Abstract
We have studied the ability of the histone (H3-H4)2 tetramer, the central part of the nucleosome of eukaryotic chromatin, to form particles on DNA minicircles of negative and positive superhelicities, and the effect of relaxing these particles with topoisomerase I. The results show that even modest positive torsional stress from the DNA, and in particular that generated by DNA thermal fluctuations, can trigger a major, reversible change in the conformation of the particle. Neither a large excess of naked DNA, nor a crosslink between the two H3s prevented the transition from one form to the other. This suggested that during the transition, the histones neither dissociated from the DNA nor were even significantly reshuffled. Moreover, the particles reconstituted on negatively and positively supercoiled minicircles look similar under electron microscopy. These data agree best with a transition involving a switch of the wrapped DNA from a left- to a right-handed superhelix. It is further proposed, based on the left-handed overall superhelical conformation of the tetramer within the octamer [Arents, G., Burlingame, R. W., Wang, B. C., Love, W. E. & Moudrianakis, E. N. (1991) Proc. Natl.Acad. Sci. USA 88, 10148-10152] that this change in DNA topology is mediated by a similar change in the topology of the tetramer itself, which may occur through a rotation (or a localized deformation) of the two H3-H4 dimers about their H3-H3 interface. Potential implications of this model for nucleosome dynamics in vivo are discussed.
Full text
PDF
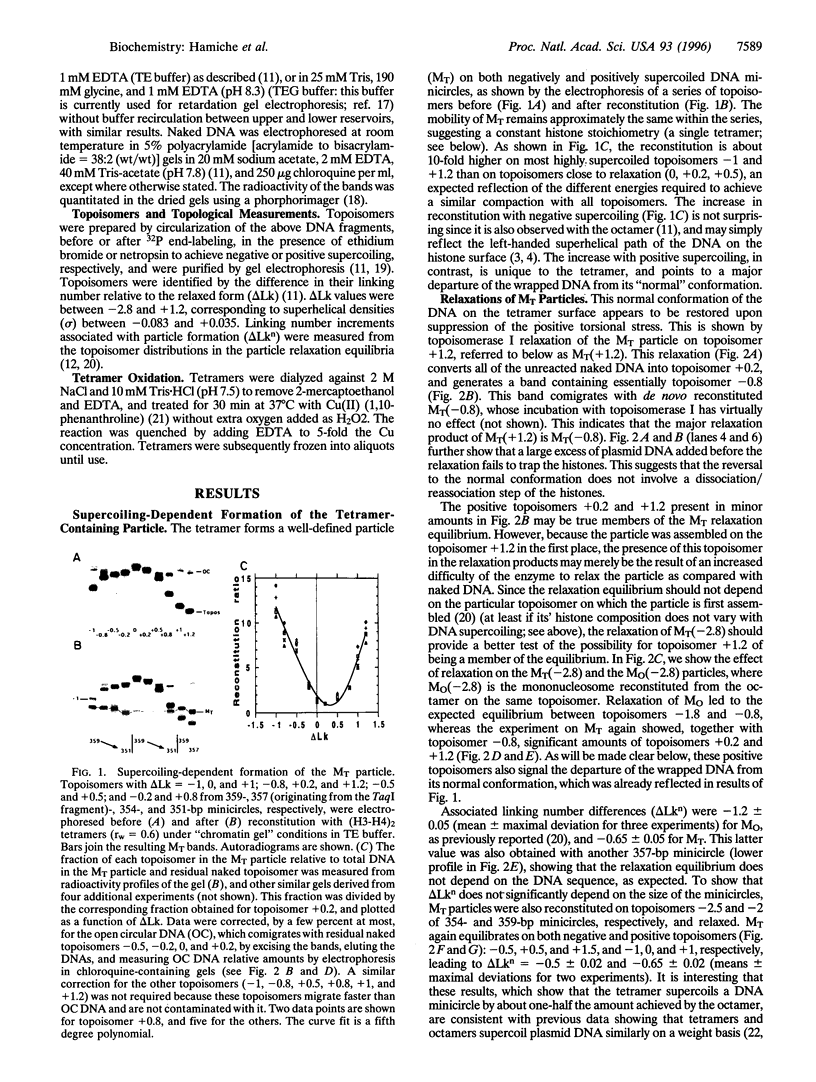



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antoniewski C., Laval M., Dahan A., Lepesant J. A. The ecdysone response enhancer of the Fbp1 gene of Drosophila melanogaster is a direct target for the EcR/USP nuclear receptor. Mol Cell Biol. 1994 Jul;14(7):4465–4474. doi: 10.1128/mcb.14.7.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G., Burlingame R. W., Wang B. C., Love W. E., Moudrianakis E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G., Moudrianakis E. N. Topography of the histone octamer surface: repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci U S A. 1993 Nov 15;90(22):10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B. W., Rhodes D. Eukaryotic RNA polymerase II binds to nucleosome cores from transcribed genes. Nature. 1983 Feb 10;301(5900):482–488. doi: 10.1038/301482a0. [DOI] [PubMed] [Google Scholar]
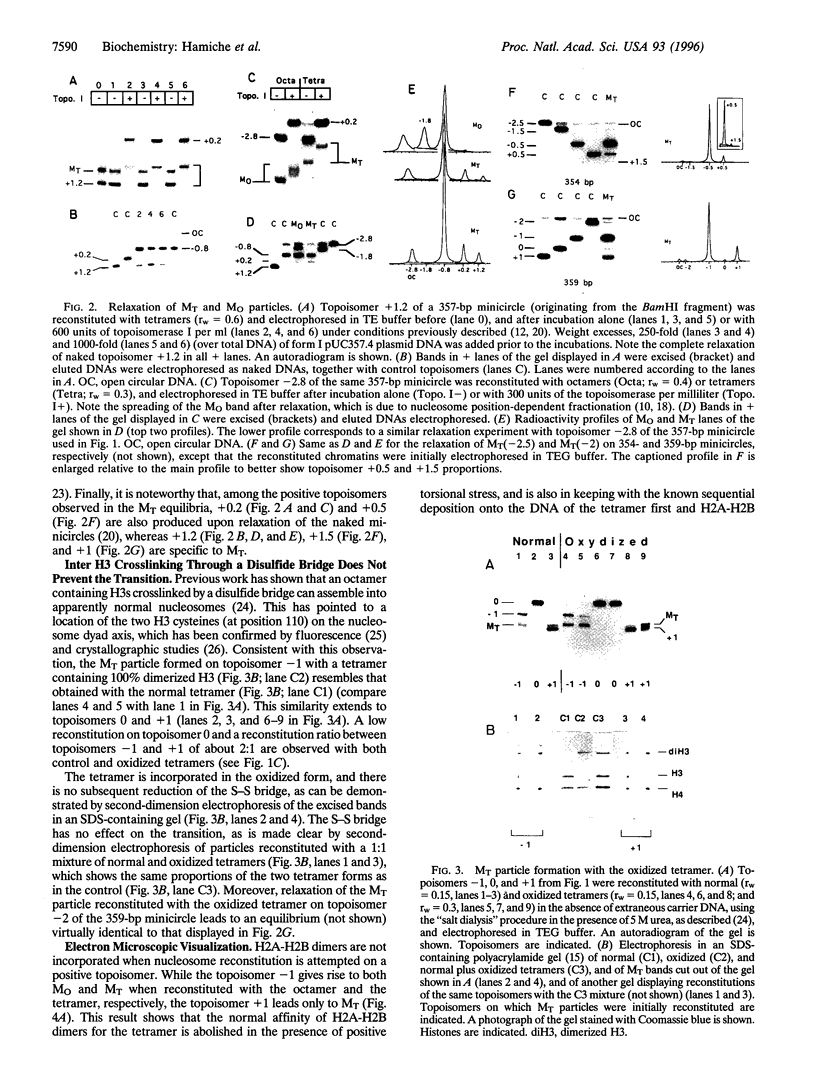
- Baxevanis A. D., Godfrey J. E., Moudrianakis E. N. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry. 1991 Sep 10;30(36):8817–8823. doi: 10.1021/bi00100a013. [DOI] [PubMed] [Google Scholar]
- Bina-Stein M., Simpson R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell. 1977 Jul;11(3):609–618. doi: 10.1016/0092-8674(77)90078-2. [DOI] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Histone H3 disulfide dimers and nucleosome structure. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5519–5523. doi: 10.1073/pnas.74.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camerini-Otero R. D., Felsenfeld G. Supercoiling energy and nucleosome formation: the role of the arginine-rich histone kernel. Nucleic Acids Res. 1977;4(5):1159–1181. doi: 10.1093/nar/4.5.1159-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Careaga C. L., Falke J. J. Thermal motions of surface alpha-helices in the D-galactose chemosensory receptor. Detection by disulfide trapping. J Mol Biol. 1992 Aug 20;226(4):1219–1235. doi: 10.1016/0022-2836(92)91063-u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Laurent B. C. The SNF/SWI family of global transcriptional activators. Curr Opin Cell Biol. 1994 Jun;6(3):396–402. doi: 10.1016/0955-0674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Crick F. H. Linking numbers and nucleosomes. Proc Natl Acad Sci U S A. 1976 Aug;73(8):2639–2643. doi: 10.1073/pnas.73.8.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daban J. R., Cantor C. R. Role of histone pairs H2A,H2B and H3,H4 in the self-assembly of nucleosome core particles. J Mol Biol. 1982 Apr 25;156(4):771–789. doi: 10.1016/0022-2836(82)90141-3. [DOI] [PubMed] [Google Scholar]
- Dong F., van Holde K. E. Nucleosome positioning is determined by the (H3-H4)2 tetramer. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duband-Goulet I., Carot V., Ulyanov A. V., Douc-Rasy S., Prunell A. Chromatin reconstitution on small DNA rings. IV. DNA supercoiling and nucleosome sequence preference. J Mol Biol. 1992 Apr 20;224(4):981–1001. doi: 10.1016/0022-2836(92)90464-u. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The histone core complex: an octamer assembled by two sets of protein-protein interactions. Biochemistry. 1978 Nov 14;17(23):4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet I., Zivanovic Y., Prunell A., Revet B. Chromatin reconstitution on small DNA rings. I. J Mol Biol. 1988 Mar 20;200(2):253–266. doi: 10.1016/0022-2836(88)90238-0. [DOI] [PubMed] [Google Scholar]
- Hamiche A., Prunell A. Chromatin reconstitution on small DNA rings. V. DNA thermal flexibility of single nucleosomes. J Mol Biol. 1992 Nov 20;228(2):327–337. doi: 10.1016/0022-2836(92)90821-z. [DOI] [PubMed] [Google Scholar]
- Hayes J. J., Clark D. J., Wolffe A. P. Histone contributions to the structure of DNA in the nucleosome. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Jackson S., Brooks W., Jackson V. Dynamics of the interactions of histones H2A,H2B and H3,H4 with torsionally stressed DNA. Biochemistry. 1994 May 10;33(18):5392–5403. doi: 10.1021/bi00184a006. [DOI] [PubMed] [Google Scholar]
- Jackson V. In vivo studies on the dynamics of histone-DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry. 1990 Jan 23;29(3):719–731. doi: 10.1021/bi00455a019. [DOI] [PubMed] [Google Scholar]
- Jackson V. Influence of positive stress on nucleosome assembly. Biochemistry. 1993 Jun 8;32(22):5901–5912. doi: 10.1021/bi00073a024. [DOI] [PubMed] [Google Scholar]
- Jackson V. Preferential binding of histones H3 and H4 to highly positively coiled DNA. Biochemistry. 1995 Aug 22;34(33):10607–10619. doi: 10.1021/bi00033a036. [DOI] [PubMed] [Google Scholar]
- Kosower E. M., Kosower N. S. Bromobimane probes for thiols. Methods Enzymol. 1995;251:133–148. doi: 10.1016/0076-6879(95)51117-2. [DOI] [PubMed] [Google Scholar]
- Kruger W., Peterson C. L., Sil A., Coburn C., Arents G., Moudrianakis E. N., Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995 Nov 15;9(22):2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- Lattman E., Burlingame R., Hatch C., Moudrianakis E. N. Crystallization of the tetramer of histones H3 and H4. Science. 1982 May 28;216(4549):1016–1018. doi: 10.1126/science.7079748. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M., Hanawalt P. C. Presence of negative torsional tension in the promoter region of the transcriptionally poised dihydrofolate reductase gene in vivo. Nucleic Acids Res. 1995 May 25;23(10):1782–1789. doi: 10.1093/nar/23.10.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louters L., Chalkley R. Exchange of histones H1, H2A, and H2B in vivo. Biochemistry. 1985 Jun 18;24(13):3080–3085. doi: 10.1021/bi00334a002. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Effect of trypsinization and histone H5 addition on DNA twist and topology in reconstituted minichromosomes. Nucleic Acids Res. 1986 Apr 25;14(8):3293–3310. doi: 10.1093/nar/14.8.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donohue M. F., Duband-Goulet I., Hamiche A., Prunell A. Octamer displacement and redistribution in transcription of single nucleosomes. Nucleic Acids Res. 1994 Mar 25;22(6):937–945. doi: 10.1093/nar/22.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn J., Fyrberg A. M., Ganster R. W., Schmidt M. C., Peterson C. L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996 Feb 29;379(6568):844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- Read C. M., Baldwin J. P., Crane-Robinson C. Structure of subnucleosomal particles. Tetrameric (H3/H4)2 146 base pair DNA and hexameric (H3/H4)2(H2A/H2B)1 146 base pair DNA complexes. Biochemistry. 1985 Jul 30;24(16):4435–4450. doi: 10.1021/bi00337a027. [DOI] [PubMed] [Google Scholar]
- Read C. M., Crane-Robinson C. The structure of sub-nucleosomal particles. The octameric (H3/H4)4--125-base-pair-DNA complex. Eur J Biochem. 1985 Oct 1;152(1):143–150. doi: 10.1111/j.1432-1033.1985.tb09174.x. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Ruiz-Carrillo A., Jorcano J. L. Nucleohistone assembly: sequential binding of histone H3-H4 tetramer and histone H2A-H2B dimer to DNA. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):165–170. doi: 10.1101/sqb.1978.042.01.018. [DOI] [PubMed] [Google Scholar]
- Simon R. H., Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979 Feb;6(2):689–696. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A., Whitlock J. P., Jr, Bina M. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5000–5004. doi: 10.1073/pnas.76.10.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. O., Butler P. J. Characterization of the octamer of histones free in solution. J Mol Biol. 1977 Nov;116(4):769–781. doi: 10.1016/0022-2836(77)90270-4. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Wang B. C., Rose J., Arents G., Moudrianakis E. N. The octameric histone core of the nucleosome. Structural issues resolved. J Mol Biol. 1994 Feb 11;236(1):179–188. doi: 10.1006/jmbi.1994.1127. [DOI] [PubMed] [Google Scholar]
- Wilhelm F. X., Wilhelm M. L., Erard M., Duane M. P. Reconstitution of chromatin: assembly of the nucleosome. Nucleic Acids Res. 1978 Feb;5(2):505–521. doi: 10.1093/nar/5.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Wolffe A. P. Transcriptional activation. Switched-on chromatin. Curr Biol. 1994 Jun 1;4(6):525–528. doi: 10.1016/s0960-9822(00)00114-7. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Duband-Goulet I., Schultz P., Stofer E., Oudet P., Prunell A. Chromatin reconstitution on small DNA rings. III. Histone H5 dependence of DNA supercoiling in the nucleosome. J Mol Biol. 1990 Jul 20;214(2):479–495. doi: 10.1016/0022-2836(90)90195-R. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Goulet I., Prunell A. Properties of supercoiled DNA in gel electrophoresis. The V-like dependence of mobility on topological constraint. DNA-matrix interactions. J Mol Biol. 1986 Dec 5;192(3):645–660. doi: 10.1016/0022-2836(86)90282-2. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y., Goulet I., Revet B., Le Bret M., Prunell A. Chromatin reconstitution on small DNA rings. II. DNA supercoiling on the nucleosome. J Mol Biol. 1988 Mar 20;200(2):267–290. doi: 10.1016/0022-2836(88)90239-2. [DOI] [PubMed] [Google Scholar]