle cross-sections from the approximate center of one-month-old littermate wildtype and knockout gastrocnemius muscle were fixed in cold 100% acetone for 5 min and then air-dried for 20 min. They were then washed with 1x PBS-T (0.1% Tween-20 in PBS) and blocked for 30 min in 2.5% horse serum in PBS-T. Sections were then incubated overnight at 4°C in primary antibody in PBS-T using the MHC type antibodies MHC1, 1:100 Sigma #M8421, Sigma-Aldrich, St. Louis, MO, USA; mouse anti-MHC Type IIA 1:50, Developmental Studies Hybridoma Bank (DSHB, Iowa City, IA, USA) #SC-71, developed by S. Schiaffino; or mouse anti-MHC Type IIB (1:50, DSHB #BF-F3, developed by S. Schiaffino), and co-stained with a rabbit anti-laminin antibody (1:100, Sigma-Aldrich #L9393) to outline the myofibers. After three 3 min washes in PBS-T, slides were incubated for 1 hr at RT with the appropriate secondary antibody in PBS-T, washed three times in PBS-T, and mounted with DAPI Vectashield (#H-1200, Vector Laboratories, Burlingame, CA, USA).
The entire section was photographed in sequential fields using a Hamamatsu Orca-ER camera (#C4742-95-12ER, Hamamatsu, Middlesex, NJ, USA) mounted on a Zeiss Axioplan 2 microscope (5× objective) with Axiovision 4.5 SP1 software. The individual fields were merged together into one image using Photoshop CS3. The numbers of MHC-positive and negative fibers in each section were manually counted using the cell counter function in ImageJ 1.4r (NIH). Significance was determined using the Student’s t-test (paired) in Excel with significance at p < 0.05. The error bars represent the standard error of measurement between samples.
Primary myoblast isolation, proliferation, and fusion assays
Limb skeletal muscles from littermate wildtype and GPR56 knockout mice were dissected, minced, and dissociated with 5 mg/mL collagenase D (Roche Applied Science #11088882001, Indianapolis, IN, USA) and 5 mg/mL dispase II (Roche #04942078001) per gram of tissue for up to an hour at 37°C. Following dissociation, filtration through 100 and 40 µm filters, and red blood cell removal (Qiagen #158904, Valencia, CA, USA), myogenic cells were isolated by FACS as previously described [42]. Myoblasts were then plated on 6 cm plastic plates coated with 5 µg/cm2 collagen type I (BD Biosciences #354236, Franklin Lakes, NJ, USA) in myoblast growth media (30% fetal bovine serum (FBS) and 1× penicillin/streptomycin/glutamine (PSG) in 1:1 F10/high-glucose DMEM) supplemented with 10 ng/mL bFGF (Atlanta Biologicals #X07995, Norcross, GA, USA).
For proliferation assays, myoblasts were plated at 1 × 104 cells on collagen-coated 12-well plates. Every 2 days for up to 10 days, duplicate wells of myoblasts were trypsinized and counted. The total number of cells was then calculated for each sample. Comparisons were made using a paired Student’s t-test. Error bars show the standard error among experimental sets. Data are represented as an average of 3 trials.
For fusion assays, 2 × 105 myoblasts were plated on gel-coated 6-well dishes in myoblast growth media + 10 ng/mL bFGF. The next day (D0), they were switched to differentiation media (2% horse serum/1x PSG in low-glucose DMEM), which was changed daily for up to 6 days. To assess fusion, cells were fixed with 4% PFA in PBS for 15 min at RT, and washed with PBS; the nuclei were then stained with DAPI in PBS. Images of cells were taken using a Photometrics CoolSNAP EZ camera (Photometrics, Tucson, AZ, USA) mounted on a Nikon Eclipse TE2000-S microscope with NIS Elements AR 2.30 SP4 software. Fusion was analyzed using the cell counter function in ImageJ 1.4r or 1.47b (Wayne Rasband, NIH). For each sample, 3 fields were taken and the results were averaged per duplicate or triplicate well. Each field contained approximately 100 to 200 nuclei, depending on the day of fusion. The fusion index was calculated as: 100 × (total number of nuclei in myotubes) / (total number of nuclei). The average myotube size was calculated as: (total number of nuclei in myotubes) / (total number of myotubes counted). For these counts, myotubes were defined as cells containing 2 or more nuclei. The final results are the averages of each experiment with 4 total experiments comprising 3 sets of littermate mouse myoblast isolations. Comparisons between WT and KO were made using a paired Student’s t-test, with significance set at p<0.05. Error bars show the standard error among experimental sets.
For immunostaining, myoblasts were fixed with 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 15 min and permeabilized with 1% Triton X-100/PBS for 3 min. After washing with PBS, cells were blocked in 10% FBS/0.1% Triton X-100 in PBS for 1 hour at RT. Cells were then treated with 1% SDS for 1 min and rinsed 3 times for 5 min in PBS. Cells were then incubated mouse anti-GPR56 (1:100, Sigma #SAB1400340) and rabbit anticaveolin-1 (1:200, Cell Signaling #3238, Danvers, MA, USA) in blocking buffer overnight at 4°C. After washing 3 times with PBS, cells were incubated with secondary antibody (Dylight 488 anti-mouse IgG and Dylight 594 anti-rabbit IgG, Jackson Immunoresearch Laboratories, West Grove, PA, USA #715-486-150, #711-516-152, respectively) in blocking buffer for 1 hour at RT. After incubation, cells were washed 3 times with PBS, and mounted with DAPI Vectashield. Cells were imaged with an Orca-ER camera mounted on a Nikon E1000 microscope with Openlab 5.5.0 software (Improvision-PerkinElmer, Waltham, MA USA).
C2C12 cell gene silencing and fusion assays
Short hairpin oligos were designed against mouse GPR56 mRNA (accession # NM_018882) and annealed into the BD RNAi-Ready pSiren-RetroQ viral vector (BD Biosciences Clontech #631526, Mountain View, CA, USA). The following sequences were used: shRNA2, 5’-TCACGTGACTACACCATCA-3’, and shRNA3, 5’-CGTTGGTGGATGTGAATAA-3’. 293GP cells were plated in 10% FBS/1x PSG in DMEM and transfected the next day with 8 µg VSV-G plasmid and 8 µg silencing or control vector using a standard calcium phosphate protocol. Twelve to 16 hours after transfection, the media was replaced with fresh media. After 65–72 hours, the media (containing virus) was collected and filtered through a 0.45 µm PES low-protein binding filter to remove cell debris.
The viral infection followed a protocol based on Springer et al. [74]. The viral suspension was supplemented with 20% FBS and 8 ng/uL polybrene (final concentrations). The viral mix was added to C2C12s (plated the previous day on gel-coated 6-well plates at 7 × 104 cells), incubated for 15 min in a cell culture incubator (37°C, 5% CO2), and spun at 1100 × g for 30 minutes at 32°C. The viral mix was then removed and replaced with fresh C2C12 growth media (20% FBS, 1x PSG in high-glucose DMEM). Cells were re-infected with this protocol at 8, 16, and 24 hours after the first infection. The efficiency of infection was verified in parallel infections with virus carrying the pQCLIN construct, which confirmed that 95–100% of C2C12 cells expressed LacZ (data not shown).
Infected C2C12 cells were switched to differentiation media 6 hours after the last infection, and assayed for fusion and mRNA and protein profiles as described below. The Day 0 (D0) timepoint was taken just prior to switching the media to differentiation. Subsequent time points were taken at D1, D2, D3, and D5 for mRNA and protein, and D2 and D5 for the fusion assay. The fusion index and myotube size were assayed as with the primary myoblasts, with the average of 10 fields taken per single well per sample per trial. The final results are the averaged results from three independent viral infections. Comparisons between GPR56-silenced samples and control samples were made using ANOVA with one-way variance from StatPlus:mac LE (Analyst Soft Inc, ver. 2009). Error bars show the standard error among experimental sets.
For immunostaining, cells were fixed and stained as with the primary myoblasts, without 1% SDS treatment. The primary antibody used was mouse anti-myosin (1:100, Developmental Studies Hybridoma Bank (DSHB) #MF-20, developed by D.A. Fischman).
RNA isolation from cells and tissue, reverse transcription and quantitative real-time PCR
For cultured cells, cells were trypsinized and pelleted. RNA was isolated using the RNeasy Mini kit (Qiagen #74104).
For tissues, RNA was isolated using the RNeasy Fibrous Tissue kit (Qiagen #74704) with modifications. Approximately 10 mg snap-frozen tissue was homogenized in 650 µL RLT + 1% β-mercaptoethanol (v/v) with Lysing Matrix D beads (MP Bio #6913, Solon, OH, USA) in a Fastprep FP120 homogenizer (Thermo Savant Qbiogene, Inc Carlsbad, CA, USA) for two cycles of 40 sec at speed 6, with a 5 min incubation on ice in between. Homogenized tissue was then spun down at 4°C at 16,100 g for 4 min, and 300 µL of the supernatant containing the RNA was transferred to an eppendorf tube. Ten microliters of proteinase K solution and 590 µL RNase-free water was then added, and the sample was incubated for 10 min at 55°C. The following RNA isolation steps with DNA removal were done as per the manufacturer’s recommendations.
cDNA was reverse-transcribed from 0.5 to 2 µg RNA using either the Quantitect Reverse Transcription kit (Qiagen #205311) or the Superscript III Reverse Transcription kit (Invitrogen Life Technologies #18080-051, Grand Island, NY, USA) with a mix of random hexamers and oligodT primers. For each experimental set, the same kit was used. From this reaction, 1/40 to 1/20 volume (0.5 to 1 µL) of cDNA was amplified using SYBR green (Invitrogen Life Technologies #4364346) and quantified in a 7900HT Fast Real-Time PCR System (Applied Biosystems Life Technologies, Grand Island, NY, USA). Relative expression was calculated using the delta delta Ct method [75]. All primers were optimized to run at the same efficiency as control primers (β-2-microglobulin (B2M) for cell and uninjured muscle assays or GAPDH for cardiotoxin injury assays) [76]. Primers were designed to span exon-intron junctions and/or large introns. All assays were run with no template and minus reverse transcriptase controls. The primers used are listed in Supplementary Table 1.
Protein isolation and Western blot analyses
Cells were lysed directly on the plate with RIPA cell lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA) with 0.1% SDS, protease inhibitor cocktail (Roche #4693159001), and phosphatase inhibitor cocktail (Roche Applied Science #4906837001). Lysates were collected and rotated for 1 hour at 4°C, then centrifuged for 25 min at 4°C, 16,100 g. The supernatant and pellet were separated.
Snap-frozen tissue samples were crushed into a powder using a liquid-nitrogen cooled ceramic mortar and pestle. An aliquot of the powder was taken and homogenized in TPER buffer (Thermo Scientific #78510, Pittsburg, PA, USA) with 0.1% SDS/ protease inhibitor cocktail/phosphatase inhibitor cocktail for 1 min at 4°C, using a hand-held homogenizer and plastic pestles. Samples were then rotated at 4°C for one hour and frozen at −20°C to complete lysis. After thawing, samples were centrifuged and the supernatant aliquoted.
One to 20 µg protein per sample was prepared with NuPage LDS loading buffer (Invitrogen Life Sciences #NP0007) and 5% β-mercaptoethanol (v/v); samples were then denatured for 10 minutes at 65°C before loading onto 4–12% Bis-Tris gels (Invitrogen Life Sciences #WG1402BOX). Proteins were transferred to nitrocellulose for 1 hour in transfer buffer (12.5 mM Tris-Glycine pH 8.3, 10% methanol). Blots were blocked in 5% BSA/1% milk in TBST (25 mM Tris, 3 mM KCl, 140 mM NaCl, 0.01% Tween-20) for one hour at RT and incubated in primary antibody in blocking buffer overnight at 4°C on a shaker. For detecting GPR56, blots were blocked and incubated with primary antibody in 5% BSA/2% milk/TBST. After 3 washes with TBST, blots were incubated with the appropriate HRP-conjugated secondary antibodies (1:10,000, Jackson Immunoresearch) in 5% milk in TBST for 40 min at RT. Bands were detected using the Western Lightning chemiluminescent detection reagent (Perkin Elmer #NEL105001EA). After detection, blots were stripped for 25 min at 80°C in stripping buffer (0.2 M glycine pH 2.5, 0.05% Tween-20), washed for 1 hour in TBST, and re-probed for other proteins. The following primary antibodies were used: mouse anti-mouse GPR56 (1:1000, H11 clone, generous gift from Xianhua Piao, Boston Children’s Hospital, Boston, MA), mouse anti-MyoD (1:1000, BD Biosciences #554130), mouse anti-myogenin (1:1000, DHSB #F5D, developed by F.W. Wright), goat anti-FHL1 (1:1000, Abcam #Ab23937, Cambridge, MA, USA), rabbit anti-GAPDH 14C10 (1:1000, Cell Signaling #2118), and rabbit anti-α/β-tubulin (1:5000, Cell Signaling #2148). For the analysis of primary mouse myoblasts, protein lysates from four sets of myoblasts isolated from littermate WT and KO mice were analyzed. For the analysis of silenced C2C12 cells, protein lysates were made from three sets of C2C12 cells that were independently infected.
For quantification of MHC types, the total protein on replicate nitrocellulose blots was stained with SYPRO Ruby Protein Blot Stain (Invitrogen Life Sciences #S-11791). Bands were imaged using the Bio-rad Chemidoc XRS+ molecular imaging system and densitometry readings were taken using the volume rectangle tool in Quantity One 4.6.2 software. The blots were destained and then blotted for either mouse anti-slow MHC (1:1000, Sigma-Aldrich #M8421), mouse anti-MHC Type IIA (1:1000, DSHB #SC-71), or mouse anti-MHC Type IIB (1:1000, DSHB #BF-F3). Multiple ECL readings of the MHC bands were taken using the Bio-rad Chemidoc XRS+ to ensure the collection of an unsaturated exposure. The densitometry measurements for MHC bands were normalized to the total protein SYPRO bands, and the results from three to five mice were averaged. Significance was determined using the Student’s t-test (unpaired) in Excel. Error bars show the standard deviation between samples.
Luciferase assays
The mouse GPR56 coding sequence (accession # NM_018882) was amplified from a V5-tagged mouse GPR56 construct kindly provided by Samir Koirala (Boston Children’s Hospital, Boston, MA) [19]. Constitutively active, truncated GPR56 (tGPR56) [26] was cloned into pCMV-XL4 by amplifying the C-terminal domain of GPR56 after the GPS cleavage site. The location of the GPS cleavage site was determined by homology to GPS sites found in other adhesion GPCRs [77]. The forward primers incorporated NotI restriction enzyme sites, a Kozak initiation sequence, and a methionine amino acid translational start site: full-length GPR56 (mGPR56) forward primer 5’-AAGCGGCCGCCACCATGGCTGTCCAGGTGCTG-3’ and tGPR56 forward primer 5’-AAGCGGCCGCCACCATGACCTACTTTGCAGTGCTGAT-3’. The reverse primer incorporated a NotI restriction enzyme site after the stop codon and was used for both mGPR56 and tGPR56: 5’-TTGCGGCCGCTGCAGAATTGCCCTAGATGC-3’.
HEK293 cells were plated in 96-well plates at a density of 6000 cells/well one day prior to transfection, or 3000 cells/well two days prior to transfection, in 10% FBS/DMEM. Cells were transfected using Lipofectamine (Invitrogen Life Sciences #18324-012) with: (A) 0–8 ng mGPR56-pCMV-XL4, tGPR56-pCMV-XL4, or pCMV-XL4 empty vector control; (B) 20 ng SRE5x-luc or NFAT-RE-luc reporter; and (C) 5 ng β-galactosidase (β-gal) pcDNA1.1 control in serum-free DMEM. Luciferase and β-gal activity were assayed as previously described [78]. Averaged triplicate data for each experiment were normalized to basal signaling from cells transfected with reporter and β-gal alone. Data from three separate experiments were averaged to produce the normalized relative luciferase activity. Significance was analyzed using two-way ANOVA with Bonferroni correction in GraphPad Prism 5. Error bars denote the standard error among experiments.
Supplementary Material
Acknowledgments
The antibodies against MHC Type IIA and MHC Type IIB, developed by S. Schiaffino, were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biology, Iowa City, IA 52242. We are grateful to Isabelle Draper for numerous helpful discussions. Thanks are also extended to Marie Torres in the Stem Cell Core Facility, Boston Children’s Hospital and Suzan Lazo-Kallanian at the Hematologic Neoplasia Core facility, Dana Farber Cancer Institute for aid with the FACS isolation of primary myoblasts, Ci Chen for cell plating at Tufts Medical Center, and Chelsea Cherenfant, Hui Meng, and Alexandra Lerch-Gaggl for help with the myofiber type analysis.
Funding This work was supported by the National Institutes of Health [2R01NS047727 and 1R01AR060317-01 to E.G., T32 GM007226 to M.P.W., K08 AR059750 and L40 AR057721 to M.W.L., and NIH-P30-HD1865 to the Boston Children’s Hospital Stem Cell Core Facility].
Abbreviations
- aGPCR
adhesion G-protein coupled receptor
- BFPP
bilateral frontoparietal polymicrogyria
- GPCR
G-protein coupled receptor
- GPR56
G-protein coupled receptor 56
- GPS
G-proteolytic site
- H&E
hematoxylin and eosin
- KO
knockout
- MHC
myosin heavy chain
- NFAT
nuclear factor of activated T-cells
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- shRNA
short-hairpin RNA
- SRE
serum response element
- SRF
serum response factor
- tGPR56
truncated G-protein coupled receptor 56
- WT
wildtype
Footnotes
Conflict of Interest Statement
The authors have no conflicts of interest to declare.
Supporting information.
Figure S1. GPR56 localization at the membrane of primary WT myoblasts undergoing differentiation (day2).
Figure S2. Myosin heavy chain type-specific isoform staining of muscle tissue sections.
Table S1. RT-qPCR primer sequences and concentrations
References
- 1.Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, Relaix F. The formation of skeletal muscle: from somite to limb. J Anat. 2003;202:59–68. doi: 10.1046/j.1469-7580.2003.00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abmayr SM, Pavlath GK. Myoblast fusion: lessons from flies and mice. Development. 2012;139:641–656. doi: 10.1242/dev.068353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lassar AB, Skapek SX, Novitch B. Regulatory mechanisms that coordinate skeletal muscle differentiation and cell cycle withdrawal. Curr Opin Cell Biol. 1994;6:788–794. doi: 10.1016/0955-0674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 4.Ott MO, Bober E, Lyons G, Arnold H, Buckingham M. Early expression of the myogenic regulatory gene, myf-5, in precursor cells of skeletal muscle in the mouse embryo. Development. 1991;111:1097–1107. doi: 10.1242/dev.111.4.1097. [DOI] [PubMed] [Google Scholar]
- 5.Halevy O, Novitch BG, Spicer DB, Skapek SX, Rhee J, Hannon GJ, Beach D, Lassar AB. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 6.Braun T, Arnold HH. Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 1996;15:310–318. [PMC free article] [PubMed] [Google Scholar]
- 7.Kablar B, Krastel K, Ying C, Asakura A, Tapscott SJ, Rudnicki MA. MyoD and Myf-5 differentially regulate the development of limb versus trunk skeletal muscle. Development. 1997;124:4729–4738. doi: 10.1242/dev.124.23.4729. [DOI] [PubMed] [Google Scholar]
- 8.Rudnicki MA, Braun T, Hinuma S, Jaenisch R. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 1992;71:383–390. doi: 10.1016/0092-8674(92)90508-a. [DOI] [PubMed] [Google Scholar]
- 9.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 10.Wright WE, Sassoon DA, Lin VK. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 11.Krauss RS. Regulation of promyogenic signal transduction by cell-cell contact and adhesion. Exp Cell Res. 2010;316:3042–3049. doi: 10.1016/j.yexcr.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerletti M, Molloy MJ, Tomczak KK, Yoon S, Ramoni MF, Kho AT, Beggs AH, Gussoni E. Melanoma cell adhesion molecule is a novel marker for human fetal myogenic cells and affects myoblast fusion. J Cell Sci. 2006;119:3117–3127. doi: 10.1242/jcs.03056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Little KD, Hemler ME, Stipp CS. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell. 2004;15:2375–2387. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tachibana I, Hemler ME. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J Cell Biol. 1999;146:893–904. doi: 10.1083/jcb.146.4.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shashidhar S, Lorente G, Nagavarapu U, Nelson A, Kuo J, Cummins J, Nikolich K, Urfer R, Foehr ED. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24:1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]
- 16.Ke N, Sundaram R, Liu G, Chionis J, Fan W, Rogers C, Awad T, Grifman M, Yu D, Wong-Staal F, Li QX. Orphan G protein-coupled receptor GPR56 plays a role in cell transformation and tumorigenesis involving the cell adhesion pathway. Mol Cancer Ther. 2007;6:1840–1850. doi: 10.1158/1535-7163.MCT-07-0066. [DOI] [PubMed] [Google Scholar]
- 17.Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N, Itoh H. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Jin Z, Koirala S, Bu L, Xu L, Hynes RO, Walsh CA, Corfas G, Piao X. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koirala S, Jin Z, Piao X, Corfas G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci. 2009;29:7439–7449. doi: 10.1523/JNEUROSCI.1182-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L, Begum S, Barry M, Crowley D, Yang L, Bronson RT, Hynes RO. GPR56 plays varying roles in endogenous cancer progression. Clin Exp Metastasis. 2010;27:241–249. doi: 10.1007/s10585-010-9322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo R, Jeong SJ, Jin Z, Strokes N, Li S, Piao X. G protein-coupled receptor 56 and collagen III, a receptor-ligand pair, regulates cortical development and lamination. Proc Natl Acad Sci U S A. 2011;108:12925–12930. doi: 10.1073/pnas.1104821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin HH, Stacey M, Yona S, Chang GW. GPS proteolytic cleavage of adhesion-GPCRs. Adv Exp Med Biol. 2010;706:49–58. doi: 10.1007/978-1-4419-7913-1_4. [DOI] [PubMed] [Google Scholar]
- 23.Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC, Brunger AT. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 2012;31:1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krasnoperov VG, Bittner MA, Beavis R, Kuang Y, Salnikow KV, Chepurny OG, Little AR, Plotnikov AN, Wu D, Holz RW, Petrenko AG. alpha-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 25.Nechiporuk T, Urness LD, Keating MT. ETL, a novel seven-transmembrane receptor that is developmentally regulated in the heart. ETL is a member of the secretin family and belongs to the epidermal growth factor-seven-transmembrane subfamily. J Biol Chem. 2001;276:4150–4157. doi: 10.1074/jbc.M004814200. [DOI] [PubMed] [Google Scholar]
- 26.Paavola KJ, Stephenson JR, Ritter SL, Alter SP, Hall RA. The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem. 2011;286:28914–28921. doi: 10.1074/jbc.M111.247973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piao X, Hill RS, Bodell A, Chang BS, Basel-Vanagaite L, Straussberg R, Dobyns WB, Qasrawi B, Winter RM, Innes AM, Voit T, Ross ME, Michaud JL, Descarie JC, Barkovich AJ, Walsh CA. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 28.Jin Z, Tietjen I, Bu L, Liu-Yesucevitz L, Gaur SK, Walsh CA, Piao X. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16:1972–1985. [Google Scholar]
- 29.Harbord MG, Boyd S, Hall-Craggs MA, Kendall B, McShane MA, Baraitser M. Ataxia, developmental delay and an extensive neuronal migration abnormality in 2 siblings. Neuropediatrics. 1990;21:218–221. doi: 10.1055/s-2008-1071501. [DOI] [PubMed] [Google Scholar]
- 30.Dobyns WB, Patton MA, Stratton RF, Mastrobattista JM, Blanton SH, Northrup H. Cobblestone lissencephaly with normal eyes and muscle. Neuropediatrics. 1996;27:70–75. doi: 10.1055/s-2007-973752. [DOI] [PubMed] [Google Scholar]
- 31.Straussberg R, Gross S, Amir J, Gadoth N. A new autosomal recessive syndrome of pachygyria. Clin Genet. 1996;50:498–501. doi: 10.1111/j.1399-0004.1996.tb02721.x. [DOI] [PubMed] [Google Scholar]
- 32.Farah S, Sabry MA, Khuraibet A, Khaffagi S, Rudwan M, Hassan M, Haseeb N, Abulhassan S, Abdel-Rasool MA, Elgamal S, Qasrawi B, Al-Busairi W, Farag TI. Lissencephaly associated with cerebellar hypoplasia and myoclonic epilepsy in a Bedouin kindred: a new syndrome? Clin Genet. 1997;51:326–330. doi: 10.1111/j.1399-0004.1997.tb02482.x. [DOI] [PubMed] [Google Scholar]
- 33.Piao X, Chang BS, Bodell A, Woods K, Benzeev B, Topcu M, Guerrini R, Goldberg-Stern H, Sztriha L, Dobyns WB, Barkovich AJ, Walsh CA. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann Neurol. 2005;58:680–687. doi: 10.1002/ana.20616. [DOI] [PubMed] [Google Scholar]
- 34.Barkovich AJ, Millen KJ, Dobyns WB. A developmental and genetic classification for midbrain-hindbrain malformations. Brain. 2009;132:3199–3230. doi: 10.1093/brain/awp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quattrocchi CC, Zanni G, Napolitano A, Longo D, Cordelli DM, Barresi S, Randisi F, Valente EM, Verdolotti T, Genovese E, Specchio N, Vitiello G, Spiegel R, Bertini E, Bernardi B. Conventional magnetic resonance imaging and diffusion tensor imaging studies in children with novel GPR56 mutations: further delineation of a cobblestone-like phenotype. Neurogenetics. 2013;14:77–83. doi: 10.1007/s10048-012-0352-7. [DOI] [PubMed] [Google Scholar]
- 36.Michele DE, Barresi R, Kanagawa M, Saito F, Cohn RD, Satz JS, Dollar J, Nishino I, Kelley RI, Somer H, Straub V, Mathews KD, Moore SA, Campbell KP. Post-translational disruption of dystroglycan-ligand interactions in congenital muscular dystrophies. Nature. 2002;418:417–422. doi: 10.1038/nature00837. [DOI] [PubMed] [Google Scholar]
- 37.Kim DS, Hayashi YK, Matsumoto H, Ogawa M, Noguchi S, Murakami N, Sakuta R, Mochizuki M, Michele DE, Campbell KP, Nonaka I, Nishino I. POMT1 mutation results in defective glycosylation and loss of laminin-binding activity in alpha-DG. Neurology. 2004;62:1009–1011. doi: 10.1212/01.wnl.0000115386.28769.65. [DOI] [PubMed] [Google Scholar]
- 38.Jimenez-Mallebrera C, Torelli S, Feng L, Kim J, Godfrey C, Clement E, Mein R, Abbs S, Brown SC, Campbell KP, Kroger S, Talim B, Topaloglu H, Quinlivan R, Roper H, Childs AM, Kinali M, Sewry CA, Muntoni F. A comparative study of alpha-dystroglycan glycosylation in dystroglycanopathies suggests that the hypoglycosylation of alpha-dystroglycan does not consistently correlate with clinical severity. Brain Pathol. 2009;19:596–611. doi: 10.1111/j.1750-3639.2008.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volonte D, Liu Y, Galbiati F. The modulation of caveolin-1 expression controls satellite cell activation during muscle repair. FASEB J. 2005;19:237–239. doi: 10.1096/fj.04-2215fje. [DOI] [PubMed] [Google Scholar]
- 40.Kim JE, Han JM, Park CR, Shin KJ, Ahn C, Seong JY, Hwang JI. Splicing variants of the orphan G-protein-coupled receptor GPR56 regulate the activity of transcription factors associated with tumorigenesis. J Cancer Res Clin Oncol. 2010;136:47–53. doi: 10.1007/s00432-009-0635-z. [DOI] [PubMed] [Google Scholar]
- 41.Taylor M, Treisman R, Garrett N, Mohun T. Muscle-specific (CArG) and serum-responsive (SRE) promoter elements are functionally interchangeable in Xenopus embryos and mouse fibroblasts. Development. 1989;106:67–78. doi: 10.1242/dev.106.1.67. [DOI] [PubMed] [Google Scholar]
- 42.Lawlor MW, Alexander MS, Viola MG, Meng H, Joubert R, Gupta V, Motohashi N, Manfready RA, Hsu CP, Huang P, Buj-Bello A, Kunkel LM, Beggs AH, Gussoni E. Myotubularin-deficient myoblasts display increased apoptosis, delayed proliferation, and poor cell engraftment. Am J Pathol. 2012;181:961–968. doi: 10.1016/j.ajpath.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gauthier-Rouviere C, Vandromme M, Tuil D, Lautredou N, Morris M, Soulez M, Kahn A, Fernandez A, Lamb N. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol Biol Cell. 1996;7:719–729. doi: 10.1091/mbc.7.5.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L'Honore A, Lamb NJ, Vandromme M, Turowski P, Carnac G, Fernandez A. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol Biol Cell. 2003;14:2151–2162. doi: 10.1091/mbc.E02-07-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Azhar G, Helms S, Burton B, Huang C, Zhong Y, Gu X, Fang H, Tong W, Wei JY. Identification of New SRF Binding Sites in Genes Modulated by SRF Over-Expression in Mouse Hearts. Gene Regul Syst Bio. 2011;5:41–59. doi: 10.4137/GRSB.S7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–338. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakuma K, Nishikawa J, Nakao R, Watanabe K, Totsuka T, Nakano H, Sano M, Yasuhara M. Calcineurin is a potent regulator for skeletal muscle regeneration by association with NFATc1 and GATA-2. Acta Neuropathol. 2003;105:271–280. doi: 10.1007/s00401-002-0647-0. [DOI] [PubMed] [Google Scholar]
- 48.Cowling BS, McGrath MJ, Nguyen MA, Cottle DL, Kee AJ, Brown S, Schessl J, Zou Y, Joya J, Bonnemann CG, Hardeman EC, Mitchell CA. Identification of FHL1 as a regulator of skeletal muscle mass: implications for human myopathy. J Cell Biol. 2008;183:1033–1048. doi: 10.1083/jcb.200804077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bahi-Buisson N, Poirier K, Boddaert N, Fallet-Bianco C, Specchio N, Bertini E, Caglayan O, Lascelles K, Elie C, Rambaud J, Baulac M, An I, Dias P, des Portes V, Moutard ML, Soufflet C, El Maleh M, Beldjord C, Villard L, Chelly J. GPR56-related bilateral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- 50.Carnac G, Primig M, Kitzmann M, Chafey P, Tuil D, Lamb N, Fernandez A. RhoA GTPase and serum response factor control selectively the expression of MyoD without affecting Myf5 in mouse myoblasts. Mol Biol Cell. 1998;9:1891–1902. doi: 10.1091/mbc.9.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delling U, Tureckova J, Lim HW, De Windt LJ, Rotwein P, Molkentin JD. A calcineurin-NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol. 2000;20:6600–6611. doi: 10.1128/mcb.20.17.6600-6611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in NFATC3(−/ −) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–126. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- 53.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 54.McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lomo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci U S A. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proc Natl Acad Sci U S A. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Parrini E, Ferrari AR, Dorn T, Walsh CA, Guerrini R. Bilateral frontoparietal polymicrogyria, Lennox-Gastaut syndrome, and GPR56 gene mutations. Epilepsia. 2009;50:1344–1353. doi: 10.1111/j.1528-1167.2008.01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Talts JF, Andac Z, Gohring W, Brancaccio A, Timpl R. Binding of the G domains of laminin alpha1 and alpha2 chains and perlecan to heparin, sulfatides, alpha-dystroglycan and several extracellular matrix proteins. EMBO J. 1999;18:863–870. doi: 10.1093/emboj/18.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito F, Blank M, Schroder J, Manya H, Shimizu T, Campbell KP, Endo T, Mizutani M, Kroger S, Matsumura K. Aberrant glycosylation of alpha-dystroglycan causes defective binding of laminin in the muscle of chicken muscular dystrophy. FEBS Lett. 2005;579:2359–2363. doi: 10.1016/j.febslet.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 59.Iyer D, Chang D, Marx J, Wei L, Olson EN, Parmacek MS, Balasubramanyam A, Schwartz RJ. Serum response factor MADS box serine-162 phosphorylation switches proliferation and myogenic gene programs. Proc Natl Acad Sci U S A. 2006;103:4516–4521. doi: 10.1073/pnas.0505338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandromme M, Gauthier-Rouviere C, Carnac G, Lamb N, Fernandez A. Serum response factor p67SRF is expressed and required during myogenic differentiation of both mouse C2 and rat L6 muscle cell lines. J Cell Biol. 1992;118:1489–1500. doi: 10.1083/jcb.118.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Megeney LA, Kablar B, Garrett K, Anderson JE, Rudnicki MA. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 1996;10:1173–1183. doi: 10.1101/gad.10.10.1173. [DOI] [PubMed] [Google Scholar]
- 62.Yablonka-Reuveni Z, Rudnicki MA, Rivera AJ, Primig M, Anderson JE, Natanson P. The transition from proliferation to differentiation is delayed in satellite cells from mice lacking MyoD. Dev Biol. 1999;210:440–455. doi: 10.1006/dbio.1999.9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pearce JM, Pennington RJ, Walton JN. Serum Enzyme Studies in Muscle Disease. Iii. Serum Creatine Kinase Activity in Relatives of Patients with the Duchenne Type of Muscular Dystrophy. J Neurol Neurosurg Psychiatry. 1964;27:181–185. doi: 10.1136/jnnp.27.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nichol CJ. Serum Creatine Phosphokinase Measurements in Muscular Dystrophy Studies. Clin Chim Acta. 1965;11:404–407. doi: 10.1016/0009-8981(65)90186-5. [DOI] [PubMed] [Google Scholar]
- 65.Chan YM, Keramaris-Vrantsis E, Lidov HG, Norton JH, Zinchenko N, Gruber HE, Thresher R, Blake DJ, Ashar J, Rosenfeld J, Lu QL. Fukutin-related protein is essential for mouse muscle, brain and eye development and mutation recapitulates the wide clinical spectrums of dystroglycanopathies. Hum Mol Genet. 2010;19:3995–4006. doi: 10.1093/hmg/ddq314. [DOI] [PubMed] [Google Scholar]
- 66.Schiaffino S, Reggiani C. Myosin isoforms in mammalian skeletal muscle. J Appl Physiol. 1994;77:493–501. doi: 10.1152/jappl.1994.77.2.493. [DOI] [PubMed] [Google Scholar]
- 67.Wigmore PM, Dunglison GF. The generation of fiber diversity during myogenesis. Int J Dev Biol. 1998;42:117–125. [PubMed] [Google Scholar]
- 68.Sanes JR. Laminin, fibronectin, and collagen in synaptic and extrasynaptic portions of muscle fiber basement membrane. J Cell Biol. 1982;93:442–451. doi: 10.1083/jcb.93.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liesi P. Do neurons in the vertebrate CNS migrate on laminin? EMBO J. 1985;4:1163–1170. doi: 10.1002/j.1460-2075.1985.tb03755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hunter DD, Llinas R, Ard M, Merlie JP, Sanes JR. Expression of s-laminin and laminin in the developing rat central nervous system. J Comp Neurol. 1992;323:238–251. doi: 10.1002/cne.903230208. [DOI] [PubMed] [Google Scholar]
- 71.Sanes JR. The basement membrane/basal lamina of skeletal muscle. J Biol Chem. 2003;278:12601–12604. doi: 10.1074/jbc.R200027200. [DOI] [PubMed] [Google Scholar]
- 72.Sasse J, von der Mark H, Kuhl U, Dessau W, von der Mark K. Origin of collagen types I, III, and V in cultures of avian skeletal muscle. Dev Biol. 1981;83:79–89. doi: 10.1016/s0012-1606(81)80010-3. [DOI] [PubMed] [Google Scholar]
- 73.Wu MP, Gussoni E. Carbamylated erythropoietin does not alleviate signs of dystrophy in mdx mice. Muscle Nerve. 2011;43:88–93. doi: 10.1002/mus.21785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Springer ML, Blau HM. High-efficiency retroviral infection of primary myoblasts. Somat Cell Mol Genet. 1997;23:203–209. doi: 10.1007/BF02721371. [DOI] [PubMed] [Google Scholar]
- 75.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 76.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 77.Silva JP, Lelianova V, Hopkins C, Volynski KE, Ushkaryov Y. Functional cross-interaction of the fragments produced by the cleavage of distinct adhesion G-protein-coupled receptors. J Biol Chem. 2009;284:6495–6506. doi: 10.1074/jbc.M806979200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doyle JR, Fortin JP, Beinborn M, Kopin AS. Selected melanocortin 1 receptor single-nucleotide polymorphisms differentially alter multiple signaling pathways. J Pharmacol Exp Ther. 2012;342:318–326. doi: 10.1124/jpet.112.194548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.