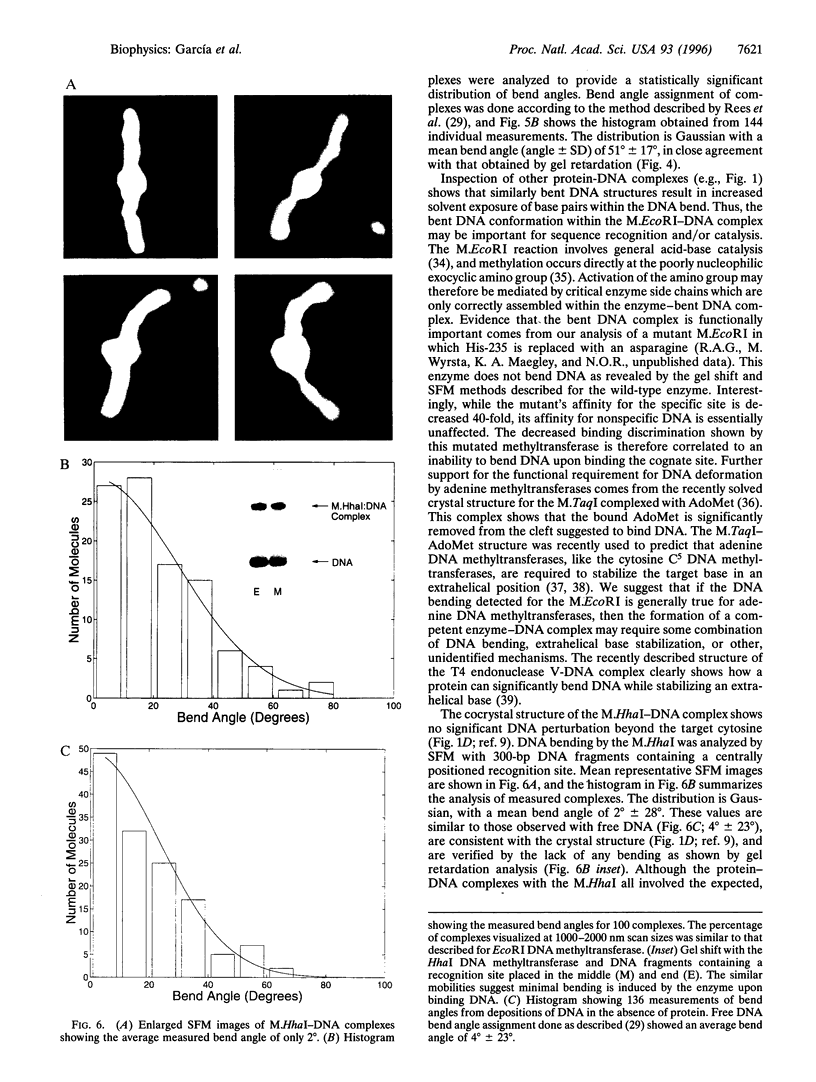
Abstract
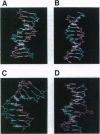
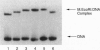
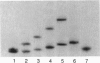
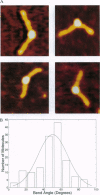
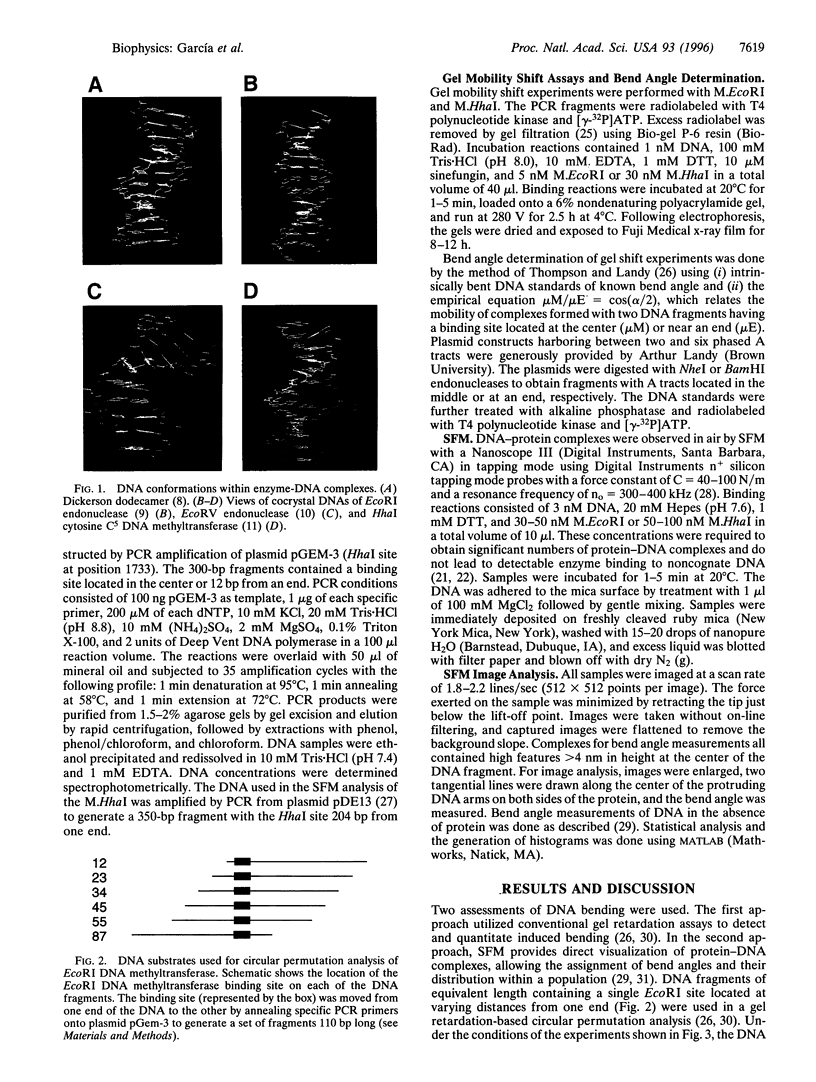
DNA methyltransferases modify specific cytosines and adenines within 2-6 bp recognition sequences. We used scanning force microscopy and gel shift analysis to show that M.HhaI, a cytosine C-5 DNA methyltransferase, causes only a 2 degree bend upon binding its recognition site. Our results are consistent with prior crystallographic analysis showing that the enzyme stabilizes an extrahelical base while leaving the DNA duplex otherwise unperturbed. In contrast, similar analysis of M.EcoRI, an adenine N6 DNA methyltransferase, shows an average bend angle of approximately 52 degrees. This distortion of DNA conformation by M.EcoRI is shown to be important for sequence-specific binding.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bustamante C., Vesenka J., Tang C. L., Rees W., Guthold M., Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992 Jan 14;31(1):22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Gartenberg M. R., Shrader T. E. DNA bending in protein-DNA complexes. Methods Enzymol. 1991;208:118–146. doi: 10.1016/0076-6879(91)08011-6. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Hajiseyedjavadi O., Young M. C., von Hippel P. H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993 Nov 5;262(5135):867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- Erie D. A., Yang G., Schultz H. C., Bustamante C. DNA bending by Cro protein in specific and nonspecific complexes: implications for protein site recognition and specificity. Science. 1994 Dec 2;266(5190):1562–1566. doi: 10.1126/science.7985026. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Ampe C., Steitz T. A., Crothers D. M. Molecular characterization of the GCN4-DNA complex. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6034–6038. doi: 10.1073/pnas.87.16.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman S. D., Nash H. A. Functional replacement of a protein-induced bend in a DNA recombination site. Nature. 1989 Sep 21;341(6239):251–254. doi: 10.1038/341251a0. [DOI] [PubMed] [Google Scholar]
- Hansma H. G., Sinsheimer R. L., Groppe J., Bruice T. C., Elings V., Gurley G., Bezanilla M., Mastrangelo I. A., Hough P. V., Hansma P. K. Recent advances in atomic force microscopy of DNA. Scanning. 1993 Sep-Oct;15(5):296–299. doi: 10.1002/sca.4950150509. [DOI] [PubMed] [Google Scholar]
- Kahn J. D., Crothers D. M. Protein-induced bending and DNA cyclization. Proc Natl Acad Sci U S A. 1992 Jul 15;89(14):6343–6347. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. L., Nikolov D. B., Burley S. K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993 Oct 7;365(6446):520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Kim Y., Geiger J. H., Hahn S., Sigler P. B. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993 Oct 7;365(6446):512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Labahn J., Granzin J., Schluckebier G., Robinson D. P., Jack W. E., Schildkraut I., Saenger W. Three-dimensional structure of the adenine-specific DNA methyltransferase M.Taq I in complex with the cofactor S-adenosylmethionine. Proc Natl Acad Sci U S A. 1994 Nov 8;91(23):10957–10961. doi: 10.1073/pnas.91.23.10957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone T., Blumenthal R. M., Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995 Nov 3;253(4):618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- Malone T., Blumenthal R. M., Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol. 1995 Nov 3;253(4):618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- Mashhoon N., Reich N. O. Investigation of ionizable residues critical for sequence-specific enzymatic DNA modification: protein modification and steady-state and pre-steady-state kinetic pH analyses of EcoRI DNA methyltransferase. Biochemistry. 1994 Jun 14;33(23):7113–7119. doi: 10.1021/bi00189a014. [DOI] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Mol C. D., Kuo C. F., Thayer M. M., Cunningham R. P., Tainer J. A. Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature. 1995 Mar 23;374(6520):381–386. doi: 10.1038/374381a0. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., McCormick R. J., Sharp P. A., Fisher D. E. Pre-bending of a promoter sequence enhances affinity for the TATA-binding factor. Nature. 1995 Feb 23;373(6516):724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- Pogolotti A. L., Jr, Ono A., Subramaniam R., Santi D. V. On the mechanism of DNA-adenine methylase. J Biol Chem. 1988 Jun 5;263(16):7461–7464. [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees W. A., Keller R. W., Vesenka J. P., Yang G., Bustamante C. Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science. 1993 Jun 11;260(5114):1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Everett E. A. Identification of peptides involved in S-adenosylmethionine binding in the EcoRI DNA methylase. Photoaffinity laveling with 8-azido-S-adenosylmethionine. J Biol Chem. 1990 May 25;265(15):8929–8934. [PubMed] [Google Scholar]
- Reich N. O., Maegley K. A., Shoemaker D. D., Everett E. Structural and functional analysis of EcoRI DNA methyltransferase by proteolysis. Biochemistry. 1991 Mar 19;30(11):2940–2946. doi: 10.1021/bi00225a030. [DOI] [PubMed] [Google Scholar]
- Reich N. O., Mashhoon N. Inhibition of EcoRI DNA methylase with cofactor analogs. J Biol Chem. 1990 May 25;265(15):8966–8970. [PubMed] [Google Scholar]
- Reich N. O., Mashhoon N. Kinetic mechanism of the EcoRI DNA methyltransferase. Biochemistry. 1991 Mar 19;30(11):2933–2939. doi: 10.1021/bi00225a029. [DOI] [PubMed] [Google Scholar]
- Renbaum P., Razin A. Mode of action of the Spiroplasma CpG methylase M.SssI. FEBS Lett. 1992 Nov 30;313(3):243–247. doi: 10.1016/0014-5793(92)81201-v. [DOI] [PubMed] [Google Scholar]
- Savva R., McAuley-Hecht K., Brown T., Pearl L. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995 Feb 9;373(6514):487–493. doi: 10.1038/373487a0. [DOI] [PubMed] [Google Scholar]
- Schluckebier G., O'Gara M., Saenger W., Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J Mol Biol. 1995 Mar 17;247(1):16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Surby M. A., Reich N. O. Contribution of facilitated diffusion and processive catalysis to enzyme efficiency: implications for the EcoRI restriction-modification system. Biochemistry. 1996 Feb 20;35(7):2201–2208. doi: 10.1021/bi951883n. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassylyev D. G., Kashiwagi T., Mikami Y., Ariyoshi M., Iwai S., Ohtsuka E., Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995 Dec 1;83(5):773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- Wing R., Drew H., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980 Oct 23;287(5784):755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Wu J. C., Santi D. V. Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4778–4786. [PubMed] [Google Scholar]