Abstract
Surface proteins in gram-positive bacteria are frequently required for virulence, and many are attached to the cell wall by sortase enzymes. Bacteria frequently encode more than one sortase enzyme and an even larger number of potential sortase substrates that possess an LPXTG-type cell wall sorting signal. In order to elucidate the sorting pathways present in gram-positive bacteria, we performed a comparative analysis of 72 sequenced microbial genomes. We show that sortase enzymes can be partitioned into five distinct subfamilies based upon their primary sequences and that most of their substrates can be predicted by making a few conservative assumptions. Most bacteria encode sortases from two or more subfamilies, which are predicted to function nonredundantly in sorting proteins to the cell surface. Only ∼20% of sortase-related proteins are most closely related to the well-characterized Staphylococcus aureus SrtA protein, but nonetheless, these proteins are responsible for anchoring the majority of surface proteins in gram-positive bacteria. In contrast, most sortase-like proteins are predicted to play a more specialized role, with each anchoring far fewer proteins that contain unusual sequence motifs. The functional sortase-substrate linkage predictions are available online (http://www.doe-mbi.ucla.edu/Services/Sortase/) in a searchable database.
Pathogenic bacteria display an array of surface proteins to adhere to a site of infection, invade host cells, and evade the immune response. Many surface proteins are covalently attached to the cell wall by membrane-associated transpeptidases, called sortases (reviewed in references 18, 45, 48, and 53). The archetype sortase is the SrtA protein from Staphylococcus aureus, which anchors proteins that contain a C-terminal cell wall sorting signal (CWS) consisting of an LPXTG motif, followed by a hydrophobic domain and a tail of mostly positively charged residues (see Fig. 1A). An N-terminal secretion signal enables the precursor surface protein to be translocated across the membrane, where SrtA cleaves it in between the threonine and glycine residues of the LPXTG motif (47). SrtA then catalyzes the formation of an amide link between the carboxyl-group of the threonine and the cell wall precursor lipid II (57, 61), which is subsequently incorporated into the peptidoglycan via the transglycosylation and transpeptidation reactions of bacterial cell wall synthesis (66). An analysis of bacterial genomes indicates that this anchoring mechanism is conserved in gram-positive bacteria, since nearly all species encode SrtA homologs and proteins bearing a CWS (34, 55). Sortases may be excellent targets for new antimicrobial agents, since pathogens deficient in these enzymes exhibit reduced virulence (11, 12, 23, 35, 43, 46).
FIG. 1.
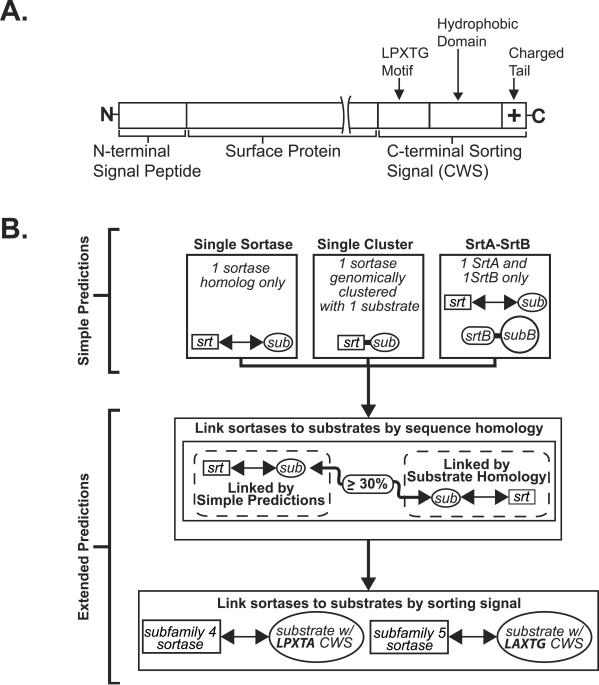
(A) Diagram illustrating a CWS-containing protein. It is composed of an N-terminal signal peptide and a C-terminal sorting signal that has a conserved LPXTG motif followed by a hydrophobic stretch of amino acids and positively charged residues at the C terminus. (B) A flowchart illustrating how functional linkages between sortase homologs and CWS-containing proteins were established. Three methods were used to make “simple” predictions: (i) single sortase, genome only contains a single sortase; (ii) single cluster, genome contains a gene cluster with a single sortase and a single CWS-containing protein; (iii) SrtA-SrtB, genome contains only SrtA and SrtB homologs. Two additional methods extended the initial predictions. With substrate sequence homology, a new functional link was made when a CWS-containing protein in one genome shared significant sequence homology (≥30%) to a previously assigned CWS-containing protein in another genome and both organisms encoded closely related sortases (see the text). With unique sorting signals, additional CWS-containing proteins were linked to their cognate sortase by examination of their sorting signal motif (subfamily-4 and subfamily-5 enzymes process the motifs LPXTA and LAXTG, respectively).
A large number of proteins are related to SrtA, but their functions have yet to be determined (55). Consistent with playing a role in surface protein chemistry, all SrtA homologs contain appropriately positioned active site residues (SrtA residues H120 and C184) (32) and transmembrane segments, and their genes are frequently clustered with genes encoding CWS-containing proteins. Moreover, several homologs have been shown to be directly involved in protein anchoring, since their elimination prevents the display of surface proteins (11, 23, 26, 54). Although the SrtA protein recognizes the sequence LPXTG within its substrates, this motif is widely varied, and a second S. aureus sortase, called SrtB, processes proteins bearing the sequence NPQTN (46). Different types of sortases may be able to attach proteins to distinct positions within the cell wall, since recent studies have shown that the cross-linked protein products of SrtA and SrtB exhibit distinct electrophoretic mobilities after cell wall digestion (44).
Many bacteria encode as many as seven sortases and 40 CWS-containing proteins. The large number of SrtA-related proteins has led to the suggestion that many perform functions other than protein anchoring and has made it difficult to predict the cognate sortase or sortases responsible for displaying many surface proteins. It is also not known whether these enzymes act nonredundantly to selectively sort proteins to the cell surface or whether they have degenerate functions. This is of major importance, because antimicrobial compounds targeted towards a particular sortase could prove ineffective if the enzymes have redundant functions or drug-resistant strains could readily evolve by horizontal gene transfer. We analyzed 72 microbial genomes and were able to conservatively predict the cognate sortase responsible for processing ∼77% of the CWS-containing proteins. Our results suggest that sortase enzymes nonredundantly sort proteins to the cell surface by selectively recognizing distinct sequence motifs within the CWS.
MATERIALS AND METHODS
Identification of sortase homologs and clustering into families.
The sequences of 241 bacterial genomes, representing 96 species of bacteria, were searched using PSI-BLAST (2) for sequence homologs of SrtA and SrtB from S. aureus (NCBI BLAST with microbial genomes: www.ncbi.nlm.nih.gov/sutils/genom_table.cgi). In addition, the genomes of Streptococcus sobrinus (The Institute for Genomic Research: tigrblast.tigr.org/ufmg/index.cgi?database=s_sobrinus|seq) and Streptomyces avermitilis (National Institute of Technology) were searched. After four iterations of PSI-BLAST, proteins that had e-values less than 0.0001 were retained. Each sequence was inspected to verify that it contained appropriately positioned catalytic cysteine and histidine residues and an N-terminal signal peptide (SignalP, version 2.0) (49, 50). Table 2 lists the completely sequenced genomes found to encode at least one sortase-related protein. The homologs were then clustered into subfamilies using a matrix of BLAST scores describing the relationship of each homolog with respect to all the other sortases. For inclusion, each member of a subfamily had to have an expectation cutoff value of at least 10−20, 30% sequence identity, and an alignment overlap length of at least 100 amino acids to every other member of the subfamily. A hidden Markov model (HMM) was then constructed for each subfamily using HMMER (20) to quantify and validate the partitioning and to classify additional sortase homologs. For inclusion into a subfamily, a sortase homolog had to have an HMM score of at least 150 with respect to the subfamily model. The HMM profiles for each subfamily are provided in the database and can be used with the program HMMER (http://hmmer.wustl.edu) to classify sortases.
TABLE 2.
Phylogenetic distribution of sortase homologs and CWS-containing proteins
| Species | Sourcea | No. of homologs in sortase subfamilyb
|
CWS substratesc
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | 3 | 4 | 5 | Unclassifiedd
|
No. in genome | No. assign. | |||
| Clustered | Not clustered | |||||||||
| Actinobacteria (high-G+C gram-positive bacteria) | ||||||||||
| Corynebacterium diphtheriae | Sanger | 5 | 1 | 16 | 13 | |||||
| Corynebacterium efficiens | 51 | 4 | 1 | 8 | 4 | |||||
| Corynebacterium glutamicum | 31 | 1 | 1 | 1 | ||||||
| Tropheryma whipplei Twist | 9 | 1 | 1 | 1 | ||||||
| Tropheryma whipplei TW08/27 | 9 | 1 | 1 | 1 | ||||||
| Streptomyces avermitilis | 30 | 4 | 13 | 13 | ||||||
| Streptomyces coelicolor | 8 | 2 | 5 | 13 | 13 | |||||
| Thermobifida fusca | DOE | 1 | 1 | 1 | ||||||
| Bifidobacterium longum DJ010A | DOE | 2 | 1 | 10 | 4 | |||||
| Bifidobacterium longum NCC2705 | 62 | 2 | 1 | 13 | 5 | |||||
| Chloroflexi (green nonSulfur bacteria) | ||||||||||
| Chloroflexus aurantiacus | DOE | 4 | 4 | 0 | ||||||
| Firmicutes (gram-positive bacteria) | ||||||||||
| Bacillus | ||||||||||
| Bacillus anthracis A2012 | 33 | 1 | 1 | 1 | 9 | 5 | ||||
| Bacillus anthracis Ames | 58 | 1 | 1 | 1 | 10 | 4 | ||||
| Bacillus anthracis KrugerB | TIGR | 1 | 1 | 1 | 11 | 4 | ||||
| Bacillus anthracis WesternNA | TIGR | 1 | 1 | 1 | 11 | 4 | ||||
| Bacillus cereus ATCC 10987 | TIGR | 1 | 1 | 4 | 1 | 19 | 11 | |||
| Bacillus cereus ATCC 14579 | 33 | 1 | 1 | 1 | 2 | 13 | 9 | |||
| Bacillus halodurans | 68 | 1 | 2 | 3 | 8 | 8 | ||||
| Bacillus subtilis | 39 | 1 | 1 | 2 | 2 | |||||
| Geobacillus stearothermophilus | OU | 1 | 1 | 1 | ||||||
| Oceanobacillus iheyensis | 69 | 2 | 3 | 3 | ||||||
| Listeria | ||||||||||
| Listeria innocua | 24 | 1 | 1 | 35 | 35 | |||||
| Listeria monocytogenes 4b | TIGR | 1 | 1 | 40 | 40 | |||||
| Listeria monocytogenes EGD-e | 24 | 1 | 1 | 38 | 38 | |||||
| Staphylococcus | ||||||||||
| Staphylococcus aureus COL | TIGR | 1 | 1 | 21 | 21 | |||||
| Staphylococcus aureus MRSA252 | Sanger | 1 | 1 | 20 | 20 | |||||
| Staphylococcus aureus MSSA476 | Sanger | 1 | 1 | 22 | 22 | |||||
| Staphylococcus aureus MW2 | 4 | 1 | 1 | 22 | 22 | |||||
| Staphylococcus aureus Mu50 | 40 | 1 | 1 | 20 | 20 | |||||
| Staphylococcus aureus N315 | 40 | 1 | 1 | 21 | 21 | |||||
| Staphylococcus aureus NCTC 8325 | OU | 1 | 1 | 18 | 18 | |||||
| Staphylococcus epidermidis RP62A | TIGR | 1 | 11 | 11 | ||||||
| Staphylococcus epidermidis ATCC 12228 | CNHGC | 1 | 1 | 11 | 8 | |||||
| Enterococcus | ||||||||||
| Enterococcus faecalis | 56 | 1 | 1 | 1 | 29 | 13 | ||||
| Enterococcus faecium | DOE | 5 | 2 | 15 | 7 | |||||
| Lactobacillaceae | ||||||||||
| Lactobacillus gasseri | DOE | 1 | 13 | 13 | ||||||
| Lactobacillus plantarum | 38 | 1 | 10 | 10 | ||||||
| Pediococcus pentosaceus | DOE | 1 | 3 | 3 | ||||||
| Leuconostocaceae | ||||||||||
| Leuconostoc mesenteroides | DOE | 2 | 3 | 0 | ||||||
| Oenococcus oeni MCW | DOE | 1 | 1 | 1 | ||||||
| Streptococcaceae | ||||||||||
| Lactococcus lactis subsp. Lactis | 13 | 1 | 1 | 8 | 3 | |||||
| Streptococcus agalactiae 2603V/R | 70 | 1 | 5 | 23 | 11 | |||||
| Streptococcus agalactiae A909 | TIGR | 1 | 4 | 19 | 9 | |||||
| Streptococcus agalactiae NEM316 | 25 | 1 | 4 | 35 | 14 | |||||
| Streptococcus equi | Sanger | 1 | 1 | 27 | 15 | |||||
| Streptococcus gordonii | TIGR | 1 | 22 | 22 | ||||||
| Streptococcus mitis | TIGR | 1 | 14 | 14 | ||||||
| Streptococcus mutans | 1 | 1 | 6 | 6 | ||||||
| Streptococcus pneumoniae R6 | 29 | 1 | 14 | 14 | ||||||
| Streptococcus pneumoniae TIGR4 | 71 | 1 | 3 | 15 | 11 | |||||
| Streptococcus pneumoniae 23F | Sanger | 1 | 13 | 13 | ||||||
| Streptococcus pneumoniae 670-6B | TIGR | 1 | 3 | 11 | 9 | |||||
| Streptococcus pyogenes MI GAS | 22 | 1 | 1 | 1 | 15 | 10 | ||||
| Streptococcus pyogenes MGAS315 | 10 | 1 | 1 | 16 | 13 | |||||
| Streptococcus pyogenes MGAS8232 | 65 | 1 | 1 | 15 | 12 | |||||
| Streptococcus pyogenes Manfredo | Sanger | 1 | 1 | 15 | 14 | |||||
| Streptococcus pyogenes SSI-I | GIRC | 1 | 1 | 15 | 13 | |||||
| Streptococcus sobrinus | TIGR | 1 | 12 | 12 | ||||||
| Streptococcus suis | Sanger | 1 | 4 | 20 | 9 | |||||
| Streptococcus uberis | Sanger | 1 | 9 | 9 | ||||||
| Clostridia | ||||||||||
| Clostridium acetobutylicum | 52 | 1 | 2 | 2 | ||||||
| Clostridium botulinum | Sanger | 1 | 1 | 1 | ||||||
| Clostridium difficile | Sanger | 1 | 7 | 7 | ||||||
| Clostridium perfringens ATCC 13124 | TIGR | 1 | 1 | 1 | 12 | 3 | ||||
| Clostridium perfringens 13 | 64 | 1 | 1 | 1 | 14 | 3 | ||||
| Clostridium tetani | 15 | 1 | 3 | 3 | ||||||
| Ruminococcus albus | TIGR | 1 | 2 | 2 | ||||||
| Gram-positive, totale | 42 | 17 | 54 | 13 | 14 | 17 | 14 | 887 | 684 | |
Either the reference for the genome sequence is given when available or the source of the preliminary sequence data: CNHGC (Chinese National Human Genome Center), DOE (U.S. Department of Energy Joint Genome Institute), GIRC (Genome Information Research Center), OU (University of Oklahoma Advanced Center for Genome Technology), Sanger (Sanger Institute), and TIGR (The Institute for Genomic Research).
Sortase homologs are clustered into subfamilies according to sequence homology using BLAST profiles and HMMs.
C-terminal sorting signal (CWS)-containing proteins. The first column is the total number of CWS-containing proteins identified as encoded in each respective genome, whereas the second column is the number of CWS-containing proteins that can be linked to a sortase homolog.
The sortase homolog is not readily classified into a subfamily based on sequence homology. The first column, “clustered", denotes that these sortases can nevertheless be linked to a CWS-containing protein by genomic positioning. The second column denotes that these unclassified sortases are not genomically adjacent to a CWS-containing protein.
For proteobacteria (purple bacteria and relatives—gram negative), each of the following has one sortase homolog, with one CWS-containing protein encoded in the genome and one CWS-containing protein that can be linked to a sortase homolog (abbreviations for sources, given in parentheses, are explained in footnote a): Bradyrhizobium japonicum (36), Colwellia psychrerythraea (TIGR), Microbulbifer degradans (DOE), Shewanella oneidensis (27), and Shewanella putrefaciens (TIGR). This brings the total for both gram-negative and gram-positive bacteria to 892 encoded CWS-containing proteins and 689 CWS- containing proteins linked to a sortase homolog.
Identification of sortase substrates.
In order to search for CWS-containing proteins, a database of protein-coding genes was constructed from the genomic data (60). First, 4,700 potential CWS-containing proteins were identified for the following reasons: (i) they had a suitable signal peptide sequence within their first 70 amino acids (49, 50) (SignalP score, >0.6), (ii) they had a potential transmembrane segment within 50 amino acids of their C terminus (TMPred) (28), and (iii) they had at least one basic residue (arginine or lysine) within their last eight residues. Each of these proteins was then examined for the presence of conserved five- to six-amino-acid motifs immediately preceding the putative transmembrane sequence. In a second complementary approach, all protein sequences were searched for the patterns [FILMPSVY][AP]X[ATS][GAKNS] (for LPXTG-like motifs) and NPX[ST][DGNS] (for SrtB substrates) positioned 17 to 45 residues from the C terminus (34). Combined these approaches yielded 892 potential sortase substrates. It should be noted that a large number of proteins contain sequences related to LPXTG elsewhere in their primary sequences, and we have elected to consider only those proteins that possess all of the known features of a sortase substrate (a cell wall sorting signal consisting of an LPXTG-like motif, hydrophobic domain, and charged tail).
Additional CWS-containing proteins were identified in 26 other species. However, the genomes of these organisms have not been completely sequenced and were therefore not used in our analysis. Bacteria with partially sequenced genomes encoding a CWS-containing protein include the following: Actinomyces naeslundii, Actinomyces viscosus, Arcanobacterium pyogenes, Arthrobacter sp., Bacillus sp., Clostridium septicum, Desulfitobacterium hafniense, Erysipelothrix rhusiopathiae, Lactobacillus leichmannii, Lactobacillus paracasei, Lactobacillus reuteri, Listeria grayi, Listeria seeligeri, Peptostreptococcus magnus (Finegoldia magna), Staphylococcus carnosus, Staphylococcus lugdunensis, Staphylococcus saprophyticus, Staphylococcus warneri, Staphylococcus xylosus, Streptococcus constellatus, Streptococcus criceti, Streptococcus downei, Streptococcus dysgalactiae, Streptococcus intermedius, Streptococcus parasanguinis, Streptococcus salivarius, and Streptococcus thermophilus.
RESULTS
Overall strategy.
We analyzed 72 sequenced microbial genomes that contained at least one sortase homolog in order to functionally link sortase enzymes to their protein substrates (the CWS-containing proteins that it presumably anchors to the extracellular surface). First, we performed a comprehensive search to identify all proteins that are related to the S. aureus SrtA and SrtB proteins, as well as all proteins harboring a CWS. We then clustered the sortase-like proteins into distinct subfamilies based upon their primary sequences and systematically analyzed how members of each subfamily and potential substrates were distributed in different microbes.
Identification of sortase homologs and clustering into subfamilies.
Sequenced microbial genomes were searched with the program PSI-BLAST using the S. aureus SrtA and SrtB genes as seeds (2). Seventy-two genomes, representing 49 bacterial species (44 gram positive and 5 gram negative), were found to encode a total of 176 sortase homologs that share greater than 21 and 32% sequence identity with the SrtA and SrtB proteins, respectively. Forty-five genomes encode two or more enzymes, with the largest number found in Bacillus cereus (strain ATCC 10987), Streptomyces coelicolor, and Enterococcus faecium, which each encode seven homologs.
The homologs were clustered into subfamilies using a matrix of BLAST scores, and then a HMM was constructed for each subfamily to quantify and validate the partitioning. A HMM is a statistical description of the consensus sequence of each subfamily and enables a rigorous evaluation of the relatedness of a particular homolog to each subfamily. As shown in Table 1, 145 of the 176 homologs can be reliably clustered into six subfamilies (one subfamily of sortases from gram-negative bacteria and five subfamilies from gram-positive bacteria). Following the convention established by Schneewind (42, 46), two of the gram-positive subfamilies are called SrtA and SrtB, since their members have primary sequences that are most closely related to the well-characterized SrtA and SrtB proteins from S. aureus. The remaining gram-positive subfamilies are numbered 3, 4, and 5, whereas the gram-negative subfamily is also known as subfamily 6. As has been previously noted, the members of a subset of SrtA family sortases (20) are distinguished by their genomic proximity to the gene encoding DNA gyrase subunit A (found in the genera Lactococcus and Streptococcus) (37, 54). Table 2 lists how members of each subfamily are distributed in the 49 species of bacteria analyzed in this study. Additional sortases have also been identified in A. naeslundii, A. viscosus (41), Streptococcus oralis, and Streptococcus sanguinis, for which complete genomic information is lacking.
TABLE 1.
Matrix of HMM scores for sortase subfamilies
| Sortase subfamilya | HMM score for sortase subfamilyb
|
|||||
|---|---|---|---|---|---|---|
| SrtA | SrtB | 3 | 4 | 5 | Gram negative | |
| SrtA | 150-610c | −110d | −79 | −56 | −56 | −146 |
| SrtB | −155 | 548-674 | −215 | −141 | −182 | −176 |
| 3 | −10 | −146 | 150-547 | −42 | −68 | −87 |
| 4 | −103 | −158 | −72 | 281-460 | −35 | −55 |
| 5 | −72 | −163 | −143 | 7 | 380-672 | −87 |
| Gram negative | −134 | −184 | −154 | −13 | −95 | 347-630 |
Six subfamilies of sortases homologs as clustered by sequence homology. The sortase subfamilies SrtA, SrtB, 3, 4, 5, and gram-negative contain 42, 17, 54, 13, 14, and 15 homologs, respectively.
A HMM for each sortase subfamily was constructed using HMMER. A HMM score was calculated for every sortase homolog to each subfamily HMM and is indicative of the similarity between a homolog and the consensus sequence of the given sortase subfamily (the higher the score is, the more similar a sortase homolog is to a subfamily HMM).
The range of HMM scores (in boldface) exhibited by sortase homologs within a subfamily to the HMM of the same subfamily. The range of scores is indicative of the similarity of these sortase homologs to the consensus sequence and hence to one another.
One score is given when a comparison is made between a set of sortase homologs within a subfamily to the HMM of a different subfamily. The score is the highest score that a sortase homolog within a given subfamily has to the HMM of a different subfamily.
Linking sortase homologs to their substrates: simple predictions.
Two different search protocols detected 892 CWS-containing proteins encoded in the sequenced genomes that also encoded a sortase (see Materials and Methods). A large number of substrate-sortase linkages could readily be made using three distinct methods (Fig. 1B). First, 27 of the 72 genomes encode only a single sortase enzyme. These organisms also encode 153 CWS-containing proteins (17.1% of the total) that can be functionally assigned to their solitary homolog (called single sortase predictions) (Table 3). Second, in many organisms a single sortase gene is positioned next to a gene encoding a CWS-containing protein (called single cluster predictions). A total of 31 CWS-containing proteins (3.5% of the total) in 16 organisms can be assigned using this approach. Finally, functional linkages were made based upon the well-characterized sorting pathways in S. aureus, which encodes two sortases, SrtA- and SrtB-like enzymes, which anchor proteins bearing the sequence LPXTG or NPQTN, respectively, within their CWSs (46). For example, the CWS-containing proteins in Listeria innocua and Listeria monocytogenes can readily be assigned, because these bacteria also encode only SrtA- and SrtB-type sortases and putative surface proteins that possess the appropriate sequence motif. Assuming behavior similar to that for S. aureus, the cognate sortase for 257 CWS-containing proteins can be predicted (28.8% of the total, called SrtA-SrtB predictions). Combined, these three straightforward approaches enabled 433 CWS-containing proteins (48.5% of the total) to be reliably assigned to their cognate sortases (Table 3).
TABLE 3.
Results of sortase-substrate predictions
| Prediction methoda or category | No. of predicted sortase-substrate linkagesb | % of total substrates |
|---|---|---|
| Single sortasec | 145 (153) | 17.1 |
| Single sortase A-single sortase Bd | 257 (257) | 28.8 |
| Single sortase-single substrate genomic clustere | 23 (31) | 3.5 |
| Single sortase and single sortase-single substrate genomic cluster | 8 (8) | <1.0 |
| Sequence homologyf | 163 (411) | 46.0 |
| Subfamily-4 sorting signal specificity—LPXTA CWSg | 14 (24) | 2.7 |
| Subfamily-5 sorting signal specificity—LAXTG CWS | 42 (46) | 5.2 |
| Subtotal | 652 | 73.0 |
| Genomic cluster with single sortase and multiple substratesh | 37 (52) | 5.8 |
| Subtotal | 689 | 77.2 |
| Unassigned substrates | 203 | 22.8 |
| Total no. of CWS-containing proteins | 892 | 100 |
General description of method used to link a CWS-containing substrate to a sortase homolog.
First number is the sum of nonredundant linkages; i.e., linkages predicted exclusively from this method. Number in parentheses is the sum total of linkages made by prediction method, which might include predictions made by more than one method.
Genome has only one sortase homolog.
Genome has only one sortase A homolog and one sortase B homolog.
Genome has one sortase homolog genomically clustered with one CWS-containing protein.
Predictions of sortase-substrate linkages are based on sequence homology between a CWS-containing protein in one species and a CWS-containing protein(s) that has been assigned by one of the above three methods.
Predictions of sortase-substrate linkages are based on the sorting signals of the CWS-containing proteins. Subfamily-4 sortases are predicted to process CWS-containing proteins with an LPXTA motif, whereas subfamily-5 sortases are predicted to process CWS-containing proteins with a LAXTG motif.
Genome has only one sortase homolog that is genomically clustered with two or more CWS-containing proteins (number of predictions excludes SrtB genomic clusters and subfamily-5 substrate in C. diphtheriae).
Extended predictions based on sequence homology between substrates.
We reasoned that sortase enzymes that are related to one another at the primary sequence level would have orthologous functions, such that they would anchor CWS-containing proteins that were also conserved at the primary sequence level. We therefore determined whether any of the remaining unassigned CWS-containing proteins shared primary sequence homology with a previously assigned protein (greater than 30% sequence identity). If a match was found, we checked if the assigned CWS-containing protein was processed by a sortase enzyme assigned to one of the subfamilies, and if so, whether the organism encoding the unassigned CWS-containing protein also encoded a single sortase from the same subfamily. When this condition was satisfied, a new functional linkage was made. Using this strategy, 163 new CWS-containing proteins were linked to their cognate sortases, extending the total number of predictions to 596 (66.8% of 892). Importantly, this method cross-validated the initial set of predictions. Specifically, 65, 16, and 173 of the linkages made using the aforementioned single-sortase, single-cluster, and SrtA-SrtB approaches were also made by analyzing substrate sequence homology.
Extended predictions based on distinct sequence motifs within the sorting signals.
We analyzed the CWSs of the previously assigned protein substrates (596 CWS-containing proteins) to ascertain whether sortases within a given subfamily processed distinct sorting signals (Fig. 2). As expected, the substrates of the SrtA subfamily contain the LPXTG motif, and SrtB subfamily substrates contain the distinct sequence NPX[TS] (24). Subfamily-5 sortases appear to process the novel sorting signal LAXTG, which is completely conserved in their predicted substrates. Since all of the remaining unassigned LAXTG CWS proteins are also present in genomes that encode a subfamily-5 sortase, an additional 16 functional linkages can be made (Table 3). The subfamily-4 sortases in bacilli are predicted to process CWS-containing proteins with the sequence motif LPXTA[ST]. This is consistent with the finding that the genes encoding subfamily-4 sortases are genomically clustered with genes containing this motif and the finding that LPXTA[ST]-containing proteins are always encoded in genomes with a subfamily-4 sortase. The subfamily-3 sortases are predicted to process a signal similar to that recognized by the SrtA subfamily, but they are distinguished by the prevalence of a glycine residue immediately following the LPXTG motif (in 83.3% of the 54 sorting signals) and by their membrane topology (discussed below). Finally, 37 additional CWS-containing proteins could be tentatively linked to their substrates because their genes were immediately adjacent to a single sortase gene (Table 3).
FIG. 2.
Sorting signals categorized by subfamily type. The figure shows the position-specific frequency of amino acids within the sorting signals of different types of sortases. The one-letter symbol for the amino acid residue is given for each position in the six-residue motif. The font size of each letter is proportional to the frequency with which an amino acid occurs. If an amino acid appears in fewer than 8% of the substrates, then the letter does not appear in the figure. When one type of amino acid is completely conserved at a particular position of the sorting signal motif or when one type of amino acid occurs in more than 92% of the CWS-containing proteins, then only one letter is present in a position. When no amino acid type is predominant in a given position of the motif, then the amino acid types found in the motif are given in brackets.
Unassigned CWS-containing proteins.
The aforementioned approaches predicted the sortase homolog responsible for processing 77.2% of the CWS-containing proteins. Functional linkages for the remaining proteins were hindered because several species encode multiple sortases that are predicted to have degenerate CWS specificities (for example, Streptococcus agalactiae, Streptococcus equi, and B. cereus encode both subfamily-3 and SrtA-type proteins). In addition, several species contain unclassifiable sortase homologs whose CWS sequence preference is not known (Enterococcus faecalis and E. faecium).
DISCUSSION
Gram-positive bacteria encode sortase-related proteins that in S. aureus and other pathogens anchor virulence determinants to the cell surface. Because many bacteria encode more than one sortase-related protein with no known function, we performed a bioinformatics analysis. First, 176 proteins that are related to the S. aureus SrtA and SrtB proteins, as well as 892 potential protein substrates harboring a CWS, were identified. Using a combination of methods, the cognate sortase responsible for processing 77% of the CWS-containing proteins was then predicted. Based upon their primary sequences, there are five subfamilies of sortases. These include the SrtA and SrtB subfamilies, which contain proteins most closely related to the S. aureus SrtA and SrtB proteins, respectively, and three previously uncharacterized groups of related homologs, called subfamilies 3, 4, and 5. The greatest number of homologs is found in the SrtA subfamily and subfamily 3, while the remaining subfamilies are of nearly equal size (Fig. 3A). Interestingly, the majority of bacteria analyzed in this study contain homologs from at least two of the five subfamilies, with several containing multiple copies of sortases from subfamilies 3, 4, and 5 (Table 2). In addition to their primary sequences, members of each subfamily are distinguished by their membrane topology, genomic positioning, and specificity for amino acid sequence motifs within the CWSs of their predicted substrates. There is no commonly agreed-upon nomenclature for sortase enzymes. We have therefore provided in the database a conversion table that lists sortase genes that have been characterized biochemically and their corresponding name in the database.
FIG. 3.
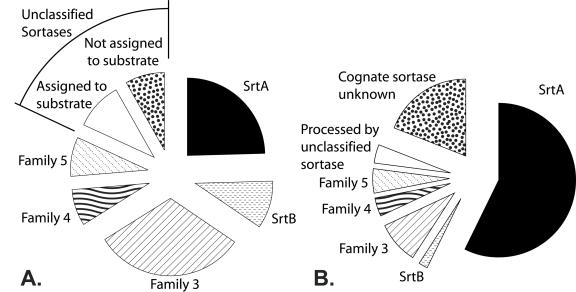
(A) Pie chart showing the distribution of sortase homologs in gram-positive bacteria. A total of 176 sortase homologs were identified in gram-positive bacteria: 42 SrtA sortases, 17 SrtB sortases, 54 subfamily-3 sortases, 13 subfamily-4 sortases, and 14 subfamily-5 sortases. (B) Pie chart showing the fraction of CWS-containing proteins that are anchored by different types of sortases. A total of 203 CWS-containing proteins were not assigned to a specific sortase. However, several lines of evidence suggest that these remaining proteins are processed by members of the SrtA subfamily (94 of the remaining 203). First, several of the unassigned substrates contain the N-terminal motif YSIRK (5). Second, they frequently contain the sequence LPXTG followed by an acidic amino acid, which appears to be an expanded recognition motif for SrtA-type enzymes (Fig. 2). Finally, most are not genomically adjacent to another sortase, similar to nearly all SrtA substrates.
The SrtA subfamily.
Several lines of evidence suggest that members of this subfamily play a housekeeping role in the cell, anchoring a large number of diverse proteins to the cell wall. First, the majority of surface proteins (a total of 511) are predicted to be anchored by SrtA-type sortases (Fig. 3B), which are distributed in a wide range of bacterial genera (Bacillus, Enterococcus, Lactobacillus, Lactococcus, Listeria, Staphylococcus, and Streptococcus). Second, bacteria always encode only a single SrtA-type homolog, which on average is predicted to anchor a large number of proteins (∼12 substrates per SrtA homolog). Third, a Pfam analysis of their predicted substrates indicates that they are functionally diverse (7). Fourth, genes encoding SrtA-type proteins are never proximal to genes encoding potential substrates. This is in contrast with other sortases, whose genes are typically clustered with a limited number of CWS-bearing substrates and thus appear to play a more specialized role in the cell. An analysis of their predicted substrates suggests that SrtA-type sortases are specific for the sequence LPXTG (Fig. 2). However, our predictions also suggest that SrtA-type proteins can process proteins in which the threonine residue is replaced by an alanine, compatible with results in recent biochemical studies (59). Assuming that members of this subfamily behave like the archetypical S. aureus SrtA protein, their substrates will be anchored to the cell wall cross-bridge.
Subfamily-3 sortases.
This is the largest subfamily, but its members play a more specialized role, anchoring far fewer proteins than the SrtA-type proteins. Interestingly, the SrtA and subfamily-3 enzymes are predicted to process proteins harboring similar sorting signals (Fig. 2), and many bacterial genomes encode both SrtA and subfamily-3 homologs (Table 2). Do these enzymes serve redundant functions in the cell, or do they differentially process proteins bearing related CWSs? Studies of Streptococcus pyogenes are consistent with the latter, because its SrtA and subfamily-3 homologs have been shown to selectively sort proteins bearing related LPXTG motifs (6). In order to account for this finding, the authors suggested that the SrtA and subfamily-3 enzymes recognized an expanded motif in which an acidic or glycine residue immediately follows the canonical LPXTG motif (6). Our results are generally consistent with this conclusion, since 53% of the predicted SrtA substrates contain the sequence LPXTG[DE], while 83% of the predicted subfamily-3 substrates contain the sequence LPXTGG. The N-terminally located sequence YSIRK has been shown to control the efficiency of export of S. aureus SrtA substrates (5), but it does not appear to be a determinant of specificity for this protein family because it is poorly conserved in the predicted SrtA-type substrates. Intriguingly, two other features of the subfamily-3 proteins may contribute to their substrate specificity. First, the expression of subfamily-3 enzymes and their potential substrates may be coordinately regulated, since their genes are always adjacent to one another. Second, the SrtA and subfamily-3 enzymes appear to be positioned in the membrane differently. Subfamily-3 enzymes contain hydrophobic amino acids at both their N and C termini, suggesting that they are type I membrane proteins (C-terminal end embedded in the membrane). In contrast, SrtA-type proteins contain only an N-terminal stretch of hydrophobic amino acids and are therefore presumably type II membrane proteins (N-terminal end embedded into the membrane). It is conceivable that their distinct membrane topology enables subfamily-3 enzymes to recognize other, as of yet undetermined features of their substrates. Many of the predicted substrates of the subfamily-3 enzymes may be involved in cell adhesion, since they contain domains associated with collagen binding, including the Cna protein B-type, DUF11, and von Willebrand factor type A domains (17, 19, 67).
The SrtB subfamily.
In contrast to the SrtA and subfamily-3 proteins, the remaining subfamilies (SrtB and subfamilies 4 and 5) are expected to process novel sorting signals (Fig. 2). Homologs most closely related to the well-characterized S. aureus SrtB protein (the SrtB subfamily) constitute a minor pathway involved in heme-iron acquisition (46). In addition to S. aureus, a single srtB gene is found in bacteria from the genera Bacillus and Listeria, and in all cases, it is proximal to a single substrate that contains an unusual sequence motif (NPQTN in S. aureus; NPKSS in Listeria; and NPKTG, NPKTD, and NPQTG in Bacillus). All SrtB proteins appear to be involved in iron metabolism, since their prospective substrates contain the NEAT domain, implicated in iron transport (3). Assuming that members of this sortase subfamily behave like the S. aureus SrtB protein, they can be expected to attach proteins to the cell surface (46).
Subfamily-4 sortases.
The subfamily-4 sortases process a unique sorting signal and constitute a specialized sorting pathway found in bacilli. This subfamily is predicted to process proteins bearing the motif LPXTA[ST] (and in B. subtilis a single protein containing the sequence LPDTSA) and is frequently found in bacteria that also contain SrtA, SrtB, and subfamily-3 proteins (Table 2). The unique placement of an alanine at position five in their recognition motif suggests that they operate nonredundantly with these other sortases (Fig. 2). The substrate selectivity of the subfamily-4 enzymes may be further enhanced by coexpression with their substrates, since their genes are typically adjacent to the genes of their predicted substrates. Although many of the predicted subfamily-4 substrates have yet to be annotated, a Pfam analysis reveals that they are predominantly enzymes (5′ nucleotidases, glycosyl hydrolase, and subtilase).
Subfamily-5 sortases.
Several high-G+C gram-positive bacteria have replaced SrtA enzymes with subfamily-5 homologs that recognize a nonstandard sorting signal, LAXTG (Fig. 2). Similar to the case with SrtA, it seems likely that the subfamily-5 proteins play a housekeeping role in the cell because their genes are never positioned adjacent to their predicted substrates and SrtA and subfamily-5 proteins are never found in the same organism (Table 2). Biochemical studies have shown that position two in the LPXTG motif is critical for protein sorting (63). The placement of an alanine at this position in the subfamily-5 substrates suggests that in Actinobacteria these enzymes and the subfamily-3 sortases nonredundantly sort proteins to the cell surface. Although the existence of an LAXTG sorting signal in Actinobacteria has been previously noted (55), our comparative genome analysis reveals that this motif is processed by subfamily-5 sortases and it predicts their cognate sortase, even in bacteria that encode more than one enzyme (Corynebacterium diphtheriae, Corynebacterium efficiens, and Bifidobacterium longum). The functions of LAXTG-containing proteins remain to be elucidated, although many appear to bind carbohydrates or to be involved in aerial hyphae formation in Streptomyces (16, 21).
Different protein sorting pathways intersect at sortase-substrate gene clusters.
Sortase genes are frequently clustered with genes encoding potential substrates (Table 4). The majority of the clusters (80%) contain a single sortase homolog and one to three genes encoding a CWS-containing protein. Although most gene clusters pair a sortase with its predicted substrates, several are points at which distinct sorting pathways intersect. These “mixed” gene clusters contain a sortase and its predicted substrates but also genes for an additional substrate(s) that is not processed by the sortase in the cluster. Schneewind and colleagues were the first to identify a mixed gene cluster in S. aureus and Listeria that contains a single srtB gene, a gene for its substrate, and two genes encoding substrates for the distantly located SrtA protein (46). In addition to this well-characterized case, our analysis reveals several other mixed clusters that have yet to be demonstrated experimentally (Table 4). For example, C. diphtheriae and C. efficiens each contain two mixed clusters that pair subfamily-3 sortases with their own substrates and substrates of a distally located subfamily-5 homolog. As in the aforementioned SrtA-SrtB mixed clusters, the ability of the subfamily-3 and -5 enzymes to discriminate between the substrates in the cluster is readily explained by the distinct CWS specificities of these enzymes (Fig. 2). An intriguing mixed cluster has recently been discovered in Streptococcus suis (54) which contains two subfamily-3 genes and three genes encoding CWS-proteins. Consistent with our predictions, one of the substrates in the cluster is attached to the cell surface by a distantly located SrtA-type protein. Interestingly, recent results suggest that the SrtA- and SrtB-type sortases attach proteins to the cell surface but that the extent of branching of these muropeptides is varied (44). The srtA-srtB mixed gene cluster may therefore enable the coordinated placement of distinctly positioned surface proteins to achieve a desired biological outcome, heme iron acquisition in this case. Further evidence that sortases are specific for acceptor groups on the cell wall comes from studies of S. aureus and S. pyogenes. In vitro, the SrtA-type sortases in these bacteria both proteolyze LPSTG peptides, but the S. pyogenes enzyme does not catalyze transpeptidation to NH2-Gly, a mimic of the S. aureus cell wall peptide that is readily used by the S. aureus protein (peptidoglycan cross-links in S. pyogenes are mediated via alanine residues) (61). By extension, the clusters identified in this study suggest that the SrtA and subfamily-3 sortases in S. suis and the subfamily-3 and subfamily-5 sortases in Actinobacteria may operate to place proteins at distinct sites within the cell wall.
TABLE 4.
Genomic clusters of sortases and CWS-containing proteins
| Species | Sortase subfamilies in genomic clustera
|
Distribution of sortases and CWS-substrates in genomic clusterb
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sortase | Substratec | 1 sortase
|
2 sortases
|
3 sortases
|
|||||||
| 1 sub. | 2 sub. | 3 sub. | 4 sub. | 1 sub. | 2 sub. | 3 sub. | 0 sub. | 3 sub. | |||
| Actinobacteria (high-G+C gram-positive bacteria) | |||||||||||
| Corynebacterium diphtheriae | 3 | 3/5d | • | ||||||||
| 3 | 3/5 | • | |||||||||
| 3 | 3 | • | |||||||||
| Corynebacterium efficiens | 3 | 3/5 | • | ||||||||
| 3 | 3/5 | • | |||||||||
| Bifidobacterium longum DJ010A | 3 | 3 | • | ||||||||
| Bifidobacterium longum NCC2705 | 3 | 3 | • | ||||||||
| Firmicutes (gram-positive bacteria) | |||||||||||
| Bacillus | |||||||||||
| Bacillus anthracis A2012 | B | B | • | ||||||||
| 4 | 4 | • | |||||||||
| Bacillus anthracis Ames | B | B | • | ||||||||
| 4 | 4 | • | |||||||||
| Bacillus anthracis KrugerB | B | B | • | ||||||||
| 4 | 4 | • | |||||||||
| Bacillus anthracis Western NA | B | B | • | ||||||||
| 4 | 4 | • | |||||||||
| Bacillus cereus ATCC 10987 | 3 | 3 | • | ||||||||
| 3 | 3 | • | |||||||||
| B | B | • | |||||||||
| 4 | 4 | • | |||||||||
| 3 | 3 | • | |||||||||
| Bacillus cereus ATCC 14579 | B | B | • | ||||||||
| 4 | 4 | • | |||||||||
| 3 | 3 | • | |||||||||
| Bacillus halodurans | Xe | X | • | ||||||||
| X | X | • | |||||||||
| X | X | • | |||||||||
| B | B | • | |||||||||
| 4 | 4 | • | |||||||||
| Bacillus subtilis | 4 | 4 | • | ||||||||
| Geobacillus stearothermophilus | 4 | 4 | • | ||||||||
| Oceanobacillus iheyensis | 4 | 4 | • | ||||||||
| Listeria | |||||||||||
| Listeria innocua | B | B/A | • | ||||||||
| Listeria monocytogenes 4b | B | B/A | • | ||||||||
| Listeria monocytogenes EGD-e | B | B/A | • | ||||||||
| Staphylococcus | |||||||||||
| Staphylococcus aureus COL | B | B/A | • | ||||||||
| Staphylococcus aureus MRSA252 | B | B/A | • | ||||||||
| Staphylococcus aureus MSSA476 | B | B/A | • | ||||||||
| Staphylococcus aureus MW2 | B | B/A | • | ||||||||
| Staphylococcus aureus Mu50 | B | B/A | • | ||||||||
| Staphylococcus aureus N315 | B | B/A | • | ||||||||
| Staphylococcus aureus NCTC 8325 | B | B/A | • | ||||||||
| Enterococcus | |||||||||||
| Enterococcus faecalis | 3 | 3 | • | ||||||||
| X | X | • | |||||||||
| Enterococcus faecium | 3 | 3 | • | ||||||||
| 3 | 3 | • | |||||||||
| 3 | 3 | • | |||||||||
| X | X | • | |||||||||
| Leuconostocaceae | |||||||||||
| Leuconostoc mesenteroides | 3 | 3 | • | ||||||||
| Streptococcaceae | |||||||||||
| Lactococcus lactis | 3 | 3 | • | ||||||||
| Streptococcus agalactiae 2603V/R | 3 | 3 | • | ||||||||
| 3 | 3 | • | |||||||||
| Streptococcus agalactiae A909 | 3 | 3 | • | ||||||||
| 3 | 3 | • | |||||||||
| Streptococcus agalactiae NEM316 | 3 | 3 | • | ||||||||
| 3 | 3 | • | |||||||||
| Streptococcus equi | 3 | 3 | • | ||||||||
| Streptococcus pneumoniae 670-6B | 3 | 3 | • | ||||||||
| Streptococcus pneumoniae TIGR4 | 3 | 3 | • | ||||||||
| Streptococcus pyogenes M1 GAS | X/3 | X | • | ||||||||
| Streptococcus pyogenes MGAS315 | X | X | • | ||||||||
| Streptococcus pyogenes MGAS8232 | X | X | • | ||||||||
| Streptococcus pyogenes Manfredo | X | X | • | ||||||||
| Streptococcus pyogenes SSI-I | X | X | • | ||||||||
| Streptococcus suis | 3 | 3/A | • | ||||||||
| Clostridia | |||||||||||
| Clostridium acetobutylicum | X | X | • | ||||||||
| Clostridium botulinum | X | X | • | ||||||||
| Clostridium perfringens ATCC 13124 | 3 | 3 | • | ||||||||
| X | X | • | |||||||||
| Clostridium perfringens 13 | 3 | 3 | • | ||||||||
| X | X | • | |||||||||
| Clostridium tetani | X | X | • | ||||||||
| Ruminococcus albus | 3 | 3 | • | ||||||||
| Proteobacteria (purple bacteria and relatives—gram-negative) | |||||||||||
| Bradyrhicobium japonicum | 6 | 6 | • | ||||||||
| Colwellia psychroerythraea | 6 | 6 | • | ||||||||
| Microbulbifer degradans | 6 | 6 | • | ||||||||
| Shewanella oneidensis | 6 | 6 | • | ||||||||
| Shewanella putrefaciens | 6 | 6 | • | ||||||||
| Total no. of clusters | 31 | 10 | 18 | 2 | 1 | 3 | 8 | 1 | 3 | ||
Sortase homologs are clustered into subfamilies according to sequence homology using BLAST profiles and HMMs.
Numbers of sortase homologs and CWS-containing proteins that are genomically adjacent in a “genomic cluster” are given in column heads. sub., substrate(s).
The subfamily of the sortase(s) and the CWS-containing protein(s) in the genomic cluster.
Shill indicates that genomic cluster encodes sortases and/or CWS-containing proteins from more than one sortase subfamily.
Sortase homolog(s) in the genomic cluster is not readily classified into a subfamily based on sequence homology and therefore is designated as belonging to “subfamily X.” There are 15 genomic clusters of these unclassified sortase homologs and CWS-containing proteins.
Five species of gram-negative bacteria encode a single sortase homolog and a single CWS-containing substrate: Colwellia psychrerythraea, Microbulbifer degradans, Bradyrhizobium japonicum, Shewanella oneidensis, and Shewanella putrefaciens. These sortases are closely related to one another (Table 1) and are positioned adjacent to a single CWS-containing substrate bearing the motif LP[QK]T[AS]T (Fig. 2). The predicted substrates for these enzymes contain a von Willebrand factor type A domain that is often associated with ligand binding in eukarya (17), and they may be attached to murein lipoproteins (14, 55). The function of these substrates is specialized, because other organisms with completely sequenced genomes from the same subphyla (α-proteobacteria and γ-proteobacteria) do not encode a sortase homolog.
In conclusion, we have shown that the majority of sortase-related proteins in gram-positive bacteria can be partitioned into five distinct subfamilies based upon their primary sequences. Most bacteria encode sortases from two or more of these subfamilies, which are predicted to function nonredundantly in sorting proteins to the cell surface. Approximately 20% of sortase homologs are most closely related to the S. aureus SrtA protein and play a housekeeping role, anchoring a large number of functionally unrelated CWS-containing proteins to the cell surface. In contrast, the majority of sortase homologs have a more specialized role, anchoring on average far fewer proteins that frequently contain unusual sequence motifs in their sorting signals. It has been suggested that many sortase-related proteins perform tasks other than cell wall protein anchoring; however, using only a few conservative assumptions, the majority of sortases are predicted to process CWS-containing proteins. The functional sortase-substrate linkages are completely compatible with all available biochemical data. They are available online (http://www.doe-mbi.ucla.edu/Services/Sortase/) in a searchable database that should prove useful in deciphering the many sorting pathways present in bacteria.
Acknowledgments
We thank Kevin Connolly, Chu Kong Liew, and Todd O. Yeates for useful discussions.
This work was supported by a grant from the National Institutes of Health to R.T.C. (AI52217).
Editor: V. J. DiRita
REFERENCES
- 1.Ajdic, D., W. M. McShan, R. E. McLaughlin, G. Savic, J. Chang, M. B. Carson, C. Primeaux, R. Tian, S. Kenton, H. Jia, S. Lin, Y. Qian, S. Li, H. Zhu, F. Najar, H. Lai, J. White, B. A. Roe, and J. J. Ferretti. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 99:14434-14439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade, M. A., F. D. Ciccarelli, C. Perez-Iratxeta, and P. Bork. 2002. NEAT: a domain duplicated in genes near the components of a putative Fe3+ siderophore transporter from Gram-positive pathogenic bacteria. Genome Biol. 3:RESEARCH0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 5.Bae, T., and O. Schneewind. 2003. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185:2910-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 9.Bentley, S. D., M. Maiwald, L. D. Murphy, M. J. Pallen, C. A. Yeats, L. G. Dover, H. T. Norbertczak, G. S. Besra, M. A. Quail, D. E. Harris, A. von Herbay, A. Goble, S. Rutter, R. Squares, S. Squares, B. G. Barrell, J. Parkhill, and D. A. Relman. 2003. Sequencing and analysis of the genome of the Whipple's disease bacterium Tropheryma whipplei. Lancet 361:637-644. [DOI] [PubMed] [Google Scholar]
- 10.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M. Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. Leung, P. M. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bierne, H., S. K. Mazmanian, M. Trost, M. G. Pucciarelli, G. Liu, P. Dehoux, L. Jansch, F. Garcia-del Portillo, O. Schneewind, and P. Cossart. 2002. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43:869-881. [DOI] [PubMed] [Google Scholar]
- 12.Bolken, T. C., C. A. Franke, K. F. Jones, G. O. Zeller, C. H. Jones, E. K. Dutton, and D. E. Hruby. 2001. Inactivation of the srtA gene in Streptococcus gordonii inhibits cell wall anchoring of surface proteins and decreases in vitro and in vivo adhesion. Infect. Immun. 69:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bost, F., M. Diarra-Mehrpour, and J. P. Martin. 1998. Inter-alpha-trypsin inhibitor proteoglycan family—a group of proteins binding and stabilizing the extracellular matrix. Eur. J. Biochem. 252:339-346. [DOI] [PubMed] [Google Scholar]
- 15.Bruggemann, H., S. Baumer, W. F. Fricke, A. Wiezer, H. Liesegang, I. Decker, C. Herzberg, R. Martinez-Arias, R. Merkl, A. Henne, and G. Gottschalk. 2003. The genome sequence of Clostridium tetani, the causative agent of tetanus disease. Proc. Natl. Acad. Sci. USA 100:1316-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Claessen, D., R. Rink, W. De Jong, J. Siebring, P. De Vreugd, F. G. Boersma, L. Dijkhuizen, and H. A. Wosten. 2003. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17:1714-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombatti, A., P. Bonaldo, and R. Doliana. 1993. Type A modules: interacting domains found in several non-fibrillar collagens and in other extracellular matrix proteins. Matrix 13:297-306. [DOI] [PubMed] [Google Scholar]
- 18.Cossart, P., and R. Jonquieres. 2000. Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc. Natl. Acad. Sci. USA 97:5013-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deivanayagam, C. C., R. L. Rich, M. Carson, R. T. Owens, S. Danthuluri, T. Bice, M. Hook, and S. V. Narayana. 2000. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Struct. Fold Des. 8:67-78. [DOI] [PubMed] [Google Scholar]
- 20.Eddy, S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755-763. [DOI] [PubMed] [Google Scholar]
- 21.Elliot, M. A., N. Karoonuthaisiri, J. Huang, M. J. Bibb, S. N. Cohen, C. M. Kao, and M. J. Buttner. 2003. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes Dev. 17:1727-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garandeau, C., H. Reglier-Poupet, L. Dubail, J. L. Beretti, P. Berche, and A. Charbit. 2002. The sortase SrtA of Listeria monocytogenes is involved in processing of internalin and in virulence. Infect. Immun. 70:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 25.Glaser, P., C. Rusniok, C. Buchrieser, F. Chevalier, L. Frangeul, T. Msadek, M. Zouine, E. Couve, L. Lalioui, C. Poyart, P. Trieu-Cuot, and F. Kunst. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499-1513. [DOI] [PubMed] [Google Scholar]
- 26.Hava, D. L., C. J. Hemsley, and A. Camilli. 2003. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166-170. [Google Scholar]
- 29.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 31.Ikeda, M., and S. Nakagawa. 2003. The Corynebacterium glutamicum genome: features and impacts on biotechnological processes. Appl. Microbiol. Biotechnol. 62:99-109. [DOI] [PubMed] [Google Scholar]
- 32.Ilangovan, U., H. Ton-That, J. Iwahara, O. Schneewind, and R. T. Clubb. 2001. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 98:6056-6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ivanova, N., A. Sorokin, I. Anderson, N. Galleron, B. Candelon, V. Kapatral, A. Bhattacharyya, G. Reznik, N. Mikhailova, A. Lapidus, L. Chu, M. Mazur, E. Goltsman, N. Larsen, M. D'Souza, T. Walunas, Y. Grechkin, G. Pusch, R. Haselkorn, M. Fonstein, S. D. Ehrlich, R. Overbeek, and N. Kyrpides. 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423:87-91. [DOI] [PubMed] [Google Scholar]
- 34.Janulczyk, R., and M. Rasmussen. 2001. Improved pattern for genome-based screening identifies novel cell wall-attached proteins in gram-positive bacteria. Infect. Immun. 69:4019-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonsson, I. M., S. K. Mazmanian, O. Schneewind, M. Verdrengh, T. Bremell, and A. Tarkowski. 2002. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J. Infect. Dis. 185:1417-1424. [DOI] [PubMed] [Google Scholar]
- 36.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110 (supplement). DNA Res. 9:225-256. [DOI] [PubMed] [Google Scholar]
- 37.Kharat, A. S., and A. Tomasz. 2003. Inactivation of the srtA gene affects localization of surface proteins and decreases adhesion of Streptococcus pneumoniae to human pharyngeal cells in vitro. Infect. Immun. 71:2758-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 40.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 41.Li, T., I. Johansson, D. I. Hay, and N. Stromberg. 1999. Strains of Actinomyces naeslundii and Actinomyces viscosus exhibit structurally variant fimbrial subunit proteins and bind to different peptide motifs in salivary proteins. Infect. Immun. 67:2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazmanian, S. K., G. Liu, T. T. Hung, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 43.Mazmanian, S. K., G. Liu, E. R. Jensen, E. Lenoy, and O. Schneewind. 2000. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc. Natl. Acad. Sci. USA 97:5510-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 45.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 46.Mazmanian, S. K., H. Ton-That, K. Su, and O. Schneewind. 2002. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. USA 99:2293-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in gram-Positive bacteria. Mol. Microbiol. 14:115-121. [DOI] [PubMed] [Google Scholar]
- 48.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 8:581-599. [DOI] [PubMed] [Google Scholar]
- 51.Nishio, Y., Y. Nakamura, Y. Kawarabayasi, Y. Usuda, E. Kimura, S. Sugimoto, K. Matsui, A. Yamagishi, H. Kikuchi, K. Ikeo, and T. Gojobori. 2003. Comparative complete genome sequence analysis of the amino acid replacements responsible for the thermostability of Corynebacterium efficiens. Genome Res. 13:1572-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nolling, J., G. Breton, M. V. Omelchenko, K. S. Makarova, Q. Zeng, R. Gibson, H. M. Lee, J. Dubois, D. Qiu, J. Hitti, Y. I. Wolf, R. L. Tatusov, F. Sabathe, L. Doucette-Stamm, P. Soucaille, M. J. Daly, G. N. Bennett, E. V. Koonin, and D. R. Smith. 2001. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 183:4823-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novick, R. P. 2000. Sortase: the surface protein anchoring transpeptidase and the LPXTG motif. Trends Microbiol. 8:148-151. [DOI] [PubMed] [Google Scholar]
- 54.Osaki, M., D. Takamatsu, Y. Shimoji, and T. Sekizaki. 2002. Characterization of Streptococcus suis genes encoding proteins homologous to sortase of gram-positive bacteria. J. Bacteriol. 184:971-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-101. [DOI] [PubMed] [Google Scholar]
- 56.Paulsen, I. T., L. Banerjei, G. S. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 57.Perry, A. M., H. Ton-That, S. K. Mazmanian, and O. Schneewind. 2002. Anchoring of surface proteins to the cell wall of Staphylococcus aureus—III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J. Biol. Chem. 277:16241-16248. [DOI] [PubMed] [Google Scholar]
- 58.Read, T. D., S. N. Peterson, N. Tourasse, L. W. Baillie, I. T. Paulsen, K. E. Nelson, H. Tettelin, D. E. Fouts, J. A. Eisen, S. R. Gill, E. K. Holtzapple, O. A. Okstad, E. Helgason, J. Rilstone, M. Wu, J. F. Kolonay, M. J. Beanan, R. J. Dodson, L. M. Brinkac, M. Gwinn, R. T. DeBoy, R. Madpu, S. C. Daugherty, A. S. Durkin, D. H. Haft, W. C. Nelson, J. D. Peterson, M. Pop, H. M. Khouri, D. Radune, J. L. Benton, Y. Mahamoud, L. Jiang, I. R. Hance, J. F. Weidman, K. J. Berry, R. D. Plaut, A. M. Wolf, K. L. Watkins, W. C. Nierman, A. Hazen, R. Cline, C. Redmond, J. E. Thwaite, O. White, S. L. Salzberg, B. Thomason, A. M. Friedlander, T. M. Koehler, P. C. Hanna, A. B. Kolsto, and C. M. Fraser. 2003. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature 423:81-86. [DOI] [PubMed] [Google Scholar]
- 59.Roche, F., R. Masey, S. Peacock, N. Day, L. Visai, P. Speziale, A. C. Lam, M. Pallen, and T. Foster. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643-654. [DOI] [PubMed] [Google Scholar]
- 60.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944-945. [DOI] [PubMed] [Google Scholar]
- 61.Ruzin, A., A. Severin, F. Ritacco, K. Tabei, G. Singh, P. A. Bradford, M. M. Siegel, S. J. Projan, and D. M. Shlaes. 2002. Further evidence that a cell wall precursor [C-55-MurNAc-(peptide)-GlcNAc] serves as an acceptor in a sorting reaction. J. Bacteriol. 184:2141-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneewind, O., P. Model, and V. A. Fischetti. 1992. Sorting of protein A to the staphylococcal cell wall. Cell 70:267-281. [DOI] [PubMed] [Google Scholar]
- 64.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasy, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strominger, J. L., K. Izaki, M. Matsuhashi, and D. J. Tipper. 1967. Peptidoglycan transpeptidase and D-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. Fed. Proc. 26:9-22. [PubMed] [Google Scholar]
- 67.Symersky, J., J. M. Patti, M. Carson, K. House-Pompeo, M. Teale, D. Moore, L. Jin, A. Schneider, L. J. DeLucas, M. Hook, and S. V. Narayana. 1997. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat. Struct. Biol. 4:833-838. [DOI] [PubMed] [Google Scholar]
- 68.Takami, H., K. Nakasone, Y. Takaki, G. Maeno, R. Sasaki, N. Masui, F. Fuji, C. Hirama, Y. Nakamura, N. Ogasawara, S. Kuhara, and K. Horikoshi. 2000. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 28:4317-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takami, H., Y. Takaki, and I. Uchiyama. 2002. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 30:3927-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tettelin, H., V. Masignani, M. J. Cieslewicz, J. A. Eisen, S. Peterson, M. R. Wessels, I. T. Paulsen, K. E. Nelson, I. Margarit, T. D. Read, L. C. Madoff, A. M. Wolf, M. J. Beanan, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, J. F. Kolonay, R. Madupu, M. R. Lewis, D. Radune, N. B. Fedorova, D. Scanlan, H. Khouri, S. Mulligan, H. A. Carty, R. T. Cline, S. E. Van Aken, J. Gill, M. Scarselli, M. Mora, E. T. Iacobini, C. Brettoni, G. Galli, M. Mariani, F. Vegni, D. Maione, D. Rinaudo, R. Rappuoli, J. L. Telford, D. L. Kasper, G. Grandi, and C. M. Fraser. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. USA 99:12391-12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]