Abstract
CheA is a multidomain histidine kinase for chemotaxis in Escherichia coli. CheA autophosphorylates through interaction of its N-terminal phosphorylation site domain (P1) with its central dimerization (P3) and ATP-binding (P4) domains. This activity is modulated through the C-terminal P5 domain, which couples CheA to chemoreceptor control. CheA phosphoryl groups are donated to two response regulators, CheB and CheY, to control swimming behavior. The phosphorylated forms of CheB and CheY turn over rapidly, enabling receptor signaling complexes to elicit fast behavioral responses by regulating the production and transmission of phosphoryl groups from CheA. To promote rapid phosphotransfer reactions, CheA contains a phosphoacceptor-binding domain (P2) that serves to increase CheB and CheY concentrations in the vicinity of the adjacent P1 phosphodonor domain. To determine whether the P2 domain is crucial to CheA's signaling specificity, we constructed CheAΔP2 deletion mutants and examined their signaling properties in vitro and in vivo. We found that CheAΔP2 autophosphorylated and responded to receptor control normally but had reduced rates of phosphotransfer to CheB and CheY. This defect lowered the frequency of tumbling episodes during swimming and impaired chemotactic ability. However, expression of additional P1 domains in the CheAΔP2 mutant raised tumbling frequency, presumably by buffering the irreversible loss of CheAΔP2-generated phosphoryl groups from CheB and CheY, and greatly improved its chemotactic ability. These findings suggest that P2 is not crucial for CheA signaling specificity and that the principal determinants that favor appropriate phosphoacceptor partners, or exclude inappropriate ones, most likely reside in the P1 domain.
CheA, a cytoplasmic kinase controlled by transmembrane receptors, plays a central role in the chemotactic signaling pathway of Escherichia coli. CheA autophosphorylates at a histidine residue by using ATP as the phosphoryl donor (11) and then donates its phosphoryl groups to two response regulators, CheB and CheY, thereby activating their signaling functions. Phospho-CheY interacts with the switching apparatus of the flagellar motors to augment clockwise (CW) rotation (5, 42); phospho-CheB regulates a subsequent sensory adaptation process (4, 20). E. coli contains many response regulator proteins whose structures and phosphotransfer chemistries are homologous to those of CheB and CheY (reviewed in references 33, 38, and 43). Although CheA can be made to donate phosphoryl groups to heterologous response regulators in vitro (9, 30, 41), such reactions are relatively inefficient and evidently do not interfere with chemotactic signaling in vivo. This report investigates a structural feature, unique to the CheA subfamily of histidine kinases, that might account for its signaling specificity.
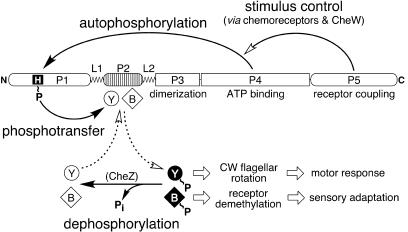
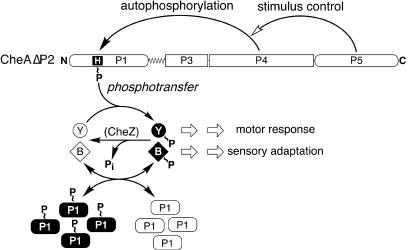
CheA has a modular organization, with different signaling functions allocated to different domains (Fig. 1). The central dimerization (P3) and ATP-binding (P4) domains are flanked by a C-terminal domain (P5) that couples autophosphorylation activity to receptor control (7) and by two N-terminal domains, P1 and P2 (27, 39), that are involved in phosphotransfer signaling. P1 contains the CheA phosphorylation site, His-48, and interacts with the P4 domain during autophosphorylation and with CheB and CheY in subsequent phosphotransfer reactions. The P2 domain binds CheY and CheB with micromolar affinity (18, 39) and increases CheA phosphotransfer rates (36, 37). Binding to P2 does not appear to induce conformational changes in CheB or CheY that appreciably influence their phosphotransfer reactions with the P1 domain, because P2 domains in trans do not affect CheA phosphotransfer rates (25, 37).
FIG. 1.
CheA domain organization and signaling activities. The relative lengths of the CheA domains (P1 to P5) and linkers (L1 and L2) are proportional to the sizes of their primary structures. The N-terminal P1 domain accepts a phosphoryl group at His-48 via interaction with the ATP-binding P4 domain. The C-terminal P5 domain enables chemoreceptors, assisted by the CheW coupling protein, to control CheA autophosphorylation activity in response to attractant or repellent stimuli. The ensuing motor response and subsequent sensory adaptation are controlled by the levels of phosphorylated CheY (Y∼P), which enhances CW flagellar rotation, and phosphorylated CheB (B∼P), which hydrolyzes glutamyl-methyl esters in the signaling domains of the chemoreceptors. The phosphorylated forms of these response regulators turn over rapidly through self-catalyzed hydrolysis. The CheZ protein further enhances dephosphorylation of Y∼P (but not B∼P). CheY and CheB acquire their phosphoryl groups from phospho-CheA. The P2 domain of CheA is not essential for these phosphotransfer reactions but rather enhances their rate by reversibly binding the unphosphorylated forms of CheY and CheB to increase phosphoacceptor concentrations in the vicinity of the P1 phosphodonor.
The P2 domain could conceivably impart specificity to CheA's signaling transactions by increasing the local concentrations of CheB and CheY, but not those of other response regulator proteins, through its binding interactions. To test this “target acquisition” model, we constructed a P2-less mutant of CheA and examined its signaling behavior in vitro and in vivo. We found that the CheAΔP2 protein had essentially wild-type autophosphorylation activity, which responded normally to chemoreceptor control. However, the mutant protein exhibited defects in phosphotransfer to both CheB and CheY, consistent with an earlier rapid kinetics study that demonstrated a substantial contribution of the P2 domain to the rate of CheA-CheY phosphotransfer (37). CheAΔP2 mutant cells, presumably owing to reduced phosphotransfer rates, exhibited predominantly counterclockwise (CCW) flagellar rotation and poor chemotactic ability. However, upward adjustment of phospho-CheB and phospho-CheY levels, by buffering the cellular pool of CheAΔP2-generated phosphoryl groups with excess P1 domains, dramatically enhanced the chemotaxis proficiency of the CheAΔP2 mutant. These results demonstrate that the CheAΔP2 protein, despite lowered phosphotransfer rates, retains sufficient signaling specificity for chemotactic responses. Thus, although the P2 domain augments the phosphotransfer process through target acquisition, it does not play a critical role in CheA signaling specificity. We conclude that the main specificity determinants that prevent excessive cross talk with heterologous response regulators reside in the P1 domain of CheA.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in this work (Table 1) were all derivatives of the E. coli K-12 strain RP437, our reference wild-type strain for chemotaxis studies (32).
TABLE 1.
Strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant properties | Reference |
|---|---|---|
| Strains | ||
| RP437 | thr-1 leuB6 his-4 metF159 eda-50 (chemotaxis wild type) | 32 |
| RP3098 | srlC::Tn10-300 Δ(flhD-flhB)4 | 35 |
| RP9005 | recD::mini-Tn10-1903 Δ(motB-cheA)m1111 | 34 |
| RP9535 | RP437 eda+ Δ(cheA)1643 | 19 |
| UU1120 | RP437 eda+ | This work |
| UU1121 | RP437 eda+ Δ(cheA)150-247ΩPA1 (cheAΔP2) | This work |
| Plasmids | ||
| pTM30 | IPTG-inducible Ptac expression vector | 27 |
| pEK46 | pUC118 motA-motB-cheA-cheW | 14 |
| pTM46 | pEK46 Δ(cheA)150-247ΩPA1 (CheAΔP2) | This work |
| pKJ9 | pTM30 (cheA) | This work |
| pKJ9-1 | pKJ9 (cheA) SacII-150 | This work |
| pKJ9-2 | pKJ9 (cheA) SacII-160 | This work |
| pKJ9-1.1 | pKJ9-1 Δ(cheA)150-247ΩPA1 (CheAΔP2) | This work |
| pKJ9-1.2 | pKJ9-1 Δ(cheA)150-247ΩPA2 | This work |
| pKJ9-2.1 | pKJ9-2 Δ(cheA)160-247ΩPA1 | This work |
| pKJ9-2.2 | pKJ9-2 Δ(cheA)160-247ΩPA2 | This work |
| pAG3 | pKJ9 cheA(1-149) | 10 |
| Oligonucleotides | ||
| kj-1 | 5′-CAGCACCAGCTGCTCCACCGCGGGCTAGCGCC-3′ (PA linker [top strand]) | This work |
| kj-2 | 5′-GGCCGGCGCTAGCCCGCGGTGGAGCAGCTGGTGCTGGC-3′ (PA linker [bottom strand]) | This work |
| kj-3 | 5′-AGTGAACCGCGGGATGAGCAG-3′ (SacII site at cheA codon 150) | This work |
| kj81 | 5′-GTGAGCATGGATATAAGC-3′ (upstream cheA primer) | This work |
| kj82 | 5′-TCAGGCGGCGGTGTTCGC-3′ (downstream cheA primer) | This work |
| kj63-3 | 5′-CGCCGCGGCGAATTATCC-3′ (SacII site at cheA codon 160) | This work |
Plasmids.
Plasmids central to this work are listed in Table 1. All are derivatives of pBR322 (6) and confer resistance to ampicillin. All except pEK46 and pTM46 carry lacI and an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible Ptac promoter driving transcription of a multiple cloning region containing the ribosome-binding site and translation start from the cheY gene of E. coli (29).
Media and culture conditions.
T broth (10 g of tryptone and 5 g of NaCl per liter) was routinely used for growth of bacterial strains, generally at 35°C. IPTG was purchased from Promega Corp. (Madison, Wis.). Ampicillin was purchased from Sigma Chemical Co. (St. Louis, Mo.) and used at a final concentration of 100 μg/ml in solid and liquid media.
Behavioral assays.
Chemotactic ability was measured on semisolid tryptone agar (T broth plus 2.6 g of agar per liter) (31). For strains harboring plasmids, ampicillin was added to a final concentration of 50 μg/ml. Flagellar rotation patterns were determined by cell tethering with antiflagellar serum (31).
Construction of pKJ9.
Pertinent features of the cheA plasmid pKJ9 have been described previously (10). It was constructed by PCR amplification of the cheA coding region in RP437 with primers kj81 and kj82 (Table 1). The PCR fragment was blunt ended by treatment with the Klenow fragment of DNA polymerase and inserted in the forward orientation at the SmaI site of pBluescript SK(+) (2). The cheA coding region was subsequently excised at the flanking PstI (upstream) and BamHI (downstream) sites in the pBluescript vector and inserted at the corresponding sites into the pTM30 vector (Table 1).
Construction of CheAΔP2 derivatives of pKJ9.
The cheA coding region was excised from pKJ9 as a PstI-BamHI fragment and inserted into the pALTER-1 phagemid (Promega Corp.). Primers kj-3 and kj63-3 (Table 1) were used to introduce a SacII site at cheA codon 150 or 160, according to the supplier's instructions for the Promega in vitro mutagenesis system. The desired cheA mutations were confirmed by DNA sequencing and then transferred back into pTM30 as a PstI-BamHI restriction fragment, yielding pKJ9-1 and pKJ9-2 (Table 1). To anneal oligonucleotides kj-1 and kj-2 (Table 1), 6 μg of each were mixed in an Eppendorf tube with annealing buffer (125 mM Tris [pH 7.6], 15 mM MgCl2, 10 mM dithiothreitol) in a total volume of 20 μl, incubated for 3 min at 95°C, and then allowed to cool to room temperature over the course of an hour. Plasmids pKJ9-1 and pKJ9-2 were digested with SacII and EagI and then ligated with 10 μl of the annealed linker mixture to produce plasmids pKJ9-1.1 and pKJ9-2.1 (Table 1). In the same manner, a second copy of the linker was inserted into these plasmids to create pKJ9-1.2 and pKJ9-2.2 (Table 1).
Construction of UU1121.
The cheAΔP2 deletion was transferred from pKJ9-1.1 to pEK46 by restriction fragment replacement, yielding plasmid pTM46. Linearized pTM46 was used to transform RP9005, and the transformation mixture was streaked onto semisolid tryptone plates. Motile transformants migrating away from the streak of nonmotile recipient cells were picked and shown to carry the cheAΔP2 deletion by PCR tests. The cheAΔP2 allele in one of these recombinants was subsequently transferred to RP437 by cotransduction with the eda locus (31). Transductants were screened by PCR to identify ones that had inherited the donor cheAΔP2 allele. One strain carrying cheAΔP2 (UU1121) and an isogenic cheA+ strain (UU1120) were kept for further analysis.
Protein purification.
CheA and CheAΔP2 (13). CheW (3), CheB (12), CheY (21), and CheZ (13) were expressed from plasmids in strain RP3098 and purified by published procedures.
Phosphorylation assays.
All reactions were carried out in phosphorylation buffer (50 mM Tris-HCl [pH 7.5], 50 mM KCl, and 5 mM MgCl2) at room temperature. CheA autophosphorylation assays were performed as described previously (3). 32P-CheA and 32P-CheAΔP2 were purified and used in CheB and CheY phosphotransfer assays as described previously (28). Receptor coupling assays were done with membranes containing the serine receptor Tsr as described previously (26). In all assays, 2 μl-reaction samples were removed at various times and added to 10 μl of sodium dodecyl sulfate protein sample buffer (15) to stop the reaction. Reaction products were separated by electrophoresis on sodium dodecyl sulfate-containing 16.5% polyacrylamide gels and quantified with a Molecular Dynamics PhosphorImager (28).
RESULTS
Construction of CheAΔP2.
We based our design of a P2-less version of CheA on the structure of FrzE, a CheA homologue of Myxococcus xanthus that lacks a P2 domain (1, 23, 24). In FrzE a proline- and alanine-rich linker, approximately 130 residues long, connects the P1 and P3/P4 domains. In the CheA proteins of E. coli and Salmonella enterica serovar Typhimurium, the P2 domain is flanked by flexible linkers, 20 to 30 residues long, whose sequences are not highly conserved (27) (Fig. 1). Accordingly, we replaced the P2 coding region, and joined the flanking L1 and L2 segments, with a proline- and alanine-rich (PA) linker that was similar to, but shorter than, the linker in FrzE. To vary the length of the connection between the P1 and ATP-binding domains, in case it proved to be critical for CheA autophosphorylation activity, the construction strategy allowed for insertion of multiple, tandem copies of the PA linker.
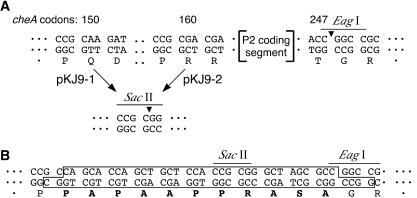
CheA constructs with deletions of P2 were made in plasmid pKJ9, which carries a wild-type cheA gene expressed from an inducible Ptac promoter (10). The construction details are summarized in Fig. 2. We first introduced a unique SacII restriction site at either codon 150 or 160 in the L1 coding region (Fig. 2A). The sequence change at codon 160 did not alter the amino acid it specified (arginine), whereas the change at codon 150 did (glutamine to arginine). Both mutant plasmids, pKJ9-1 and pKJ9-2, complemented a Δ(cheA) host strain (RP9535) as proficiently as did the parent plasmid (data not shown), demonstrating that neither change had any discernible effect on CheA function. The cheA coding segment between the introduced SacII sites in L1 and a unique EagI restriction site in L2 was then replaced with the double-stranded oligonucleotide shown in Fig. 2B to introduce the PA linker. The linker oligonucleotide contained an internal SacII site to permit introduction of additional copies of the PA linker in the same manner. In all, we constructed four P2-less CheA plasmids (pKJ9-1.1, pKJ9-1.2, pKJ9-2.1, and pKJ9-2.2) which had either one or two tandem copies of the PA linker beginning at either codon 150 or 160 in the L1 segment (Table 1).
FIG. 2.
Strategy for constructing cheA genes with deletions of P2. (A) Features of cheA flanking the P2 coding segment. A SacII restriction site was introduced at codon 150 (pKJ9-1) or codon 160 (pKJ9-2) to permit excision of the P2 coding segment by double digestion with SacII and EagI enzymes. (B) The PA linker. The boxed sequence depicts the double-stranded linker made by annealing the complementary single-stranded oligonucleotides kj-1 and kj-2 (Table 1). The PA linker carried a SacII-compatible overhang at one end and an EagI-compatible overhang at the other for joining the P2-excised coding segments. An internal SacII site in the PA linker permitted additional excision-replacement cycles for creating tandem PA linker inserts.
All four mutant plasmids behaved similarly in preliminary tests. Their CheA expression levels were identical to that of pKJ9 over a range of IPTG concentrations (data not shown), indicating that the proteins with deletions of P2 were similar in synthesis and stability to the wild type. Moreover, at IPTG concentrations above 10 μM, both the parental and mutant plasmids caused episodes of CW flagellar rotation in tethered cells of RP9535 [Δ(cheA)] (data not shown), indicating that the mutant proteins were capable of autophosphorylation and subsequent phosphotransfer to CheY. However, unlike pKJ9, the mutant plasmids failed to complement RP9535 for chemotaxis at any level of induction (data not shown), suggesting that one or more of the CheA signaling reactions was impaired. To examine the activities of CheA with a deletion of P2 in more detail, we purified and characterized the mutant protein made by plasmid pKJ9-1.1, which carries the Δ(cheA)150-247ΩPA1 alteration, in which one copy of the PA linker replaces cheA codons 150 to 247. Hereafter, we refer to this cheA allele as cheAΔP2 and to its gene product as CheAΔP2.
In vitro assays of CheA function.
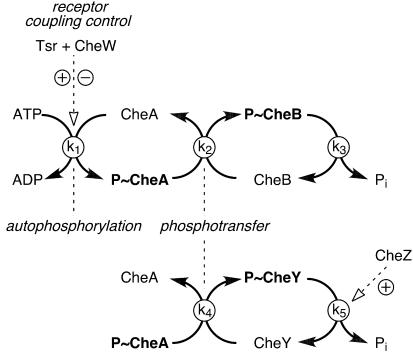
The CheA reactions involved in phosphorelay signaling are summarized in Fig. 3. CheA catalyzes its own phosphorylation, using ATP, and then serves as a phosphodonor in transfer reactions catalyzed by CheB or CheY. Finally, the phosphoryl groups on CheB and CheY are released as inorganic phosphate. In the case of phospho-CheY, the dephosphorylation step can be accelerated by CheZ. In chemoreceptor signaling complexes, CheA autokinase activity responds to the signaling state of the receptor. CW-signaling (e.g., unliganded) receptors stimulate the CheA autophosphorylation rate more than 100-fold, whereas CCW-signaling (e.g., attractant-bound) receptors inhibit CheA to approximately its uncoupled level of activity.
FIG. 3.
The phosphorelay signaling reactions of chemotaxis. Only the net forward rates are depicted: CheA autophosphorylation (which is subject to modulation in signaling complexes with chemoreceptors and CheW), phosphotransfer from phospho-CheA (P∼CheA) to CheB and CheY, and the subsequent dephosphorylation of phospho-CheB (P∼CheB) and phospho-CheY (P∼CheY). Note that dephosphorylation of P∼CheY (but not that of P∼CheB) is accelerated by CheZ.
Based on the reaction scheme depicted in Fig. 3, we devised assays to compare the autophosphorylation, receptor control, and phosphotransfer abilities of CheA and CheAΔP2. Two general considerations influenced our experimental designs. (i) The autophosphorylation and phosphotransfer reactions are readily reversible (10, 40). Accordingly, we chose reactant concentrations and stoichiometries that minimized the reverse reactions to facilitate comparison of their net forward rates, which are the ones depicted in Fig. 3. (ii) At physiological component concentrations, the phosphotransfer and receptor-stimulated autophosphorylation reactions are exceedingly fast and cannot be directly measured by manual methods (17, 36, 37). To evaluate those activities, we chose conditions that either slowed the reactions to directly measurable rates or enabled us to determine the steady-state levels of reaction intermediates and thereby infer their relative turnover rates. The rationale, design, and analysis of those experiments are detailed in the relevant sections below.
Autophosphorylation of CheAΔP2.
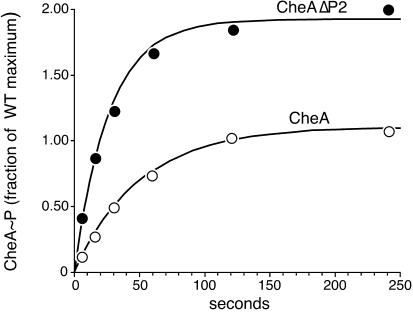
When CheA is not coupled to receptor control, its autophosphorylation rate (Fig. 3, k1) depends mainly on ATP concentration (Km ≈ 200 μM) and is low enough to measure by manual methods (half-life of ∼15 s). Thus, CheA and CheAΔP2 were mixed with a saturating level of [γ-32P]ATP (1 μM), and the formation of radiolabeled protein was monitored over time (Fig. 4). Both reactions followed pseudo-first-order kinetics, but the autophosphorylation activity of CheAΔP2 differed from that of CheA in two respects. First, the rate constant was about twofold greater for the mutant protein (0.024 ± 0.005 s−1 for CheA and 0.046 ± 0.011 s−1 for CheAΔP2 [averages and standard deviations of five determinations]). The higher specific activity of CheAΔP2 is most likely a consequence of changing the length and nature of the segment connecting the substrate histidine to the ATP-binding site in the catalytic center, which could allow more frequent productive encounters between them. Second, the steady-state phosphorylation level of CheAΔP2 was consistently severalfold higher than that of wild-type CheA (Fig. 4). We have not investigated the basis for this difference, but it may be that the forward (ATP-mediated) and backward (ADP-mediated) reactions are differentially affected by the removal of the P2 domain or that a greater proportion of the His-48 substrate sites are accessible to catalysis in the mutant protein. Alternatively, CheAΔP2 might be phosphorylated at one or more sites in addition to His-48, but this seems unlikely in view of the fact that all of its phosphoryl groups are available for transfer to CheB and CheY (see below).
FIG. 4.
Receptor-uncoupled autophosphorylation activity ofCheAΔP2. Reaction mixtures contained purified CheA and CheAΔP2 (5 μM) and [γ-32P]ATP (1 mM). The data points were fitted to an exponential function to determine the pseudo-first-order rate constant and steady-state phosphorylation level of each reaction (10).
Receptor control of CheAΔP2 autophosphorylation.
CheA and CheAΔP2 were mixed with the coupling protein CheW and membranes containing the serine chemoreceptor, Tsr, to form ternary signaling complexes. The autophosphorylation rate of CheA in such complexes is typically too high to measure directly without rapid-reaction instruments (40). To slow the receptor-stimulated autophosphorylation reaction, we used a limiting ATP concentration (45 nM) and determined the initial rate of the reactions (before ADP product levels had risen sufficiently to cause a significant reverse reaction). We fitted the CheA and CheAΔP2 data points to single exponentials to obtain a pseudo-first-order rate constant for each reaction. Under these conditions, the autophosphorylation activities of both CheA and CheAΔP2 were stimulated about 150-fold (data not shown). The formation of phospho-CheAΔP2 was about twofold faster and its extrapolated steady-state level was about twofold higher than those for phospho-CheA (data not shown), reminiscent of the activity differences seen in the absence of receptor coupling. No stimulation was seen for either CheA protein if CheW was omitted from the coupling reactions, implying that CheAΔP2 forms ternary signaling complexes in the same manner as CheA (data not shown). Moreover, saturating levels of the attractant serine reduced autophosphorylation activity to approximately the same extent in both CheA and CheAΔP2 signaling complexes (2.7-fold versus 4.5-fold [averages of seven determinations]). In summary, the receptor coupling experiments demonstrated that CheAΔP2 can form receptor signaling complexes that regulate its autophosphorylation ability in the same manner as wild-type CheA.
Phosphotransfer by CheAΔP2.
Phosphotransfer from phospho-CheA to CheY occurs on a millisecond time scale at physiological reactant concentrations (kcat of ∼750 s−1) (22, 36). In a study that used rapid-reaction instruments, Stewart et al. found that phosphotransfer from phospho-CheAΔP2 to CheY was about 25-fold slower than that from wild-type CheA but still quite fast under physiological conditions (37). To extend these studies to CheB, we devised two experimental paradigms for comparing the phosphotransfer abilities of CheA and CheAΔP2 without the use of rapid-reaction instruments. In describing those experiments, we refer to the reactions and rate constants summarized in Fig. 3.
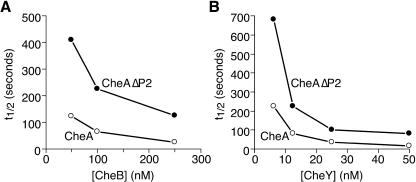
In the first experiment, CheA and CheAΔP2 were prelabeled by autophosphorylation with [γ-32P]ATP. We then monitored the dephosphorylation of purified phospho-CheA and phospho-CheAΔP2 upon addition of CheB (k2) or CheY (k4). The reactant stoichiometries were adjusted so that only a small fraction (<20%) of the phosphoacceptor molecules would be phosphorylated at steady state (data not shown), thereby ensuring that hydrolysis of phospho-CheB (k3) or phospho-CheY (k5, assisted by CheZ) was not rate limiting in the overall dephosphorylation reaction. To slow the phosphotransfer reaction, these experiments were done at protein concentrations well below reported physiological levels. Under these conditions, we determined the half-times for the phospho-CheA and phospho-CheAΔP2 dephosphorylation reactions at several concentrations of CheB and CheY (Fig. 5). At the lowest phosphoacceptor concentrations, dephosphorylation of phospho-CheAΔP2 by both CheB (Fig. 5A) and CheY (Fig. 5B) was approximately fourfold slower than that for phospho-CheA (4.0 ± 0.9 for CheB and 3.5 ± 1.3 for CheY). At higher phosphoacceptor concentrations, dephosphorylation was faster and the difference in phosphotransfer ability was less apparent (Fig. 5). At nominally physiological phosphoacceptor concentrations (∼1 μM and above), CheA and CheAΔP2 were indistinguishable as phosphodonors in this assay (data not shown).
FIG. 5.
Receptor-uncoupled phosphotransfer from phospho-CheAΔP2 to CheB (A) and CheY (B). Reaction mixtures contained CheB or CheY at the indicated concentrations and, in the case of CheY, 0.5 μM CheZ to augment turnover of phospho-CheY. Phosphotransfer was initiated by addition of 0.5 μM 32P-CheA or 32P-CheAΔP2, and the phosphodonor levels were monitored for 600 s (14 time points) for CheB and for 120 s (seven time points) for CheY to determine their half-lives (t1/2) at each phosphoacceptor concentration tested.
To compare the phosphodonor abilities of phospho-CheA and phospho-CheAΔP2 at more physiologically relevant concentrations, we set up a coupled reaction sequence (Fig. 3) beginning with CheA or CheAΔP2 autophosphorylation (k1), followed by phosphotransfer to CheB (k2) or CheY (k4), and ending with dephosphorylation of phospho-CheB (k3) or phospho-CheY (k5). We devised conditions in which phosphotransfer was the rate-limiting step in the overall flow of phosphate groups from ATP to Pi and examined the effect of phosphoacceptor concentration on the steady-state levels of phospho-CheB and phospho-CheY. To ensure that the phosphodonor supply was not rate limiting in the overall reaction scheme, we assembled stimulatory chemoreceptor signaling complexes and used a saturating concentration of ATP to accelerate the autophosphorylation step. Because CheA and CheAΔP2 have similar receptor-stimulated autophosphorylation rates at subsaturating ATP levels (see above), we assumed that their rates would also be comparable at ATP excess. Moreover, we assumed that those rates would not be influenced by changes in the phosphoacceptor concentration. At an initial concentration of 1 mM ATP, the level of phosphodonors (phospho-CheA or phospho-CheAΔP2) reached steady state by 10 s and remained at steady state for at least 30 s (data not shown). Over that interval, less than 5% of the ATP was consumed in the course of the reactions (data not shown). Under these conditions, we found that the proportion of CheA or CheAΔP2 molecules in the phosphorylated form at steady state dwindled with increasing amounts of phosphoacceptor in the reaction mixtures (data not shown), indicating that until the concentration-dependent phosphotransfer reaction outpaced the concentration-independent autophosphorylation reaction, phosphodonor production was not rate limiting. The phosphoacceptor concentrations that produced half-maximal steady-state levels of phospho-CheA were similar to their in vivo levels (∼2 μM for CheB and ∼1 μM for CheY [data not shown]). In this concentration range, the steady-state level of phospho-CheB or phospho-CheY in the reactions should reflect the ratios of their phosphotransfer to dephosphorylation rates (k2/k3 or k4/k5). We assumed that the rates of the dephosphorylation reactions (k3 and k5) were independent of phosphoacceptor concentration and the nature of the phosphodonor (phospho-CheA or phospho-CheAΔP2). Thus, any phosphodonor-dependent differences in the steady-state levels of phospho-CheB or phospho-CheY should reflect differences in their respective phosphotransfer rates (k2 or k4).
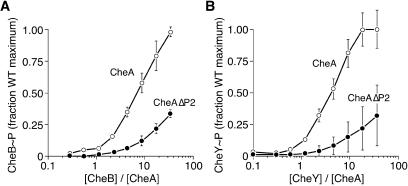
In the CheB titration experiment, the steady-state level of phospho-CheB rose substantially more slowly with phospho-CheAΔP2 as the phosphodonor than with phospho-CheA (Fig. 6A). The difference in the CheY titration was equally dramatic (Fig. 6B). (Note, as detailed in the legend to Fig. 6, that CheZ was added to the CheY titration to ensure that dephosphorylation of phospho-CheY was not rate limiting.) These results imply that phosphotransfer to both CheB and CheY was substantially slower from phospho-CheAΔP2 than from phospho-CheA. The reduction in the phosphotransfer rate of CheAΔP2 appeared to be similar for both phosphoacceptors. At physiological reactant concentrations (e.g., 1 μM phosphodonor and 1 to 10 μM phosphoacceptor), phospho-CheAΔP2 supported approximately fivefold-lower steady-state levels of phospho-CheB or phospho-CheY than did phospho-CheA. We conclude that the P2 domain of CheA enhances the CheB and CheY phosphotransfer reactions to similar extents.
FIG. 6.
Steady-state levels of phospho-CheB (A) and phospho-CheY (B) in chemoreceptor-activated reaction systems. Reaction mixtures contained 0.5 μM CheA and CheAΔP2, 4 μM membrane-embedded Tsr (serine chemoreceptor), 4 μM CheW, and variable amounts of CheB or CheY. The CheY reaction mixtures also contained 0.5 μM CheZ to augment phospho-CheY turnover. The phosphorylation cascade was initiated by addition of 1 mM [γ-32P]ATP, and the levels of phospho-CheB and phospho-CheY were measured after 10 and 20 s, when the reactions had reached steady-state.
Behavior of a cheAΔP2 mutant.
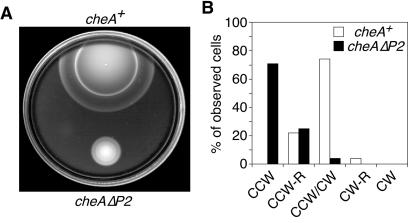
The cheAΔP2 allele was transferred from pKJ9-1.1 into the E. coli chromosome by homologous recombination, as detailed in Materials and Methods. The resulting cheAΔP2 mutant (UU1121) showed reduced chemotactic ability on semisolid tryptone agar, spreading at about 25% the rate of UU1120, an isogenic cheA+ control strain (Fig. 7A). However, the mutant colonies still exhibited cell bands characteristic of chemotactic movements, indicating that the behavioral defect of UU1121 was not complete. Conceivably, the normal receptor-mediated regulation of CheAΔP2 autophosphorylation activity permits the mutant cells some stimulus control over flagellar rotation despite a reduced rate of phosphotransfer to CheB and CheY. We compared the unstimulated flagellar rotation patterns of cheAΔP2 and cheA+ strains by cell tethering (Fig. 7B). Whereas most of the wild-type cells exhibited frequent reversals of motor rotation, more than two-thirds of the mutant cells rotated exclusively in the CCW direction during the 15-s observation period. The low frequency of CW rotation in UU1121 is consistent with a reduced rate of phosphotransfer from CheAΔP2 to CheY and most likely accounts for its chemotactic disability.
FIG. 7.
Chemotaxis phenotypes of UU1121 (cheAΔP2). (A) Colony morphology on semisolid agar. Colonies of UU1120 (cheA+) and UU1121 (cheAΔP2) were transferred with a toothpick to a semisolid tryptone agar plate and incubated at 35°C for 8 h. (B) Flagellar rotation pattern. Strains UU1120 (cheA+) and UU1121 (cheAΔP2) were analyzed by cell tethering as described in Materials and Methods. Rotating cells were assigned to five categories: exclusively CCW or CW, predominantly CCW or CW with some reversals (CCW-R and CW-R, respectively), or with frequent reversals and no significant directional bias (CCW/CW).
Phenotypic rescue of a cheAΔP2 mutant by free P1 domains.
Upon prolonged incubation on semisolid agar, UU1121 gave rise to pseudorevertants with enhanced chemotactic ability (data not shown). If its CCW-biased flagellar rotation and poor chemotactic ability are due to the reduced phosphotransfer activity of CheAΔP2, mutations that either augment the steady-state pool of phospho-CheY or otherwise increase the CW bias of the flagellar motors could conceivably alleviate those behavioral defects. To test this idea, we examined the behavior of UU1121 cells containing high levels of CheA[1-149], the P1 domain of CheA. At high stoichiometries, P1 fragments can efficiently exchange phosphoryl groups with CheB and CheY, in effect creating a reservoir of CheA-generated phosphoryl groups (10). Because phospho-P1 has a much longer half-life than phospho-CheB and phospho-CheY, CheA[1-149] fragments should serve to buffer the loss through dephosphorylation of signaling phosphates from CheB and CheY and thereby raise the steady-state levels of phospho-CheY and phospho-CheB.
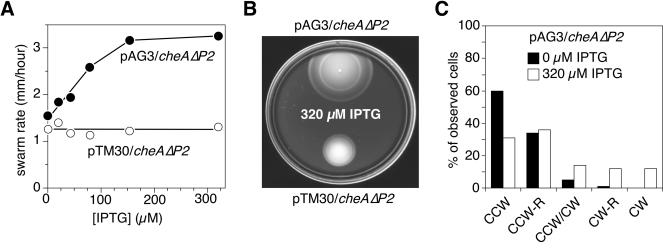
We found that pAG3, an IPTG-inducible P1 expression plasmid, produced a dramatic improvement in the chemotactic ability of UU1121 (Fig. 8). In the absence of IPTG inducer, pAG3-containing cells spread on soft agar plates at essentially the same rate as vector-containing control cells (Fig. 8A). However, in the presence of IPTG, pAG3-containing cells spread much faster than the control cells. At maximal induction (320 μM IPTG), UU1121/pAG3 spread at ∼75% of the wild-type rate, about threefold faster than control cells (Fig. 8B). This improvement in chemotactic ability was accompanied by an increase in the proportion of cells with frequent episodes of CW flagellar rotation (Fig. 8C), suggesting that the aberrant steady-state swimming pattern of UU1121, rather than a defect in stimulus detection per se, is primarily responsible for its poor chemotactic ability.
FIG. 8.
Suppression of CheAΔP2 chemotaxis defects by free P1 domains. The chemotaxis phenotypes of strain UU1121 (cheAΔP2) carrying pAG3 (expressing CheA[1-149] under IPTG-inducible control) are compared to those of UU1121 carrying pTM30 (vector control). (A) Rates of colony expansion on semisolid tryptone agar containing various IPTG concentrations. The diameters of colonies incubated at 35°C were measured at three or more time points to calculate an expansion rate. (B) Colony sizes on semisolid tryptone agar containing 320 μM IPTG. The plate was incubated for 8 h at 35°C. (C) Flagellar rotation patterns. The rotation patterns of tethered UU1121/pAG3 cells, grown without inducer or with 320 μM IPTG, were classified as described in the legend to Fig. 7B.
DISCUSSION
Signaling defects of CheAΔP2.
To investigate the contribution of the P2 domain to chemotactic signal processing by CheA, we constructed several mutant proteins in which the P2 domain had been replaced by a flexible linker and characterized one of them in detail. We found that extirpation of P2 had no deleterious effect on the CheA autophosphorylation reaction. In fact, the mutant protein autophosphorylated about twice as fast as wild-type CheA and reached roughly twofold-higher steady-state levels. CheA autophosphorylation is a trans reaction in which the phosphorylation site from one subunit of the dimer interacts with the dimerization and ATP-binding domains from the other subunit. At saturating ATP concentrations, the rate of this reaction is determined mainly by the frequency of collisional encounters between the P1 domain and the P3/P4 domain of the other subunit. However, the P1 phosphorylation sites in a wild-type dimer are probably in steric conflict (16). The absence of the P2 domain in CheAΔP2 might be expected to enhance the flexibility of the P1-P3/P4 connection, which could facilitate the autophosphorylation reaction by alleviating steric clashes between the two subunits.
In ternary complexes with CheW and the serine receptor (Tsr), CheAΔP2 responded like wild-type CheA to chemoreceptor control: the rate of autophosphorylation activity was approximately 100-fold greater than the receptor-uncoupled rate and was subject to down-regulation by serine stimuli. Evidently, the P2 domain of CheA plays no important role in the assembly or subsequent autophosphorylation behavior of receptor signaling complexes.
The principal biochemical difference between CheAΔP2 and wild-type CheA was the speed of their phosphotransfer reactions with CheB and CheY. We showed with several qualitative reaction schemes that CheAΔP2 had reduced rates of phosphotransfer to both CheB and CheY, consistent with the findings of a more precise kinetic analysis of the CheY reaction (37). Most importantly, CheAΔP2 phosphotransfer was at least 10-fold slower than that of wild-type CheA in ternary signaling complexes, which would most likely reduce the steady-state phosphorylation levels of the CheB and CheY response regulators in vivo.
Chemotaxis without P2.
CheAΔP2 cells exhibited predominantly CCW flagellar rotation, consistent with a reduced intracellular level of phospho-CheY. If phospho-CheB levels are also reduced in the CheAΔP2 mutant, we would expect the cells to have elevated receptor methylation states owing to feedback control of receptor signals by the sensory adaptation system. Although we did not examine this prediction in the present work, the low tumbling frequency of the mutant cells demonstrates that their adaptation system was not able to fully compensate for the decreased flux of phosphates to CheB and CheY. Nevertheless, CheAΔP2 cells exhibited slow chemotactic spreading on semisolid medium, indicative of functional chemoreceptor control of CheAΔP2 activity.
If the aberrant tumbling pattern of the CheAΔP2 mutant is responsible for its low rate of chemotactic spreading, mutations that raise the tumbling frequency should improve its chemotactic ability. Indeed, the CheAΔP2 mutant readily acquires mutations that enhance its rate of chemotactic spreading, suggesting that it may be amenable to a number of bypass-type suppression mechanisms. Preliminary genetic analyses of such pseudorevertants has revealed a variety of suppressor mutations located in or near the main cluster of chemotaxis and flagellar genes at min 42 on the E. coli chromosome (A. Garzón and C. Jensen, unpublished results). These suppressors might include flagellar switch mutations, partial cheZ lesions, cheY overproducers, signal-biased receptor mutations, and other alterations that promote CW flagellar rotation without compromising phosphorelay signaling.
To show more directly that the CheAΔP2 mutant was proficient at chemotactic signaling, we contrived to elevate its steady-state intracellular levels of phospho-CheB and phospho-CheY by expressing additional P1 domains in the cells, a tactic previously shown to restore chemotactic ability to a CheAΔP1P2 mutant lacking both the P1 and P2 domains (10). Indeed, elevated P1 levels improved the chemotactic ability of the CheAΔP2 mutant to about 75% of that of the wild type. The probable signaling mechanism for the P1 helping effect is shown in Fig. 9. In the CheAΔP1P2 system described previously (10), the free P1 domains have to acquire phosphoryl groups through trans interactions with the ATP-binding domains of the CheAΔP1P2 molecules before relaying them to CheB and CheY. In the CheAΔP2 system, the P1 domain resident in the CheAΔP2 molecule most likely excludes free P1 domains from interacting with the ATP-binding and catalytic center, as demonstrated in several analogous cases (10, 16). Thus, essentially all phosphoryl groups transmitted to CheB and CheY probably pass through the P1 domain tethered to the CheAΔP2 molecules (Fig. 9). The free P1 domains most likely influence the flagellar bias of CheAΔP2 by participating in reversible exchange reactions with phospho-CheB and phospho-CheY (10), as demonstrated previously (10). Because phospho-P1 is much less susceptible to hydrolysis, the exchange reactions should slow the irreversible loss of phosphoryl groups from CheB and CheY, thereby serving to buffer the phosphoryl pool. Using an empirical relationship between cellular phospho-CheY levels and CW rotational bias derived by Cluzel et al. (8), we estimate that P1 buffering would need to increase intracellular phospho-CheY levels only about 15% (from ∼2.4 to ∼2.8 μM) to produce the observed CW shift in CheAΔP2 flagellar rotation pattern. This modest increase is consistent with the extent of P1 buffering seen in our previous in vitro study (10). In summary, our model proposes that the free P1 domains create a phosphoryl group reservoir that serves to augment the steady-state phosphorylation levels of CheB and CheY.
FIG. 9.
Model of chemotactic signaling in a CheAΔP2 mutant containing free P1 domains. CheAΔP2 autophosphorylates at essentially normal rates and responds normally to chemoreceptor coupling control. However, without the target-acquiring P2 domain, the reduced rate of phosphotransfer to CheB and CheY cannot support efficient chemotaxis. A stoichiometric excess of free P1 domains may enhance chemotactic ability by buffering the CheAΔP2-generated pool of signaling phosphates through reversible exchange reactions with B∼P and Y∼P. See text for additional details of the model.
This signaling model predicts that P1-assisted CheAΔP2 cells might show delayed motor responses to stimuli, such as attractant increases, that suppress CheA autophosphorylation because the ensuing drop in the phospho-CheY level would be slowed by replenishment from the phospho-P1 buffer. This delay could be largely offset by an accompanying increase in CheZ activity, but the case for stimulus control of CheZ remains controversial. Thus, an increased response latency may be one of the factors that limits the chemotactic efficiency of P1-assisted CheAΔP2. However, on semisolid media, where metabolism-generated attractant gradients are steep, response latency is probably a less critical factor than tumbling rate. Under such conditions, the cells might show more profound defects in their response to CheA-activating stimuli, such as repellent increases, because the model provides no obvious way to accelerate the flux of phosphoryl groups to CheY. This analysis assumes that the relatively slow P2-independent phosphotransfer reaction is the rate-limiting step, as indicted by our steady-state measurements with receptor-coupled CheAΔP2. Again, this predicted signaling defect could be offset by a concomitant reduction in CheZ activity, allowing phospho-CheY levels to rise through slowed dephosphorylation. These considerations suggest that the P1-assisted CheAΔP2 signaling system might prove valuable in exploring the possible stimulus control of CheZ activity.
The signaling role of P2.
This study and a previous study (37) demonstrate that the principal signaling role of the P2 domain is to facilitate the transfer of CheA phosphoryl groups to CheB and CheY through reversible binding reactions. Despite the specificity of P2 binding behavior, two observations indicate that it is not the primary determinant of signaling specificity in the chemotaxis phosphorelay. First, the unassisted P1 domain is capable of rapid phosphotransfer to CheB and CheY, suggesting that it can discriminate among potential phosphoacceptor partners. If a discrete binding interaction is involved, it must have very low affinity, because the P1 phosphotransfer reactions to CheB and CheY are not readily saturable (37). Alternatively, P1 residues might promote phosphotransfer specificity by controlling access to the donor phosphohistidine or by participating with target residues in the phosphotransfer chemistry. Second, although the reactivity of P1 with heterologous response regulators has not been examined in detail, the proficient chemotaxis of the P1-assisted CheAΔP2 mutant might mean that P1 is not a highly promiscuous phosphodonor.
The P2 domain seems to be a relatively recent evolutionary embellishment to CheA. It was probably acquired not because it enhanced signaling specificity, but rather because it accelerated signaling reactions that were already quite specific. For example, a P2-promoted increase in the cell's phospho-CheY level might have increased turning episodes while swimming, which was possibly an improved foraging pattern. Whatever the evolutionary scenario, in the right genetic background the P2 domain is largely dispensable, at least for chemotactic movements on semisolid media. Moreover, some CheA homologues, such as FrzE of M. xanthus, lack a P2 domain and may have never had one. Instead, their signaling partner is covalently tethered to the phosphodonor molecule, ensuring a rapid phosphotransfer reaction. The P1 domain, a more ancient and essential component of the CheA kinase, probably carries the critical (and still poorly understood) determinants of signaling specificity in the chemotaxis phosphorelay.
Acknowledgments
We thank Bill McCleary (Brigham Young University) and Rick Stewart (University of Maryland) for helpful comments on the manuscript.
This work was supported by research grant GM19559 from the National Institutes of Health. K.J. was supported by a Feodor Lynen postdoctoral fellowship from the Alexander von Humboldt Foundation. A.G. was supported by a postdoctoral fellowship from the Spanish Ministry of Education under the auspices of the Fulbright Program. The Protein-DNA Core Facility at the University of Utah receives support from National Cancer Institute grant CA42014 to the Huntsman Cancer Institute.
REFERENCES
- 1.Acuna, G., W. Shi, K. Trudeau, and D. R. Zusman. 1995. The ′CheA' and ′CheY' domains of Myxococcus xanthus FrzE function independently in vitro as an autokinase and a phosphate acceptor, respectively. FEBS Lett. 358:31-33. [DOI] [PubMed] [Google Scholar]
- 2.Alting-Mees, M. A., and J. M. Short. 1989. pBluescript II: gene mapping vectors. Nucleic Acids Res. 17:9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, P., and J. S. Parkinson. 1994. Constitutively signaling fragments of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 176:6340-6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand, G. S., P. N. Goudreau, and A. M. Stock. 1998. Activation of methylesterase CheB: evidence of a dual role for the regulatory domain. Biochemistry 37:14038-14047. [DOI] [PubMed] [Google Scholar]
- 5.Barak, R., and M. Eisenbach. 1992. Correlation between phosphorylation of the chemotaxis protein CheY and its activity at the flagellar motor. Biochemistry 31:1821-1826. [DOI] [PubMed] [Google Scholar]
- 6.Bolivar, F., R. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. Gene 2:95-113. [PubMed] [Google Scholar]
- 7.Bourret, R. B., J. Davagnino, and M. I. Simon. 1993. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J. Bacteriol. 175:2097-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cluzel, P., M. Surette, and S. Leibler. 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287:1652-1655. [DOI] [PubMed] [Google Scholar]
- 9.Deretic, V., J. H. J. Leveau, C. D. Mohr, and N. S. Hibler. 1992. In vitro phosphorylation of AlgR, a regulator of mucoidy in Pseudomonas aeruginosa, by a histidine protein kinase and effects of small phospho-donor molecules. Mol. Microbiol. 6:2761-2767. [DOI] [PubMed] [Google Scholar]
- 10.Garzon, A., and J. S. Parkinson. 1996. Chemotactic signaling by the P1 phosphorylation domain liberated from the CheA histidine kinase of Escherichia coli. J. Bacteriol. 178:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hess, J. F., R. B. Bourret, and M. I. Simon. 1988. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature 336:139-143. [DOI] [PubMed] [Google Scholar]
- 12.Hess, J. F., K. Oosawa, N. Kaplan, and M. I. Simon. 1988. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53:79-87. [DOI] [PubMed] [Google Scholar]
- 13.Hess, J. F., K. Oosawa, P. Matsumura, and M. I. Simon. 1987. Protein phosphorylation is involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 84:7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kofoid, E. C., and J. S. Parkinson. 1991. Tandem translation starts in the cheA locus of Escherichia coli. J. Bacteriol. 173:2116-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Levit, M., Y. Liu, M. Surette, and J. Stock. 1996. Active site interference and asymmetric activation in the chemotaxis protein histidine kinase CheA. J. Biol. Chem. 271:32057-32063. [DOI] [PubMed] [Google Scholar]
- 17.Levit, M. N., and J. B. Stock. 2002. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 277:36760-36765. [DOI] [PubMed] [Google Scholar]
- 18.Li, J. Y., R. V. Swanson, M. I. Simon, and R. M. Weis. 1995. The response regulators CheB and CheY exhibit competitive binding to the kinase CheA. Biochemistry 34:14626-14636. [DOI] [PubMed] [Google Scholar]
- 19.Liu, J. 1990. Molecular genetics of the chemotactic signaling pathway in Escherichia coli. Ph.D. thesis. University of Utah, Salt Lake City.
- 20.Lupas, A., and J. Stock. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264:17337-17342. [PubMed] [Google Scholar]
- 21.Matsumura, P., J. J. Rydel, R. Linzmeier, and D. Vacante. 1984. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J. Bacteriol. 160:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayover, T. L., C. J. Halkides, and R. C. Stewart. 1999. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry 38:2259-22571. [DOI] [PubMed] [Google Scholar]
- 23.McCleary, W. R., and D. R. Zusman. 1990. FrzE of Myxococcus xanthus is homologous to both CheA and CheY of Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 87:5898-8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCleary, W. R., and D. R. Zusman. 1990. Purification and characterization of the Myxococcus xanthus FrzE protein shows that it has autophosphorylation activity. J. Bacteriol. 172:6661-6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrison, T. B. 1995. Structure/function studies of the chemotaxis kinase CheA of Escherichia coli. Ph.D. thesis. University of Utah, Salt Lake City.
- 26.Morrison, T. B., and J. S. Parkinson. 1997. A fragment liberated from the Escherichia coli CheA kinase that blocks stimulatory, but not inhibitory, chemoreceptor signaling. J. Bacteriol. 179:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morrison, T. B., and J. S. Parkinson. 1994. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5485-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morrison, T. B., and J. S. Parkinson. 1994. Quantifying radiolabeled macromolecules and small molecules on a single gel. BioTechniques 17:922-926. [PubMed] [Google Scholar]
- 29.Mutoh, N., and M. I. Simon. 1986. Nucleotide sequence corresponding to five chemotaxis genes in Escherichia coli. J. Bacteriol. 165:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninfa, A. J., E. G. Ninfa, A. N. Lupas, A. Stock, B. Magasanik, and J. Stock. 1988. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc. Natl. Acad. Sci. USA 85:5492-5496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkinson, J. S. 1976. cheA, cheB, and cheC genes of Escherichia coli and their role in chemotaxis. J. Bacteriol. 126:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson, V. L., D. R. Buckler, and A. M. Stock. 2000. A tale of two components: a novel kinase and a regulatory switch. Nat. Struct. Biol. 7:626-633. [DOI] [PubMed] [Google Scholar]
- 34.Sanatinia, H., E. C. Kofoid, T. B. Morrison, and J. S. Parkinson. 1995. The smaller of two overlapping cheA gene products in not essential for chemotaxis in Escherichia coli. J. Bacteriol. 177:2713-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, R. A., and J. S. Parkinson. 1980. Overlapping genes at the cheA locus of E. coli. Proc. Natl. Acad. Sci. USA 77:5370-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart, R. C. 1997. Kinetic characterization of phosphotransfer between CheA and CheY in the bacterial chemotaxis signal transduction pathway. Biochemistry 36:2030-2040. [DOI] [PubMed] [Google Scholar]
- 37.Stewart, R. C., K. Jahreis, and J. S. Parkinson. 2000. Rapid phosphotransfer to CheY from a CheA protein lacking the CheY-binding domain. Biochemistry 39:13157-13165. [DOI] [PubMed] [Google Scholar]
- 38.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 39.Swanson, R. V., S. C. Schuster, and M. I. Simon. 1993. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry 32:7623-7629. [DOI] [PubMed] [Google Scholar]
- 40.Tawa, P., and R. C. Stewart. 1994. Kinetics of CheA autophosphorylation and dephosphorylation reactions. Biochemistry 33:7917-7924. [DOI] [PubMed] [Google Scholar]
- 41.Via, L., R. Curcic, M. Mudd, S. Dhandayuthapani, R. Ulmer, and V. Deretic. 1996. Elements of signal transduction in Mycobacterium tuberculosis: in vitro phosphorylation and in vivo expression of the response regulator MtrA. J. Bacteriol. 178:3314-3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch, M., K. Oosawa, S.-I. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. USA 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]